Abstract
Food security has become a pressing issue in the modern world. The ever-increasing world population, ongoing COVID-19 pandemic, and political conflicts together with climate change issues make the problem very challenging. Therefore, fundamental changes to the current food system and new sources of alternative food are required. Recently, the exploration of alternative food sources has been supported by numerous governmental and research organizations, as well as by small and large commercial ventures. Microalgae are gaining momentum as an effective source of alternative laboratory-based nutritional proteins as they are easy to grow under variable environmental conditions, with the added advantage of absorbing carbon dioxide. Despite their attractiveness, the utilization of microalgae faces several practical limitations. Here, we discuss both the potential and challenges of microalgae in food sustainability and their possible long-term contribution to the circular economy of converting food waste into feed via modern methods. We also argue that systems biology and artificial intelligence can play a role in overcoming some of the challenges and limitations; through data-guided metabolic flux optimization, and by systematically increasing the growth of the microalgae strains without negative outcomes, such as toxicity. This requires microalgae databases rich in omics data and further developments on its mining and analytics methods.
Keywords: microalgae, omics, machine learning, alternative proteins, systems biology
Statements of significance.
Microalgae can be a promising platform for alternative protein production. However, there are several challenges related to cultivation costs, protein extraction, and processing, as well as taste and sensory properties. The employment of modern omics techniques, artificial intelligence (AI), and advanced data analytics can help unleash the full potential of microalgae in alternative protein production.
Introduction
The world is currently facing food security and cost inflation challenges, where millions of people are struggling for physical and economic access to sufficient, safe, and nutritious food [1]. By 2050, it is estimated that the world will have ∼10 billion people to feed [2]. In times of exceptional events, such as the current climate changes affecting large parts of the world with severe drought and floods, together with recurrent pandemics and the ongoing Russia-Ukraine conflict, the food security problem severely manifests itself with added challenges in the food supply chain and reduced food production over the coming seasons. Moreover, the recent volatility in oil prices also adds immense stress to common people worldwide, notably in the developing world, as the overall cost of food is significantly increasing [3]. Collectively, these issues pose an unprecedented threat, as current agricultural approaches and food distribution systems are both unable to cover the current need and cope with the increased nutritional demands for the future [4].
Proteins, which are vital components of food, are getting more traction recently as any forthcoming food output should be nutritional. The current major source of protein is plant-based, and increasing its production to match the demand with traditional agricultural approaches is highly resource-intensive with an overall negative environmental impact [5]. The production of animal-based proteins, on top of concerns regarding animal welfare and healthy diets, is also a burden to agricultural land use and water resources and, thus, increases greenhouse gas emissions [6]. The current food production system accounts for between 20% and 30% of the total environmental impact and for almost 30% of global greenhouse gas emissions [5]. Therefore, there is an urgent need for a transformation in the food system that supports its availability, accessibility, affordability, and desirability [7].
Novel plant-based and laboratory-grown meat protein alternatives have recently been developed [8,9]. Several plant-based products are successfully commercialized, and the first laboratory-grown meat product has recently been approved by the authorities in Singapore [10]. Although this is a positive new direction toward the alternative food initiative, especially imminent in current food challenges [3], such novel food products usually involve additives, genetic modifications, or synthetic modifications that may improve the product safety and quality; however, they do not yet meet the increasing consumer demand for healthy, vegan, or natural food products [[11], [12], [13]]. Furthermore, although plant proteins are typically insufficient in essential amino acids compared to animal proteins, laboratory-grown meat faces religious and cultural challenges [14]. A new solution is needed to provide protein without causing climate change, deforestation, and depletion of water resources.
Alternative Proteins from Single-Cell Organisms
Compared with plant or meat proteins, producing protein from single-cell organisms is thought to be a good alternative for several reasons: 1) they require less water; 2) wastewater can be recycled within the process; 3) they need little land area; 4) they are less harmful to the environment with minimal contribution to climate change (as they have a smaller net carbon footprint, through releasing oxygen and sequestrating carbon dioxide); 5) unlike plants, they do not require herbicides, fertilizers, or antibiotics; 6) they can be cultured on non-agricultural land; and 7) their positive impact on human health is highlighted as many of these organisms are already used as supplements for human or in traditional medications [[15], [16], [17], [18], [19]].
Therefore, single-cell organisms, such as bacteria, yeasts, or algae, might be promising sources of alternative proteins [20]. They were reported to have high protein concentration and contain amounts of indispensable amino acids (IAAs) that are important for health, growth, and pregnancy, and they are usually obtained from meat-based proteins or plant alternatives [21]. Additionally, the amount of protein in some of these organisms was shown to be relatively similar to oilseed plants, and they can be mass-produced using standard bioreactors [22].
There already are several examples of single-cell organisms being included in human diets either as food or ingredients (e.g., mushrooms and yeast). Fungi, such as Fusarium venenatum which grows on carbohydrate substrates, eliminating the need for an exogenous protein input, produces mycoproteins that are used as meat and chicken analogs, and are available in markets of many countries [23]. Bacteria, which are a very rich source of protein, have long been included in human diets, mainly for fermenting food and as probiotic drinks [24,25]. However, the high contents of nucleic acids in bacteria, which can be harmful, require further technological development for neutralization [20].
Microalgae-based protein products can be classified into 5 groups based on their protein content and the degree of purification. A whole-cell protein levels range from 60% to 89% with ∼40%–50% comprising of protein components, protein concentrates, isolates, hydrolysates, and bioactive peptides [26]. The microaglae protein components are characterized by their slightly denatured protein state compared with other types because of their preserved intact rigid cell wall and membranes during processing; however, it has been reported to be poorly digestible [27]. To produce protein concentrates and isolates, the host-cell protein is subjected to several steps of extraction and purification using chemicals and heat. Subsequently, the extracted protein is subjected to enzymatic degradation into smaller peptides to form protein hydrolysates and bioactive peptides that correlate with positive contributions to human physiology functions [26,28].
Microalgae as Potential Biofactory Platform
Various microorganisms, including bacteria, mushrooms, yeasts, and microalgae, have been explored as sources for alternative proteins [20,29]. Microalgae form a diverse group containing ∼200,000 species of photosynthetic and heterotrophic microorganisms with morphological, physiological, and genetic diversity [30] (Table 1). Some microalgae species such as Arthrospira platensis, Spirulina maxima, Chlorella vulgaris, and Chlorella pyrenoidosa contain up to 70% of the dry cell weight (DCW) as protein [31]. Such high protein concentration exceeds the protein levels found in many fungi including several Aspergillus, Fusarium, and Saccharomyces species that were reported to have concentrations ranging from 40% to 50% (DCW) [32,33] and are comparable with some types of bacteria [34]. In addition, microalgae have less nucleic acid and their amino acid profile is closer to conventional protein sources such as eggs and oyster larvae [35,36]. Yet, they differ widely in terms of cellular composition and metabolic characteristics [24].
TABLE 1.
Microalgae classification and examples of some key metabolites
| Class | Species | Metabolite | Biological activities |
|---|---|---|---|
| Cyanophyceae (blue-green algae) | Arthrospira platensis | c-phycocyanin | Antitumor, antioxidant, antibacterial, anti-inflammatory, hepatoprotective |
| Arthrospira maxima | |||
| Chlorophyceae (green algae) | Chlorella sp. | β-1,3-glucan-peptides | Immune-stimulating, antioxidant, blood lipid reducing, antitumor |
| Haematococcus pluvialis | Astaxanthin | Antioxidant, antihypertensive, anti-inflammatory, anticancer, photoprotective | |
| Dunaliella salina | β-carotene | Antioxidant, anticancer, eye protective | |
| Porphyridiophyceae (red algae) | Porphyridium sp. | Phycoerythrin-polyunsaturated fatty acids | Antiviral (herpes), antibacterial, antioxidant, immunomodulatory |
| Bacillariophyceae | Phaeodactylum tricornutum | Fucuxanthin | Insulin resistance improving, anticancer, anti-inflammatory, eye- and cardiovascular-protective |
In response to external conditions, microalgae synthesize different metabolites (primary and secondary), many of which are of significant health, nutritional, and industrial value [37]. Some microalgae can produce compounds with anticarcinogenic, anti-inflammatory, immunomodulatory, antimicrobial, antioxidative, antihypertensive, and anticoagulant activities [38]. They can also produce environmentally friendly agricultural compounds such as plant biostimulants [39]. In addition, microalgae are known to grow effectively using CO2 as a carbon source, making them play significant roles in carbon capture while producing alternative proteins for food production. On scaling up efforts, they are promising for environmental and sustainable biomanufacturing. Therefore, microalgae present a promising platform for pharmaceutical and industrial applications, as the cultivation processes can be optimized in a controlled culture. A better understanding of microalgae biology, genetics, and cultivation processes will lead to a bioproduction platform that is environmentally friendly and economically viable [37].
