Abstract
Diribonucleotides arise from two sources: turnover of RNA transcripts (rRNA, tRNA, mRNA, and others) and linearization of cyclic-di-nucleotide signaling molecules. In both cases, there appears to be a requirement for a dedicated set of enzymes that will cleave these diribonucleotides into mononucleotides. The first enzyme discovered to mediate this activity is oligoribonuclease (Orn) from Escherichia coli. In addition to being the enzyme that cleaves dinucleotides and potentially other short oligoribonucleotides, Orn is also the only known exoribonuclease enzyme that is essential for E. coli, suggesting that removal of the shortest RNAs is an essential cellular function. Organisms naturally lacking the orn gene encode other nanoRNases (nrn) that can complement the conditional E. coli orn mutant. This review covers the history and recent advances in our understanding of these enzymes and their substrates. In particular, we focus on (i) the sources of diribonucleotides; (ii) the discovery of exoribonucleases; (iii) the structural features of Orn, NrnA/NrnB, and NrnC; (iv) the enzymatic activity of these enzymes against diribonucleotides versus other substrates; (v) the known physiological consequences of accumulation of linear dinucleotides; and (vi) outstanding biological questions for diribonucleotides and diribonucleases.
Keywords: RNA degradation, diribonucleotides, oligoribonuclease, NanoRNases, diribonuclease
Specialized enzymes, including oligoribonuclease (orn), nanornase A (NrnA), NrnB, and NrnC, share structural features that allow them to cleave dinucleotides that arise from RNA turnover and cyclic-di-nucleotide signaling.
Sources of oligoribonucleotides/diribonucleotides
RNA polymerization produces RNA for ribosomes (rRNA), transfer RNA (tRNA), messenger RNA (mRNA) and a number of regulatory RNAs. However, these molecules have a limited lifespan inside the cell and are turned over by the action of specific subsets of enzymes (Fig. 1). Turnover ensues when RNAs are internally cleaved by endonucleases. These long RNA fragments are acted on by exoribonucleases which remove one nucleotide at a time from the resulting RNA fragments leading to the release of mononucleotides and the accumulation of short RNAs, from 2–7 nucleotides in length, presumably because they are not likely to be good substrates for general exoribonucleases. The process through which these short RNA oligonucleotides are recycled to mononucleotides was first attributed to an enzyme called oligoribonuclease (Orn). The discovery of a number of cyclic dinucleotides acting as second messengers, including cyclic-di-GMP (c-di-GMP), cyclic-di-AMP (c-di-AMP), and cyclic-GMP-AMP (cGAMP), indicated the existence of a separate pool of diribonucleotides that is not generated by RNA polymerase (reviewed in (Krasteva and Sondermann 2017)). These cyclic nucleotide signals are removed by a two-step process: linearization into linear RNA dinucleotides and cleavage of diribonucleotides into mononucleotides. Studies into cleavage of dinucleotides into mononucleotides revealed a specific subset of ribonucleases responsible for recycling (Fig. 2). The greater implication of these studies is that the terminal step in RNA degradation, i.e. the processing of dinucleotides into mononucleotides, could comprise a distinct step. This final step appears to require a specific set of diribonucleases. Thus, diribonucleotides could represent a point of convergence between RNA degradation and cellular signaling.
Figure 1.

Overview of RNA degradation. In a general way, RNAs become unstable upon endolytic cleavage of unstructured and structured regions by endoribonucleases. A combination of exoribonucleases successively remove single nucleotides from the termini either to continue the degradation or to mature functional RNAs. In addition to mononucleotides, the penultimate step of RNA degradation creates diribonucleotides. A second source of ribonucleotides comes from the cleavage of specific signaling nucleotides, the cyclic diribonucleotides. The diribonucleotide species has been linked to feedback inhibition of c-di-GMP-specific phosphodiesterases and RNases, transcription control through nanopriming, and potentially the binding of protein receptors. One or more of these functions appear to be detrimental to cellular growth when linear dinucleotide levels rise above a certain level.
Figure 2.
Distribution of Orn and nanoRNases. Shown is a taxonomic distribution at the class level. Bacterial taxa are shown with purple lines, eukaryotic taxa with green lines, and archaeal taxa with red lines. The presence of each RNase homolog as a proportion of the total proteins in that taxonomic group is shown as either a filled square (>50% presence of a homolog per genome) or an empty square (<50% presence of a homolog per genome). Lack of a square indicates no homologs for that family were present in genomes of that class. The tree focuses on Orn, NrnC, and related enzymes RNase T and RNase D, as well as NrnA/B. The illustration is based on (Lormand et al. 2021).
Identification and discovery of RNases
In the history of characterizing the enzymes that degrade RNA molecules, the process has been enabled by the identification of substrates, biochemical purification of enzymatic activities that act on the substrates, and identification of the genes encoding each of the enzymes. Below is a brief history that led to our current understanding of RNA degradation.
Early characterization of enzymes that degrade RNA molecules was limited by the substrates available for detection of biochemical activity. The first RNase was identified before the structure of nucleic acids were even fully characterized. RNase A (also known as RNase I (Spahr and Schlessinger 1963)) was characterized by its ability to convert acid-precipitable yeast nucleic acids to a non-precipitable form (nucleotides) (Jones 1920). Purification and crystallization of thermostable and acid-resistant RNase A from beef pancreas revealed that it acted on yeast nucleic acid (RNA), but not thymus nucleic acid (DNA) (Kunitz 1939, Kunitz 1940). Even at this time, there was recognition that RNase A did not release mononucleotides that would be diffusible through cellophane (Schmidt and Levene 1938). Characterization of this RNase protein provided insight into nuclease activity specifically and protein folding and protein primary and secondary structures in general (Anfinsen 1973, Moore and Stein 1973). The crystal structure of RNase A (Avey et al. 1967, Kartha et al. 1967, Wyckoff et al. 1967) represents one of the first high-resolution protein structures. An additional feature of RNase A is the two-step catalytic mechanism with a cyclic 2′-3′ intermediate for the newly released 3′ ribose, which is subsequently hydrolyzed (Findlay et al. 1961). The characterization of RNase A revealed it as an endonuclease that cleaved internally in RNA polymers, which therefore suggested the presence of other enzymes—exoribonucleases that could cleave mononucleotides from the ends of the RNA fragments.
While a few ribonucleases were subsequently purified and characterized, their functions in the cellular context remained elusive at these early times. The discovery of the structure of DNA (Watson and Crick 1953), the genetic code (Barondes and Nirenberg 1962, Nirenberg 2004, Nirenberg and Matthaei 1961) and the characterization of ribosomal RNA (rRNA) transcripts (Fellner and Sanger 1968) led to a realization that there are many unique RNA substrates in the cell. For example, rRNA maturation required endo- and exo-nucleolytic RNase processing to produce mature rRNA (Deutscher 2009). In another example, characterization of transfer RNA (tRNA) revealed that pre-tRNAs are processed through 3′ exoribonuclease activity (Deutscher 2015, Deutscher et al. 1984, Zhang and Deutscher 1988). Through analyses of these new RNA substrates, several other new RNases were identified. RNase II was the first exoribonuclease discovered for degrading polyA RNA (Singer and Tolbert 1965, Spahr and Schlessinger 1963). Using short RNA oligos, Orn was identified as an enzyme that preferentially cleaves short RNA (Datta and Niyogi 1975, Niyogi and Datta 1975, Stevens and Niyogi 1965). Using tRNA altered at the 3′ end, RNase D was identified as the enzyme responsible for degrading damaged tRNA (Ghosh and Deutscher 1978). Using precursor tRNA, RNase PH was identified as the maturation exoribonuclease required to trim back the 3′ end of tRNA to the CCA sequence (Deutscher et al. 1988).
Once the unique biochemical activity was discovered and the responsible enzyme purified, the gene encoding this enzyme was identified by screening for mutants lacking the biochemical activity. The gene encoding RNase II was isolated from a screen for mutants lacking activity against poly-U substrate (Nikolaev et al. 1976). Using an E. coli mutant lacking RNase I (rna) and RNase II (rnb), the mutant defective for RNase D was identified, allowing mapping of the rnd gene in the E. coli genome (Zaniewski and Deutscher 1982). In a separate line of investigation into resistance to phage infection, E. coli mutant strains resistant to T4 phage exhibited a defect in processing of a phage-specific ser-tRNA, suggesting that there exists an RNase responsible for this reaction (Seidman et al. 1975). Using mutants lacking known RNases (rnb– and rnd–) and the assay for measuring tRNA processing, the RNase BN protein was purified (Asha et al. 1983). Similarly, mutagenesis of this E. coli strain lacking rnb, rnd and rbn identified mutants in the gene encoding RNase T (Deutscher et al. 1984, Deutscher et al. 1985). Together, the iterative approach of identifying biochemical activity and the genes encoding for these enzymes led to discovery of the various endo- and exo-nucleases in E. coli including RNase II, PNPase, RNase D, RNase BN, RNase T, RNase PH (Deutscher et al. 1988), RNase R (Cheng et al. 1998, Cheng and Deutscher 2002), and oligoribonuclease (Yu and Deutscher 1995, Zhang et al. 1998). Biochemical characterization of these exoribonucleases revealed that, after processively removing mononucleotides of the 3′ ends, the enzymes produce short 5′ oligoribonucleotides that are not processed further (Frazao et al. 2006). For example, Rnase R leaves 5′ tetranucleotides and dinucleotides (Matos et al. 2009); whereas RNase II leaves 5′ tetranucleotides and pentanucleotides (Matos et al. 2011). The explanation for the accumulation of 5′ oligonucleotide is that the substrate binding pocket of the enzyme is located at a distance from the catalytic site (Frazao et al. 2006). Once the product length is shorter than the distance between the binding site and catalytic site, the enzyme is unable to cleave these short substrates any further.
Upon identification of the genes encoding ribonucleases, the contribution of these genes in RNA maturation and degradation could be assessed. Analysis of triple, quadruple, and quintuple mutants indicated that many exoribonucleases display similar and overlapping activities suggesting that exoribonucleases have redundant functions (Cheng et al. 1998, Kelly and Deutscher 1992, Li and Deutscher 1996, Zaniewski et al. 1984). Interestingly, there is an exception to these general observations. Orn was specifically attributed to the degradation of short RNA, thereby exhibiting a unique function not shared with other exoribonucleases (Yu and Deutscher 1995). N-terminal Edman degradation of the purified Orn revealed the possible gene sequence, subsequently leading to the cloning of the gene from an E. coli genomic library (Zhang et al. 1998). Orn is essential in E. coli as mutants cannot be generated (Ghosh and Deutscher 1999). Similarly, the orn gene appears to be essential in Vibrio cholerae (Kamp et al. 2013) and Yersinia pestis (Palace et al. 2014). These results suggest that degradation of short oligo RNA is a process that is distinct from the reactions carried out by other exoribonucleases.
