Abstract
We studied the role of two members of the 100-kDa heat shock protein family, the ClpC and ClpE ATPases, in cell adhesion and invasion of the intracellular pathogen Listeria monocytogenes. During the early phase of infection, a clpC mutant failed to disseminate to hepatocytes in the livers of infected mice whereas the invasive capacity of a clpE mutant remained unchanged. This was confirmed by a confocal microscopy study on infected cultured hepatocyte and epithelial cell lines, showing a strong reduction of cell invasion only by the clpC mutant. Western blot analysis with specific antisera showed that the absence of ClpC, but not that of ClpE, reduced expression of the virulence factors InlA, InlB, and ActA. ClpC-dependent modulation of these factors occurs at the transcriptional level with a reduction in the transcription of inlA, inlB, and actA in the clpC mutant, in contrast to the clpE mutant. This work provides the first evidence that, in addition to promoting escape from the phagosomes, ClpC is required for adhesion and invasion and modulates the expression of InlA, InlB, and ActA, further supporting the major role of the Clp chaperones in the virulence of intracellular pathogens.
Listeria monocytogenes is a gram-positive bacterium that is widespread in nature and responsible for severe infections in humans and most animal species (19). It is easy to reproduce the natural disease in animal models, especially in the mouse (26). The virulence of this ubiquitous pathogen is due to its capacity to invade and multiply within macrophages (26) and nonprofessional phagocytes, including epithelial cells and hepatocytes (9, 12, 13, 15, 35, 45). This well-adapted facultative intracellular pathogen induces its own internalization by cultured mammalian cells (13). Several surface proteins are involved in this process, including InlA (internalin) and InlB, both of which are required for entry into various cultured cell lines, each with its own specificity (3, 10, 11, 28). ActA also plays a role in entry (1). After phagocytosis, bacteria rapidly disrupt the phagosomal membrane, a process requiring the secretion of listeriolysin O and phospholipases (7, 13, 39, 44), and grow within the cytoplasm of host cells (13, 30). Bacteria can spread from cell to cell within tissues using an actin-based motility process due to ActA (8, 24), thus taking advantage of the host cell machinery (30, 44). These virulence genes are transcribed under heat or nutrient stress conditions and controlled by PrfA, a transcriptional activator (2, 25, 41, 42).
Like any other bacterium, L. monocytogenes rapidly adapts to sudden changes in the environment during its saprophytic life by synthesizing a group of proteins acting as chaperones and proteases, allowing its survival under adverse conditions, including low and high temperatures (4 to 44°C), starvation, variations in pH and osmolarity, chemical stresses, and competition with other microorganisms (19). In living cells, chaperones assist the proper folding, refolding, or assembly of proteins while the proteases process those that cannot be refolded. In host tissues, L. monocytogenes is also exposed to hostile conditions induced by the immune response during the infectious process mimicking the environmental conditions. Following bacterial uptake by macrophages, a set of proteins are produced (21). Several stress proteins of L. monocytogenes are involved in the fate of intracellular bacteria in macrophages. The ClpC ATPase belongs to the Clp 100-kDa heat shock protein family, a class of highly conserved proteins implicated in the stress tolerance of many prokaryotic and eukaryotic organisms (17, 18, 38, 43), and is implicated in the virulence of L. monocytogenes by promoting early bacterial escape from the phagosomal compartment of macrophages (36, 37). Clp ATPases have also been shown to play a role in the survival and virulence of other bacterial pathogens, including Salmonella typhimurium (22) and Staphylococcus aureus (27). Another 100-kDa heat shock protein, ClpE, is also involved in the virulence of L. monocytogenes, acting synergistically with ClpC in cell division under conditions of nutrient and energy deprivation at elevated temperatures (32). ClpE displays the typical structural organization of the Clp ATPases; however, this protein is smaller (726 amino acids) than ClpC (826 amino acids), with a smaller spacer region and a conserved shorter N-terminal region with a potential zinc finger motif. As opposed to clpC, clpE expression is not stimulated by various stresses, including elevated temperatures and salt stress. Transcription of clpE is strongly up-regulated in the absence of ClpC, whereas transcription of clpC remains unchanged in a clpE mutant (32). It has also been shown that a stress-induced ClpP is required for the intracellular survival of L. monocytogenes in macrophages (16). Transcription of clpC, clpE, and clpP is regulated by CtsR, a negative transcriptional regulator of the stress response in L. monocytogenes (31).
