Abstract
Aim
Left ventricular systolic dysfunction (LVSD) carries an increased risk for overt heart failure and mortality, yet treatable to mitigate disease progression. An artificial intelligence (AI)-enabled 12-lead electrocardiogram (ECG) model demonstrated promise in LVSD screening, but the performance dropped unexpectedly in external validation. We thus sought to train de novo models for LVSD detection and investigated their performance across multiple institutions and across a broader set of patient strata.
Methods and results
ECGs taken within 14 days of an echocardiogram were obtained from four academic hospitals (three in the United States and one in Japan). Four AI models were trained to detect patients with ejection fraction (EF) <40% using ECGs from each of the four institutions. All the models were then evaluated on the held-out test data set from the same institution and data from the three external institutions. Subgroup analyses stratified by patient characteristics and common ECG abnormalities were performed. A total of 221 846 ECGs were identified from the 4 institutions. While the Brigham and Women’s Hospital (BWH)-trained and Keio-trained models yielded similar accuracy on their internal test data [area under the receiver operating curve (AUROC) 0.913 and 0.914, respectively], external validity was worse for the Keio-trained model (AUROC: 0.905–0.915 for BWH trained and 0.849–0.877 for Keio-trained model). Although ECG abnormalities including atrial fibrillation, left bundle branch block, and paced rhythm-reduced detection, the models performed robustly across patient characteristics and other ECG features.
Conclusion
While using the same model architecture, different data sets produced models with different performances for detecting low-EF highlighting the importance of external validation and extensive stratification analysis.
Keywords: Electrocardiogram, Neural network, External validation, Ejection fraction
Graphical Abstract
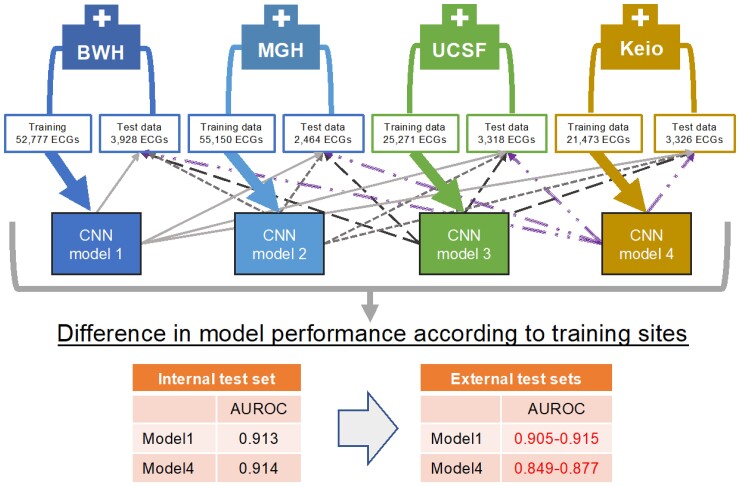
Graphical Abstract.
Convolutional neural network models were trained using training data at each institution. Then the models were tested in the held-out test set from the institution and in three external test sets from the remaining three institutions, and their performances were compared. AUROC, area under the receiver operating curve; BWH, Brigham and Women’s Hospital; CNN, convolutional neural network; Keio, Keio University Hospital; MGH, Massachusetts General Hospital; UCSF, University of California San Francisco.
Introduction
Left ventricular systolic dysfunction (LVSD) is a frequently observed finding that carries an increased risk for overt heart failure and mortality.1 If detected early, LVSD can be treated to mitigate disease progression. An artificial intelligence (AI)-enabled 12-lead electrocardiogram (ECG) model has demonstrated promise in LVSD screening2 but an unexpected drop in performance was observed in external validation.3 In addition to problems with generalizability, AI models have also shown uneven performance in distinct subpopulations,4 which has important implications for downstream decision-making should these models be applied to general practice. We thus sought to train a de novo model for LVSD detection from ECG data and investigate its performance across multiple institutions and across a broader set of patient strata.
Methods
ECGs taken within 14 days of an echocardiogram in patients aged ≥20 were obtained from 4 academic hospitals from the USA [Brigham and Women’s Hospital (BWH), Massachusetts General Hospital (MGH), University of California San Francisco (UCSF)] and Japan (Keio University Hospital). No patients were excluded based on the presence or absence of symptoms. A convolutional neural network (CNN) was trained to detect patients with left ventricular ejection fraction (EF) <40% from ECG alone. The details of the model architecture are described at https://github.com/obi-ml-public/ECG-LV-Dysfunction. In brief, the model consisted of a layer of 2D-CNN followed by 20 layers of a multi-2D-CNN module and a fully connected layer. The data set for each institution was randomly divided into three groups (derivation, validation, and test) in a 5:2:3 ratio without overlaps of patients across groups. Models were trained on the derivation data set and those with the highest performance on the validation data set across 50 epochs were chosen as the final model. These models were then evaluated on the test data set from the same institution and all data from the three external institutions. While the training was performed using all ECGs expecting a similar effect as data augmentation, testing was done using a single ECG–echocardiogram pair with the closest dates for each patient to prevent exaggeration of the model performance. Subgroup analyses stratified by patient characteristics and common overt ECG abnormalities were performed. The model performance was evaluated by area under the receiver operating characteristics curve (AUROC) analysis.