Bacillus subtilis and other Firmicutes lack genes encoding Orn. However, heterologous expression of B. subtilis YtqI (renamed NrnA) led to complementation of the growth phenotype exhibited by a conditional E. coli ∆orn strain, suggesting that NrnA might be functionally equivalent to E. coli Orn (Mechold et al. 2007). When nrnA was deleted from B. subtilis, the resulting ∆nrnA mutant cells were viable, suggesting that an additional enzyme might serve to hydrolyze short chain RNA fragments. Using this genetic strategy, a few other B. subtilis genes (NrnB and YhaM) were discovered to complement growth of E. coli ∆orn, suggesting that they too can degrade short RNA oligonucleotides (Fang et al. 2009). When all three genes were deleted, the B. subtilis ∆nrnA ∆nrnB ∆yhaM triple mutant had a similar doubling time as the parental strain suggesting either that still a-yet-unknown additional RNase awaits discovery or the requirement of these enzymes in Firmicutes is substantially different from γ-proteobacteria (Fang et al.2009).
Cyclic-di-GMP and pGpG—diribonucleotides with specific physiological functions
A line of studies into bacterial cellulose revealed that a unique nucleotide, cyclic di-guanylate (c-di-GMP), acted as an allosteric activator of the bacterial cellulose synthase (Bcs) complex (Ross et al. 1987). This finding led to the identification of diguanylate cyclases, which synthesize c-di-GMP, and phosphodiesterase-A (PDE-A) enzymes, which cleave c-di-GMP to linear di-GMP (abbreviated as pGpG or pGG) (Tal et al. 1998). In addition to these two classes of enzymes, the earliest description of c-di-GMP synthesis and degradation postulated that a second enzyme is required for the degradation of the linear pGpG to two guanine monophosphates (GMP) (Ross et al. 1987). Thus, linear pGpG represented a physiologically relevant dinucleotide within the bacterial cells. The identity of the enzyme that degraded pGpG was later revealed in two separate studies on Orn (Cohen et al. 2015, Orr et al. 2015). Both groups made this discovery for Pseudomonas aeruginosa as the ∆orn mutant is impaired but viable. Using lysates from the parental and isogenic ∆orn strains, activity for the cleavage of pGpG was shown to be largely absent in the ∆orn strain indicating that Orn is the primary enzyme for hydrolysis of this dinucleotide (Cohen et al. 2015, Orr et al. 2015).
While the PA14 ∆orn strain was viable, it demonstrates a small colony variant phenotype on agar plates and an accelerated sedimentation phenotype in broth culture (Orr et al. 2015). One mechanism for increased cell aggregation is due to feedback inhibition of pGpG on c-di-GMP phosphodiesterases resulting in elevated c-di-GMP levels in the cells triggering biofilm-related processes (Cohen et al. 2015, Orr et al. 2015). When genes encoding proteins containing known RNase domains from Vibrio cholerae and B. subtilis were tested for their ability to restore wild-type colony morphology to P. aeruginosa ∆orn, only four genes could be identified; this included V. cholerae orn, B. subtilis nrnA, B. subtilis nrnB and nrnC from Caulobacter crescentus (Fang et al. 2009, Liu et al. 2012, Orr et al. 2018) (Table 1). These findings support the idea that the ability to hydrolyze pGpG is performed by a few discrete enzymes that are distinct from other exoribonucleases.
Table 1.
Experimental evidence for bacterial enzymes with potential dinuclease activity. For details, please refer to text.
| Enzyme | Organisms | References | Experimental data |
|---|---|---|---|
| Orn | Escherichia coli | Datta and Niyogi 1975 | Biochemical |
| (DnaQ- | Niyogi and Datta 1975 | Biochemical | |
| DEDDh) | Yu and Deutscher 1995 | Genetic, Biochemical | |
| Zhang et al. 1998 | Biochemical | ||
| Ghosh and Deutscher 1999 | Genetic, Biochemical | ||
| Vibrio cholerae | Kim et al. 2019 | Structural, Biochemical | |
| Pseudomonas aeruginosa | Cohen et al. 2015 | Genetic, Biochemical | |
| Orr et al. 2015 | Genetic, Biochemical | ||
| Colwellia psychrerythraea | Lee et al. 2019 | Structural, Biochemical | |
| NrnC | Bartonella birtlesii | Liu et al. 2012 | Genetic, Biochemical |
| (DnaQ- | Brucella melitensis | Lormand et al. 2021 | Structural, Biochemical |
| DEDDy) | Bartonella henselae | Lormand et al. 2021 | Structural, Biochemical |
| Caulobacter cresentus | Orr et al. 2018 | Genetic, Biochemical | |
| NrnA | Bacillus subtilis | Mechold et al. 2007 | Genetic, Biochemical |
| (DHH- | Wakamatsu et al. 2011 | Biochemical | |
| DHHA1) | Schmier et al. 2017 | Structural, Biochemical | |
| Thermus thermophilus | Wakamatsu et al. 2011 | Biochemical | |
| Staphylococcus aureus | Bowman et al. 2016 | Genetic, Biochemical | |
| Mycobacterium tuberculosis | Postic et al. 2012 | Genetic, Biochemical | |
| Yang et al. 2014 | Genetic, Biochemical | ||
| He et al. 2016 | Structural, Genetic, Biochemical | ||
| Mycoplasma pneumoniae | Postic et al. 2012 | Genetic, Biochemical | |
| Streptococcus mutans | Postic et al. 2012 | Genetic | |
| Vibrio cholerae | Heo et al., 2022 | Structural, Biochemical, Genetic | |
| NrnB | Bacillus subtilis | Fang et al. 2009 | Genetic, Biochemical |
| (DHH- | |||
| DHHA1) |
Enzymatic activity of Orn—a historical perspective
Orn was initially identified through biochemical purification of enzymes that can cleave 14C-labeled oligoribonucleotides of various lengths, from 2 to 6 nucleotides (Datta and Niyogi 1975, Niyogi and Datta 1975) in reactions buffers containing 5 mM Mn2+ as catalysis-supporting cations. The authors concluded that when substrate concentration were saturating, the rate of hydrolysis scaled linearly with time and was inversely proportional to chain length (Datta and Niyogi 1975). Under substrate limiting conditions, dinucleotides were cleaved quickly, whereas processing of substrates with higher chain length was slower at the beginning, but accelerated over time (Datta and Niyogi 1975), suggesting shorter substrates were preferred by Orn over longer substrates. Because this enzyme preparation had activity against ribonucleotides of various length generated by hydroxide treatment of synthesized polyuridine followed by paper-chromatography separation, the authors named the enzyme oligoribonuclease.
Subsequent studies of various E. coli mutants lacking known RNases (pnp, rnd, rnt, rnb, and rph) revealed that oligoribonuclease activity remained in all of these mutants suggesting that oligoribonuclease activity is encoded by a separate gene (Yu and Deutscher 1995). Here, the oligoribonucleotide substrate was prepared in a similar manner—hydroxide hydrolysis of polyuridine. Oligoribonucleotides were separated from uridine and UMP and oligonucleotides of specific lengths were not further separated (Yu and Deutscher 1995). The authors tested this substrate against purified RNase D, RNase T, RNase PH, RNase II and PNPase in buffer containing 5 mM Mn2+. While RNase D, T and PH had minimal activity against the oligouridine mixture, purified RNase II or PNPase cleaved 42% and 95% of this substrate, respectively (Yu and Deutscher 1995). Later studies demonstrated that rnb and pnp expression did not restore cleavage of pGpG (Orr et al. 2018), which confirmed the oligoribonuclease activity against short oligoribonucleotides is distinct from known exoribonucleases.
The orn gene is essential as attempts to delete the gene were unsuccessful (Ghosh and Deutscher 1999, Zhang et al. 1998). Using a different strategy, the authors showed that the chromosomal orn gene can be disrupted by insertion with the kanamycin gene if the cell has an additional plasmid expressing orn. To study a mutant lacking orn, the authors placed the orn gene on a temperature sensitive (ts) plasmid. After heat inactivation of the plasmid, cell lysates were assessed for oligoribonuclease activity using oligonucleotide substrate ApCpC[32P]pC prepared by ligating [5′-32P]pCp to ApCpC (Ghosh and Deutscher 1999). This substrate is a major advance over previous studies since this substrate has a more defined polymer length (i.e. 4 nucleotides) and was only enabled by the commercial availability of ApCpC. The results for activity assays performed in buffer with 5 mM Mn2+ showed that non-permissive conditions inhibited growth, reduced plasmid copy number, and reduced oligoribonuclease activity against this 4-nucleotide substrate. While there was less oligoribonuclease activity, growth of the ts-conditional orn strain at the non-permissive temperature did not grossly alter RNA degradation as assessed by a pulse-chase experiment. Nonetheless, the oligoribonucleotides collected from wild-type and the conditional orn mutant grown at non-permissive temperature showed large difference in the appearance of labeled mononucleotide after a pulse with 3H-uridine as assessed by acid soluble label (Ghosh and Deutscher 1999). When the oligonucleotide fraction was separated by chromatography, the authors concluded that there was an increase in the accumulation of 2-, 3-, 4-, and 5-mer RNAs (Ghosh and Deutscher 1999). These results support the idea that Orn cleaves oligonucleotide RNA of various lengths in the cell.
In 2006, a key study utilized a different substrate and assay format to assess the substrate preference of Orn (Mechold et al. 2006). The authors used a 5′ Cy5-labeled pentacytosine ribonucleotide as a substrate, assayed in the presence of 5 mM Mn2+, and monitored the degradation by a 22% polyacrylamide gel. This fluorescently labeled substrate provides sensitivity and the improved separation technique provided clear and unambiguous distinction of 2-, 3-, 4-, and 5-mer from each other and mononucleotides (Mechold et al. 2006). Substrate preference by comparing degradation of nanoRNAs of different length was not examined in this study and would likely be affected by the presence of the Cy5 group. A potential preference for dinucleotides might not be detectable using Cy5-labeled substrates. However, such a preference was observed for an NrnA homolog from Mycobacterium tuberculosis (Rv2837C) using the same modified nanoRNA substrates and resulted in the absence of dinucleotides from the pattern of degraded nanoRNA substrate (Postic et al. 2012). Whether the Cy5 group on the substrate affects Orn and NrnA in different ways is currently unknown. Whether the 5′ modification renders RNA suboptimal substrates for Orn will be discussed below.