In this work, we studied the role of ClpC and ClpE of L. monocytogenes in the process of cell invasion in vivo and in vitro. We found that ClpC, but not ClpE, is involved in the invasion of hepatocytes in vivo during infection. This is due to ClpC-dependent modulation of the invasion virulence factors InlA, InlB, and ActA.
MATERIALS AND METHODS
Bacterial strains and culture media.
We used L. monocytogenes reference strain LO28 and several allelic mutants of this strain: a clpC mutant (36), a clpE mutant, and a clpC clpE double mutant (32). All bacteria were grown in brain heart infusion (BHI) media. For virulence assays, overnight bacterial cultures were harvested (5 × 109/ml) in 1-ml aliquots and stored at −80°C until required. For each experiment, a vial was thawed and diluted appropriately in saline (0.15 M NaCl) for intravenous inoculation (0.5 ml) into a lateral tail vein as described previously (32).
Infection of mice and histology.
Specific-pathogen-free Swiss mice were challenged intravenously (i.v.) with 108 bacteria (0.5 ml) and killed by cervical dislocation 1 and 8 h after infection. Small pieces of liver were removed and processed for Gram staining and semithin sections. Samples for Gram staining were fixed in 10% formalin and embedded in paraffin; 2- to 3-μm sections were cut and stained using the Gram-Weigert procedure. Tissue samples were also fixed in 2% glutaraldehyde (Sigma Chemical Co.) and embedded in Epon 812 (TAA-Jamming) as previously described (14). Semithin sections were cut and stained with 1% toluidine blue for light microscopy.
Culture of cell lines and invasion assays.
We used the murine embryonic hepatocyte cell line TIB73 (ATCC TIB73) and the human colon carcinoma cell line Caco-2 (ATCC HTB37) from the American Type Culture Collection (Manassas, Va.). Cells were cultured in Dulbecco's modified eagle medium (DMEM) containing glutamax (Gibco Laboratories, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (Gibco Laboratories). TIB73 cells and Caco-2 cells were grown without antibiotics as previously described (9, 14) and used between passages 12 and 20 and passages 25 and 35, respectively. Cells were maintained in 10% CO2 at 37°C. TIB73 cells were seeded at a density of 105/cm2 in 24-well tissue culture plates (Falcon Labware, Becton Dickinson & Co., Lincoln Park, N.J.) for invasion assays and onto 12-mm-diameter glass coverslips in 24-well tissue culture plates (Falcon Labware) for fluorescence microscopy. Monolayers were used 24 to 48 h after seeding.
Invasivity assays were performed with 24-well plates using the gentamicin killing assay as previously described (29). Bacteria grown overnight at 37°C in BHI broth (optical density at 600 nm, 1.2 to 1.4) were pelleted by centrifugation, washed once, and diluted appropriately in DMEM. Cells were exposed to bacteria at a ratio of 100 bacteria per cell for 1 h at 37°C. After extensive washings, cells were incubated with fresh DMEM; gentamicin (10 mg/liter) was added to kill extracellular bacteria. At intervals, cells were washed twice and lysed by adding cold water. Released intracellular bacteria were counted by plating on BHI agar. Each determination was made in triplicate and expressed as the mean ± the standard deviation.
Immunofluorescence and confocal microscopy.
Cells were infected with bacteria at a multiplicity of infection of 100 bacteria per cell as described above and then incubated with fresh DMEM containing gentamicin (10 mg/liter). At 1, 3, and 8 h, cells were washed twice with phosphate-buffered saline (PBS), fixed with 3% (wt/vol) paraformaldehyde in PBS for 30 min at room temperature, washed three times with PBS, and permeabilized for 5 min in 0.1% Triton X-100 (Sigma Chemical Co.) in PBS and processed for immunolabeling and actin staining. For immunolabeling of listeriae, cells were incubated sequentially with appropriate dilutions of polyclonal rabbit anti-Listeria serovar 1/2a (1/1,000 dilution) immunoglobulin and of goat anti-rabbit immunoglobulin (1/1,000 dilution) coupled to CY3 (Jackson ImmunoResearch Laboratories Inc., Bio/Can Scientific, Mississauga, Ontario, Canada) in 1% bovine serum albumin–PBS; incubations were carried out for 30 min at room temperature. For F-actin staining, cells were incubated with Oregon green phalloidin (Molecular Probes, Inc., Eugene, Oreg.) diluted 1/40 for 30 min at room temperature. Coverslips were mounted on slides and examined by fluorescence microscopy using a confocal microscope (AxiosLop; Carl Zeiss, Inc., Thornwood, N.Y.).
