Abstract
The mountainous areas of Ethiopia represent one of the most extreme environmental challenges in Africa faced by humans and other inhabitants. Selection for high-altitude adaptation is expected to have imprinted the genomes of livestock living in these areas. Here we assess the genomic signatures of positive selection for high altitude adaptation in three cattle populations from the Ethiopian mountainous areas (Semien, Choke, and Bale mountains) compared to three Ethiopian lowland cattle populations (Afar, Ogaden, and Boran), using whole-genome resequencing and three genome scan approaches for signature of selection (iHS, XP-CLR, and PBS). We identified several candidate selection signature regions and several high-altitude adaptation genes. These include genes such as ITPR2, MB, and ARNT previously reported in the human population inhabiting the Ethiopian highlands. Furthermore, we present evidence of strong selection and high divergence between Ethiopian high- and low-altitude cattle populations at three new candidate genes (CLCA2, SLC26A2, and CBFA2T3), putatively linked to high-altitude adaptation in cattle. Our findings provide possible examples of convergent selection between cattle and humans as well as unique African cattle signature to the challenges of living in the Ethiopian mountainous regions.
Keywords: high altitude, hypoxia, Ethiopian cattle, adaptation, candidate gene, convergent evolution
Introduction
Ethiopia is endowed with diverse agro-climatic regions and altitudes that range from the lowest Afar depression (−160 m above sea level, masl) to the highest Semien Mountain (4,600 masl). The Ethiopian highlands are commonly found in the central part of the country, on both sides of the Rift Valley, extending from the Semien Mountain in the North to the Bale Mountain in the Southeast. Cold temperatures and humid weather are characteristics of the high-altitude plateaus in Ethiopia. Mixed livestock farming and crop cultivation are major agricultural activities for livelihoods. These environments are characterized by some unique agricultural production activities and food sources, providing cash income to the local communities (Asresie and Zemedu, 2015). Likewise, they have contributed to the diversity of Ethiopian livestock.
Human populations started to occupy the Ethiopian high plateau by migrating from the Rift Valley to the Bale Mountain in the Middle Stone Age around 50–30,000 years ago (Ossendorf et al., 2019). Overtime, people living in such environments have become adapted to high-altitude stressors, including hypobaric hypoxia, ultraviolet light, cold temperature, and oxidative stress (Beall et al., 2002; San et al., 2013; Debevec et al., 2017; Yang et al., 2017; Storz, 2021). The possible genetic basis of human adaptation to high altitudes in Ethiopia has been previously reported (Beall et al., 2002; Alkorta-Aranburu et al., 2012; Scheinfeldt et al., 2012; Huerta-Sánchez et al., 2013; Edea et al., 2019; Wiener et al., 2021). Likewise, evolutionary adaptations of livestock exposed to high altitudes are expected to be associated with major changes in the anatomy and physiological functions following a long period of acclimatization. For example, the yak Bos grunniens possesses a larger heart and lungs as compared to cattle (Wang et al., 2016), leading to a high amount of inhaled air to supply sufficient oxygen to the respiratory cell system. Uteroplacental oxygen flow at the fetal stage (Simonson, 2015), higher hemoglobin concentration in blood (Alkorta-Aranburu et al., 2012; Scheinfeldt et al., 2012), pulmonary vasoconstriction (Wang et al., 2016), ability to avoid altitude sickness (Dolt et al., 2007), calcium metabolism (Wang et al., 2015), and better foraging ability and energy metabolism (Qiu et al., 2012; Edea et al., 2014) may all contribute to the high-altitude adaptation in cattle and other livestock species.
High-altitude adaptation in animals relies on their genetic background attained through natural selection. Hypoxia induced factors such as HIF-1a and its paralogs of HIF-2a and HIF-3a are oxygen regulating factors in a hypobaric hypoxia environment and are thus considered candidate genes for high-altitude adaptation. The HIF-1a gene is over-expressed in cattle, yak, humans, and the Tibetan gray wolf living at high altitudes (Newman et al., 2011; Bigham and Lee, 2014; Zhang et al., 2014; Wang et al., 2016; Verma et al., 2018; Werhahn et al., 2018). The HIF-1a pathways include the endothelial PAS domain 1 (EPAS1), vascular endothelial growth factor-A (VEGF-A), endothelial converting enzyme-1 (ECE1), glucose transport members 1 (GLUT-1), hexokinase 2 (HK2), and nitric oxide synthesis (NOS2) genes. These are all expressed in cattle adapted to high altitudes (Verma et al., 2018), and they play an important role in maintaining oxygen homeostasis and glucose metabolism in mammals (Hu et al., 2006; Majmundar et al., 2010). Hypoxia-related genes, including EPAS1, RYR2, and ANGPT1, were identified in high-altitude Tibetan gray wolves, and they were associated with the HIF signaling pathway, ATP binding, and response to oxygen-containing compounds (Zhang et al., 2014). Using the Illumina bovine low-density 50K SNP array, a study on the Ethiopian cattle population living at an altitude of 2,400 masl identified energy metabolism (ATP2A3, CA2, MYO18B, SIK3, INPP4A, and IREB2) and response to hypoxia (BDNF, TFRC, and PML) genes as candidate genes to the adaptation of cattle to the high-altitude environments (Edea et al., 2014).
Physiological and genomic landscape studies have revealed convergent evolution in several species to independently adapt to high altitudes in different geographic locations across the world (Scheinfeldt et al., 2012; Huerta-Sánchez et al., 2013; Azad et al., 2017; Witt and Huerta-Sánchez, 2019; Friedrich and Wiener, 2020). For example, human populations in the Tibetan, Andean, and Ethiopian highlands shared common candidates selected regions and genes linked to high-altitude adaptation, such as PPARA and EDNRA (Scheinfeldt et al., 2012; Simonson, 2015; Witt and Huerta-Sánchez, 2019). However, ARNT2 and THRB were uniquely identified in the Ethiopian population (Scheinfeldt et al., 2012).
Only a few studies have reported the environmental adaptations of Ethiopian cattle at the full autosomal genome level (Kim et al., 2017; Kim et al., 2020). Though the bovine low-density SNP array (Edea et al., 2014) was the first to investigate Ethiopian cattle adaptation to different environments including hypoxia. However, it did not include high altitude adaptation of cattle population living at an altitude of >3,000 masl. Therefore, this study aimed to identify signatures of positive selection for high altitude adaptation in Ethiopian cattle using whole-genome resequencing. For this purpose, we selected three cattle populations from the highest mountainous areas (>3,000 masl) of the country (Bale, Choke, and Semien Mountain areas) (Table 1), and three Ethiopian cattle populations living at low altitudes.
TABLE 1.
Sampling location and cattle population description in the high and low altitudes.
| Category | Breed | Region (district) | Location | Altitude (masl) | GPS | Climate | |
|---|---|---|---|---|---|---|---|
| Latitude (degree) | Longitude (degree) | ||||||
| High altitude | Bale | Oromia (Bale) | Bale Mountain | 3,586 | 6.77 | 39.75 | Cold humid, highland |
| Choke | Amhara (East Gojam) | Choke Mountain | 3,410 | 10.60 | 37.84 | Cold humid, highland | |
| Semien | Amhara (North Goder) | Semien Mountain | 3,732 | 13.23 | 38.13 | Cold humid, highland | |
| Low altitude | Afar | Afar | Melka Were/Asayta | 729 | 9.34 | 40.17 | Hot and dry lowland |
| Boran | Oromia (Borena) | Dubulk | 1,368 | 4.55 | 38.10 | Hot and dry lowland | |
| Ogaden | Ogaden | Jigjiga | 1,200 | 09.58 | 41.85 | Hot and dry lowland | |
Materials and methods
Cattle populations and whole-genome resequencing
The study involved a comparative analysis of indigenous cattle distributed at high altitudes (>3,000 masl) and low altitudes (<1,500 masl) in Ethiopia (Table 1). The high-altitude populations included Bale (n = 10) sampled in the Bale district (Bale Mountain, ∼3,586 masl), Semien (n = 10) in the Gondar district (Semien Mountain, ∼3,732 masl), and Choke (n = 10) in the Gojam district (Choke Mountain, ∼3,410 masl). The low-land populations included Afar (n = 11) sampled in the Afar district (∼729 masl), Ethiopian Boran (n = 10) from the Borana district (∼1,368 masl), and Ogaden (n = 9) from the Ogaden district (∼1,200 masl) (Figure 1).
FIGURE 1.
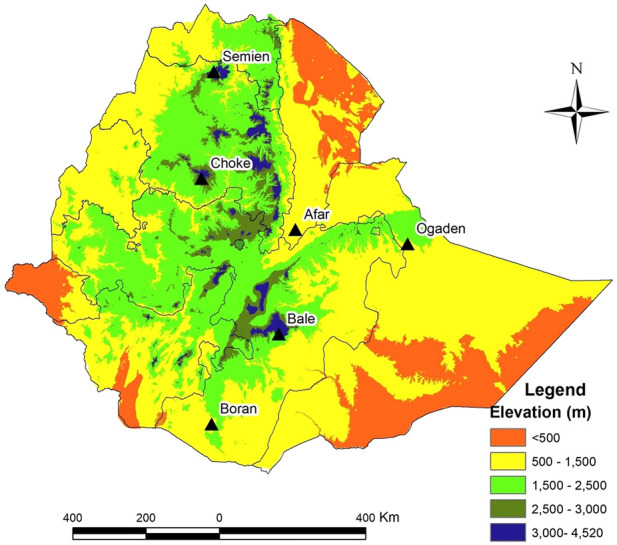
The elevation map of Ethiopia and sampling locations for high and low-altitude cattle populations.
Whole blood samples were collected aseptically from the jugular veins of unrelated individuals into 10 ml vacuum tubes containing EDTA. Genomic DNA was extracted using the QIAGEN DNeasy Blood and Tissue Kit (https://www.qiagen.com/us/) following the manufacturer’s protocol. The DNA integrity was checked by a 1% agarose gel electrophoresis and observed under a UV light-based gel viewer. The concentration and quality of DNA for each sample were checked by using a DeNovix DS-11 FX Series Spectrophotometer/Fluorometer (DeNovix Inc., Wilmington, DE, United States). DNA samples (>50 μg/μl) were then shipped to Novogene, China (https://en.novogene.com/services/research-services/genome-sequencing/whole-genome-sequencing/animal-plant-whole-genome-sequencing-wgs/), where whole-genome sequencing was performed on an Illumina NovaSeq 6000 Platform (Illumina, San Diego, CA, United States) to generate 150 bp of paired-end reads. We included Gir (GenBank accession no. PRJNA343262), Angus (PRJNA318087), Muturu (PRJNA386202), and Butana (PRJNA574857) cattle for comparative analyses, following the same sequence quality control and variant calling procedures.
Read mapping and variant calling
The quality control of raw sequencing reads was performed using the FastQC v0.11.9 program (https://github.com/s-andrews/FastQC/releases/tag/v0.11.9). Qualified raw reads were processed for initial trimming and filtering of the low-quality reads by removing adapters, short reads (sequence length <35 bp), and reads with low sequence base quality (quality score <20) using the Trimmomatic v0.38 tool (Bolger et al., 2014). Clean reads were mapped to the latest taurine cattle reference genome of ARS-UCD1.2 (Shamimuzzaman et al., 2020) using the BWA-MEM algorithm of Burrows-Wheeler Aligner (bwa v0.7.17) (Li and Durbin, 2010).