A different approach for understanding the substrate preference of Orn was also ascertained through high-throughput sequencing. Short oligoribonucleotides are capable of initiating transcription as nano-primers (Nickels and Dove 2011). To understand the contribution of Orn in this process, a depletion strategy was developed to induce protein degradation (Goldman et al. 2011). Total RNA was isolated and the RNA with 5′ phosphate, which are products of RNA degradation, was selectively sequenced as they can be ligated at the 5′ end with an oligonucleotide adapter that is compatible with modern sequencing technologies. Sequencing and comparing transcripts from Orn-depleted and untreated controls allowed determination of the transcripts that exhibited nanopriming by oligoribonucleotides. These analyses revealed that the majority of the nanoprimed transcripts were primed by diribonucleotides (Druzhinin et al. 2015, Goldman et al. 2011, Vvedenskaya et al. 2012). This preferential accumulation of dinucleotides is reversed by treatment of these oligonucleotides with purified Orn enzyme (Goldman et al. 2011). Together, these results support the idea that Orn might be an enzyme with preference for dinucleotides.
Substrate preference of Orn remains a key issue for our general understanding of RNA degradation and needs to be further investigated. The outcome of these studies will potentially depend on: (i) the particular enzyme homolog used, (ii) the composition of the reaction buffer, (iii) the method and resolution of substrate and product detection, and (iv) the concentrations and ratios of enzymes and substrates in the reactions.
Is Orn a diribonuclease?
Because of the overlap between the enzymes that cleave pGpG and the enzymes that are thought to cleave short RNA oligonucleotides (previously called ‘nano-RNAs’ (Mechold et al. 2007)), a key question is how do these enzymes distinguish between diribonucleotides, short oligoribonucleotides, and longer oligoribonucleotides? Moreover, do certain RNase enzymes recognize discrete subclasses of short RNAs or is there a broader range of substrates that they prefer? While Orn was previously described as having a preference for ‘short RNAs’, a high-resolution three-dimensional structure of Orn bound to different diribonucleotides unexpectedly revealed a highly constrained active site (Kim et al. 2019). Indeed, the substrate binding pocket appeared to be unable to accommodate substrates larger than a dinucleotide. This observation was further supported by biochemical analyses, which showed that Orn exhibits an exceptionally strong preference for 5′ 32P-labeled diribonucleotide substrates over 5′ 32P-labeled 3-, 4-, 5-, 6- or 7-mers (Kim et al. 2019). These findings were also supported by ex-vivo experiments with lysates from the P. aeruginosa ∆orn strain in buffer with Mg2+ (Fig. 3). When 5′-radiolabeled 7-mers were added to wild-type lysates it resulted in a progression in the degradation of the 7-mer to shorter lengths over time; by 30 minutes, the labeled RNA was fully processed to individual nucleoside monophosphates (Kim et al. 2019). In contrast, degradation of the 7-mer RNA substrate in ∆orn lysates resulted in processing of the 7-mer but with robust accumulation of dinucleotides with some residual trinucleotide, with no apparent production of nucleoside monophosphates (Kim et al. 2019). The residual trinucleotide is likely due to feedback inhibition in manner similar to accumulation of c-di-GMP through the inhibition of c-di-GMP phosphodiesterases by pGpG (Cohen et al. 2015, Orr et al. 2015). These results suggest that cellular exoribonucleases can act on the 7-mer by cleaving successive nucleotides from the 3′ terminus until a 5′ diribonucleotide remains. This diribonucleotide is then specifically cleaved into mononucleotides by Orn. Because 5′ 32P-labeled oligoribonucleotides of various length are chemically identical to the endogenous oligoribonucleotides, these results likely reflect more accurately the substrate preference of the enzyme for cellular substrates than 5′ Cy5 labeled substrate. If this interpretation is correct, diribonucleotide processing appears to represent both a connection to cyclic dinucleotide signaling pathways and a key step in the general RNA degradation pathway. These results generally support the idea that Orn might be a specific diribonuclease rather than the historical prospective that Orn acts on oligonucleotides. Additional biochemical and structural studies for the E. coli Orn enzyme should settle this important issue.
Figure 3.
Orn is the primary dinuclease in P. aeruginosa. Degradation of (A) 32P-GG or (B) 32P-AAAAAGG into mononucleotides by whole cell lysates of PA14, orn mutant, orn mutant complemented with ornVc, or ornVc D12A in buffer with Mg2+. Reactions were stopped at indicated times and separated on 20% PAGE, exposed to phosphorimager to reveal the location of the radiolabel. For these studies, purified V. cholerae Orn was used, which is 72% identical to P. aeruginosa Orn with a strictly conserved active site. The image is modified from (Kim et al. 2019).
Structural features of Orn
A sequence and phylogenetic survey classified Orn, the founding member of the nano-RNases, and corresponding eukaryotic enzymes as DEDD-superfamily nucleases that also contains other RNases such as RNase D and RNase T, but also many nucleases that act on DNA (Zuo and Deutscher 2001, Yang 2011). The DEDD denotation refers to four strictly conserved, acidic residues in the DnaQ fold of the nucleases, which are involved in the coordination of two metal ions at the enzyme's active site (Zuo and Deutscher 2001). A sub-classification can be made into DEDDy- and DEDDh-type enzymes, highlighting the conservation of an additional active-site residue, a tyrosine or histidine, respectively. This residue acts on the nucleophilic water crucial for the hydrolytic cleavage of the substrate's phosphodiesterase bond. Orn and its eukaryotic orthologs (e.g. human Rexo2) belong to the DEDDh group containing a histidine residue as the general base (Zuo and Deutscher 2001). The alignments also revealed four conserved stretches of sequence, unique motifs that distinguish Orn from other DEDD nucleases. Their functional relevance was unknown at the time. Furthermore, early studies established Orn as a homodimer in its purified form (Zhang et al. 1998).
The first high-resolution, molecular views of Orn came from crystal structures of orthologs from Haemophilus influenzae (PDB 1j9a; unpublished, 2003), E. coli (PDBs 1yta and 2igi; unpublished, 2006, 2007), and Xanthomonas campestris (PDB 2gbz; (Chin et al. 2006)). They confirmed that Orn adopts the canonical DnaQ fold characteristic for DEDDh exoribonucleases, with the structural similarity indicating a conserved catalytic mechanism within this superfamily (Hamdan et al. 2002, Steitz 1999, Yang 2011). The crystal structures also support a homodimeric assembly as the biologically active unit of Orn-type enzymes with two seemingly independent active sites. In X. campestris Orn, a disulfide bond bridges the two monomers in the dimer (Chin et al. 2006). The disulfide bond is also present in the crystal structure of a metagenomic Orn from an arctic marine bacterium and mutating the involved cysteine residue to an alanine or glycine severely impacted enzyme activity (Piotrowski et al. 2019). Covalent dimerization, however, does not appear to be mandatory since the involved cysteine residue is not strictly conserved in the Orn family. On the other hand, several of the conserved, Orn-specific motifs first described by Zhu and Deutscher stabilize the dimeric assembly, presenting a general dimer interface in the Orn family (Zuo and Deutscher 2001, Chin et al. 2006). These features were also observed in the crystal structures of Coxiella burnetii Orn (PDB 3tr8; (Franklin et al. 2015)) and Acinetobacter baumannii Orn (PDB 5cy4; unpublished, 2015).
Up to this point, substrate-bound states were only modeled based on the corresponding crystallized apo states of the proteins (Franklin et al. 2015). The models suggested a path for nano-RNAs with up to 5 residues in length, with an Orn-specific helix (‘helix H’ in X. campestris Orn) or loop containing an HYR motif proposed to be involved in the processing of RNAs with more than two nucleotide residues. One question at the heart of Orn's nano-RNase mechanism remained unanswered by these early structural studies: What molecular features determine the narrow substrate range of Orn and its eukaryotic orthologs, i.e. what prevents longer (>5–7 nucleotides of length), single-stranded RNAs to be degraded by these enzymes? Associated with this question, how is processivity achieved for the degradation of nano-RNAs?
A first glimpse at ligand-bound states came from structures of Colwellia psychrerythraea Orn (cpsOrn) (Lee et al. 2019). In addition to the apo state, structures bound to a substrate analog (pNP-TMP), two separate uridine molecules, or a di-uridine molecule from a penta-uridine fragment used during crystallization were determined. From these structures, it is apparent that several conserved motifs together form a narrow active site: (i) a leucine residue that splays apart two most 3′ residues of the substrate; (ii) two serine residues, a tyrosine and an arginine residue that coordinate the phosphate moiety of pNP-TNP; and (iii) a tryptophan residue in a small, helical lobe that buttresses the 3′ base of the substrate. In addition, a loop harboring the histidine of the DEDDh motif undergoes a conformational change between the apo and substrate-engaged states, with the histidine residue moving into the active site to engage with the RNA ligands. A structure-guided mutagenesis experiment showed that the conserved catalytic residues responsible for metal coordination or activation of the attacking water molecule are critically important for enzyme activity towards the substrate analog pNP-TMP. Mutation of other active site residues affected cleavage of the substrate proxy much less. Most notably though, substrate-bound structures only resolved two nucleotide residues at the active site, even when longer substrates were used, failing to explain how substrate-length preference towards nano-RNAs is achieved.
A contemporary structure-function study on Vibrio cholerae Orn (and human Rexo2) resulted in structures of the enzymes bound with 5′-phosphorylated diribonucleotides of various sequence, all revealing a narrow active site formed by the same structural features described for cpsOrn (Kim et al. 2019, Lee et al. 2019) (Fig. 4A). A loop segment carrying the histidine residue of the DEDDh motif narrows the active site further in the catalytically competent state (Fig. 4B). The consensus features that contribute to the narrow and specialized active site—the leucine wedge, the 5′-phosphate cap, and a lobe that blocks off the 3′ end of the RNA substrate—are strictly conserved in the Orn family of enzymes, including the human ortholog Rexo2, but are divergent in the structural homolog RNase T and other DEDDh-type enzymes (Fig. 4C and D) (Kim et al. 2019). The structures suggested an optimization of the active site for 5′-phosphorylated diribonucleotides. Quantitative binding and activity studies confirmed a much more stringent substrate preference towards diribonucleotides than reported earlier and a strict requirement of 5′ phosphorylation of diribonucleotides for substrate binding. Most strikingly, P. aeruginosa cell lysates lacking Orn were capable of degrading nano-RNAs down to diribonucleotides with kinetics comparable to those observed in lysates from wild-type cells (Kim et al. 2019). Discrepancies to earlier kinetic studies that showed nano-RNase activity towards longer substrates by Orn likely stems from the 5′ labeling with bulky fluorophores used for detection. Considering that the 5′ phosphate is required for dinucleotide cleavage likely due to its observed tight coordination by conserved residues at Orn's narrow active site, any bulky modification at that site would render substrates suboptimal (Kim et al. 2019, Lee et al. 2019). The negative impact of fluorescent labels at the 5′ phosphate of substrates would be more pronounced for dinucleotides, as longer substrates already fit poorly into Orn's restricted active site. The use of 5′ 32P-labeled substrates that is chemically identical to the native substrates in the more recent studies eliminates any convoluting effects that may have arisen due to substrate modification (Kim et al.2019).
