Abstract
We have previously isolated 32 mutants of Legionella pneumophila that are defective in the infection of mammalian cells but not protozoa. The mutated loci have been designated macrophage-specific infectivity (mil) loci. In this study we characterized the mil mutant GK11. This mutant was incapable of growth within U937 macrophage-like cells and WI-26 alveolar epithelial cells. This defect in intracellular replication correlated with a defect in cytopathogenicity to these cells. Sequence analysis of the GK11 locus revealed it to be highly similar to rep helicase genes of other bacteria. Since helicase mutants of Escherichia coli are hypersensitive to thymine starvation, we examined the sensitivity of GK11 to thymineless death (TLD). In the absence of thymine and thymidine, mutant GK11 did not undergo TLD but was defective for in vitro growth, and the defect was partially restored when these compounds were added to the growth medium. In addition, supplementation with thymidine or thymine partially restored the ability of GK11 to grow within and kill U937 macrophage-like cells. The data suggested that the low levels of thymine or thymidine in the L. pneumophila phagosome contributed to the defect of GK11 within macrophages. Using confocal laser scanning microscopy, we determined the effect of the mutation in the Rep helicase homologue on the intracellular trafficking of GK11 within macrophages. In contrast to the wild-type strain, phagosomes harboring GK11 colocalized with several late endosomal/lysosomal markers, including LAMP-1, LAMP-2, and cathepsin D. In addition, only 50% of the GK11 phagosomes colocalized with the endoplasmic reticulum marker BiP 4 h postinfection. Colocalization of BiP with GK11 phagosomes was absent 6 h postinfection, while 90% of the wild-type phagosomes colocalized with this marker at both time points. We propose that the low level of thymine within the L. pneumophila phagosome in combination with simultaneous exposure to multiple stress stimuli results in deleterious mutations that cannot be repaired in the rep helicase homologue mutant, rendering it defective in intracellular replication.
The Legionnaires' disease bacterium, Legionella pneumophila, is a gram-negative facultative intracellular bacterial pathogen (15, 49). Legionnaires' disease is a common pneumonia that occurs in both nosocomial and community settings (14, 19, 41). In the lungs of affected individuals, L. pneumophila replicates in phagocytic and possibly alveolar epithelial cells, within a specialized compartment that is surrounded by the host-cell rough endoplasmic reticulum (RER) and mitochondria (1, 24, 39, 51, 62). Furthermore, the L. pneumophila phagosome has been characterized as an endosome maturation-blocked phagosome due to its inability to mature along the classical endocytic trafficking pathway into a phagolysosome (1). More specifically, this phagosome has been shown to be devoid of the late endosomal/lysosomal markers LAMP-1, LAMP-2, and cathepsin D (17, 40, 61, 63).
The ability of L. pneumophila to cause disease has been linked to its ability to replicate within its environmental protozoan hosts (8, 21, 65). Within protozoa, L. pneumophila replicates within a phagosome that appears similar to its mammalian counterpart in that it is surrounded by the RER and mitochondria (3, 64, 65). In addition, the L. pneumophila phagosome is blocked from maturation into a phagolysosome (13). Interestingly, we have isolated 89 distinct mutants of L. pneumophila (pmi mutants) that are defective for growth within both mammalian and protozoan cells (30, 31, 60). Furthermore, 12 of the pmi mutants contain insertions in the dot/icm gene clusters (reference 57 and our unpublished data), indicating the requirement of the Dot/Icm secretion system for infectivity of both mammalian and protozoan cells (30, 57, 66). This observation has recently been supported by the finding that other dot/icm genes are required for intracellular growth within both host cells (58). In addition to these genes, the pilD product, a component of the type II secretion apparatus of L. pneumophila, is also required for growth within mammalian cells and protozoa (45, 46). The above findings support the idea that L. pneumophila may utilize similar mechanisms to infect its two evolutionarily distant hosts (1, 7, 8).
L. pneumophila, however, appears to have evolved distinct mechanisms to infect and replicate within its various host cells. We have isolated 32 mutants (mil mutants) of L. pneumophila that are defective for growth within macrophages but not within Acanthamoeba polyphaga (29, 31). None of the mil mutants contain insertions in the dot/icm loci (29). The identity of the mil genes is currently under investigation and will likely yield clues about the nature of the interaction between L. pneumophila and macrophages. One of the mil loci (milA) has been recently characterized and found to encode a potential transporter with homology to sugar transport proteins from other bacteria (33). In this study we characterized the mil mutant GK11.
MATERIALS AND METHODS
Bacterial strains and vectors.
The virulent strain of L. pneumophila (AA100) is a clinical isolate and has been described previously (4). Plasmid pBC-SK+ (Stratagene, La Jolla, Calif.) was used to subclone L. pneumophila DNA. Cloning experiments were performed with Escherichia coli DH5α as a host strain (Bethesda Research Laboratories [BRL], Gaithersburg, Md.). The L. pneumophila chromosomal cosmid DNA library that contains a chloramphenicol resistance marker has been previously described (35, 53, 59).
DNA manipulations and sequence analysis.
L. pneumophila chromosomal DNA was prepared using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.). Transfections, restriction enzyme digestions, and DNA ligations were performed as described elsewhere (56) unless specified. Restriction enzymes were purchased from Promega (Madison, Wis.), and T4 DNA ligase was obtained from BRL. Plasmid and cosmid DNA preparations were performed using Qiagen mini and midi plasmid kits, respectively (Qiagen Inc., Chatsworth, Calif.). Transformations were done using a Gene Pulser as recommended by the manufacturer (Bio-Rad, Hercules, Calif.). Purification of DNA fragments from agarose gels for subcloning or labeling for Southern hybridization was carried out using a QIAquick gel purification kit (Qiagen). Fluorescein labeling of DNA probes for Southern hybridization was done using the ECL random prime labeling system, version II (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Oligonucleotides for sequencing were purchased from Integrated DNA Technologies Inc. (Coralville, Calif.). Initial sequencing was performed using a chromosomal HindIII subclone of the kanamycin insertion region of GK11. This subclone included half of the kanamycin cassette and the adjacent L. pneumophila DNA, which allowed sequencing using a primer, designated Tn10.2 (5′-TCATTAGGGGATTCATCA-3′), homologous to the inverted repeat of the IS10 elements of the mini-Tn10::Kan. Subsequent sequencing was performed directly from cosmid clone K9 that complemented the GK11 defect. Sequence comparisons and alignments were performed using the BlastX and Blast2 programs, respectively.
Bacterial cultures.
For all experiments described, L. pneumophila AA100 and GK11 were grown to the postexponential phase in the following manner. Bacteria were grown from frozen stocks on buffered charcoal-yeast extract (BCYE) agar (supplemented with kanamycin [20 μg/ml] for strain GK11) for 48 h at 37°C. The bacteria were resuspended to an optical density at 550 nm (OD550) of 0.1 to 0.4 in buffered yeast extract (BYE) broth and grown at 37°C in a shaking incubator for 15 to 18 h to an OD550 of 2.1 to 2.2, when bacterial replication completely stopped (post-exponential growth phase).
