Abstract
Methyl mercaptan production by oral bacteria is thought to be one of the main causes of oral malodor. We examined the ability of periodontopathic Porphyromonas gingivalis to produce methyl mercaptan from l-methionine and found that the invasive strains W83 and W50 produced large amounts of methyl mercaptan. We cloned and sequenced the mgl gene encoding l-methionine-α-deamino-γ-mercaptomethane-lyase (METase) from P. gingivalis W83. The structural mgl gene consisted of 1,200 bp and encoded a 43.3-kDa protein. To examine the role of methyl mercaptan in the pathogenesis of P. gingivalis, a METase-deficient mutant of P. gingivalis W83 was constructed. The methionine degradation activity and virulence of the mutant (M1217) and the parent strain (W83) in mice were compared. M1217 showed a marked decrease in the formation of methyl mercaptan from l-methionine and decreased virulence compared with the wild-type strain W83. These results suggest that methyl mercaptan not only is one of the sources of oral malodor, but may also play a role in the pathogenicity of P. gingivalis.
Oral malodor is caused mainly by volatile sulfur compounds (VSCs), such as hydrogen sulfide, methyl mercaptan, and dimethyl sulfide (21). Of these VSCs, hydrogen sulfide and methyl mercaptan are predominant in mouth air. Both compounds are highly toxic, especially methyl mercaptan (21). VSCs can increase the permeability of the oral mucosa (16) and decrease protein or collagen synthesis (6, 8). It is possible that the presence of methyl mercaptan within a periodontal pocket is involved in the induction or progression of periodontal disease. Coil et al. (2) reported that the increase in the ratio of methyl mercaptan to hydrogen sulfide in human gingival crevicular sites is correlated with deeper pockets or bleeding pockets. Exposure to methyl mercaptan alters protein synthesis in human gingival fibroblasts (7) and inhibits cell migration in periodontal ligament cells (11). These findings suggest that methyl mercaptan not only may be responsible for oral malodor but also may contribute to the pathogenesis of periodontal disease.
Methyl mercaptan is produced from l-methionine by the enzymatic action of l-methionine-α-deamino-γ-mercaptomethane-lyase (METase), which catalyzes the α,γ elimination of l-methionine to produce α-ketobutyrate, methyl mercaptan, and ammonia. This enzyme is detected in anaerobic, nonoral microorganisms, such as Pseudomonas, Trichomonas, and Clostridium (4, 5, 9, 13). In addition, Porphyromonas gingivalis, a black-pigmented anaerobe, which is implicated as a major pathogen in adult periodontitis, is known to produce large amounts of methyl mercaptan in human serum (18). However, little is known about P. gingivalis METase and the role of methyl mercaptan in the pathogenesis of this organism. In this study, we cloned the mgl gene homolog encoding METase from the virulent strain P. gingivalis W83 and confirmed its nucleotide sequence. To determine the role of METase in the pathogenicity of P. gingivalis, we constructed a METase-deficient mutant from P. gingivalis strain W83 and compared the virulence of the mutant and the parent strain W83 in a mouse model.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. gingivalis strains W83 and W50 and Fusobacterium nucleatum ATCC 10953 were grown anaerobically (10% CO2, 10% H2, 80% N2) in GAM broth (Nissui Medical Co., Tokyo, Japan) supplemented with hemin (5 μg/ml), menadione (1 μg/ml), and enriched tryptic soy agar (per liter: 40 g of tryptic soy agar [Difco, Detroit, Mich.], 5 g of brain heart infusion, 1 g of cysteine, 5 mg of hemin, and 1 mg of menadione) at 37°C. Actinobacillus actinomycetemcomitans Y4 was grown in THY broth (per liter: 30 g of Trypticase soy broth [BBL Microbiology Systems, Cockeysville, Md.], and 10 g of yeast extract [Difco]) at 37°C in a 5% CO2 atmosphere. Escherichia coli strains DH5α and BL21 were grown aerobically in 2× TY broth (per liter: 16 g of Bacto tryptone [Difco], 10 g of yeast extract [Difco], and 5 g of sodium chloride) at 37°C. For transformants of P. gingivalis, erythromycin at a final concentration of 10 μg/ml was added.
Gas chromatography of VSCs.
