Abstract
Typical enteropathogenic Escherichia coli (EPEC) strains produce bundle-forming pili (BFP), type IVB fimbriae that have been implicated in EPEC virulence, antigenicity, autoaggregation, and localized adherence to epithelial cells (LA). BFP are polymers of bundlin, a pilin protein that is encoded by the bfpA gene found on a large EPEC plasmid. Striking sequence variation has previously been observed among type IV pilin genes of other gram-negative bacterial pathogens (e.g., Pseudomonas and Neisseria spp.). In contrast, the established sequences of bfpA genes from two distantly related prototype EPEC strains vary by only a single base pair. To determine whether bundlin sequences vary more extensively, we used PCR to amplify the bfpA genes from 19 EPEC strains chosen for their various serotypes and sites and years of isolation. Eight different bfpA alleles were identified by sequencing of the PCR products. These alleles can be classified into two major groups. The α group contains three alleles derived from strains carrying O55, O86, O111, O119, O127, or O128 somatic antigens. The β group contains five alleles derived from strains carrying O55, O110, O128ab, O142, or nontypeable antigens. Sequence comparisons show that bundlin has highly conserved and variable regions, with most of the variation occurring in the C-terminal two-thirds of the protein. The results of multilocus enzyme electrophoresis support the hypothesis that bfpA sequences have spread horizontally across distantly related clonal lineages. Strains with divergent bundlin sequences express bundlin protein, produce BFP, and carry out autoaggregation and LA. However, four strains lack most or all of these phenotypes despite having an intact bfpA gene. These results have important implications for our understanding of bundlin structure, transmission of the bfp gene cluster among EPEC strains, and the role of bundlin variation in the evasion of host immune system responses.
Enteropathogenic Escherichia coli (EPEC) is one of several pathovars of E. coli capable of causing diarrhea (45). While human EPEC infections, which are manifest primarily in infants, were once commonplace in industrialized nations (56), they are now identified primarily in developing countries (10). EPEC strains possess distinct virulence factors not found in most E. coli strains (45). Typical EPEC strains produce bundle-forming pili (BFP), long, flexible, rope-like structures composed of intertwining fibers (19). Based on protein sequence analysis and morphology, BFP belong to the type IV group of fimbriae or pili found on a variety of gram-negative bacteria, many of which are human, animal, or plant pathogens (67, 70, 77). BFP have recently been shown to elicit an antibody response in natural infections (37, 38, 53) and a modest response in experimentally infected adults (13). BFP are required for two phenotypes of EPEC that can be studied in vitro and probably play a role in colonization in vivo. The first is autoaggregation, which is the ability of EPEC to reversibly form multicellular clusters in liquid culture (5, 76). The second is localized adherence (LA), which is the capacity of EPEC to form defined multicellular clusters (microcolonies) upon epithelial cells (7, 59) and upon human intestinal tissue (26). Both of these phenomena seem to be manifestations of the ability of BFP to intertwine, creating a fibrous network connecting individual bacteria. BFP also appear to adhere directly to epithelial cells (19, 31), although no BFP receptor on epithelial cells has been identified. During the course of tissue culture infection, BFP bundles undergo a transformation to a longer and thicker form that is correlated with dispersal of bacteria from microcolonies (31). In a recent clinical study, volunteers fed mutant EPEC strains lacking BFP developed diarrhea at a much lower frequency and of less severity than did volunteers receiving equivalent doses of an isogenic strain producing BFP (5). Thus, BFP are dynamic fimbriae that contribute to EPEC pathogenesis.
BFP biogenesis is specified by a cluster of 14 bfp genes (65, 66) located on large (∼50- to 70-MDa) EPEC adherence factor (EAF) plasmids (3, 47, 48). The first gene of this cluster is bfpA, which encodes bundlin, a pilin protein that is the only known structural component of BFP. Like other type IV pilins, bundlin is synthesized with a leader peptide removed by a prepilin peptidase, has a hydrophobic region at its mature N terminus, and has two cysteine residues forming a disulfide bond near the C terminus. Bundlin is membrane associated (2, 76). It seems likely that the ultimate function of most of the bfp gene products is to remove bundlin subunits from the bacterial membrane and assemble them into pili (2).
Little is known about the structure of bundlin monomers or how they assemble into BFP. However, some information on these topics can be inferred from studies of other type IV pili. The crystal structures of type IV pilin monomers from Neisseria gonorrhoeae and Pseudomonas aeruginosa have been solved (15, 25, 52). These pilins are ladle shaped with a long N-terminal α-helical handle and a globular head. Models of the way in which pilin monomers pack together into the pilus fiber have been proposed for the gonococcal and Pseudomonas pili (16, 25, 52), as well as the toxin-coregulated pili (TCP) of Vibrio cholerae (R. Chattopadhyaya and A. C. Ghose, unpublished data; Protein Data Bank accession number 1QQZ).
Extensive sequence variability has been observed in the type IV pilins of Dichelobacter nodosus, Eikenella corrodens, Moraxella bovis, P. aeruginosa (reviewed in reference 70), N. gonorrhoeae (52), N. meningitidis (1), and Vibrio cholerae (27, 43, 50, 55). In some of these species, pilin variability is clearly linked to changes in antigenicity. The host immune response may have played a major role in selecting for pilin variants with altered sequences. In the N. gonorrhoeae genome, extra copies of partial pilin genes are also present. These normally silent copies can be exchanged with the active copy via recombination, leading to antigenic variation that can aid the gonococcus in evading the host immune response (reviewed in references 41 and 62). A comparison of many gonococcal pilin gene products shows that extensive sequence variation occurs in amino acid residues of the “head” region but hardly any occurs in residues of the “handle” region (52). A hypervariable region of gonococcal pilin occurs between the disulfide-bonded cysteine residues. This region, as well as a disaccharide moiety and the extreme C terminus of the pilin, is predicted to be exposed on the outside of the pilus fiber (16, 52). In P. aeruginosa pilins, the disulfide region is also variable in sequence and serves as a binding domain for cell surface receptors (reviewed in reference 23).
In contrast to the considerable variation noted in other type IV pilins, the bundlins encoded by the two known bfpA alleles contain only a single amino acid difference (11, 64). Bundlin proteins from diverse EPEC strains might be expected to exhibit more significant sequence variation than has previously been noted. The current study was undertaken to assess the level of sequence diversity in bundlin proteins among EPEC strains and to gain insight into structural and functional constraints on different regions of the molecule.
MATERIALS AND METHODS
EPEC strains.
The origins of the EPEC strains examined in this study are listed in Table 1. To analyze the motility of each strain, a loopful of culture was stabbed into a motility agar plate (1% Bacto-Tryptone, 0.5% NaCl, 0.35% agar) and examined after overnight incubation at 37°C. Based on this analysis, strains lacking H serotype information were designated H+ (motile) or NM (nonmotile) (Table 1).
TABLE 1.
EPEC strains used in this study
| Strain | Serotype | Isolation:
|
Reference(s) | Source | ||
|---|---|---|---|---|---|---|
| Yr | Location | Circumstances | ||||
| 10 | O119:H+ | 1986 | Santiago, Chile | Health center-based study | 36, 46 | CVDa |
| 009-271082 | O111:NM | 1982 | Lima, Peru | Infant cohort study | 44 | CVD |
| 010-311082 | O128:H+ | 1982 | Lima, Peru | Infant cohort study | 44 | CVD |
| 012-050982 | O142:H+ | 1982 | Lima, Peru | Infant cohort study | 44 | CVD |
| 0659-79 | O119:H6 | 1979 | Florida | Infant diarrhea | 18, 20, 35, 47 | CDCb |
| 2309-77 | O111ab:H2 | 1977 | California | Infant stool | 47 | CDC |
| Beta | O55:H6 | 1947 | Aberdeen, Scotland | Infant diarrhea outbreak | 17, 57 | R. M. Robins-Browne |
| C771 | O142:H6 | Pre-1960 | Djakarta, Indonesia | Infant diarrhea | 51, 57 | R. M. Robins-Browne |
| CA89-4221 | NTc:H+ | 1989 | Montreal, Quebec, Canada | Canine diarrhea | 4, 14 | J. Harel |
| DIF043256 | O111:H2 | 1986 | Morelos, Mexico | Acute infant diarrhea | 9 | A. Cravioto |
| E56/54 | O128ab:H2 | 1953 | England | Infant diarrhea outbreak | 57, 69 | R. M. Robins-Browne |
| E851/71 | O142:H6 | 1971 | Glasgow, Scotland | Infant diarrhea outbreak | 7, 29, 34, 35, 47 | B. Rowe |
| E990 | O86:NM | 1950 | England | Infant diarrhea outbreak | 57, 68 | R. M. Robins-Browne |
| E2348/69 | O127:H6 | 1969 | Taunton, England | Infant diarrhea outbreak | 7, 11, 34, 35, 47 | CVD |
| HSP19/4 | O86:H+ | 1995 | São Paulo, Brazil | Acute infant diarrhea | I. C. A. Scaletsky | |
| MA551/1 | O55:NM | 1998 | São Luiz, Brazil | Acute infant diarrhea | I. C. A. Scaletsky | |
| RN191/1 | O111:H+ | 1996 | Natal, Brazil | Acute infant diarrhea | I. C. A. Scaletsky | |
| RN410/1 | O119:H+ | 1998 | Natal, Brazil | Pediatric control | I. C. A. Scaletsky | |
| Stoke W | O111ab:NM | 1947 | Aberdeen, Scotland | Pediatric control | 17, 57 | R. M. Robins-Browne |
| Z188-93 | 0110:H6 | 1993 | Germany | Avian diarrhea | 60 | J. E. Lohr |
CVD, Center for Vaccine Development, University of Maryland School of Medicine, Baltimore.