To compare the growth of mutant GK11 to that of the wild-type strain in BYE medium, bacteria were grown on BCYE agar plates for 48 h at 37°C. Bacteria were then resuspended in BYE medium to an OD550 of 0.1 to 0.4, and growth was measured spectrophotometrically. The effect of thymine or thymidine supplementation on the in vitro growth of mutant GK11 was determined by monitoring growth in a semidefined medium (CAA medium) as described previously (50). Briefly, bacteria were grown on BCYE agar plates for 48 h at 37°C, washed three times in sterile water, and resuspended to an OD550 of 0.1 to 0.4 in CAA medium supplemented with thymine or thymidine (200 μg/ml), and cultures were grown at 37°C with shaking at 220 rpm. Bacterial growth was monitored spectophotometrically at an OD of 550 nm, and viability of GK11 in the absence or presence of thymine or thymidine in CAA medium was determined by colony enumeration on BCYE agar plates. Note that supplementation with either thymine or thymidine yielded identical results in all described experiments; thus, only results from thymidine supplementation are presented.
Tissue culture and infections.
U937 macrophage-like cells were maintained at 37°C and 5% CO2 in RPMI 1640 tissue culture medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% heat-inactivated fetal bovine serum (Gibco BRL). For infections, cells (105 cells/well in 96- or 24-well plates) were differentiated for 48 h using phorbol 12-myristate 13-acetate as described previously (5).
WI-26 human type I alveolar epithelial cells were maintained in minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum at 37°C and 5% CO2 (31). For infections, 104 cells/well were seeded in 96-well plates and incubated at 37°C and 5% CO2 overnight.
Infection protocol.
A multiplicity of infection (MOI) of 10 was used to determine bacterial growth kinetics in U937 macrophage-like cells or WI-26 alveolar epithelial cells, and an MOI of 1 was used to determine cytopathogenicity. An MOI of 1 was also used in laser scanning confocal microscopy experiments (see below).
Infections were performed as described previously (29, 30). Briefly, bacteria were diluted to the appropriate concentrations in tissue culture medium and added to monolayers in 96-well plates (to determine intracellular growth kinetics and cytopathogenicity) and 24-well plates (for microscopy experiments). Plates were then spun at 1,000 × g for 5 min in order to synchronize the infection. For cytopathogenicity and microscopy experiments, plates were then incubated at 37°C for several time intervals. To examine intracellular growth and survival, extracellular bacteria were killed using gentamicin (50 μg/ml) for 1 h following an initial 1-h infection at 37°C. Infections were then incubated at 37°C for several time intervals, and the number of intracellular bacteria was determined following growth on BCYE plates.
Cytopathogenicity was determined by treatment of infected or noninfected monolayers with 10% Alamar blue dye (Alamar Bioscience Inc., Sacramento, Calif.). Viability of monolayers was measured optically at a wavelength of 570 nm and corrected for background at 600 nm, using a VMAX kinetic microplate reader (Molecular Devices, Menlo Park, Calif.) (6). The relative degree of cytopathogenicity was expressed as percent survival compared to noninfected cells, using the formula (mean OD value of infected/mean OD value of noninfected) × 100 as described previously (30). Noninfected cells were considered 100% viable.
To determine the effect of thymine or thymidine on the in vivo growth kinetics and cytopathogenicity of GK11, U937 macrophage-like cells were preincubated with thymidine or thymine (200 μg/ml) for 24 h and then infected in the presence or absence of thymine or thymidine (200 μg/ml). Growth kinetics and cytopathogenicity were determined exactly as described above.
Antibodies, stains, and laser scanning confocal microscopy.
U937 cells were differentiated on 12-mm-diameter (0.13 to 0.17 mm thick) circular glass coverslips (Fisher Scientific, Pittsburgh, Pa.) in 24-well culture plates. Following infections, cells were processed for confocal laser scanning microscopy exactly as described previously (28). Briefly, to differentiate between intracellular and extracellular bacteria, a polyclonal anti-L. pneumophila antibody (26) was used to label extracellular bacteria; cells were then permeabilized, and intracellular and extracellular bacteria were stained with Toto-3 (Molecular Probes Inc., Eugene, Oreg.). The primary antibodies anti-LAMP-1, anti-LAMP-2, anti-cathepsin D, and anti-BiP, as well as conjugated secondary antibodies, were used exactly as described previously (28). Samples were analyzed using a Leica TCS SP laser scanning confocal microscope (Leica Microscopy and Scientific Instruments Group, Heerburg, Switzerland). On average, 15 serial 0.2-μm sections of each image were captured, compiled, and stored for further analyses, using Adobe Photoshop 5.0 (Adobe, Inc.). Duplicate samples from at least three independent experiments were analyzed, and results of a representative experiment are provided in Results. Colocalization was assessed by counting 100 to 150 intracellular bacteria per experiment. Fluorescence intensity was measured using Leica TCS SP software.
Hemolysis assays.
Hemolysis of sheep red blood cells (sRBCs) was performed as previously described (43). Bacteria were mixed with sRBCs at an MOI of 25, pelleted for 2 min at 13,000 × g, and incubated at 37°C for 60 min. Hemolysis of sRBCs was assessed spectrophotometrically at an OD of 415 nm.
Nucleotide sequence accession number.
Partial sequence of the L. pneumophila rep helicase gene was submitted to GenBank and assigned accession number AF173009.
RESULTS
Intracellular growth of GK11 within macrophages and alveolar epithelial cells.
L. pneumophila has been shown to undergo growth phase-dependent expression of several virulence-associated phenotypes that are triggered at the post-exponential growth phase (16). Thus, it is crucial to take special note of the bacterial growth phase when characterizing potential mutants. For our initial characterization of the mil and pmi mutants, we used bacteria grown to the mid-late logarithmic growth phase (29, 30).
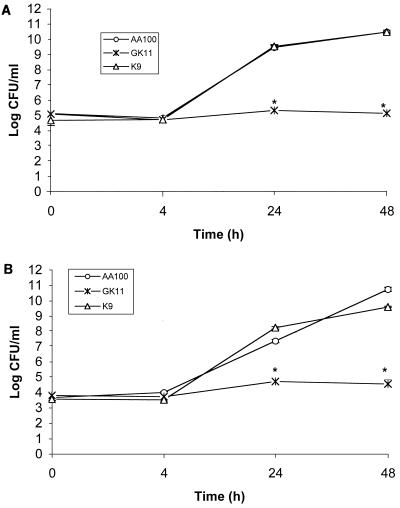
In this study, bacteria were grown into the post-exponential phase in BYE broth and used for infection studies. The intracellular growth of GK11 was severely defective compared to the parental strain AA100 within both U937 macrophages (Fig. 1A) and WI-26 alveolar epithelial cells (Fig. 1B). Similar numbers of GK11 and AA100 were present within these cells at 0 and 4 h postinfection after gentamicin treatment (Fig. 1). Therefore, GK11 was not defective for entry into these cells. Interestingly, the number of intracellular GK11 remained constant throughout the time course of the experiment (48 h) within both cell types, while AA100 counts increased ∼10,000-fold (Fig. 1). These results showed that GK11 was severely defective in intracellular replication within both macrophages and alveolar epithelial cells.