Over the last 2 decades, microalgae research has attracted great interest for the potential to produce high-value compounds, biofuels, and waste effluent remediation [40]. Previously, in addition to the cost of harvesting, extraction, and refinement, most of the research was geared toward biofuel application where the photosynthesis requirement was a significant cost barrier [41]. It currently is estimated that the cost of producing 1 L of microalgae-based biodiesel in photobioreactors with 60% oil content is $3.96–$10.56. This is 10-fold greater than the cost of producing 1 L of soybean-based biodiesel [42]. From 70%–80% of the total cost is attributed to the downstream processing of microalgal biomass where energy consumption represents the most expensive factor in the process [43]. Microalgae-based waste removal of chemicals or nutrients might be achieved by either conversion to different forms or accumulation in biomass. However, harvesting at low cost and dealing with contaminants are still potential challenges for its wider applications [[44], [45], [46]].
Microalgae Utilization in Food and Health Industries
Microalgae can be used as a key platform for alternative protein production due to several positive reasons: 1) Some microalgae contain relatively high concentrations of essential amino acids including lysine, tryptophan, methionine, threonine, histidine, valine, and isoleucine that are comparable with those reported in oyster larvae and eggs [35,36]; 2) they also contain polyunsaturated fatty acids including omega-3 fatty acids, such as docosahexaenoic acid and eicosapentaenoic acid, carotenoids, chlorophyll, and pigments; 3) they can be used as a source of vitamins such as vitamin A, ascorbic acid (vitamin C), nicotinic acid, tocopherol (vitamin E), thiamin (vitamin B1), riboflavin (vitamin B2), pyridoxin (vitamin B-6), cobalamin (vitamin B-12), folic acid, pantothenic acid, and biotin (vitamin H), in addition to fiber and other valuable nutrients [47]; 4) some microalgae are heterotrophic where they can grow on both light and carbon source at comparable rates to other microbes; and 6) they require simple and defined media to grow [24,48,49]. Furthermore, many microalgae strains are high-protein producers with up to 70% dry weight (DW) protein contents. Additionally, compared to bacteria, microalgae have a relatively low content of nucleic acids, which makes them more suitable for edible applications [50]. They can be grown using traditional fermentation approaches or bioreactors; although growing microalgae in open ponds is also common, it risks biological and chemical contaminations [51].
In recent years, employing microalgae as an innovative platform for food, feed, and health products is generating immense interest. The primary focus of these attempts was to use microalgae to produce compounds that are used as food supplements, additives, or ingredients [48]. Currently, microalgae products are available in several pharmaceutical forms such as tablets, liquid or capsules, and baking ingredients for pasta and snacks [24,48]. These products include β-glucan content, food ingredients, and whole-cell products produced using food-grade microalgae species such as Arthrospira, Arthrospira, Chlorella, Dunaliella, Aphanizomenon, Euglena [[52], [53], [54]]. Microalgae are also used to produce feed for aquaculture, where they mainly provide livestock with fatty acids (e.g., omega-3 fatty acids), pigments, and carotenoids, while their high protein content contributed to fish nutrition [55]. Because of their high nutrient contents, several microalgae species have been used in aquaculture as a live feed for bivalve molluscs (i.e., mussels, clams oysters, and clams) of all growth stages [56].
In addition to utilization for producing health products, food ingredients, and feed, microalgae are a promising source of alternative proteins mainly due to their generally high protein content ranging from 6% to 71 % of dry matter [49]; for example, A. platensis was reported with 630 g protein/kg dry mass [20]. Furthermore, the composition of IAA in A. platensis is nearly the same as that of animal protein and exceeds that of most plant-derived protein [57]. Similarly, Chlorella and Arthrospira species are promising due to their best protein quality [49]. According to the recommended requirements of essential amino acids by the World Health Organization and the Food and Agriculture Organization, these 2 species have proteins with balanced amino acid profiles suitable for human requirements [58,59]. The amino acid profiles of these 2 species are also similar to that of animal-based protein foods such as eggs [60]. By making use of the ability of these species to grow heterotrophically, i.e., in the absence of light, and of utilizing an external carbon source, Sophie’s Bionutrients, a food-based startup, plans to generate microalgae-based food protein as fast as yeast; their first prototype contains 50% protein by dry biomass weight. Microalgae are also unique in its seafood umami flavor, and hence, they are a potential alternative to conventional seafood [61]. Nevertheless, the microalgae contribution to global food security is still limited due to several challenges faced by the new field.
Challenges to Microalgae Utilization for Protein Production
Conventional protein sources including plant proteins are widely consumed worldwide due to their relatively high protein digestibility [62,63]. In contrast, microalgae represent an age-old high-protein product for human consumption [49], and its commercial application has not virtually materialized and is restricted to only a few species such as Chlorella vulgaris, Scenedesmus obliquus, Dunaliella sp., and Arthrospira platensis [59,64,65]. This limitation, for instance, might correlate with their safety as some microalgae species belonging to diatoms, dinoflagellates, and cyanobacteria were reported to produce toxins [66]. In addition, allergenicity was also recorded with consumption of microalga species such as Arthrospira platensis [67,68]. Overall, the safety data for existing microalgae are still very limited, and the future selected candidates will have to undergo several toxicological and allergen tests to confirm their safety, which introduces restrictions to their commercial applications.
Currently, only Chlorella vulgaris and Arthrospira sp. are sold on the market shelves, usually as health supplements in tablets or powdered form. These are of dried cell biomass without further cellular extraction [69]. Bioactive compounds of much higher price also are being extracted and purified from other algae species (e.g., Dunaliella salina and Haematococcus pluvialis), but food protein ingredients from microalgae are still at the premarket stage [70]. The low level of exploitation of these highly nutritious microbes is linked to the missing data on the algal biology, toxicity, food safety, and optimal growth conditions and feed [71].
Compared to animal source proteins, microalgae have characteristic strong cellulosic indigestible cell walls that are associated with lower digestibility and further limit their acceptability [63,72]. Thus, post-harvesting treatments are required to disrupt the cell wall and make the extracted protein more accessible for digestive proteases. This requires a downstream process such as enzymatic hydrolysis using pancreatin enzymes, which was reported to improve the in vitro hydrolysate digestibility of Chlorella vulgaris and Arthrospira platensis by 70% and 97%, respectively [73].
In addition to digestibility, the microalgae sensory quality presents another challenge. Although the desirable sensory qualities of the conventional protein sources represent a major cause of wider consumer satisfaction, in contrast, the green color, odor, and fishy smell associated with microalgae are considered undesirable features for consumer choice [28,74]. Volatile sulfur and diketone compounds were reported to have fishy odors in microalgae [75]. Unpleasant odors might be mitigated by various routes such as changing medium components, altering harvest time, and cooking [76,77]. Overall, the organoleptic assessment of microalgae is important for its use as a source of alternative protein and further research is required to improve their digestibility and sensory quality.
Microalga cultivation is expensive and the possibilities to decrease its production cost remain a major challenge [78]. Culture conditions of microalgae such as temperature, pH, light, aeration, and nutrients potentially influence their overall production cost and biomass yield. Cheap carbon sources were successfully used to produce single cell protein, including Kluyveromyces marxianus on whey, Fusarium graminearum on molasses, Candida utilis on starch, and Cellulomonas sp. on bagasse [50]. Moreover, more than fifty types of media are used for microalgae cultivation [79], and some of them such as f/2 medium (enriched seawater medium, National Centre for Marine Algae and Microbiota, Maine) cost ∼$55/L. Similarly, cost-effective storage techniques are well established and reported to maintain the microorganism viable and free of contamination and genetic alterations by bacteria and fungi, including cryopreservation and lyophilization that are highly utilized [80,81]. Common ways to conserve microalgae are either by routine serial subculturing or cryopreservation [79]. The former method is a labor- and consumable-intensive process. In addition, the long continuous subculturing may result in culture degeneration and loss of essential morphological and physiological features with the high possibility of cross contaminations [79,82]. On the other hand, cryopreservation provides an alternative approach that overcomes such drawbacks, and nevertheless, it has also been reported to cause culture death of some algae strains [83].
For large-scale cultivation, a few studies have been conducted on cost-effective cultivation of algae using several systems, such as photobioreactors, polyethylene sleeve, and open ponds [84]. Norsker et al. [65] reported costs of €4.15, €4.95, and €5.96/kg of algae for horizontal tubular photobioreactors, open ponds, and flat panel photobioreactors, respectively. The authors believed that through optimizing the cost factors, such as mixing, irradiation, photosynthesis, and medium components, the cost declined to €0.68/kg. Another economic study reported the overall costs ranging €1.4–€2.5 [85]. Furthermore, the cultivation process is a species-specific and non-generic process due to the variations of species nutrients, light, temperature, pH, and other requirements of culture conditions [86]. Similarly, downstream processing is also species-specific. This process includes harvesting, drying, and by-product purification [87]. Furthermore, the variation among alga species in geometric, cell weight, and mucilage secretion also makes such processes species-specific and requires considerable research to be optimized [88,89].