CDC, Centers for Disease Control and Prevention, Atlanta, Ga.
NT, nontypeable.
Isolation and sequencing of bfpA genes.
Partial and complete bfpA genes were amplified by PCR using DeepVent DNA polymerase in accordance with the manufacturer's (New England Biolabs) instructions. EPEC colonies were picked from Luria agar plates and boiled for 10 min in ThermoPol buffer (New England Biolabs) containing 2 to 6 mM MgSO4. DeepVent DNA polymerase (2 U), oligonucleotide primers (0.35 to 1.2 μM), and deoxynucleoside triphosphates (400 μM each) were added to bring the total reaction volume to 100 μl before beginning the PCR. Primer pairs used for PCR were Donne-28 and -29, -362 and -363, -423 and -363, -6 and -382, and -423 and -382 (Table 2). The precise reaction conditions varied with the strain and primers used. The PCR products were examined by agarose gel electrophoresis. If the PCR mixture contained a single DNA species of the expected size, it was purified directly using the Wizard PCR Preps DNA Purification System (Promega). If additional DNA species were present, the desired PCR product was isolated from a preparative agarose gel. Automated DNA sequencing of PCR products was carried out by the staff of The Biopolymer Laboratory (University Of Maryland School of Medicine) using two or more of the primers listed in Table 2. For some strains, the entire double-stranded sequence of the bfpA gene could be obtained using only primers Donne-423 and Donne-363.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequencea | Annealing sitea |
|---|---|---|
| Coding strand | ||
| Donne-6 | 5′-TCTTGGTGCTTGCGTGTC-3′ | 19–208 |
| Donne-28 | 5′-CGCGGATCCATGGTTTCTAAAATCATGAAT-3′ | 274–294 |
| Donne-362 | 5′-AGGTCTGTCTTTGATTGAA-3′ | 309–327 |
| Donne-423 | 5′-GATTATTCCGTGACCTATT-3′ | 223–241 |
| Noncoding strand | ||
| Donne-26 | 5′-GTAAGCGTCAGATAGTAACCAA-3′ | 638–617 |
| Donne-29 | 5′-GCGAAGCTTTTACTTCATAAAATATGTAAC-3′ | 855–835 |
| Donne-363 | 5′-CCTGAGTAAAACAGGATAC-3′ | 953–935 |
| Donne-382 | 5′-TCCTTCGGGTGATTGTGTA-3′ | 1300–1282 |
| Donne-416 | 5′-GTTGCGCTCATTACTTCT-3′ | 443–426 |
The underlined portion of each primer corresponds to the annealing site indicated within the bfp cluster nucleotide sequence (EMBL accession number Z68186).
Sequence analysis.
DNASIS (Hitachi Software) was used to align bfpA nucleotide sequences from each strain and determine a consensus sequence. Multiple sequence alignment was performed on the bfpA gene sequences and the deduced bundlin amino acid sequences using DNASIS and CLUSTAL W (71). Gene trees were constructed with the computer program MEGA (Molecular Evolutionary Genetics Analysis, version 1.0; S. Kumar, K. Tamura, and M. Nei, Institute of Molecular Evolutionary Genetics, The Pennsylvania State University, 1993 [http://www.bio.psu.edu/People/Faculty/Nei/Lab/megaform.txt]). The proportions of polymorphic synonymous (pS) and nonsynonymous (pN) sites were calculated by the method of Nei and Gojobori (49). To examine variation in the functional constraints of different parts of the molecule, these statistics were tabulated in a sliding-window analysis of 30 codons along the gene by the program PSWIN. The theoretical three-dimensional structure of bundlin was analyzed and colored using RASMOL (58) (http://www.umass.edu/microbio/rasmol/).
MLEE.
Every strain was characterized by its profile of electromorphs for 20 enzymes by multilocus enzyme electrophoresis (MLEE) (63, 78, 79). Genetic relationships between strains were determined based on a matrix of genetic distances between all pairs constructed by comparison of the allelic arrays. A dendrogram was constructed with MEGA using the neighbor-joining algorithm.
Immunoblotting.
Overnight cultures of EPEC in LB were diluted 1:100 into 20 ml of Dulbecco's modified Eagle medium–nutrient mixture F-12 (DMEM–F-12) containing 15 mM HEPES buffer and lacking phenol red (Gibco-BRL Life Technologies catalog no. 11039-021). After 6 h of growth at 37°C with shaking, the bacteria were pelleted by centrifugation at 2,500 × g for 5 min and then resuspended in 1.2 ml of cell lysis buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 0.1 mM EDTA, 0.1% Triton X-100). The optical density at 600 nm of a portion of these samples was measured on a spectrophotometer. Aliquots (75 μl or less) of these samples were mixed with 25 μl of 4× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer (2) and then adjusted to a total volume of 100 μl with additional cell lysis buffer such that the resulting gel samples were derived from roughly equivalent densities of bacteria. The gel samples were boiled for 10 min, and then 5 μl of each was loaded per lane of a sodium dodecyl sulfate–15% polyacrylamide gel. After electrophoretic separation, the protein in the gel was electrotransferred to a polyvinylidene difluoride membrane. The membrane was blocked with phosphate-buffered saline containing 0.1% Tween 20 plus 5% nonfat dry milk and then incubated sequentially with a polyclonal antibody (1:5,000 dilution) directed against six-His-tagged prebundlin (81), an anti-rabbit horseradish peroxidase conjugate (1:30,000 dilution), and ECL Western blotting detection reagents (Amersham Pharmacia Biotech).
Transmission electron microscopy.
To prepare samples for electron microscopy, 1-ml aliquots were removed from EPEC cultures grown in DMEM–F-12 for 5 h and the bacteria were pelleted by brief centrifugation. Most of the medium was decanted, and the bacterial pellet was gently resuspended in the remainder. Aliquots (10 μl) were spotted onto electron microscopy grids, which were dried for 10 min and then washed, stained, and examined for BFP as described previously (66).
Autoaggregation assay.
EPEC strains were cultured overnight at 37°C in LB. The resulting stationary-phase cultures were diluted 1:250 (making appropriate adjustments for optical density at 600 nm) into 20 ml of DMEM–F-12. These cultures were shaken at 250 rpm for 5 h in 50-ml conical polypropylene tubes at 37°C. Autoaggregation was initially gauged by visually inspecting the cultures for bacterial aggregates and sedimentation. For some strains, it was necessary to use light microscopy in order to detect aggregates. Culture aliquots (5 μl) were examined in hanging-drop slides at ×40 magnification.
LA assay.
LA to HeLa epithelial cells was assayed as described previously (12) using the eight-well chamber slide modification.
Nucleotide sequence accession numbers.
The nucleotide sequences of the bfpA genes isolated from all of the strains listed in Table 1 (except prototype strain E2348/69) have been deposited in GenBank under accession numbers AF304468 through AF304486.
RESULTS
Identification of novel bfpA alleles.