Mapped reads were sorted and indexed using the samtools v1.8 (Li et al., 2009) to produce a BAM file. Alignment sorting by coordinate and marking of potential PCR and optical duplicates were carried out using the MarkDuplicates tool in Picard v2.18.2 package (http://picard.sourceforge.net). Base quality score recalibration (BQSR) and haplotype caller analysis were performed using the GATK v3.8-1-0-gf15c1c3ef according to its best practice workflows (McKenna et al., 2010). The known variants of ARS1.2PlusY_BQSR_v3.vcf.gz provided by the 1,000 Bull Genomes project were used for masking known sites for all cattle samples. The GATK PrintReads was run to adjust the base quality scores in the data based on information from the table and to produce a recalibrated bam file.
Then, the genomic variant call format (gVCF) file for each sample was created using the GATK HaplotypeCaller command from the recalibrated bam file, and all samples were combined to obtain a joint genotype file using the GATK CombineGVCFs. Finally, the variants were processed for variant recalibration with a 99.9 truth sensitivity filter level using the GATK to reduce false discovery rates that minimize the noise created by low standard variants. After all quality checking, approximately 36.6 million biallelic autosomal SNPs were identified and used for downstream analyses.
Population genetic structure
Principal component analysis (PCA) and admixture modeling were performed, based on the SNP genotypes, to determine the population genetic structure of indigenous Ethiopian cattle living in high and low altitudes. We used Angus, Gir, Muturu, and Butana cattle as reference breeds (European taurine, Asian zebu, African taurine, and non-Ethiopian zebu cattle). The dataset in the vcf file was first converted to a plink format (map, ped, and fam) after pruning the SNPs in linkage disequilibrium (LD) (r2 ≥ 0.5), minor allele frequency (MAF) (< 0.05), and missing genotype (call rate > 10%) based on a step-wise procedure using 50 SNPs windows and 10 SNPs steps. After the stringent variant quality check, 5.1 million autosomal SNPs with an average of 98.1% genotyping rate were used for admixture modeling with the Admixture v1.3.0 software (Alexander et al., 2009) to estimate the ancestry proportion of individual samples. The ancestral proportions in the hierarchical clustering of individual samples were optimized at the K values ranging from 2 to 10 and the admixture plot was visualized using the R package. For PCA, we removed SNPs with MAF < 0.01, SNPs with missing genotypes > 10%, and SNP calling rate < 90%. After this filtering, 25.5 million SNPs were used for PCA. The eigenvectors of each sample were calculated using PLINK 1.9 (Purcell et al., 2007) and the result was plotted with the ggplot2 in the R package.
Integrated haplotype homozygosity analysis
The phasing and imputation of missed genotypes were estimated per chromosome for each population using the Beagle v5.1 software (Browning and Browning, 2007). The length of homozygous haplotypes along a chromosome was used to estimate the LD decay. The extended haplotype homozygosity (EHH) from each SNP, which is the probability that two randomly chosen homologous chromosomes carrying the core haplotype of interest are identical by descent (Sabeti et al., 2002), was then calculated. The integrated haplotype score (iHS) compares the integrated EHH between the ancestral allele relative to the derived allele in a population (Voight et al., 2006). It detects selective sweeps when alleles are near fixation. The iHS analysis was done using the REHH v2.0 R package (Gautier et al., 2017) for alleles with MAF within a population > 0.05. A genomic window of 100 kb and a step size of 50 kb were used to identify candidate regions of selection signatures.
Cross-population composite likelihood ratio (XP-CLR)
XP-CLR test was done between cattle living at high and low altitudes using the haplotype phased data of each chromosomal window of 100 kb and a step size of 50 kb. The test is based on local allele frequency changes in a genomic region between the two groups. The method is most sensitive to recent selection and detects departures from neutrality that could be compatible with hard or soft selection sweeps (Chen et al., 2010).
Population branch statistic (PBS)
PBS analysis was employed to detect genomic regions under selection with highly divergent haplotypes (Yi et al., 2010). We run population differentiation (F ST) analysis using the vcftools v0.1.15 (Danecek et al., 2011) between the Semien population for the highest altitude representative compared to Afar, Boran, and Ogaden cattle for the low altitude one, and the Sudanese Butana cattle as an outgroup. The top 0.5% of the candidate regions detected by the iHS and XP-CLR tests were used for PBS analysis. We estimated divergence time using a log-transformation of one minus the F ST value for each comparison and calculated the PBS value using the method described in Huerta-Sánchez et al. (2013).
Functional annotation and haplotype structure of candidate genes
The candidate genomic regions were annotated using the taurine cattle reference genome ARS-UCD1.2 (Shamimuzzaman et al., 2020) in the Ensembl database (http://www.ensembl.org/index.html). The Database for Annotation, Visualization, and Integrated Discovery (DAVID, v6.8, https://david.ncifcrf.gov/home.jsp) was used to understand the biological functions and molecular pathways of the candidate genes (Huang et al., 2009), according to the minimum similarity thresholds for enrichment scores at 1.0 and p values ≤ 0.05. Further functional annotations were done from the literature published for humans and other vertebrates. Additional structural and functional analyses of the genomic regions of the candidate genes were evaluated using haplotype structure, LD, F ST, nucleotide diversity, and STRING protein-protein interaction network database. The haplotype structure was estimated based on pairwise LD heatmap of the SNPs using the LDBlockShow (Dong et al., 2020). The F ST and nucleotide diversity of the candidate gene regions were estimated using the vcftools v 0.1.15 (Danecek et al., 2011) in a 10 kb window and 5 kb step size to determine the strength and pattern of selection signatures between the high- and low-altitude cattle. Protein-protein interaction analyses were done using the STRING online platform for the cattle genome reference database (https://string-db.org/).
Results
Population genetic structure
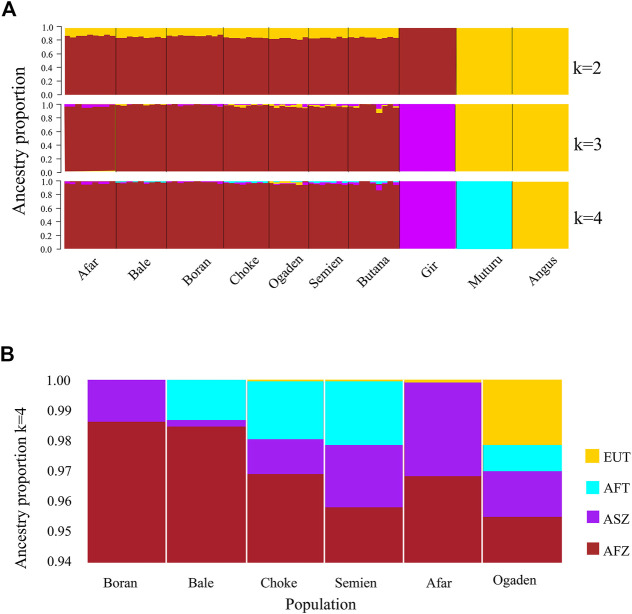
The PCA and admixture plots describe the population genetic structure of the indigenous Ethiopian cattle living in the high and low altitudes compared with European taurine cattle (Angus), African taurine (Muturu), Asian zebu cattle (Gir), and African zebu (Butana) (Figure 2A). The first and second principal components (PC) represent 57.8% of the total variation. The first PC (PC1, 41.4%) separates the zebu (Gir, Butana, and Ethiopian cattle) from the taurine (Angus and Muturu), while the second PC (PC2, 16.4%) differentiate the African zebu (Ethiopian cattle Butana) and Muturu from all non-African cattle (Gir and Angus) (Figure 2A). Next, a second PCA was conducted, excluding the reference cattle. The PC1 (11.8%) and PC2 (10.3%) differentiate the Ethiopian high-altitude (HA) from low-altitude (LA) cattle (Figure 2B). The population genetic admixture plot supports the PCA result, which separated cattle populations in the whole dataset into four ancestry clusters (Figure 3A). At K = 4, the genetic ancestry of the Ethiopian cattle was inferred to be 97.0% of African zebu, 1.6% Asian zebu (represented here by the Gir), 1.0% African taurine (Muturu), and 0.4% European taurine (Angus) (Figure 3B). A small shared European taurine component is observed in Bale, Choke, Semien, and Boran cattle. It is, however, higher (2.2%) in Ogaden cattle. The Afar and Boran populations have very little African taurine ancestry proportion. Butana cattle share a similar genetic background to other Ethiopian cattle. To explore potential genome-wide selection signatures for high-altitude adaptation, we analyzed the HA and LA cattle populations separately, following the Ethiopian cattle PCA results (Figure 2B).
FIGURE 2.
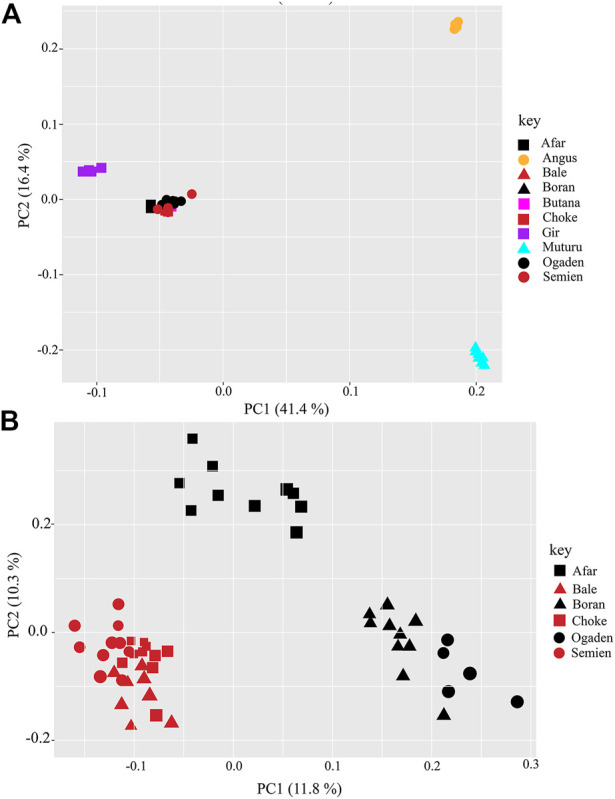
A plot of the first and second principal components (PC1 and PC2) of (A) the whole population dataset and (B) Ethiopian high altitude (brown color: Bale, Choke, and Semien) and low altitude (black color: Afar, Boran, and Ogaden) cattle populations.
FIGURE 3.
The plot of population genetic admixture analysis. (A) Cattle population in the whole dataset and (B) Ethiopian cattle ancestry proportion.
