Pairtree is a computational method that uses many samples from a cancer to build clone trees reconstructing the cancer's evolutionary history, thereby offering insight into the cancer's development.
Abstract
Cancers are composed of genetically distinct subpopulations of malignant cells. DNA-sequencing data can be used to determine the somatic point mutations specific to each population and build clone trees describing the evolutionary relationships between them. These clone trees can reveal critical points in disease development and inform treatment. Pairtree is a new method that constructs more accurate and detailed clone trees than previously possible using variant allele frequency data from one or more bulk cancer samples. It does so by first building a Pairs Tensor that captures the evolutionary relationships between pairs of subpopulations, and then it uses these relations to constrain clone trees and infer violations of the infinite sites assumption. Pairtree can accurately build clone trees using up to 100 samples per cancer that contain 30 or more subclonal populations. On 14 B-progenitor acute lymphoblastic leukemias, Pairtree replicates or improves upon expert-derived clone tree reconstructions.
Significance:
Clone trees illustrate the evolutionary history of a cancer and can provide insights into how the disease changed through time (e.g., between diagnosis and relapse). Pairtree uses DNA-sequencing data from many samples of the same cancer to build more detailed and accurate clone trees than previously possible.
See related commentary by Miller, p. 176.
This article is highlighted in the In This Issue feature, p. 171.
INTRODUCTION
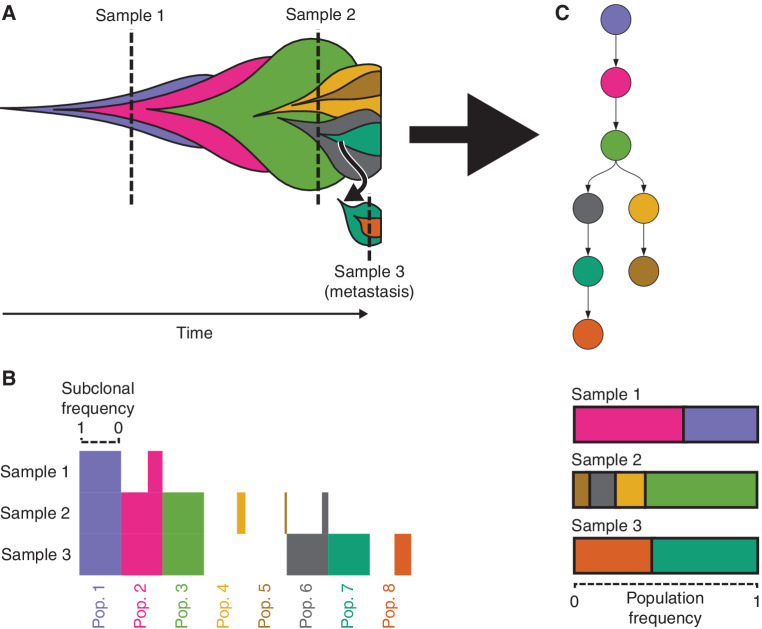
Individual cancers contain substantial genetic heterogeneity arising from an ongoing evolutionary process of random somatic mutation and selection (1). Cancers typically arise from a small number of founder mutations that confer a growth advantage (2). Over time, additional somatic mutations accrue, and their frequency and distribution are shaped by evolutionary forces such as selection and genetic drift, resulting in the emergence of multiple genetically distinct cell subpopulations (ref. 3; Fig. 1A). A clone tree is the evolutionary tree delineating the cell subpopulations in a cancer, the genetic mutations specific to each, and the proportions of cells in each sample that arose from each subpopulation (Fig. 1). Within the tree, subclones correspond to a cell subpopulation and all its descendants.
Figure 1.
Construction of clone trees from multiple cancer samples. A, Schematic illustrates cancer development under the clonal evolution model. Each color represents a genetically distinct subpopulation. Each subpopulation emerges within the mass of its parent. The leftmost point for a subpopulation denotes the cell that was its most recent common ancestor. Dashed vertical lines indicate when and where cancer samples were taken. The relative abundance of each subpopulation in a cancer sample, including any nested descendent subpopulations composing a subclone, is represented by the height of that subpopulation or subclone along the sample's dashed line. B, Horizontal bar plot showing idealized input to clone tree reconstruction algorithms. Bar length indicates the subclonal frequency of each subpopulation and its descendants (column) in each sequenced sample (row). The clonal evolution model asserts that a subpopulation's point mutations are inherited by its descendants. Consequently, mutation VAFs in DNA sequencing data provide estimates of subclonal frequencies, corresponding to the proportion of cells that originated from a subclonal population and its descendants. C, Clone tree representing the ancestry of subpopulations (top). Nodes indicate subpopulations. Arrows extend from each subpopulation to its direct descendants. Inferred frequencies of each subpopulation in each sample are based on the clone tree and mutation frequency data (bottom).
Clone trees built from bulk cancer samples have important biomedical applications. Those built from single samples already reveal important genomic events in evolution (3–5) and provide insights into heterogeneity (1). But as sequencing costs continue to drop, sequencing different regions of the same tumor (6), multiple tumors of the same cancer (7), or longitudinal samples from different timepoints (8) will become more common. When bulk samples have different mixtures of subpopulations, each sample can provide unique information about the single clone tree that characterizes the cancer's evolutionary history. This can include revealing new subpopulations or deconvolving large subpopulations into smaller constituents. Clone trees built from multiple samples of the same cancer have helped identify factors associated with metastasis (9) and probed how treatment (10–12) or tumor microenvironment (13, 14) shaped evolution. This, in turn, can inform strategies to counteract treatment resistance (15). Beyond cancer, clone trees have applications in other studies of somatic genetic heterogeneity (16, 17).
Current subclonal reconstruction methods (18–24) are severely limited in their ability to build clone trees based on large multi-sample studies. Most of these methods were designed for single cancer samples from which no more than three subclones can be discerned at typical whole-genome sequencing depths (1). Recent studies with greater sequencing depth and multiple cancer samples have revealed that a single cancer can have dozens of resolvable subclones (6, 11). Here we show that existing clone tree reconstruction methods become highly inaccurate on datasets with many subclones or many cancer samples, necessitating a new approach.
We introduce Pairtree, a new method that can accurately construct clone trees containing as many as 30 subclones. Pairtree outperforms a representative set of state-of-the-art clone tree reconstruction packages on simulated benchmark datasets of variable complexity. Pairtree is also the only method tested that can recover or improve upon expert reconstructions of clone trees for 14 B-progenitor acute lymphoblastic leukemias (B-ALLs) containing up to 90 samples and 26 subclones per cancer. The Pairtree method, along with an interactive visual interface for exploring the clone tree posterior, is available at https://github.com/morrislab/pairtree.