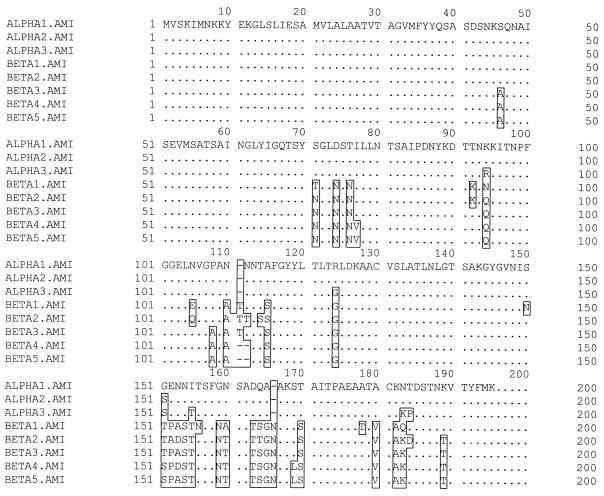
The goal of this study was to assess the amount of sequence variation among bfpA genes found in EPEC strains. To this end, we initially analyzed 13 strains from our laboratory collection (a subset of those listed in Table 1) representing a variety of serotypes and well-documented outbreaks (or sporadic cases) of human diarrhea. These strains were originally isolated from diverse geographical locations between 1947 and 1986. They belong to classical EPEC O serogroups O55, O86, O111, O119, O127, O128, and O142. Three of them (C771, E990, and Stoke W) are type strains for the preparation of O antisera. Canine EPEC strain CA89-4221 and avian EPEC strain Z188-93, which are both probe positive for the bfpA gene, were also examined. PCR was used to amplify the bfpA gene from each EPEC strain. The DNA sequences of both strands of the PCR products were determined, and the resulting bfpA coding sequences were compared to each other and to the known bfpA sequences from prototypic strains E2348/69 and B171 (GenBank accession numbers Z12295 and U27184). Eight different bfpA alleles were identified among the 15 strains. Six of these alleles have not been previously described. Each bfpA allele specifies a prebundlin protein carrying a variant amino acid at one or more positions (Fig. 1).
FIG. 1.
Variable amino acids are clustered in prebundlin. An alignment of the prebundlin amino acid sequences encoded by the eight bfpA alleles described in this study is shown. Note that amino acids 1 to 13 comprise the cleaved leader peptide. The α1 (ALPHA1.AMI) prebundlin prototype sequence is listed on the top line. In the remaining seven sequences (ALPHA2.AMI through ALPHA3.AMI and BETA1.AMI through BETA5.AMI), invariant amino acids are indicated by dots. Variant amino acids are boxed and indicated by single-letter abbreviations.
Genetic relationships among bfpA alleles and correspondence to O serogroups.
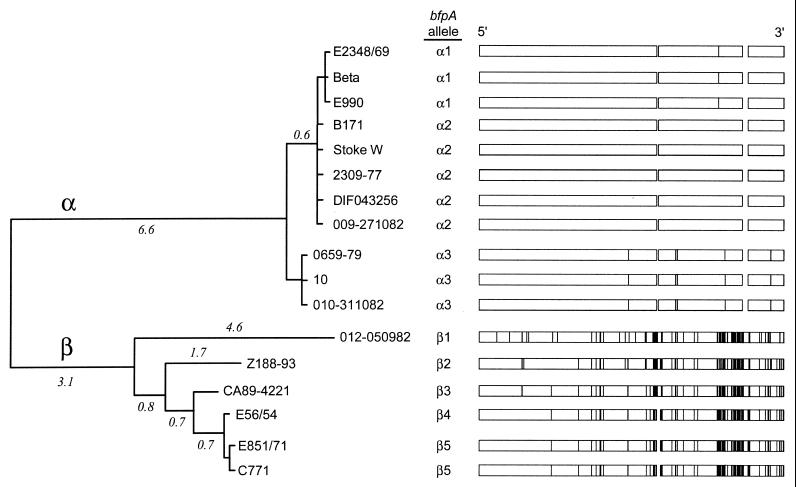
Sequence comparisons of the bfpA alleles show that they fall into two distinct clonal groups, which we term bfpA alpha (α) and bfpA beta (β) (Fig. 2 and Table 3). The three α alleles show polymorphism at only 7 (1.2%) of 579 nucleotides. The α prebundlins differ at only 6 (3.1%) of 193 amino acid residues. The previously reported α1 allele is found in prototype O127 strain E2348/69 (11), as well as in the single O86 and O55 strains initially examined in the current study. The previously reported α2 allele is carried by prototype O111 strain B171 (64) and each of the other O111 strains examined. The α1 allele varies from α2 at a single nucleotide, resulting in an amino acid substitution. The α3 allele of bfpA is found in both of the O119 strains but in only one of the two O128 strains we initially examined. This allele varies from α1 and α2 at six nucleotides located in the 3′ half of the gene. Five of these six nucleotide changes specify altered amino acids relative to α2 prebundlin. Three of the sequence changes are identical to those found throughout the β group, suggesting a closer relationship to a common α/β ancestor.
FIG. 2.
Eight bfpA alleles cluster into two major groups. To the left is a phylogenetic tree for the bfpA gene from 17 EPEC strains constructed by the neighbor-joining algorithm based on the gamma distance with α = 2. Branch lengths in terms of nucleotide substitutions per 100 sites are given above the major branches. Allele designations are listed for each strain. To the right is a graph of the locations of polymorphic nucleotide sites, which are marked as vertical lines that indicate nucleotides differing from those of the α2 allele at each position. Gaps indicate codons absent from specific alleles.
TABLE 3.
Expression of BFP-related phenotypes by EPEC strains
| bfpA allele | Strain | Serotype | Bundlina | BFPb | LAc | Autoaggregationd |
|---|---|---|---|---|---|---|
| α1 | Beta | O55:H6 | + | NDe | + | + |
| α1 | E990 | O86:NM | + | + | + | + |
| α1 | E2348/69 | O127:H6 | + | + | + | + |
| α1 | HSP19/4 | O86:H+ | + | ND | + | + |
| α2 | 009-271082 | O111:NM | − | − | − | − |
| α2 | 2309-77 | O111ab:H2 | − | − | − | − |
| α2 | DIF043256 | O111:H2 | + | ND | + | + |
| α2 | Stoke W | O111ab:NM | + | ND | + | + |
| α2 | RN191/1 | O111:H+ | + | ND | + | + |
| α3 | 10 | O119:H+ | + | ND | + | + |
| α3 | 0659-79 | O119:H6 | + | ND | + | + |
| α3 | 010-311082 | O128:H+ | + | + | + | + |
| α3 | RN410/1 | O119:H+ | + | ND | + | + |
| β1 | 012-050982 | O142:H+ | + | + | + | + |
| β2 | Z188-93 | O110:H6 | + | ND | + | + |
| β3 | CA89-4221 | NTf:H+ | + | + | + | + |
| β4 | E56/54 | O128ab:H2 | − | − | + | − |
| β5 | C771 | O142:H6 | − | − | + | − |
| β5 | E851/71 | O142:H6 | + | + | + | + |
| β5 | MA551/1 | O55:NM | + | ND | + | + |
Antigenic detection of bundlin protein by immunoblotting. +, presence of bundlin; −, absence of bundlin.
Visual detection of BFP by electron microscopy. +, pili present; −, pili absent.
LA of EPEC to cultured epithelial cells. +, presence of LA; −, absence of LA.
Autoaggregation of EPEC in tissue culture medium. +, presence of autoaggregation; −, absence of autoaggregation.
ND, not determined.
NT, nontypeable.
Among the human-derived strains, β alleles were identified in each of the three strains of the O142 serogroup and in one of the two strains of the O128 serogroup. Unique β alleles were also found in the avian and canine EPEC strains. The five bfpA β alleles are more diverse in sequence than the α alleles, with the exception of β4 and β5, which differ from one another by only one nucleotide. At the nucleotide level, β alleles are polymorphic at 51 (8.7%) of 585 nucleotides. The β prebundlins vary at 22 (11.3%) of 195 amino acids. Relative to α alleles, β alleles of bfpA carry either a net insertion (β1/β2/β3) or a net deletion (β4/β5) of one codon directly after codon 110 plus a one-codon insertion between codons 164 and 165 (Fig. 1 and 2). When both α and β sequences are considered, bfpA exhibits polymorphism in 94 (16.1%) of 585 nucleotides and prebundlin varies at 36 (18.5%) of 195 amino acids.