Selection signatures within Ethiopian high- and low-altitude cattle populations
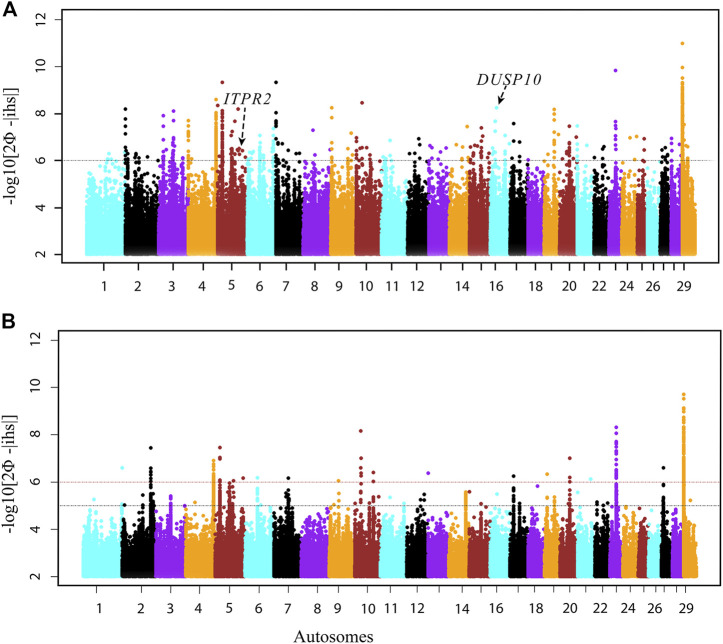
We performed a genomic scan combining the three HA, as well as combining the three LA populations, using the within-population iHS test to identify recent and/or ongoing footprints of natural selection (Vatsiou et al., 2016). Using the REHH v2.0 R package, we calculated genome-wide iHS for each focal SNP from the phased data (Gautier et al., 2017). Subsequently, we summarized the selection statistics across a sliding 100 kb genomic window with a 50 kb step size. From the empirical distribution of iHS statistics, we applied a p-value threshold < 1.0E-6, equivalent to -log10 iHS ≥ 6, to select the candidate regions under selection (Figure 4A, Supplementary Table S1).
FIGURE 4.
Manhattan plots of genome-wide scans based on the iHS test. (A) HA, high-altitude and (B) LA, low-altitude Ethiopian cattle populations.
There are 144 candidate selected regions across 29 autosomes within the Ethiopian zebu populations living in high altitudes (Supplementary Table S1). These regions vary in size from 150 to 750 kb. They overlap with 264 protein-coding genes based on the Ensembl taurine cattle assembly (ARS-UCD1.2) (Supplementary Table S1). Of these, 28 protein-coding genes were identified within the top 10 iHS regions. Most of these genes remain uncharacterized with the exception of ITPR2 on BTA5 (5:83.45–83.60 Mb, iHS −log10 p = 8.19), DUSP10 on BTA16 (16:25.05–25.20 Mb, iHS −log10 p = 8.24), and GTPase IMAP family members 4–7 genes (GIMAP4–7) on BTA4 (4:112.95–113.35 Mb, iHS −log10 p = 8.60). These annotated genes are possibly involved in high-altitude adaptation, especially the former two genes with functions linked to the response to hypoxia (ITPR2) and oxygen-containing compounds (DUSP10), respectively. GIMAP4, GIMAP5, and GIMAP7 functions are related to the immune response and hematopoiesis (Chen et al., 2011; Schwefel et al., 2013). They play a significant role in modulating immune functions by controlling cell death and the activation of T cells (Ho and Tsai, 2017).
To contrast the selection signatures of Ethiopian cattle living in high altitudes with the ones living at low altitudes, we performed an additional genomic scan based on the iHS in the three LA cattle populations at the same threshold (p-value < 1.0E-6). We identified only 20 candidate regions under selection (Figure 4B, Supplementary Table S2). These regions vary in size from 150 kb to 650 kb. They overlap with 50 protein-coding genes. Twenty-three (∼45%) were common with those identified in the HA populations. The common genes include 14 genes, mostly uncharacterized, present in the top 10 iHS regions, except for the three GIMAP family members, GIMAP 4, 5, and 7 genes. Among the remaining nine common genes, VEGFC and EP300 are possibly linked to the adaptation to high altitudes due to their functions in the vascular system (Herbert and Stainier, 2012; Huerta-Sánchez et al., 2013; Bigham and Lee, 2014; Azad et al., 2017; Zheng et al., 2017). However, due to the fewer candidate regions identified in the LA populations, we decided to increase the iHS threshold to p value < 1.0E-7, equivalent to -log10 iHS ≥ 5, which added 113 protein-coding genes, of which 63 were common in both HA and LA populations (Figure 4B, Supplementary Table S2).
Comparative genomic signatures between Ethiopian high- and low-altitude cattle populations
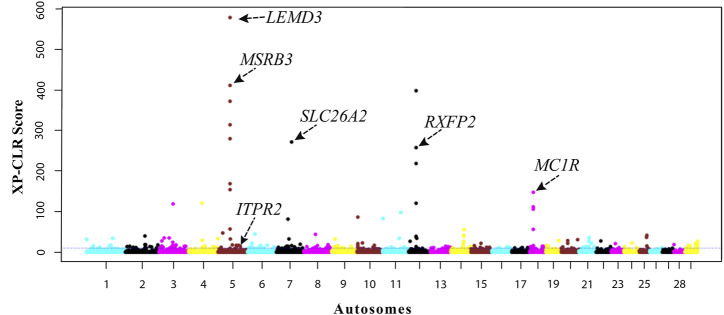
We further investigate the genomic footprints of natural selection in indigenous Ethiopian cattle by contrasting the allele frequency profiles between the HA and LA populations using the XP-CLR test. The top 0.5% XP-CLR scores (XP-CLR > 10) include 216 candidate windows of 100 kb size regions, from which 251 protein-coding genes were annotated (Supplementary Table S3). Unlike many uncharacterized genes within the iHS regions, the top 10 signals identified by the XP-CLR test include seven annotated genes (MSRB3, LEMD3, and WIF1 on BTA5, SLC26A2, HMGXB3, and CSF1R on BTA7, and RXFP2 on BTA12) (Figure 5). These are not found within the top high altitude iHS signals. Some have functions that may be related to high-altitude adaptation. For instance, MSRB3 is involved in cold tolerance in Arabidopsis and high-altitude adaptation in Tibetan dogs and sheep (Kwon et al., 2007; Vaysse et al., 2011; Wei et al., 2016; Witt and Huerta-Sánchez, 2019). This gene was also reported to protect cells from oxidative stress caused by hypoxia (Hansel et al., 2005) as well as from cold and heat stress (Lim et al., 2012). RXFP2 was reported to control horn type, development, and morphology (Pan et al., 2018; Ahbara et al., 2019; Liu et al., 2020) and linked to high-altitude adaptation to hypoxia in sheep (Guo et al., 2021).
FIGURE 5.
Manhattan plot of genome-wide XP-CLR scores by contrasting the high- and low-altitude cattle populations.
There are 14 genes in common to both XP-CLR and iHS (HA) tests, (CLCA4, CLCA1, CLCA2, ITPR2, ABCB10, NUP133, GDA, GALNT13, ENSBTAG00000050002, COL14A1, AXDND1, B3GLCT, SOAT1, and ENSBTAG00000034225) (Table 2; Figure 7). These genes could be regarded as promising candidates subjected to natural selection for high-altitude adaptation in Ethiopian HA cattle. On the other hand, no shared candidate gene was found between the LA iHS cattle and the XP-CLR test.
TABLE 2.
List of overlapping regions and candidate genes identified using the iHS and XPCLR selection scan methods including, gene functions in reported species.
| BAT | Region start | Region end | XPCLR score | iHS value | Gene name | Gene function/phenotype | Species | References |
|---|---|---|---|---|---|---|---|---|
| 2 | 42.25 | 42.45 | 10.99 | 6.28 | GALNT13 | |||
| 3 | 17.55 | 17.6 | 11.54 | 6.15 | ENSBTAG0 | Novel gene/uncharacterized protein | — | — |
| 0000050002 | ||||||||
| 3 | 57.5 | 57.6 | 16.01 | 6.75 | CLCA4 | Rennin secretion, ion channel activity | Palubiski et al. (2020); Weir and Olschewski, (2006) | |
| CLCA1 | “ | “ | ||||||
| 3 | 57.55 | 57.65 | 12.65 | 6.75 | CLCA2 | “ | “ | |
| 5 | 83.45 | 83.55 | 16.65 | 8.19 | ITPR2 | Response to hypoxia | Human | Huerta-Sánchez et al. (2013); Jurkovicova et al. (2008); Manalo et al. (2005); Qu et al. (2015) |
| 8 | 48.4 | 48.5 | 43.83 | 6.23 | GDA | |||
| 12 | 29.6 | 29.7 | 11.17 | 6.31 | B3GLCT | Carbohydrate metabolic process, protein glycosylation, horn development, environment adaptation | Cattle, sheep | Ahbara et al. (2019); Flori et al. (2019); Pan et al. (2018) |
| 14 | 81.45 | 81.55 | 13.63 | 6.04 | COL14A1 | Protein binding, angiogenesis | Mice, rats, sheep, human | Chai et al. (2004); Copple et al. (2011); Wiener et al. (2021); Zhang et al. (2018) |
| 16 | 60.65 | 60.75 | 12.94 | 7.05 | SOAT1 | Cholesterol metabolic process | Insects, mice | Guan et al. (2020); Miron and Tirosh, (2019); Zuniga-Hertz and Patel, (2019) |
| AXDND1 | Response to bone fracture/bone synthesis | human | Pettersson-Kymmer et al. (2013) | |||||
| 28 | 14.5 | 15.5 | 18.81 | 6.94 | NUP133 | Regulate mitochondrial function and oxidative stress response | Mouse | Sunny et al. (2020) |
| ABCB10 | ATPase-coupled transmembrane transporter activity; regulates heme synthesis; Iron metabolism; reactive oxygen species | Mouse, zebrafish, human cell culture | Bayeva et al. (2013); Liesa et al. (2012); Seguin and Ward, (2018); Valverde et al. (2015); Yamamoto et al. (2014) | |||||
| ENSBTAG0 | Basal transcription, coactivators, and promotor recognition. | |||||||
| 0000034225 |
FIGURE 7.
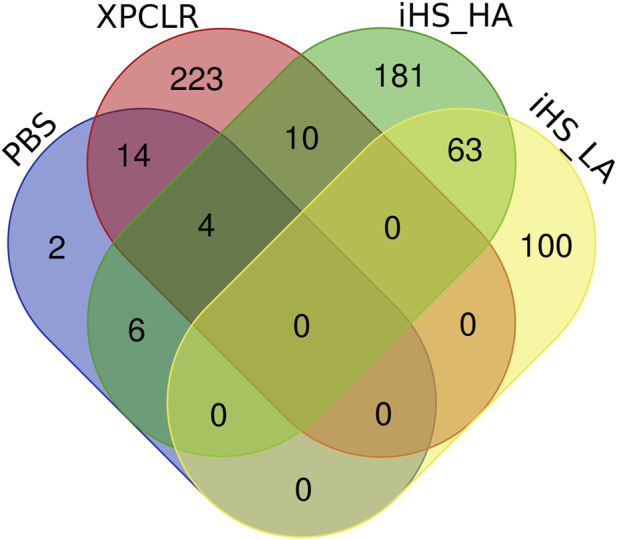
Candidate genes supported by the iHS, XP-CLR, and PBS analyse in high-altitude (HA) and low-altitude (LA) cattle populations.