Western blot analysis.
Protein extracts were prepared from cultures of wild-type LO28, clpC mutant, clpE mutant, and clpC clpE double-mutant bacteria grown in BHI broth at 37°C (optical density at 600 nm, 0.6) as previously described (32). Proteins in culture supernatants of bacterial strains were precipitated with 10% (vol/vol) trichloroacetic acid. Bacterial whole-cell extracts were prepared by boiling the cells for 5 min in 100 mM Tris (pH 6.8)–200 mM dithiothreitol–4% (wt/vol) sodium dodecyl sulfate–0.2% (wt/vol) bromophenol blue–20% (vol/vol) glycerol. The concentration of protein was measured using the Bradford assay, and equal amounts were loaded in each lane. The membrane was stained with Ponceau red, confirming the efficacy of the transfer before hybridization with the antibodies. Gels were stained with Coomassie blue or subjected to immunoblot analysis with mouse monoclonal antibodies, directed against purified ActA, InlA, and InlB, obtained from P. Cossart (Pasteur Institute, Paris, France). Anti-mouse immunoglobulin-horseradish peroxidase conjugate and the ECL kit were used for immunodetection (Amersham). A rabbit anti-PrfA antibody was obtained by immunizing rabbits with purified PrfA. Briefly, the entire prfA gene from L. monocytogenes was amplified by PCR and cloned into the pET-20b expression vector (Novagen), allowing the fusion of a six-His tag to the C-terminal coding sequence. The expressed PrfA protein was purified by affinity chromatography with an Ni-nitrilotriacetic acid column. The sequence of the first 10 N-terminal amino acid residues was verified by Edman degradation (Laboratory of Microsequencing of Proteins, Department of Biotechnologies, Pasteur Institute, Paris, France). Anti-rabbit-horseradish peroxidase conjugate (Amersham) was used to reveal PrfA.
RNA slot blot analysis.
RNA slot blot analysis and hybridizations of total RNA extracted during exponential phase at 37°C from wild-type LO28 and the mutants were done as previously described (37). Specific probes used for hybridizations were generated with the following primers: inlA, 5′-CCTGTGGCACCACCAAC-3′ and 5′-CTATTTACTAGCACGTGC-3′; inlB, 5′-GTACGCAGTATTTAAAGCGG-3′ and 5′-TTATTTCTGTGCCCTTAA-3′; actA, 5′-GTGGGATTAAACAGA-3′ and 5′-ATTTTTTCTTAATTGAA-3′; prfA, 5′-ATGAACGCTCAAGCAGAATTC-3′ and 5′-TAATTTTCCCCAAGTAGCAGG-3′. A probe (867 bp) corresponding to the 16S RNA from L. monocytogenes was amplified using primers 5′-AGGCCCGGGAACGTATTCAC-3′ and 5′-GTGCCAGCAGCCGCGGTAAT-3′.
RESULTS
ClpC ATPase is required for in vivo cell invasion in infected mice.