Figure 4.
Structure of substrate-bound Orn and Rexo2. (A) Crystal structure of V. cholerae Orn bound to pGpG. A homodimer is shown composed of two monomers colored in grey and purple (the red coordinate axis highlights the rotational symmetry axis of the dimer). pGpG bound to the active site is shown in yellow (Kim et al. 2019). The diagram on the right illustrates the structural features around the metal-coordinating DEDD motif, which are characteristic for Orn and restrict substrate specificity. (B) An activation loop associated with the catalytic histidine residue of the DEDDh motif is shown in green, indicating a catalytically competent state. (C)Crystal structures of Rexo2 bound to the diribonucleotide pGpG and an oligoT DNA are shown. In the pGpG -bound state, both active sites of the dimeric enzyme are occupied with substrate (Kim et al. 2019). In contrast, oligoT fragments were observed only at one of the two active sites (Chu et al. 2019). In addition, nucleotides marked with ‘X’ are poorly resolved resulting in discontinuous electron density for the DNA. (D) The structure of the related RNase T, another 3′-5′ exoribonuclease in the DEDDh family is shown with bound dTAGG substrate. A wider active site that accommodates longer substrates correlates with the enzymes activity in vitro (Hsiao et al. 2011). All structural figures in this review were prepared using ChimeraX (Pettersen et al. 2021).
Structure-function relationship in Rexo2, a eukaryotic Orn ortholog
Similar mechanistic questions have been raised concerning Rexo2’s substrate preference. The majority of Rexo2 resides in mitochondria with a smaller fraction localizing to the cytosol and nuclei of mammalian cells (Bruni et al. 2013, Szewczyk et al. 2020). Rexo2 is also called small fragment nuclease (Sfn) based on the early observation that the enzyme degrades both short RNA and DNA fragments (Nguyen et al. 2000). Orn was initially described as a dedicated RNase (Niyogi and Datta 1975); later studies showed that it is also capable of degrading small DNA fragments, albeit with much lower efficiency (Mechold et al. 2006). In a direct comparison, human Rexo2 degraded nano-RNAs ∼50 fold slower compared to E. coli Orn, with a preference for substrates with less than 5 residues, although longer oligoribonucleotides were processed as well, albeit much slower (Chu et al. 2019). Rexo2 processed small fragments of RNAs ∼4-fold faster than DNA fragments of similar length (Chu et al. 2019, Nguyen et al. 2000). Substrates to assess Rexo2 activity were labeled at the 5′ end with a bulky fluorescent group, possibly preventing native coordination of the terminal 5′ phosphate of dinucleotides. As a result, it is possible that labeled dinucleotides could act as suboptimal substrates, leading to an underestimation of Rexo2’s activity on these specific RNA fragments. Another study reported severe accumulation of mitochondrial non-coding transcripts and short RNA species in cells when Rexo2 expression was silenced (Szewczyk et al. 2020). Some of these effects, especially those on structured RNAs, may be indirect by affecting the function of the mitochondrial degradosome comprising the helicase SUV3 and PNPase; others may indicate direct Rexo2 targets. While structured RNAs were poor substrates for Rexo2 in vitro, single-stranded, unstructured RNA of various length (< 40 nucleotides in length) were degraded with some sequence specificity. How length- and sequence-dependent RNA degradation by Rexo2 is achieved on a molecular level is not fully understood.
Crystal structures of the human enzyme have been reported bound to nano-DNAs and -RNAs ranging from two to twelve residues in length (Chu et al. 2019, Kim et al. 2019, Nicholls et al. 2019, Szewczyk et al. 2020) (Fig. 4C). With one exception, only the 3′ dinucleotide is well resolved in those structures. Additional, partially discontinuous density was only modeled for a nano-DNA with seven residues (Chu et al. 2019). Accommodation of the longer DNA fragment required a sharp turn after the penultimate base, preventing some of the ‘phosphate cap’ residues (especially the conserved serine residues) to engage with the substrate. These residues were crucial for Orn's activity and tightly coordinate the 5′ phosphate of diribonucleotide substrates bound to Orn and Rexo2 (Kim et al. 2019). It also remains to be seen if the additional hydroxyl in the RNA backbone would allow a similar kinking of the substrate as seen with nano-DNAs. Furthermore, affinity measurements using the fluorescently labeled RNA and DNA fragments of different length showed only marginal difference (Chu et al. 2019), an indication that the additional contacts do not result in a tighter binding of the longer substrate. An independent study reported that Rexo2 crystallized in the presence of a longer RNA (7–11 nucleotides) (Szewczyk et al. 2020). In agreement with the previous study that used single-stranded DNA fragments, the last and penultimate 3′ nucleotides were well resolved at the active site. A sharp turn connects to the third-most 3′ nucleotide that is poorly resolved. The catalytic histidine residue of the DEDDh motif is also poorly resolved, with its Cβ position pointing away from the active site. Hence, it is not clear whether the model is representative of a catalytically competent state.
A third study on human Rexo2 reports structures that are identical to those reported previously alongside those of V. cholerae Orn (Kim et al. 2019, Nicholls et al. 2019). Hallmarks of these structures are the tight coordination of the entire 5′-phosphorylated diribonucleotide at a narrow active site. All catalytic residues for processing diribonucleotides, including divalent ion coordination and activation of the attacking water molecule, are in place. Biochemical experiments supported the strong preference of diribonucleotides that had also been observed for bacterial Orn.
In summary, the substrate-length preference of Rexo2 remains controversial. However, a common theme in kinetic studies is the extremely rapid degradation of diribonucleotides compared to the longer species, especially when native-like substrates are used. Structurally, the tight coordination of 5′-phosphorylated diribonucleotides at an optimally configured active site in a catalytically competent state would argue for a higher preference towards the shortest RNAs with only two nucleotide residues (Fig. 4).
Cleavage of dinucleotides by DHH-DHHA1 proteins
The DHH family of phosphoesterases is broadly distributed among diverse organisms and includes multiple major subfamilies of enzymes, such as Drosophila prune protein, pyrophosphatase, polyphosphatase, cyclic di-NMP phosphodiesterases, and RecJ exonucleases (Aravind and Koonin 1998, Srivastav et al. 2019). All of these proteins are defined by a unique N-terminal domain (‘DHH’; Pfam: PF01368). Within this domain is a conserved DHH sequence, which, along with neighboring acidic residues, coordinates two divalent metals that participate in the cleavage of phosphoester bonds (Commichau et al. 2019). For many of the bacterial subclasses of DHH family proteins, a second domain (‘DHHA1’; Pfam: PF02272) is connected by a short flexible linker (Aravind and Koonin 1998, Srivastav et al. 2019). While some subfamilies of proteins only include one of these two domains, such as alanyl-tRNA synthetase, which includes the DHHA1 domain but lacks DHH (Aravind and Koonin 1998), most protein architectures feature both domains (Fig. 5A). In addition to their core DHH-DHHA1 regions, some members of this overall protein family utilize additional domains, including but not limited to PAS, polyA polymerase, cystathionine-beta-synthase (CBS) and GGDEF domains. However, not all DHH-DHHA1 proteins contain these additional domains; indeed, many bacteria encode for standalone proteins consisting only of the DHH-DHHA1 domains. In total, DHH-DHHA1 proteins act on a wide variety of substrates (Commichau et al. 2019). For instance, some DHH-DHHA1 subfamilies act on linear RNAs while others process signaling nucleotides. For these reasons, some DHH-DHHA1 proteins have been found to exhibit cellular roles that may partially or fully overlap with short RNA-degrading enzymes, such as Orn.
Figure 5.
Overview of NrnA/B-type proteins. (A) Schematic architectures of certain DHH and DHHA1 proteins. (B) Representative alignment of NrnA-like proteins. Strictly conserved and similar residues are highlighted in red and orange, respectively. (C) Crystal structure of pAp-bound NrnA. The crystal structure of an enzyme dimer is shown in two nearly orthogonal views (monomer 1 shown in shades of pink, monomer 2 shown in shades of grey), highlighting the difference in inter-domain spacing (Schmier et al. 2017). Only the closed-state monomer carries a pAp molecule (yellow) at the active site. (D) Schematic view of the open and closed state of NrnA observed in the crystal structure.
NrnA, NrnA-Like Proteins, and NrnB
The initial discovery of NrnA resulted from a series of pulldown assays in which researchers were attempting to identify novel proteins that associate with 3-phosphoadenosine 5′-phosphate (pAp). A common source of pAp is from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) as a result of sulfate assimilation and coenzyme A metabolism. One of the enzymes that exhibits 3′,5′-bisphosphate (pAp) nucleotidase activity, which removes the 3′ phosphate to yield AMP, is CysQ. CysQ was shown to associate with pAp-coated agarose resin when it was incubated with E. coli lysates, consistent with its role in acting on pAp (Mechold et al. 2006). Orn was also found to associate with the pAp resin; however, purified Orn does not exhibit pAp phosphatase activity in vitro, although pAp can act as a competitive inhibitor to reduce Orn's processing of short RNAs (Mechold et al. 2006). Given the results of the pAp-agarose pulldown assays, researchers sought to identify proteins that bind or process pAp for the Gram-positive model organism Bacillus subtilis, which lacks both cysQ and orn. The incubation of B. subtilis lysates with pAp-bound agarose led to enrichment of HisIE, GuaC and YtqI (Mechold et al. 2007). YtqI is a standalone DHH-DHHA1 protein, lacking any additional domains, and was later renamed NrnA (as it will be referred to herein). Deletion of E. coli cysQ results in a requirement for supplemental cysteine and expression of B. subtilis nrnA could complement this phenotype, suggesting that NrnA might be capable of affecting pAp homeostasis, although other proteins may be involved (Mechold et al. 2007). However, the diminished growth phenotype of an E. coli strain that contained a conditional copy of orn was fully restored to wild-type levels by heterologous expression of B. subtilis nrnA, demonstrating that B. subtilis NrnA shares cellular functions with Orn. B. subtilis NrnA was purified and incubated with RNA substrates of varying lengths and the reaction products were analyzed by denaturing PAGE. Although only a pilot set of RNA substrates were analyzed in the initial study, it suggested that B. subtilis NrnA preferentially acts on RNAs less than 5 nucleotides in length, with a greatest preference for 3-mers. To expand upon this observation, B. subtilis NrnA protein was again purified in a subsequent study (Wakamatsu et al. 2011) and analyzed against a wider array of RNA substrates. It was also assayed alongside homologous proteins that had been purified from Thermus thermophilus (TTHA0118, herein referred to as NrnA) and Mycoplasma pneumoniae (MPN140) (Fig. 5B). Kinetic analyses of oligonucleotide cleavages confirmed that short nucleic acid substrates were strongly favored over longer substrates. For example, the kcat/Km values for 3-mer and 21-mer RNA substrates varied five orders of magnitude, strongly suggesting that shorter RNA substrates comprise the physiological targets of these enzymes. Interestingly, the T. thermophilus and M. pneumoniae proteins could also efficiently process pAp in addition to short oligonucleotides (i.e. less than 6 nucleotides in length). Using Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR), the reaction products were analyzed for these reactions. This revealed that the T. thermophilus and B. subtilis NrnA proteins both acted at the 5′ terminus of both RNA and DNA substrates. This contrasts with other published data, which argue that NrnA acts from the 3′ terminus for short substrates (Mechold et al. 2007, Schmier et al. 2017) and from the 5′ terminus for longer substrates (Schmier et al. 2017). Although the presence of a large 5′ fluorophore may have confounded the interpretation of some of the published data, the orientation of NrnA's exonucleolytic activity still remains to be conclusively resolved. Similarly, it is still unclear how much the individual activities and substrate preferences will vary between DHH-DHHA1 proteins from different bacteria and whether NrnA-like proteins represent a cohesive subfamily of proteins that display similar activities.