FIG. 1.
Intracellular growth kinetics of GK11 and AA100 (wild-type L. pneumophila) in U937 macrophage-like cells (A) and WI-26 alveolar epithelial cells (B). Strain K9 is a cosmid-complemented clone of GK11. Experiments were performed in triplicate and repeated at least three times. The data are from a representative experiment. Error bars represent standard deviation of the mean; some of the error bars are too small to be seen. ∗, significantly fewer intracellular bacteria than in wild-type (AA100)-infected cells, based on Student's t test (P < 0.05).
Cytopathogenicity of GK11 to U937 macrophages and WI-26 alveolar epithelial cells.
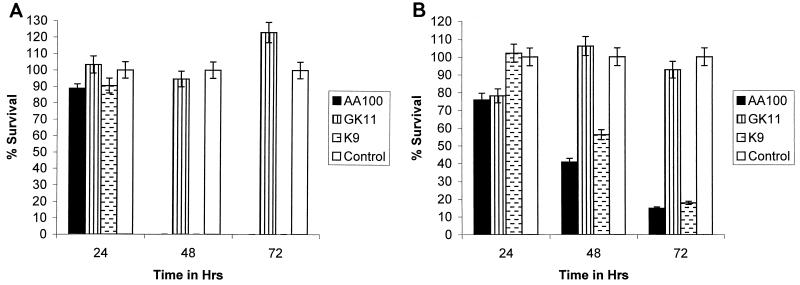
Cytopathogenicity of L. pneumophila to its host cells correlates with its intracellular growth. Furthermore, all of the previously described mutants of L. pneumophila that are defective for intracellular growth are also defective for cytopathogenicity (29, 30, 54, 55, 66). GK11 was less cytopathogenic to both macrophages and WI-26 cells than AA100 (Fig. 2). Furthermore, the defect in cytopathogenicity remained relatively constant throughout the course of the infection (Fig. 2). Thus, the defect of GK11 in intracellular growth correlated with its defect in cytopathogenicity to both macrophages and epithelial cells.
FIG. 2.
Cytopathogenicity of GK11 and AA100 to U937 macrophage-like cells (A) and WI-26 alveolar epithelial cells (B) as determined by Alamar blue dye staining. K9 is a cosmid-complemented clone of GK11. Percent survival of cells was normalized to uninfected cells, which were considered 100% viable. Experiments were performed in triplicate and repeated at least three times. The data are from a representative experiment. Error bars represent standard deviation of the mean. ∗, significantly fewer intracellular bacteria than in wild-type (AA100)-infected cells, based on Student's t test (P < 0.05).
The ability of L. pneumophila to kill macrophages and epithelial cells has been shown to occur via a biphasic mechanism (25, 26) that involves apoptosis early during the first few hours of infection (25, 26, 52), followed by a rapid and independent necrosis prior to escape of the bacteria from the host cell (16). Examination of the DNA of infected macrophages by agarose gel electrophoresis (26) showed that mutant GK11 induced apoptosis in macrophages similarly to the wild-type strain (data not shown). Thus, it is possible that its defect in cytopathogenicity is due to a defect in induction of necrosis. L. pneumophila-mediated necrotic damage to the host cell is mediated by a pore-forming activity that can be tested for by determining hemolysis of sRBCs (43). However, post-exponential-growth-phase GK11 lysed sRBCs similarly to strain AA100 (data not shown). Since GK11 is not defective in induction of apoptosis or necrosis, its defect in cytopathogenicity is most likely due to its defect in intracellular replication.
Sequence analysis of the GK11 locus and complementation of the defect.
A 2.7-kb chromosomal fragment from GK11 that contained the kanamycin cassette and flanking L. pneumophila sequences was subcloned and used as a probe to screen an L. pneumophila genomic library. Two cosmid clones that hybridized to the insert were isolated and introduced into GK11. One of these cosmids (clone K9) complemented the GK11 defect in intracellular replication in and cytopathogenicity to U937 macrophage-like cells and WI-26 alveolar epithelial cells (Fig. 1 and 2). The second cosmid clone did not complement the GK11 defects, presumably because it contained only part of the complementing element or was a false-positive clone.
Sequence analysis showed that the kanamycin cassette was inserted 300 bp upstream of the carboxy terminus of a 1.4-kb open reading frame (ORF). Comparison of the predicted amino acid sequence of the 1.4-kb ORF to other sequences using the BlastX program revealed it to contain a high degree of similarity to the Rep helicase of several bacterial species. The highest degree of similarity was to the Rep helicase of E. coli (49% identity and 66% similarity) (data not shown). In addition, helicases in general contain six conserved motifs, five of which (motifs II, III, IV, V, and VI) were identical to those of E. coli (data not shown). Furthermore, the location of the kanamycin insertion was between two of the conserved motifs (motif V and VI) (44, 67). Thus, the GK11 ORF most likely encoded the Rep helicase of L. pneumophila, and the kanamycin cassette disrupted a helicase function. Sequencing of the region downstream of the termination codon of the rep gene showed that there were no ORFs present in all six possible reading frames at least up to 600 bp downstream of the termination codon. Thus, the GK11 ORF may either be monocistronic or a terminal gene in an operon. In either case, the kanamycin insertion is not expected to result in polar effects on other genes. Furthermore, the rep gene is monocistronic in other bacterial species such as E. coli and Haemophilus influenzae (22, 32).
TLD of GK11.