RESULTS
Pairtree Algorithm
Figure 1 outlines the process of constructing a clone tree to represent the evolutionary history of a cancer. Pairtree takes as input allele frequency data for point mutations detected in one or more samples from a single cancer. These data can be derived from whole-genome sequencing (WGS), whole-exome sequencing (WES), or more targeted sequencing. Each bulk cancer sample is a mixture of genetically heterogeneous cells (Fig. 1A). For each mutation, Pairtree uses counts of variant and reference reads in each sample to estimate the variant allele frequency (VAF), that is, the proportion of reads at a mutation's locus that contain the mutation. By correcting a mutation's VAF for copy-number aberrations (CNA) affecting the locus, Pairtree computes an estimate of the proportion of cells in each sample carrying the mutation, termed the mutation's subclonal frequency (ref. 25; Fig. 1B).
Pairtree outputs a set of possible clone trees explaining evolutionary relationships between the input mutations. Clone tree nodes correspond to cancerous subpopulations, while arrows (i.e., directed edges) extend from a subpopulation's node to the nodes representing its direct descendants (Fig. 1C). We define a subpopulation as those cells containing exactly the same subset of the somatic mutations input into Pairtree. In each cancer sample, each subpopulation is assigned a population frequency, representing what proportion of cells in that sample share the same mutation subset. Note that many, if not most, of a cancer's mutations will not be provided in the input because of incomplete genome coverage or because the mutations are too low in frequency to be detected.
Each subpopulation and its descendant subpopulations (both direct and indirect) form a subclone (Fig. 1A). Pairtree assigns a tree-constrained subclonal frequency to each subclone in each cancer sample, which is equal to the sum of the population frequencies of all the subpopulations contained within the subclone (Fig. 1A and B). This relationship follows from the infinite sites assumption (ISA), which states that no site is mutated more than once during cancer evolution. The ISA implies that subpopulations inherit all the mutations of their parent populations, and that each mutation appears only once in the evolutionary history of the cancer. Although violations of the ISA occur (26), it remains broadly valid (27), and if the input dataset includes ISA-violating mutations, Pairtree can detect and discard them before starting to build a clone tree (see Supplementary Information). Like most other clone tree reconstruction methods, Pairtree assumes the ISA when building trees. Other methods permit some, but not all, types of ISA violations (28–31).
Pairtree identifies which mutations belong to each subclone based on the estimated subclonal frequencies provided by the VAF data (Fig. 1B), then searches for clone trees whose structures allow subclonal frequencies that best match these estimates (Fig. 1C). This search is performed by inferring the evolutionary relationships between subclone pairs (Fig. 2A) and then using these to inform overall tree construction (Fig. 2B). Pairtree's output consists of a set of clone trees, each scored by a likelihood indicating how well the tree-constrained subclonal frequencies match the frequency estimates given by the VAF data (Fig. 2C). Although there is a single true clone tree explaining how subpopulations are related, this tree is not observed directly, and the input data often permit multiple solutions.
Figure 2.
The Pairtree algorithm. A, Pairtree uses VAF data to compute the Pairs Tensor. This tensor denotes the probability of every possible pairwise ancestral relationship between subclones (left). An initial clone tree is built using relationships scored by the Pairs Tensor. B, Pairtree samples trees using Markov Chain Monte Carlo. The method proposes tree modifications by identifying a subclone whose ancestral relationships in the current tree are assigned low probability by the Pairs Tensor (top), then ascertaining where that subclone can be moved within the tree to increase its ancestral relationship probabilities (bottom right). Proposed trees are then accepted or rejected based on their likelihoods that reflect how well they fit the VAF data (bottom left). C, Sampled clone trees are returned along with posterior probability estimates proportional to the likelihood of each tree.
Grouping mutations into subclones is not necessary—algorithms can instead build clone trees in which each mutation is assigned to a unique subclone, yielding a mutation tree. However, because of limited resolution in the data's estimated subclonal frequencies, sets of mutations often have subclonal frequency estimates that are too similar to separate the mutations into distinct subclones. As such, the first step in clone tree reconstruction is often clustering mutations with similar estimated subclonal frequencies across all input samples, and associating subclones with these clusters. Mutation clustering can be performed with Pairtree (see Supplementary Information) or by another method (32–34) and input into Pairtree. This step simplifies clone tree reconstruction by reducing the number of subclones. In addition, this approach permits more precise estimates of each subclone's subclonal frequency by combining data from the subclone's mutations (see Supplementary Information), at the risk of grouping together mutations from different subclones. Increasing the number of cancer samples provides more subclonal frequency estimates for each mutation, thereby reducing the risk of improper mutation grouping.
Pairtree Outperformed the State-of-the-art on Simulated Data
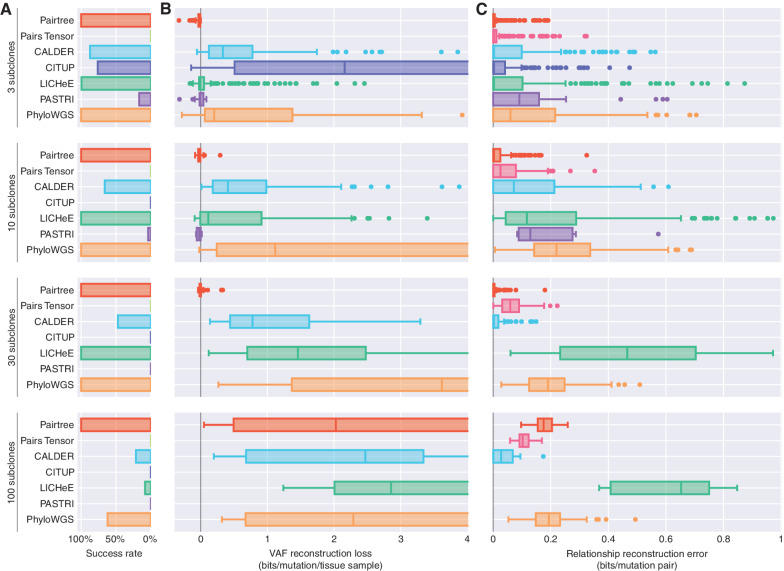
Figure 3 summarizes how Pairtree and alternative methods performed on simulated data, with an method's scores reflecting its performance on only the datasets for which it produced output (Supplementary Fig. S1 shows untruncated distributions). See Methods for how comparison methods were chosen and evaluation metrics were established. Pairtree was the only method that produced results for all 576 simulations (Fig. 3A). Nevertheless, Pairtree fared better than comparison methods on trees with 30 or fewer subclones, succeeding on all datasets while achieving negative median VAF losses (Fig. 3B and C). In fact, Pairtree always produced lower error than other methods for every such dataset (Supplementary Fig. S2), except for two datasets with three subclones and a single cancer sample where CALDER had negligibly better VAF losses (i.e., 0.002 bits lower or less). Pairtree also performed better than comparison methods with respect to relationship error. In general, for 30 subclones or fewer, relationship error was almost zero when the number of cancer samples exceeded the number of subclones (Supplementary Fig. S3 and S3B). For these cases, only one clone tree fit the ground-truth subclonal frequencies (Supplementary Fig. S4A) and Pairtree achieved low error by finding that tree or a close approximation thereof (Supplementary Fig. S4B and S4C). When applied to datasets with 100 subclones, Pairtree had higher VAF losses (Fig. 3B) and relationship errors (Fig. 3C) than with fewer subclones. Pairtree outperformed other methods for 100-subclone trees with respect to VAF loss, except for 16 datasets (15%) where PhyloWGS performed better (Supplementary Fig. S2) and 22 where CALDER was better. As a complement to relationship error, we also evaluated the methods with the tree error metric defined in other studies (23, 35), where Pairtree again showed good performance (Supplementary Fig. S5A and S5B). Furthermore, in evaluations limited to low-depth datasets where observations of mutation VAFs were less precise, Pairtree continued to perform well (Supplementary Figs. S6 and S7).