However, some clues have been recently obtained through high-resolution structural data of the B. subtilis NrnA protein, resolved in its apo form and bound to pAp or a nonhydrolyzable 3-mer RNA (Schmier et al. 2017) (Fig. 5C). The high-resolution structure of B. subtilis NrnA (PDB 5j21, 5izo, 5iuf) revealed a two-lobed structure that resembled structures of other DHH proteins (Commichau et al. 2019, Schmier et al. 2017). The overall architecture of the B. subtilis NrnA DHH domain consists of a ß-sheet, comprised of five ß-strands, enveloped by a layer of α-helices. Similarly, the neighboring DHHA1 domain also consists of a core β-sheet flanked by α-helices on either side. The protein crystallized as a dimer of dimers, which is consistent with the observation that NrnA purifies as a tetramer by size-exclusion chromatography (Wakamatsu et al. 2011). The catalytic site is located at the surface of the DHH domain, oriented toward the DHHA1 lobe, and the interface between the DHH and DHHA1 domains within individual monomers faces away from the dimerization interface. The DHHA1 domain is thought to aid in recognition of substrate(s); therefore, signature residues that are directly involved in substrate selection are likely to be located in this domain. It is in the region between the DHH and DHHA1 lobes where the substrate is thought to enter. This process is potentially impacted by mobility of the DHHA1 domain, which swings away or towards the DHH domain in open and closed states, respectively (Fig. 5C and D). While the apo form of the protein showed heterogeneity in the distances between the two domains, the structure of NrnA bound to a 3-mer RNA was fully locked in the closed conformation, with positively charged residues lining the surface of the mobile region. Two sequence motifs, RxRxR and GGGH, are conserved among NrnA-like proteins (e.g. Fig. 5B) and appear to participate in binding of substrate. The RxRxR residues interact directly with the phosphodiester backbone of co-crystallized RNA substrates, including recognition of the 5′ phosphate, while glycines from the GGGH further stabilize the substrate. While the GGGH motif is common amongst all DHH-DHHA1 proteins, the RxRxR motif may be more restricted to the NrnA subfamily. However, the full structural basis of NrnA's preference for shorter RNAs has not yet been fully resolved and it is not yet clear whether additional sequence motifs influence substrate recognition by other NrnA-like proteins. Nor has it been fully resolved whether subclasses of NrnA-like proteins might exhibit substrate preferences that differ from B. subtilis NrnA, although some clues have been obtained through biochemical and structural analyses of NrnA-like proteins from Mycobacterium tuberculosis (Rv2837c/CnpB) and Thermotoga maritima TM1595.
The M. tuberculosis Rv2837c protein (renamed CnpB (Yang et al. 2014)) shows 25% identity (and 41% similarity) to B. subtilis NrnA (Postic et al. 2012). Similarly, the high-resolution, three-dimensional structures of Rv2837c/CnpB revealed a global fold that closely resembles B. subtilis NrnA (He et al. 2016). Furthermore, expression of the M. tuberculosis cnpB gene within a conditional orn strain of E. coli resulted in partial complementation of growth, suggesting overlapping functions (Postic et al. 2012). Indeed, the cnpB gene was also able to complement E. coli cysQ as well. In vitro assays of RNA cleavage also showed that CnpB was capable of processing RNAs of varying lengths, although it exhibited a clear preference for shorter RNAs and a particular preference for dinucleotides (Postic et al. 2012). However, deletion of the M. tuberculosis cnpB gene also resulted in accumulation of the signaling nucleotide c-di-AMP and led to reduced virulence in a manner that is consistent with elevated c-di-AMP (Yang et al. 2014). This raised the hypothesis that CnpB might act on c-di-AMP directly. Biochemical assays further supported this claim by showing cleavage of c-di-AMP (Yang et al. 2014) and, to a lesser extent, c-di-GMP (He et al. 2016). CnpB was also shown to be capable of cleaving the linear dinucleotides pApA and pGpG (He et al. 2016, Yang et al. 2014), suggesting that cyclic di-NMPs might be processed fully to their nucleoside monophosphate constituents by CnpB. However, a separate structural and biochemical analysis of a close homolog from Thermotoga maritima (TM1595) supported the argument that the enzymes can cleave linear dinucleotides but cautioned that c-di-NMP cleavage may only occur under non-physiological concentrations (Drexler et al. 2017).
Similar observations were made for a close homolog of NrnA from Staphylococcus aureus, referred to as Pde2 (Bowman et al. 2016). Cellular data revealed that Pde2 affected S. aureus c-di-AMP signaling in vivo while biochemical data suggested that it preferentially hydrolyzed the linear diribonucleotide pApA in vitro, although limited cleavage of c-di-AMP was also observed after longer incubations. A relationship between c-di-AMP signaling and NrnA-like proteins was further observed in Streptococcal strains. For example, mutation of a gene encoding for Streptococcus pyogenes Pde2 triggered a variety of phenotypes that together are consistent with perturbation of c-di-AMP signaling (Fahmi et al. 2019), although it is currently unknown if S. pyogenes Pde2 acts directly on c-di-AMP. Similarly, two Streptococcus pneumoniae DHH-DHHA1 proteins, SPD_2032 (Pde1) and SPD_1153 (Pde2) were discovered to display c-di-AMP phosphodiesterase activity (Bai et al. 2013). This revealed that Pde1 preferentially processes c-di-AMP to its linear dinucleotide, while Pde2 can process c-di-AMP to AMP. Interestingly, Mycoplasma pneumoniae is also thought to encode two DHH-DHHA1 proteins that might exhibit different activities. M. pneumoniae MPN549 (PdeM) has been proposed to cleave c-di-AMP, while MPN140 (NrnA) was proposed to preferentially process short, linear RNAs (Blotz et al. 2017, Postic et al. 2012, Wakamatsu et al. 2011). Finally, a Streptococcus mutans NrnA-like homolog SMU1297 could complement E. coli strains containing either a cysQ mutant or a conditional orn (Postic et al. 2012, Zhang and Biswas 2009), confirming a role in cleavage of short RNAs.
There is yet another standalone DHH-DHHA1 protein, NrnB, that has been implicated in cleavage of short RNAs, including pGpG (Fang et al. 2009, Orr et al. 2018). Structural data are currently lacking for NrnB-like proteins; therefore, it is not yet clear what roles NrnB may serve in the bacteria that encode for it. Similar to NrnA, NrnB can complement an E. coli strain containing a conditional copy of orn (Fang et al. 2009, Orr et al. 2018). Moreover, the gene encoding NrnB is present in the genomes of some bacteria that lack both Orn and NrnA, suggesting that NrnB may exhibit Orn-like function.
GdpP
GdpP proteins also include DHH and DHHA1 domains but they occur alongside transmembrane spanning helices, a PAS domain and a degenerate GGDEF domain (Fig. 5A). GdpP proteins act as phosphodiesterases that specifically cleave c-di-AMP to release pApA (Corrigan et al. 2011, Huynh and Woodward 2016). This activity can be modulated by binding of b-type heme to the PAS domain (Rao et al. 2010, Rao et al. 2011, Tan et al. 2013). Their c-di-AMP phosphodiesterase activity can also be subjected to competitive inhibition by binding of (p)ppGpp (Bowman et al. 2016, Corrigan et al. 2015, Wang et al. 2017). These proteins are widespread in Firmicutes and their deletion results in elevated c-di-AMP levels, which results in a wide variety of c-di-AMP-related phenotypes (reviewed in (Commichau et al. 2019, Huynh and Woodward 2016). However, while some standalone DHH-DHHA1 proteins, such as NrnA, can act on short RNAs, GdpP proteins do not serve in this capacity; therefore, they are not considered in depth herein. Instead, the role(s) of these proteins in c-di-AMP signaling have been reviewed thoroughly elsewhere (Commichau et al. 2019, Stulke and Kruger 2020, Yin et al. 2020).
Together, the investigations of standalone DHH-DHHA1 proteins have shown that there is a common need in bacteria for proteins that act specifically on short RNAs and that without these proteins there is a bottleneck that deleteriously affects nucleotide recycling and signaling. Furthermore, these aggregate data demonstrate that some NrnA/B-like proteins fulfill this cellular role when orn is absent from the genome. However, the analyses of these proteins have also highlighted some of the challenges in annotating and predicting the substrate preferences of DHH-DHHA1 proteins. Some proteins exhibit a preference for linear oligonucleotides, while others may specialize in processing of cyclic dinucleotides. And perhaps some uncharacterized DHH-DHHA1 proteins might exhibit a preference for yet-to-be-described nucleic acid substrates. Because the linker region between DHH and DHHA1 domains allows great flexibility and access to bulk solvent, the active sites of different DHH-DHHA1 proteins might have evolved to accommodate a more diverse than expected range of nucleic acid substrates. Therefore, much more biochemical and structural data are needed for additional representatives of DHH-DHHA1 family proteins. Only then will the sequence and structural elements be discovered that are diagnostic for certain nucleic acid substrates.