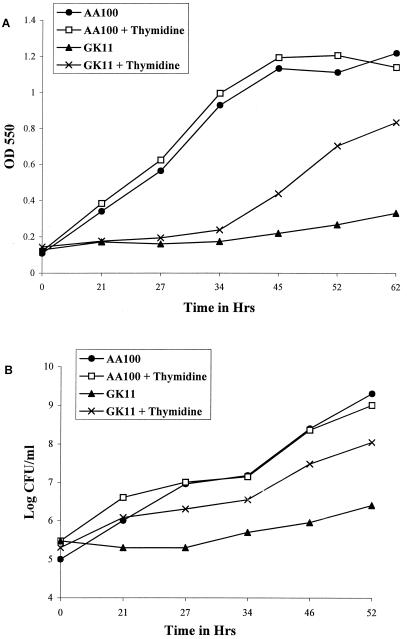
Several DNA repair mutants of E. coli, including Rep helicase mutants, are also hypersensitive to thymineless death (TLD) (10, 18). Thymine starvation has been shown to result in enhanced DNA damage such as double-strand breaks and incorporation of dUMP in place of dTMP in newly synthesized DNA, which could potentially become lethal if not properly repaired. In addition, thymine-starved bacteria exhibit inefficient assembly of Okazaki fragments during DNA replication (for a recent review, see reference 10). Thus, an inability to repair thymine starvation-induced DNA damage due to a helicase defect could be detrimental to the bacterium. Mutant GK11 grew as well as AA100 in BYE rich medium (data not shown). However, to determine whether GK11 is susceptible to TLD, we examined its growth in the semidefined CAA medium (lacking thymine and thymidine) in the presence or absence of thymidine supplementation. In the absence of thymidine supplementation, GK11 did not grow in CAA medium or grew very slightly (Fig. 3A). In the presence of thymidine (200 μg/ml), GK11 grew in CAA medium (following a lag phase of >20 h) with a generation time of 9 h, compared to 5.5 h for AA100 (Fig. 3A). The slower growth of GK11 was not likely due to insufficient thymidine in the medium since increasing the concentration of supplied thymidine to 400 μg/ml did not enhance its growth (data not shown). Furthermore, clone K9 (cosmid-complemented GK11) exhibited growth kinetics identical to those of AA100 (data not shown).
FIG. 3.
Effect of thymidine starvation on growth and survival of GK11 and AA100. (A) Growth of GK11 and AA100 in CAA medium with or without supplementation with thymidine (200 μg/ml), measured at an OD of 550. (B) CFU counts of GK11 and AA100 in CAA medium in the presence or absence of thymidine. Data are from one of three experiments with identical results.
To determine whether the absence of thymidine also resulted in increased death of GK11, we examined the viability of GK11 grown in CAA medium in the absence or presence of thymidine. Thymidine supplementation did not enhance the growth of AA100 (Fig. 3B). The number of GK11 CFU increased only eightfold in the absence of thymidine in CAA medium during 52 h of incubation (Fig. 3B). In contrast, GK11 grown in CAA medium in the presence of thymidine exhibited a more than 500-fold increase in CFU (Fig. 3B). In addition, clone K9 grew as well as AA100 in the absence of a thymidine supplement (data not shown). Thus, the mutation in the putative rep gene of L. pneumophila resulted in an in vitro growth defect in CAA semidefined medium in the absence of thymidine. This is similar to observations that the E. coli Rep helicase mutant is sensitive to thymine starvation (10, 18).
Thymine supplementation complements the intracellular growth and cytopathogenicity defects of GK11.
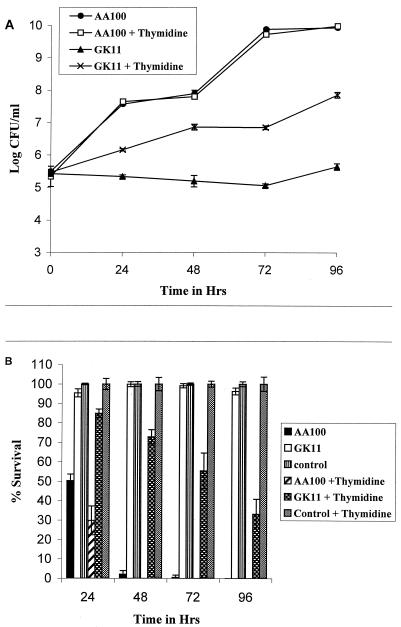
Thymine auxotroph mutants of L. pneumophila have been shown to be defective for intracellular growth within macrophages, which indicates that the L. pneumophila phagosome is deficient in thymine or thymidine (50). Thus, we determined whether supplementation of thymidine to the tissue culture media could complement the GK11 defect in intracellular replication in and cytopathogenicity to U937 macrophages. In the presence or absence of a thymidine supplement, strain AA100 grew 10,000-fold by 72 h postinfection (Fig. 4A). In contrast, the number of intracellular GK11 remained relatively constant (or slightly declined) in the absence of thymidine over the time course of the experiment (Fig. 4A). However, the intracellular numbers of GK11 increased 100-fold when thymidine was added to the tissue culture medium (Fig. 4A). It may not be surprising that in the presence of thymidine, GK11 did not grow intracellularly as well as AA100, since in vitro growth of GK11 in the presence of thymidine in CAA medium was also less than that of AA100 (Fig. 3A). The lower intracellular growth of GK11 was not likely due to insufficient thymidine supplementation to the tissue culture medium, since increasing the concentration of the supplement to 400 μg/ml did not increase growth (data not shown).
FIG. 4.
Effect of thymidine supplementation on intracellular replication (A) and cytopathogenicity (B) of GK11 in U937 macrophage-like cells in the presence or absence of thymidine. Experiments were performed in triplicate and repeated at least three times. The data are from a representative experiment. Error bars represent standard deviation of the mean; some of the error bars are too small to be seen.
The enhanced growth of GK11 within macrophages in the presence of thymidine correlated with increased cytopathogenicity to these cells (Fig. 4B). Although GK11 was not as cytopathogenic to U937 cells as AA100, over 60% of the macrophages infected with GK11 in the presence of thymidine were killed by 96 h postinfection (Fig. 4B). In stark contrast, macrophages infected with GK11 in the absence of thymidine remained completely viable throughout these experiments (Fig. 4B). Thus, the Rep helicase defect of GK11 resulted in a defect in intracellular replication and cytopathogenicity that was likely contributed to by the lack (or low concentration) of thymine or thymidine in the L. pneumophila phagosome.
Colocalization of the GK11 phagosome with late endosomal/lysosomal markers.
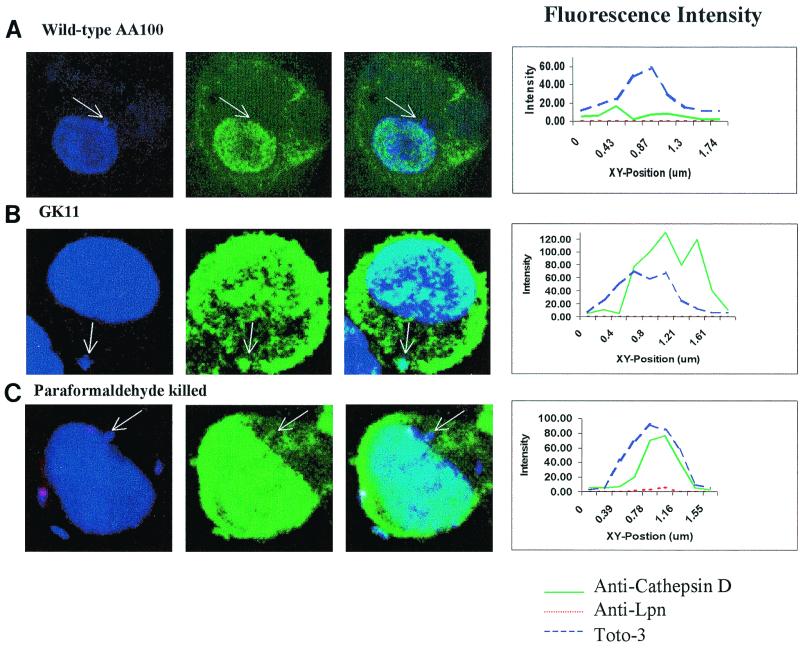
Since GK11 was incapable of intracellular growth, we determined whether it exhibited aberrant intracellular trafficking of its phagosome as well. Using confocal laser scanning microscopy, we examined colocalization of the GK11 phagosome with the late endosomal/lysosomal markers LAMP-1, LAMP-2, and cathepsin D. Visual assessment of colocalization was corroborated by measurement of fluorescence intensity across the phagosome. Colocalization was determined by the presence of two fluorescence peaks in the same area (one green for late endosomal/lysosomal markers and one blue for the bacteria) (Fig. 5 and 6) as we described previously (33).