Microalgae are generally slow-growing compared to bacterial or yeast culture in nutrient media, and growth is limited by the daylight cycle, CO2 availability, and photoinhibition during the peak of day. For photosynthetic modes of cell production, open pond systems of cultivation remain the cheaper option, but productivity and contamination risks by bacteria, protozoa, and other algae are typical limitations [90]. Furthermore, less than a meter of culture depth poses light penetration limitation to the cells [91]. Several photobioreactors are designed to reduce the light penetration depth problem but, often, performance increases with the increase of both equipment capital and operational cost [92]. Fouling, aeration, and light-source direction are some of the operational challenges involved. Hence, several commercial approaches have focused on heterotrophic species capable of higher productivity when supplied with nutrient media [93].
Systems Biology and Machine Learning Approaches Help Tackle the Challenges
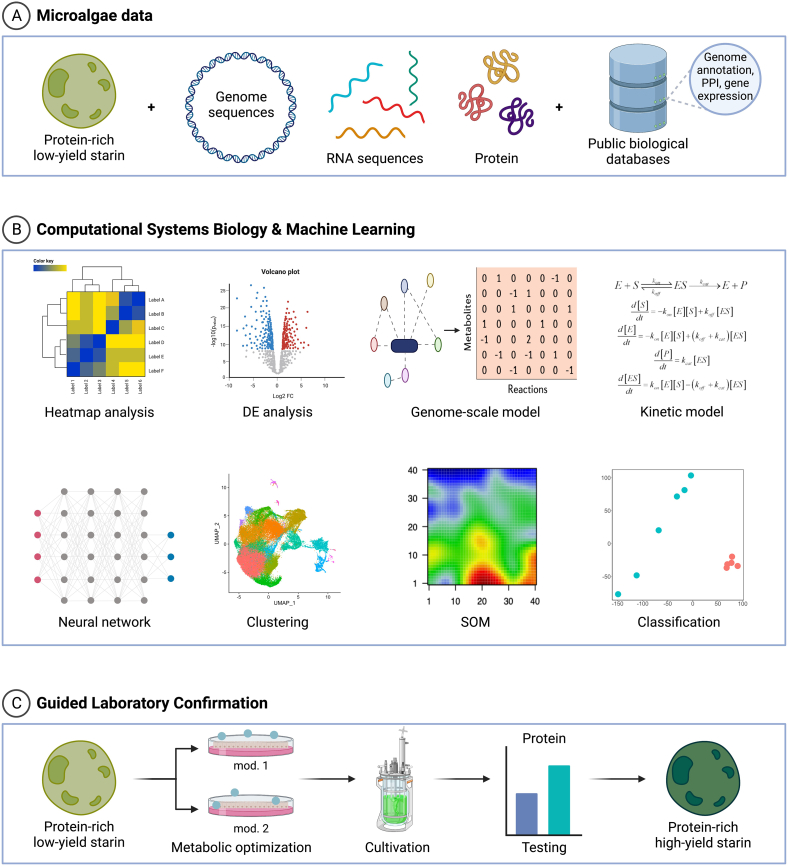
Most of the challenges faced in the utilization of microalgae are centralized around the understanding of their biology and genetics. For instance, the process of growing and optimizing microalgae in the laboratory as a source of alternative proteins requires an adequate understanding of microalga biology that involves many variables and multiple steps. This, therefore, makes it a highly complex process that requires long time investment and high-cost and labor-intensive experiments. The processes face multiple levels of challenges starting with the identification of food-safe strains, evaluating their protein production capacities, and assessing their abilities to grow under laboratory/bioreactor conditions [94,95]. These steps also produce large volumes of data, such as multiomics data, that require advanced data mining and analytics to generate a holistic view of the microalga biology at the systems level. Thus, data analytics and machine learning approaches are required for the success of the research and process. Alternatively, systems biology approaches, which include biosimulation using mathematical models (e.g., nonparametric steady-state or dynamic in silico models) [96], can provide mechanistic understanding and predictive opportunity of microalgae research (Figure 1).
FIGURE 1.
Systems biology and machine learning pipeline. (A) Microalgae data can be stored or retrieved from private/public databases. They can contain gene/protein sequences and structures and omics data (transcriptomics, proteomics, and metabolomics). (B) The data from (A) can be used to perform biostatistics, data analytics, and machine learning to identify expression correlations; co-regulated clusters; and differentially activated genes, proteins, or metabolites between samples. Metabolites and enzyme data can be used to develop and test a genome-scale model to identify growth constraints, as well as kinetic models, which could highlight bottlenecks in metabolic fluxes. (C) The model-optimized microalga strains can then be experimentally tested for better yield, especially at a commercial scale. DE, deferential expression; SOM, self-organization map.
Due to the overwhelming interest in using microalgae to produce biofuel and high-value products (i.e., pharmaceuticals, food supplements, and pigments), there is a large amount of previously published genome and transcriptome data of different microalga species under different growth conditions [[97], [98], [99]]. Genomic (DNA-sequencing) and transcriptomic (RNA-sequencing) studies play a crucial role in the qualitative and quantitative improvement of microalgal biomass [98,100]. Currently, the Gene Expression Omnibus database has over 6350 gene expression datasets for algae, and the Genome Online Database has 230 alga genome sequencing projects [101,102]. The genomic and transcriptomic information can be used to assess metabolic pathways and their respective genes of the enzymes that are involved. Furthermore, a good annotation of the whole genome, alongside a comparative transcriptomic approach, not only allows gaining insight into the metabolic pathways and their key-enzymes but also enables identification of regulatory factors, transcriptional factors, and promoters of gene expression [[103], [104], [105]].
Computational models developed for biological applications require actual experimental data to be used in the building, training, and testing processes of these models [106]. Data are usually obtained through well-defined experiments. Quantitative high-throughput transcriptomic data measuring the dynamic gene expression profiles of the microalgae, under multiple growth conditions, can be analyzed to identify differentially expressed (DE) genes between the conditions. These can be subsequently clustered into distinct expression patterns. Such specific clusters with the aid of bioinformatic databases, such as UniProt databases, Gene Ontology Resource [107], Kyoto Encyclopaedia of Genes and Genomes (KEGG) [108], PathwaysCommons [109], and Reactome [110], can rapidly highlight genes and their pathways that are involved in alternative protein production [111]. For example, KEGG databases highlight genes that are differentially present between different strains and species, and these can be used to focus on alternative protein pathways [112].
To illustrate the utility of mathematical approaches, there are several extrinsic environmental factors reported to potentially affect the tubular photobioreactors and open pond large-scale culturing processes of microalgae, including fluctuation of daily temperatures, dissolved oxygen, and light intensity [113,114]. Applying predictive microbiology mathematical models to correlate such changes and predict the microalgae growth performance, one could optimize the culturing process and decrease the overall costs. For example, the Geeraerd and Van Impe Inactivation Model Fitting Tool has been successfully applied to bacteria, yeast, and filamentous fungi growth performance covering both classical log-linear and nonlinear performance curves of microbial survivors [115,116]. These tools and equations were reported to evaluate the growth performance of microorganisms under several variables including pH, inoculum size, temperature, mixed culture, and incubation time to determine the actual growth behavior in a particular or desirable environment [[117], [118], [119], [120]]. Overall, the authors of these studies used their observations to develop a model describing the impact of several variables and their combined effects on the growth of the target microorganism. For example, Ram et al. [120] illustrated a possible way to overcome the laborious and expensive traditional way of evaluating the microbial fitness of different stains in a mixed culture. In this study, a computational approach was created from growth curves of mono- and mixed cultures, which replaced the traditional way of measuring the occurrences of single isolates within a mixed culture. Theoretical models can also be used to predict, control, and optimize the culture and growth conditions of microalgae and to direct their metabolism. For example, it was postulated that lipid yield can be increased by 5 times to reach a realistic maximum of 0.5 g triacylglycerol (TAG)/mol photons [121].
Another aspect of theoretical or computational models is based on metabolic network stoichiometry. Here, thousands of metabolites and their reactions can be simulated with reasonable computational cost and prediction outcome, especially when biological restrictions, such as growth constraints, are added [77]. In one study, Betenbaugh et al. [99] developed such a constraint-based genome-scale model and optimized metabolic fluxes for sustainable growth with increased nutrient supply and lipid productivity in Chlorella vulgaris.
Another area of mathematical models that have been widely used for microbial metabolic optimization research is kinetic models. These models are based on ordinary differential equations applied to each biochemical reaction in a metabolic network [96]. Although sparingly applied in microalga research so far, they have been used to simulate dynamic lipid metabolisms, TAG synthesis, and growth under multiple cultivation conditions and have led to novel insights. Lenka et al. [122] have provided a succinct review of several modeling applications in the microalga lipid and TAG yield increase.
The culture, harvesting, and extraction costs are a major limitation to the commercialization of microalgae. The organic carbon sources are indispensable for microalga growth, and they account for >80% of the overall cost of the used culture medium [123]. Cheap carbon sources such as CO2, lignocellulosic biomass, organic acids, and other wastes might be explored to reduce the overall production cost [124]. Screening of genes and enzymes that can degrade such substrates among microalga species using available databases, such as the KEGG database, might potentially save time, cost, and resources in experimental conformation compared to the traditional laboratory screening process, which is expensive and labor- and time-consuming [96]. Furthermore, metabolic engineering might also be used to encode the desired hydrolytic enzymes into selected microalgae. Similar approaches have been successfully applied to improve the capability of industrial microbes to break down substrates that cannot be degraded naturally [125,126].