Cleavage of dinucleotides by NrnC
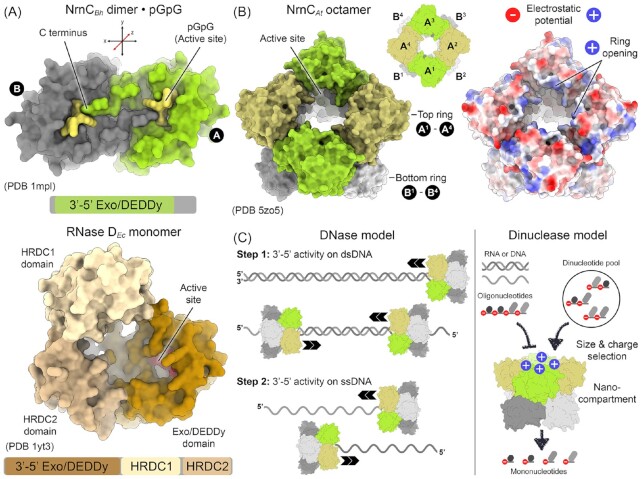
From the enzyme families that can substitute for Orn in P. aeruginosa and E. coli (Orr et al. 2018), NrnC belongs to one of the least characterized entities, both structurally and functionally. NrnC was discovered in a genome-wide screen using a library from the alphaproteobacterium Bartonella birtlesii. Specifically, expression of NrnC could rescue the growth defect associated with the conditional knock-down of orn in E. coli, suggesting overlapping biological activities of the two enzymes (Liu et al. 2012). In vitro, and similar to Orn, NrnC was able to successively degrade a 5′ fluorescently labeled oligoribonucleotide down to mononucleotides. Another similarity to Orn pertains to the essentiality of NrnC in several bacteria, including Brucella abortus, Bartonella henselae, and Caulobacter crescentus (Christen et al. 2011, Liu et al. 2012), indicating that accumulation of the substrates of these RNases is deleterious for cellular fitness. NrnC has been classified as a DEDDy-type exonuclease. Its closet relative is RNase D, a protein involved in the 3′ processing of structured RNAs (Zhang and Deutscher 1988). Unlike NrnC, which comprises a single DEDDy domain, RNase D contains two additional domains proposed to aid in substrate selection (Zuo et al. 2005) (Fig. 6A). The respective catalytic domains of RNase D and NrnC are similar but the proteins differ in their quaternary structure (Yuan et al. 2018, Zuo et al. 2005) (Fig. 6B). While RNase D is a monomer, NrnC from Agrobacterium tumefaciens forms an octamer with identical subunits. In this arrangement, four copies of NrnC form a ring-like structure, and two such rings stack in a head-to-head fashion (Yuan et al. 2018). The resulting octamer has a central cavity that is lined at the ring opening with positively charged residues (Fig. 6B). The active sites are located at the inside of the channel, at the midpoint of each ring. The central gateway of the NrnC octamer has a diameter that could accommodate double-stranded DNA and single-stranded DNA or RNA, but not double-stranded RNA. Such an activity profile was also observed in a purified system, leading to the proposal that NrnC can act as a processive DNase powered by nucleolytic events on the substrate (Fig. 6C, left panel). Where such an activity would come into play remains unknown.
Figure 6.
Structure of NrnC. (A) Dimer unit from a NrnC octamer. A homodimer is shown composed of two monomers colored in grey and green (the red coordinate axis highlights the rotation symmetry axis of the dimer) (Yuan et al. 2018). The C-terminal tail of one monomer reaches into the active site of an adjacent monomer. The bottom panel shows a RNase D monomer in comparison, with the DEDDy domain colored in dark brown, and the additional HRDC1 and HrDC2 domains colored in lighter hues of brown. (B) Octameric NrnC assembly. Four NrnC dimers assemble into a homo-octamer. The subunits colored in green form the top ring, the grey-colored subunits the bottom ring via tail-to-tail packing (see panel A). The active sites are midway through each ring, lining the central tunnel. Positively charged residues line the ring opening leading into the tunnel. (C) Models for NrnC function. Based on the geometrical constraints, A. tumefaciens apo-NrnC structures were interpreted to accommodate single-stranded RNA and DNA as well as double stranded DNA. Under certain experimental circumstances, DNase activity could be detected with purified enzyme, which led to the model presented in the left panel (Yuan et al. 2018). Another study showed narrow substrate specificity of B. heneselae NrnC with a strong preference for dinucleotides (Lormand et al. 2021). Here, an octameric assembly could serve as a nano-compartment that selects for 2-mer nucleotides (right panel).
A contemporary study presents the crystal structures of substrate-bound forms of B. henseleae and B. melitensis NrnC (Lormand et al. 2021). The overall architecture including an octameric assembly is conserved in NrnC orthologs characterized to date. (Lormand et al. 2021, Yuan et al. 2018). Structural analyses of the substrate complexes coupled with the biochemical approaches that had been established to re-evaluate Orn's substrate specificity revealed many parallels between NrnC and Orn, respectively (Kim et al. 2019, Lormand et al. 2021). Strikingly, the active site of both enzymes appears to be optimized for 5′-phosphorylated dinucleotides. In both cases, the region around the metal-coordinating DEDD motif is lined by 5′ phosphate-coordinating residues, a leucine wedge splaying the two substrate bases apart, a structural feature blocking off the 3′ end of the substrate, and an activation loop that coordinates the catalytic tyrosine or histidine residue, completing a catalytically competent state (Lormand et al. 2021). Longer substrates bind NrnC less tightly and appear to prevent the enzyme to adopt an active conformation, correlating with poor activity on RNA or DNA with more than two nucleotides. Since no general hydrolytic activity on double-stranded DNA was observed with these orthologs, it is possible that the peculiar NrnC octamer acts as a nano-compartment that attracts the smallest RNA (or DNA) fragments, i.e. dinucleotides, through a positive electrostatic potential at the outer-ring surface, with the narrow ring opening and optimized active site selecting against longer substrates (Fig. 6C, right panel). Notably, NrnC- and Orn-type enzymes usually do not co-occur and have evolved independently, indicating again the necessity to preserve dinuclease activity in an organism to support cellular fitness.
Possible physiological functions of linear dinucleotides
There are several instances in which linear dinucleotides have demonstrated functions in cells (Table 2). One function is in feedback inhibition of phosphodiesterase enzymes that cleave cyclic dinucleotides. For P. aeruginosa, a number of individual EAL- and HD-GYP-domain-containing phosphodiesterases have been shown to cleave c-di-GMP into pGpG (Kulasakara et al. 2006). The pGpG product is then cleaved primarily by Orn since the ∆orn mutant accumulates pGpG as compared to the parental strain (Orr et al. 2015). Not only is pGpG elevated in ∆orn, c-di-GMP levels are also elevated suggesting that pGpG is actively causing feedback inhibition on EAL and HD-GYP phosphodiesterases (Cohen et al. 2015, Orr et al. 2015). In an analogous scenario, c-di-AMP degradation in S. aureus also occurs by a two-step process involving two enzymes. C-di-AMP is linearized by GdpP into pApA which is subsequently hydrolyzed into AMP by Pde2 (NrnA) (Bowman et al. 2016). In ∆pde2 strains, pApA accumulates and causes feedback inhibition of GdpP. As a consequence, there is also elevated c-di-AMP in ∆pde2 mutant strain (Bowman et al. 2016). A third cyclic dinucleotide is c-GMP-AMP (cGAMP) made by related dinucleotide cyclases (DncV in V. cholerae (Davies et al. 2012), DncE in E. coli (Li et al. 2019, Whiteley et al. 2019) and Cdn in Geobacter sulfurreducens (Hallberg et al. 2016). In V. cholerae, cGAMP is cleaved by three proteins termed V-cGAPs (VCA0681, VCA0210 and VCA0933) (Gao et al. 2015). Since orn is essential in V. cholerae, it is unclear whether degradation of cGAMP occurs in a two-step process similar to c-di-GMP and c-di-AMP. Testing of purified proteins in vitro suggest that the primary product of V-cGAPs is pApG, and only VCA0681 can cleave the 5′ phosphate after cGAMP is depleted (Gao et al. 2015). Whether feedback inhibition by linearized cyclic dinucleotides on the phosphodiesterases is generalizable within cells remains to be determined.
Table 2.
Biological functions of diribonucleotides.
| Nucleotide | Function | References |
|---|---|---|
| pGpG | 1. Feedback inhibition of c-di-GMP phosphodiesterases 2. Nanopriming of transcripts |
(Cohen et al. 2015, Orr et al. 2015) (Druzhinin et al. 2015) |
| pApA | 1. Feedback inhibition of c-di-AMP phosphodiesterases | (Bowman et al. 2016) |
| pUpA | 1. Nanopriming of transcripts | (Druzhinin et al. 2015, Vvedenskaya et al. 2012) |
| NAD/NAD+ | 1. Cellular metabolism 2. Nanopriming of transcripts |
(Bird et al. 2016) (Vvedenskaya and Nickels 2020) |
| FAD | 1. Cellular metabolism 2. Nanopriming of transcripts |
(Vvedenskaya and Nickels 2020) |
Dinucleotides also serve as nano primers in cells (Nickels and Dove 2011). Elegant sequencing-based experiments revealed that depletion of Orn in P. aeruginosa led to a large proportion (∼40%) of transcripts to initiate at the −1 position (Goldman et al. 2011). This accumulation is restored upon expression of Orn or NrnB (Goldman et al. 2011). This shift in transcription start site led to a large number of genes (>1000) that is changed by more than 2-fold (Goldman et al. 2011). This large dysregulation of gene expression was attributed to the essentiality of Orn. The effect of dinucleotide priming was also observed in E. coli cells with endogenous levels of orn expression, but only in stationary phase, not exponential phase (Vvedenskaya et al. 2012). Further analysis in E. coli and V. cholerae revealed that two transcriptional start site sequences at −1/+1 consisting of TA and GG are primarily affected by expression of the NrnB dinuclease (Druzhinin et al. 2015, Orr et al. 2018). While the source of UA and GG dinucleotides that lead to the observed nanoprimer-dependent initiation is unknown, these dinucleotides are likely produced during the transition from exponential growth to stationary growth phase.
