Recurrent tandem duplications of UBTF exon 13 are associated with poor outcome in pediatric AML and likely define a unique molecular category.
Abstract
The genetics of relapsed pediatric acute myeloid leukemia (AML) has yet to be comprehensively defined. Here, we present the spectrum of genomic alterations in 136 relapsed pediatric AMLs. We identified recurrent exon 13 tandem duplications (TD) in upstream binding transcription factor (UBTF) in 9% of relapsed AML cases. UBTF-TD AMLs commonly have normal karyotype or trisomy 8 with cooccurring WT1 mutations or FLT3-ITD but not other known oncogenic fusions. These UBTF-TD events are stable during disease progression and are present in the founding clone. In addition, we observed that UBTF-TD AMLs account for approximately 4% of all de novo pediatric AMLs, are less common in adults, and are associated with poor outcomes and MRD positivity. Expression of UBTF-TD in primary hematopoietic cells is sufficient to enhance serial clonogenic activity and to drive a similar transcriptional program to UBTF-TD AMLs. Collectively, these clinical, genomic, and functional data establish UBTF-TD as a new recurrent mutation in AML.
Significance:
We defined the spectrum of mutations in relapsed pediatric AML and identified UBTF-TDs as a new recurrent genetic alteration. These duplications are more common in children and define a group of AMLs with intermediate-risk cytogenetic abnormalities, FLT3-ITD and WT1 alterations, and are associated with poor outcomes.
See related commentary by Hasserjian and Nardi, p. 173.
This article is highlighted in the In This Issue feature, p. 171.
INTRODUCTION
Children with acute myeloid leukemia (AML) have a long-term survival rate that is still below 70% (1). Most children with AML respond to initial treatment and achieve complete remission, but many experience a relapse. Relapsed AML is typically treated with chemotherapy or a wide variety of novel agents, followed by allogeneic hematopoietic stem cell transplantation (2). Unfortunately, AML after relapse is often refractory, and many patients eventually succumb to the relapsed disease (1–3).
Cytogenetic and molecular studies on pediatric AMLs have led to the identification of genetic alterations that are used to risk-stratify patients at diagnosis. For example, RUNX1-RUNX1T1 and CBFB-MYH11 are favorable-risk lesions, whereas DEK-NUP214, CBFA2T3-GLIS2, and NUP98-NSD1 fusions are regarded as high-risk factors (4–6). The TARGET pediatric AML study revealed that mutations in CEBPA, FLT3, and WT1 are prognostic in pediatric AML (7) while also illuminating significant genetic differences between adult and pediatric AML, including more KMT2A and NUP98 fusions in pediatric AML, along with fewer somatic mutations in NPM1, DNMT3A, and TET2 (7–9).
To date, most genetic studies on pediatric AML have focused on disease at diagnosis, despite relapse serving as a primary driver of poor outcome (8, 10). Here we performed genetic and transcriptional profiling of 136 relapsed pediatric AML cases to define the spectrum of alterations, and we demonstrated an overrepresentation of WT1, KMT2A, and NUP98 alterations in relapsed AML relative to the diagnosis cohort of the TARGET study. Notably, we identified tandem duplications in exon 13 of upstream binding transcription factor UBTF (UBTF-TD) as a recurrent alteration in pediatric AML associated with poor prognosis that is frequently present with FLT3-ITD and WT1 mutations but is mutually exclusive with known subtype-defining fusion oncoproteins.
RESULTS
Comprehensive Genetic Background of Relapsed Pediatric AML
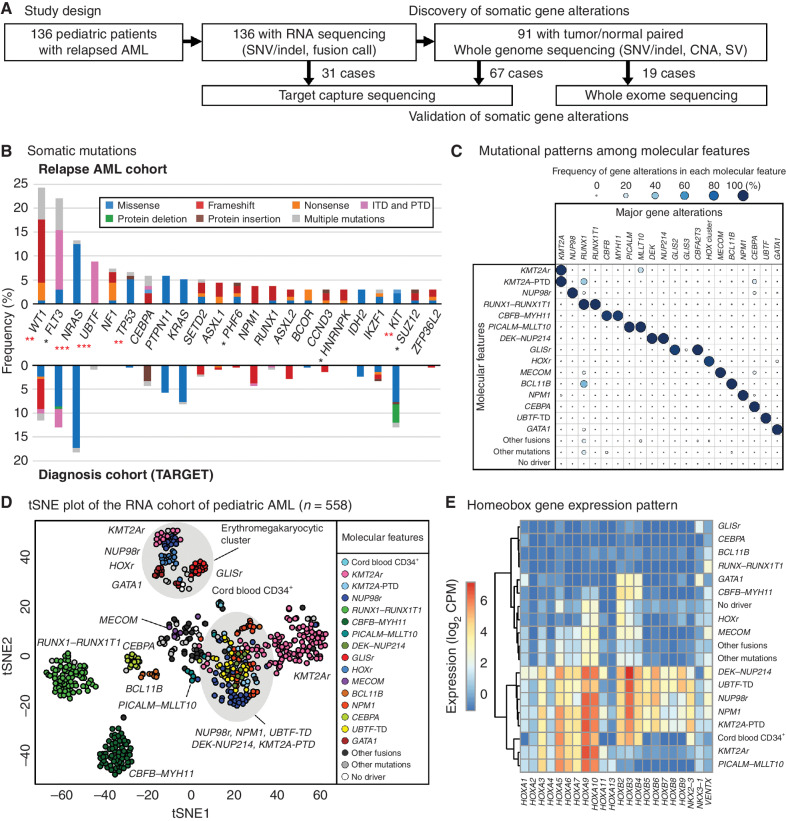
We investigated the genomic profile of relapsed pediatric AML from 136 patients (median age of 9.2) utilizing whole-genome sequencing (WGS), whole-exome sequencing (WES), target capture sequencing (TCS), and RNA sequencing (RNA-seq; Fig. 1A; Supplementary Fig. S1A and S1B; Supplementary Tables S1–S7, see Methods for details). These analyses identified gene fusions in 106 patients (77.9%; Supplementary Figs. S2A–S2C and S3; Supplementary Tables S8–S10). The most common in-frame fusions involved KMT2A (n = 36, 26.5%) or NUP98 (n = 18, 13.2%). We also found rare fusions associated with poor prognosis, including PICALM-MLLT10 (ref. 11; n = 5, 3.7%), FUS-ERG (ref. 12; n = 4, 2.9%), DEK-NUP214 (ref. 6; n = 4, 2.9%), and GLIS family fusions (ref. 5; n = 3, 2.2%). Structural variants leading to outlier high expression and allele-specific expression (ASE) of oncogenic genes were detected by cis-X (13), most notably involving MECOM (refs. 14, 15; n = 3, 2.2%), BCL11B (ref. 16; n = 2, 1.5%), or MNX1 (ref. 17; n = 1, 0.7%; Supplementary Figs. S2A and S4A–S4C; Supplementary Table S11). In comparison to a patient cohort at diagnosis in the TARGET AML study (7, 18), this relapse cohort was enriched for NUP98 rearrangements (P = 0.02) along with fewer CBFB-MYH11 fusions (P < 0.001). We also identified recurrent somatic mutations, including single-nucleotide variant (SNV), insertion and deletion (Indel), tandem duplications, copy-number alterations (CNA), and copy neutral loss of heterozygosity (CN-LOH; Fig. 1B; Supplementary Fig. S5A; Supplementary Tables S12–S14). Overall, 14.1 ± 13.3 (mean ± SD) somatic coding mutations and CNA/CN-LOH were identified per patient with WGS data, many of which are in cell signaling genes and transcription factors (Supplementary Fig. S5B–S5D). Using the genomic random interval (GRIN) model (19), we identified 39 significantly mutated genes, including genes associated with poor prognosis, such as WT1 (n = 33, 24.3%), FLT3 (n = 30, 22.1%), and TP53 (n = 9, 6.6%; Fig. 1B; Supplementary Tables S12 and S15). Recurrent somatic and heterozygous tandem duplications in UBTF (herein referred to as UBTF-TD) were identified in nearly 9% of the relapse AML cohort. The genome-wide mutation pattern of relapsed AML is significantly different (P < 0.001) from the TARGET cohort (7, 18), with more FLT3, WT1, and UBTF mutations. We also identified 9 pathogenic or likely pathogenic germline alterations from 8 patients (8/91, 8.8%), including one germline RUNX1 mutation (Supplementary Table S16).
Figure 1.
Molecular landscape of relapsed pediatric acute myeloid leukemia (AML). A, The study design. Tumor samples from 136 pediatric patients with relapsed AML were subjected to RNA-seq followed by WGS, WES, and TCS when patient samples were available. B, The ratio of patients with recurrent somatic coding mutations in the relapsed AML cohort. The color in each bar represents the type of mutation. Asterisks denote the significance of the difference with the TARGET cohort calculated by Fisher exact test (*, P < 0.05; **, P < 0.01; ***, P < 0.001) and red asterisks denote q < 0.05 after adjustment for multiple testing by the Benjamini-Hochberg method. C, Mutually exclusive gene alteration patterns. Each dot's color and size denote the ratio of patients with the gene alteration. D, t-Distributed Stochastic Neighbor Embedding (tSNE) of expression profiles of the pediatric AML cohort (n = 558) performed with the top 250 most variably expressed genes. The color of each dot denotes the molecular feature of the sample. E, An expression heat map of representative homeobox genes in each molecular feature. The colors denote averaged log2 CPM (counts per million) within each molecular feature.
Molecular Features of Pediatric AML and UBTF-TD Defined by Integrated Molecular Profiling
These data suggest many gene alterations are more common in relapsed pediatric AML than at diagnosis, most notably UBTF-TD (Fig. 1B; Supplementary Fig. S2A). To further investigate these molecular alterations, we established a transcriptomic extension cohort with 417 additional pediatric AMLs from previous studies (5, 7, 10, 20–24), including 36 cases of a therapy-related AML cohort (ref. 15; Supplementary Table S17), as well as samples sequenced through our clinical service (23, 25). We detected fusion transcripts and somatic mutations with the same in-house pipeline and collectively evaluated the mutational and expression patterns (Supplementary Fig. S6; Supplementary Tables S18–S21). These analyses demonstrated that UBTF-TDs are mutually exclusive with other known subtype-defining alterations in pediatric AML, including both the common subtypes like NUP98r, KMT2Ar, NPM1, and CBF-AMLs (CBFB-MYH11 and RUNX1-RUNX1T1), as well as rare subtypes like DEK-NUP214, PICALM-MLLT10, CBFA2T3-GLIS2, and BCL11B (Fig. 1C; Supplementary Figs. S7A, S7B, S8A, and S8B).