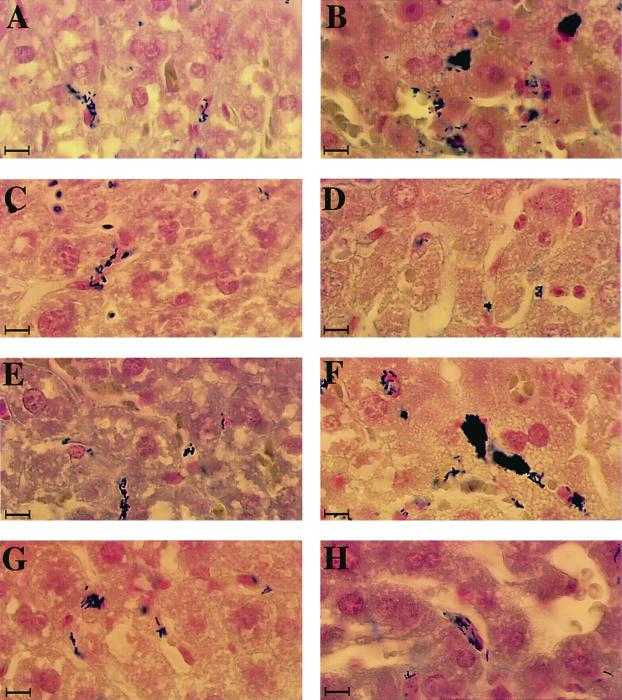
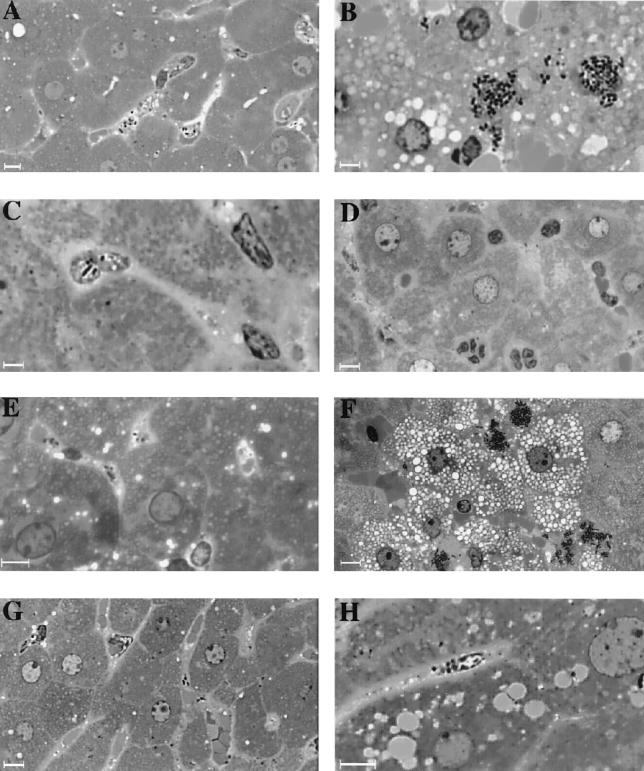
We previously demonstrated that the virulence of clpC-, clpE-, and clpC clpE-inactivated mutants of L. monocytogenes is strongly reduced in the mouse model (32, 36, 37). A reduction of bacterial survival was observed in the livers of infected mice during the first 3 days, suggesting that ClpC and ClpE are involved in the invasion process in the liver during acute infection. In this work, bacterial invasion of hepatocytes in vivo was directly visualized by inoculating mice i.v. (108 bacteria) with four strains of L. monocytogenes (wild-type LO28 and clpE, clpC, and clpC clpE mutants). Mice were sacrificed 1 and 8 h after inoculation, and a histological study of the liver sections stained with Gram-Weigert stain or toluidine blue (semithin sections) was performed. One hour after inoculation, bacteria of all of the strains were visible exclusively in the Kupffer cells, without visible spreading to the hepatocytes at this early stage of infection (Fig. 1). The fate of these bacterial strains was very different in the liver 8 h after infection. In mice infected with either the wild type or the clpE mutant, bacteria were packed in the Kupffer cells and often invaded adjacent hepatocytes with vacuolization and necrosis of surrounding cells (Fig. 1 and 2). In contrast, rare bacteria were visible in the Kupffer cells of mice infected with either the clpC or the clpC clpE mutant. These mutant bacteria showed no visible invasion of hepatocytes and infiltrated polymorphonuclear cells in tissues (Fig. 2). Higher magnification (not shown) showed that the morphology of these intracellular bacteria was altered. This correlates with the results of bacterial counts obtained by monitoring bacterial survival in the livers of mice 1 and 8 h after infection. No difference in bacterial counts was found among the four strains 1 h after inoculation. Eight hours after inoculation, a significant 1- to 1.5-log drop in bacteria was observed in the liver for the clpC and clpC clpE mutants, whereas this reduction was very weak for wild-type LO28 and the clpE mutant (data not shown). These results indicate that, in contrast to ClpE, ClpC plays a crucial role in bacterial survival and invasion of hepatocytes in vivo during the early phase of infection.
FIG. 1.