In addition to linear dinucleotides that can lead to altered priming and altered transcript stability, other dinucleotides in the cell, including nicotinamide adenine dinucleotide (NAD+) and reduced nicotinamide adenine dinucleotide (NADH), can also serve as initiating base in vivo for transcription start sites in which the + 1 position is an adenine (or + 1A promoters) (Bird et al. 2016). The addition of NAD + and NADH to the 5′ termini of RNAs, so called 5′ capping of the mRNA, increases the size of the transcript by 1 base. These NAD+/NADH 5′ caps are sensitive to NudC that removes the 5′ cap of the mRNA, but the triphosphate mRNAs are resistant. The mRNA capped with NAD+/NADH have a 3–4 increase in half-life indicating another important mechanism for regulating gene expression (Bird et al. 2016). Flavine adenine dinucleotide (FAD) is another dinucleotide that can cap + 1A promoters in vitro (Julius and Yuzenkova 2017). Not only have these caps been identified in bacteria, capping with NAD + and NADH is observed in human and yeast mitochondrial RNA indicating that 5′ capping is likely a widespread and conserved process that is responsive to cellular metabolism (Bird et al. 2018, Jiao et al. 2017). Since + 1A promoters are fairly ubiquitous, there are likely additional determinants for selecting transcripts to be capped. Using a high-throughput sequencing approaching in combination with decapping enzymes (Vvedenskaya and Nickels 2020), termed CapZyme-seq, revealed promoters that are subject to non- canonical initiating nucleotides (Vvedenskaya et al. 2018). These results suggest that there may be a subset of genes that is responsive to NAD + capping depending on the metabolic changes in the cell.
Another mechanism whereby linear dinucleotides can act to alter cell physiology is to directly target proteins either by competition for active site or binding to allosteric sites. In analogy to cyclic dinucleotide binding proteins (Cohen et al. 2015, Lacey et al. 2010, Orr et al. 2015), linear dinucleotides may bind competitively at the active site (feedback inhibition), bind at allosteric sites (Chan et al. 2004, Christen et al. 2006, De et al. 2009, Lee et al. 2007, Morgan et al. 2014) or binding at sites known to bind related molecules (Chin et al. 2010, Leduc and Roberts 2009, Tao et al. 2010). In addition to pGpG, pApA, pApG binding to the active site of phosphodiesterases that linearize c-di-GMP, c-di-AMP, and c-GAMP, respectively, exoribonucleases that can produce dinucleotides may also be subject to competition at the active site. These exoribonucleases include RNase R (Rnr) (Matos et al. 2009, Matos et al. 2011) and possibly other exonuleases. As for allosteric binding, dinucleotides bind catalytically inactive EAL and HD-GYP proteins that no longer have activity against c-di-GMP (Orr et al. 2015) and may serve as a dinucleotide binding protein. Lastly, proteins that bind NAD, FAD and other natural dinucleotides may be susceptible to competition by linear dinucleotides. Additional investigation in the future can lead to the identification of dinucleotide binding proteins that may explain the essentiality of dinucleases.
Currently, there is little known regarding the function of longer oligonucleotides. However, in analogy to the cellular effects of linear dinucleotides, it is conceivable that there may be cellular targets or pathways impacted by trinucleotides and longer oligonucleotides awaiting discovery.
Conclusions
The differences in the biochemical activities of diribonucleases and oligoribonuclease observed in various studies is a key issue that should be resolved with further studies to determine the function of these enzymes in a physiologically context. With recent advances in RNA sequencing and genetic methods, as well as deep structural and mechanistic insight into enzyme function, it is timely to revisit the role of each and every RNases in a cellular context. These studies should be guided by questions regarding substrate specificity and enzymatic mechanism, but also taking into account the multitude of modifications of the RNA backbone, termini and bases that make RNA such a versatile biopolymer. Other levels of complexity arise from the regulation of RNA turnover, the biological function of its intermediates, and the crosstalk between cell signaling and housekeeping pathways. The recent discoveries of a multitude of novel signaling nucleotides, including a broader range of cyclic dinucleotides and cyclic trinucleotides opens up questions of their biological function and regulation (Lau et al. 2020, Whiteley et al. 2019). Future studies will have to incorporate these emerging links for an integrated view of the biological function of nano-RNAs.
Acknowledgements
This work was supported by funding from National Institutes of Health (NIH) R01-AI110740 (V.T.L, H.S. and W.W). We thank members of our labs for critical reading of the manuscript.
Contributor Information
Vincent T Lee, Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD 20742 United States.
Holger Sondermann, CSSB Centre for Structural Systems Biology, Deutsches Elektronen-Synchrotron DESY, Notkestr. 85, 22607 Hamburg, Germany.
Wade C Winkler, Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD 20742 United States.
Conflict of interest
None declared.
References
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–30. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem Sci. 1998;23:17–19. [DOI] [PubMed] [Google Scholar]
- Asha PK, Blouin RT, Zaniewski Ret al. Ribonuclease BN: identification and partial characterization of a new tRNA processing enzyme. Proc Natl Acad Sci. 1983;80:3301–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avey HP, Boles MO, Carlisle CHet al. Structure of ribonuclease. Nature. 1967;213:557–62. [DOI] [PubMed] [Google Scholar]
- Bai Y, Yang J, Eisele LEet al. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol. 2013;195:5123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barondes SH, Nirenberg MW. Fate of a synthetic polynucleotide directing cell-free protein synthesis I. Characteristics of degradation. Science. 1962;138:810–3. [DOI] [PubMed] [Google Scholar]
- Bird JG, Basu U, Kuster Det al. Highly efficient 5′ capping of mitochondrial RNA with NAD(+) and NADH by yeast and human mitochondrial RNA polymerase. Elife. 2018;7:e42179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird JG, Zhang Y, Tian Yet al. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature. 2016;535:444–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blotz C, Treffon K, Kaever Vet al. Identification of the components involved in cyclic Di-AMP signaling in Mycoplasma pneumoniae. Front Microbiol. 2017;8:1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman L, Zeden MS, Schuster CFet al. New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus. J Biol Chem. 2016;291:26970–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni F, Gramegna P, Oliveira JMet al. REXO2 is an oligoribonuclease active in human mitochondria. PLoS One. 2013;8:e64670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray Det al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A. 2004;101:17084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–9. [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Zuo Y, Li Zet al. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–80. [DOI] [PubMed] [Google Scholar]
- Chin KH, Yang CY, Chou CCet al. The crystal structure of XC847 from Xanthomonas campestris: a 3′-5′ oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix. Proteins. 2006;65:1036–40. [DOI] [PubMed] [Google Scholar]
- Chin KH, Lee YC, Tu ZLet al. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396:646–62. [DOI] [PubMed] [Google Scholar]
- Christen B, Christen M, Paul Ret al. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281:32015–24. [DOI] [PubMed] [Google Scholar]
- Christen B, Abeliuk E, Collier JMet al. The essential genome of a bacterium. Mol Syst Biol. 2011;7:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu LY, Agrawal S, Chen YPet al. Structural insights into nanoRNA degradation by human Rexo2. RNA. 2019;25:737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Mechold U, Nevenzal Het al. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2015;112:11359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Heidemann JL, Ficner Ret al. Making and breaking of an essential poison: the cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in bacteria. J Bacteriol. 2019;201:e00462–00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne Het al. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Bowman L, Willis ARet al. Cross-talk between two nucleotide-signaling pathways in Staphylococcus aureus. J Biol Chem. 2015;290:5826–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta AK, Niyogi K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J Biol Chem. 1975;250:7313–9. [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TSet al. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V . cholerae virulence. Cell. 2012;149:358–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De N, Navarro MV, Raghavan RVet al. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J Mol Biol. 2009;393:619–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–91. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Twenty years of bacterial RNases and RNA processing: how we've matured. RNA. 2015;21:597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Marlor CW, Zaniewski R. Ribonuclease T: new exoribonuclease possibly involved in end-turnover of tRNA. Proc Natl Acad Sci U S A. 1984;81:4290–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Marlor CW, Zaniewski R. RNase T is responsible for the end-turnover of tRNA in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:6427–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Marshall GT, Cudny H. RNase PH: an Escherichia coli phosphate-dependent nuclease distinct from polynucleotide phosphorylase. Proc Natl Acad Sci U S A. 1988;85:4710–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler DJ, Muller M, Rojas-Cordova CAet al. Structural and biophysical analysis of the soluble DHH/DHHA1-Type phosphodiesterase TM1595 from Thermotoga maritima. Structure. 2017;25:1887–97. e1884. [DOI] [PubMed] [Google Scholar]
- Druzhinin SY, Tran NT, Skalenko KSet al. A conserved pattern of primer-dependent transcription initiation in Escherichia coli and Vibrio cholerae revealed by 5′ RNA-seq. PLos Genet. 2015;11:e1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmi T, Faozia S, Port GCet al. The second messenger c-di-AMP regulates diverse cellular pathways involved in stress response, biofilm formation, cell wall homeostasis, SpeB expression, and virulence in Streptococcus pyogenes. Infect Immun. 2019;87:e00147–00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Zeisberg WM, Condon Cet al. Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis. Nucleic Acids Res. 2009;37:5114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellner P, Sanger F. Sequence analysis of specific areas of the 16S and 23S ribosomal RNAs. Nature. 1968;219:236–8. [DOI] [PubMed] [Google Scholar]
- Findlay D, Herries DG, Mathias APet al. The active site and mechanism of action of bovine pancreatic ribonuclease. Nature. 1961;190:781–4. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Cheung J, Rudolph MJet al. Structural genomics for drug design against the pathogen Coxiella burnetii. Proteins. 2015;83:2124–36. [DOI] [PubMed] [Google Scholar]
- Frazao C, McVey CE, Amblar Met al. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–4. [DOI] [PubMed] [Google Scholar]
- Gao J, Tao J, Liang Wet al. Identification and characterization of phosphodiesterases that specifically degrade 3′3′-cyclic GMP-AMP. Cell Res. 2015;25:539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh RK, Deutscher MP. Identification of an Escherichia coli nuclease acting on structurally altered transfer RNA molecules. J Biol Chem. 1978;253:997–1000. [PubMed] [Google Scholar]
- Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci U S A. 1999;96:4372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SR, Sharp JS, Vvedenskaya IOet al. NanoRNAs prime transcription initiation in vivo. Mol Cell. 2011;42:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg ZF, Wang XC, Wright TAet al. Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3′, 3′-cGAMP). Proc Natl Acad Sci USA. 2016;113:1790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan S, Carr PD, Brown SEet al. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10:535–46. [DOI] [PubMed] [Google Scholar]