At a transcriptional level, UBTF-TD AMLs showed the highest expression of UBTF, although UBTF expression is generally high among AML cases, and are globally similar to NPM1 and NUP98-NSD1 subtypes, with expression of PRDM16, NKX2–3, and HOX cluster genes (Fig. 1D and E; Supplementary Figs. S7A and S9A–S9D; Supplementary Table S22). Although HOXA cluster genes are broadly expressed across different molecular features, HOXB5–9 expression is specific to AMLs with UBTF-TD, NPM1, NUP98r, DEK-NUP214, and KMT2A-PTD (Fig. 1E). In addition, PRDM16, which is required for maintenance of hematopoietic stem cells (26), is homogenously expressed in UBTF-TD and NUP98r subtypes, whereas the expression in NPM1 subtype is more heterogeneous (Supplementary Fig. S9A).
These data on a larger cohort of pediatric AML samples further demonstrate unique cooperating mutation patterns across all cases and within relapse cases (Supplementary Fig. S10A–S10C). For example, UBTF-TD–positive AMLs showed significant enrichment of WT1 and FLT3 mutations across the entire cohort, like NPM1 and NUP98r AMLs. It has also been recognized that a subset of FLT3 or WT1-positive pediatric AMLs lacked a known initiating event (e.g., DEK-NUP214, BCL11B, NUP98-NSD1; refs. 7, 20). Here we show that many of these cases harbor TDs in exon 13 of UBTF. For example, in the relapse cohort, 7 of the 25 FLT3-ITD+ AMLs lacked a concurrent subtype-defining lesion, and 6 of these 7 cases (85.7%) were found to have a UBTF-TD. Likewise, all 9 of the 29 WT1+ AMLs without a known driver alteration were found to harbor a UBTF-TD. Collectively, these data indicate that UBTF-TD is a unique genetic alteration that shares transcriptional and mutational backgrounds with NPM1 and NUP98r AMLs.
UBTF-TD in Pediatric AML
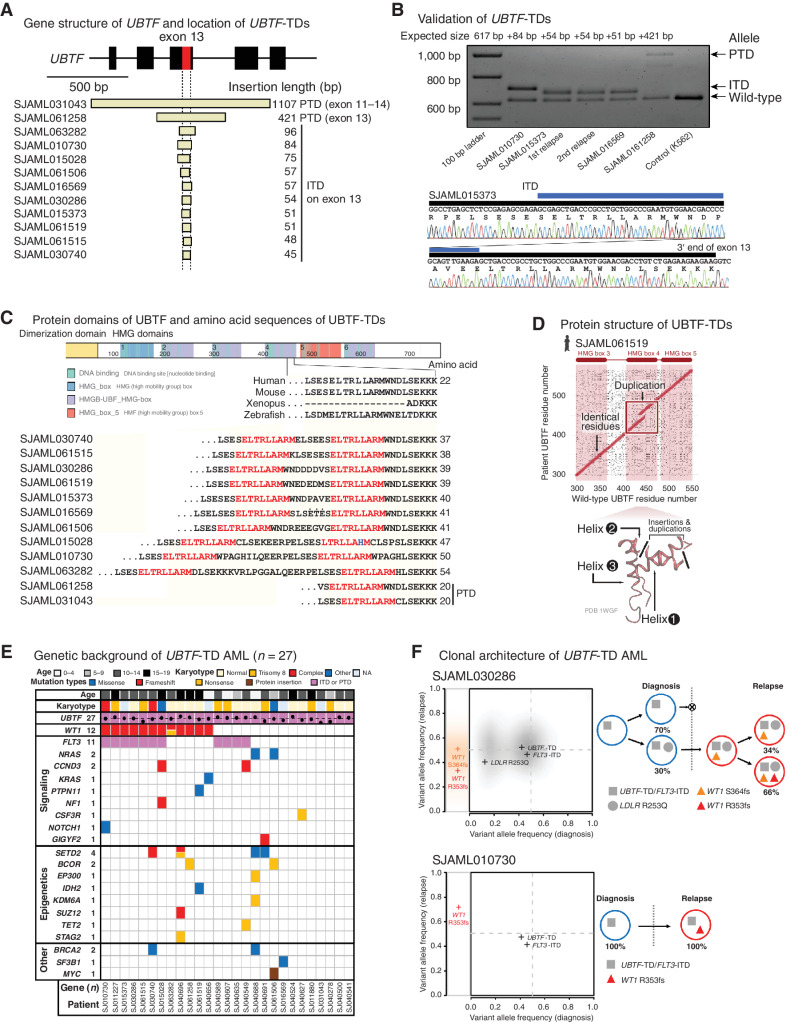
UBTF is a nucleolar protein that regulates the epigenetic status of ribosomal DNA (rDNA) and ribosomal RNA (rRNA) transcription (27, 28). Because UBTF mutations have rarely been observed in myeloid malignancies (8, 10, 18, 21), the frequency in relapsed pediatric AML (n = 12, 8.8%) is surprising. These UBTF mutations all involved either in-frame insertions on the 3′ end of exon 13 of UBTF (internal tandem duplication: ITD) or in-frame duplication of exon 13 (partial tandem duplication: PTD), collectively referred to as UBTF-TD (Fig. 2A). We validated these heterozygous UBTF-TDs by Sanger sequencing and long-read sequencing when patient tumor DNA was available (Fig. 2B; Supplementary Fig. S11A–S11E).
Figure 2.
UBTF-TDs in pediatric AML. A, Exons 11–15 of UBTF gene and the location of UBTF-TDs (n = 12) identified in the relapse AML cohort. B, The results of validation of UBTF-TDs in the relapsed AML cohort by polymerase chain reaction (PCR) and Sanger sequencing. Blue bars denote duplicated sequences. C, Illustrative schema of UBTF protein and amino acid sequences within the HMG domain 4 of both UBTF-wild-type and UBTF-TDs. A part of UBTF-TDs encoded on exon 13 of UBTF genes is shown in comparison with UBTF-wild-type of human and other vertebrates. Amino acid sequences highlighted in red denote leucine-rich sequences duplicated in all UBTF-TDs. D, Comparisons of amino acid sequences of UBTF-wild-type and UBTF-TD at the likely insertion site in helix 2 of HMB box 4 for observed UBTF-TDs. E, Mutational landscape of UBTF-TD AML. F, Clonal dynamics of WT1 mutations in UBTF-TD AMLs. Comparison of variant allele frequency between diagnosis (x-axis) and relapse (y-axis) tumors for cases SJAML030286 and SJAML010730 (left). SNVs/Indels detected from SJAML030286 WGS were drawn as density clouds, and representative mutations for each subclone were marked by crosses. Relapse-specific mutations are shown to the left. The clonal evolution scheme for the patients imputed from bulk WGS data (SJAML030286) or RNA-seq and TCS (SJAML010730; right).
We noted that complex secondary indels frequently occur alongside the duplication, which can limit the detection by CICERO (29). To enhance our ability to detect all UBTF-TD events, we performed an integrative screening of UBTF-TD by CICERO, RNAIndel (30, 31), and a novel soft-clip read approach (see Methods). This screening identified 15 additional UBTF-TDs in the extension cohort, many of which have not been reported in previously published studies (Supplementary Tables S23 and S24). These include 7 newly identified cases from the TARGET cohort that were missed by prior CICERO analysis because of the complex secondary indels mentioned above and the omission of UBTF from the required candidate gene list for ITD detection due to not being previously recognized as an oncogene. These duplications varied in size, but all yielded in-frame insertions of exon 13 of UBTF. At the amino acid level, UBTF-TDs caused amino acid insertions of variable sizes (15–181 amino acids), duplicating a portion of high mobility group domains 4 (HMG4) of UBTF protein, which contains short leucine-rich sequences (Fig. 2C and D; Supplementary Table S24).
The 27 UBTF-TD AMLs (12 in the relapse AML cohort and 15 in the extension cohort) mostly occurred in early adolescence (median age: 12.6, range: 2.4–19.6), and 19 of 27 cases had either normal karyotype (n = 12) or trisomy 8 (n = 7; Fig. 2E; Supplementary Table S25). In patients with documented relapse, the median time from diagnosis to relapse was 1.1 years (Supplementary Table S26). Although UBTF-TD frequently occurred with FLT3-ITD (44.4%) or WT1 mutations (40.7%; Fig. 2E; Supplementary Fig. S10A), mutations in other signaling genes frequently observed in AMLs such as NRAS, KRAS, and PTPN11 were also observed in cases that lacked a FLT3-ITD, suggesting a cooperating signaling alteration contributes to the leukemic phenotype. Across the 27 cases, the median variant allele fraction (VAF) of UBTF-TD was 48.0% (range: 9.7%–66.7%). The three cases with a VAF below 25% had either a paucity of other somatic variants or a low tumor purity before sampling, and the low VAF can likely be attributed to contamination of normal cells together with an overall difficulty with establishing accurate VAFs from complex indels (Supplementary Table S24). These findings suggest that UBTF-TD is predominantly a clonal alteration.
To gain further insights into the clonal dynamics of UBTF-TD AMLs, we studied four cases with data at multiple disease time points. In every case, UBTF-TD was present with a high VAF at both timepoints (Fig. 2F; Supplementary Fig. S12A and S12B; Supplementary Table S27). In contrast, WT1 mutations were commonly enriched in the relapse samples, indicating that WT1 alterations are late cooperating events. Although FLT3-ITD is imputed to cooccur with UBTF-TD in the founding clone in some cases, UBTF-TD AMLs can occur in the absence of FLT3-ITDs (e.g., SJAML016569 in Supplementary Fig. S12B), showing that UBTF-TD AMLs do not require FLT3-ITD. These cumulative data suggest that the recurrent alterations of UBTF-TD in pediatric AML are a subgroup-defining lesion, and we hypothesized that UBTF-TD could drive the leukemic phenotype.