Liver sections of mice infected with L. monocytogenes. Liver sections were prepared 1 (A, C, E, and G) and 8 (B, D, E, and H) h after infection of mice inoculated i.v. with 108 bacteria and examined by light microscopy after Gram-Weigert staining (magnification, ×500). Panels: A and B, wild-type LO28; C and D, clpC mutant; E and F, clpE mutant; G and F, clpC clpE mutant. After 1 h, all of the strains of bacteria were found exclusively in Kupffer cells. After 8 h, the wild type and the clpE mutant replicated massively in Kupffer cells, with large clusters of bacteria packed in sinusoid capillaries (B and F). In contrast, clpC and clpC clpE bacteria remained confined to Kupffer cells without visible invasion of hepatocytes (D and H). Bars, 2.5 μm.
FIG. 2.
Livers of mice infected with wild-type and clp mutant L. monocytogenes. Semithin liver sections were prepared 1 (A, C, E, and G) and 8 (B, D, E, and H) h after infection of mice inoculated i.v. with 108 bacteria and examined by light microscopy after toluidine blue staining (magnification, ×1,200). Panels: A and B, wild-type LO28; C and D, clpC mutant; E and F, clpE mutant; G and H, clpE/clpC mutant. After 1 h, bacteria of all of the strains were found exclusively in Kupffer cells. After 8 h, wild-type and clpE mutant bacteria formed typical infectious foci, consisting of large areas of necrosis with bacteria packed in Kupffer cells and spreading to adjacent hepatocytes. clpC and clpC clpE bacteria remained confined to Kupffer cells without visible invasion of hepatocytes and there were few inflammatory cells. Bars, 2.5 μm.
ClpC ATPase is required for invasion of hepatocytes.
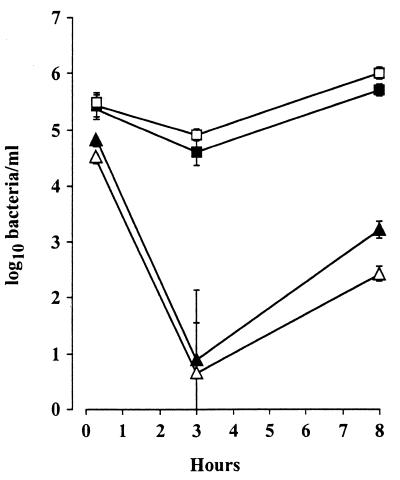
We further investigated the involvement of the ClpC and ClpE ATPases of L. monocytogenes during the in vitro invasion of hepatocyte and epithelial cell lines. Murine TIB73 hepatocytes were exposed to either wild-type strain LO28 or clpC, clpE, or clpC clpE mutant bacteria. Cells were then washed and incubated in the presence of gentamicin (10 mg/liter) to eliminate extracellular bacteria. Intracellular bacteria were counted at 1, 3, and 8 h after cell lysis. The curves of bacterial intracellular survival in TIB73 cells are illustrated in Fig. 3. We found that both wild-type and clpE mutant bacteria could adhere to and penetrate hepatocytes. In contrast, the rate of adhesion was approximately 1 log lower for the clpC and clpC clpE mutant bacteria. After 3 h, the growth rates of wild-type and clpE mutant bacteria were similar. We observed a strong reduction in cell invasion with a 4-log decrease in bacteria at 3 h for the clpC and clpC clpE mutants compared to the wild type (Fig. 3), corresponding to the killing of membrane-associated extracellular bacteria by the antibiotic (13). Regrowth of the clpC and clpC clpE mutants was observed after 8 h of incubation, but the amount of intracellular bacteria remained significantly lower than that of wild-type bacteria (Fig. 3). Similar results were obtained with Caco-2 cells (data not shown).
FIG. 3.
Entry of L. monocytogenes and clp mutants into TIB73 hepatocytes. Cells were exposed for 1 h to bacteria (100 bacteria per cell) (time zero), extensively washed, and further incubated with gentamicin (10 mg/liter). Cells were washed after 3 and 8 h and lysed by addition of cold water. Released intracellular bacteria were plated on BHI agar (each time point represents triplicate measurements). Symbols: □, LO28; ▴, clpC; ■, clpE; ▵, clpC clpE. The values are means ± standard deviations.