FIG. 5.
Colocalization of phagosomes with cathepsin D at 4 h after infection with wild-type L. pneumophila (AA100), GK11, and paraformaldehyde-killed AA100. Nucleic acids were stained with Toto-3 (blue pseudocolor), which labels both intracellular and extracellular bacteria and the cell nucleus (first column); cathepsin D was visualized using secondary antibodies conjugated to Oregon green (green pseudocolor; second column); extracellular bacteria were visualized using secondary antibody conjugated to Alexa red (red pseudocolor). Colocalization is shown in the third column. Intensity of fluorescence across the indicated phagosomes (arrows) is shown at the right.
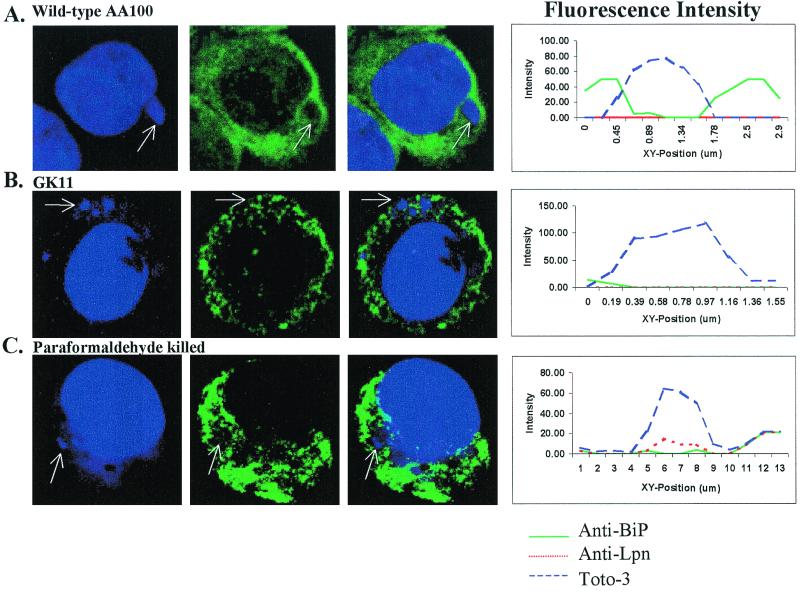
FIG. 6.
Colocalization of BiP with phagosomes of wild-type L. pneumophila (AA100), GK11, and paraformaldehyde-killed AA100 at 6 h postinfection. Nucleic acids were stained with Toto-3 (blue pseudocolor), which labels both intracellular and extracellular bacteria and the cell nucleus (first column); BiP was visualized using secondary antibodies conjugated to Oregon green (green pseudocolor; second column). Extracellular bacteria were visualized using secondary antibody conjugated to Alexa red (red pseudocolor). Colocalization is shown in the third column. Intensity of fluorescence across the indicated phagosomes (arrows) is shown at the right.
At 2 h postinfection, 60% of the GK11 phagosomes and 80% of the paraformaldehyde-killed AA100 phagosomes, but only 18% of the AA100 phagosomes, colocalized with LAMP-1 (Table 1). Following 4 h of infection, 80% of both the GK11 and paraformaldehyde-killed AA100 phagosomes colocalized with LAMP-1 (Table 1). In contrast, only 5% of the AA100 phagosomes colocalized with LAMP-1 at 4 h postinfection (Table 1). Patterns of colocalization of the GK11 and paraformaldehyde-killed AA100 phagosomes with LAMP-1 and cathepsin D were similar at 2 and 4 h postinfection (75 to 85%) (Table 1 and Fig. 5). In contrast, <15% of the AA100 phagosomes colocalized with these markers at 2 and 4 h post infection (Table 1 and Fig. 5). Thus, the defect in GK11 resulted in an aberrant trafficking of its phagosome along the endosomal/lysosomal maturation pathway.
TABLE 1.
Intracellular trafficking of AA100, GK11, and paraformaldehyde-killed AA100 phagosomes in U937 macrophage-like cells
| Marker | Strain | % Colocalization aftera:
|
||
|---|---|---|---|---|
| 2 h | 4 h | 6 h | ||
| LAMP-1 | AA100 | 17 ± 2.6 | 4.3 ± 4 | ND |
| GK11 | 61.3 ± 7.1 | 83.9 ± 7.9 | ND | |
| Pf-AA100b | 80 ± 5 | 83.9 ± 7.6 | ND | |
| LAMP-2 | AA100 | 5.5 ± 2.3 | 10.7 ± 4 | ND |
| GK11 | 87.3 ± 6.4 | 81.7 ± 7.6 | ND | |
| Pf-AA100 | 88.3 ± 7 | 90.6 ± 9.3 | ND | |
| Cathepsin D | AA100 | 8 ± 1 | 13.5 ± 4.3 | ND |
| GK11 | 72 ± 10.6 | 69.4 ± 9.5 | ND | |
| Pf-AA100 | 84 ± 7.9 | 82.2 ± 6.6 | ND | |
| BiP | AA100 | NDc | 85 ± 5.1 | 88.7 ± 5.5 |
| GK11 | ND | 48.3 ± 4.2 | 12.3 ± 3.5 | |
| Pf-AA100 | ND | 4.2 ± 2.6 | 3.7 ± 1.5 | |
Mean percentage of colocalization of bacterial phagosomes with LAMP-1, LAMP-2, cathepsin D, or BiP at the indicated time points from a representative experiment. The standard error of each mean is shown.
Pf-AA100, paraformaldehyde-killed AA100.
ND, not determined.
Colocalization of the GK11 phagosome with the RER.