Results
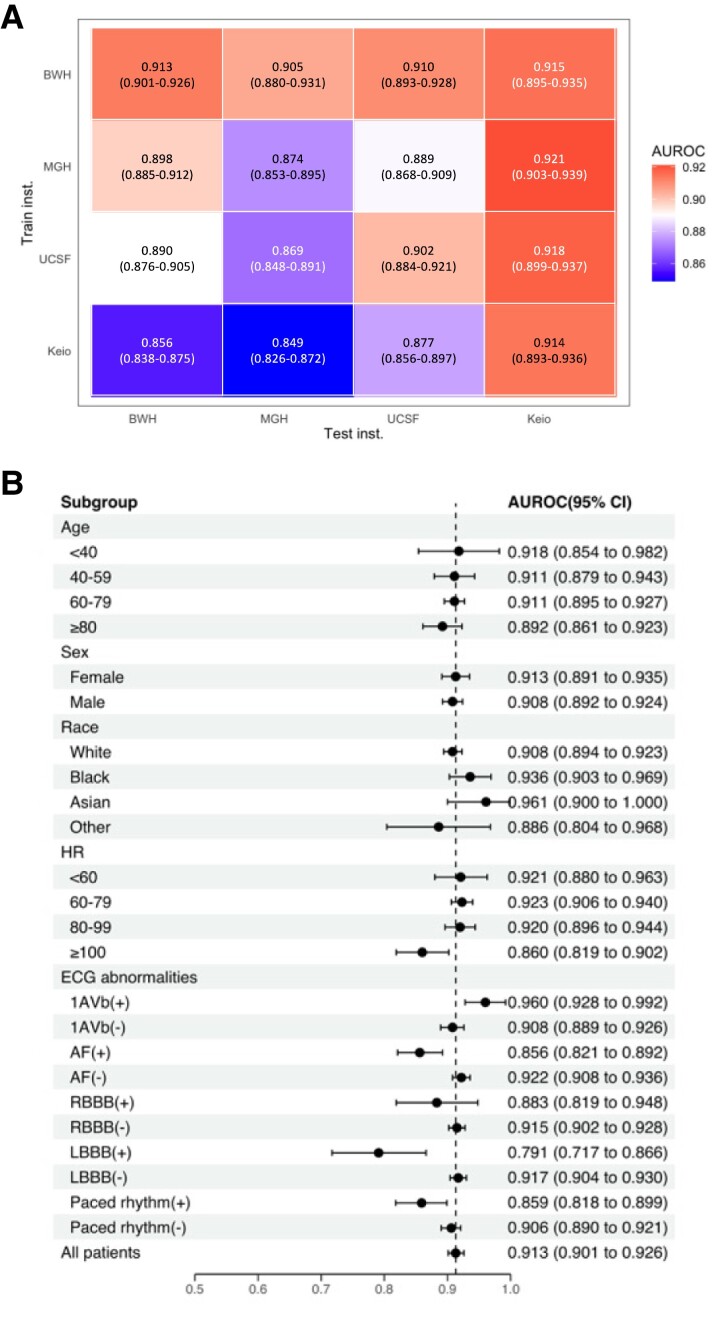
There were 75 033, 79 663, 36 314, and 30 836 ECGs for BWH, MGH, UCSF, and Keio, respectively. While the BWH-trained model yielded excellent accuracy on internal test data [AUROC 0.913, 95% confidence internal (CI), 0.901–0.926] and good external validity (AUROC 0.905, 95% CI 0.880–0.905; 0.910, 95% CI 0.893–0.928; and 0.915, 95% CI 0.895–0.935, respectively, for MGH, UCSF and Keio), the Keio-trained model, having a similar performance on its test set (AUROC 0.914, 95% CI 0.893–0.936), showed poor external validity (AUROC 0.856, 95% CI 0.838–0.875; 0.849, 95% CI 0.826–0.872; and 0.877, 95% CI 0.856–0.897, respectively, for BWH, MGH, and UCSF; Figure 1A). This resulted in higher positive predictive value (PPV) when the cutoffs were chosen to have the same sensitivity n the external institutions for the BWH-trained model compared with the Keio-trained model (PPVs for BWH- vs. Keio-trained model: 0.41 vs. 0.27 for the MGH data set and 0.33 vs. 0.26 for USCF data set at sensitivity 0.80). The stratified analysis, using the BWH-trained model, demonstrated consistent performance across patient age, sex, race, and common ECG abnormalities: first-degree atrio-ventricular block and right bundle brunch block (AUROC 0.960, 95% CI 0.928–0.992 and 0.883, 95% CI 0.819–0.948, respectively). However, the model showed lower accuracy in individuals with atrial fibrillation (AF), left bundle branch block (LBBB), or a paced rhythm (AUROC 0.856, 95% CI 0.821–0.892; 0.791, 95% CI 0.717–0.866; and 0.859, 95% CI 0.818–0.899, respectively; Figure 1B). These findings were consistent upon external validation.