Functional annotation of genes under selection in Ethiopian high-altitude cattle populations
We conducted a functional annotation using the DAVID visualization tools, based on the Ensembl taurine cattle assembly (ARS-UCD1.2, to identify GO terms and KEGG pathways for the candidate genes that we detected in the Ethiopian HA cattle populations following iHS and XP-CLR analyses. Genes with fold enrichment >1.2 and p-value ≤ 0.05 were considered to be significant (Table 3). Several top candidate genes related to environmental stress such as hypobaric hypoxia, temperature, and UV radiation were clustered into important GO terms, including response to hypoxia (GO:0001666; p-value: 9.3E-06), response to oxygen-containing compound (GO:1901700; p-value: 1.0E-04), ion channel activity (GO: 0005216; p-value: 4.8E-06), glucose homeostasis (GO:0042593; p-value: 5.1E-03), and ATPase activity (GO:0016887; p-value: 1.3E-03), which are biological processes potentially relevant to high altitude adaptation. These findings are in line with previous reports on cattle and other species adapted to high altitudes (Remillard and Yuan, 2006; Edwards et al., 2007; Shimoda and Polak, 2011; Ge et al., 2013; Veith et al., 2016; Moore, 2017; Hu et al., 2019; Friedrich and Wiener, 2020).
TABLE 3.
Gene ontology (GO) clustering and enrichment analyses of candidate genes identified by genome-wide iHS and XP-CLR scans in the high-altitude cattle populations.
| GO term | Count | p value | Fold change | Gene |
|---|---|---|---|---|
| GO: 0005216∼ion channel activity | 15 | 4.8E-06 | 4.5 | ITPR2, KCNJ8, GPR89A, TRPM3, CLCA1, NOX5, GRIK3, CACNG2, SLC26A, CLCA4, GABRB2, ABCC9, KCNQ3, CLCA2, TPC3 |
| GO: 0001666∼response to hypoxia | 7 | 9.3E-04 | 6.1 | ITPR2, MB, CBFA2T3, SRF, VEGFC, HMOX1, ARNT |
| GO: 0034101∼erythrocyte homeostasis | 6 | 1.0E-03 | 7.7 | MAFB, MB, FOXO3, SRF, HMOX1, ARID4A |
| bta04924: Renin secretion | 6 | 2.9E-04 | 9.9 | ITPR2, CLCA1, PRKACB, CLCA4, CLCA2, GUCY1A2 |
| GO: 1901700∼response to oxygen-containing compound | 19 | 1.0E-04 | 2.8 | DUSP10, ITPR2, KCNJ8, IMPACT, NDUFS4, SESN3, AVPR1A, STAT1, PDX1, PTK7, SLC11A1, ADH5, EFNA5, SSTR2, NOX4, HRH4, CRY2, ENPP1, MZB1 |
| GO: 0008217∼regulation of blood pressure | 6 | 2.1E-03 | 6.6 | AVPR1A, POMC, GRIP2, ERAP1, ADH5, MYH6 |
| GO: 0020037∼heme binding | 5 | 2.1E-02 | 4.7 | MB, HMOX1, CYP4F2, GUCY1A2, ENSBTAG00000048257 |
| GO: 0016887∼ATPase activity | 8 | 1.3E-02 | 3.1 | ABCB10, ATP6V0A1, YTHDC2, MYO10, ABCC9, ATP6V1G1, MYH7, MYH6 |
| GO: 0042593∼glucose homeostasis | 6 | 5.1E-03 | 5.3 | SESN3, POMC, PDX1, FOXO3, CRY2, NOX4 |
Low atmospheric oxygen concentration in the inhaled air causes low oxygen levels in the arterial blood reducing cellular energy production, which leads to cellular stress and then induces several factors to increase oxygen availability to cell mitochondria for energy homeostasis. Physiological homeostasis is established through increasing tissue oxygen supply by mounting vascular smooth muscle tone to withstand fast blood flow pressure by inducing the formation of additional blood vessels (angiogenesis), increasing the number of erythrocytes, and improving heme-binding affinity. Supporting these adaptive mechanisms, we identified candidate genes in the biological processes of erythrocyte homeostasis (GO:0034101; p-value: 1.0E-03), heme-binding (GO:0020037; p-value: 2.1E-02), and the regulation of blood pressure (GO:0008217; p-value: 2.1E-03) enhancement. The increases in erythrocyte, hemoglobin concentration, and heme-binding affinity enable more oxygen transportation to tissues in hypoxia-adapted animals (Storz, 2007; Zhang et al., 2007; Storz and Moriyama, 2008; Storz et al., 2010; Yalcin and Cabrales, 2012; Storz, 2016).
Identification of candidate genes associated with high-altitude adaptation
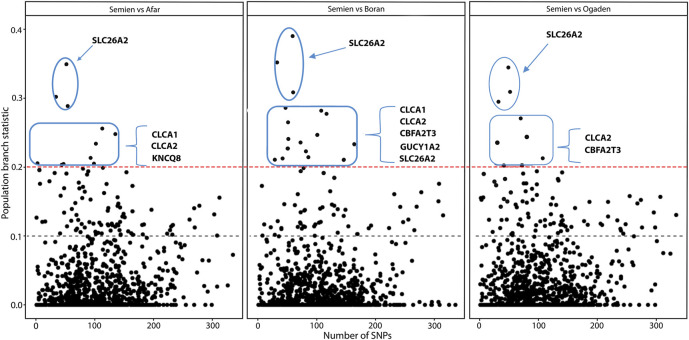
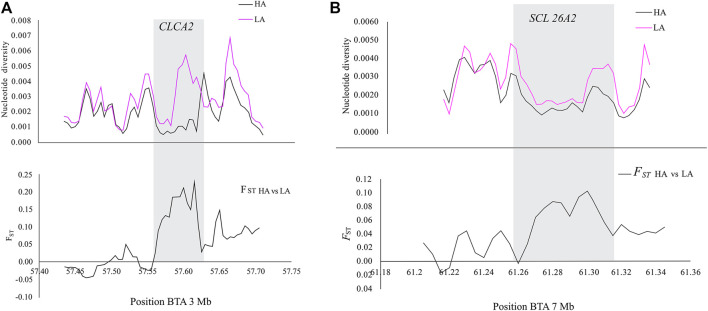
We further analyzed the candidate genes clustered into biological processes relevant to high altitude adaptation (Table 3) using the population branch statistics (PBS). We compared Semien cattle from the highest Ethiopian mountain area to each of the LA cattle populations (Afar, Boran, and Ogaden) using Butana cattle from the Sudanese arid region as an outgroup (see Materials and methods). The 10 kb window outliers from the PBS analysis represent the most differentiated genomic regions (PBS value ≥ 0.2). They overlap with SLC26A2, CLCA1, CLCA2, KCNJ8, GUCY1A2, and CBFA2T3 (Figure 6, Supplementary Table S4). These genes have possible roles in ion channel activity, renin secretion, response to hypoxia, response to oxygen-containing compounds, and heme-binding (Table 3). The genomic region within the SLC26A2 gene was the most differentiated in the PBS scans (Figure 6). SLC26A2 is a ubiquitously expressed SO42− transporter with high expression levels in cartilage and several epithelia (Ohana et al., 2012; Park et al., 2014). This gene is involved in body size and male fertility in humans (Kujala et al., 2007; Touré, 2019), and its mutations have been implicated in dwarfism (Yang and Liang, 2021) and dysplasia (Pineda et al., 2013; Zheng et al., 2019; Heidari et al., 2021).
FIGURE 6.
The distribution of the population branch statistic (PBS) values in 10 kb genomic regions as a function of the number of SNPs.
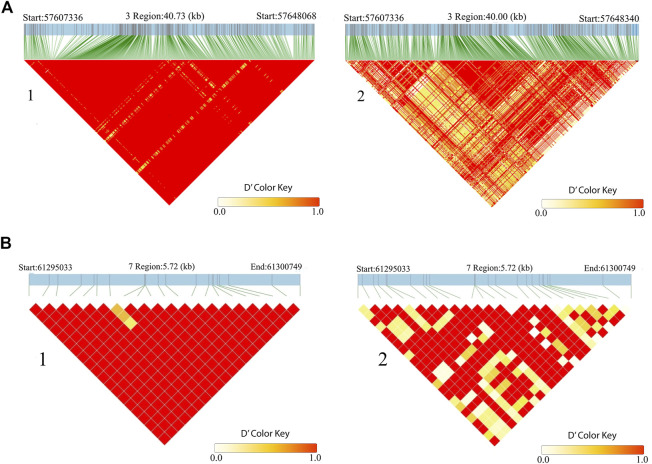
In addition, CLCA2 and two other paralogs, CLCA1 and CLCA4, and ITPR2 were the only four candidate genes detected by the three genomic scans (Figure 7, Supplementary Table S5). Moreover, the variants within the CLCA2 in the HA populations showed a higher level of linkage disequilibrium (LD) compared to the LA populations (Figure 8). Similarly, the nucleotide diversity and population differentiation plot show the CLCA2 gene region with significant variation compared to regions of the two paralog genes (Figure 9). Therefore, we considered CLCA2, ITPR2, SCL26A2, and CBFA2T3 as strong candidate genes putatively linked to high-altitude adaptation in Ethiopian cattle.
FIGURE 8.
LD block heatmap of the candidate genes of CLCA2 (A) and SLC26A2 (B) in the high-altitude (1) and low-altitude (2) cattle populations.
FIGURE 9.
Plots of the nucleotide diversity within and F ST values between the genomic regions of CLCA2 (A) and SCL26A2 (B) in high-altitude (HA) and low-altitude (LA) cattle populations.
Other genes of interest include GO terms linked to the response to hypoxia (Table 3). These include MB (BTA5: 73.81–73.82 Mb), CBFA2T3 (BTA18: 14.05–14.1 Mb), and SRF (BTA23: 16.77–16.78 Mb) from XP-CLR scans results, ARNT (BTA3: 19.8–19.9 Mb) and VEGFC (BTA27: 8.0–8.1 Mb) from iHS scans results, and ITPR2 from both XP-CLR and iHS scan results. ARNT is involved in the positive regulation of vascular endothelial growth factor (VEGF) activation. VEGFC, a VEGF homolog, is involved in regulating endothelial cell proliferation and angiogenesis in response to the low oxygen concentration in the arterial blood (Kumar et al., 2011; Herbert and Stainier, 2012; Ramakrishnan et al., 2014).
ARNT also enhances endothelial cell growth in the vascular line and it is expressed during the early phase of the growth of new blood vessels (angiogenesis) (Scheinfeldt et al., 2012; Geng et al., 2014; Graham and Presnell, 2017). Protein-protein interaction network analysis shows that ARNT interacts with hypoxia-inducible factors such as HIF1a, EPAS1, EP300, and its paralog CREBBP (Figure 10). CBFA2T3 is clustered in response to hypoxia and functions as a transcription regulator of HIF1a through interaction with EGLN1 and promoting the HIF1a prolyl hydroxylation-dependent ubiquitination and proteasomal degradation pathways (Kumar et al., 2015). It also contributes to the inhibition of glycolysis and the stimulation of mitochondrial respiration by down-regulating the expression of glycolytic genes as direct targets of HIF1a (Kumar et al., 2013).
FIGURE 10.