Figure 3.
Benchmark performance on 576 simulated datasets. Simulations are grouped by number of subclones (rows). A, Bar plots show each method's success rate in the group. Successes are reconstruction problems for which the method produced at least one tree in 24 hours (wall-clock time) and did not crash. B, Boxplots show distributions of VAF reconstruction losses for a method on a problem group. Scores reflect only datasets where a method ran successfully. VAF reconstruction loss is the decrease in average, per-mutation log likelihood of VAF data using subclonal frequencies assigned by the method, when compared with the true frequencies used to generate the data. Negative loss indicates better VAF reconstructions than true trees, while high loss indicates inaccurate tree structures. Midlines in box plots indicate medians. Plots are truncated at four bits. C, Boxplots show distributions of relationship reconstruction error in each group for each method's successful runs. Relationship reconstruction error is measured as the average Jensen–Shannon divergence per subclone pair between the true distributions over pairwise relations, and empirical distributions computed from the trees output by a method. Errors can range between zero bits (perfect match) and one bit (complete mismatch).
CITUP failed on all datasets with ten or more subclones, and on 32% of three-subclone cases (Fig. 3A). All failures on three-subclone datasets and 71% of failures on ten subclone datasets occurred because CITUP crashed (see Supplementary Information). The remaining 29% of ten subclone failures occurred because CITUP ran out of time. On the three-subclone cases where it ran successfully, its VAF loss was poor (Fig. 3B), perhaps because of a mismatch between its sequencing error model and the model used for computing VAF loss. Conversely, the method exhibited better relationship error than other non-Pairtree methods (Fig. 3C), suggesting its tree structures were more accurate.
PASTRI, which cannot run on datasets with more than 15 subclones, failed for 83% of three-subclone cases and 96% of ten-subclone cases (Fig. 3). For datasets with three or ten subclones, PASTRI produced output on 10%, terminated without producing a result on 84%, and ran out of time on the remaining 6% (see Supplementary Information). When it produced solutions, PASTRI generally performed well, reaching negative median VAF losses for three- and ten-subclone datasets, and relatively low relationship errors.
LICHeE fared better, producing results on all cases with 3, 10, or 30 subclones (Fig. 3). However, the method ran out of time for 92% of 100-subclone datasets. After Pairtree, LICHeE was the next-best performing method, with low VAF losses and moderate relationship errors on datasets with three or ten subclones, beating PhyloWGS on both measures. LICHeE performed less well on 30-subclone cases, where it exhibited lower VAF losses than PhyloWGS but higher relationship errors.
PhyloWGS produced clone trees for all datasets with 30 or fewer subclones (Fig. 3). In these cases, PhyloWGS generally had worse VAF losses and relationship errors than Pairtree or LICHeE, except for the 30-subclone datasets, where it had better relationship error than LICHeE but worse VAF loss. PhyloWGS performed better than other non-Pairtree methods on 100-subclone cases, where it finished within 24 hours for 62% of such datasets, but usually had higher VAF losses than Pairtree (Supplementary Fig. S2).
CALDER in its nonlongitudinal mode failed on 13% of three-subclone cases, 34% of ten-subclone cases, 53% of 30-subclone cases, and 79% of 100-subclone cases (Fig. 3). On datasets with 30 or fewer subclones where it succeeded, CALDER generally produced VAF losses lower than PhyloWGS and on par with LICHeE, and relationship errors that were better than all non-Pairtree methods. On the 21% of 100-subclone cases where it produced a result, CALDER exhibited performance that was generally the best of all methods, achieving lower VAF loss than Pairtree on 22 of the 108 datasets with 100 subclones.
Relationship error can also be measured for the Pairs Tensor alone, without requiring trees. The Pairs Tensor estimates pairwise relationships well (Fig. 3C), requiring only a fraction of the computational resources of the full Pairtree method (Supplementary Fig. S8). Although the Pairs Tensor does slightly worse than Pairtree on trees with 30 or fewer subclones, it has less relationship error than any other method. On datasets with 100 subclones, the Pairs Tensor was better able to delineate pairwise relationships between subclones than the full Pairtree method (Fig. 3C).
With respect to computational resources, Pairtree was competitive with other methods (Supplementary Figs. S9 and S10), particularly when compared on only the subset of datasets where other methods could produce answers (Supplementary Figs. S8 and S11).