Natural mutations were reported among alga isolates that produced the generally desired lighter-colored green biomass. To investigate such positive attributes systematically, computational genomics approaches might be used to identify novel secondary metabolite-related genes, such as that of carotenoids, that can be biochemically determined using the available databases with the well-known Escherichia coli expression systems. Such approaches could be aimed to modulate and improve the sensory quality of green microalgae [127,128]. For example, computational techniques unleashed the phylogenetics of the carotenoid pathways and helped study their gene expression and mutations, thereby providing better opportunities for increasing production rates [129].
High-throughput data analytics also play a major role in recent microalga research. A transcriptome-wide analysis by Azaman et al. [130] on Chlorella sorokiniana showed 2000 upregulated and downregulated genes related to lutein biosynthesis, fatty acid biosynthesis, TAG accumulation, starch biosynthesis, sucrose biosynthesis, etc. under the mixotrophic condition. In another work, by studying differentially activated genes from transcriptome-wide data, Kobayashi et al. [131] illustrated a safe way to produce green Chlamydomonas reinhardtii with a chlorophyll-deficient mutant by the deletion of phytoene synthase. Here, an understanding of the gene regulation mechanism might increase consumer acceptance of microalgae as an alternative protein source, since the dark green–colored biomass generally decreases customer desire, as discussed above.
Machine learning (ML) algorithms can also be used to analyze the transcriptome data and protein structure data of many available microalgae datasets to predict the connection between genes and pathways/networks to reveal novel gene regulation mechanisms [132,133]. Some of the data analytic and AI methods, such as PCA, random forest, k-means clustering, and support vector machine, can be utilized to predict key pathways and rank them based on their expression levels and statistical scores [96]. This will subsequently allow for testing and validating the best ranking pathways biosynthetically on stable microalgae for enhanced production of alternative protein [134,135]. Thus, advanced data mining and analytics methods (e.g., AI) can provide breakthroughs through analyses and predictions that cannot be achieved otherwise [96].
Another area of ML models under current development is related to those attempting to integrate complex dynamic omics data [136]. Here, the models are first developed to learn between high-throughput omics “training” data linking an input to an output (e.g., proteomics with metabolomics in wildtype). Next, the trained model is to be tested on a different “test” data (e.g., under mutant or different growth condition) to finalize a machine-learned connectivity model that, without knowing the detailed mechanistic understanding of the cell system, will be able to guide synthetic biology applications [[137], [138], [139]]. However, such models are yet to be developed or tested in microalga research, partly due to the lack of expertise or domain knowledge in the cross-disciplinary area. Table 2 lists current examples of systems biology and machine learning contributions to microalga research.
TABLE 2.
Overview of some key contributions of systems biology and AI in microalga research
| Methodology | Key contributions | Reference |
|---|---|---|
| Computational analysis of MFA data: using mathematical and statistical approaches to process and analyze MFA data | Identify suppressor proteins of methionine and cysteine biosynthesis | [140] |
| Increase carbon fixation and biomass through the identification of an alternative pathway for isoleucine synthesis | [141] | |
| Investigate and increase the astaxanthin synthesis | [142] | |
| Control the hydrogen production through the regulation of hydrogenase and poly-β-hydroxybutyrate synthase | [143] | |
| Genome-scale metabolic model: a mathematical approach for simulating metabolism in genome-scale reconstructions. It uses steady-state assumptions and stoichiometry of all known reactions of the metabolic pathway | Predict alternative metabolic routes for fixed carbon through an analysis of all possible double reaction knockouts | [144] |
| Dynamic or kinetic modeling: computational modeling of metabolic pathways using enzymes kinetics and rate laws | Control the growth of the microalgae in the bioreactors with artificial lights | [145] |
| Predict the photosynthetic apparatus status (open, closed, and damaged reaction centers) under different lighting conditions | [146] | |
| Optimize the growth of microalgae for increased biomass through the simulation of the production in a virtual system | [147] | |
| Machine learning: data analytics, bioinformatics, and deep learning using artificial neural networks | Analyze microscopic imaging for the classification, identification, and growth stage estimation of microalgae | [148] |
| Analyze microscopic imaging for the identification of individual microalgae in a free or symbiosis state | [149] | |
| Improve the design of a semicontinuous algal cultivation to overcome the mutual shading that limited the growth | [150] | |
| Define and diagnose algal cultures under stress conditions to save them from imminent crashing by utilizing 4 biomarkers | [151] | |
| Predict phosphorylation site from mass spectrometry–based proteomics data | [152] | |
| Bioinformatics analysis of gene expression data | Identify the phytoene synthase gene of microalgae | [153] |
| Identify the sequence encoding P-type ATPases from RNA-Seq transcriptomic data | [154] |
MFA, metabolic flux analysis
In conclusion, overpopulation, the COVID-19 pandemic, and ongoing geopolitical conflicts are some of the major challenges faced in today’s world that are negatively impacting global food security. Increasing food production through most of the currently available approaches (agriculture, livestock, and fishing) to match the increasing demand is not possible due to limited resources, environmental impact, and increased awareness of healthier diets. This raises the need for unconventional solutions and fundamental changes in the current food system. Alternative food and proteins sources are 2 potential solutions to close the gap between food supply and demand. Microalgae can be a potential platform for alternative proteins production. They are known for their high protein content and producing proteins rich in essential amino acids, and can grow in different environments using cheap and recyclable feed, such as wastes. They are also being successfully used as a live feed in aquaculture.
Utilizing microalgae to produce alternative proteins require overcoming of several limitations, such as finding the most suitable strains, reducing the cost of cultivation, harvesting and extraction, increasing the yield from the proteins, eliminating the green color, and improving the sensory properties of the final products. Overcoming these challenges requires employing modern systems biology approaches (such as multiomics) and advanced computational data analytics (such as AI) to provide guidance to the downstream experimental work. Modern high-throughput techniques (e.g., genomics, transcriptomics, proteomics, and metabolomics) can provide better details on the biology of the microalgae to reveal key molecules and pathways that are important for the quality and quantity of the produced proteins. However, the immense amount of the data requires advanced computational and statistical methods to be employed and used. Coupling AI and mathematical modeling is a promising systems approach to processing and analyzing microalga data in the future. Such an approach can provide novel insights into their biology, such as identifying key pathways for growth, protein production, and improvement of their taste and sensory properties. Thus, multidisciplinary and integrated approaches are potentially the way forward to overcome the challenges faced by microalgae research and unleash their potential to contribute to food sustainability and circular economy.
Funding
This project was supported by the Agency for Science, Technology and Research under the Singapore Food Story R&D Programme (Theme 2 – 1st Alternative Protein Seed Challenge; W20W2D0017). Any opinions, findings and conclusions, or recommendations expressed in this material are those of the authors and do not reflect the views of the Agency for Science, Technology and Research.
Author disclosures
The authors report no conflicts of interest.
Acknowledgments
We thank Derek Smith for critical reading of the manuscript.
Author contribution
The authors’ responsibilities were as follows – MH: contributed to every section of the manuscript and wrote the first draft; HE, LY, and YC: contributed to the sections on microalgae on food and health and challenges to microalga utilization for protein production; KS: conceptualized the idea, contributed to writing, and supervised the entire study; and all authors: read and approved the final manuscript.