Methyl mercaptan was formed by using a modification of the method of Persson et al. (18). Briefly, bacterial strains were grown at 37°C until an optical density at 550 nm (OD550) of about 0.6 was attained. Culture supernatants were obtained by centrifugation (8,000 × g for 10 min). The cells were harvested and washed with a buffered salt solution (40 mM potassium phosphate buffer [pH 7.7] and 50 mM sodium chloride). The cells were resuspended in the salt solution to an OD550 of 0.3. To determine the formation of methyl mercaptan, a reaction mixture consisting of 100 μl of the cell suspension or culture supernatant and 870 μl of buffered salt solution was added in a sterile 15-ml polypropylene tube sealed with a silicon plug. The reaction was initiated by adding 30 μl of 33 mM l-methionine. The reaction mixtures were kept at 37°C. After 90 min of incubation, the reaction was stopped by adding 500 μl of 3 M phosphoric acid. Ten minutes later, a sample (1 ml) of the vapor above the reaction mixture in the tube was removed with a gastight syringe and analyzed by gas chromatography (model GC-14B; Shimadzu Works, Tokyo, Japan) using a glass column packed with 25% β, β′-oxydipropionitrile on a 60–80 mesh Chromosorb W AW-DMCS-ST device (Shimadzu Works) fitted with a flame photometric detector at 70°C. The concentration of each VSC was determined with standard hydrogen sulfide, methyl mercaptan, or dimethyl sulfide gas prepared with Permeater PD-1B (GL Science, Tokyo, Japan).
The VSC contents in the culture supernatants of P. gingivalis were also determined. P. gingivalis cells were grown until an OD550 of 0.7 was attained. The cultures were immediately centrifuged at 8,000 × g for 10 min, and the culture supernatants were obtained. A reaction mixture consisting of 100 μl of the culture supernatant and 900 μl of buffered salt solution was added to a tube and sealed with a silicon plug. Ten minutes later, a sample (1 ml) of the vapor above the reaction mixture in the tube was analyzed by gas chromatography as described above.
Electrotransformation of P. gingivalis.
Electroporation of P. gingivalis cells was performed by the method of Nakayama et al. (14). Briefly, P. gingivalis cells grown in 10 ml of GAM broth containing hemin (5 μg/ml) and menadione (1 μg/ml) were resuspended in 1 ml of the electroporation solution (300 mM sucrose). Fifteen microliters of plasmid DNA solution (270 μg/ml in distilled water) was added to 400 μl of the cell suspension. The mixtures were pulsed with a Gene Pulser apparatus (Bio-Rad Laboratories, Hercules, Calif.) at 2.5 kV with a time constant of 5 ms. The cell suspension was then diluted with 10 ml of prewarmed GAM broth containing 0.1% α-ketobutyrate and incubated anaerobically at 37°C for 15 h. Aliquots of the cell suspension were plated on enriched tryptic soy agar containing erythromycin (10 μg/ml) and 0.1% α-ketobutyrate and incubated anaerobically at 37°C for 7 days.
DNA manipulation.
Standard DNA recombinant procedures such as DNA isolation, restriction endonuclease digestion, ligation, transformation of competent E. coli cells, and agarose gel electrophoresis were carried out as described by Sambrook et al. (20). Chromosomal DNA was isolated from P. gingivalis cells by the guanidine isothiocyanate method (12) with an IsoQuick DNA extraction kit (MicroProbe, Garden Grove, Calif.). DNA sequencing was performed by the dideoxy-chain technique with a Taq Dye Primer Cycle Sequencing kit and an ABI 373A DNA sequencer (Applied Biosystems, Foster City, Calif.).
Southern hybridization and colony hybridization.
Southern hybridization and colony hybridization were performed with digoxigenin-labeled PCR probes using a nonradioactive digoxigenin DNA labeling and detection kit (Rosche Diagnostics GmbH, Mannheim, Germany) according to the supplier's instructions. To detect the mgl gene of P. gingivalis, an mgl probe was amplified with primers designed from the nucleotide sequence of the mgl gene homolog found in data from the P. gingivalis Genome Project (http://www.forsyth.org/pggp/) and the Institute for Genomic Research (http://www.tigr.org/). The primers used were 5′-GCTATCGAGAACGCCTTC-3′ (sense), located 40 to 57 bases downstream of the initiation codon of the mgl gene homolog of P. gingivalis, and 5′-GCAGTGCCATCTGCTTCT-3′ (antisense), located 895 to 913 bases downstream of the initiation codon of the mgl gene homolog of P. gingivalis, and genomic DNA of P. gingivalis W83 was used as the template. To confirm the appropriate insertional inactivation of the mgl gene, the ermF-ermAM cassette probe was constructed with a kit based on random hexanucleotide primers (Rosche Diagnostics GmbH) with a 2.1-kb EcoRI-PstI fragment isolated from pVA2198 as a template.