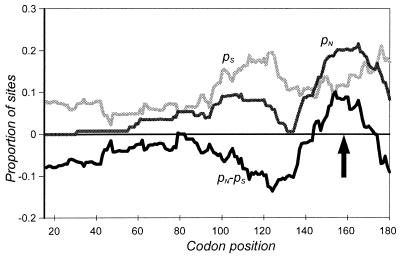
Evidence of selection for sequence diversification near the 3′ end of the bfpA gene.
To determine how the level of selective constraint varies along the length of the bfpA gene, the proportions of nonsynonymous and synonymous codon changes (pN and pS, respectively) were calculated within a 30-codon sliding window (Fig. 3). Nonsynonymous codon changes result in amino acid substitution; synonymous codon changes do not. The difference, pN − pS, is a measure of the degree of selective constraint. The zero-difference line indicates selectively neutral variation, where pS and pN are equal. The more positive the value, the less the contribution of synonymous substitutions and the greater the contribution of replacement substitutions. A positive difference in a particular region suggests that some selective pressure is driving diversification in that region, whereas a negative difference indicates selection against mutations resulting in amino acid variation. The results shown in Fig. 3 suggest that most bundlin is under selective constraint. However, a single region of bfpA, located between codons 143 and 174, exhibits a positive value of pN − pS. This region corresponds to amino acids 130 through 161 of mature bundlin, which are located between the disulfide-bonded cysteines. An attractive possibility is that this region of bundlin is located on the surface of the BFP filament and that the host immune system has selected for antigenic diversity at this site.
FIG. 3.
Evidence for diversifying selection near the 3′ end of the bfpA gene. A sliding-window plot of the proportions of synonymous and nonsynonymous sites (pS and pN, respectively) that have mutated among eight bfpA alleles is shown. The difference (pN − pS) is a measure of the level of selective constraint on various parts of the molecule. Note that pN − pS > 0 for codons 143 to 174 (indicated by the arrow), a pattern consistent with the effect of diversifying selection.
Genetic relationships between EPEC strains.
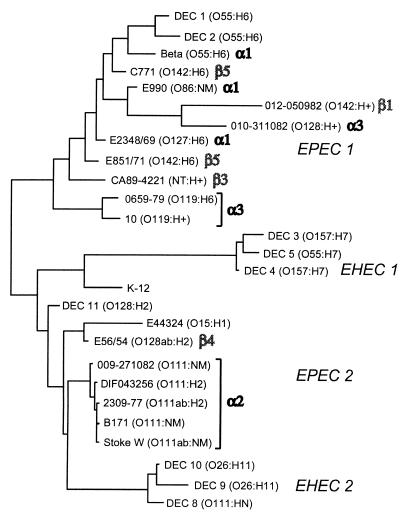
MLEE was carried out to define the overall phylogenetic relationships between selected EPEC strains. A dendrogram resulting from this analysis (Fig. 4) shows that all of the α2 alleles are found in a closely related cluster of O111 strains. Among the remaining EPEC strains, however, there is a curious lack of correspondence between the deduced clonal pattern of the strains and the pattern of bfpA alleles they possess. Alleles of both the α and β types are found among strains in either of the two major clonal EPEC groups, EPEC 1 and EPEC 2 (54, 78). In the EPEC 1 group, the α1, α3, β1, β3, and β5 alleles are interspersed among each other in a way that obviously does not correlate with the overall strain lineage. These results strongly suggest that the bfpA gene has been recently transferred between EPEC strains through multiple horizontal transfer events.
FIG. 4.
The alleles of bfpA identified in EPEC strains do not correlate precisely with the overall clonal lineage. Shown is a dendrogram of genetic relationships of EPEC strains from this study and representative DEC strains (79). The genetic distance was estimated in terms of electrophoretically detectable codon differences per enzyme locus for 20 enzymes. Previously identified DEC clusters (EPEC 1 and 2 and EHEC 1 and 2) are indicated (54, 78). Strain serotypes are in parentheses. Alleles of bfpA identified in specific EPEC strains are displayed in large, shaded type.
Examination of bfpA alleles in recent EPEC isolates.
To determine whether bfpA α alleles have remained stable over recent decades, we examined a set of four EAF+ EPEC strains of very recent Brazilian origin (Table 1). The bfpA alleles derived from these strains were isolated by PCR and sequenced. As expected from their serogroups, the O86, O111, and O119 strains (HSP19/4, RN191/1, and RN410/1) possessed bfpA genes of the α1, α2, and α3 varieties, respectively. These results indicate that bfpA α sequences have been retained unchanged in current EPEC strains. The O55 strain (MA551/1), however, did not carry the α1 allele found in the previously examined O55 EPEC (strain β). Rather, it carried a β5 allele identical to that carried by O142 strains C771 and E851/71. This unexpected finding provides a further suggestion of the horizontal spread of a particular bfpA allele.
Expression and function of BFP variants.
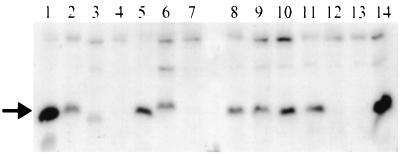
Selected EPEC strains were examined to determine whether they produced detectable bundlin and BFP and whether they exhibited the BFP-dependent phenotypes of autoaggregation and LA. Some of these characteristics have been previously reported for a subset of these strains (see references in Table 1). Prototype EPEC strain E2348/69 served as a positive control for all of our studies. To identify bundlin, a polyclonal antiserum was used to probe whole-cell extracts from each of the EPEC strains (Fig. 5 and Table 3). Bundlin protein was readily detected in extracts from 10 out of 12 strains tested carrying α alleles of bfpA and 5 of 7 strains carrying β alleles. Bundlin was not identified in strain 009-271082 or 2309-77 carrying the α2 allele or in strain E56/54 or C771 carrying the β4 or β5 allele. Particular β bundlins migrated more slowly (β1 and β2) or more quickly (β5) than did α bundlins (Fig. 5 and data not shown). The relative migration correlates well with the lengths (180 amino acids for α1 and β5 and 182 amino acids for β1 and β2) and molecular masses (19,986 Da for β5, 20,269 Da for α1, 20,306 Da for β2, and 20,328 Da for β1) predicted for these bundlin types. Such a migration anomaly has been noted for bundlin from other strains in previous studies (18, 20, 38).
FIG. 5.
Immunoblot of bundlin protein from EPEC whole-cell extracts. The arrow indicates the position of bundlin. The extracts are from the following strains: lane 1, E2348/69; lane 2, 0659-79; lane 3, E851/71; lane 4, 2309-77; lane 5, 10; lane 6, 012-050982; lane 7, 009-271082; lane 8, 010-311082; lane 9, DIF043256; lane 10, β; lane 11, Stoke W; lane 12, E56/54; lane 13, C771; lane 14, E990. The remaining strains exhibited detectable bundlin but are not shown here.
Ten of the strains were examined by electron microscopy for the presence of BFP fimbriae. Fimbrial bundles were readily detected in six strains (Table 3). No BFP was seen in samples of the remaining four strains, which had also failed to express detectable bundlin. Noticeably more BFP was produced by α1 strains E990 and E2348/69 than the remaining strains. All of the strains were tested for LA and autoaggregation (Table 3). The α2 allele strains that failed to produce both bundlin and BFP (009-271082 and 2309-77) exhibited neither autoaggregation nor LA. The β allele strains E56/54 and C771, which failed to produce both bundlin and BFP, also did not exhibit autoaggregation. However, they did exhibit LA, as has been previously noted (57). All of the remaining strains exhibited both phenotypes. In the LA assay, no conspicuous differences were noted in the size, pattern, or relative numbers of adherent clusters produced by strains carrying different bfpA alleles. While some strains consistently produced aggregates in the autoaggregation assay that could be readily seen by the naked eye, the aggregates of other strains (e.g., E851/71) could be detected only by microscopy. To summarize, most strains expressed BFP that had normal structure and function regardless of the bfpA allele they carried. Four strains, however, were defective for BFP expression and function in most or all of the assays. In each of these cases, other strains carrying an identical or closely related bfpA allele produced functional BFP, suggesting that the allele sequence itself is not the cause of the defect.