- He Q, Wang F, Liu Set al. Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase. J Biol Chem. 2016;291:3668–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao YY, Yang CC, Lin CLet al. Structural basis for RNA trimming by RNase T in stable RNA 3′-end maturation. Nat Chem Biol. 2011;7:236–43. [DOI] [PubMed] [Google Scholar]
- Huynh TN, Woodward JJ. Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol. 2016;30:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Doamekpor SK, Bird JGet al. 5′ End nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-Mediated deNADding. Cell. 2017;168:1015–27. e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. The action of boiled pancreas extract on yeast nucleic acid. Am J Physiol. 1920;52:203–7. [Google Scholar]
- Julius C, Yuzenkova Y. Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic Acids Res. 2017;45:8282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp HD, Patimalla-Dipali B, Lazinski DWet al. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 2013;9:e1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartha G, Bello J, Harker D. Tertiary structure of ribonuclease. Nature. 1967;213:862–5. [DOI] [PubMed] [Google Scholar]
- Kelly KO, Deutscher MP. The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli. J Bacteriol. 1992;174:6682–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lormand JD, Weiss CAet al. A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation. Elife. 2019;8:e46313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Sondermann H. Versatile modes of cellular regulation via cyclic dinucleotides. Nat Chem Biol. 2017;13:350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasakara H, Lee V, Brencic Aet al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci USA. 2006;103:2839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitz M. Isolation from beef pancreas of a crystalline protein possessing ribonuclease activity. Science. 1939;90:112–3. [DOI] [PubMed] [Google Scholar]
- Kunitz M. Crystalline Ribonuclease. J Gen Physiol. 1940;24:15–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey MM, Partridge JD, Green J. Escherichia coli K-12 YfgF is an anaerobic cyclic di-GMP phosphodiesterase with roles in cell surface remodelling and the oxidative stress response. Microbiology. 2010;156:2873–86. [DOI] [PubMed] [Google Scholar]
- Lau RK, Ye Q, Birkholz EAet al. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol Cell. 2020;77:723–33. e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191:7121–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Park SH, Jeong CSet al. Structural basis of small RNA hydrolysis by oligoribonuclease (CpsORN) from Colwellia psychrerythraea strain 34H. Sci Rep. 2019;9:2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Matewish JM, Kessler JLet al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Cimdins A, Rohde Met al. DncV synthesizes cyclic GMP-AMP and regulates biofilm formation and motility in Escherichia coli ECOR31. Mbio. 2019;10:e02492–02418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. Maturation pathways for E . coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–12. [DOI] [PubMed] [Google Scholar]
- Liu MF, Cescau S, Mechold Uet al. Identification of a novel nanoRNase in Bartonell a. Microbiology. 2012;158:886–95. [DOI] [PubMed] [Google Scholar]
- Lormand JD, Kim S-K, Walters-Marrah GAet al. Structural characterization of NrnC identifies unifying features of dinucleotidases. Elife. 2021;10:e70146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos RG, Barbas A, Arraiano CM. RNase R mutants elucidate the catalysis of structured RNA: RNA-binding domains select the RNAs targeted for degradation. Biochem J. 2009;423:291–301. [DOI] [PubMed] [Google Scholar]
- Matos RG, Barbas A, Gomez-Puertas Pet al. Swapping the domains of exoribonucleases RNase II and RNase R: conferring upon RNase II the ability to degrade ds RNA. Proteins. 2011;79:1853–67. [DOI] [PubMed] [Google Scholar]
- Mechold U, Ogryzko V, Ngo Set al. Oligoribonuclease is a common downstream target of lithium-induced pAp accumulation in Escherichia coli and human cells. Nucleic Acids Res. 2006;34:2364–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Fang G, Ngo Set al. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007;35:4552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S, Stein WH. Chemical structures of pancreatic ribonuclease and deoxyribonuclease. Science. 1973;180:458–64. [DOI] [PubMed] [Google Scholar]
- Morgan JL, McNamara JT, Zimmer J. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol. 2014;21:489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LH, Erzberger JP, Root Jet al. The human homolog of Escherichia coli Orn degrades small single-stranded RNA and DNA oligomers. J Biol Chem. 2000;275:25900–6. [DOI] [PubMed] [Google Scholar]
- Nicholls TJ, Spahr H, Jiang Set al. Dinucleotide degradation by REXO2 maintains promoter specificity in mammalian mitochondria. Mol Cell. 2019;76:784–796 e786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels BE, Dove SL. NanoRNAs: a class of small RNAs that can prime transcription initiation in bacteria. J Mol Biol. 2011;412:772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev N, Folsom V, Schlessinger D. Escherichia coli mutants deficient in exoribonucleases. Biochem Biophys Res Commun. 1976;70:920–4. [DOI] [PubMed] [Google Scholar]
- Nirenberg M. Historical review: deciphering the genetic code–a personal account. Trends Biochem Sci. 2004;29:46–54. [DOI] [PubMed] [Google Scholar]
- Nirenberg MW, Matthaei JH. The dependence of cell-free protein synthesis in e . coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci USA. 1961;47:1588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi SK, Datta AK. A novel oligoribonuclease of Escherichia coli. I. Isolation and properties. J Biol Chem. 1975;250:7307–12. [PubMed] [Google Scholar]
- Orr MW, Donaldson GP, Severin GBet al. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci USA. 2015;112:E5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Weiss CA, Severin GBet al. A subset of exoribonucleases serve as degradative enzymes for pGpG in c-di-GMP signaling. J Bacteriol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palace SG, Proulx MK, Lu Set al. Genome-wide mutant fitness profiling identifies nutritional requirements for optimal growth of Yersinia pestis in deep tissue. MBio. 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CCet al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski Y, Berg K, Klebl DPet al. Characterization of an intertidal zone metagenome oligoribonuclease and the role of the intermolecular disulfide bond for homodimer formation and nuclease activity. FEBS Open Bio. 2019;9:1674–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic G, Danchin A, Mechold U. Characterization of NrnA homologs from Mycobacterium tuberculosis and Mycoplasma pneumoniae. RNA. 2012;18:155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, Ji Q, Soehano Iet al. Unusual heme-binding PAS domain from YybT family proteins. J Bacteriol. 2011;193:1543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, See RY, Zhang Det al. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Yet al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–81. [DOI] [PubMed] [Google Scholar]
- Schmidt G, Levene PA. The effect of nucleophosphatase on “Native” and depolymerized thymonucleic acid. Science. 1938;88:172–3. [DOI] [PubMed] [Google Scholar]
- Schmier BJ, Nelersa CM, Malhotra A. Structural basis for the bidirectional activity of Bacillus nanoRNase NrnA. Sci Rep. 2017;7:11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman JG, Schmidt FJ, Foss Ket al. A mutant of Escherichia coli defective in removing 3′ terminal nucleotides from some transfer RNA precursor molecules. Cell. 1975;5:389–400. [DOI] [PubMed] [Google Scholar]
- Singer MF, Tolbert G. Purification and properties of a potassium-activated phosphodiesterase (RNAase II) from Escherichia coli. Biochemistry. 1965;4:1319–30. [DOI] [PubMed] [Google Scholar]
- Spahr PF, Schlessinger D. Breakdown of messenger ribonucleic acid by a Potassium-activated phosphodiesterase from Escherichia coli. J Biol Chem. 1963;238:PC2251–2253. [PubMed] [Google Scholar]
- Srivastav R, Sharma R, Tandon Set al. Role of DHH superfamily proteins in nucleic acids metabolism and stress tolerance in prokaryotes and eukaryotes. Int J Biol Macromol. 2019;127:66–75. [DOI] [PubMed] [Google Scholar]
- Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–8. [DOI] [PubMed] [Google Scholar]
- Stevens A, Niyogi SK. Hydrolysis of oligoribonucleotides by an enzyme fraction from Escherichia coli. Biochem Biophys Res Commun. 1967;29:550–5. [DOI] [PubMed] [Google Scholar]
- Stulke J, Kruger L. Cyclic di-AMP signaling in Bacteria. Annu Rev Microbiol. 2020;74:159–79. [DOI] [PubMed] [Google Scholar]
- Szewczyk M, Malik D, Borowski LSet al. Human REXO2 controls short mitochondrial RNAs generated by mtRNA processing and decay machinery to prevent accumulation of double-stranded RNA. Nucleic Acids Res. 2020;48:5572–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal R, Wong HC, Calhoon Ret al. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E, Rao F, Pasunooti Set al. Solution structure of the PAS domain of a thermophilic YybT protein homolog reveals a potential ligand-binding site. J Biol Chem. 2013;288:11949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao F, He YW, Wu DHet al. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol. 2010;192:1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Nickels BE. CapZyme-Seq: a 5′-RNA-Seq method for differential detection and quantitation of NAD-Capped and uncapped 5′-Triphosphate RNA. STAR Protoc. 2020;1:100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Sharp JS, Goldman SRet al. Growth phase-dependent control of transcription start site selection and gene expression by nanoRNAs. Genes Dev. 2012;26:1498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Bird JG, Zhang Yet al. CapZyme-Seq comprehensively defines promoter-sequence determinants for RNA 5′ capping with NAD<sup/>. Mol Cell. 2018;70:553–64. e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu T, Kim K, Uemura Yet al. Role of RecJ-like protein with 5′-3′ exonuclease activity in oligo(deoxy)nucleotide degradation. J Biol Chem. 2011;286:2807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Davlieva M, Reyes Jet al. A novel phosphodiesterase of the GdpP family modulates cyclic di-AMP levels in response to cell membrane stress in daptomycin-resistant enterococci. Antimicrob Agents Chemother. 2017;61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–8. [DOI] [PubMed] [Google Scholar]
- Whiteley AT, Eaglesham JB, de Oliveira Mann CCet al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff HW, Hardman KD, Allewell NMet al. 1967; The structure of ribonuclease-S at 3.5 A resolution. J Biol Chem. 2019;242:3984–8. [PubMed] [Google Scholar]
- Yang J, Bai Y, Zhang Yet al. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol Microbiol. 2014;93:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Cai X, Ma Het al. A decade of research on the second messenger c-di-AMP. FEMS Microbiol Rev. 2020;44:701–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Deutscher MP. Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli. J Bacteriol. 1995;177:4137–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Gao F, Yin Ket al. NrnC, an RNase D-Like protein from Agrobacterium, is a novel octameric nuclease that specifically degrades dsDNA but leaves dsRNA intact. Front Microbiol. 2018;9:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaniewski R, Deutscher MP. Genetic mapping of mutation in Escherichia coli leading to a temperature-sensitive RNase D. Mol Gen Genet. 1982;185:142–7. [DOI] [PubMed] [Google Scholar]
- Zaniewski R, Petkaitis E, Deutscher MP. A multiple mutant of Escherichia coli lacking the exoribonucleases RNase II, RNase D, and RNase BN. J Biol Chem. 1984;259:11651–3. [PubMed] [Google Scholar]
- Zhang J, Biswas I. 3′-Phosphoadenosine-5′-phosphate phosphatase activity is required for superoxide stress tolerance in Streptococcus mutans. J Bacteriol. 2009;191:4330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JR, Deutscher MP. Transfer RNA is a substrate for RNase D in vivo. J Biol Chem. 1988;263:17909–12. [PubMed] [Google Scholar]
- Zhang X, Zhu L, Deutscher MP. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J Bacteriol. 1998;180:2779–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Wang Y, Malhotra A. Crystal structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure. 2005;13:973–84. [DOI] [PubMed] [Google Scholar]