Cloning the mgl gene from P. gingivalis.
Southern blotting with the mgl gene-specific probe suggested that a 4.2-kb ClaI chromosome fragment of P. gingivalis W83 contained an mgl homolog. Based on this result, the complete ClaI digests of chromosomal DNA of P. gingivalis W83 were electrophoresed on 0.8% agarose gels, and the 4.2-kb ClaI fragment was extracted from the gel with a QIAEX II agarose gel extraction kit (Qiagen, Hilden, Germany). The 4.2-kb ClaI fragment was ligated with ClaI-digested pBluescriptII KS+. E. coli DH5α-competent cells were then transformed with the resultant plasmid. Positive colonies hybridized with the mgl probe were selected. Plasmid DNA was purified from one of these colonies and designated pME220.
Plasmid construction.
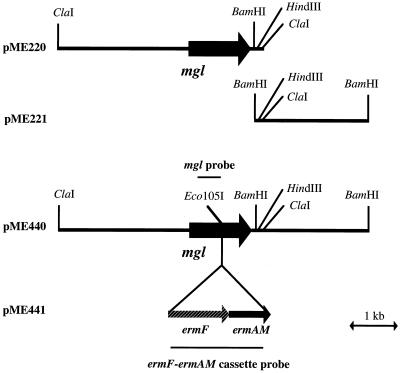
The BamHI fragment located downstream from the mgl gene of P. gingivalis W83 was amplified and isolated by PCR (Fig. 1). The primers used were 5′-GACAATATGAACCCCGAA-3′ (sense), located 1,216 to 1,233 bases downstream of the initiation codon of the mgl gene homolog of P. gingivalis, and 5′-GACCCGATATCCTCACTT-3′ (antisense), located 3,539 to 3,556 bases downstream of the initiation codon of the mgl gene homolog of P. gingivalis. The PCR conditions were as follows: an initial denaturation at 95°C for 2 min and then 25 cycles of denaturation at 96°C for 15 s, annealing at 50°C for 30 s, and extension at 72°C for 2 min. The amplified fragment was cloned with pGEM-T Easy (Promega, Madison, Wis.), and the resultant plasmid was designated pME221. pME221 was digested with HindIII, and the 2.0-kb HindIII-BamHI fragment was isolated. The isolated fragment was ligated into pME220 to replace the 73-bp HindIII-ClaI fragment. The plasmid obtained was designated pME440. The mgl gene of plasmid pME440 was then disrupted by inserting the 2.1-kb ermF-ermAM cassette from pVA2198 (3) into the Eco105I site within the mgl gene. This plasmid, designated pME441, was linearized with NotI-XhoI and used for electroporation. The resultant plasmids were purified, and the nucleotide sequences of the inserted fragments were determined to confirm that the fragments did not contain nucleotide substitutions or deletions resulting from PCR with Taq polymerase.
FIG. 1.
Restriction map and construction of plasmids containing the mgl gene and its adjacent region. pME220 contains the entire mgl gene and its upstream region. pME221 contains the flanking region of the mgl gene. pMF440 contains the 6.2-kb ClaI-BamHI fragment reconstructed from pME220 and pME221. pME441 contains the mgl gene interrupted by an ermF-ermAM cassette. pME441 was linearized with NotI and XhoI and introduced into P. gingivalis W83 by electroporation.
Preparation of recombinant METase.
The mgl gene homolog of P. gingivalis W83 was amplified by PCR with the primers 5′-AAAAGGATCCCGTAGTGGCTTTGCCACA-3′ (sense) and 5′-AAAAGTCGACTTAGATCAGGCTGTCCAG-3′ (antisense). These primers were designed to create BamHI and SalI restriction sites (underlined) in the PCR product. The PCR product was double digested with BamHI and SalI and ligated to BamHI-SalI double-digested pGEX-6P-1 expression vector (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) to produce pRM83. The nucleotide sequence of the insert was determined to confirm that no mutations had been introduced. To obtain the mgl products of P. gingivalis W83, E. coli BL21 was transformed with pRM83. The transformant was grown in 2 × TY broth with ampicillin (50 μg/ml) at 37°C until an OD550 of 0.7 was attained. Isopropyl-β-thiogalactopyranoside was added to the culture at a final concentration of 1 mM, and the culture was grown for 4 h. The cells were harvested by centrifugation at 1,600 × g for 20 min at 4°C and lysed by ultrasonication. The cell extract was obtained by centrifugation at 12,000 × g for 10 min at 4°C. Binding to glutathione-Sepharose 4B medium (Amersham Pharmacia Biotech), cleavage of the fusion protein by PreScission protease, and elution of the product were done according to the manufacturer's instructions.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as follows. Purified recombinant METase (10 μg) was suspended in 100 μl of 10 mM Tris-HCl (pH 6.8) containing 1% SDS, 1% mercaptoethanol, and 20% glycerol and heated for 3 min at 100°C. After the sample (10 μl) was electrophoresed at 25 mA per gel with 4% stacking and 10% resolving acrylamide gels containing 0.1% SDS (10), proteins were stained with Coomassie brilliant blue R-250. The LMW electrophoresis calibration kit (Amersham Pharmacia Biotech) was used as a molecular weight standard.