Other mutants of L. pneumophila that exhibit aberrant intracellular trafficking are also defective in recruitment of the RER (25, 31, 61, 63). We determined whether the inability of GK11 to inhibit colocalization with late endosomal/lysosomal markers correlated with a defect in recruitment of the RER. Using anti-BiP antibody (luminal ER protein) and laser scanning confocal microscopy, recruitment of the RER was determined by an accumulation of BiP around the L. pneumophila phagosome (Fig. 6). Intensity of fluorescence around the bacterial phagosome was measured and corresponded with the visual assessment of RER recruitment (Fig. 6), as we described previously (33). Since the RER surrounded the phagosome as a ring, green fluorescence (used to visualize BiP) peaked on either ends of a line drawn across the phagosome, while blue fluorescence (used to visualize bacteria) peaked only in the center of the phagosome (Fig. 6). Lack of colocalization was determined by the absence of green fluorescence peaks around the bacterial phagosome. Recruitment of the RER was scored in AA100, GK11, and paraformaldehyde-killed AA100-infected U937 macrophages. Following 4 h of infection, BiP colocalized with 48% of the GK11 phagosomes, while 85% of the AA100 phagosomes colocalized with this marker (Table 1). Colocalization of BiP with the GK11 phagosomes decreased to 12% after 6 h of infection, while colocalization with the AA100 phagosomes remained relatively constant (Table 1). More than 95% of the paraformaldehyde-killed AA100 phagosomes did not colocalize with BiP (Table 1). Thus, GK11 was defective in recruitment of the RER or at least was not able to sustain the RER-phagosome interaction.
DISCUSSION
Several intracellular bacterial pathogens have evolved mechanisms to modulate various host cell processes in order to survive and proliferate (9, 34, 37, 38). Among the most fascinating aspects of the intracellular life cycle of L. pneumophila are its mechanisms of phagosomal modulation and escape from its host cells via heterogeneous and specific mechanisms such as apoptosis and/or necrosis (11, 23, 27, 28). Such functions are likely to require specific bacterial products. For example, mutations in several of the dot/icm loci have been shown to alter the intracellular trafficking of L. pneumophila (42, 57, 66). In addition to the identification of these genes, understanding the nature of the intracellular microenvironment within which bacterial pathogens reside is essential for the understanding of the nature of host-pathogen interactions (9, 36). Such an understanding would also allow for the identification of new targets for therapeutic compounds. For example, the use of auxotrophic mutants of L. pneumophila has revealed that the phagosomal environment of L. pneumophila is devoid of thymine or thymidine and is not accessible to diaminopimelic acid (35, 50). Tryptophan, on the other hand, is readily available to auxotrophs of this amino acid within macrophages (50).
To identify genes that are required for survival within its host cells, Gao et al. have isolated 121 mutants of L. pneumophila (29, 30). A subset of these mutants (mil mutants) is specifically defective for intracellular growth within macrophages but not protozoa (29). Here we show that the mil mutant GK11 exhibits a severe defect in intracellular replication within U937 macrophage-like cells and WI-26 alveolar epithelial cells. Interestingly, the numbers of intracellular bacteria remain constant throughout the time course of the infection within both cell types. The defect in intracellular replication of GK11 cannot be attributed to a defect in attachment or entry since equal numbers of GK11 and AA100 cells are present within macrophages and epithelial cells at 0 and 4 h postinfection.
Sequence analysis of the gene affected by the kanamycin insertion has revealed it to be most likely a rep helicase gene of L. pneumophila. Furthermore, the insertion site is localized between two conserved motifs (motifs V and VI) found in most helicase genes (44, 67). In general, helicases are enzymes that catalyze the unwinding of DNA during replication, recombination, conjugation, and DNA repair (47, 48). The Rep helicase is a 3′-5′ helicase that is involved in unwinding of the replication fork in E. coli and in recombination and may play a role in DNA repair (18, 47, 48). In addition, the Rep helicase of E. coli has been shown to oligodimerize with other helicases such as DnaB and UvrD (48). Interestingly, E. coli Rep helicase mutants are not lethal, but rep/uvrD double mutants are lethal (47). The Rep helicase mutation in GK11 has no effect on growth when the bacteria are grown in BYE rich medium. However, several E. coli helicase mutants including rep mutants are hypersensitive to TLD (10). This is thought to occur due to double-strand breaks and to an increase in deleterious mutations due to misincorporation of dUMP in place of dTMP in newly synthesized DNA that cannot be repaired (10). In this regard, we determined if GK11 exhibits increased sensitivity to thymine or thymidine starvation. The data indicate that mutant GK11 does not grow or grows extremely slowly in CAA semidefined medium in the absence of thymine or thymidine. Furthermore, supplementation with thymine or thymidine restores the growth of GK11, although to a rate still lower than that of the wild-type strain. Thus, thymine or thymidine alone is not sufficient to completely complement the rep mutation in GK11. Additional sequencing of 600 bp downstream of the L. pneumophila rep gene does not reveal any ORFs, suggesting that it is monocistronic or a terminal gene in an operon, in which case the kanamycin insertion is not expected to result in polar effects on other genes.
Thymidine auxotrophs of L. pneumophila are attenuated for growth within macrophages, but growth can be restored by adding thymidine to the culture media (50). This indicates that the L. pneumophila phagosome is devoid of or contains low concentrations of thymine or thymidine but that these compounds can freely access this compartment. This prompted us to determine the effects of thymidine supplementation on the intracellular growth and cytopathogenicity of mutant GK11. Addition of thymidine to the growth medium for GK11 restores (at least partially) intracellular growth within and cytopathogenicity to U937 macrophage-like cells. Thus, the Rep helicase of L. pneumophila serves an essential function in intracellular replication that becomes more pronounced in the absence of thymidine. To our knowledge, this is the first example of the requirement of a helicase function for the intracellular replication of an intracellular bacterial pathogen. Since thymine starvation results in an increased mutation rate (10), it is possible that the putative Rep helicase of L. pneumophila plays an essential role in the repair of these mutations.
Berger and Isberg have used a thymidine auxotroph of L. pneumophila to isolate attenuated strains in a TLD-enrichment assay (12). Their studies have indicated that mutations arise spontaneously at a high rate in the thymidine auxotroph of L. pneumophila grown in macrophages in the absence of thymidine (12). However, thymine or thymidine starvation is not the only source of DNA damage for intracellular L. pneumophila. Previous studies have revealed that the intracellular milieu of the phagosome inhabited by L. pneumophila within mammalian cells induces the expression of several stress-induced bacterial proteins such as GroEL, GroES, and GspA (2, 4, 9, 20, 36). Furthermore, L. pneumophila is constitutively exposed to stress stimuli within its intracellular environment, since the global stress protein GspA is constitutively expressed throughout intracellular growth (6). Since thymine starvation results in an increased mutation rate (10, 12), we postulate that exposure of intracellular L. pneumophila to stress stimuli within mammalian cells results in enhanced DNA damage that cannot be repaired in the Rep helicase mutant, particularly in combination with thymine deprivation within the phagosome. This hypothesis is supported by our finding of the defect in intracellular trafficking of the Rep helicase mutant. While thymine auxotrophs of L. pneumophila are transported similarly to the parental strain (12), the Rep helicase mutant moves through the existing endosomal/lysosomal degradation pathway. The GK11 phagosome colocalizes with the three late endosomal/lysosomal markers LAMP-1, LAMP-2, and cathepsin D and is not surrounded by the RER. It is unlikely that the Rep helicase plays a direct role in intracellular trafficking. It is likely that the exposure of L. pneumophila to stress stimuli combined with the starvation for thymine or thymidine within mammalian cells results in high rates of DNA damage in some of the genes involved in modulating the biogenesis of the phagosome (e.g., dot/icm genes), and this DNA damage cannot be repaired in the rep helicase mutant. To our knowledge, this is the first example of the requirement of a Rep helicase function for the intracellular replication of an intracellular bacterial pathogen. This may not be surprising, since these pathogens inhabit idiosyncratic niches, the nature of which is likely unique for each pathogen (9, 36).