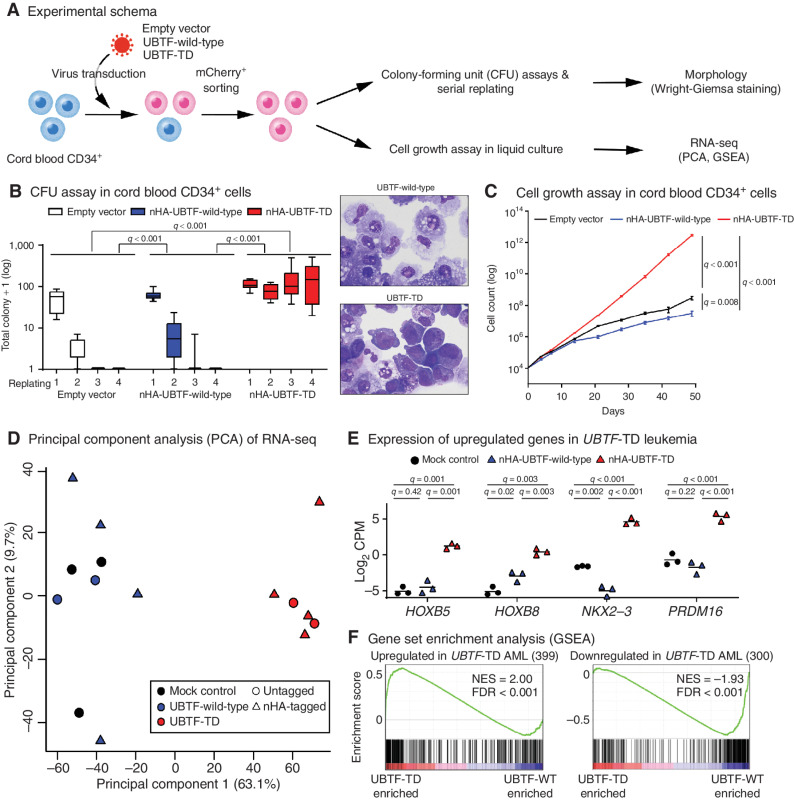
Functional Assessment of UBTF-TD
The impact of exogenous UBTF-TD expression on differentiation and growth in primary hematopoietic cells was assessed in cord blood CD34+ cells (Fig. 3A; Supplementary Fig. S13A–S13C). Colony-forming assays revealed that UBTF-TD expression promotes colony-forming activity over several rounds of replating and yields cells with a persistent blast-like morphology, in contrast to macrophage-like differentiation in UBTF-wild-type and empty controls (Fig. 3B; Supplementary Fig. S14A). UBTF-TD expression also resulted in a growth advantage over controls (Fig. 3C; Supplementary Fig. S14B–S14D). Furthermore, transcriptional profiling of these cell cultures demonstrated an expression program similar to what we observe in patients with UBTF-TD AMLs, including expression of HOXB genes, NKX2–3, and PRDM16 (Fig. 3D–F), implying that UBTF-TD expression is sufficient for this phenotype. We next investigated the impact of UBTF-TD on the known function of UBTF in regulating rDNA activity (28). Overexpression of both UBTF-wild-type and UBTF-TD decreased inactive rDNA sites compared with mock control with no consistent difference between UBTF-wild-type and UBTF-TD (Supplementary Fig. S15A and S15B), suggesting that the duplication does not alter UBTF function on rDNA. Finally, we interrogated the subcellular localization of UBTF-TD in a variety of human cell lines and primary patient samples, which showed no consistent changes in the localization of UBTF-TD compared with wild-type (Supplementary Fig. S16A–S16C).
Figure 3.
In vitro modeling of UBTF-TD. A, Experimental design of in vitro modeling of UBTF-TD in cord blood (CB) CD34+ cells. B, The effects of UBTF-wild-type and UBTF-TD overexpression in colony-forming potential of cord blood CD34+ cells. Boxplots of logged colony count from technical replicates (Empty vector: n = 7, UBTF-wild-type: n = 12, UBTF-TD: n = 12) from five independent experiments are shown. A box represents quartiles, and whiskers represents max and minimal values. Statistical significances were calculated by ANOVA test followed by pairwise comparisons and adjustment with Tukey method (left). Wright-Giemsa staining of cells derived from the second replating. Both images are at equal magnification (60×; right). C, The effects of UBTF-wild-type and UBTF-TD overexpression on cell growth of CD34+ cord blood in liquid culture. Experimental design and error bars are the same in Fig. 3B. Statistical significances were calculated at day 49 by Student t test followed by adjustment for multiple testing by the Benjamini-Hochberg method. D, Principal Component Analysis (PCA) of transcriptional profiles of transduced cord blood CD34+ cells at day 32 (Empty vector: n = 3, UBTF-wild-type: n = 6, UBTF-TD: n = 6). E, Expression of representative genes upregulated in UBTF-TD AMLs. Bars denote mean from biological triplicates from which data for all conditions were available. Statistical significances were calculated as in Fig. 3C. F, Gene Set Enrichment Analysis (GSEA) between nHA-UBTF-WT (n = 3) and nHA-UBTF-TD (n = 3) transduced conditions using gene sets identified in patient samples (Supplementary Table S21).
Prevalence and Clinical Impact of UBTF-TD in De Novo AML Cohorts
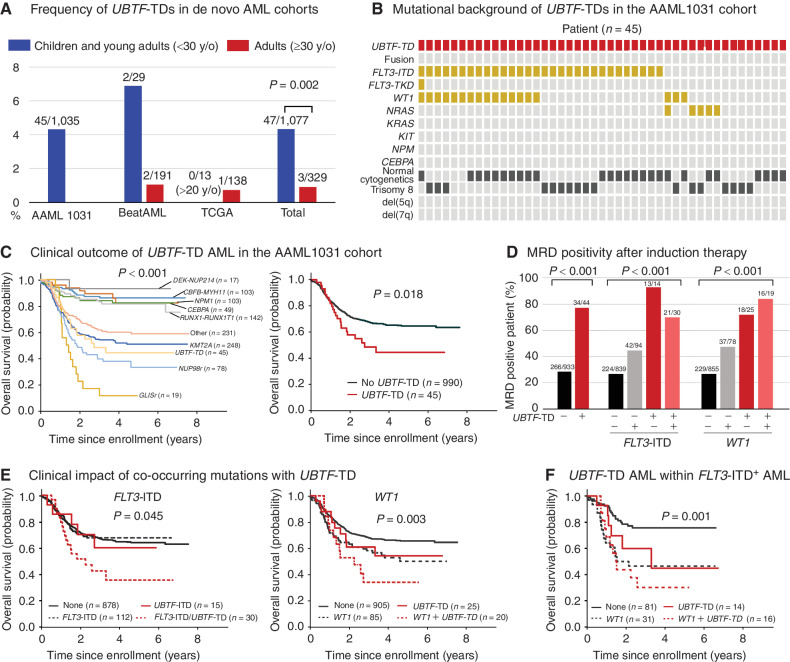
To investigate the prevalence of UBTF-TDs in de novo pediatric and adult AML, we applied the above UBTF-TD screening method to the available large de novo AML cohorts of TCGA (n = 151, adult; ref. 32), BeatAML (n = 220, pediatric and adult; ref. 33), and AAML1031 (n = 1,035, pediatric; ref. 34). We identified UBTF-TDs in 4.3% (45/1,035) of the pediatric AAML1031 cohort (Fig. 4A; Supplementary Tables S28–S30), and all of these cases lacked a cooccurring recurrent fusion oncoprotein (Fig. 4B). We also confirmed frequent cooccurring FLT3-ITD (30/45: 66.7%) and WT1 mutations (14/45: 31.1%), as well as rare NRAS mutations in UBTF-TD AMLs, and the enrichment in AML in adolescence (median: 14.3, range: 6.3–27.3) with normal karyotype or trisomy 8. In contrast, UBTF alterations are less common (P = 0.002) in the adult AML cohorts, occurring in only 0.9% (3/329) of patients (Fig. 4A; Supplementary Tables S28 and S29).
Figure 4.
Prevalence and clinical outcome of UBTF-TDs in de novo AML cohorts. A, Frequencies of UBTF-TDs in published de novo AML cohorts in Supplementary Table S25. Statistical significance was calculated between the total pediatric and adult cohorts by Fisher exact test. B, Cytogenetic and genetic background of UBTF-TD cases in the AAML1031 cohort. C, Clinical outcomes of UBTF-TD cases and AML with major molecular features in the AAML1031 cohort. D, Minimal residual disease (MRD) positivity of UBTF-TD case with cooperating mutations of FLT3-ITD or WT1. E, Clinical outcomes of UBTF-TD cases with cooperating mutations of FLT3-ITD or WT1. F, Subgroup analysis of outcomes of UBTF-TDs with or without WT1 mutations within FLT3+ AMLs. In C, E, and F, the statistical significance of variables was tested with the log-rank test. In D, the statistical significance was calculated by Pearson χ2 test.
The enrichment of UBTF-TDs in the relapse cohort suggests an association with poor outcomes. Furthermore, all 9 patients with UBTF-TDs in the TARGET cohort (7) experienced relapse, and an additional 3 patients were part of the induction failure cohort (ref. 10; Supplementary Fig. S17; Supplementary Table S31). To test this hypothesis, we investigated the clinical outcomes within the AAML1031 cohort. Overall, patients with UBTF-TD have a 5-year overall survival (OS) of 44%, which is less than patients without a UBTF-TD (64%, P = 0.018), and tend to have lower 5-year event-free survival (30% vs. 45%, P = 0.078; Fig. 4C; Supplementary Fig. S18A). The clinical course of UBTF-TD AML is similar to those of NUP98r or KMT2Ar among major molecular features (Fig. 4C). Notably, UBTF-TD was strongly associated with minimal residual disease (MRD) positivity at the end of the first induction, independent of FLT3-ITD or WT1 status (Fig. 4D). Furthermore, the cooccurrence of FLT3-ITD with UBTF-TD resulted in a poor clinical outcome (Fig. 4E; Supplementary Fig. S18B). Also, UBTF-TD and WT1 demonstrated additive impact on the clinical outcomes in the entire cohort and within FLT3-ITD+ AMLs (Fig. 4E and F; Supplementary Fig. S18C). Univariate and multivariate analyses with genetic alterations revealed that UBTF-TD, WT1, and NUP98 fusions are independent risk factors for overall survival within FLT3-ITD+ AMLs, whereas the age of the patients had only mild impacts on the clinical outcome in univariate analysis (Supplementary Table S32). These clinical data show that UBTF-TD is a new risk factor for pediatric AML with a poor prognosis and high rates of MRD positivity.