Enzyme assay.
METase activity was assayed as described by Tan et al. (22). Briefly, the assay was carried out with 1 ml of 50 mM potassium phosphate buffer (pH 8.0) containing 10 μM pyridoxal 5′-phosphate, 10 mM l-methionine, and various amounts of enzyme solution. The enzyme was added to start the reaction. After incubation for 10 min at 37°C, the reaction was terminated by adding 0.5 ml of 4.5% trichloroacetic acid. The suspension was centrifuged, and 0.5 ml of the supernatant was added to 0.5 ml of 0.05% 3-methyl-2-benzothiazolinone hydrazone in 1 ml of 1 M sodium acetate (pH 5.2) and then incubated at 50°C for 30 min. The amount of α-ketobutyrate produced was determined by spectrophotometry with A335. The amount of protein was determined by using the Bio-Rad protein assay with bovine serum albumin as a standard.
Mouse virulence assays.
The virulence of P. gingivalis strains W83 and M1217 was determined as described by Neiders et al. (15). Each bacterial strain was grown in GAM broth supplemented with hemin and menadione to an OD550 of 1.0. The cells were harvested, resuspended and adjusted to a concentration of 3 × 1011 CFU/ml in the same medium. Female BALB/c mice (8 to 10 weeks of age) were challenged with subcutaneous injections of 0.1 ml of bacterial suspension at two sites on the depilated dorsal surface (0.2 ml per mouse). Injected mice were examined daily for the size, consistency, and ulceration of each local lesion, the presence and locations of secondary lesions, and general health status. Three sets of experiments were carried out (61 mice in total).
Statistical analysis.
Student's t-test was used to evaluate the difference in the mean value of the survival rate of three experiments between mice infected with W83 and those infected with M1217.
RESULTS
Formation of methyl mercaptan by P. gingivalis, F. nucleatum, A. actinomycetemcomitans, and E. coli.
P. gingivalis, F. nucleatum, A. actinomycetemcomitans, and E. coli were tested for their ability to form methyl mercaptan from l-methionine. The amount of methyl mercaptan in the vapor above reaction mixtures containing the cell suspension in sealed tubes was determined by gas chromatography. P. gingivalis and F. nucleatum produced methyl mercaptan from l-methionine. Interestingly, the two invasive strains, W83 and W50, produced a large amount of methyl mercaptan. The content of methyl mercaptan in the vapor above the reaction mixture containing the culture supernatant did not change after addition of l-methionine (data not shown). On the other hand, A. actinomycetemcomitans and E. coli could not form methyl mercaptan (Table 1).
TABLE 1.
Production of methyl mercaptan from l-methionine by P. gingivalis, F. nucleatum, A. actinomycetemcomitans, and E. coli
| Strain | Production of methyl mercaptan (nmol/liter)a |
|---|---|
| P. gingivalis | |
| W83 | 79.44 ± 10.87 |
| W50 | 66.77 ± 7.78 |
| F. nucleatum ATCC 10953 | 54.31 ± 4.00 |
| A. actinomycetemcomitans Y4 | NDb |
| E. coli DH5α | ND |
The production of methyl mercaptan by each strain was estimated as described in the text. The values are expressed as the means ± standard deviations of triplicate assays.
ND, not detected.
Cloning the P. gingivalis mgl gene.