Figure 1.
(A) Performance and external validity of models trained at individual institution. (B) Stratification analyses on patient characteristics and electrocardiogram abnormalities in Brigham and Women’s Hospital. AF, atrial fibrillation; CLBBB, complete left bundle branch block; HR, heart rate; PM, pacemaker.
Discussion
Although a previously published model reported high performance on an internal data set, retrospectively and prospectively,2,5 external validation unexpectedly revealed lower performance.3 Our neural network models trained at BWH and Keio; both displayed similar performance for the detection of low-EF on internal test sets but varied substantially upon external validation. If only internal testing had been performed, one could have concluded these models were equally useful. While our results confirmed the robustness of the models across patient demographics as reported previously,2,6 the subgroup analysis by ECG abnormalities demonstrated settings in which the model had a lower performance, which was confirmed upon external validation. The patterns of performance in each subgroup were similar for all four models trained at different institutions. Given that patients with newly detected AF and LBBB are likely to be referred for echocardiography, regardless, the models’ high accuracy for other subgroups implies excellent utility for screening. Given the similar performance patterns across models trained at different institutions, our analysis could not identify the cause of the difference in performance by the training institution. The performance drop could be attributable to the differences in the internal processes of different vendors (e.g. noise reduction), but this could not be assessed due to the lack of availability of multiple vendors within one institution.
Conclusion
Our findings highlight the importance of extensive stratification analysis and external validation to establish model applicability.
Acknowledgements
This study complies with all ethical regulations and guidelines. Approval was obtained from the Institutional Review Boards of all institutions.
Contributor Information
Ryuichiro Yagi, One Brave Idea and Division of Cardiovascular Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA.
Shinichi Goto, One Brave Idea and Division of Cardiovascular Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA; Department of Cardiology, Keio University School of Medicine, Shinjuku, Tokyo, Japan.
Yoshinori Katsumata, Department of Cardiology, Keio University School of Medicine, Shinjuku, Tokyo, Japan.
Calum A MacRae, One Brave Idea and Division of Cardiovascular Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA.
Rahul C Deo, One Brave Idea and Division of Cardiovascular Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA.
Funding
This work was supported by One Brave Idea, co-funded by the American Heart Association and Verily with significant support from AstraZeneca and pillar support from Quest Diagnostics.
Data availability
The code for training and testing the model is provided at https://github.com/obi-ml-public/ECG-LV-Dysfunction. The model weights may contain personal information from patients and thus, are not shared. We provide a web-interface to run our model and generate predictions at http://onebraveideaml.org. All data are included in the manuscript.
References
- 1. Echouffo-Tcheugui JB, Erqou S, Butler J, Yancy CW, Fonarow GC. Assessing the risk of progression from asymptomatic left ventricular dysfunction to overt heart failure a systematic overview and meta-analysis. J Am Coll Cardiol HF 2016;4:237–248. [DOI] [PubMed] [Google Scholar]
- 2. Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez-Sarano M, Noseworthy PA, Munger TM, Asirvatham SJ, Scott CG, Carter RE, Friedman PA. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 3. Attia ZI, Tseng AS, Benavente ED, Medina-Inojosa JR, Clark TG, Malyutina S, Kapa S, Schirmer H, Kudryavtsev AV, Noseworthy PA, Carter RE, Ryabikov A, Perel P, Friedman PA, Leon DA, Lopez-Jimenez F. External validation of a deep learning electrocardiogram algorithm to detect ventricular dysfunction. Int J Cardiol 2021;329:130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seyyed-Kalantari L, Zhang H, McDermott MBA, Chen IY, Ghassemi M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat Med 2021;27:2176–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yao X, Rushlow DR, Inselman JW, McCoy RG, Thacher TD, Behnken EM, Bernard ME, Rosas SL, Akfaly A, Misra A, Molling PE, Krien JS, Foss RM, Barry BA, Siontis KC, Kapa S, Pellikka PA, Lopez-Jimenez F, Attia ZI, Shah ND, Friedman PA, Noseworthy PA. Artificial intelligence–enabled electrocardiograms for identification of patients with low ejection fraction: a pragmatic, randomized clinical trial. Nat Med 2021;27:815–819. [DOI] [PubMed] [Google Scholar]
- 6. Noseworthy PA, Attia ZI, Brewer LC, Hayes SN, Yao X, Kapa S, Friedman PA, Lopez-Jimenez F. Assessing and mitigating bias in medical artificial intelligence: the effects of race and ethnicity on a deep learning model for ECG analysis. Circulation Arrhythmia Electrophysiol 2020;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The code for training and testing the model is provided at https://github.com/obi-ml-public/ECG-LV-Dysfunction. The model weights may contain personal information from patients and thus, are not shared. We provide a web-interface to run our model and generate predictions at http://onebraveideaml.org. All data are included in the manuscript.