DISCUSSION
The outcome of pediatric AML has improved over the past few decades (1) largely due to intensification of chemotherapy and enhancements in supportive care, rather than new therapies guided by molecular alterations (35). This has partly resulted from the lack of understanding of the genetic features of childhood AML at relapse, which drives the poor outcome in this disease (3, 35). To address this deficiency, we comprehensively characterized 136 relapsed pediatric AMLs. This relapsed AML cohort was enriched for KMT2A and NUP98 rearrangements and WT1 mutations compared with de novo pediatric AML (7, 18). Notably, the third most common molecular feature in our relapsed AML cohort was tandem duplications in exon 13 of UBTF. UBTF-TDs have rarely been reported in the literature (10, 18), most recently in 3 of 25 cases of relapsed pediatric AML (8). Here we report for the first time that UBTF-TDs involving exon 13 are a common gene alteration in pediatric AML and are present only in cases lacking known subtypes defining molecular alterations. In addition to the recognition of UBTF-TD as a recurrent alteration, this cohort of 136 relapse AMLs also identified a number of rare genetic alterations that may also be associated with high-risk disease (e.g., structural variants involving MNX1) or poor response to chemotherapy (e.g., SETD2 mutations).
Both FLT3-ITD and WT1 mutations are among the most common mutations in pediatric AML and typically cooccur with NUP98-NSD1 and other fusion oncoproteins, such as DEK-NUP214, and less commonly KMT2A rearrangements (4, 7). However, a subset of FLT3-ITD or WT1-mutant AMLs have previously lacked a known initiating event (7, 20), and this study confirms that many of these cases have an exon 13 duplication in UBTF. Our data also show that UBTF-TD is predominantly present in the founding clone and is stable during disease progression, whereas WT1 alterations are cooperating events acquired at later stages of the disease. Although our data also show that FLT3-ITD is commonly present with UBTF-TD, it has been well established that FLT3-ITD is not a stable marker during disease progression or patient-derived xenograft (PDX) propagation (36–38). Likewise, not all UBTF-TD AMLs cooccur with FLT3 or WT1 mutations, suggesting that mutations involving the Ras pathway or other signaling pathways can contribute to the development of UBTF-TD AML. The clonal nature of UBTF-TDs and the mutual exclusivity with other oncogenic drivers of pediatric AML across multiple cohorts suggests that UBTF-TDs may represent a new subtype-defining lesion in pediatric AML.
At a transcriptional level, UBTF-TD AMLs are most similar to NUP98-NSD1 or NPM1 subtypes, partly driven by the expression of HOX cluster genes and variable patterns of PRDM16 expression. The similarity to NUP98-NSD1 is intriguing as they share mutational features (e.g., cooccurrence of WT1 and FLT3-ITD) and a refractory nature. On the other hand, NPM1-mutant AMLs, which are enriched in adults and not associated with poor prognosis, have a more heterogenous PRDM16 expression pattern than UBTF-TD AMLs. PRDM16 encodes for a protein with histone methyltransferase activity (39) and is a known regulator of hematopoietic stem cells (26). Previous studies have shown that elevated expression of PRDM16 in pediatric AMLs are associated with poor outcomes (40), and the expression pattern of PRDM16 may account for some of the differences in outcomes between NPM1 and UBTF-TD AMLs. At the protein level, UBTF and NPM1 localize in nucleoli, whereas mutant NPM1 is known to mislocalize to the cytoplasm (41). However, our data suggest that mislocalization is likely not a major consequence of UBTF-TDs, and additional mechanistic studies are needed to understand how UBTF-TD leads to leukemia. Importantly, our data show that UBTF-TD expression in CD34+ cells is sufficient to induce a proliferative advantage, increased clonogenic activity, and a similar transcriptional program to AML in vivo, demonstrating clear hematopoietic phenotypes.
By combining data from additional AML cohorts, we demonstrated that UBTF-TD AMLs are uniquely associated with adolescent age with a median age of 13.4 years of all patients presented in this study, including those from AAML1031. AMLs with this alteration commonly have either normal karyotype or trisomy 8 in addition to FLT3-ITD and WT1 mutations. The initial identification of UBTF-TDs as a recurrent alteration in relapsed disease suggests a link to poor outcome, and we confirmed this using data from the AAML1031 trial. Our findings show a clear association with poor outcome (5-year OS, 44% vs. 64%, P = 0.018), which is similar to KMT2Ar and NUP98r leukemias. Likewise, UBTF-TDs appear to have a poor response to conventional therapy, as shown by the high rates of MRD positivity. These biological and clinical features have been commonly ascribed to FLT3-ITD and WT1 mutations (7); however, our data confirmed that these can be partly attributed to UBTF-TD, showing that WT1 and UBTF-TD are independent risk factors in FLT3-ITD–positive AMLs with an especially dismal outcome (5-year OS, 30.0%) in patients with WT1 and UBTF-TD alterations.
The finding that UBTF-TD is a recurrent alteration in pediatric AML with prognostic significance justifies the need to ascertain its mutational status in pediatric AMLs without a known driver, especially in those with either FLT3-ITD or WT1 mutations, or that are MRD-positive at the end of the first induction. Furthermore, our data also suggest that it should be regarded as a high-risk molecular alteration in future pediatric AML trials. Currently, UBTF is likely not covered by most commercially available panels, and updated approaches are needed to ensure accurate detection. Moreover, the fact that UBTF-TDs have been underrecognized so far suggests that these alterations have been frequently missed by many current bioinformatic approaches even when the sequencing modality effectively covers UBTF. Our findings importantly imply that additional recurrent lesions can still be detected with improved technical and bioinformatic strategies despite the major emphasis on genomic profiling of tumors in the past decade or more.
METHODS
Subject Cohorts and Sample Details
Tumor samples from 136 patients with relapsed AML from the St. Jude Children's Research Hospital tissue resource core facility were obtained with written informed consent using a protocol approved by the St. Jude Children's Research Hospital institutional review board (IRB). Studies were conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects. These relapsed AML cases were part of multiple clinical studies, and detailed information of these patients and clinical trials are provided in Supplementary Table S1. CD3+ T cells were first depleted from all AML samples by magnetic beads (EasySep Human CD3 Positive Selection Kit II, 17851, StemCell Technologies). For samples with low-tumor purity (<60%), additional enrichment was performed by flow cytometry using a combination of mouse anti-human CD45 PerCP-Cy5.5 (eBioscience, #8045–9459–120, clone:2D1, RRID:AB_1907397) and mouse anti-human CD33 APC (eBioscience, #17–0338–42, clone: WM-53, RRID:AB_10667747), along with negative selection of T cells with mouse anti-human CD3 APC-Cy7 (BD Biosciences, #557832, clone:SK7, RRID:AB_396890). Matched germline DNA was obtained from purified T cells (n = 18), skin or bone marrow fibroblasts (n = 70), or remission samples (n = 3). Twenty-eight samples were sequenced through our clinical pipeline (n = 28; refs. 23, 25).
Library Preparation and Sequencing
Patient gDNA was extracted from tumor samples using Quick-gDNA Miniprep Kit (D3024, Zymo Research) followed by WGS as described previously (24). Tumor RNA was extracted from tumor samples using RNeasy Mini kit (74104, Qiagen) followed by library preparation for RNA-seq as described previously (24). RNA from freshly thawed cord blood CD34+ cells [purchased from Lonza (catalog no. 2C-101)] were subjected to RNA-seq as normal controls.
Whole-Genome Sequencing Data Analysis
WGS analysis (n = 91) were performed using standard methods as described (15, 24). Briefly, DNA reads were mapped using BWA (WGS: v0.7.15-r1140, RRID:SCR_010910) to the GRCh37-lite/hg19 human genome assembly. Aligned files were merged, sorted, and deduplicated using Picard tools 1.65 (broadinstitute.github.io/picard/, RRID:SCR_006525). Single-nucleotide variant (SNV) and Insertion and deletion (Indels) were called using Bambino (42). Structural variations (SV) were analyzed by using CREST (43) (v1.0). Copy-number alterations (CNA) were analyzed by using CONSERTING (44). For the tumor purity inference, SNVs with at least 20× total coverage and in the diploid region of the genome were subjected to unsupervised clustering using R mclust package, and the tumor purity was estimated by twice the highest cluster center value among all cluster centers ≤0.5.
RNA Sequencing, Mapping, and Fusion Detection
RNA reads were mapped using our StrongARM pipeline (45). Chimeric fusion detection was carried out using CICERO (ref. 29; v1.7.0) and ChimeraScan (ref. 46; v0.4.5). DNA sequences of NUP98-ZFX1 fusions were confirmed by polymerase chain reaction (PCR) and Sanger sequencing (primers can be found in Supplementary Table S10). Illustrative schemas of structural variants were drawn by ProteinPaint (https://proteinpaint.stjude.org/). Tumor purity was estimated from RNA-seq data using ESTIMATE (47) algorithm.
Somatic Mutation Calling from RNA-Seq
We applied the following approach to simultaneously account for germline polymorphisms (without germline control) and sequencing artifacts specific to RNA-seq, on a panel of 75 predefined genes previously reported to be significantly mutated in pediatric AML (7) and myelodysplastic syndrome (MDS; Supplementary Table S4). Briefly, candidate SNVs/Indels were called by Bambino (42) or RNAindel (30, 31), annotated by VEP (48), and in turn classified for putative pathogenicity with PeCanPie/MedalCeremony (49). Candidate variants (n = 83,765 SNVs and 44,987 Indels) with putative pathogenicity were considered germline or artifacts if present in >5% of the cases. Candidate variants were further filtered if the number of supporting reads was ≤5 or if the variant allele fraction (VAF) was ≤5%. All called variants were manually confirmed for pathogenicity. This resulted in 1,039 (627 SNVs + 412 Indel) high confidence variants.