We have recently shown that like the rep gene, the stress-induced htrA homologue of L. pneumophila is a mil gene that is indispensable for growth within mammalian cells but is dispensable in protozoa (L. Pedersen et al., submitted for publication). This stress-induced chaperon/protease is required under extreme stress conditions in other gram-negative bacteria, which is a further testimony to the stressful phagosomal microenvironment inhabited by L. pneumophila within mammalian cells (4, 6, 9, 36). It is interesting that the rep gene that is involved in DNA damage, which occurs under certain stress conditions, is also a mil locus that is not required for intracellular replication within protozoa. Taken together, the data suggest that the L. pneumophila phagosome faces a harsher microenvironment in mammalian cells than in the protozoan host. However, we cannot exclude the possibility that the relative deficiency of thymidine does not occur in amoebae. Further elucidation of this microenvironment will yield interesting knowledge about the adaptation of L. pneumophila to the two evolutionarily distant hosts.
ACKNOWLEDGMENTS
Y.A.K. was supported by Public Health Service grants RO1AI43965 and R29AI38410. O.S.H. was supported by NIH training grant 5T32CA09509.
We thank Bruce Maley, Richard Watson, and Mary Gail Engle, University of Kentucky microscopy imaging suite, for technical assistance with confocal microscopy imaging.
REFERENCES
- 1.Abu Kwaik Y. Fatal attraction of mammalian cells to Legionella pneumophila. Mol Microbiol. 1998;30:689–696. doi: 10.1046/j.1365-2958.1998.01092.x. [DOI] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection. Infect Immun. 1998;66:203–212. doi: 10.1128/iai.66.1.203-212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y. The phagosome containing Legionella pneumophila within the protozoan Hartmanella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, Eisenstein B I, Engleberg N C. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect Immun. 1993;61:1320–1329. doi: 10.1128/iai.61.4.1320-1329.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu Kwaik Y, Engleberg N C. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress stimuli. Mol Microbiol. 1994;13:243–251. doi: 10.1111/j.1365-2958.1994.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 6.Abu Kwaik Y, Gao L-Y, Harb O S, Stone B J. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol Microbiol. 1997;24:629–642. doi: 10.1046/j.1365-2958.1997.3661739.x. [DOI] [PubMed] [Google Scholar]
- 7.Abu Kwaik Y, Gao L-Y, Stone B J, Harb O S. Invasion of mammalian and protozoan cells by Legionella pneumophila. Bull Inst Pasteur. 1998;96:237–247. [Google Scholar]
- 8.Abu Kwaik Y, Gao L-Y, Stone B J, Venkataraman C, Harb O S. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64:3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abu Kwaik Y, Harb O S. Phenotypic modulation by intracellular bacterial pathogens. Electrophoresis. 1999;20:2248–2258. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2248::AID-ELPS2248>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad S I, Kirk S H, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol. 1998;152:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 11.Alli O A T, Gao L-Y, Pedersen L, Zink S, Radulic M, Doric M, Abu Kwaik Y. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect Immun. 2000;68:6431–6440. doi: 10.1128/iai.68.11.6431-6440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger K H, Isberg R R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 13.Bozue J A, Johnson W. Interaction of Legionella pneumophila with Acanthamoeba catellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozzoni M, Radice L, Frosi A, Vezzoli S, Cuboni A, Vezzoli F. Prevalence of pneumonia due to Legionella pneumophila and Mycoplasma pneumoniae in a population admitted to a department of internal medicine. Respiration. 1995;62:331–335. doi: 10.1159/000196475. [DOI] [PubMed] [Google Scholar]
- 15.Brenner D J, Steigerwalt A G, McDade J E. Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann Intern Med. 1979;90:656–658. doi: 10.7326/0003-4819-90-4-656. [DOI] [PubMed] [Google Scholar]
- 16.Byrne B, Swanson M S. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemens D L, Horwitz M A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagolysosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denhardt D T, Iwaya M, Larison L L. The rep mutation. II. Its effect on Escherichia coli and on the replication of bacteriophage phi X174. Virology. 1972;49:486–496. doi: 10.1016/0042-6822(72)90500-4. [DOI] [PubMed] [Google Scholar]
- 19.Edelstein P H. Legionnaires' disease. Clin Infect Dis. 1993;16:741–747. doi: 10.1093/clind/16.6.741. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez R C, Logan S, Lee S H S, Hoffman P S. Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlated with virulence. Infect Immun. 1996;64:1968–1976. doi: 10.1128/iai.64.6.1968-1976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 22.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 23.Gao L Y, Kwaik Y A. The modulation of host cell apoptosis by intracellular bacterial pathogens. Trends Microbiol. 2000;8:306–313. doi: 10.1016/s0966-842x(00)01784-4. [DOI] [PubMed] [Google Scholar]
- 24.Gao L Y, Susa M, Ticac B, Abu Kwaik Y. Heterogeneity in intracellular replication and cytopathogenicity of Legionella pneumophila and Legionella micdadei in mammalian and protozoan cells. Microb Pathog. 1999;27:273–287. doi: 10.1006/mpat.1999.0308. [DOI] [PubMed] [Google Scholar]
- 25.Gao L-Y, Abu Kwaik Y. Activation of caspase-3 in Legionella pneumophila-induced apoptosis in macrophages. Infect Immun. 1999;67:4886–4894. doi: 10.1128/iai.67.9.4886-4894.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao L-Y, Abu Kwaik Y. Apoptosis in macrophages and alveolar epithelial cells during early stages of infection by Legionella pneumophila and its role in cytopathogenicity. Infect Immun. 1999;67:862–870. doi: 10.1128/iai.67.2.862-870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, L.-Y., and Y. Abu Kwaik. Hijacking the apoptotic pathways of the host cell by bacterial pathogens. Microbes Infect., in press. [DOI] [PubMed]
- 28.Gao L-Y, Abu Kwaik Y. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ Microbiol. 2000;2:79–90. doi: 10.1046/j.1462-2920.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- 29.Gao L-Y, Harb O S, Abu Kwaik Y. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect Immun. 1998;66:883–892. doi: 10.1128/iai.66.3.883-892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao L-Y, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao L-Y, Stone B J, Brieland J K, Abu Kwaik Y. Different fates of Legionella pneumophila pmi and mil mutants within human-derived macrophages and alveolar epithelial cells. Microb Pathog. 1998;25:291–306. doi: 10.1006/mpat.1998.0237. [DOI] [PubMed] [Google Scholar]