The mgl genes encoding METase have been cloned from Pseudomonas putida (4, 5) and Trichomonas vaginalis (13), and their nucleotide sequences have been determined. A database search was performed to detect homologs of the P. gingivalis mgl gene. Although both P. putida and T. vaginalis have two genes reported to encode METase (4, 5, 13), only one homolog of the mgl gene was found in the preliminary search of the P. gingivalis genome database obtained from websites. From this, a probe for the P. gingivalis mgl gene was synthesized by PCR with chromosomal DNA of strain W83 as a template (Fig. 1) and was used to isolate the 4.2-kb ClaI fragment cloned into the plasmid vector pBluescript KS+. The segment coding for the METase gene, mgl, of P. gingivalis contained a 1,200-bp open reading frame. The deduced peptide sequence has 399 amino acid residues and a molecular mass of 43.3 kDa. The predicted amino acid sequence of METase of P. gingivalis W83 was compared with the amino acid sequence of the METases of P. putida (4, 5) and T. vaginalis (13). The sequence identities of P. gingivalis METase with the gene products of mgl1 and mgl2 of P. putida were 43.9 and 43.2%, respectively. Those of P. gingivalis METase with the gene products of mgl1 and mgl2 of T. vaginalis were 42.9 and 39.6%, respectively.
Characterization of recombinant METase of P. gingivalis W83.
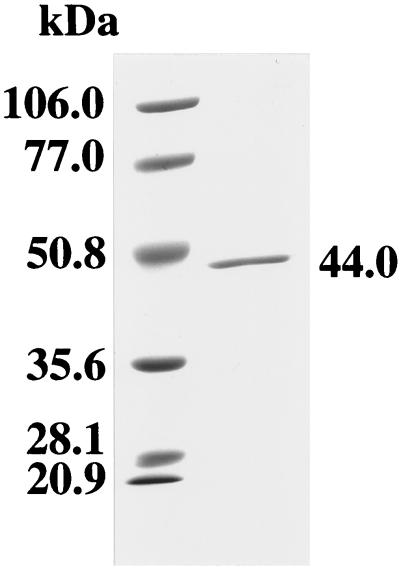
To evaluate the enzymatic activity of P. gingivalis METase, the mgl gene of P. gingivalis W83 was cloned into pGEX-6P Expression Vector. The glutathione S-transferase GST-fusion protein was cleaved with PreScission protease and purified by affinity chromatography with glutathione-Sepharose 4B medium. A single protein band was obtained in SDS-PAGE analysis of recombinant METase. The enzyme migrated at approximately 44 kDa on SDS-PAGE gels (Fig. 2). Approximately 1.2 mg of purified recombinant METase was obtained from each liter of E. coli culture. The breakdown of l-methionine by the recombinant enzyme was determined by assaying the production of α-ketobutyrate, a by-product of methyl mercaptan formation, as described previously (22). The Km and Vmax of W83 recombinant enzyme were 23.1 mM and 21.9 μmol/min/mg of protein, respectively.
FIG. 2.
SDS-PAGE analysis of purified recombinant METase of P. gingivalis W83. The purified recombinant protein was suspended in SDS-PAGE loading buffer and heated at 100°C for 3 min. A sample (1.0 μg) was then subjected to SDS-PAGE (10% polyacrylamide), and the gel was stained with Coomassie brilliant blue R-250. The positions of molecular size markers are indicated to the left.
Construction of a METase-deficient mutant.
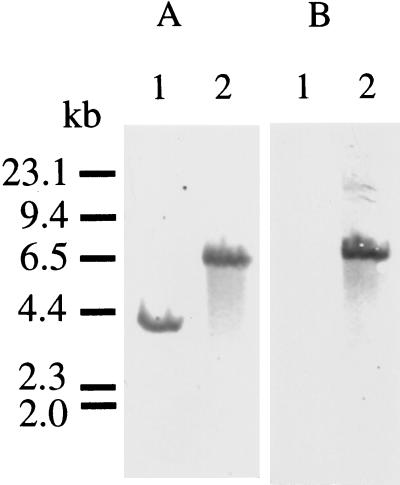
To examine the role of methyl mercaptan in the pathogenicity of P. gingivalis, we constructed a METase-deficient mutant. To construct the METase-deficient mutant, the mgl gene of plasmid pME440 was disrupted by inserting the 2.1-kb ermF-ermAM cassette from pVA2198 into the Eco105I site within the mgl gene. This construct, pME441, was linearized with NotI-XhoI and electroporated into P. gingivalis W83, and erythromycin-resistant colonies were isolated. To confirm the presence of the ermF-ermAM cassette in the predicted location, Southern blot analysis was carried out with the mgl gene as a probe. Upon digestion of the DNA with ClaI, a 4.2-kb fragment hybridized with the mgl probe was observed in strain W83. A 6.3-kb fragment was present in the erythromycin-resistant mutant of W83 (Fig. 3A). Southern blots probed with the ermF-ermAM cassette revealed the presence of an identical 6.3-kb fragment in the erythromycin-resistant mutants, but not in W83 (Fig. 3B). These data indicate that the ermF-ermAM cassette caused the predicted recombination. One of the erythromycin-resistant mutants of W83 was chosen and designated M1217. The growth rate of transformant M1217 was similar to that of the parent strain W83.