We focused on 98 samples with both RNA-seq and target capture sequencing (TCS) data available to investigate the accuracy of our RNA-seq based calling method. Of these, 167 variants (93 SNVs, 74 Indels) were called from RNA-seq, and 164 variants (91 SNVs, 73 Indels) were validated by TCS (98.2%), indicating a high specificity of our RNA-based variant calling method. Furthermore, variants in the 75 genes called by TCS were first subjected to the following filtering criteria: present in ≤10% cases, RNA-seq FPKM (fragments per kilobase of exon per million reads mapped) ≥0.5 for the gene, and reads supporting mutant allele ≥15 (SNV) or ≥25 (Indel). Of the 102 SNVs further filtered with the same pathogenicity filtering as mentioned above, 91 SNVs (89.2%) were called from RNA-seq. Of the 82 Indels, 69 Indels (84.1%) were called from RNA-seq. Overall, of the 184 variants called by TCS, 160 were called from RNA-seq (87%), indicating a high sensitivity of our RNA-based variant calling method. The lower call rate of indels from RNA-Seq is concordant with previous observation (18) where frameshifting variants tend to have reduced expression and therefore lower mutant alleles read counts for detection, possibly due to nonsense-mediated decay.
Validation of Somatic Variants with TCS and WES
Mutations were validated using WES (n = 19) or TCS (n = 98; TWIST Biosciences) designed to cover genes frequently mutated in pediatric AML as well as recurrent mutations found in this current study. A total target region of 2,320,524 bp was directly covered (Supplementary Table S7). BWA (v0.7.12) MEM algorithm was used to map the target capture sequencing reads to the GRCh37-lite/hg19 human genome assembly. Variant detection was done using VarScan 2 (50) (v2.3.5, RRID:SCR_006849) on the target capture sequencing data with the following criteria: MAPQ ≥1; base quality Phred (RRID:SCR_001017) score ≥20; VAF ≥ 0.001 and variant call p-value ≤ 0.05. Among the 91 patients with WGS, TCS data were obtained for 67 patients. 176 somatic SNV/Indel calls by the DNA variant-calling pipeline from 58 patients were covered by the custom capture panel, and 173 variants (98.3%) were validated (Supplementary Fig. S1). WES data was used for validation in 19 patients sequenced through our clinical service and 349 of 359 SNV/Indel calls (97.2%) were validated. For the 31 RNA-seq only cases with TCS data, 45 of 46 (97.8%) somatic SNV/Indel calls from RNA-seq data were validated by TCS. In total, 567 of 581 (97.6%) somatic SNV/Indel calls from DNA and RNA variant-calling pipelines were validated, indicating a high accuracy of our pipeline including RNA-based variant calling approach.
Germline Variant Curation Methods
All nonsynonymous variants with a VAF ≥0.3 were comprehensively reviewed and classified as pathogenic, likely pathogenic, of uncertain significance, likely benign, or benign based on recommendations from the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (51) and the Clinical Genome Resource (52–55) by a variant scientist. Missense variants in well-established genes (e.g., DDX41, RUNX1, SAMD9L) were evaluated regardless of in silico predictions. Missense variants in all other genes were filtered on the basis of a REVEL score ≥0.5 and a CADD score ≥15. Population frequency of all variants were evaluated on the basis of their prevalence in gnomAD (v2.1.1, RRID: SCR_014964; ref. 56).
GRIN Analysis for Significantly Mutated Genes
For the 91 cases with WGS data, the genomic random interval (GRIN) model (19) was used to evaluate the statistical significance of the number of subjects with each type of lesion [fusions, CNAs (amplifications and deletions), copy neutral loss of heterozygosity (CN-LOH), SNV/indels, and tandem duplications] in each gene. For each type of lesion, robust false discovery estimates were computed from P values using Storey method (57) with Pounds and Cheng (58) estimator of the proportion of hypothesis tests with a true null hypothesis. In addition, P values for the number of subjects with any one type of lesion in each gene were computed using the beta distribution derived as order statistics of uniform random variables (59). Thirty-nine genes were considered significantly mutated genes at a FDR <0.01, of which 14 genes were involved.
Analysis of Clonal Evolution For UBTF-TD AMLs
Clonal evolution analysis was performed similar to previous study (60, 61). Briefly, allele fractions of somatic SNVs were compared between time points to detect subclones that are shared or specific to diagnosis, relapse, or subsequent relapses. A representative mutation (i.e., being known driver or being protein-coding) is used to label each subclone. Non-representative mutations were drawn as a background cloud to facilitate visual comparison, as done previously (60). The detected subclones were then ordered to generate schematic evolutionary trees as done previously (61).
cis-X Analysis for Outlier and Allele-Specific Expression
To detect noncoding variants that lead to dysregulation of cancer driver genes such as MECOM and BCL11B, cis-X (ref. 13; version 1.4.0) was used to detect outlier high expression (OHE) and allele-specific expression (ASE) with default parameters (cis-X run … -w 10 -r 10 -f 5). OHE with outlier P < 0.05 and ASE with binominal P < 0.01 were considered significant.
UBTF-TD Screening and Validation
Based on findings in the previous report (18) that UBTF-TDs can be accompanied with secondary small indels that could hamper the detection by CICERO (29), we developed a novel approach that integrates following three features to comprehensively detect UBTF-TDs.
Detection by CICERO (v1.7.0) focusing ITD or PTD with supporting reads ≥3 on exon 13 of UBTF gene or adjacent introns and CICERO score >10.
Detection of Indels (30, 31) on exon 13 of the UBTF gene, which were found to be recurrent in UBTF-TD cases.
Counting reads with 10 or more soft-clipped nucleotide sequences and total reads on the 3′ end of exon 13 that contains a hotspot of ITD and PTD (GRCh37-lite, chr17:42288162–42288192; GRCh38, chr17: 44210794–44210824).
We first performed this approach on the RNA cohort followed by manual examination of RNA-seq BAM files to validate the efficacy of the method. CICERO detected 21 UBTF-TDs in the RNA cohort with 6 UBTF-TDs undetected due to secondary alterations on duplications. However, the combination of these methods detected UBTF-TDs efficiently with a sensitivity of 100% (27/27) and specificity of 96.6% (508/526) when we put the threshold of soft-clipped count ratio at 21% in total count on the hotspot (Supplementary Table S23). To account for the possible large variance of tumor cellularity, and by extensive evaluation of sensitivity and specificity through cross-comparison with CICERO and RNAIndel as well as manual verification, we concluded that 10% of soft-clipped read count is a reasonable threshold for screening of external cohorts.
This screening approach was applied to de novo AML cohorts [TCGA (32), BeatAML (33), and AAML1031 (34); Supplementary Table S28]. For cases in the relapse cohort with enough DNA sample (n = 4), heterozygous ITD or PTD allele was confirmed by PCR, and DNA sequence of insertion was confirmed by Sanger sequencing (primers are listed in Supplementary Table S10).
We also validated UBTF-TD with long-read sequencing. Briefly, high molecular weight (HMW) DNA from cryopreserved xenograft samples from UBTF-TD AML patient SJ015373 were extracted using Monarch HMW DNA Extraction Kit for Cells & Blood (T3050L, NEB). About 5 μg HMW DNA was sheared to about 20 Kb using Covaris g-TUBE (520079, Covaris). Sheared DNA was size selected using BluePippin (BLU0001, Sage Science) to obtain DNA fragments that were >10 Kb. Library for sequencing was prepared following the protocol of PacBio HiFi library Preparation using SMRTbell Express Template Prep Kit 2.0 (100–938–900, Pacific Bioscience) and was sequenced with 12 SMRT cells (101–531–001, SMRT Cell 1M v3 LR, Pacific Bioscience) using the Sequel system. Sequences in the subread.bam format were converted into CCS reads using smrttools (version 8.0) with default parameters. PacBio CCS fastq file was generated from CCS ubam files using bam2fastx (version 1.3.0; https://github.com/pacificbiosciences/bam2fastx/). Then minimap2 (ref. 62; version 2.18) was used to map the CCS reads to human GRCh37-lite genome with parameters (-ax asm20). The 3 insertion sequences are aligned using CLUSTAL Omega (ref. 63; version 1.2.4, RRID: SCR_014964).
Gene Expression Data Summarization, Batch Correction, and Dimension Reduction
Reads from aligned BAM files were assigned to genes and counted using HTSeq (ref. 64; v0.11.2, RRID: SCR_005514) with the GENCODE (RRID: SCR_014966) human release 19 gene annotation. The gene count matrix was generated, and the Log2 CPM (counts per million) values were used for downstream analysis. For a gene to be considered as expressed, we required that at least 5 samples (equal to the smallest group in the RNA cohort) should have ≥10 read counts per million reads sequenced. The batch effect between St. Jude and the TARGET cases was corrected using the ComBat method available from R package SVA (ref. 65; v3.36.0). R package Limma (ref. 66; version 3.32.10, RRID: SCR_010943) was used for differential gene expression analysis and we set Log2 CPM = –1 if it is < –1 based on the Log2 CPM data distribution. P values were adjusted by the Benjamini–Hochberg method to calculate FDR. Genes with absolute fold change >2 and FDR <0.05 were regarded as significantly differentially expressed. Dimension reduction was done using t-Distributed Stochastic Neighbor Embedding (tSNE; ref. 67) to visualize the clustering of cases from different molecular features. Clustering visualization was done using top 250 by MAD (median absolute deviation, sex-specific genes were excluded) and the following tSNE parameters: perplexity = 21, max_iter = 10,000 (R version 3.4.2, package Rtsne v0.13, RRID: SCR_016342). Dimension reduction was also performed using UMAP (Uniform Manifold Approximation and Projection; ref. 68) with the top 125 genes by MAD and the following UMAP parameters: n_components = 3, n_neighbors = 6, min_dist = 0.4 and n_epochs = 500 (R version 3.6.1, UMAP version 0.2.7.0). For hierarchical clustering of homeobox genes, genes were selected from ANTP and PRD family genes and averaged within each molecular feature. Clustering and visualization were performed by R package pheatmap (version 1.0.12, RRID: SCR_016418) with Euclidian distance and Ward.D2 linkage. Principal Component Analysis (PCA) was performed by R prcomp function. Gene Set Enrichment Analysis (GSEA; ref. 69) was performed by GSEA (v4.1.0, RRID: SCR_003199) using MSigDB gene sets c2.all (v7.4) for UBTF-TD AMLs in patients. For GSEA with transduced CD34+ cells, gene sets were made from differentially expressed genes in UBTF-TD patients (Supplementary Table S22).