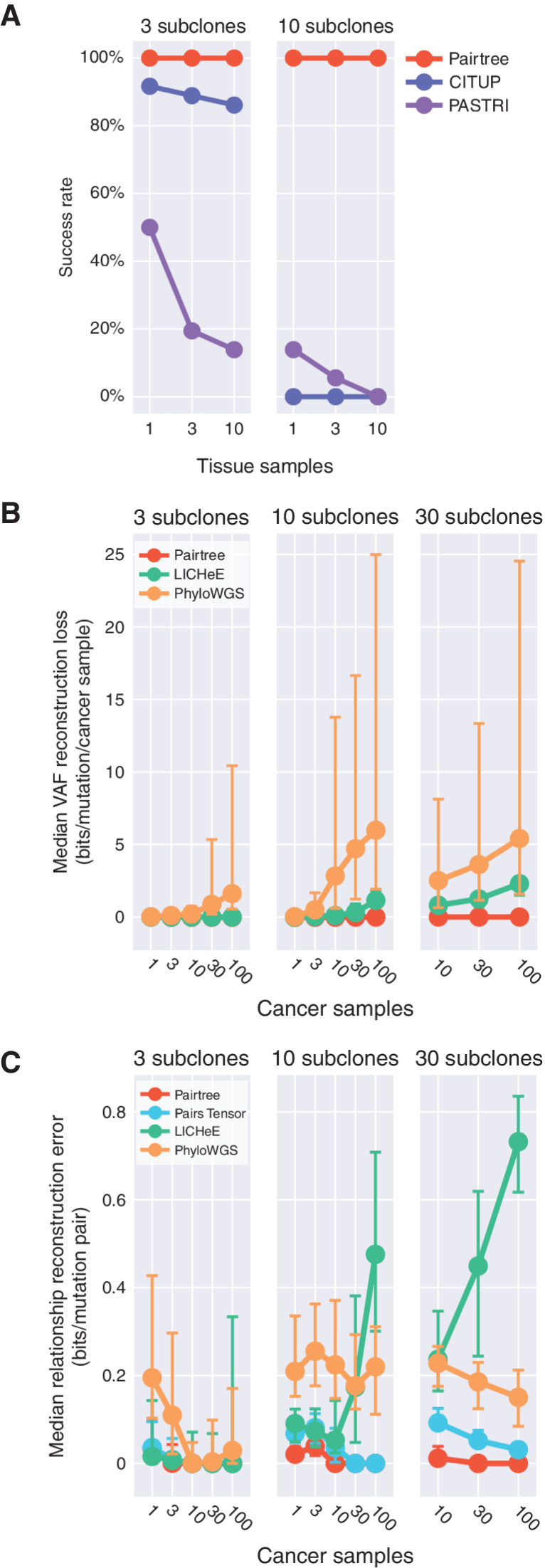
Pairtree Improved with More Cancer Samples; Other Methods Worsened
After controlling for other variables, all methods except Pairtree performed worse when provided more cancer samples. CITUP and PASTRI's failure rates increased with the number of cancer samples (Fig. 4A). Although LICHeE and PhyloWGS produced output for all cases with 30 subclones or fewer, they had higher VAF losses with more cancer samples (Fig. 4B). By contrast, Pairtree never failed and had nearly zero median VAF loss regardless of the number of simulated cancer samples on datasets with 30 subclones or fewer (Fig. 4A and B). Relationship errors decreased for both full Pairtree and the Pairs Tensor with more samples (Fig. 4C). LICHeE, conversely, exhibited rapidly increasing error with more samples, while PhyloWGS’ performance fluctuated. Because CALDER failed on subsets of all datasets when partitioned by number of subclones, we consider it separately. CALDER generally failed more frequently as the number of subclones or number of cancer samples increased (Supplementary Fig. S12), while its VAF loss was largely independent of the number of subclones (Supplementary Fig. S13).
Figure 4.
Performance on simulated datasets as a function of number of subclones and cancer samples. CALDER is not shown because it succeeded only on subsets of the different dataset groups shown, while the methods represented here succeeded on all datasets in the depicted groups. A, Method success rate. For CITUP and PASTRI, success rate depended on the number of subclones and/or cancer samples in datasets. Pairtree, LICHeE, and PhyloWGS succeeded on all datasets depicted. B, Median VAF reconstruction loss as a function of number of samples. For LICHeE and PhyloWGS, VAF loss increases with more cancer samples. C, Median relationship reconstruction error as a function of number of samples. LICHeE's error generally increased with more cancer samples, while other methods showed the opposite effect. Error bars represent the first and third quartiles in (B and C).
Pairtree Detected Mutations that Violate the Infinite Sites Assumption
Pairtree's Pairs Tensor can be used to identify mutations that violate the ISA (see Supplementary Information). We evaluated Pairtree's ISA violation detection algorithm under four different scenarios—technical noise (i.e., sequencing artifacts), homoplasy, back mutation, and miscalled CNAs—for trees with 10 subclones (Supplementary Fig. S14A and S14C) or 30 subclones (Supplementary Fig. S14B and S14D). This evaluation was performed under two different strengths of evidence, encompassing strong support (i.e., a 5% difference in VAF, implying a 10% difference in subclonal frequency; Supplementary Fig. S14C and S14D) and weak support (i.e., a 0.025% difference in VAF, implying a 0.05% difference in subclonal frequency; Supplementary Fig. S14A and S14B). Pairtree had 100% precision and recall for simulated sequencing artifacts in all scenarios, and its precision for finding ISA-violating mutations did not drop below 99% for the other three cases. Its recall exceeded 97% for all cases within these three except for the weak-support case on 30-subclone trees, where its recall dropped to 88% for homoplasy and back mutation. This demonstrated that Pairtree could detect ISA violations nearly perfectly in most scenarios, save for two where its detection was still excellent. Moreover, this performance was insensitive to hyperparameters used in the algorithm (Supplementary Fig. S15). Pairtree also provides an alternative means of detecting mutations affected by putative loss of heterozygosity (LOH) events without computing pairwise relations, where it again showed strong performance (Supplementary Fig. S16A–S16D).
Pairtree Met or Exceeded Expert Baselines on Real Data
We applied Pairtree, CALDER, CITUP, LICHeE, PASTRI, and PhyloWGS to genomic data from 14 B-ALL patients (11). Samples were obtained at diagnosis and relapse for each patient. In addition, each sample was transplanted into immunodeficient mice, generating multiple patient-derived xenografts (PDX). The patient samples were profiled using WES, while the PDXs were used targeted sequencing based on leukemic variants found in the patient WES data. There were 16 to 509 mutations called per patient (median 40), clustered into 5 to 26 subclones per patient (median 8). By combining patient and PDX samples, we obtained between 13 and 90 tissue samples per cancer (median 42). Across cancers, the median read depth was 212 reads.
To define an expert-derived baseline for these datasets, we first built high-quality clone trees for each dataset manually, subjecting them to extensive review and refinement before evaluating them for biological plausibility (11). Then we used the same technique as Pairtree to fit tree-constrained subclonal frequency estimates to the VAF data. The data fit of these estimates, as computed by likelihood, yielded the expert-derived baseline. As a reminder, methods that improve on the baseline achieve negative VAF losses.