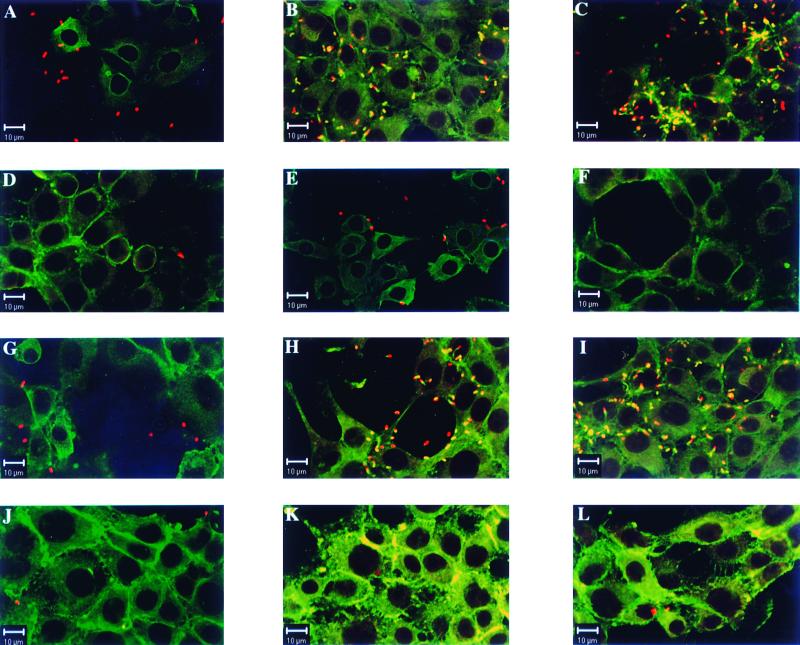
TIB73 and Caco-2 cells were further examined under these conditions by immunofluorescence assay with a confocal microscope at 1, 3, and 8 h after infection. Cells on glass coverslips were first incubated with rabbit anti-Listeria serovar 1/2a antibodies, with goat anti-rabbit antibodies coupled to CY3, and then with Oregon green phalloidin. The results obtained with the TIB73 cell line are illustrated in Fig. 4, and similar results (not shown) were obtained with Caco-2 cells. As expected, wild-type and clpE mutant bacteria adhered to and rapidly invaded the cell monolayers (Fig. 4). As early as 3 h, actin comets were observed with these bacteria (Fig. 4B and H) and massive intracytoplasmic multiplication was visible by 8 h, with multiple comets of actin polymerization (Fig. 4C and I). Under the same conditions of infection, clpC and clpC clpE mutants adhered very weakly to cells, even at time zero (Fig. 4D and J), and remained located extracellularly. Bacteria were scarce, with very rare actin comets visible after 3 to 8 h. We can estimate that at 8 h postinfection, 90% of the cells infected with the wild type had at least 50 bacteria per cell, in contrast to the mutants, where <10% of the cells infected contained intracellular bacteria, again correlating with the bacterial survival in Fig. 3. These results confirm the in vivo data indicating that ClpC plays a crucial role in cell adhesion and invasion of L. monocytogenes.
FIG. 4.
Confocal microscopy of TIB73 hepatocytes infected with L. monocytogenes and clp mutants. Cells were exposed to bacteria for 1 h (100 bacteria/cell), extensively washed, and further incubated in the presence of gentamicin (10 mg/liter) for 1, 3, and 8 h. Wild-type LO28 (A, B, and C), the clpC mutant (D, E, and F), the clpE mutant (G, H, and I), and the clpC clpE mutant (J, K, and L) are shown. F-actin was stained with phalloidin (green), and bacteria were labeled with anti-Listeria antibodies (red). F-actin sheaths associated with bacteria are indicated by the overlapping of green and red light (orange-yellow). After 1 h, the bacterial uptake is similar for both the wild type (A) and the clpE mutant (G) with cell-associated bacteria whereas only very rare clpC and clpC clpE mutant bacteria were visible at that time (D and J). After 3 h (B, E, H, and K) and 8 h (C, F, I, and L), many replicating wild type (B and C) and clpE mutant (H and I) bacteria were associated with the F-actin sheaths. In contrast, few clpC (E and F) and clpC clpE (K and L) mutant bacteria were seen under the same conditions, with no evidence of cell division or actin polymerization.
ClpC ATPase is required for expression of invasion virulence factors.