FIG. 3.
Southern blot analysis of chromosomal DNA from W83 and M1217. (A) Southern blot hybridization with the mgl probe. (B) Southern blot hybridization with the ermF-ermAM cassette probe. Lanes: 1, ClaI-digested W83 chromosomal DNA; 2, ClaI-digested M1217 chromosomal DNA. HindIII-digested λ DNA size markers are indicated to the left.
Formation of methyl mercaptan by the METase-deficient mutant and the wild-type strain.
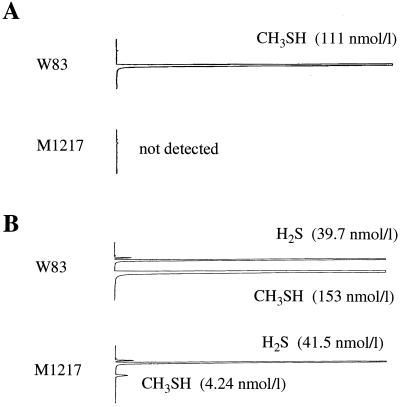
Whole cells of P. gingivalis W83 and M1217 were tested for their ability to form methyl mercaptan from l-methionine. The bacterial cells were incubated in the buffered salt solution containing l-methionine at 37°C for 90 min, and then the amount of methyl mercaptan produced was determined by gas chromatography. A large amount of methyl mercaptan was detected in the reaction mixture containing W83 cells (Fig. 4A). On the other hand, no methyl mercaptan peak was detected in the reaction mixture containing M1217. The VSCs in the culture supernatants of W83 and M1217 were also analyzed by gas chromatography. The hydrogen sulfide contents in the culture supernatants of M1217 and W83 were similar (Fig. 4B). On the other hand, the amount of methyl mercaptan in the culture supernatant of M1217 was approximately 2.8% of that in the culture supernatant of W83.
FIG. 4.
Gas chromatographic patterns of VSCs produced by P. gingivalis W83 and M1217. (A) To measure the formation of methyl mercaptan, a reaction mixture consisting of 100 μl of the cell suspension of P. gingivalis (OD550 of 0.3), 870 μl of the buffered salt solution, and 30 μl of 33 mM l-methionine was added to a tube, which was sealed with a silicon plug. The reaction mixtures were kept at 37°C for 90 min. The reaction was stopped by adding 500μl of 3 M phosphoric acid. Ten minutes later, the sample (1 ml) of the vapor above the reaction mixture in the tube was analyzed by gas chromatography. (B) VSCs in the vapor above 100 μl of the supernatants of the cultures of P. gingivalis strains in a sealed tube were determined by gas chromatography.
Pathogenicity of P. gingivalis in mice.
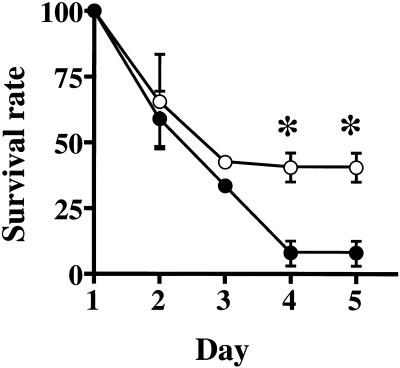
To examine the pathogenicity of P. gingivalis W83 and M1217, BALB/c mice were challenged with subcutaneous injections of bacterial suspension at two sites on the depilated dorsal surface. Within 24 h, strains M1217 and W83 (6 × 1010 CFU per animal) induced spreading, ulcerative lesions on the abdomens of all mice tested. The mice did not have any lesions at the dorsal site of injection. Four days after the subcutaneous injections, about 7.7% of mice challenged with W83 at a dose of 6 × 1010 CFU per mouse survived (Fig. 5). In contrast, 36% of mice challenged with M1217 at the same dose per animal survived. Student's t test showed a significant difference between the survival rates of mice infected with W83 and with M1217 (P = 0.01) at days 4 and 5.
FIG. 5.