References
- 1.United Nations World Population Prospects. https://www.un.org/development/desa/publications/world-population-prospects-the-2017-revision.html The 2017 Revision | Multimedia Library - United Nations Department of Economic and Social Affairs [Internet]. 2017 [cited 2021 Mar 5]. Available from:
- 2.FAO http://www.fao.org/3/a0607e/a0607e00.htm World agriculture: towards 2030/2050 [Internet]. 2006 [cited 2021 Feb 25]. Available from:
- 3.Global Food Crisis https://www.bloomberg.com/news/articles/2022-03-13/how-russia-s-invasion-of-ukraine-is-tearing-apart-the-global-food-system Latest News and Analysis With Ukraine-Russia War - Bloomberg [Internet]. [cited 2022 Apr 13]. Available from:
- 4.Wu G., Fanzo J., Miller D.D., Pingali P., Post M., Steiner J.L., Thalacker-Mercer A.E. Production and supply of high-quality food protein for human consumption: sustainability, challenges, and innovations. Ann N Y Acad Sci. 2014;1321:1–19. doi: 10.1111/nyas.12500. [DOI] [PubMed] [Google Scholar]
- 5.Poore J., Nemecek T. Reducing food’s environmental impacts through producers and consumers. Science (80- ) [Internet] American Association for the Advancement of Science. 2018 doi: 10.1126/science.aaq0216. http://science.sciencemag.org/ [cited 2021 Jul 5];360:987–92. Available from: [DOI] [PubMed] [Google Scholar]
- 6.Buller H., Blokhuis H., Jensen P., Keeling L. Towards farm animal welfare and sustainability. Animals (Basel) 2018;8(6):81. doi: 10.3390/ani8060081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb P., Benton T.G., Beddington J., Flynn D., Kelly N.M., Thomas S.M. The urgency of food system transformation is now irrefutable. Nat Food. 2021;1:584–585. doi: 10.1038/s43016-020-00161-0. [DOI] [PubMed] [Google Scholar]
- 8.Post M.J., Levenberg S., Kaplan D.L., Genovese N., Fu J., Bryant C.J., et al. Scientific, sustainability and regulatory challenges of cultured meat. Nat Food. 2020;1:403–415. [Google Scholar]
- 9.Lynch H., Johnston C., Wharton C. Plant-based diets: considerations for environmental impact, protein quality, and exercise performance. Nutrients. 2018;10:1841. doi: 10.3390/nu10121841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravindan A., Geddie J. 2020. Singapore approves sale of lab-grown meat in world first | Reuters.https://www.reuters.com/article/uk-eat-just-singapore/singapore-becomes-first-country-to-approve-sale-of-lab-grown-meat-idUKKBN28C06Q?edition-redirect=ca [Internet] [cited 2021 Feb 25]. Available from: [Google Scholar]
- 11.Bakhsh A., Lee S.J., Lee E.Y., Hwang Y.H., Joo S.T. Evaluation of rheological and sensory characteristics of plant-based meat analog with comparison to beef and pork. Food Sci Anim Resour. 2021;41:983–996. doi: 10.5851/kosfa.2021.e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood P., Tavan M. A review of the alternative protein industry. Curr Opin Food Sci. 2022;47 [Google Scholar]
- 13.Boukid F. Plant-based meat analogues: from niche to mainstream. Eur Food Res Technol. 2020;247:297–308. [Google Scholar]
- 14.Pakseresht A., Ahmadi Kaliji S., Canavari M. Review of factors affecting consumer acceptance of cultured meat. Appetite. 2022;170 doi: 10.1016/j.appet.2021.105829. [DOI] [PubMed] [Google Scholar]
- 15.Katiyar R., Gurjar B.R., Biswas S., Pruthi V., Kumar N., Kumar P. Microalgae: An emerging source of energy based bio-products and a solution for environmental issues. Renew Sustain Energy Rev Pergamon. 2017;72:1083–1093. [Google Scholar]
- 16.Jach M.E., Serefko A., Ziaja M., Kieliszek M. Yeast protein as an easily accessible food source. Metabolites. 2022;12:63. doi: 10.3390/metabo12010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdel-Azeem A.M., Abdel-Azeem M.A., Khalil W.F. Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Diseases (2nd Ed. Interv Arthritis Relat Inflamm Dis.; 2019. Endophytic fungi as a new source of antirheumatoid metabolites; pp. 355–384. [Google Scholar]
- 18.Vethathirri R.S., Santillan E., Wuertz S. Microbial community-based protein production from wastewater for animal feed applications. Bioresour Technol. 2021;341 doi: 10.1016/j.biortech.2021.125723. [DOI] [PubMed] [Google Scholar]
- 19.Gervasi T., Pellizzeri V., Calabrese G., Di Bella G., Cicero N., Dugo G. Production of single cell protein (SCP) from food and agricultural waste by using Saccharomyces cerevisiae. Nat Prod Res. 2018;32:648–653. doi: 10.1080/14786419.2017.1332617. [DOI] [PubMed] [Google Scholar]
- 20.Salter A.M., Lopez-Viso C. Role of novel protein sources in sustainably meeting future global requirements. Proc Nutr Soc. 2021;80:186–194. doi: 10.1017/S0029665121000513. [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization . 2007. Protein and amino acid requirements in human nutrition : report of a joint FAO/WHO/UNU expert consultation.https://apps.who.int/iris/handle/10665/43411 [Internet] Available from: [PubMed] [Google Scholar]
- 22.Westendorf M.L., Wohlt J.E. Brewing by-products: their use as animal feeds. Vet Clin North Am Food Anim Pract. 2002;18:233–252. doi: 10.1016/s0749-0720(02)00016-6. [DOI] [PubMed] [Google Scholar]
- 23.TJA Finnigan. Handbook of Food Proteins. Woodhead Publishing; 2011. Mycoprotein: origins, production and properties; pp. 335–352. [Google Scholar]
- 24.Ritala A., Häkkinen S.T., Toivari M., Wiebe M.G. Single cell protein-state-of-the-art, industrial landscape and patents 2001-2016. Front Microbiol. 2017;8:2009. doi: 10.3389/fmicb.2017.02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nangul A., Bhatia R. 15–8. 2013. (Microorganisms: a marvelous source of single cell proteins). [Google Scholar]
- 26.Klamczynska B., Mooney W.D. Academic Press; 2017. Heterotrophic microalgae: a scalable and sustainable protein source. Sustainable Protein Sources; pp. 327–339. [Google Scholar]
- 27.Barka A., Blecker C. Microalgae as a potential source of single-cell proteins. A review. 2016;20:427–436. https://popups.uliege.be/1780-4507 https://popups.uliege.be/1780-4507/index.php?id=13132 [Internet] FAC UNIV SCIENCES AGRONOMIQUES GEMBLOUX. Available from: [Google Scholar]
- 28.Morris H.J., Almarales A., Carrillo O., Bermúdez R.C. Utilisation of Chlorella vulgaris cell biomass for the production of enzymatic protein hydrolysates. Bioresour Technol. 2008;99:7723–7729. doi: 10.1016/j.biortech.2008.01.080. [DOI] [PubMed] [Google Scholar]
- 29.Wu J. Emerging sources and applications of alternative proteins: An introduction. Adv Food Nutr Res. 2022;101:1–15. doi: 10.1016/bs.afnr.2022.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Torres-Tiji Y., Fields F.J., Mayfield S.P. Microalgae as a future food source. Biotechnol Adv. 2020:41. doi: 10.1016/j.biotechadv.2020.107536. [DOI] [PubMed] [Google Scholar]
- 31.Koyande A.K., Chew K.W., Rambabu K., Tao Y., Chu D.T., Show P.L. Microalgae: a potential alternative to health supplementation for humans. Food Sci Hum Wellness. 2019;8:16–24. [Google Scholar]
- 32.Christias C., Couvaraki C., Georgopoulos S.G., Macris B., Vomvoyanni V. Protein content and amino acid composition of certain fungi evaluated for microbial protein production. Appl Microbiol. 1975;29:250. doi: 10.1128/am.29.2.250-254.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada E.A., Sgarbieri V.C. Yeast (Saccharomyces cerevisiae) protein concentrate: preparation, chemical composition, and nutritional and functional properties. J Agric Food Chem. 2005;53:3931–3936. doi: 10.1021/jf0400821. [DOI] [PubMed] [Google Scholar]
- 34.Matassa S., Boon N., Pikaar I., Verstraete W. Microbial protein: future sustainable food supply route with low environmental footprint. Microb Biotechnol. 2016;9:568–575. doi: 10.1111/1751-7915.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christaki E., Florou-Paneri P., Bonos E. Microalgae: a novel ingredient in nutrition. Int J Food Sci Nutr. 2011;62:794–799. doi: 10.3109/09637486.2011.582460. [DOI] [PubMed] [Google Scholar]
- 36.Brown M.R., Jeffrey S.W., Volkman J.K., Dunstan G.A. Nutritional properties of microalgae for mariculture. Aquaculture. 1997;151:315–331. [Google Scholar]
- 37.De Morais M.G., Vaz B.D.S., De Morais E.G., Costa J.A.V. Biologically active metabolites synthesized by microalgae. Biomed Res Int. 2015 doi: 10.1155/2015/835761. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vieira M.V., Pastrana L.M., Fuciños P. Microalgae encapsulation systems for food, pharmaceutical and cosmetics applications. Mar Drugs. 2020;18:644. doi: 10.3390/md18120644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chanda M., Merghoub N., El Arroussi H. Microalgae polysaccharides: the new sustainable bioactive products for the development of plant bio-stimulants? World J Microbiol Biotechnol. 2019;35 doi: 10.1007/s11274-019-2745-3. [DOI] [PubMed] [Google Scholar]