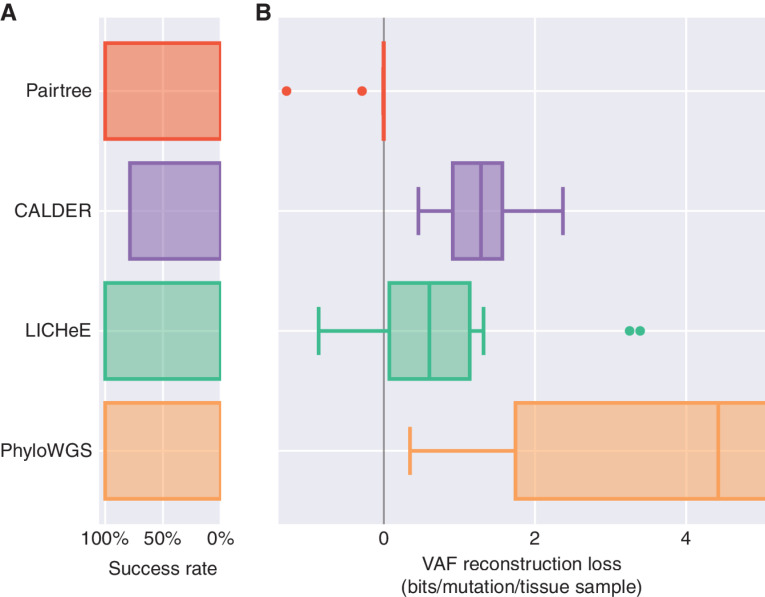
CITUP and PASTRI failed on 13 of the 14 cancers, and so we excluded these methods from the comparison. Pairtree, LICHeE, and PhyloWGS produced results for all 14 cancers (Fig. 5A; Supplementary Fig. S17), while CALDER failed on three of them. Pairtree found trees that fit the sequencing data as well as, or slighter better than, the expert baseline for 12 of 14 cancers (Fig. 5B), achieving VAF losses between 0 and −0.05 bits. On two cancers, Pairtree inferred clone trees that fit the VAF data substantially better than the expert baseline, resulting in negative losses of −0.32 bits and −1.42 bits. LICHeE beat the baseline for one cancer, reaching a negative loss of -0.86 bits; (nearly) matched the baseline for four other patients, incurring between 0 and 0.11 bits of loss; and had substantially worse VAF losses for the remaining nine patients. PhyloWGS suffered at least 0.35 bits of loss on all patients, reaching a median VAF loss of 4.42 bits. As PhyloWGS could not adhere to the expert-derived clustering, unlike other methods, it often merged clusters incorrectly, causing high VAF loss. CALDER failed on 3 of the 14 cancers (Fig. 5A), was worse than Pairtree on all the other 11, and worse than LICHeE on 9 of the 11.
Figure 5.
Method performance loss for 14 B-ALL patient datasets. The number of cancer samples for each dataset ranged from 13 to 90. A, Pairtree, LICHeE, and PhyloWGS succeeded on all 14 datasets. CALDER succeeded on only 11 of the 14 (79%). CITUP and PASTRI each failed on 13 of 14 datasets and so are not shown. B, VAF loss on the subset of datasets where each method succeeded. VAF reconstruction losses are reported as a negative log likelihood normalized to the number of mutations and cancer samples, relative to the MAP subclonal frequencies for expert-derived trees. Lower loss indicates better performance, while negative loss corresponds to performance better than human experts. Mid-lines in box plots indicate medians. The axis is truncated at 5 bits.
Consensus Graphs Illustrate Uncertainties in Clone Tree Reconstructions
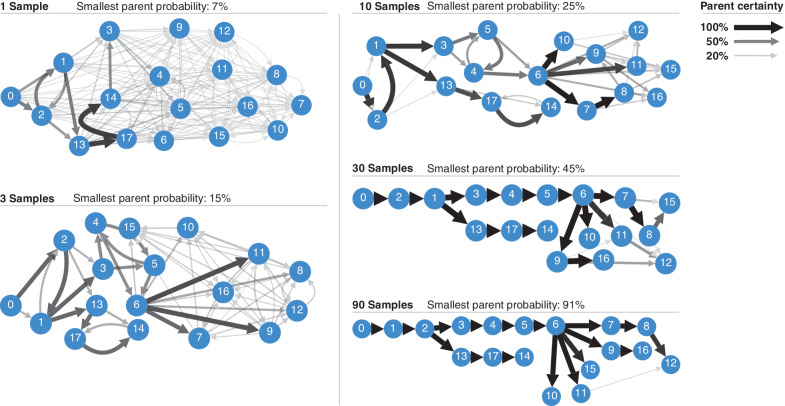
Pairtree provides interactive visualizations to help navigate the multiple clone tree solutions that it produces for each dataset (Fig. 6). By using the likelihoods associated with each solution as weights, Pairtree produces a weighted consensus graph, in which the nodes represent subclones, and each directed edge is assigned a weight equal to the marginal probability that it appears in a clone tree drawn from the empirical clone tree distribution produced by Pairtree. Thus, the consensus graph summarizes the estimated posterior probability of each parental relationship between subclones. These summaries are useful for interpreting Pairtree's results, as they provide a concise representation of the evolutionary relationships supported by the data, alongside the confidence underlying each. By taking the maximum-weight spanning tree of this graph, the user can generate a single consensus tree. To demonstrate the consensus graph's utility, we ran Pairtree multiple times on one of the B-ALL cases from Fig. 5, using variable numbers of cancer samples (Fig. 6). As we provided more cancer samples, confidence in evolutionary relationships increased, until all parents were resolved with near certainty. Providing more samples also corrected erroneous inferences—with 30 samples, population 8 appeared to be the likely parent of population 15, but with 90 samples, it became clear that population 15's parent is population 6.
Figure 6.
Consensus graph visualization of posterior tree distributions. These consensus graph visualizations are based on one of the 14 B-ALL cancers analyzed with Pairtree, for which 90 cancer samples were available. Consensus graphs are shown for variable numbers of samples, ranging from a single sample to all 90. All edges with less than 5% posterior probability are hidden. The minimum spanning tree certainty is the minimum of the maximum parent probabilities of each subclone.
DISCUSSION
Pairtree is the first automated method that reliably recovers large, complex clone trees from bulk DNA-sequencing data. For simulated clone trees with up to 30 subclones, Pairtree's reconstructed clone trees almost always fit the VAF data as well as or better than the original clone trees used to generate the data. On 14 B-ALL cancers, with up to 26 subclones and 90 samples per cancer, Pairtree's clone trees fit the VAF data as well as, or better than, those constructed by experts. No other tested method was consistently accurate on real or simulated benchmarks containing ten subclones or more. Pairtree was also the only method whose clone trees reliably became more accurate when more samples were used in the reconstructions. This is surprising—as each cancer sample provides additional information about evolutionary relationships between subpopulations, subclonal reconstruction problems should become easier with more cancer samples, not more difficult.
Identifying the correct clone tree for a given dataset may not be possible, and so Pairtree is specifically designed to identify and report ambiguities in the clone tree reconstruction. For example, the relationships among subclones with low VAFs in all samples may not be possible to resolve because, depending on the read coverage, the low VAFs might be consistent with multiple ancestral relationships between the subclone pair. In these circumstances, Pairtree is designed to capture this uncertainty in both its Pairs Tensor and through its MCMC-derived samples from the clone tree posterior. In addition, due to incomplete genome coverage or the inherent sequencing limits of mutation detection, all clone trees provide an incomplete view of a cancer's evolution. Accounting for uncertainty under these conditions becomes even more important because of the difficulty of resolving the true tree.