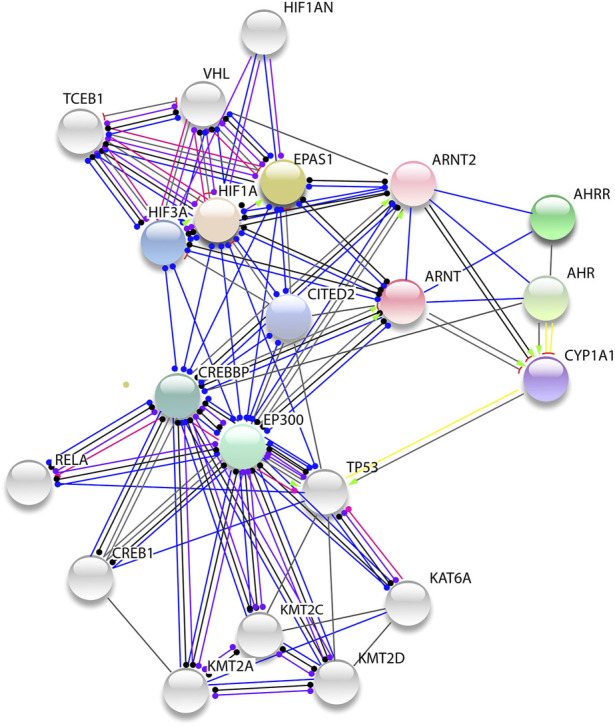
Protein-protein network of ARNT. The genes show several protein interaction networks interacting with hypoxia induced factors (HIF1a, EPAS1, CREBBP, and EP300).
Discussion
This study aimed to unravel at the autosomal genome level the adaptation of Ethiopian indigenous cattle to the extreme environmental conditions of its mountainous areas. We studied specifically three cattle populations of Semien, Bale, and Choke living in a mountainous area of more than 3,000 masl by contrasting them with the indigenous cattle population from the Ethiopian lowlands. Population genetic structure validated the African zebu admixture of indicine and taurine status of all the studied indigenous Ethiopian cattle (Figure 1), while the PCA result shows some level of genetic differentiation between the high-altitude Ethiopian cattle populations from those originating from the low altitude locations (Figure 2B). We then applied the iHS, XP-CLR, and PBS methods to detect selection signatures within and between the Ethiopian cattle populations living at high and low altitudes. Finally, additional comparisons of the candidate genomic regions between the high- and low-altitude cattle populations were carried out based on their nucleotide diversity, population differentiation, and haplotype LD heatmap differences for a detailed exploration of the high altitude adaptation.
Novel candidate gene identified in this study
We identified three novel strong candidate genes (CLCA2, SLC26A2, and CBFA2T3) (Figure 6) for high altitude adaptation along with the previously reported ITPR2 gene. The functional analysis clustered the candidate genes into ion channel activity (CLCA2, SLC26A2, and ITPR2), response to hypoxia (ITPR2 and CBFA2T3), and renin secretion KEGG pathway (CLCA2) (Table 3). The renin pathway and ion channel activity regulate smooth muscle tone and epithelial secretion in response to hypoxia (Al-Hashem et al., 2012; Palubiski et al., 2020) by controlling arterial blood flow pressure (Shimoda and Polak, 2011). Hypoxia induces the expression of CLCA2 in the pulmonary artery smooth muscle of rats and controls cell proliferation and apoptosis in the ERK1/2-MAPK signaling pathway (Huang et al., 2017; Zhao et al., 2017). Similarly, the renin secretion pathways maintain the amount of plasma renin and aldosterone concentration by modulating the normal relationship between plasma osmolality and plasma vasopressin concentration in humans (Bestle et al., 2002; Savoia et al., 2011). The renin-angiotensin and vasopressin function is stimulated by increased blood pressure caused by vesicular smooth muscle tone (Chassagne et al., 2000) to regulate high blood flow to balance cellular oxygen demand.
SLC26A2 showed the highest PBS value in the high-altitude cattle populations (Figure 6). The haplotype LD heatmap, nucleotide diversity, and population differentiation index all supporting positive selection at the genome region overlapping with the gene (Figures 8, 9). The function of this gene is related to ion transport, and it plays a role in chondrocyte proliferation, differentiation, and growth in endochondral bone formation (Park et al., 2014). In humans, it regulates body size, and its recessive allele contributes to the dwarfism phenotype (Yang and Liang, 2021) and dysplasia (Pineda et al., 2013; Zheng et al., 2019; Heidari et al., 2021). A previous study reported a dominant allele at SLC26A2 linked to higher heels and stronger claws in dairy cattle, while mutation at the gene causes dysplasia (Brenig et al., 2003). Considering the rugged and rocky terrain of the Ethiopian highlands, strong claws and high heels may prove advantageous. Further phenotypic characterization of the Ethiopian highland cattle may support this interpretation. The short stature and small body size of cattle observed in the Ethiopian high-altitude cattle confer the evidence. Though confirmatory analysis is required to differentiate the nature of short stature and small body size for HA adaptation in Ethiopian cattle, it could be a possible mechanism of the cold and high-altitude adaptation as it was reported in humans adapted to high altitude (Ma et al., 2019).
Candidate convergent genome evolution between cattle and humans living in the Ethiopian highlands
The human population in Ethiopia occupied the high altitudes thousands of years ago, expanding from the lower Rift Valley in the early Pleistocene age (Aldenderfer, 2006). Archaeological evidence suggests that humans inhabited Bale Mountain approximately 50–30 thousand years ago (Ossendorf et al., 2019). Today, the human communities occupying the high altitude areas where the cattle samples were collected are the Oromo (Bale Mountain) and the Amhara (Semien and Choke mountains). The beginning of the settlement of the Amhara to these high-altitude regions is thought to have started around 5,000 years (Alkorta-Aranburu et al., 2012), while the settlement of the Oromo people was since early 1500s as reported by Hassen (1990) (cited in Alkorta-Aranburu et al., 2012; Huerta-Sánchez et al., 2013). The settlers in these territories were agrarian and had close interaction with their animals as sources of food and means of food production. Both humans and cattle living in the Ethiopian high altitudes share similar environmental challenges. Humans and ruminants living at high altitudes can be exposed to extended hypoxia stress and develop high-altitude sicknesses that may lead to high-altitude pulmonary hypertension (Friedrich and Wiener, 2020). For example, reports have indicated that cattle exposed to high altitudes may develop brisket disease caused by hypoxia (Newman et al., 2011; Wuletaw et al., 2011). Besides hypoxia, UV light and cold temperatures have been reported as major risk factors that challenge the survival of humans and other species in high altitude environments. Through a long-term evolutionary process, these risk factors may have induced positive selection pressures for physiological and morphological features that contribute to the evolutionary adaptation to high altitude environments (Witt and Huerta-Sánchez, 2019). Candidate genes detected in Ethiopian people living at high altitudes (Huerta-Sánchez et al., 2013), including ITPR2, ARNT, EP300, MB, and HMOX1, were also detected in Ethiopian cattle living in similar environments, supporting a convergent evolution between these two mammalian species.
Previous studies on the Ethiopian human population adapted to high altitudes have reported the ITPR2 gene (Huerta-Sánchez et al., 2013) as a candidate gene. The ITPR2 is also one of the candidate genes detected in HA cattle populations. It regulates vascular endothelial cells and intracellular calcium ion channel activity (Manalo et al., 2005; Jurkovicova et al., 2008). Following hypoxia, the cardiovascular system will increase blood flow by increasing pressure through vasoconstriction, increased heart rate, and myocardial contractility (Parati et al., 2015). These adaptive physiological mechanisms will enhance the supply of blood oxygen to tissues. ITPR2 increases intracellular calcium concentration in vascular smooth muscle and it controls vasoconstriction avoiding pulmonary hypertension (Remillard and Yuan, 2006; Newman et al., 2011; Lai et al., 2015). ITPR2, as part of the calcium gated channel activities, also enhances endothelial cell proliferation lining and it triggers the vasculature and remodeling of the arterial tone to control the high blood pressure following hypoxic exposure (Makino et al., 2011; Hübner et al., 2015).
High altitude adaptation also depends on the concentration of hemoglobin in red blood cells and its affinity to oxygen in tissues (Alkorta-Aranburu et al., 2012). Also, increasing the number of erythrocytes will lead to higher hemoglobin concentration at the tissue level (Siebenmann et al., 2015). The candidate MB gene (Table 3) has been reported to play a role in increasing the hemoglobin concentration in muscle (Fraser et al., 2006; Jaspers et al., 2014) and increasing oxygen storage and binding affinity in hypoxic conditions (Hoppeler and Vogt, 2001; Li et al., 2018). This myoglobin gene was also reported under selection in the Ethiopian and Tibetan human populations living in highlands (Beall et al., 2002; Moore et al., 2002; Alkorta-Aranburu et al., 2012; Scheinfeldt et al., 2012). The gene is involved in erythrocyte homeostasis and regulates the level of hemoglobin (Avivi et al., 2010) in response to high altitude adaptation.
Selection signatures overlap between Ethiopia cattle and other species adapted to high altitudes
Several studies have reported candidate positive selection signatures for high-altitude adaptation in different species. Here, besides the overlap with human candidate selected regions, we identified several candidate regions which aligned with genes reported under selection in other species adapted to high altitudes. They include MSRB3 with the highest XP-CLR score in our study and MC1R previously reported under selection in Ethiopian highland sheep (Edea et al., 2019). The MSRB3 gene was also reported in Tibetan dogs and sheep (Vaysse et al., 2011; Wei et al., 2016; Witt and Huerta-Sánchez, 2019). It has a pleiotropic effect in being involved in the ossification and adipose tissue development in cattle (Saatchi et al., 2014). It has also been linked to ear size in Tibetan sheep (Wei et al., 2016), pigs (Zhang et al., 2015), and dogs (Vaysse et al., 2011). The MSRB3 gene also protects cells from oxidative stress caused in mammals by hypoxia (Hansel et al., 2005), while it is linked to cold and heat tolerance in Drosophila (Lim et al., 2012) and cold tolerance in Arabidopsis (Kwon et al., 2007). Cold temperature is one of the environmental stressors that trigger animal cells to transduce energy to adapt to cold temperatures.
Last but not least, among the genes present within candidate genomic regions detected by both XP-CLR and iHS analyses, we do have ABCB10 (Table 3). This gene was previously reported in candidate selected regions in humans, and several other species, including cattle (Bayeva et al., 2013; Martinez et al., 2020). Its function is related to iron metabolism and heme biosynthesis (Haase, 2010; Shah et al., 2013; Yamamoto et al., 2014; Seguin and Ward, 2018). ABCB10 is also involved in the transport of heme out of the mitochondria, before hemoglobinization of erythropoietic cells (Liesa et al., 2012; Bayeva et al., 2013).
Conclusion
Despite the particularly challenging environmental conditions of the high-altitude Ethiopian highlands and the relatively recent arrival of African indicine cattle in these areas, we identified several genomic regions with evidence of positive selection for high-altitude environment adaptations at the autosomal level. These include genes previously reported in other mammalian species, including humans, living in high altitude areas in Ethiopia or other parts of the world, as well as in Ethiopian-specific cattle genomic regions. Our results show that these indigenous livestock populations are locally adapted, and they have developed a physiological mechanism to cope with the environmental challenges of hypoxia, UV radiation, and cold temperature. It calls for the conservation of these indigenous cattle adaptations as well as for their utilization in breeding programs combining the improvement of productivity with adaptability.