Construction of UBTF-TD–Expressing Vectors and Virus Production
UBTF-wild-type and UBTF-TD cDNA were amplified from patient cDNA using High-Capacity RNA-to-cDNA Kit (#4387406, Thermo Fisher) and cloned into pENTR/D-TOPO Gateway entry vector (K240020, Thermo Fisher) then transferred to Gateway-compatible lentivirus vector (MND-mPGK-mCherry). N- and C-terminal HA and tags were introduced by amplification during cloning. pCDNA3.1(+)-N-eGFP based UBTF-wild-type and UBTF-TD were synthesized in full by Genscript. cDNA sequence, schema of the vectors, primers used for cloning and Sanger sequence are found in Supplementary Fig. S13 and Supplementary Table S10. The above lentivirus vectors were cotransfected with packaging vectors (pHDM-G, pCAGG-HIVgpco, and pCAG4-RTR2, provided from the St. Jude Vector Laboratory) into 50%–60% confluent low-passage HEK293T cells (CVCL_0063, ATCC #CRL-3216, obtained from ATCC in 2016) using FuGene HD Transfection Reagent (E2311, Promega). Supernatant containing lentiviral particles was harvested at 48 hours after transfection and concentrated for cord blood CD34+ transduction. For MOLM-13 (CVCL_2119, DSMZ #ACC 554, obtained from DSMZ in 2016), virus-containing supernatant was used without concentration. HEK293T cells are maintained in DMEM (#11965, Invitrogen) supplemented with 10% FBS (S11550H, Atlanta Bio) and 100 U/mL penicillin–streptomycin (15140122, Invitrogen), and MOLM-13 cells are maintained in RPMI1640 media (11875, Invitrogen) supplemented with 10% FBS and 100 U/mL penicillin–streptomycin. MOLM-13 cells and HEK293T cells were validated by short tandem repeat (STR) analysis and were generally used within 15 passages after thawing. Mycoplasma testing was performed on cell lines, including cord blood CD34+ cells, using MycoAlert Mycoplasma Detection Kit (#LT08–118, Lonza).
Functional Assays of Cord Blood CD34+ Cells
Commercially available cord blood CD34+ cells were purchased from Lonza (catalog no. 2C-101, Lot# 18TL248959) or the Carolinas Cord Blood Bank/Duke University. After thawing, cells were cultured for 24 hours in StemSpan SFEM II media (#09655, STEMCELL Technologies) supplemented with penicillin–streptomycin, l-glutamine, and recombinant human SCF, FLT-3, TPO, and IL6 (all 50 ng/mL, HHSC6, PeproTech), UM171 (35 nmol/L, 72914, STEMCELL Technologies), and SR-1 (1 μmol/L, 72344, STEMCELL Technologies). Cells were transduced with MND-PGK-mCherry lentivirus expressing untagged or nHA-tagged UBTF-wild-type, untagged or nHA-tagged UBTF-TD, or mock controls at an MOI (multiplicity of infection) of ≥50. Transduced mCherry+ cells were enriched by flow cytometric sorting at day 7 of transduction and plated for colony-forming unit (CFU) assay in MethoCult H4435 (#04435, STEMCELL Technologies) at a plating concentration of 1 × 103 cells per dish, at 1 × 104 per well for growth curve cultures on StemSpan media with cytokines, or 2 × 103 per well for cell growth assay by IncuCyte (4647, SARTORIUS). Colonies were counted after 10 days of growth at 37°C, harvested, and serially replated at a concentration of 1 × 104 cells per dish. Cells in culture were counted every 7 days and growth curve was generated. Blinding to the groups was not used.
Western Blotting
Ten micrograms of purified proteins were separated by SDS-PAGE on a 10% Protein gel (Bio-Rad, #4561033) and transferred onto nitrocellulose membranes (0.2 μmol/L, Bio-Rad, 1620252). Membranes were incubated overnight at 4°C with rabbit anti-HA-tag (mAb #3724, Cell Signaling Technology, clone: C29F4, RRID:AB_1549585, 1:1,000 dilution) or Mouse anti-β-actin (mAb #3700, Cell Signaling Technology, clone: 8H10D10, RRID:AB_2242334, 1:2,000 dilution). IRDye 800CW Donkey anti-Rabbit IgG (LI-COR Biosciences, #926–32213, RRID:AB_2715510, 1:15,000) and IRDye 680RD Goat anti-Mouse IgG (LI-COR Biosciences, #926–68070, RRID:AB_2651128, 1:15,000 dilution) were used as secondary antibody. Imaging was performed on the Odyssey CLx Imaging System (LI-COR Biosciences, 9140, RRID:SCR_014579).
Fluorescence Microscopy
U-2 OS cells (CVCL_0042, originally purchased from ATCC (catalog no. HTB-9) and kindly provided by the lab of Paul Taylor at St. Jude Children's Research Hospital) and HEK293T cells, cultured in DMEM supplemented with 10% FBS and 100 U/mL penicillin–streptomycin, were transfected with expressing GFP-tagged UBTF-WT and UBTF-TD constructs using FuGene HD Transfection Reagent (E2311, Promega) were fixed with 4% paraformaldehyde (43368, Alfa Aesar), permeabilized first with methanol (A412–1, Fisher Chemical) then in 0.3% triton X-100 (X100–500 mL, Sigma) blocked with 5% goat serum (ab7481, Abcam), and stained with Alexa Fluor 568 Anti-Fibrillarin (ab202540, Abcam, clone: EPR10823(B)), Alexa Fluor 647 anti-alpha tubulin (ab190573, Abcam, clone: EP1332Y), and DAPI (564907, BD Biosciences). Coverslips were mounted using ProLong Diamond Antifade (P36980, Invitrogen). Images were acquired on a Nikon C2 laser scanning confocal microscope using a 60X oil-objective lens controlled by NIS-Elements software (Nikon, RRID: SCR_014329). MOLM-13 cells transduced with lentivirus expressing cHA-tagged UBTF-WT and UBTF-TD were also analyzed for HA, Fibrillarin, and DAPI as described above. U-2 OS cells were used within 15 passages after thawing.
Immunohistochemistry (IHC)
Photomicrographs of bone marrow core biopsy of 4 cases with UBTF-TDs and 2 cases with either KMT2A rearrangement or NPM1c mutation using anti-UBF (sc-13125, Santa Cruz Biotechnology, clone: F-9, RRID: AB_671403, 1:500 dilution). All images are at equal magnification (60×).
Sequential Ribosomal RNA and DNA Fluorescent In Situ Hybridization (FISH)
MOLM-13 cells that had been transduced with either cHA-tagged UBTF-wild-type, cHA-tagged UBTF-TD, or an empty vector were subjected to HA immunofluorescence staining and ribosomal RNA FISH, followed by RNase treatment and ribosomal DNA FISH on the same cells to visualize the location of cHA-tagged UBTF and its relationship to ribosomal RNA and ribosomal DNA and to identify rDNA gene clusters that are outside of the nucleolus and not being expressed. Unfixed MOLM-13 cells were applied to glass slides by cytocentrifugation at 300,000 cells/slide. Slides were fixed in 4% PFA (sc-281692, Santa Cruz Biotechnology) containing 0.5% Tween 20 (P9416–50ML, Sigma-Aldrich) and 0.5% Nonidet P-40 (N6507, Sigma-Aldrich) for 10 minutes. Following fixation, slides were stored in 70% ethanol at −20°C until ready to use. To remove cells from storage, slides were first placed in room temperature PBS for 1 minute followed by application of a solution composed of 1% BSA and 2X SSC [saline-sodium citrate, diluted from 20× SSC containing 3 mol/L NaCl and 300 mmol/L trisodium citrate (#S6639–1L, Sigma-Aldrich)] for 10 minutes under a glass coverslip. Primary HA antibody (11583816001, Sigma-Aldrich, clone: 12CA5, RRID: AB_514505) was then applied in the same 1% BSA and 2X SSC solution under a coverslip for 45 minutes. Slides were then washed in PBS for 5 minutes followed by detection with an Alexa Fluor 488 labeled secondary antibody (A-11001, Invitrogen, polyclonal, RRID: AB_2534069) for 45 minutes followed by washing in PBS for 5 minutes. After immunostaining, the slides were then dehydrated in a graded ethanol series (70%, 80%, and 100%) for 2 minutes each followed by application of an Alexa Fluor 594 labeled fosmid clone (WI2–3042B13, BACPAC Genomics) that was suspended in 50% formamide (4610-OP, Calbiochem. OmniPur Formamide, Deionized), 10% dextran (4610-OP, Sigma-Aldrich), and 2X SSC and denatured for 5 minutes at 70°C to detect ribosomal RNA. Following overnight hybridization of the probe to RNA at 37°C, slides were washed in 50% formamide, 2X SSC for 5 minutes and then mounted in Vectashield Antifade Mounting Medium (H-1000–10, Vector Laboratories) with DAPI. Mounted slides were then imaged in 3D using widefield fluorescence microscopy with Nikon NIS-Elements software (all imaged fields coordinates were recorded). Images were captured at 0.15 μmol/L plane spacing, and subsequently subjected to 3D deconvolution. Enough fields were imaged so that at least 200 cells could be analyzed for each condition. Following imaging the coverslips were removed and the slides were treated with RNase A (R4642–5MG, Sigma-Aldrich) in 2X SSC for 45 minutes at 37°C and then briefly rinsed in PBS. The slides were then denatured in 70% formamide, 2X SSC at 80°C for 10 minutes followed by a second hybridization using the same fosmid clone that had previously been used to identify ribosomal RNA, this time to identify rDNA. Following overnight hybridization at 37°C, the slides were washed in 50% formamide, 2X SSC at 37°C and imaged for a second time using the same coordinates as the first set of images.