DISCUSSION
In this study, we have identified eight alleles of the bfpA gene by surveying 19 EPEC strains of diverse origins. Previous studies using prototype EPEC strains E2348/69 and B171 had identified two of these alleles, α1 and α2, that differ by only a single nucleotide (11, 64). The eight bfpA alleles fall into two distinct groups based upon sequence analysis. By analogy to the intimins, we call these groups α and β. The α group, isolated from 12 of the EPEC strains examined here plus prototype strains B171 and E2348/69, contains three different alleles showing little sequence variation. The β group, represented by seven strains, exhibits more significant sequence variation, both internally and when compared to α alleles. We note that additional alleles of bfpA have recently been deposited in sequence databases. Bortolini and colleages have described a nonfunctional bfpA allele (GenBank accession number AF119170) that is prematurely truncated by an insertion sequence and is found in certain clonally related O128ab:H2 and O119:H2 EPEC strains (6). We did not identify this allele in our study; however, the residual 5′ bfpA sequence of this allele is identical to that of the β3 allele, with the exception of a single nucleotide. Incomplete bfpA sequences can be found in GenBank records with accession numbers AF233895 through AF233899 (S. Subpasu, M. Yamasaki, J. Yatsuyanagi, O.-A. Ratchatrachenchai, and K. Ito, unpublished data). One of these sequences appears to be from an α1 allele, but the remaining four differ from our α3, β1, and β4/β5 alleles at 1 to 10 nucleotide positions. These sequences support our division of bfpA alleles into two groups and attest that there is further sequence diversity to be identified in the bfpA gene. The existence of multiple bfpA alleles has important implications for the study of BFP structure and function, for our understanding of the evolution of EPEC as a pathogen, and for EPEC diagnosis and vaccine development as detailed below.
Implications for the structure of bundlin and BFP.
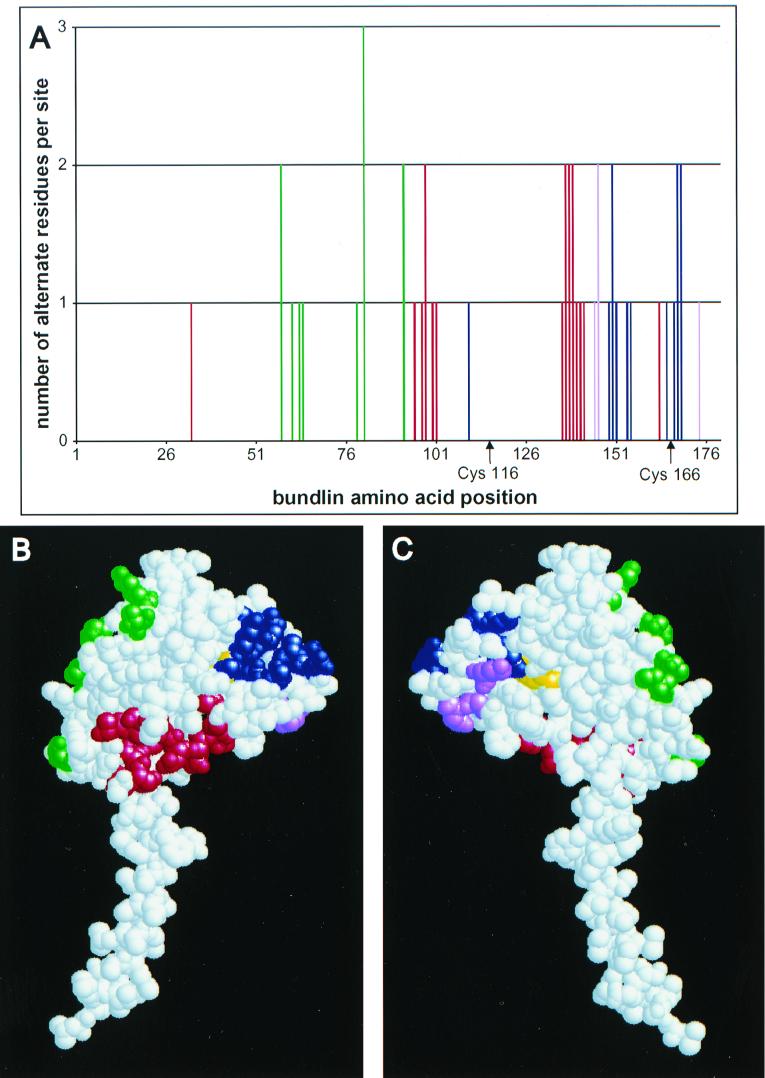
To better understand the structural significance of amino acid polymorphism in bundlin, we carried out a graphical analysis of variability in the sequence of the protein (Fig. 6A). We also mapped the variable residues onto a theoretical three-dimensional (3D) structure model of α1 bundlin (Fig. 6B and C) that has been recently deposited in the Protein Data Bank (http://www.rcsb.org/pdb) under accession number 1QT2 (R. Chattopadhyaya and A. C. Ghose, unpublished data). The variable amino acids of bundlin are generally clustered, within both the linear sequence and the theoretical 3D structure. Variable residues which group together in the 3D structure are sometimes derived from different segments of the linear bundlin sequence (Fig. 6). Most of the highly variable amino acids are located in the C-terminal third of bundlin. Many of the variable residues are located on the pilin surface in the theoretical 3D model. Variable residues 58, 61, 63 and 64, 79, 81, and 92 (numbered in accordance with the 180-residue mature α1 bundlin prototype) occur along one side of the pilin head (shown in green in Fig. 6B and C). Variable residues 33, 95, 97 and 98, 100 and 101, 136 to 142, and 163 (shown in red) are located in a cleft on the lower convex face of the pilin head. Residues 110, 149 to 151, 154 and 155, 165, and 167 to 169 (shown in blue) are found on one side of the upper convex face of the pilin head. Residues 145 and 146 and residue 174 (shown in violet) are located on the flattened reverse face of the pilin. One possibility is that these variable positions define regions of molecular interactions either between pilin monomers or between BFP filaments. A more appealing possibility is that these variable positions define regions on the surface of BFP that are directly exposed to the host immune system. Interestingly, other regions of bundlin show little or no variation. These include the cleaved leader peptide, the α-helical handle of the pilin, and five beta strands that probably serve to create the structure of the pilin head. Such regions may lack variation due to constraints inherent in critical structural domains.
FIG. 6.
Variable amino acids may be clustered on the surface of bundlin. (A) Graphical analysis of bundlin variation. The bars represent amino acid residues that vary among the eight bundlin types identified in this study. The heights of the bars are proportional to the numbers of alternate residues per site. The positions of conserved cysteine residues are indicated below the x axis. (B and C) Theoretical 3D structure of bundlin (Chattopadhyaya and Ghose, unpublished). Colored amino acid residues are those that vary between bundlin types. These residues have been assigned to four color groups (green, red, blue, and violet) in accordance with their relative locations in the molecule. These colors correspond to those displayed in the graph (A). The disulfide-bonded cysteines are yellow. Panel B shows the convex face of bundlin that is likely to be facing outward on the surface of the BPF filament. Panel C shows the flattened face on the opposite side of the molecule that is likely to be facing toward the center of the filament, based on existing models of gonococcal and V. cholerae pili.
Like other type IV pilins, bundlin contains two disulfide-bonded cysteine residues near its C terminus. These residues are completely conserved in all bundlin variants, consistent with the previous finding that replacement of either cysteine with serine greatly reduces bundlin stability (81). Replacement of the entire disulfide region of bundlin with the corresponding region of the V. cholerae TcpA pilin also leads to instability of the hybrid pilin (40). Studies of multiple type IV pili have shown the importance of variation in the disulfide region. The hypervariable disulfide region of the gonococcal pilin participates in antigenic variation, while that of the P. aeruginosa pilin is involved in host cell interaction. A detailed mutational analysis of the TcpA pilin indicates that an intact disulfide region is critical for both proper structure and function of TCP (30). Based on these findings and on the 3D model of Chattopadhyaya and Ghose, it was suggested that this region is surface exposed on TCP and defines surfaces of interaction both between pilin monomers and between individual pilus fibers within a bundle. Interestingly, there is minimal variation in a subregion of ∼20 to 25 residues surrounding the more N-terminal cysteine of both BfpA (Fig. 6A) and TcpA (43), corresponding in part to the structural domain defined by Kirn and colleagues (30). Furthermore, there is evidence of constraint against nonsynonymous substitutions in this subregion (Fig. 3), indicating selection against diversity. In contrast, much of the remaining disulfide region is highly variable in both pilins. In fact, our analysis (Fig. 3) suggests that it is under positive selection for diversification. Thus, the disulfide region of type IV pilins may be caught in a struggle to maintain structural integrity, as well as pilin-pilin and filament-filament interactions, while at the same time presenting a changing antigenic face to the host immune system.