Acknowledgments
The authors would like to acknowledge the following institutions and personnel for funding and facilitating the research. The International Livestock Research Institute (ILRI) LiveGne program, supported by the CGIAR Research Program on Livestock (CRP livestock project) sponsored by the CGIAR funding contributors to the Trust Fund (http://www.cgiar.org/about-us/our-funders/). The Bill and Melinda Gates Foundation and UK aid from the UK Foreign, Commonwealth, and Development Office (Grant Agreement OPP1127286) under the auspices of the Centre for Tropical Livestock Genetics and Health (CTLGH), established jointly by the University of Edinburgh, SRUC (Scotland’s Rural College), and The Chinese Government contribution to CAAS-ILRI Joint Laboratory on Livestock and Forage Genetic Resources in Beijing (2018-GJHZ-01). ET is a post-graduate fellow registered at Addis Ababa University (Ethiopia) and he was granted an ILRI post-graduate fellowship for his Ph.D. study.
Data availability statement
The original contributions presented in the study are publicly available. The WGS data of Bale and Semien cattle samples are available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA698721, while the Choke cattle samples WGS data is available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA841948/.
Ethics statement
Ethical review and approval were not required for the animal study because they were not applicable in the country. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
ET collected samples and carried out laboratory analysis. ET and AT performed analysis and data interpretation. OH reviewed critically the draft manuscript input from JH who assisted in the acquisition of the grant. OH and JH sequenced the data. ET wrote the draft manuscript, which was critically reviewed by AT, GB, OH, and JH. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.960234/full#supplementary-material
References
- Ahbara A., Bahbahani H., Almathen F., Abri M. A., Agoub M. O., Abeba A., et al. (2019). Genome-wide variation, candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous sheep. Front. Genet. 10, 699–721. 10.3389/fgene.2018.00699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashem F. H., Alkhateeb M. A., Shatoor A. S., Khalil M. A., Sakr H. F. (2012). Chronic exposure of rats to native high altitude increases in blood pressure via activation of the renin-angiotensin-aldosterone system. Saudi Med. J. 33, 1169–1176. [PubMed] [Google Scholar]
- Aldenderfer M. (2006). Modelling plateau peoples: The early human use of the world’s high plateaux. World Archaeol. 38, 357–370. 10.1080/00438240600813285 [DOI] [Google Scholar]
- Alexander D. H., Novembre J., Lange K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkorta-Aranburu G., Beall C. M., Witonsky D. B., Gebremedhin A., Pritchard J. K., Di Rienzo A. (2012). The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 8, e1003110. 10.1371/journal.pgen.1003110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asresie A., Zemedu L. (2015). Contribution of livestock sector in Ethiopian economy : A review. Adv. Life Sci. Technol. 29, 79–91. [Google Scholar]
- Avivi A., Gerlach F., Joel A., Reuss S., Burmester T., Nevo E., et al. (2010). Neuroglobin, cytoglobin, and myoglobin contribute to hypoxia adaptation of the subterranean mole rat Spalax. Proc. Natl. Acad. Sci. U. S. A. 107, 21570–21575. 10.1073/pnas.1015379107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad P., Stobdan T., Zhou D., Hartley I., Akbari A., Bafna V., et al. (2017). High-altitude adaptation in humans: From genomics to integrative physiology. J. Mol. Med. 95, 1269–1282. 10.1007/s00109-017-1584-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayeva M., Khechaduri A., Wu R., Burke M. A., Wasserstrom J. A., Singh N., et al. (2013). ATP-binding cassette B10 regulates early steps of heme synthesis. Circ. Res. 113, 279–287. 10.1161/CIRCRESAHA.113.301552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall C. M., Decker M. J., Brittenham G. M., Kushner I., Gebremedhin A., Strohl K. P. (2002). An Ethiopian pattern of human adaptation to high-altitude hypoxia. Proc. Natl. Acad. Sci. U. S. A. 99, 17215–17218. 10.1073/pnas.252649199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle M. H., Olsen N. V., Poulsen T. D., Roach R., Fogh-Andersen N., Bie P. (2002). Prolonged hypobaric hypoxemia attenuates vasopressin secretion and renal response to osmostimulation in men. J. Appl. Physiol. 92, 1911–1922. 10.1152/japplphysiol.00936.2001 [DOI] [PubMed] [Google Scholar]
- Bigham A. W., Lee F. S. (2014). Human high-altitude adaptation: Forward genetics meets the HIF pathway. Genes Dev. 28, 2189–2204. 10.1101/gad.250167.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenig B., Baumgartner B. G., Kriegesmann B., Habermann F., Fries R., Swalve H. H. (2003). Molecular cloning, mapping, and functional analysis of the bovine sulfate transporter SLC26a2 gene. Gene 319, 161–166. 10.1016/S0378-1119(03)00806-0 [DOI] [PubMed] [Google Scholar]
- Browning S. R., Browning B. L. (2007). Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097. 10.1086/521987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J., Jones M. K., Tarnawski A. S. (2004). Serum response factor is a critical requirement for VEGF signaling in endothelial cells and VEGF-induced angiogenesis. FASEB J. 18, 1264–1266. 10.1096/fj.03-1232fje [DOI] [PubMed] [Google Scholar]
- Chassagne C., Eddahibi S., Adamy C., Rideau D., Marotte F., Dubois-Rande J.-L., et al. (2000). Modulation of angiotensin II receptor expression during development and regression of hypoxic pulmonary hypertension. Am. J. Respir. Cell Mol. Biol. 22, 323–332. 10.1165/ajrcmb.22.3.3701 [DOI] [PubMed] [Google Scholar]
- Chen H., Patterson N., Reich D. (2010). Population differentiation as a test for selective sweeps. Genome Res. 20, 393–402. 10.1101/gr.100545.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yu M., Dai X., Zogg M., Wen R., Weiler H., et al. (2011). Critical role for Gimap5 in the survival of mouse hematopoietic stem and progenitor cells. J. Exp. Med. 208, 923–935. 10.1084/jem.20101192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copple B. L., Bai S., Burgoon L. D., Moon J. O. (2011). Hypoxia-inducible factor-1α regulates the expression of genes in hypoxic hepatic stellate cells important for collagen deposition and angiogenesis. Liver Int. 31, 230–244. 10.1111/j.1478-3231.2010.02347.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debevec T., Millet G. P., Pialoux V. (2017). Hypoxia-induced oxidative stress modulation with physical activity. Front. Physiol. 8, 84–89. 10.3389/fphys.2017.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolt K. S., Mishra M. K., Karar J., Baig M. A., Ahmed Z., Pasha M. A. Q. (2007). cDNA cloning, gene organization and variant specific expression of HIF-1α in high altitude yak (Bos grunniens). Gene 386, 73–80. 10.1016/j.gene.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Dong S.-S., He W.-M., Ji J.-J., Zhang C., Guo Y., Yang T.-L. (2020). LDBlockShow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. bioRxiv, 151332. 06.14. 10.1101/2020.06.14.151332 [DOI] [PubMed] [Google Scholar]
- Edea Z., Dadi H., Dessie T., Kim K.-S. (2019). Genomic signatures of high-altitude adaptation in Ethiopian sheep populations. Genes Genomics 41, 973–981. 10.1007/s13258-019-00820-y [DOI] [PubMed] [Google Scholar]
- Edea Z., Dadi H., Kim S. W., Park J. H., Shin G. H., Dessie T., et al. (2014). Linkage disequilibrium and genomic scan to detect selective loci in cattle populations adapted to different ecological conditions in Ethiopia. J. Anim. Breed. Genet. 131, 358–366. 10.1111/jbg.12083 [DOI] [PubMed] [Google Scholar]
- Edwards C. J., Bollongino R., Scheu A., Chamberlain A., Tresset A., Vigne J. D., et al. (2007). Mitochondrial DNA analysis shows a Near Eastern Neolithic origin for domestic cattle and no indication of domestication of European aurochs. Proc. Biol. Sci. 274, 1377–1385. 10.1098/rspb.2007.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flori L., Moazami-Goudarzi K., Alary V., Araba A., Boujenane I., Boushaba N., et al. (2019). A genomic map of climate adaptation in Mediterranean cattle breeds. Mol. Ecol. 28, 1009–1029. 10.1111/mec.15004 [DOI] [PubMed] [Google Scholar]
- Fraser J., De Mello L. V., Ward D., Rees H. H., Williams D. R., Fang Y., et al. (2006). Hypoxia-inducible myoglobin expression in nonmuscle tissues. Proc. Natl. Acad. Sci. U. S. A. 103, 2977–2981. 10.1073/pnas.0508270103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J., Wiener P. (2020). Selection signatures for high-altitude adaptation in ruminants. Anim. Genet. 51, 157–165. 10.1111/age.12900 [DOI] [PubMed] [Google Scholar]
- Gautier M., Klassmann A., Vitalis R. (2017). Rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol. Ecol. Resour. 17, 78–90. 10.1111/1755-0998.12634 [DOI] [PubMed] [Google Scholar]
- Ge R. L., Cai Q., Shen Y. Y., San A., Ma L., Zhang Y., et al. (2013). Draft genome sequence of the Tibetan antelope. Nat. Commun. 4, 1858–1867. 10.1038/ncomms2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X., Feng J., Liu S., Wang Y., Arias C., Liu Z. (2014). Transcriptional regulation of hypoxia inducible factors alpha (HIF-α) and their inhibiting factor (FIH-1) of channel catfish (ictalurus punctatus) under hypoxia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 169, 38–50. 10.1016/j.cbpb.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Graham A. M., Presnell J. S. (2017). Hypoxia Inducible Factor (HIF) transcription factor family expansion, diversification, divergence and selection in eukaryotes. PLoS One 12, e0179545. 10.1371/journal.pone.0179545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C., Niu Y., Chen S. C., Kang Y., Wu J. X., Nishi K., et al. (2020). Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat. Commun. 11, 2478–2511. 10.1038/s41467-020-16288-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Zhao H., Yuan C., Huang S., Zhou S., Lu Z., et al. (2021). Selective sweeps uncovering the genetic basis of horn and adaptability traits on fine-wool sheep in China. Front. Genet. 12, 604235. 10.3389/fgene.2021.604235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase V. H. (2010). Hypoxic regulation of erythropoiesis and iron metabolism. Am. J. Physiol. Ren. Physiol. 299, F1–F13. 10.1152/ajprenal.00174.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel A., Heinemann S. H., Hoshi T. (2005). Heterogeneity and function of mammalian MSRs: Enzymes for repair, protection and regulation. Biochim. Biophys. Acta 1703, 239–247. 10.1016/j.bbapap.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Heidari M., Soleyman-Nejad M., Isazadeh A., Taskiri M. H., Bolhassani M., Sadighi N., et al. (2021). Identification of a novel homozygous mutation in the DDR2 gene from a patient with spondylo-meta-epiphyseal dysplasia by whole exome sequencing. Iran. J. Basic Med. Sci. 24, 191–195. 10.22038/IJBMS.2020.44487.10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S. P., Stainier D. Y. R. (2012). Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat. Rev. Mol. Cell Biol. 12, 551–564. 10.1038/nrm3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. H., Tsai S. F. (2017). Functional and biochemical characterization of a T cell-associated anti-apoptotic protein, GIMAP6. J. Biol. Chem. 292, 9305–9319. 10.1074/jbc.M116.768689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppeler H., Vogt M. (2001). Muscle tissue adaptations to hypoxia. J. Exp. Biol. 204, 3133–3139. 10.1242/jeb.204.18.3133 [DOI] [PubMed] [Google Scholar]