- 40.Cuellar-Bermudez S.P., Aguilar-Hernandez I., Cardenas-Chavez D.L., Ornelas-Soto N., Romero-Ogawa M.A., Parra-Saldivar R. Extraction and purification of high-value metabolites from microalgae: essential lipids, astaxanthin and phycobiliproteins. Microb Biotechnol. 2015;8:190–209. doi: 10.1111/1751-7915.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapoore R.V., Butler T.O., Pandhal J., Vaidyanathan S. Microwave-assisted extraction for microalgae: from biofuels to biorefinery. Biology (Basel) 2018;7:1–25. doi: 10.3390/biology7010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.González-Delgado Á -D., Kafarov V. SciELO Anal; 2011. Microalgae based biorefinery: issues to consider.http://www.scielo.org.co/scielo.php?script=sci_arttext&pid=S0122-53832011000200001 [cited 2022 Apr 13]; Available from: [Google Scholar]
- 43.Chisti Y. Biodiesel from microalgae. Biotechnol Adv. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Hoffmann J.P. Wastewater treatment with suspended and nonsuspended algae. J Phycol. 1998;34:757–763. [Google Scholar]
- 45.Richmond A. Asian Pacific Phycology in the 21st Century: Prospects and Challenges. 33–7. 2004. Principles for attaining maximal microalgal productivity in photobioreactors: an overview. [Google Scholar]
- 46.Ruiz J., Álvarez P., Arbib Z., Garrido C., Barragán J., Perales J.A. Effect of nitrogen and phosphorus concentration on their removal kinetic in treated urban wastewater by Chlorella vulgaris. Int J Phytoremediation. 2011;13:884–896. doi: 10.1080/15226514.2011.573823. [DOI] [PubMed] [Google Scholar]
- 47.Fabregas J., Herrero C. Vitamin content of four marine microalgae. Potential use as source of vitamins in nutrition. J Ind Microbiol. 1990;5:259–263. [Google Scholar]
- 48.Gouveia L., Batista A.P., Sousa I., Raymundo A., Bandarra N.M. Food Chemistry Research Developments. 2008. https://www.repository.utl.pt/bitstream/10400.5/2434/1/REP-I.Sousa-CapLivro algasGouveia.pdf Available from. [Google Scholar]
- 49.Caporgno M.P., Mathys A. Trends in microalgae incorporation into innovative food products with potential health benefits. Front Nutr. 2018;5:58. doi: 10.3389/fnut.2018.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nasseri A.T., Rasoul-Amini S., Morowvat M.H., Ghasemi Y. Single cell protein: production and process. Am J Food Technol. 2011;6:103–116. [Google Scholar]
- 51.Mahmoud R., Ibrahim M., Ali G. Closed photobioreactor for microalgae biomass production under indoor growth conditions. J Algal Biomass Utln. 2016;7:86–92. [Google Scholar]
- 52.Enzing C., Ploeg M., Barbosa M., Sijtsma L., Vigani M., Parisi C., et al. Microalgae-based products for the food and feed sector: an outlook for Europe. JRC Scientific and policy reports. 2014 https://publications.jrc.ec.europa.eu/repository/handle/JRC85709 Available from: [Google Scholar]
- 53.Vigani M., Parisi C., Rodríguez-Cerezo E., Barbosa M.J., Sijtsma L., Ploeg M., et al. Food and feed products from micro-algae: Market opportunities and challenges for the EU. Trends Food Sci Technol. 2015;42:81–92. [Google Scholar]
- 54.Food and Drug Administration . 2018. Microorganisms & Microbial-Derived Ingredients Used in Food (Partial List) | FDA.https://www.fda.gov/food/generally-recognized-safe-gras/microorganisms-microbial-derived-ingredients-used-food-partial-list [Internet] Available from: [Google Scholar]
- 55.Muller-Feuga A. The role of microalgae in aquaculture: situation and trends. J Appl Phycol. 2000;12:527–534. [Google Scholar]
- 56.M.R. Brown, L.E. Cruz-Suárez, D. Ricque-Marie, M. Tapia-Salazar, M.G. Gaxiola-Cortés, Nutritional value of microalgae for aquaculture, Handbook of Microalgal Culture: Biotechnology and Applied Phycology, 2003, pp. 380–391.
- 57.Lupatini A.L., Colla L.M., Canan C., Colla E. Potential application of microalga Spirulina platensis as a protein source. J Sci Food Agric. 2017;97:724–732. doi: 10.1002/jsfa.7987. [DOI] [PubMed] [Google Scholar]
- 58.World Health Organization, Food and Agriculture Organization, United Nations University . FAO/WHO/UNU expert consultation; 2007. Protein and amino acid requirements in human nutrition : report of a joint.https://apps.who.int/iris/handle/10665/43411 [Internet] Available from: [Google Scholar]
- 59.Becker E.W. Micro-algae as a source of protein. Biotechnol Adv. 2007;25:207–210. doi: 10.1016/j.biotechadv.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Chronakis I.S., Madsen M. Woodhead Publishing; 2011. Algal proteins. Handbook of Food Proteins; pp. 353–394. [Google Scholar]
- 61.Southey F. Food Navigator. [Internet; 2021. Sophie’s BIoNutrients: Protein made from microalgae fed with food waste comes to Europe; p. 12. 02. [Google Scholar]
- 62.Tulbek M.C., Lam R.S.H., Wang Y.C., Asavajaru P., Lam A. Academic Press; 2017. Pea: a sustainable vegetable protein crop. Sustainable Protein Sources; pp. 145–164. [Google Scholar]
- 63.Day L. Proteins from land plants – Potential resources for human nutrition and food security. Trends Food Sci Technol. 2013;32:25–42. [Google Scholar]
- 64.Jensen A. Fourteenth International Seaweed Symposium. Springer; Dordrecht: 1993. Present and future needs for algae and algal products; pp. 15–23. [Google Scholar]
- 65.Pulz O., Gross W. Valuable products from biotechnology of microalgae. Appl Microbiol Biotechnol. 2004;65:635–648. doi: 10.1007/s00253-004-1647-x. [DOI] [PubMed] [Google Scholar]
- 66.Caruso G., Caruso G., Laganà P.L., Santi Delia A., Parisi S., Barone C., et al. Springer International Publishing; Cham: 2015. Microbial toxins and related contamination in the food industry.http://link.springer.com/10.1007/978-3-319-20559-5 Available from. [Google Scholar]
- 67.Szabo N.J., Matulka R.A., Chan T. Safety evaluation of whole algalin protein (WAP) from Chlorella protothecoides. Food Chem Toxicol. 2013;59:34–45. doi: 10.1016/j.fct.2013.05.035. [DOI] [PubMed] [Google Scholar]
- 68.Le T.M., Knulst A.C., Röckmann H. Anaphylaxis to Spirulina confirmed by skin prick test with ingredients of Spirulina tablets. Food Chem Toxicol. 2014;74:309–310. doi: 10.1016/j.fct.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 69.Lafarga T. Effect of microalgal biomass incorporation into foods: Nutritional and sensorial attributes of the end products. Algal Res. 2019;41 [Google Scholar]
- 70.Borowitzka M.A. High-value products from microalgae—their development and commercialisation. J Appl Phycol. 2013;25:743–756. [Google Scholar]
- 71.Fabris M., Abbriano R.M., Pernice M., Sutherland D.L., Commault A.S., Hall C.C., et al. Emerging technologies in algal biotechnology: toward the establishment of a sustainable, algae-based bioeconomy. Front Plant Sci. 2020;11:279. doi: 10.3389/fpls.2020.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tchorbanov B., Bozhkova M. Enzymatic hydrolysis of cell proteins in green algae Chlorella and Scenedesmus after extraction with organic solvents. Enzyme Microb Technol. 1988;10:233–238. [Google Scholar]
- 73.Kose A., Ozen M.O., Elibol M., Oncel S.S. Investigation of in vitro digestibility of dietary microalga Chlorella vulgaris and cyanobacterium Spirulina platensis as a nutritional supplement. 3 Biotech. 2017;7:1–7. doi: 10.1007/s13205-017-0832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ellies-Oury M.P., Cantalapiedra-Hijar G., Durand D., Gruffat D., Listrat A., Micol D., et al. An innovative approach combining Animal Performances, nutritional value and sensory quality of meat. Meat Sci. 2016;122:163–172. doi: 10.1016/j.meatsci.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 75.Van Durme J., Goiris K., De Winne A., De Cooman L., Muylaert K. Evaluation of the volatile composition and sensory properties of five species of microalgae. J Agric Food Chem. 2013;61:10881–10890. doi: 10.1021/jf403112k. [DOI] [PubMed] [Google Scholar]
- 76.Isleten Hosoglu M. Aroma characterization of five microalgae species using solid-phase microextraction and gas chromatography–mass spectrometry/olfactometry. Food Chem. 2018;240:1210–1218. doi: 10.1016/j.foodchem.2017.08.052. [DOI] [PubMed] [Google Scholar]
- 77.Milovanović I., Mišan A., Simeunović J., Kovač D., Jambrec D., Mandić A. Determination of volatile organic compounds in selected strains of cyanobacteria. J Chem. 2015 2015. [Google Scholar]
- 78.Chen L., Liu T., Zhang W., Chen X., Wang J. Biodiesel production from algae oil high in free fatty acids by two-step catalytic conversion. Bioresour Technol. 2012;111:208–214. doi: 10.1016/j.biortech.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 79.Andersen R.A. Elsevier; Amsterdam: 2004. Algal Culturing Techniques. 1st Ed.https://www.elsevier.com/books/algal-culturing-techniques/andersen/978-0-12-088426-1 [Internet] Available from: [Google Scholar]
- 80.Tedeschi R., De Paoli P. Collection and preservation of frozen microorganisms. Methods Mol Biol. 2011;675:313–326. doi: 10.1007/978-1-59745-423-0_18. [DOI] [PubMed] [Google Scholar]
- 81.Gorman R., Adley C.C. An evaluation of five preservation techniques and conventional freezing temperatures of −20°C and −85°C for long-term preservation of Campylobacter jejuni. Lett Appl Microbiol. 2004;38:306–310. doi: 10.1111/j.1472-765x.2004.01490.x. [DOI] [PubMed] [Google Scholar]