Implications for the role of BFP in the evolution of EPEC as a pathogen.
EPEC strains are defined as diarrheagenic E. coli (DEC) strains producing attaching and effacing histopathology but lacking Shiga-like toxins (28). Typical EPEC strains also carry the EAF plasmid and adhere in a BFP-dependent localized fashion to cultured epithelial cells. The importance of the EAF plasmid and plasmid-encoded BFP in pathogenicity has been demonstrated in volunteer studies (5, 35). Further support is provided by some case-control studies, where EAF+, but not EAF−, strains have been significantly associated with diarrhea (reviewed in reference 28). As bundlin is a virulence factor and a potentially important antigen of EPEC, it is useful to understand whether bfpA alleles are capable of being mobilized between different clonal groups of EPEC strains and how this might take place.
The majority of EPEC strains form two clonal groups, EPEC 1 and EPEC 2 (54, 78), with EPEC 1 exhibiting greater strain diversity. The low G+C content of the 11.5-kb bfp gene cluster on the EAF plasmid suggests that it has been derived from a non-E. coli bacterium (72). The 14 bfp cluster genes from EPEC 1 strain E2348/69 (66) and EPEC 2 strain B171 (65) are more similar to each other in nucleotide sequence than would be expected from the distance between their clonal frames, based on MLEE analysis. Therefore, Whittam and McGraw (78) hypothesized that the EAF plasmid carrying the bfp cluster was acquired independently by EPEC 1 and then by EPEC 2 after the divergence of these two groups. The results of the current study indicate that the evolutionary picture is more complex. There has been a radiation of bundlin alleles into at least two major types, α and β, both of which appear to have an ancient origin. In addition, bfpA alleles appear to have been exchanged multiple times among divergent EPEC strains, presumably as a result of conjugation (see below). Domesticated animals are a potential reservoir of β alleles, as EPEC strains isolated from a pet dog and a pet bird were both found to carry unique β alleles in this study. The complicated intermingling of both α and β alleles of bfpA in EPEC 1 (Fig. 4) suggests that, in addition to mutation, bfpA sequences have diverged by multiple recombination, horizontal transfer, and positive-selection events. At least two transfers of bfpA into the EPEC 2 group have also taken place.
Since the bfpA gene is located on the EAF plasmid, the most likely mechanism for its movement between EPEC strains is plasmid transfer by conjugation. The available evidence seems to indicate that EAF plasmids from some EPEC strains are capable of being mobilized for conjugation while others are not (24, 32, 39). The EAF plasmid of O111 strain B171, which has been completely sequenced, appears to lack plasmid transfer genes (72). It does, however, contain many remnants of insertion elements, suggesting an alternative method of bfp cluster transfer via a transposon. The lack of transfer genes on the O111 EAF plasmid may explain why the α2 allele of bfpA was identified only among a closely related cluster of O111 strains.
An unexpected finding of this study was the identification of four strains that contained an intact bfpA gene yet did not produce detectable bundlin or BFP or exhibit autoaggregation. Two of the strains (E56/54 and C771) still exhibited LA, suggesting that they either produce sufficient amounts of BFP under the conditions of the adherence assay or that they produce an alternative adherence factor. In such strains, bfpA may be regulated differently than in prototype strains. Alternatively, expression of bundlin may have been abrogated during the course of the original infection or subsequently during multiple passages through various laboratories. It is possible that BFP is not strictly required for adherence to cells and infection of the host by some EPEC strains (6, 26).
Implications for EPEC diagnosis.
The presence of multiple bfpA alleles in nature requires a re-evaluation of the methods used to identify typical EPEC strains. Molecular diagnosis of bfpA+ EPEC has previously been carried out with hybridization probes consisting of restriction digest fragments, PCR products, or labeled oligonucleotides (4, 18, 42, 45, 64). PCR amplification has also been described for the identification of bfpA from E. coli (22, 74, 75, 80). The current results indicate that oligonucleotides for bfpA identification should be designed such that the annealing regions represent sequences conserved in all known bfpA alleles. Use of a poorly conserved oligonucleotide sequence might result in the incorrect designation of bonafide bfpA+ strains as BFP negative. Results from the functional studies we conducted, along with the findings of others (38), show the limitations of using probes or PCR for diagnosis—some strains carrying an intact bfpA gene may not actually produce readily detectable levels of bundlin or BFP. Therefore, accurate designation of an E. coli strain as BFP positive ultimately requires one or more tests of BFP expression or function. Our results have shown for the first time that diverse EPEC strains exhibit autoaggregation, a phenotype that is BFP dependent. While the autoaggregation assay is not yet in common use, it appears to be an acceptable and facile substitute for LA in those laboratories lacking tissue culture facilities but having a simple microscope.
Antigenic variation of bundlin and implications for vaccine development.
BFP has been proposed as a potential component of an EPEC vaccine (33, 61). This concept is supported by the detection of antibundlin antibodies in children with natural EPEC infections (38) and in healthy children and mothers living in areas where EPEC is endemic (37, 53). Further support is provided by the finding that antisera raised against BFP blocks LA (19). Studies of EPEC infections in adult volunteers have demonstrated a correlation between the level of antibundlin antibodies and the propensity for loss of the plasmid encoding BFP (13). Since the plasmid-cured strain is less pathogenic than the plasmid-containing strain (35), this association establishes a link between antibundlin antibodies and virulence. The results of the current study have a direct bearing on considerations for vaccine formulations. Strains of serogroups O55, O111, and O119 have the highest prevalence in areas where EPEC is endemic (8, 21, 36, 73). Our results, which demonstrate that O55 strains, at least, may possess either α or β bfpA alleles, suggest that successful vaccines may need to include both classes of antigen. Further studies are needed to test the hypotheses that immune responses discriminate between these classes of bundlin proteins and that the immune response is a significant factor driving the evolution of bfpA allelic variants of EPEC.
In summary, our analysis of 19 EPEC strains indicates that there is heretofore unappreciated allelic diversity in the bfpA gene. Although some strains with intact bfpA alleles failed to produce BFP, most of the strains produced BFP having readily detectable expression and function. The bfpA gene contains both variable and invariable regions, whose locations are likely to correlate with their role in bundlin and BFP filament structure. In particular, the most highly variable stretch of bundlin is located in a region of the molecule that is predicted to be surface exposed and under diversifying selection from a host immune response. This study additionally establishes evidence for the horizontal transfer of bfpA across diverse EPEC lineages, which likely contributed to the evolution of this important pathogen.
ACKNOWLEDGMENTS
We are grateful to each of the researchers who previously provided EPEC strains that were used in this study, including Alejandro Cravioto, Josée Harel, J. E. Lohr, Roy Robins-Browne, Bernard Rowe, Isabel Scaletsky, and Nancy Strockbine. We thank Jorge Girón for critical review of the manuscript.
This investigation was supported by a Public Health Service grant (AI37606) to M.S.D. and a National Research Service Award postdoctoral training fellowship (AI10191) to T.E.B.