- Hu C.-J., Iyer S., Sataur A., Covello K. L., Chodosh L. A., Simon M. C. (2006). Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol. Cell. Biol. 26, 3514–3526. 10.1128/MCB.26.9.3514-3526.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X. J., Yang J., Xie X. L., Lv F. H., Cao Y. H., Li W. R., et al. (2019). The genome landscape of Tibetan sheep reveals adaptive introgression from Argali and the history of early human settlements on the Qinghai-Tibetan Plateau. Mol. Biol. Evol. 36, 283–303. 10.1093/molbev/msy208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang L. J., Zhang C. C., Zhao M. P., Zheng M. X., Ying L., Chen X. W., et al. (2017). The regulation of MAPK signaling pathway on cell proliferation and apoptosis in hypoxic PASMCs of rats. Chin. J. Appl. Physiol. 33, 226–230. 10.12047/j.cjap.5422.2017.056 [DOI] [PubMed] [Google Scholar]
- Hübner C. A., Schroeder B. C., Ehmke H. (2015). Regulation of vascular tone and arterial blood pressure: Role of chloride transport in vascular smooth muscle. Pflugers Arch. 467, 605–614. 10.1007/s00424-014-1684-y [DOI] [PubMed] [Google Scholar]
- Huerta-Sánchez E., DeGiorgio M., Pagani L., Tarekegn A., Ekong R., Antao T., et al. (2013). Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol. Biol. Evol. 30, 1877–1888. 10.1093/molbev/mst089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers R. T., Testerink J., Gaspera B. D., Chanoine C., Bagowski C. P., Van Der Laarse W. J. (2014). Increased oxidative metabolism and myoglobin expression in zebrafish muscle during chronic hypoxia. Biol. Open 3, 718–727. 10.1242/bio.20149167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkovicova D., Sedlakova B., Lacinova L., Kopacek J., Sulova Z., Sedlak J., et al. (2008). Hypoxia differently modulates gene expression of inositol 1, 4, 5-trisphosphate receptors in mouse kidney and HEK 293 cell line. Ann. N. Y. Acad. Sci. 1148, 421–427. 10.1196/ANNALS.1410.034 [DOI] [PubMed] [Google Scholar]
- Kim J., Hanotte O., Mwai O. A., Dessie T., Bashir S., Diallo B., et al. (2017). The genome landscape of indigenous African cattle. Genome Biol. 18, 34–14. 10.1186/s13059-017-1153-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kwon T., Dessie T., Yoo D. A., Mwai O. A., Jang J., et al. (2020). The mosaic genome of indigenous African cattle as a unique genetic resource for African pastoralism. Nat. Genet. 52, 1099–1110. 10.1038/s41588-020-0694-2 [DOI] [PubMed] [Google Scholar]
- Kujala M., Hihnala S., Tienari J., Kaunisto K., Hästbacka J., Holmberg C., et al. (2007). Expression of ion transport-associated proteins in human efferent and epididymal ducts. Reproduction 133, 775–784. 10.1530/rep.1.00964 [DOI] [PubMed] [Google Scholar]
- Kumar B., Chile S. A., Ray K. B., Vidyadhar Reddy G. E. C., Addepalli M. K., Manoj Kumar A. S., et al. (2011). VEGF-C differentially regulates VEGF-A expression in ocular and cancer cells; Promotes angiogenesis via RhoA mediated pathway. Angiogenesis 14, 371–380. 10.1007/s10456-011-9221-5 [DOI] [PubMed] [Google Scholar]
- Kumar P., Gullberg U., Olsson I., Ajore R. (2015). Myeloid translocation gene-16 co-repressor promotes degradation of hypoxia-inducible factor 1. PLoS One 10, e0123725. 10.1371/journal.pone.0123725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Sharoyko V. V., Spégel P., Gullberg U., Mulder H., Olsson I., et al. (2013). The transcriptional Co-repressor myeloid translocation gene 16 inhibits glycolysis and stimulates mitochondrial respiration. PLoS One 8, e68502. 10.1371/journal.pone.0068502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. J., Kwon S. Il, Bae M. S., Cho E. J., Park O. K. (2007). Role of the methionine sulfoxide reductase MsrB3 in cold acclimation in Arabidopsis. Plant Cell Physiol. 48, 1713–1723. 10.1093/pcp/pcm143 [DOI] [PubMed] [Google Scholar]
- Lai N., Lu W., Wang J. (2015). Ca2+ and ion channels in hypoxia-mediated pulmonary hypertension. Int. J. Clin. Exp. Pathol. 8, 1081–1092. [PMC free article] [PubMed] [Google Scholar]
- Li C., Li X., Liu J., Fan X., You G., Zhao L., et al. (2018). Investigation of the differences between the Tibetan and Han populations in the hemoglobin–oxygen affinity of red blood cells and in the adaptation to high-altitude environments. Hematology 23, 309–313. 10.1080/10245332.2017.1396046 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M., Qiu W., Shirihai O. S. (2012). Mitochondrial ABC transporters function: The role of ABCB10 (ABC-me) as a novel player in cellular handling of reactive oxygen species. Biochim. Biophys. Acta 1823, 1945–1957. 10.1016/j.bbamcr.2012.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D. H., Han J. Y., Kim J. R., Lee Y. S., Kim H. Y. (2012). Methionine sulfoxide reductase B in the endoplasmic reticulum is critical for stress resistance and aging in Drosophila. Biochem. Biophys. Res. Commun. 419, 20–26. 10.1016/j.bbrc.2012.01.099 [DOI] [PubMed] [Google Scholar]
- Liu J., Yuan C., Guo T., Wang F., Zeng Y., Ding X., et al. (2020). Genetic signatures of high-altitude adaptation and geographic distribution in Tibetan sheep. Sci. Rep. 10, 18332–18413. 10.1038/s41598-020-75428-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Jia, Zhang Z., Niu W., Chen J., Guo S., Liu S., et al. (2019). Education, altitude, and humidity can interactively explain spatial discrepancy and predict short stature in 213, 795 Chinese school children. Front. Pediatr. 7, 425–510. 10.3389/fped.2019.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar A. J., Wong W. J., Simon M. C. (2010). Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294–309. 10.1016/j.molcel.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A., Firth A. L., Yuan J. X.-J. (2011). “Endothelial and smooth muscle cell ion channels in pulmonary vasoconstriction and vascular remodeling,” in Comprehensive physiology (Hoboken, NJ, USA: John Wiley & Sons; ), 139–148. 10.1002/cphy.c100023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manalo D. J., Rowan A., Lavoie T., Natarajan L., Kelly B. D., Ye S. Q., et al. (2005). Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105, 659–669. 10.1182/blood-2004-07-2958 [DOI] [PubMed] [Google Scholar]
- Martinez M., Fendley G. A., Saxberg A. D., Zoghbi M. E. (2020). Stimulation of the human mitochondrial transporter ABCB10 by zinc-mesoporphrin. PLoS One 15, e0238754. 10.1371/journal.pone.0238754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron N., Tirosh O. (2019). Cholesterol prevents hypoxia-induced hypoglycemia by regulation of a metabolic ketogenic shift. Oxid. Med. Cell. Longev. 2019, 5829357. 10.1155/2019/5829357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. G. (2017). Measuring high-altitude adaptation. J. Appl. Physiol. 123, 1371–1385. 10.1152/japplphysiol.00321.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. G., Zamudio S., Zhuang J., Droma T., Shohet R. V. (2002). Analysis of the myoglobin gene in tibetans living at high altitude. High. Alt. Med. Biol. 3, 39–47. 10.1089/152702902753639531 [DOI] [PubMed] [Google Scholar]
- Newman J. H., Holt T. N., Hedges L. K., Womack B., Memon S. S., Willers E. D., et al. (2011). High-altitude pulmonary hypertension in cattle (brisket disease): Candidate genes and gene expression profiling of peripheral blood mononuclear cells. Pulm. Circ. 1, 462–469. 10.4103/2045-8932.93545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohana E., Shcheynikov N., Park M., Muallem S. (2012). Solute carrier family 26 member a2 (Slc26a2) protein functions as an electroneutral SOFormula/OH-/Cl- exchanger regulated by extracellular Cl-. J. Biol. Chem. 287, 5122–5132. 10.1074/jbc.M111.297192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossendorf G., Groos A. R., Bromm T., Tekelemariam M. G., Glaser B., Lesur J., et al. (2019). Middle Stone Age foragers resided in high elevations of the glaciated Bale Mountains, Ethiopia. Science 365, 583–587. 10.1126/science.aaw8942 [DOI] [PubMed] [Google Scholar]
- Palubiski L. M., O’Halloran K. D., O’Neill J. (2020). Renal physiological adaptation to high altitude: A systematic review. Front. Physiol. 11, 756. 10.3389/fphys.2020.00756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z., Li S., Liu Q., Wang Z., Zhou Z., Di R., et al. (2018). Whole-genome sequences of 89 Chinese sheep suggest role of RXFP2 in the development of unique horn phenotype as response to semi-feralization. Gigascience 7, giy019–15. 10.1093/gigascience/giy019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parati G., Ochoa J. E., Torlasco C., Salvi P., Lombardi C., Bilo G. (2015). Aging, high altitude, and blood pressure: A complex relationship. High. Alt. Med. Biol. 16, 97–109. 10.1089/ham.2015.0010 [DOI] [PubMed] [Google Scholar]
- Park M., Ohana E., Choi S. Y., Lee M. S., Park J. H., Muallem S. (2014). Multiple roles of the SO4(2-)/Cl-/OH- exchanger protein Slc26a2 in chondrocyte functions. J. Biol. Chem. 289, 1993–2001. 10.1074/jbc.M113.503466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson-Kymmer U., Lacroix A., Eriksson J., Bergstrom U., Melin B., Wibom C., et al. (2013). Genome-wide association study meta-analysis identifies the SOAT1/AXDND1 locus to be associated with hip and forearm fracture risk. Bone Abstr. 10.1530/boneabs.2.1 [DOI] [Google Scholar]
- Pineda T., Rossi A., Bonafè L., Superti-Furga A., Velasco H. M. (2013). Report of a novel mutation in the SLC26A2 gene found in a Colombian adult patient with diastrophic dysplasia. Rev. Fac. Med. 61, 255–259. [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., et al. (2007). Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q., Zhang G., Ma T., Qian W., Ye Z., Cao C., et al. (2012). The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–949. 10.1038/ng.2343 [DOI] [PubMed] [Google Scholar]
- Qu Y., Tian S., Han N., Zhao H., Gao B., Fu J., et al. (2015). Genetic responses to seasonal variation in altitudinal stress : Whole-genome resequencing of great tit in eastern himalayas. Sci. Rep. 5, 14256–14310. 10.1038/srep14256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan S., Anand V., Roy S. (2014). Vascular endothelial growth factor signaling in hypoxia and inflammation. J. Neuroimmune Pharmacol. 9, 142–160. 10.1007/s11481-014-9531-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remillard C. V., Yuan J. X.-J. (2006). High altitude pulmonary hypertension: Role of K+ and Ca2+ channels. High. Alt. Med. Biol. 6, 133–146. 10.1089/ham.2005.6.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., Schnabel R. D., Taylor J. F., Garrick D. J. (2014). Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics 15, 442. 10.1186/1471-2164-15-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C. C., Reich D. E. E., Higgins J. M. M., Levine H. Z. P. Z. P., Richter D. J. J., Schaffner S. F. F., et al. (2002). Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837. 10.1038/nature01140 [DOI] [PubMed] [Google Scholar]