A key factor in Pairtree's success is its efficient search through the space of clone trees. Beyond ten subclones, this tree space quickly becomes too large for exhaustive enumeration (CITUP) or unguided stochastic search (PhyloWGS). Even methods that reduce the search space by applying hard constraints to exclude some parent–child relationships (LICHeE, PASTRI, CALDER) can fail to recover clone trees with more subclones because as the number of samples increases, these hard constraints become more likely to be incorrect and thus exclude the correct solution (see Supplementary Information). By contrast, Pairtree's stochastic tree search is guided by the Pairs Tensor, which provides soft constraints defined by a well-motivated probability model. Consequently, Pairtree's constraints become more precise as more cancer samples are provided, without excluding the true clone tree.
As Pairtree's performance degrades on the 100-subclone benchmarks, alternative search strategies may be necessary for very large clone trees. While Pairtree almost always fails to correctly resolve a subclone's parent (Supplementary Fig. S4C), it achieves relatively low relationship error (Supplementary Fig. S4D), suggesting it may be capturing the coarse tree structure. If so, Pairtree may fare better using a tiered approach, in which it would group together subclones with similar pairwise relations to others, build subtrees for each group separately, and then connect the subtrees using the groups’ pairwise relations to compose the full clone tree. Given 100 subclones with 10 or more cancer samples, the Pairs Tensor is already better than Pairtree itself at capturing the correct evolutionary relationships between subclones (Supplementary Fig. S3A–S3C). Future work should focus on understanding what conditions (e.g., high read depth or many cancer samples) under which the Pairs Tensor converges to a partial clone tree (36) that succinctly summarizes all clone trees with nonnegligible posterior probability.
Future extensions of Pairtree could incorporate alternative models of cancer evolution by introducing nonuniform priors on clone tree structure or subclonal frequencies. Any evolutionary model that can assign a likelihood to a given clone tree structure and set of subclonal frequencies can be immediately incorporated in the Metropolis–Hastings scoring, though some subclonal frequency priors may make the subclonal frequency optimization nonconvex or reduce the accuracy of the MAP approximation to the marginal likelihood of the tree (see Supplementary Information). In addition, nonuniform priors may decrease the value of the Pairs Tensor as a proposal distribution for tree inference, but, encouragingly, any prior that permits tractable computation of their marginal distributions over subclone pairs can also be incorporated into the Pairs Tensor. For example, CALDER's longitudinal constraints (i.e., once a subclone goes extinct at a given timepoint, it never returns; ref. 35) can be incorporated as time-dependent priors on subclonal frequencies for individual subclones and subclone pairs where one is the ancestor of the other. The Supplementary Information describes some possible alternative evolutionary models in detail.
Throughout this work, we have stressed performance metrics that recognize there are often many solutions consistent with observed data (see Supplementary Information), extending previous ones that compare single clone trees to one another (see (23, 35) and others). We developed new metrics that extend ones we previously developed (24) to score multiple candidate solutions from a method against a single ground-truth tree. Our new metrics permit the ground truth to be uncertain, with multiple potential truths equally consistent with noise-free observations. In general, characterizing uncertainty in clone tree reconstructions is critical. Even when methods produce multiple solutions, users typically want a single answer, and so select the highest-scoring tree while neglecting other credible candidates that fit their data nearly as well. Consequently, they lose information about which evolutionary relationships between subclones are well defined by the data, and which are uncertain because they have multiple equally likely possibilities. If users are to benefit from a method's ability to produce multiple solutions, the method must provide tools for interpreting this uncertainty. Pairtree's weighted consensus graph characterizes the uncertainty present in each evolutionary relationship, depicting all credible possibilities and the confidence underlying each (Fig. 6). This allows users to make informed conclusions about their data.
In summary, Pairtree can reconstruct highly accurate trees representing the evolutionary relationships among up to 30 subclones based on sequencing data from up to 100 samples from a cancer. Using pairwise mutation relationships, Pairtree can detect mutations that violate the ISA (see Supplementary Information) or have technical issues corrupting their observed data. By scaling to many more subclones and cancer samples than past approaches, and by illustrating the uncertainty present in solutions, Pairtree can address questions in many cancer research domains. These include understanding the origin and progression of tumors, measuring tumor age and heterogeneity, mapping out mechanisms of tumor adaptation to therapy, and understanding the relationship between primaries and metastases. Pairtree also has applications beyond cancer, where it can be used to examine somatic evolution in noncancerous tissues for any asexually dividing cell population. In the future, the Pairtree framework can be extended to scale to even more complex trees, integrate single-cell sequencing data, and permit violations of the infinite sites assumption (see Supplementary Information).
METHODS
Here we provide a nontechnical description of the Pairtree methods. The Supplementary Information contains a concise, formal description of the algorithms and the remaining Supplementary Data sections expand on this concise summary, provide motivation for some of our design choices, and provide some analysis of the solution space of the simulations.
Delineating Ancestral Relationships between Pairs of Subclones using the Pairs Tensor
Pairtree uses the estimated subclonal frequencies to predict the ancestral relationship between every subclone pair. These pairwise relationships then serve as a guide when Pairtree searches for clone trees that best fit the VAF data. Under the ISA, one of three mutually exclusive ancestral relationships exist between an ordered pair of subclones A and B (23, 37).
Ancestor A is ancestral to B. Here, the subpopulation associated with A contains A's mutations but not B's. No cell subpopulation has B's mutations without also inheriting A's.
Descendant B is ancestral to A. As above but with the roles of A and B switched.
Branching neither A nor B is the ancestor of the other. That is, they occur on different branches of the clone tree. Consequently, no subpopulations have both A's and B's mutations.
Each relationship constrains the frequencies that can be assigned to the two subclones (see Supplementary Information). For a given subclone pair, Pairtree combines a prior probability distribution incorporating these constraints with a likelihood distribution based on the VAF data for each subclone's mutations, then uses Bayesian inference to compute the probability of each relationship type for the pair (see Supplementary Information). This yields a data structure termed the Pairs Tensor (Fig. 2A), the elements of which are the marginal posterior probability distributions over the three possible ancestral relationships for every subclone pair.
Using Pairwise Ancestry to Guide the Search for Clone Trees
Pairtree uses the Pairs Tensor to define a proposal distribution for a Markov Chain Monte Carlo (MCMC) algorithm (38) that samples from the posterior distribution over clone trees (Fig. 2B). The algorithm's Metropolis–Hastings scheme generates proposal trees using two distributions over subclones derived from the Pairs Tensor (see Supplementary Information). The first distribution helps choose a poorly placed subclone to move within the tree, with each subclone's selection probability determined by the degree of discordance between the data-implied pairwise relationships and those imposed by its present position within the tree. The second distribution guides the choice of new parent for the selected subclone, evaluating potential destinations based on how much this discordance is decreased. Though other MCMC-based subclonal reconstruction methods also modify trees by moving subclones (18, 20, 39) or mutations (40, 41), Pairtree is the first to guide this decision with data, allowing the algorithm to rapidly navigate to and explore high-probability regions of the clone-tree posterior.