REFERENCES
- 1.Aho E L, Botten J W, Hall R J, Larson M K, Ness J K. Characterization of a class II pilin expression locus from Neisseria meningitidis: evidence for increased diversity among pilin genes in pathogenic Neisseria species. Infect Immun. 1997;65:2613–2620. doi: 10.1128/iai.65.7.2613-2620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anantha R P, Stone K D, Donnenberg M S. Effects of bfp mutations on biogenesis of functional enteropathogenic Escherichia coli type IV pili. J Bacteriol. 2000;182:2498–2506. doi: 10.1128/jb.182.9.2498-2506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldini M M, Kaper J B, Levine M M, Candy D C, Moon H W. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J Pediatr Gastroenterol Nutr. 1983;2:534–538. doi: 10.1097/00005176-198302030-00023. [DOI] [PubMed] [Google Scholar]
- 4.Beaudry M, Zhu C, Fairbrother J M, Harel J. Genotypic and phenotypic characterization of Escherichia coli isolates from dogs manifesting attaching and effacing lesions. J Clin Microbiol. 1996;34:144–148. doi: 10.1128/jcm.34.1.144-148.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 6.Bortolini M R, Trabulsi L R, Keller R, Frankel G, Sperandio V. Lack of expression of bundle-forming pili in some clinical isolates of enteropathogenic Escherichia coli (EPEC) is due to a conserved large deletion in the bfp operon. FEMS Microbiol Lett. 1999;179:169–174. doi: 10.1111/j.1574-6968.1999.tb08723.x. [DOI] [PubMed] [Google Scholar]
- 7.Cravioto A, Gross R J, Scotland S M, Rowe B. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol. 1979;3:95–99. [Google Scholar]
- 8.Cravioto A, Reyes R E, Trujillo F, Uribe F, Navarro A, De La Roca J M, Hernández J M, Pérez G, Vázquez V. Risk of diarrhea during the first year of life associated with initial and subsequent colonization by specific enteropathogens. Am J Epidemiol. 1990;131:886–904. doi: 10.1093/oxfordjournals.aje.a115579. [DOI] [PubMed] [Google Scholar]
- 9.Cravioto A, Tello A, Navarro A, Ruiz J, Villafán H, Uribe F, Eslava C. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet. 1991;337:262–264. doi: 10.1016/0140-6736(91)90868-p. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg M S. Enteropathogenic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press, Ltd.; 1995. pp. 709–726. [Google Scholar]
- 11.Donnenberg M S, Girón J A, Nataro J P, Kaper J B. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 12.Donnenberg M S, Nataro J P. Methods for studying adhesion of diarrheagenic Escherichia coli. Methods Enzymol. 1995;253:324–336. doi: 10.1016/s0076-6879(95)53028-2. [DOI] [PubMed] [Google Scholar]
- 13.Donnenberg M S, Tacket C O, Losonsky G, Frankel G, Nataro J P, Dougan G, Levine M M. Effect of prior experimental human enteropathogenic Escherichia coli infection on illness following homologous and heterologous rechallenge. Infect Immun. 1998;66:52–58. doi: 10.1128/iai.66.1.52-58.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drolet R, Fairbrother J M, Harel J, Hélie P. Attaching and effacing and enterotoxigenic Escherichia coli associated with enteric colibacillosis in the dog. Can J Vet Res. 1994;58:87–92. [PMC free article] [PubMed] [Google Scholar]
- 15.Forest K T, Dunham S A, Koomey M, Tainer J A. Crystallographic structure reveals phosphorylated pilin from Neisseria: phosphoserine sites modify type IV pilus surface chemistry and fibre morphology. Mol Microbiol. 1999;31:743–752. doi: 10.1046/j.1365-2958.1999.01184.x. [DOI] [PubMed] [Google Scholar]
- 16.Forest K T, Tainer J A. Type-4 pilus-structure: outside to inside and top to bottom—a minireview. Gene. 1997;192:165–169. doi: 10.1016/s0378-1119(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 17.Giles C, Sangster G, Smith J. Epidemic gastroenteritis of infants in Aberdeen during 1947. Arch Dis Child. 1949;24:45–53. doi: 10.1136/adc.24.117.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girón J A, Donnenberg M S, Martin W C, Jarvis K G, Kaper J B. Distribution of the bundle-forming pilus structural gene (bfpA) among enteropathogenic Escherichia coli. J Infect Dis. 1993;168:1037–1041. doi: 10.1093/infdis/168.4.1037. [DOI] [PubMed] [Google Scholar]
- 19.Girón J A, Ho A S Y, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 20.Girón J A, Qadri F, Azim T, Jarvis K J, Kaper J B, Albert M J. Monoclonal antibodies specific for the bundle-forming pilus of enteropathogenic Escherichia coli. Infect Immun. 1995;63:4949–4952. doi: 10.1128/iai.63.12.4949-4952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes T A T, Vieira M A M, Wachsmuth I K, Blake P A, Trabulsi L R. Serotype-specific prevalence of Escherichia coli strains with EPEC adherence factor genes in infants with and without diarrhea in São Paulo, Brazil. J Infect Dis. 1989;160:131–135. doi: 10.1093/infdis/160.1.131. [DOI] [PubMed] [Google Scholar]
- 22.Gunzburg S T, Tornieporth N G, Riley L W. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol. 1995;33:1375–1377. doi: 10.1128/jcm.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn H P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 24.Hales B A, Hart C A, Batt R M, Saunders J R. The large plasmids found in enterohemorrhagic and enteropathogenic Escherichia coli constitute a related series of transfer-defective Inc F-IIA replicons. Plasmid. 1992;28:183–193. doi: 10.1016/0147-619x(92)90050-k. [DOI] [PubMed] [Google Scholar]
- 25.Hazes B, Sastry P A, Hayakawa K, Read R J, Irvin R T. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. J Mol Biol. 2000;299:1005–1017. doi: 10.1006/jmbi.2000.3801. [DOI] [PubMed] [Google Scholar]
- 26.Hicks S, Frankel G, Kaper J B, Dougan G, Phillips A D. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iredell J R, Manning P A. Biotype-specific tcpA genes in Vibrio cholerae. FEMS Microbiol Lett. 1994;121:47–54. doi: 10.1111/j.1574-6968.1994.tb07074.x. [DOI] [PubMed] [Google Scholar]
- 28.Kaper J B. Defining EPEC. Rev Microbiol São Paulo. 1996;27(Suppl. 1):130–133. [Google Scholar]
- 29.Kennedy D H, Walker G H, Fallon R J, Boyd J F, Gross R J, Rowe B. An outbreak of infantile gastroenteritis due to E. coli 0142. J Clin Pathol. 1973;26:731–737. doi: 10.1136/jcp.26.10.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirn T J, Lafferty M J, Sandoe C M, Taylor R K. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- 31.Knutton S, Shaw R K, Anantha R P, Donnenberg M S, Zorgani A A. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999;33:499–509. doi: 10.1046/j.1365-2958.1999.01495.x. [DOI] [PubMed] [Google Scholar]
- 32.Laporta M Z, Silva M L M, Scaletsky I C A, Trabulsi L R. Plasmids coding for drug resistance and localized adherence to HeLa cells in enteropathogenic Escherichia coli O55:H− and O55:H6. Infect Immun. 1986;51:715–717. doi: 10.1128/iai.51.2.715-717.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine M M. Vaccines against enteropathogenic Escherichia coli. Rev Microbiol São Paulo. 1996;27(Suppl. 1):126–129. [Google Scholar]
- 34.Levine M M, Bergquist E J, Nalin D R, Waterman D H, Hornick R B, Young C R, Sotman S, Rowe B. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;i:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 35.Levine M M, Nataro J P, Karch H, Baldini M M, Kaper J B, Black R E, Clements M L, O'Brien A D. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 1985;152:550–559. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 36.Levine M M, Prado V, Robins-Browne R, Lior H, Kaper J B, Moseley S L, Gicquelais K, Nataro J P, Vial P, Tall B. Use of DNA probes and HEp-2 cell adherence assay to detect diarrheagenic Escherichia coli. J Infect Dis. 1988;158:224–228. doi: 10.1093/infdis/158.1.224. [DOI] [PubMed] [Google Scholar]
- 37.Loureiro I, Frankel G, Adu-Bobie J, Dougan G, Trabulsi L R, Carneiro-Sampaio M M. Human colostrum contains IgA antibodies reactive to enteropathogenic Escherichia coli virulence-associated proteins: intimin, BfpA, EspA, and EspB. J Pediatr Gastroenterol Nutr. 1998;27:166–171. doi: 10.1097/00005176-199808000-00007. [DOI] [PubMed] [Google Scholar]
- 38.Martinez M B, Taddei C R, Ruiz-Tagle A, Trabulsi L R, Girón J A. Antibody response of children with enteropathogenic Escherichia coli infection to the bundle-forming pilus and locus of enterocyte effacement-encoded virulence determinants. J Infect Dis. 1999;179:269–274. doi: 10.1086/314549. [DOI] [PubMed] [Google Scholar]