Statistical Analysis
For discrete values of the molecular feature and the mutation frequency in cohorts, statistical significance was calculated by Fisher exact test. For functional assays, statistical significance was calculated by two-tailed Student t test. The sample size and statistical methods were determined according to the SD and distribution of the results of preliminary experiments. Adjustment of multiple testing was performed by the Benjamini–Hochberg method using p.adjust function on R. For the statistical analysis of multiple rounds of colony assays with technical and biological replicates, generalized linear mixed effects model with Poisson distribution and the random effect defined from nested data in each condition was applied. A global test was first performed with ANOVA test for two nested models with or without the condition followed by pairwise comparisons and adjustment with the Tukey method.
For survival data, Kaplan–Meier curves for the probability of overall survival (pOS), and event-free survival (pEFS) were constructed using R package survival, IBM SPSS (20.0, RRID: SCR_019096; AAML1031) and GraphPad Prism (9.1.0, RRID: SCR_002798; TARGET). The significance of predictor variables was tested with the log-rank statistic for pOS, pEFS, and Pearson χ2 test for minimal residual disease (MRD) positivity. Summary statistics for each group are presented in Supplementary Tables S30–S32. Events in pEFS calculations were defined as relapse, death in remission by any cause, and nonresponse, which was included as an event at the date of diagnosis. For univariable and multivariate analysis, the Cox proportional hazards model was used to obtain the estimates and the 95% confidence interval of the relative risk for prognostic factors. Computations were performed using SAS (Statistical Analysis System Version 9.3; SAS Institute, RRID: SCR_008567), GraphPad Prism and R statistical environment.
Data Availability
The genomic data and expression data of the relapse AML cohort generated in this study have been deposited in the European Genome-Phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI), under accession EGAS00001005760. The dataset is also available through St. Jude Cloud Genomics Platform under accession SJC-DS-1015 at https://permalinks.stjude.cloud/permalinks/rpaml. The results published here are in whole or part based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative, phs000218, managed by the NCI. Data for TARGET AML (n = 159) and TARGET AML-IF (n = 29) are available as a part of phs000218.v24.p8 under dbGaP accession number phs000465.v21.p8. Information about TARGET can be found at http://ocg.cancer.gov/programs/target. Additional RNA-seq data in the extension RNA cohort were obtained from St. Jude Cloud (n = 159), and other published studies (5, 7, 10, 15, 20–24). Expression data of normal cord blood CD34+ cells (n = 5) generated in this study as controls in the RNA cohort are publicly available in Gene Expression Omnibus (GEO) at GSE190269. RNA-seq data of adult AML cohort for UBTF-TD screening were obtained from the GDC Data Portal under accession phs000178 (TCGA-LAML; ref. 32, n = 151) and phs001657 (BEATAML1.0-COHORT; ref. 33, n = 220). Expression data of transduced cord blood CD34+ cells generated in this study are publicly available in GEO at GSE189901. Other data generated in this study are available in the Supplementary tables or upon request to the corresponding author.
Supplementary Material
Acknowledgments
We thank all the patients and their families at St. Jude Children's Research Hospital (SJCRH) for their contribution of the biological specimens used in this study. We also thank the Biorepository, the Flow Cytometry and Cell Sorting Core, and the Hartwell Center for Bioinformatics and Biotechnology at SJCRH for their essential services. Julie Justice in the Anatomic Pathology lab established the IHC for UBTF. This work was funded by the American Lebanese and Syrian Associated Charities of St. Jude Children's Research Hospital and grants from the NIH (P30 CA021765, Cancer Center Support Grant and a Developmental Fund Award, to J.M. Klco and X. Ma). This work was also supported in part by the Fund for Innovation in Cancer Informatics (www.the-ici-fund.org, to X. Ma and J.M. Klco), St. Baldrick's Consortium Grant (to S. Meshinchi), Target Pediatric AML (to S. Meshinchi), Leukemia and Lymphoma Society (6558-18, to S. Meshinchi), National Institutes of Health (R01-CA114563-10 and HHSN-261200800001E, to S. Meshinchi), COG Chair's Grant U10-CA098543 (to S. Meshinchi), Andrew McDonough B+ Foundation (to S. Meshinchi), Hyundai Hope on Wheels (to S. Meshinchi), NCTN Statistics & Data Center U10-CA180899 (to S. Meshinchi and T.A. Alonzo), NCTN Operations Center Grant U10CA180886 (to S. Meshinchi), and Project Stella (to S. Meshinchi). J.M. Klco holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is a previous recipient of the V Foundation Scholar Award (Pediatric).
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
I. Iacobucci reports other support from Amgen and other support from Mission Bio outside the submitted work. J. Miller reports personal fees from Janssen Research & Development outside the submitted work. H. Inaba reports grants and personal fees from Amgen and Servier; personal fees from Jazz Pharmaceuticals, and personal fees from Chugai pharmaceuticals outside the submitted work. C.G. Mullighan reports personal fees from Illumina during the conduct of the study; grants from Pfizer and grants from AbbVie outside the submitted work. S. Pounds reports other support from ALSAC and grants from NIH during the conduct of the study; in addition, S. Pounds has a patent for PCT/US2020/051961 pending and a patent for US Provisional Application no. 63/233,673 pending. J.E. Rubnitz reports personal fees from Kura Oncology, Biomea Fusion, Kronos Bio, Inc, Geron Corporation, and personal fees from PinotBio, Inc outside the submitted work. No disclosures were reported by the other authors.
Disclaimer
This research content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Authors’ Contributions
M. Umeda: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Ma: Conceptualization, resources, data curation, software, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. B.J. Huang: Data curation, formal analysis, validation, investigation, visualization, writing–review and editing. K. Hagiwara: Data curation, software, formal analysis, investigation, methodology, writing–review and editing. T. Westover: Conceptualization, resources, data curation, investigation, visualization, writing–review and editing. S. Abdelhamed: Supervision, investigation, writing–review and editing. J.M. Barajas: Validation, investigation, writing–review and editing. M.E. Thomas: Validation, investigation, writing–review and editing. M.P. Walsh: Resources, data curation, formal analysis, investigation, writing–review and editing. G. Song: Resources, data curation, formal analysis, investigation, visualization, writing–review and editing. L. Tian: Resources, data curation, software, formal analysis, visualization, writing–review and editing. Y. Liu: Resources, data curation, software, formal analysis, visualization, writing–review and editing. X. Chen: Resources, data curation, software, formal analysis, visualization, writing–review and editing. P. Kolekar: Resources, data curation. Q. Tran: Resources, data curation. S.G. Foy: Resources, data curation. J.L. Maciaszek: Data curation, formal analysis, investigation, writing–review and editing. A.B. Kleist: Investigation, visualization. A.R. Leonti: Resources, data curation, formal analysis. B. Ju: Validation, visualization, methodology, writing–review and editing. J. Easton: Supervision, investigation, visualization, methodology, writing–review and editing. H. Wu: Software, formal analysis, investigation, writing–review and editing. V. Valentine: Investigation, visualization. M.B. Valentine: Formal analysis, supervision, investigation, visualization, writing–review and editing. Y. Liu: Investigation, visualization. R.E. Ries: Resources, data curation, formal analysis. J.L. Smith: Resources, data curation, formal analysis. E. Parganas: Resources, data curation, investigation. I. Iacobucci: Writing–review and editing. R. Hiltenbrand: Investigation, writing–review and editing. J. Miller: Data curation, investigation, writing–review and editing. J.R. Myers: Data curation, investigation, writing–review and editing. E. Rampersaud: Resources, data curation, writing–review and editing. D. Rahbarinia: Resources, data curation, writing–review and editing. M. Rusch: Resources, data curation, writing–review and editing. G. Wu: Resources, data curation, supervision, writing–review and editing. H. Inaba: Supervision, writing–review and editing. Y. Wang: Resources, data curation, formal analysis, validation, investigation, writing–original draft, writing–review and editing. T.A. Alonzo: Resources, data curation, formal analysis, supervision, validation, investigation, writing–review and editing. J.R. Downing: Supervision, writing–review and editing. C.G. Mullighan: Supervision, writing–review and editing. S. Pounds: Data curation, software, formal analysis, supervision, validation, writing–original draft, writing–review and editing. M. Babu: Supervision, visualization, writing–review and editing. J. Zhang: Resources, supervision, writing–review and editing. J.E. Rubnitz: Supervision, writing–review and editing. S. Meshinchi: Resources, supervision, funding acquisition, validation, investigation, project administration, writing–review and editing. X. Ma: Resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. J.M. Klco: Conceptualization, resources, data curation, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Rubnitz JE. How I treat pediatric acute myeloid leukemia. Blood 2012;119:5980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cornelissen JJ, Gratwohl A, Schlenk RF, Sierra J, Bornhauser M, Juliusson Get al. The European LeukemiaNet AML Working Party consensus statement on allogeneic HSCT for patients with AML in remission: an integrated-risk adapted approach. Nat Rev Clin Oncol 2012;9:579–90. [DOI] [PubMed] [Google Scholar]
- 3. Kaspers GJ, Zimmermann M, Reinhardt D, Gibson BE, Tamminga RY, Aleinikova Oet al. Improved outcome in pediatric relapsed acute myeloid leukemia: results of a randomized trial on liposomal daunorubicin by the International BFM Study Group. J Clin Oncol 2013;31:599–607. [DOI] [PubMed] [Google Scholar]
- 4. Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JEet al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 2011;118:3645–56. [DOI] [PubMed] [Google Scholar]
- 5. de Rooij JD, Branstetter C, Ma J, Li Y, Walsh MP, Cheng Jet al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet 2017;49:451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrison CJ, Hills RK, Moorman AV, Grimwade DJ, Hann I, Webb DKet al. Cytogenetics of childhood acute myeloid leukemia: United Kingdom Medical Research Council Treatment trials AML 10 and 12. J Clin Oncol 2010;28:2674–81. [DOI] [PubMed] [Google Scholar]