Pairtree uses a maximum a posteriori (MAP) approximation of the clone tree's marginal likelihood, both for the Metropolis–Hastings accept–reject decision and for estimating the tree's posterior probability (Fig. 2C). The Bayesian prior enforces tree constraints but is otherwise uninformative. By this constraint, the root subclone must have a subclonal frequency of 1 in every sample, as it corresponds to the germline and all subclones are descended from it. In addition, the prior requires that every subclone has a frequency greater than or equal to the sum of its direct descendants’ subclonal frequencies. Pairtree can compute the MAP estimate either using a fast approximate scheme (42) or a slower exact one (see Supplementary Information). A clone tree's likelihood scores how well the variant and reference read counts for each mutation match the MAP subclonal frequencies under a binomial sequencing noise model that includes the provided CNA correction for the mutation.
Benchmarking Pairtree Performance using Novel Scoring Metrics
Evaluating Pairtree against other common subclonal reconstruction methods required developing new metrics, as existing metrics are limited to datasets with single cancer samples (24), do not consider uncertainty about the best-fitting clone tree (23), or both. Below, we introduce two novel metrics well-suited for the multisample domain that also permit uncertainty about the best-fitting clone tree.
The first, termed VAF reconstruction loss, uses likelihood to compare the data fit of a tree's subclonal frequencies to a baseline (see Supplementary Information). For simulated data, the baseline frequencies are the ground-truth frequencies used to generate the VAF data. For real data with an unknown ground truth, the baseline is MAP subclonal frequencies computed for an expert-constructed clone tree. If a method outputs multiple clone trees, the VAF reconstruction loss of this solution set is the average loss of each clone tree, weighted by the likelihood the method associated to the tree. Negative VAF losses indicate the evaluated frequencies have better data fit than the baseline. Importantly, this is an unbiased metric can be used even when the ground-truth is unknown, or when the simulated data supports a better-fitting clone tree than the one to generate it in the first place.
The second evaluation metric, termed relationship reconstruction error, compares the structure of candidate clone trees to the ground truth (see Supplementary Information) using the evolutionary relationships between subclone pairs. This metric is a generalization of previous pairwise-relation-dependent metrics (23, 24) to permit the comparison of distributions over clone trees to one another. The metric permits uncertainty in the ground truth clone tree while also rewarding methods that report multiple clone trees when the correct solution is indeed uncertain. To compute it, we construct an empirical Pairs Tensor from the clone tree solutions found by a method, then compare it via the Jensen–Shannon divergence (JSD) to a tensor based on the ground truth. As multiple clone trees may be consistent with the ground-truth subclonal frequencies, we construct the ground-truth Pairs Tensor by enumerating all trees consistent with these frequencies (36) and denoting the pairwise relationships between subclones that each expresses. Building the ground-truth collection of clone trees requires knowing the ground-truth subclonal frequencies with no measurement error, so this metric is best suited to simulated data.
Selecting Comparison Methods and Generating Simulated Data
Clone tree reconstruction methods use one of two approaches: exhaustive enumeration or stochastic search. To evaluate Pairtree, a stochastic search method, we compared it against four exhaustive enumeration methods [CALDER (35), PASTRI (23), CITUP (19), and LICHeE (22)) and one stochastic search method (PhyloWGS (41).] All methods produce multiple candidate clone trees that are scored based on how well their tree-constrained subclonal frequencies fit the observed VAF data (see also ref. 43).
We assessed method performance on 576 simulated datasets with variable read depths and numbers of subclones, cancer samples, and mutations. These included trees with 3, 10, 30, and 100 subclones. Three subclones are the most that can typically be resolved at WGS read depths of 50x (1). In multi-sample datasets, ten subclones are often discernible (6), while 30 was the approximate maximum we could resolve in the high-depth, many-sample B-ALL data evaluated here (11). We also included trees with 100 subclones to probe the methods’ limits, anticipating challenges presented by future datasets. The number of simulated cancer samples ranged from 1 to 100.
We designed the simulation process to generate realistic, diverse, and resolvable clone trees (see Supplementary Information). In this simulation framework, most large trees consist of subclones comprised of only a few subpopulations (Supplementary Fig. S18). As trees grow larger, they are dominated by subpopulations whose frequency becomes small (Supplementary Fig. S19A), such that the populations become difficult to place in the tree, while the variance in subclone frequency across cancer samples also becomes less (Supplementary Fig. S19B), reducing the value provided by multiple samples. We did not include one- or three-sample datasets in the 30- and 100-subclone simulations, as resolving so many subclones from so few samples would be unrealistic—in these cases, 39% or more of subpopulations would have frequencies less than 1% across all cancer samples (Supplementary Fig. S20). Building trees from such subpopulations is also difficult, as many subtrees within the larger tree will be comprised solely of such small-frequency subpopulations (Supplementary Fig. S21), so that any arrangement of subpopulations within that subtree would be nearly equally consistent with the data.
Methods were allowed up to 24 hours of wall-clock time to produce results. Some caveats must be noted. LICHeE does not report subclonal frequencies for its solutions, so we used Pairtree to fit MAP frequencies to LICHeE's trees. Although LICHeE does not produce a likelihood, unlike the other methods here, it reports an error score for each tree that we interpreted as a likelihood when weighting its solutions. PhyloWGS, unlike other methods, could not use a fixed mutation clustering. This led to the method incorrectly merging clusters, causing artificially high VAF loss and relationship error. More generally, all methods except Pairtree failed to produce output on some simulated datasets. These failures stemmed from methods terminating without producing output, crashing outright, or failing to finish within 24 hours (see Supplementary Information).
Data and Source Code Availability
Real and simulated data used to evaluate the methods are available at https://github.com/morrislab/pairtree-experiments. The framework used to generate simulated data is available at https://github.com/morrislab/pearsim. Pairtree is available at https://github.com/morrislab/pairtree.
Supplementary Material
Acknowledgments
This research was partially supported by NIH/NCI Cancer Center Support Grant P30 CA008748, a CIFAR Catalyst AI grant to Q.M., and a Vector Institute Research Grant. Q. Morris holds a Canada CIFAR AI chair. Experiments were run using computational resources provided by SciNet and Compute Canada. The authors gratefully acknowledge Bei Jia and José Bento for extending their method for computing subclonal frequencies. Alexandre Bouchard-Côté, Kieran Campbell, and Jared Simpson provided invaluable feedback on this project as members of J.A. Wintersinger's PhD committee.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
No disclosures were reported.