- 39.McConnell M M, Chart H, Scotland S M, Smith H R, Willshaw G A, Rowe B. Properties of adherence factor plasmids of enteropathogenic Escherichia coli and the effect of host strain on expression of adherence to HEp-2 cells. J Gen Microbiol. 1989;135:1123–1134. doi: 10.1099/00221287-135-5-1123. [DOI] [PubMed] [Google Scholar]
- 40.McNamara B P, Donnenberg M S. Evidence for specificity in type 4 pilus biogenesis by enteropathogenic Escherichia coli. Microbiology. 2000;146:719–729. doi: 10.1099/00221287-146-3-719. [DOI] [PubMed] [Google Scholar]
- 41.Meyer T F, Gibbs C P, Haas R. Variation and control of protein expression in Neisseria. Annu Rev Microbiol. 1990;44:451–477. doi: 10.1146/annurev.mi.44.100190.002315. [DOI] [PubMed] [Google Scholar]
- 42.Nagayama K, Bi Z, Oguchi T, Takarada Y, Shibata S, Honda T. Use of an alkaline phosphatase-conjugated oligonucleotide probe for the gene encoding the bundle-forming pilus of enteropathogenic Escherichia coli. J Clin Microbiol. 1996;34:2819–2821. doi: 10.1128/jcm.34.11.2819-2821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nandi B, Nandy R K, Vicente A C, Ghose A C. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect Immun. 2000;68:948–952. doi: 10.1128/iai.68.2.948-952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nataro J P, Baldini M M, Kaper J B, Black R E, Bravo N, Levine M M. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J Infect Dis. 1985;152:560–565. doi: 10.1093/infdis/152.3.560. [DOI] [PubMed] [Google Scholar]
- 45.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nataro J P, Kaper J B, Robins-Browne R, Prado V, Vial P, Levine M M. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J. 1987;6:829–831. doi: 10.1097/00006454-198709000-00008. [DOI] [PubMed] [Google Scholar]
- 47.Nataro J P, Maher K O, Mackie P, Kaper J B. Characterization of plasmids encoding the adherence factor of enteropathogenic Escherichia coli. Infect Immun. 1987;55:2370–2377. doi: 10.1128/iai.55.10.2370-2377.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nataro J P, Scaletsky I C A, Kaper J B, Levine M M, Trabulsi L R. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect Immun. 1985;48:378–383. doi: 10.1128/iai.48.2.378-383.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 50.Novais R C, Coelho A, Salles C A, Vicente A C. Toxin-co-regulated pilus cluster in non-O1, non-toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol Lett. 1999;171:49–55. doi: 10.1111/j.1574-6968.1999.tb13411.x. [DOI] [PubMed] [Google Scholar]
- 51.Ørskov F, Ørskov I, Rees T A, Sahab K. Two new E. coli O-antigens: O141 and O142 and two new coli K-antigens: K85 and K86. Acta Pathol Microbiol Scand. 1960;48:48–50. doi: 10.1111/j.1699-0463.1960.tb04736.x. [DOI] [PubMed] [Google Scholar]
- 52.Parge H E, Forest K T, Hickey M J, Christensen D A, Getzoff E D, Tainer J A. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 53.Parissi-Crivelli A, Parissi-Crivelli J M, Girón J A. Recognition of enteropathogenic Escherichia coli virulence determinants by human colostrum and serum antibodies. J Clin Microbiol. 2000;38:2696–2700. doi: 10.1128/jcm.38.7.2696-2700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reid S D, Herbelin C J, Bumbaugh A C, Selander R K, Whittam T S. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 55.Rhine J A, Taylor R K. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol Microbiol. 1994;13:1013–1020. doi: 10.1111/j.1365-2958.1994.tb00492.x. [DOI] [PubMed] [Google Scholar]
- 56.Robins-Browne R M. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev Infect Dis. 1987;9:28–53. doi: 10.1093/clinids/9.1.28. [DOI] [PubMed] [Google Scholar]
- 57.Robins-Browne R M, Yam W C, O'Gorman L E, Bettelheim K A. Examination of archetypal strains of enteropathogenic Escherichia coli for properties associated with bacterial virulence. J Med Microbiol. 1993;38:222–226. doi: 10.1099/00222615-38-3-222. [DOI] [PubMed] [Google Scholar]
- 58.Sayle R A, Milner-White E J. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 59.Scaletsky I C A, Silva M L M, Trabulsi L R. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–536. doi: 10.1128/iai.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schremmer C, Lohr J E, Wastlhuber U, Kösters J, Ravelshofer K, Steinrück H, Wieler L H. Enteropathogenic Escherichia coli in Psittaciformes. Avian Pathol. 1999;28:349–354. doi: 10.1080/03079459994605. [DOI] [PubMed] [Google Scholar]
- 61.Schriefer A, Maltez J R, Silva N, Stoeckle M Y, Barral-Netto M, Riley L W. Expression of a pilin subunit BfpA of the bundle-forming pilus of enteropathogenic Escherichia coli in an aroA live Salmonella vaccine strain. Vaccine. 1999;17:770–778. doi: 10.1016/s0264-410x(98)00261-8. [DOI] [PubMed] [Google Scholar]
- 62.Seifert H S. Questions about gonococcal pilus phase and antigenic variation. Mol Microbiol. 1996;21:433–440. doi: 10.1111/j.1365-2958.1996.tb02552.x. [DOI] [PubMed] [Google Scholar]
- 63.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sohel I, Puente J L, Murray W J, Vuopio-Varkila J, Schoolnik G K. Cloning and characterization of the bundle-forming pilin gene of enteropathogenic Escherichia coli and its distribution in Salmonella serotypes. Mol Microbiol. 1993;7:563–575. doi: 10.1111/j.1365-2958.1993.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 65.Sohel I, Puente J L, Ramer S W, Bieber D, Wu C-Y, Schoolnik G K. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J Bacteriol. 1996;178:2613–2628. doi: 10.1128/jb.178.9.2613-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stone K D, Zhang H-Z, Carlson L K, Donnenberg M S. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for biogenesis of a type IV pilus. Mol Microbiol. 1996;20:325–337. doi: 10.1111/j.1365-2958.1996.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 67.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 68.Taylor J, Charter R E. The isolation of serological types of Bact. coli in two residential nurseries and their relation to infantile gastroenteritis. J Pathol Bacteriol. 1952;64:715–728. doi: 10.1002/path.1700640405. [DOI] [PubMed] [Google Scholar]
- 69.Taylor J, Charter R E. Escherichia coli O.128 causing gastroenteritis of infants. J Clin Pathol. 1955;8:276–281. doi: 10.1136/jcp.8.4.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tennent J M, Mattick J S. Type 4 fimbriae. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 127–146. [Google Scholar]
- 71.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tobe T, Hayashi T, Han C G, Schoolnik G K, Ohtsubo E, Sasakawa C. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect Immun. 1999;67:5455–5462. doi: 10.1128/iai.67.10.5455-5462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toledo M R, Alvariza M, Murahovschi J, Ramos S R, Trabulsi L R. Enteropathogenic Escherichia coli serotypes and endemic diarrhea in infants. Infect Immun. 1983;39:586–589. doi: 10.1128/iai.39.2.586-589.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tornieporth N G, John J, Salgado K, de Jesus P, Latham E, Melo M C, Gunzburg S T, Riley L W. Differentiation of pathogenic Escherichia coli strains in Brazilian children by PCR. J Clin Microbiol. 1995;33:1371–1374. doi: 10.1128/jcm.33.5.1371-1374.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsukamoto T. PCR methods for detection of enteropathogenic Escherichia coli (localized adherence) and enteroaggregative Escherichia coli. Kansenshogaku Zasshi. 1996;70:569–573. doi: 10.11150/kansenshogakuzasshi1970.70.569. [DOI] [PubMed] [Google Scholar]
- 76.Vuopio-Varkila J, Schoolnik G K. Localized adherence by enteropathogenic Escherichia coli is an inducible phenotype associated with the expression of new outer membrane proteins. J Exp Med. 1991;174:1167–1177. doi: 10.1084/jem.174.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 78.Whittam T S, McGraw E A. Clonal analysis of EPEC serogroups. Rev Microbiol São Paulo. 1996;27(Suppl. 1):7–16. [Google Scholar]
- 79.Whittam T S, Wolfe M L, Wachsmuth I K, Ørskov F, Ørskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wieler L H, Vieler E, Erpenstein C, Schlapp T, Steinruck H, Bauerfeind R, Byomi A, Baljer G. Shiga toxin-producing Escherichia coli strains from bovines: association of adhesion with carriage of eae and other genes. J Clin Microbiol. 1996;34:2980–2984. doi: 10.1128/jcm.34.12.2980-2984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang H-Z, Donnenberg M S. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–797. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]