- San T., Polat S., Cingi C., Eskiizmir G., Oghan F., Cakir B. (2013). Effects of high altitude on sleep and respiratory system and theirs adaptations. ScientificWorldJournal. 2013, 241569. 10.1155/2013/241569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoia C., Burger D., Nishigaki N., Montezano A., Touyz R. M. (2011). Angiotensin II and the vascular phenotype in hypertension. Expert Rev. Mol. Med. 13, e11–e25. 10.1017/S1462399411001815 [DOI] [PubMed] [Google Scholar]
- Scheinfeldt L. B., Soi S., Thompson S., Ranciaro A., Woldemeskel D., Beggs W., et al. (2012). Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 13, R1. 10.1186/gb-2012-13-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwefel D., Arasu B. S., Marino S. F., Lamprecht B., Köchert K., Rosenbaum E., et al. (2013). Structural insights into the mechanism of GTPase activation in the GIMAP family. Structure 21, 550–559. 10.1016/J.STR.2013.01.014 [DOI] [PubMed] [Google Scholar]
- Seguin A., Ward D. M. (2018). Mitochondrial ABC transporters and iron metabolism. J. Clin. Exp. Pathol. 08, 6–10. 10.4172/2161-0681.1000338 [DOI] [Google Scholar]
- Shah D. I., Takahashi-makise N., Cooney J. D., Li L., Iman J., Pierce E. L., et al. (2013). Mitochondrial Atpif1 regulates heme synthesis in developing erythroblasts. Nature. 491, 608–612. 10.1038/nature11536.Mitochondrial [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamimuzzaman M., Le Tourneau J. J., Unni D. R., Diesh C. M., Triant D. A., Walsh A. T., et al. (2020). Bovine genome database: New annotation tools for a new reference genome. Nucleic Acids Res. 48, D676–D681. 10.1093/nar/gkz944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda L. A., Polak J. (2011). Hypoxia. 4. Hypoxia and ion channel function. Am. J. Physiol. Cell Physiol. 300, 951–967. 10.1152/ajpcell.00512.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenmann C., Cathomen A., Hug M., Keiser S., Lundby A. K., Hilty M. P., et al. (2015). Hemoglobin mass and intravascular volume kinetics during and after exposure to 3, 454-m altitude. J. Appl. Physiol. 119, 1194–1201. 10.1152/japplphysiol.01121.2014 [DOI] [PubMed] [Google Scholar]
- Simonson T. S. (2015). Altitude adaptation: A glimpse through various lenses. High. Alt. Med. Biol. 16, 125–137. 10.1089/ham.2015.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F. (2007). Hemoglobin function and physiological adaptation to hypoxia in high-altitude mammals. J. Mammal. 88, 24–31. 10.1644/06-MAMM-S-199R1.1 [DOI] [Google Scholar]
- Storz J. F. (2016). Hemoglobin–oxygen affinity in high-altitude vertebrates: Is there evidence for an adaptive trend? J. Exp. Biol. 219, 3190–3203. 10.1242/jeb.127134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F. (2021). High-altitude adaptation: Mechanistic insights from integrated genomics and physiology. Mol. Biol. Evol. 38, 2677–2691. 10.1093/molbev/msab064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F., Moriyama H. (2008). Mechanisms of hemoglobin adaptation to high altitude hypoxia. High. Alt. Med. Biol. 9, 148–157. 10.1089/ham.2007.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J. F., Scott G. R., Cheviron Z. A. (2010). Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 213, 4125–4136. 10.1242/jeb.048181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunny D. E., Hammer E., Strempel S., Joseph C., Manchanda H., Ittermann T., et al. (2020). Nup133 and ERα mediate the differential effects of hyperoxia-induced damage in male and female OPCs. Mol. Cell. Pediatr. 7, 10. 10.1186/s40348-020-00102-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré A. (2019). Importance of slc26 transmembrane anion exchangers in sperm post-testicular maturation and fertilization potential. Front. Cell Dev. Biol. 7, 230. 10.3389/fcell.2019.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde G., Zhou H., Lippold S., De Filippo C., Tang K., Herráez D. L., et al. (2015). A novel candidate region for genetic adaptation to high altitude in Andean populations. PLoS One 10, 01254444–e125522. 10.1371/journal.pone.0125444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsiou A. I., Bazin E., Gaggiotti O. E. (2016). Detection of selective sweeps in structured populations: A comparison of recent methods. Mol. Ecol. 25, 89–103. 10.1111/mec.13360 [DOI] [PubMed] [Google Scholar]
- Vaysse A., Ratnakumar A., Derrien T., Axelsson E., Pielberg G. R., Sigurdsson S., et al. (2011). Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 7, e1002316. 10.1371/journal.pgen.1002316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith C., Schermuly R. T., Brandes R. P., Weissmann N. (2016). Molecular mechanisms of hypoxia-inducible factor-induced pulmonary arterial smooth muscle cell alterations in pulmonary hypertension. J. Physiol. 594, 1167–1177. 10.1113/JP270689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P., Sharma A., Sodhi M., Thakur K., Bharti V. K., Kumar P., et al. (2018). Overexpression of genes associated with hypoxia in cattle adapted to Trans Himalayan region of Ladakh. Cell Biol. Int. 9999, 1141–1148. 10.1002/cbin.10981 [DOI] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K. (2006). A map of recent positive selection in the human genome. PLoS Biol. 4, e72–e0458. 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Yang Y., Wang L., Ma T., Shang H., Ding L., et al. (2016). Different gene expressions between cattle and yak provide insights into high-altitude adaptation. Anim. Genet. 47, 28–35. 10.1111/age.12377 [DOI] [PubMed] [Google Scholar]
- Wang M. D., Dzama K., Rees D. J. G., Muchadeyi F. C. (2015). Tropically adapted cattle of Africa: Perspectives on potential role of copy number variations. Anim. Genet. 47, 154–164. 10.1111/age.12391 [DOI] [PubMed] [Google Scholar]
- Wei C., Wang H., Liu G., Zhao F., Kijas J. W., Ma Y., et al. (2016). Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci. Rep. 6, 26770–26811. 10.1038/srep26770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir E. K., Olschewski A. (2006). Role of ion channels in acute and chronic responses of the pulmonary vasculature to hypoxia. Cardiovasc. Res. 71, 630–641. 10.1016/j.cardiores.2006.04.014 [DOI] [PubMed] [Google Scholar]
- Werhahn G., Senn H., Ghazali M., Karmacharya D., Sherchan A. M., Joshi J., et al. (2018). The unique genetic adaptation of the Himalayan wolf to high-altitudes and consequences for conservation. Glob. Ecol. Conserv. 16, e00455. 10.1016/J.GECCO.2018.E00455 [DOI] [Google Scholar]
- Wiener P., Robert C., Ahbara A., Salavati M., Abebe A., Kebede A., et al. (2021). Whole-genome sequence data suggest environmental adaptation of Ethiopian sheep populations. Genome Biol. Evol. 13, 1–18. 10.1093/gbe/evab014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt K. E., Huerta-Sánchez E. (2019). Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180235. 10.1098/rstb.2018.0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuletaw Z., Wurzinger M., Holt T., Dessie T., Sölkner J. (2011). Assessment of physiological adaptation of indigenous and crossbred cattle to hypoxic environment in Ethiopia. Livest. Sci. 138, 96–104. 10.1016/j.livsci.2010.12.005 [DOI] [Google Scholar]
- Yalcin O., Cabrales P. (2012). Increased hemoglobin O2 affinity protects during acute hypoxia. Am. J. Physiol. Heart Circ. Physiol. 303, H271–H281. 10.1152/ajpheart.00078.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Arimura H., Fukushige T., Minami K., Nishizawa Y., Tanimoto A., et al. (2014). Abcb10 role in heme biosynthesis in vivo : Abcb10 knockout in mice causes anemia with protoporphyrin IX and iron accumulation. Mol. Cell. Biol. 34, 1077–1084. 10.1128/MCB.00865-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Jin Z.-B., Chen J., Huang X.-F., Li X.-M., Liang Y.-B., et al. (2017). Genetic signatures of high-altitude adaptation in Tibetans. Proc. Natl. Acad. Sci. U. S. A. 114, 4189–4194. 10.1073/pnas.1617042114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. L., Liang S. S. (2021). Study on pathogenic genes of dwarfism disease by next-generation sequencing. World J. Clin. Cases 9, 1600–1609. 10.12998/wjcc.v9.i7.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z. X. P., Pool J. E., et al. (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78. 10.1126/science.1190371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Niu W., Dong H. Y., Liu M. L., Luo Y., Li Z. C. (2018). Hypoxia induces endothelial-mesenchymal transition in pulmonary vascular remodeling. Int. J. Mol. Med. 42, 270–278. 10.3892/ijmm.2018.3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu C. X., Chamba Y., Ling Y. (2007). Blood characteristics for high altitude adaptation in Tibetan chickens. Poult. Sci. 86, 1384–1389. 10.1093/ps/86.7.1384 [DOI] [PubMed] [Google Scholar]
- Zhang W., Fan Z., Han E., Hou R., Zhang L., Galaverni M., et al. (2014). Hypoxia adaptations in the grey wolf (Canis lupus chanco) from qinghai-tibet plateau. PLoS Genet. 10, e1004466. 10.1371/journal.pgen.1004466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liang J., Zhang L., Wang Ligang, Liu X., Yan H., et al. (2015). Porcine methionine sulfoxide reductase B3: Molecular cloning, tissue-specific expression profiles, and polymorphisms associated with ear size in Sus scrofa . J. Anim. Sci. Biotechnol. 6, 60–69. 10.1186/s40104-015-0060-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M. P., Ma Y. C., Zhang C. C., Huang L. J., Zheng M. X., Li G. L., et al. (2017). The effects of ERK1/2 pathway on the expression of calcium activated chloride channel in hypoxia in PASMCs rat model. Zhongguo Ying Yong Sheng Li Xue Za Zhi 33, 47–50. 10.12047/j.cjap.5448.2017.011 [DOI] [PubMed] [Google Scholar]
- Zheng C., Lin X., Xu X., Wang C., Zhou J., Gao B., et al. (2019). Suppressing UPR-dependent overactivation of FGFR3 signaling ameliorates SLC26A2-deficient chondrodysplasias. EBioMedicine 40, 695–709. 10.1016/j.ebiom.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W. S., He Y. X., Cui C. Y., Ouzhu L., Deji Q., Peng Y., et al. (2017). EP300 contributes to high-altitude adaptation in Tibetans by regulating nitric oxide production. Zool. Res. 38, 163–170. 10.24272/j.issn.2095-8137.2017.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga-Hertz J. P., Patel H. H. (2019). The evolution of cholesterol-rich membrane in oxygen adaption: The respiratory system as a model. Front. Physiol. 10, 1340–1418. 10.3389/fphys.2019.01340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. The WGS data of Bale and Semien cattle samples are available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA698721, while the Choke cattle samples WGS data is available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA841948/.