- 32.Gilchrist C A, Denhardt D T. Escherichia coli rep gene: sequence of the gene, the encoded helicase, and its homology with uvrD. Nucleic Acids Res. 1987;15:465–475. doi: 10.1093/nar/15.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harb O S, Abu Kwaik Y. Characterization of a macrophage-specific infectivity locus (milA) of Legionella pneumophila. Infect Immun. 1999;68:368–376. doi: 10.1128/iai.68.1.368-376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harb O S, Abu Kwaik Y. Interaction of Legionella pneumophila with protozoa provides lessons. ASM News. 2000;66:609–616. [Google Scholar]
- 35.Harb O S, Abu Kwaik Y. Identification of the aspartate-β-semiadehyde dehydrogenase gene of Legionella pneumophila and characterization of a null mutant. Infect Immun. 1998;66:1898–1903. doi: 10.1128/iai.66.5.1898-1903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harb O S, Abu Kwaik Y. Probing the microenvironment of intracellular bacterial pathogens. Microbes Infect. 1999;1:445–453. doi: 10.1016/s1286-4579(99)80048-3. [DOI] [PubMed] [Google Scholar]
- 37.Harb O S, Gao L-Y, Abu Kwaik Y. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ Microbiol. 2000;2:251–265. doi: 10.1046/j.1462-2920.2000.00112.x. [DOI] [PubMed] [Google Scholar]
- 38.Harb O S, Venkataraman C, Haack B J, Gao L-Y, Abu Kwaik Y. Heterogeneity in the attachment and uptake mechanisms of the Legionnaires' disease bacterium, Legionella pneumophila, by protozoan hosts. Appl Environ Microbiol. 1998;64:126–132. doi: 10.1128/aem.64.1.126-132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horwitz M A. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horwitz M A. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller D W, Hajjeh R, DeMaria A, Fields B S, Pruckler J M, Benson R S, Kludt P E, Lett S M, Mermel L A, Giorgio C, Breiman R F. Community outbreak of legionnaires' disease: an investigation confirming the potential for cooling towers to transmit Legionella species. Clin Infect Dis. 1995;22:257–261. doi: 10.1093/clinids/22.2.257. [DOI] [PubMed] [Google Scholar]
- 42.Kirby J E, Isberg R R. Legionnaires' disease: the pore macrophage and the legion of terror within. Trends Microbiol. 1998;6:256–258. doi: 10.1016/s0966-842x(98)01310-9. [DOI] [PubMed] [Google Scholar]
- 43.Kirby J E, Vogel J P, Andrews H L, Isberg R R. Evidence for pore-forming ability by Legionella pneumophila. Mol Microbiol. 1998;27:323–336. doi: 10.1046/j.1365-2958.1998.00680.x. [DOI] [PubMed] [Google Scholar]
- 44.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 45.Liles M R, Edelstein P H, Cianciotto N P. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol Microbiol. 1999;31:959–970. doi: 10.1046/j.1365-2958.1999.01239.x. [DOI] [PubMed] [Google Scholar]
- 46.Liles M R, Viswanathan V K, Cianciotto N P. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohman T M. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 48.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 49.McDade J E, Shepard C C, Fraser D W, Tsai T R, Redus M A, Dowdle W R. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 50.Mintz C S, Chen J X, Shuman H A. Isolation and characterization of auxotrophic mutants of Legionella pneumophila that fail to multiply in human monocytes. Infect Immun. 1988;56:1449–1455. doi: 10.1128/iai.56.6.1449-1455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mody C H, Paine R I, Shahrabadi M S, Simon R H, Pearlman E, Eisenstein B I, Toews G B. Legionella pneumophila replicates within rat alveolar epithelial cells. J Infect Dis. 1993;167:1138–1145. doi: 10.1093/infdis/167.5.1138. [DOI] [PubMed] [Google Scholar]
- 52.Muller A, Hacker J, Brand B C. Evidence for apoptosis of human macrophage-like HL-60 cells by Legionella pneumophila infection. Infect Immun. 1996;64:4900–4906. doi: 10.1128/iai.64.12.4900-4906.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pérez E, Munoz M D L, Ortega A. Entamoeba histolytica: involvement of pp125FAK in collagen-induced signal transduction. Exp Parasitol. 1996;82:164–170. doi: 10.1006/expr.1996.0021. [DOI] [PubMed] [Google Scholar]
- 54.Purcell M, Shuman H A. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect Immun. 1998;66:2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadosky A B, Wiater L A, Shuman H A. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Segal G, Shuman H A. How is the intracellular fate of the Legionella pneumophila phagosome determined. Trends Microbiol. 1998;6:253–255. doi: 10.1016/s0966-842x(98)01308-0. [DOI] [PubMed] [Google Scholar]
- 58.Segal G, Shuman H A. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stone B J, Abu Kwaik Y. Natural competency for DNA uptake by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181:1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stone B J, Brier A, Kwaik Y A. The Legionella pneumophila prp locus; required during infection of macrophages and amoebae. Microb Pathog. 1999;27:369–376. doi: 10.1006/mpat.1999.0311. [DOI] [PubMed] [Google Scholar]
- 61.Swanson M S, Isberg R R. Analysis of the intracellular fate of Legionella pneumophila mutants. Ann NY Acad Sci. 1996;797:8–18. doi: 10.1111/j.1749-6632.1996.tb52944.x. [DOI] [PubMed] [Google Scholar]
- 62.Swanson M S, Isberg R R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swanson M S, Isberg R R. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun. 1996;64:2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venkataraman C, Gao L-Y, Bondada S, Abu Kwaik Y. Identification of putative cytoskeletal protein homologues in the protozoan Hartmannella vermiformis as substrates for induced tyrosine phosphatase activity upon attachment to the Legionnaires' disease bacterium, Legionella pneumophila. J Exp Med. 1998;188:505–514. doi: 10.1084/jem.188.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venkataraman C, Haack B J, Bondada S, Abu Kwaik Y. Identification of a Gal/GalNAc lectin in the protozoan Hartmannella vermiformis as a potential receptor for attachment and invasion by the Legionnaires' disease bacterium, Legionella pneumophila. J Exp Med. 1997;186:537–547. doi: 10.1084/jem.186.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogel J P, Andrews H L, Wong S K, Isberg R R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 67.Zhang G, Deng E, Baugh L R, Hamilton C M, Maples V F, Kushner S R. Conserved motifs II to VI of DNA helicase II from Escherichia coli are all required for biological activity. J Bacteriol. 1997;179:7544–7550. doi: 10.1128/jb.179.23.7544-7550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]