Authors’ Contributions
J.A. Wintersinger: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. S.M. Dobson: Resources, data curation, validation, investigation, writing–review and editing. E. Kulman: Software, visualization, writing–review and editing. L.D. Stein: Resources, supervision, funding acquisition, project administration, writing–review and editing. J.E. Dick: Conceptualization, resources, data curation, supervision, validation, investigation, project administration, writing–review and editing. Q. Morris: Conceptualization, resources, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Dentro SC, Leshchiner I, Haase K, Tarabichi M, Wintersinger J, Deshwar AG, et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell 2021;184:2239–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 3. Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Espiritu SMG, Liu LY, Rubanova Y, Bhandari V, Holgersen EM, Szyca LM, et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell 2018;173:1003–13. [DOI] [PubMed] [Google Scholar]
- 5. Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell 2012;149:994–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TB, Veeriah S, et al. Tracking the evolution of non–small-cell lung cancer. N Engl J Med 2017;376:2109–21. [DOI] [PubMed] [Google Scholar]
- 7. Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015;520:353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sakamoto H, Attiyeh MA, Gerold JM, Makohon-Moore AP, Hayashi A, Hong J, et al. Evolutionary origins of recurrent pancreatic cancer. bioRxiv 2019;811133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alves JM, Prado-López S, Cameselle-Teijeiro JM, Posada D. Rapid evolution and biogeographic spread in a colorectal cancer. Nat Commun 2019;10:5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Z, Ding J, Ma Z, Sun R, Seoane JA, Shaffer JS, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 2019;51:1113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dobson SM, García-Prat L, Vanner RJ, Wintersinger J, Waanders E, Gu Z, et al. Relapse fated latent diagnosis subclones in acute B lineage leukemia are drug tolerant and possess distinct metabolic programs. Cancer Discov 2020;10:568–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Z, Li Z, Ma Z, Curtis C. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat Genet 2020;52:701–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zahir N, Sun R, Gallahan D, Gatenby RA, Curtis C. Characterizing the ecological and evolutionary dynamics of cancer. Nat Genet 2020;52:759–67. [DOI] [PubMed] [Google Scholar]
- 14. Williams MJ, Werner B, Heide T, Curtis C, Barnes CP, Sottoriva A, et al. Quantification of subclonal selection in cancer from bulk sequencing data. Nat Genet 2018;50:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pogrebniak KL, Curtis C. Harnessing tumor evolution to circumvent resistance. Trends Genet 2018;34:639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coorens THH, Oliver TRW, Sanghvi R, Sovio U, Cook E, Vento-Tormo R, et al. Inherent mosaicism and extensive mutation of human placentas. Nature 2021;592.80–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bizzotto S, Dou Y, Ganz J, Doan RN, Kwon M, Bohrson CL, et al. Landmarks of human embryonic development inscribed in somatic mutations. Science 2021;371:1249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang Y, Qiu Y, Minn AJ, Zhang NR. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc Natl Acad Sci U S A 2016;113:E5528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malikic S, McPherson AW, Donmez N, Sahinalp CS. Clonality inference in multiple tumor samples using phylogeny. Bioinformatics 2015;31:1349–56. [DOI] [PubMed] [Google Scholar]
- 20. Malikic S, Jahn K, Kuipers J, Sahinalp SC, Beerenwinkel N. Integrative inference of subclonal tumour evolution from single-cell and bulk sequencing data. Nat Commun 2019;10:2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deshwar AG, Vembu S, Morris Q. Comparing nonparametric Bayesian tree priors for clonal reconstruction of tumors. Pac Symp Biocomput 2015;20–31. [PubMed] [Google Scholar]
- 22. Popic V, Salari R, Hajirasouliha I, Kashef-Haghighi D, West RB, Batzoglou S. Fast and scalable inference of multi-sample cancer lineages. Genome Biol 2015;16:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Satas G, Raphael BJ. Tumor phylogeny inference using tree-constrained importance sampling. Bioinformatics 2017;33:i152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salcedo A, Tarabichi M, Espiritu SMG, Deshwar AG, David M, Wilson NM, et al. A community effort to create standards for evaluating tumor subclonal reconstruction. Nat Biotechnol 2020;38.97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dentro SC, Wedge DC, Van Loo P. Principles of reconstructing the subclonal architecture of cancers. Cold Spring Harb Perspect Med 2017;7:a026625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuipers J, Jahn K, Raphael BJ, Beerenwinkel N. Single-cell sequencing data reveal widespread recurrence and loss of mutational hits in the life histories of tumors. Genome Res 2017;27:1885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singer J, Kuipers J, Jahn K, Beerenwinkel N. Single-cell mutation identification via phylogenetic inference. Nat Commun 2018;9:5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McPherson A, Roth A, Laks E, Masud T, Bashashati A, Zhang AW, et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat Genet 2016;48:758–67. [DOI] [PubMed] [Google Scholar]
- 29. Bonizzoni P, Ciccolella S, Della Vedova G, Soto M. Does relaxing the infinite sites assumption give better tumor phylogenies? An ILP-based comparative approach. bioRxiv 2017. [DOI] [PubMed] [Google Scholar]
- 30. Ciccolella S, Soto Gomez M, Patterson M, Della Vedova G, Hajirasouliha I, Bonizzoni P. Inferring cancer progression from single-cell sequencing while allowing mutation losses. bioRxiv 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Satas G, Zaccaria S, Mon G, Raphael BJ. SCARLET: single-cell tumor phylogeny inference with copy-number constrained mutation losses. Cell Syst 2020;10:323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014;11:396–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 2014;10:e1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gillis S, Roth A. PyClone-VI: scalable inference of clonal population structures using whole genome data. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Myers MA, Satas G, Raphael BJ. CALDER: inferring phylogenetic trees from longitudinal tumor samples. Cell Syst 2019;8.514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sundermann LK, Wintersinger J, Rätsch G, Stoye J, Morris Q. Reconstructing tumor evolutionary histories and clone trees in polynomial-time with SubMARine. PLoS Comput Biol 2021;17.1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tarabichi M, Salcedo A, Deshwar AG, Ni Leathlobhair M, Wintersinger J, Wedge DC, et al. A practical guide to cancer subclonal reconstruction from DNA sequencing. Nat Methods 2021;18:144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hastings WK. Monte carlo sampling methods using markov chains and their applications. Biometrika 1970;57:97–109. [Google Scholar]
- 39. Jahn K, Kuipers J, Beerenwinkel N. Tree inference for single-cell data. Genome Biol 2016;17:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiao W, Vembu S, Deshwar AG, Stein L, Morris Q. Inferring clonal evolution of tumors from single nucleotide somatic mutations. BMC Bioinf 2014;15:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deshwar AG, Vembu S, Yung CK, Jang GH, Stein L, Morris Q. PhyloWGS: reconstructing subclonal composition and evolution from whole genome sequencing of tumors. Genome Biol 2015;16:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jia B, Ray S, Safavi S, Bento J. Efficient projection onto the perfect phylogeny model. arXiv 2018. [Google Scholar]
- 43. Strino F, Parisi F, Micsinai M, Kluger Y. TrAp: a tree approach for fingerprinting subclonal tumor composition. Nucleic Acids Res 2013;41:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.