- 7. Bolouri H, Farrar JE, Triche T Jr, Ries RE, Lim EL, Alonzo TAet al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018;24:103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stratmann S, Yones SA, Mayrhofer M, Norgren N, Skaftason A, Sun Jet al. Genomic characterization of relapsed acute myeloid leukemia reveals novel putative therapeutic targets. Blood Adv 2021;5:900–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LRet al. Genomic profiling of pediatric acute myeloid leukemia reveals a changing mutational landscape from disease diagnosis to relapse. Cancer Res 2016;76:2197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McNeer NA, Philip J, Geiger H, Ries RE, Lavallee VP, Walsh Met al. Genetic mechanisms of primary chemotherapy resistance in pediatric acute myeloid leukemia. Leukemia 2019;33:1934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borel C, Dastugue N, Cances-Lauwers V, Mozziconacci MJ, Prebet T, Vey Net al. PICALM-MLLT10 acute myeloid leukemia: a French cohort of 18 patients. Leuk Res 2012;36:1365–9. [DOI] [PubMed] [Google Scholar]
- 12. Noort S, Zimmermann M, Reinhardt D, Cuccuini W, Pigazzi M, Smith Jet al. Prognostic impact of t(16;21)(p11;q22) and t(16;21)(q24;q22) in pediatric AML: a retrospective study by the I-BFM Study Group. Blood 2018;132:1584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y, Li C, Shen S, Chen X, Szlachta K, Edmonson MNet al. Discovery of regulatory noncoding variants in individual cancer genomes by using cis-X. Nat Genet 2020;52:811–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck Cet al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 2014;157:369–81. [DOI] [PubMed] [Google Scholar]
- 15. Schwartz JR, Ma J, Kamens J, Westover T, Walsh MP, Brady SWet al. The acquisition of molecular drivers in pediatric therapy-related myeloid neoplasms. Nat Commun 2021;12:985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Montefiori LE, Bendig S, Gu Z, Chen X, Polonen P, Ma Xet al. Enhancer hijacking drives oncogenic BCL11B expression in lineage-ambiguous stem cell leukemia. Cancer Discov 2021;11:2846–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tosi S, Mostafa Kamel Y, Owoka T, Federico C, Truong TH, Saccone S. Paediatric acute myeloid leukaemia with the t(7;12)(q36;p13) rearrangement: a review of the biological and clinical management aspects. Biomark Res 2015;3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad Cet al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pounds S, Cheng C, Li S, Liu Z, Zhang J, Mullighan C. A genomic random interval model for statistical analysis of genomic lesion data. Bioinformatics 2013;29:2088–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buelow DR, Pounds SB, Wang YD, Shi L, Li Y, Finkelstein Det al. Uncovering the genomic landscape in newly diagnosed and relapsed pediatric cytogenetically normal FLT3-ITD AML. Clin Transl Sci 2019;12:641–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iacobucci I, Wen J, Meggendorfer M, Choi JK, Shi L, Pounds SBet al. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat Genet 2019;51:694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwartz JR, Ma J, Lamprecht T, Walsh M, Wang S, Bryant Vet al. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun 2017;8:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rusch M, Nakitandwe J, Shurtleff S, Newman S, Zhang Z, Edmonson MNet al. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat Commun 2018;9:3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faber ZJ, Chen X, Gedman AL, Boggs K, Cheng J, Ma Jet al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet 2016;48:1551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Newman S, Nakitandwe J, Kesserwan CA, Azzato EM, Wheeler DA, Rusch Met al. Genomes for kids: the scope of pathogenic mutations in pediatric cancer revealed by comprehensive DNA and RNA sequencing. Cancer Discov 2021;11:3008–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corrigan DJ, Luchsinger LL, Justino de Almeida M, Williams LJ, Strikoudis A, Snoeck HW. PRDM16 isoforms differentially regulate normal and leukemic hematopoiesis and inflammatory gene signature. J Clin Invest 2018;128:3250–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 2009;4:374–82. [DOI] [PubMed] [Google Scholar]
- 28. Maiser A, Dillinger S, Langst G, Schermelleh L, Leonhardt H, Nemeth A. Super-resolution in situ analysis of active ribosomal DNA chromatin organization in the nucleolus. Sci Rep 2020;10:7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tian L, Li Y, Edmonson MN, Zhou X, Newman S, McLeod Cet al. CICERO: a versatile method for detecting complex and diverse driver fusions using cancer RNA sequencing data. Genome Biol 2020;21:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hagiwara K, Ding L, Edmonson MN, Rice SV, Newman S, Easton Jet al. RNAIndel: discovering somatic coding indels from tumor RNA-Seq data. Bioinformatics 2020;36:1382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hagiwara K, Edmonson MN, Wheeler DA, Zhang J. indelPost: harmonizing ambiguities in simple and complex indel alignments. Bioinformatics 2022;38:549–51. [DOI] [PubMed] [Google Scholar]
- 32. Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis Jet al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012;366:1079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SLet al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aplenc R, Meshinchi S, Sung L, Alonzo T, Choi J, Fisher Bet al. Bortezomib with standard chemotherapy for children with acute myeloid leukemia does not improve treatment outcomes: a report from the Children's Oncology Group. Haematologica 2020;105:1879–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rasche M, Zimmermann M, Borschel L, Bourquin JP, Dworzak M, Klingebiel Tet al. Successes and challenges in the treatment of pediatric acute myeloid leukemia: a retrospective analysis of the AML-BFM trials from 1987 to 2012. Leukemia 2018;32:2167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin Met al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell 2014;25:379–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Boer B, Prick J, Pruis MG, Keane P, Imperato MR, Jaques Jet al. Prospective isolation and characterization of genetically and functionally distinct AML subclones. Cancer Cell 2018;34:674–89. [DOI] [PubMed] [Google Scholar]
- 38. Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy Ret al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou B, Wang J, Lee SY, Xiong J, Bhanu N, Guo Qet al. PRDM16 suppresses MLL1r leukemia via intrinsic histone methyltransferase activity. Mol Cell 2016;62:222–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shiba N, Ohki K, Kobayashi T, Hara Y, Yamato G, Tanoshima Ret al. High PRDM16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT3-ITD, KMT2A-PTD, and NUP98-NSD1: the results of the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 trial. Br J Haematol 2016;172:581–91. [DOI] [PubMed] [Google Scholar]
- 41. Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran Ret al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell 2018;34:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edmonson MN, Zhang J, Yan C, Finney RP, Meerzaman DM, Buetow KH. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics 2011;27:865–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang J, Mullighan CG, Easton J, Roberts S, Heatley SL, Ma Jet al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods 2011;8:652–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen X, Gupta P, Wang J, Nakitandwe J, Roberts K, Dalton JDet al. CONSERTING: integrating copy-number analysis with structural-variation detection. Nat Methods 2015;12:527–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Yet al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 2014;46:444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 2011;27:2903–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia Wet al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann Aet al. The ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Edmonson MN, Patel AN, Hedges DJ, Wang Z, Rampersaud E, Kesserwan CAet al. Pediatric Cancer Variant Pathogenicity Information Exchange (PeCanPIE): a cloud-based platform for curating and classifying germline variants. Genome Res 2019;29:1555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin Let al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster Jet al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LGet al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat 2018;39:1517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee K, Krempely K, Roberts ME, Anderson MJ, Carneiro F, Chao Eet al. Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline CDH1 sequence variants. Hum Mutat 2018;39:1553–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luo X, Feurstein S, Mohan S, Porter CC, Jackson SA, Keel Set al. ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants. Blood Adv 2019;3:2962–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gelb BD, Cave H, Dillon MW, Gripp KW, Lee JA, Mason-Suares Het al. ClinGen's RASopathy Expert Panel consensus methods for variant interpretation. Genet Med 2018;20:1334–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Qet al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol 2002;64:479–98. [Google Scholar]
- 58. Pounds S, Cheng C. Robust estimation of the false discovery rate. Bioinformatics 2006;22:1979–87. [DOI] [PubMed] [Google Scholar]
- 59. Casella G, Berger RL. Statistical Inference Vol. 70. Belmont, CA: Duxbury Press;1990. [Google Scholar]
- 60. Li B, Brady SW, Ma X, Shen S, Zhang Y, Li Yet al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood 2020;135:41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch Met al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 2015;6:6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li Wet al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi Wet al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maaten LVD, Hinton GE. Visualizing Data using t-SNE. J Mach Learn Res 2008;9:2579–605. [Google Scholar]
- 68. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LGet al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2019;37:38–44. [DOI] [PubMed] [Google Scholar]
- 69. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MAet al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic data and expression data of the relapse AML cohort generated in this study have been deposited in the European Genome-Phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI), under accession EGAS00001005760. The dataset is also available through St. Jude Cloud Genomics Platform under accession SJC-DS-1015 at https://permalinks.stjude.cloud/permalinks/rpaml. The results published here are in whole or part based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative, phs000218, managed by the NCI. Data for TARGET AML (n = 159) and TARGET AML-IF (n = 29) are available as a part of phs000218.v24.p8 under dbGaP accession number phs000465.v21.p8. Information about TARGET can be found at http://ocg.cancer.gov/programs/target. Additional RNA-seq data in the extension RNA cohort were obtained from St. Jude Cloud (n = 159), and other published studies (5, 7, 10, 15, 20–24). Expression data of normal cord blood CD34+ cells (n = 5) generated in this study as controls in the RNA cohort are publicly available in Gene Expression Omnibus (GEO) at GSE190269. RNA-seq data of adult AML cohort for UBTF-TD screening were obtained from the GDC Data Portal under accession phs000178 (TCGA-LAML; ref. 32, n = 151) and phs001657 (BEATAML1.0-COHORT; ref. 33, n = 220). Expression data of transduced cord blood CD34+ cells generated in this study are publicly available in GEO at GSE189901. Other data generated in this study are available in the Supplementary tables or upon request to the corresponding author.