- 82.Wang H., Zhang W., Chen L., Wang J., Liu T. The contamination and control of biological pollutants in mass cultivation of microalgae. Bioresour Technol. 2013;128:745–750. doi: 10.1016/j.biortech.2012.10.158. [DOI] [PubMed] [Google Scholar]
- 83.Day J.G., Harding K. Plant Cryopreservation: A Practical Guide. 95–116. Springer; New York, NY: 2008. Cryopreservation of algae. [Internet] [Google Scholar]
- 84.Ugwu C.U., Aoyagi H., Uchiyama H. Photobioreactors for mass cultivation of algae. Bioresour Technol. 2008;99:4021–4028. doi: 10.1016/j.biortech.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 85.Sari Y.W., Sanders J.P.M., Bruins M. Techno-economical evaluation of protein extraction for microalgae biorefinery. IOP Conference Series: Earth and Environmental Science. IOP Publishing. 2016;31 [Google Scholar]
- 86.Laurens L.M.L., Van Wychen S., McAllister J.P., Arrowsmith S., Dempster T.A., McGowen J., et al. Strain, biochemistry, and cultivation-dependent measurement variability of algal biomass composition. Anal Biochem. 2014;452:86–95. doi: 10.1016/j.ab.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 87.Tan J Sen, Lee S.Y., Chew K.W., Lam M.K., Lim J.W., Ho S.H., et al. A review on microalgae cultivation and harvesting, and their biomass extraction processing using ionic liquids. Bioengineered. 2020;11:116–129. doi: 10.1080/21655979.2020.1711626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rhodes C.J. Oil from algae; salvation from peak oil? Sci Prog. 2009;92:39–90. doi: 10.3184/003685009X440281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oey M., Sawyer A.L., Ross I.L., Hankamer B. Challenges and opportunities for hydrogen production from microalgae. Plant Biotechnol J. 2016;14:1487–1499. doi: 10.1111/pbi.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borowitzka M.A., Reza Moheimani N. Algae for Biofuels and Energy. Springer; Dordrecht: 2013. Open pond culture systems; pp. 133–152. [Google Scholar]
- 91.Torzillo G., Pushparaj B., Masojidek J., Vonshak A. Biological constraints in algal biotechnology. Biotechnol Bioprocess Eng. 2003;8:338–348. [Google Scholar]
- 92.Posten C. Design principles of photo-bioreactors for cultivation of microalgae. Eng Life Sci. 2009;9:165–177. [Google Scholar]
- 93.Morales-Sánchez D., Martinez-Rodriguez O.A., Martinez A. Heterotrophic cultivation of microalgae: production of metabolites of commercial interest. J Chem Technol Biotechnol. 2017;92:925–936. [Google Scholar]
- 94.Shukal S., Chen X., Zhang C. Systematic engineering for high-yield production of viridiflorol and amorphadiene in auxotrophic. Escherichia coli. 2019;55:170–178. doi: 10.1016/j.ymben.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 95.Curran K.A., Crook N.C., Karim A.S., Gupta A., Wagman A.M., Alper H.S. Design of synthetic yeast promoters via tuning of nucleosome architecture. Nat Commun. 2014;5:1–8. doi: 10.1038/ncomms5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Helmy M., Smith D., Selvarajoo K. Systems biology approaches integrated with artificial intelligence for optimized metabolic engineering. Metab Eng Commun. 2020;11 doi: 10.1016/j.mec.2020.e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Villanova V., Spetea C. Mixotrophy in diatoms: Molecular mechanism and industrial potential. Physiol Plant. 2021;173:603–611. doi: 10.1111/ppl.13471. [DOI] [PubMed] [Google Scholar]
- 98.Kumar G., Shekh A., Jakhu S., Sharma Y., Kapoor R., Sharma T.R. Bioengineering of microalgae: recent advances, perspectives, and regulatory challenges for industrial application. Front Bioeng Biotechnol. 2020;8:914. doi: 10.3389/fbioe.2020.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ng I.S., Tan S.I., Kao P.H., Chang Y.K., Chang J.S. Recent developments on genetic engineering of microalgae for biofuels and bio-based chemicals. Biotechnol J. 2017;12 doi: 10.1002/biot.201600644. [DOI] [PubMed] [Google Scholar]
- 100.Jagadevan S., Banerjee A., Banerjee C., Guria C., Tiwari R., Baweja M., et al. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnol Biofuels. 2018;11:1–21. doi: 10.1186/s13068-018-1181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clough E., Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Sundaramurthi J.C., Lee J., et al. Genomes OnLine Database (GOLD) v.8: overview and updates. Nucleic Acids Res. 2021;49:D723–D733. doi: 10.1093/nar/gkaa983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Banerjee C., Dubey K.K., Shukla P. Metabolic engineering of microalgal based biofuel production: Prospects and challenges. Front Microbiol. 2016;7:432. doi: 10.3389/fmicb.2016.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu W.L., Ansari W., Schoepp N.G., Hannon M.J., Mayfield S.P., Burkart M.D. Modifications of the metabolic pathways of lipid and triacylglycerol production in microalgae. Microb Cell Fact. 2011;10:1–11. doi: 10.1186/1475-2859-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shahid A., Rehman A ur, Usman M., Ashraf M.U.F., Javed M.R., Khan A.Z., et al. Engineering the metabolic pathways of lipid biosynthesis to develop robust microalgal strains for biodiesel production. Biotechnol Appl Biochem. 2020;67:41–51. doi: 10.1002/bab.1812. [DOI] [PubMed] [Google Scholar]
- 106.Helmy M., Gohda J., Inoue J., Tomita M., Tsuchiya M., Selvarajoo K. In: PLoS One. Unutmaz D., editor. Vol. 4. 2009. Predicting novel features of Toll-like receptor 3 signaling in macrophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carbon S., Douglass E., Good B.M., Unni D.R., Harris N.L., Mungall C.J., et al. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kanehisa M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:277D. doi: 10.1093/nar/gkh063. 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rodchenkov I., Babur O., Luna A., Aksoy B.A., Wong J.V., Fong D., et al. Pathway Commons 2019 Update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 2020;48:D489–D497. doi: 10.1093/nar/gkz946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fabregat A., Sidiropoulos K., Viteri G., Forner O., Marin-Garcia P., Arnau V., et al. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18:1–9. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Helmy M., Agrawal R., Ali J., Soudy M., Bui T.T., Selvarajoo K. GeneCloudOmics: a data analytic cloud platform for high-throughput gene expression analysis. Front Bioinformatics. 2021:63. doi: 10.3389/fbinf.2021.693836. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Longworth J., Wu D., Huete-Ortega M., Wright P.C., Vaidyanathan S. Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Res. 2016;18:213–224. doi: 10.1016/j.algal.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grivalský T., Ranglová K., da Câmara Manoel J.A., Lakatos G.E., Lhotský R., Masojídek J. Development of thin-layer cascades for microalgae cultivation: milestones (review) Folia Microbiol. 2019;64:603–614. doi: 10.1007/s12223-019-00739-7. [DOI] [PubMed] [Google Scholar]
- 114.Gupta P.L., Lee S.M., Choi H.J. A mini review: photobioreactors for large scale algal cultivation. World J Microbiol Biotechnol. 2015;31:1409–1417. doi: 10.1007/s11274-015-1892-4. [DOI] [PubMed] [Google Scholar]
- 115.Geeraerd A.H., Valdramidis V.P., Van Impe J.F. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int J Food Microbiol. 2005;102:95–105. doi: 10.1016/j.ijfoodmicro.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 116.Nielsen L., Rolighed M., Buehler A., Knøchel S., Wiedmann M., Marvig C. Development of predictive models evaluating the spoilage-delaying effect of a bioprotective culture on different yeast species in yogurt. J Dairy Sci. 2021;104:9570–9582. doi: 10.3168/jds.2020-20076. [DOI] [PubMed] [Google Scholar]
- 117.Baker D.A., Genigeorgis C. Predicting the safe storage of fresh fish under modified atmospheres with respect to Clostridium botulinum toxigenesis by modeling length of the lag phase of growth. J Food Prot. 1990;53 doi: 10.4315/0362-028X-53.2.131. [DOI] [PubMed] [Google Scholar]
- 118.Genigeorgis C., Savoukidis, And M., Martin S. Initiation of Staphylococcal growth in processed meat environments. Appl Microbiol. 1971;21:940–942. doi: 10.1128/am.21.5.940-942.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Skinner G.E., Larkin J.W., Rhodehamel E.J. Mathematical modeling of microbial growth: a review. J Food Saf. 1994;14:175–217. [Google Scholar]
- 120.Ram Y., Dellus-Gur E., Bibi M., Karkare K., Obolski U., Feldman M.W., et al. Predicting microbial growth in a mixed culture from growth curve data. Proc Natl Acad Sci U S A. 2019;116:14698–14707. doi: 10.1073/pnas.1902217116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Remmers I.M., Wijffels R.H., Barbosa M.J., Lamers P.P. Can we approach theoretical lipid yields in microalgae? Trends Biotechnol. 2018;36:265–276. doi: 10.1016/j.tibtech.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 122.Lenka S.K., Carbonaro N., Park R., Miller S.M., Thorpe I., Li Y. Current advances in molecular, biochemical, and computational modeling analysis of microalgal triacylglycerol biosynthesis. Biotechnol Adv. 2016;34:1046–1063. doi: 10.1016/j.biotechadv.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 123.Mata T.M., Martins A.A., Caetano N.S. Microalgae for biodiesel production and other applications: a review. Renew Sustain energy Rev. 2010;14:217–232. [Google Scholar]