Survival rates of mice challenged with P. gingivalis W83 (●) and M1217 (○). Female BALB/c mice (five mice per group) were subcutaneously injected with 0.1 ml of bacterial suspension (3 × 1011 CFU/ml) of P. gingivalis at two sites on the depilated dorsal surface (0.2 ml per mouse). The survival rate in each group was calculated as the number of survivors/the total number of mice. Data are the mean survival rate ± standard error of three different experiments. On days 4 and 5 after injection, the values were significantly higher for M1217 than for W83. ∗, P = 0.01 (Student's t test).
DISCUSSION
Although several studies have reported on the formation of methyl mercaptan by various oral bacteria (1, 17, 18), the mechanism and pathogenic role of the formation of methyl mercaptan by oral bacteria are not well defined. We tested the capacity of A. actinomycetemcomitans, F. nucleatum, E. coli, and two strains of P. gingivalis to form methyl mercaptan from l-methionine. Of these strains, P. gingivalis strain W83 produced the most methyl mercaptan (Table 1). This strain is also known as an invasive virulent strain (15). Therefore, it is possible that methyl mercaptan may be one of the virulence factors of P. gingivalis.
To determine whether the mgl gene participates in the formation of methyl mercaptan by P. gingivalis, we constructed an mgl mutant of P. gingivalis W83. Whole cells of the mgl-defective mutant M1217 did not form methyl mercaptan from l-methionine (Fig. 4A), indicating that the mgl gene product participates in the formation of methyl mercaptan from l-methionine in P. gingivalis. Gas chromatography detected methyl mercaptan in the supernatant of cultures of both W83 and M1217, although the amount of methyl mercaptan produced by M1217 was approximately 2.8% of that of W83 (Fig. 4B). It is possible that an enzyme or enzymes different from METase may produce methyl mercaptan from substrates other than l-methionine.
Ng and Tonzetich (16) reported that exposure of sublingual porcine mucosa to methyl mercaptan increases the permeability of the tissue. Methyl mercaptan has also been shown to induce interleukin-1β secretion from mononuclear cells (19). To evaluate whether the mgl gene is associated with the pathogenicity of P. gingivalis, the virulence of the parent strain W83 and the mgl-deficient mutant M1217 was tested in a mouse abscess model. Four days after injection, a significant difference (P = 0.01) between the survival rate of mice infected with W83 and that of those infected with M1217 was observed (Fig. 5). These results suggest that methyl mercaptan may play a role in the pathogenicity of P. gingivalis. However, we should note that the mouse abscess model used in this study is not periodontitis. Further work is needed to determine a role of methyl mercaptan in induction and/or progress of periodontitis.
In this regard, several studies have examined the relationship between oral malodor and periodontitis from an epidemiological perspective (2, 23). Yaegaki and Sanada (23) found that the total amount of VSCs and the ratio of methyl mercaptan to hydrogen sulfide were greater in the mouth air from patients with periodontal disease than in that of control subjects. Coil et al. (2) found a significantly higher methyl mercaptan/hydrogen sulfide ratio in deep or inflamed crevicular sites than in shallow or noninflamed sites. These findings suggest that methyl mercaptan from periodontal pockets may be associated with the oral malodor of patients with periodontitis. Understanding the mechanism of methyl mercaptan production in periodontal pockets not only may help us elucidate the source of oral malodor, but may also provide a clue to understanding the pathology in periodontal disease.
Since METase has not been found in mammals, inhibition of this enzyme should have little effect on humans. Therefore, we hypothesize that this enzyme may be a potentially exploitable target for designing chemotherapeutic drugs. An inhibitor of P. gingivalis METase may represent a new class of compounds with the potential for preventing and treating oral malodor.
ACKNOWLEDGMENTS
We thank Masahiro Yoneda for helpful advice on the murine virulence assay. Katsuji Okuda kindly supplied strains of P. gingivalis.
This work was supported in part by Grant-in-Aid for Scientific Research no. 12877338 (T.K.) from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Claesson R, Edlund M B, Persson S, Carlsson J. Production of volatile sulfur compounds by various Fusobacterium species. Oral Microbiol Immunol. 1990;5:137–142. doi: 10.1111/j.1399-302x.1990.tb00411.x. [DOI] [PubMed] [Google Scholar]
- 2.Coil J M, Tonzetich J. Characterization of volatile sulphur compounds production at individual gingival crevicular sites in humans. J Clin Dent. 1992;3:97–103. [PubMed] [Google Scholar]
- 3.Fletcher H M, Schenkein H A, Morgan R M, Bailey K A, Berry C R, Macrina F L. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hori H, Takabayashi K, Orvis L, Carson D A, Nobori T. Gene cloning and characterization of Pseudomonas putidal-methionine-α-deamino-γ-mercaptomethane-lyase. Cancer Res. 1996;56:2116–2122. [PubMed] [Google Scholar]
- 5.Inoue H, Inagaki K, Sugimoto M, Esaki N, Soda K, Tanaka H. Structural analysis of the l-methionine γ-lyase gene from Pseudomonas putida. J Biochem (Tokyo) 1995;117:1120–1125. doi: 10.1093/oxfordjournals.jbchem.a124816. [DOI] [PubMed] [Google Scholar]
- 6.Johnson P, Yaegaki K, Tonzetich J. Effect of methyl mercaptan on synthesis and degradation of collagen. J Periodontal Res. 1996;31:323–329. doi: 10.1111/j.1600-0765.1996.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 7.Johnson P W, Ng W, Tonzetich J. Modulation of human gingival fibroblast cell metabolism by methyl mercaptan. J Periodontal Res. 1992;27:476–483. doi: 10.1111/j.1600-0765.1992.tb01820.x. [DOI] [PubMed] [Google Scholar]
- 8.Johnson P W, Yaegaki K, Tonzetich J. Effect of volatile thiol compounds on protein metabolism by human gingival fibroblasts. J Periodontal Res. 1992;27:553–561. doi: 10.1111/j.1600-0765.1992.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 9.Kreis W, Hession C. Isolation and purification of l-methionine-α-deamino-γ-mercaptomethane-lyase (l-methioninase) from Clostridium sporogenes. Cancer Res. 1973;33:1862–1865. [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Lancero H, Niu J, Johnson P W. Exposure of periodontal ligament cells to methyl mercaptan reduces intracellular pH and inhibits cell migration. J Dent Res. 1996;75:1994–2002. doi: 10.1177/00220345960750121201. [DOI] [PubMed] [Google Scholar]
- 12.Lippke J A, Strzempko M N, Raia F F, Simon S L, French C K. Isolation of intact high-molecular-weight DNA by using guanidine isothiocyanate. Appl Environ Microbiol. 1987;53:2588–2589. doi: 10.1128/aem.53.10.2588-2589.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKie A E, Edlind T, Walker J, Mottram J C, Coombs G H. The primitive protozoon Trichomonas vaginalis contains two methionine γ-lyase genes that encode members of the γ-family of pyridoxal 5′-phosphate-dependent enzymes. J Biol Chem. 1998;273:5549–5556. doi: 10.1074/jbc.273.10.5549. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J Biol Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 15.Neiders M E, Chen P B, Suido H, Reynolds H S, Zambon J J, Shlossman M, Genco R J. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 1989;24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 16.Ng W, Tonzetich J. Effect of hydrogen sulfide and methyl mercaptan on the permeability of oral mucosa. J Dent Res. 1984;63:994–997. doi: 10.1177/00220345840630071701. [DOI] [PubMed] [Google Scholar]
- 17.Persson S, Claesson R, Carlsson J. The capacity of subgingival microbiotas to produce volatile sulfur compounds in human serum. Oral Microbiol Immunol. 1989;4:169–172. doi: 10.1111/j.1399-302x.1989.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 18.Persson S, Edlund M B, Claesson R, Carlsson J. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol. 1990;5:195–201. doi: 10.1111/j.1399-302x.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 19.Ratkay L G, Waterfield J D, Tonzetich J. Stimulation of enzyme and cytokine production by methyl mercaptan in human gingival fibroblast and monocyte cell cultures. Arch Oral Biol. 1995;40:337–344. doi: 10.1016/0003-9969(94)00165-8. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Scully C, El-Maaytah M, Porter S R, Greenman J. Breath odor: etiopathogenesis, assessment and management. Eur J Oral Sci. 1997;105:287–293. doi: 10.1111/j.1600-0722.1997.tb00242.x. [DOI] [PubMed] [Google Scholar]
- 22.Tan Y, Xu M, Tan X, Wang X, Saikawa Y, Nagahama T, Sun X, Lenz M, Hoffman R M. Overexpression and large-scale production of recombinant l-methionine-α-deamino-γ-mercaptomethane-lyase for novel anticancer therapy. Protein Expr Purif. 1997;9:233–245. doi: 10.1006/prep.1996.0700. [DOI] [PubMed] [Google Scholar]
- 23.Yaegaki K, Sanada K. Biochemical and clinical factors influencing oral malodor in periodontal patients. J Periodontol. 1992;63:783–789. doi: 10.1902/jop.1992.63.9.783. [DOI] [PubMed] [Google Scholar]