- 124.Xu J., Du W., Zhao X., Zhang G., Liu D. Microbial oil production from various carbon sources and its use for biodiesel preparation. Biofuels Bioprod Biorefining. 2013;7:65–77. [Google Scholar]
- 125.Blazeck J., Hill A., Liu L., Knight R., Miller J., Pan A., et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun. 2014;5:1–10. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- 126.Ledesma-Amaro R., Dulermo T., Nicaud J.M. Engineering Yarrowia lipolytica to produce biodiesel from raw starch. Biotechnol Biofuels. 2015;8:1–12. doi: 10.1186/s13068-015-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schüler L., Greque de Morais E., Trovão M., Machado A., Carvalho B., Carneiro M., et al. Isolation and characterization of novel Chlorella vulgaris mutants with low chlorophyll and improved protein contents for food applications. Front Bioeng Biotechnol. 2020;8:469. doi: 10.3389/fbioe.2020.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dall’Osto L., Cazzaniga S., Guardini Z., Barera S., Benedetti M., Mannino G., et al. Combined resistance to oxidative stress and reduced antenna size enhance light-to-biomass conversion efficiency in Chlorella vulgaris cultures. Biotechnol Biofuels. 2019;12:1–17. doi: 10.1186/s13068-019-1566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sankari M., Rao P.R., Hemachandran H., Pullela P.K., Doss C.G.P., Tayubi I.A., et al. Prospects and progress in the production of valuable carotenoids: Insights from metabolic engineering, synthetic biology, and computational approaches. J Biotechnol. 2018;266:89–101. doi: 10.1016/j.jbiotec.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 130.Azaman S.N.A., Wong D.C.J., Tan S.W., Yusoff F.M., Nagao N., Yeap S.K. De novo transcriptome analysis of Chlorella sorokiniana: effect of glucose assimilation, and moderate light intensity. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-74410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McCarthy S.S., Kobayashi M.C., Niyogi K.K. White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics. 2004;168:1249–1257. doi: 10.1534/genetics.104.030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mochida K., Koda S., Inoue K., Nishii R. Statistical and machine learning approaches to predict gene regulatory networks from transcriptome datasets. Front Plant Sci. 2018;9 doi: 10.3389/fpls.2018.01770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Anuntakarun S., Lertampaiporn S., Laomettachit T., Wattanapornprom W., Ruengjitchatchawalya M. mSRFR: a machine learning model using microalgal signature features for ncRNA classification. BioData Min. 2022;15 doi: 10.1186/s13040-022-00291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Osterloff J., Nilssen I., Eide I., de Oliveira Figueiredo M.A., de Souza Tâmega F.T., Nattkemper T.W. Computational visual stress level analysis of calcareous algae exposed to sedimentation. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Asplund-Samuelsson J., Hudson E.P. Wide range of metabolic adaptations to the acquisition of the Calvin cycle revealed by comparison of microbial genomes. PLoS Comput Biol. 2021:17. doi: 10.1371/journal.pcbi.1008742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lawson C.E., Martí J.M., Radivojevic T., Jonnalagadda S.V.R., Gentz R., Hillson N.J., et al. Machine learning for metabolic engineering: A review. Metab Eng. 2021;63:34–60. doi: 10.1016/j.ymben.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 137.Jervis A.J., Carbonell P., Vinaixa M., Dunstan M.S., Hollywood K.A., Robinson C.J., et al. Machine learning of designed translational control allows predictive pathway optimization in Escherichia coli. ACS Synth Biol. 2019;8:127–136. doi: 10.1021/acssynbio.8b00398. [DOI] [PubMed] [Google Scholar]
- 138.Liebal U.W., Phan A.N.T., Sudhakar M., Raman K., Blank L.M. Machine learning applications for mass spectrometry-based metabolomics. Metabolites. 2020;10:243. doi: 10.3390/metabo10060243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sanchez-Lengeling B., Aspuru-Guzik A. Inverse molecular design using machine learning: Generative models for matter engineering. Science. 2018;361:360–365. doi: 10.1126/science.aat2663. [DOI] [PubMed] [Google Scholar]
- 140.Krömer J.O., Heinzle E., Schröder H., Wittmann C. Accumulation of homolanthionine and activation of a novel pathway for isoleucine biosynthesis in Corynebacterium glutamicum McbR deletion strains. J Bacteriol. 2006;188:609–618. doi: 10.1128/JB.188.2.609-618.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Feng X., Tang K.H., Blankenship R.E., Tang Y.J. Metabolic flux analysis of the mixotrophic metabolisms in the green sulfur bacterium Chlorobaculum tepidum. J Biol Chem. 2010;285:39544–39550. doi: 10.1074/jbc.M110.162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dong Q.L., Zhao X.M., Ma H.W., Xing X.Y., Sun N.X. Metabolic flux analysis of the two astaxanthin-producing microorganisms Haematococcus pluvialis and Phaffia rhodozyma in the pure and mixed cultures. Biotechnol J. 2006;1:1283–1292. doi: 10.1002/biot.200600060. [DOI] [PubMed] [Google Scholar]
- 143.Tao Y., Liu D., Yan X., Zhou Z., Lee J.K., Yang C. Network identification and flux quantification of glucose metabolism in Rhodobacter sphaeroides under photoheterotrophic H(2)-producing conditions. J Bacteriol. 2012;194:274–283. doi: 10.1128/JB.05624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hendry J.I., Prasannan C.B., Joshi A., Dasgupta S., Wangikar P.P. Metabolic model of Synechococcus sp. PCC 7002: Prediction of flux distribution and network modification for enhanced biofuel production. Bioresour Technol. 2016;213:190–197. doi: 10.1016/j.biortech.2016.02.128. [DOI] [PubMed] [Google Scholar]
- 145.Minzu V., Ifrim G., Arama I. Control of microalgae growth in artificially lighted photobioreactors using metaheuristic-based predictions. Sensors (Basel). 2021:21. doi: 10.3390/s21238065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nikolaou A., Bernardi A., Meneghesso A., Bezzo F., Morosinotto T., Chachuat B. A model of chlorophyll fluorescence in microalgae integrating photoproduction, photoinhibition and photoregulation. J Biotechnol. 2015;194:91–99. doi: 10.1016/j.jbiotec.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 147.Kenny P., Flynn K.J. In silico optimization for production of biomass and biofuel feedstocks from microalgae. J Appl Phycol. 2015;27:33–48. doi: 10.1007/s10811-014-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xu Z., Jiang Y., Ji J., Forsberg E., Li Y., He S. Classification, identification, and growth stage estimation of microalgae based on transmission hyperspectral microscopic imaging and machine learning. Opt Express. 2020;28 doi: 10.1364/OE.406036. [DOI] [PubMed] [Google Scholar]
- 149.Luo J., Zhang H., Forsberg E., Hou S., Li S., Xu Z., et al. Confocal hyperspectral microscopic imager for the detection and classification of individual microalgae. Opt Express. 2021;29 doi: 10.1364/OE.438253. [DOI] [PubMed] [Google Scholar]
- 150.Long B., Fischer B., Zeng Y., Amerigian Z., Li Q., Bryant H., et al. Machine learning-informed and synthetic biology-enabled semi-continuous algal cultivation to unleash renewable fuel productivity. Nat Commun. 2022;13 doi: 10.1038/s41467-021-27665-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fisher C.L., Lane P.D., Russell M., Maddalena R., Lane T.W. Low molecular weight volatile organic compounds indicate grazing by the marine rotifer Brachionus plicatilis on the microalgae Microchloropsis salina. Metabolites. 2020;10:1–20. doi: 10.3390/metabo10090361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Thapa N., Chaudhari M., Iannetta A.A., White C., Roy K., Newman R.H., et al. A deep learning based approach for prediction of Chlamydomonas reinhardtii phosphorylation sites. Sci Rep. 2021:11. doi: 10.1038/s41598-021-91840-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shaker S., Morowvat M.H., Ghasemi Y. Bioinformatics analysis and identification of phytoene synthase gene in microalgae. Recent Pat Biotechnol. 2021;15:216–226. doi: 10.2174/1872208315666210712121951. [DOI] [PubMed] [Google Scholar]
- 154.Popova L.G., Belyaev D.V., Shuvalov A.V., Yurchenko A.A., Matalin D.A., Khramov D.E., et al. In silico analyses of transcriptomes of the marine green microalga Dunaliella tertiolecta: identification of sequences encoding P-type ATPases. Mol Biol. 2018;52:601–615. doi: 10.1134/S002689841804016X. [DOI] [PubMed] [Google Scholar]