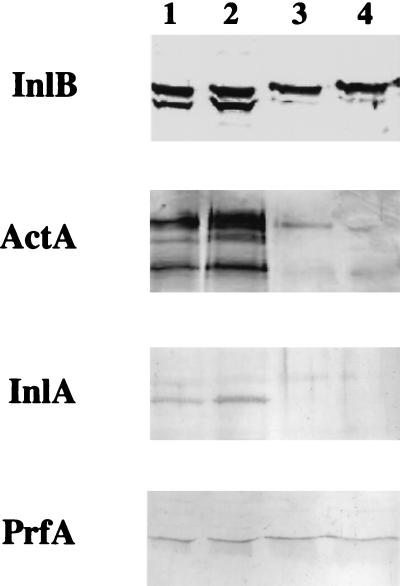
It is known that InlA, InlB, and ActA play a role in cell adhesion and invasion of L. monocytogenes. The expression of these virulence factors in the clpC, clpE, and clpC clpE mutants, compared to the wild type, was studied by Western blot analysis. Whole-cell extracts and culture supernatants from the wild type and the mutants (see Materials and Methods) were prepared and exposed to specific antibodies raised against InlA, InlB, and ActA. We also used an antibody directed against PrfA, a cytoplasmic transcriptional activator. In contrast to the clpC and clpC clpE mutants, bands corresponding to InlA and ActA were detected only in bacterial extracts from the wild type and the clpE mutant (Fig. 5). As previously described for the anti-InlB antibody used, two major bands were recognized in extracts of wild-type and clpE mutant bacteria, the lower band corresponding to InlB (60 kDa) and the second band cross-reacting with an undefined 67-kDa protein (11). The 60-kDa InlB protein almost disappeared in the clpC and clpC clpE mutants (Fig. 5). These results obtained with InlB were confirmed in the supernatants of bacteria, where many bands, presumably corresponding to digested polypeptides, were observed in the wild-type and clpE mutant bacteria; in contrast, InlB was weakly expressed in the clpC and clpC clpE mutants (data not shown). The same amount of PrfA was detected in bacterial extracts of all of the strains (Fig. 5). These results show that the expression of InlA, InlB, and ActA is strongly reduced in the absence of ClpC, indicating that ClpC is required for the expression of these virulence factors at the surface and in the supernatant of bacteria.
FIG. 5.
Western blot analysis of whole-cell extracts of wild-type and clp mutant L. monocytogenes revealed with anti-InlB, anti-ActA, anti-InlA, and anti-PrfA antibodies. Lanes: 1, LO28; 2, clpE mutant; 3, clpC mutant; 4, clpC clpE mutant. The adhesion factors were hardly detected in the clpC and clpC clpE mutants, in contrast to the wild type and the clpE mutant. No difference in the expression of PrfA was found.
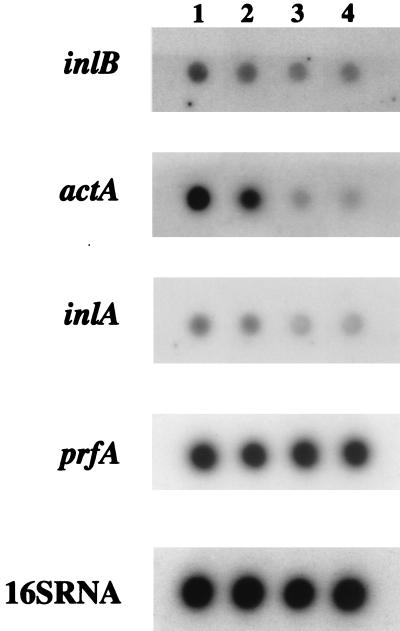
A transcriptional study of inlA, inB, actA, and prfA was then performed on RNA extracted from the wild-type and mutant strains during the exponential growth phase. A calibrated amount of RNA was then tested by dot blot analysis using specific probes for these genes, compared to the 16S rRNA used as a control. Whereas the amounts of prfA transcript were similar in all of the strains tested, the amounts of inlA, inlB, and actA transcripts were similar in wild-type LO28 and the clpE mutant but partly reduced in the clpC and clpC clpE mutants (Fig. 6). These results suggest that the chaperone ClpC is involved at the level of transcription of genes implicated in cell adhesion and invasion.
FIG. 6.
RNA slot blot analysis of wild-type strain LO28 and clp mutant L. monocytogenes. Calibrated amounts of RNA extracted during exponential growth phase were tested using specific probes for inlA, inlB, actA, and prfA compared to the 16S RNA control. Lanes: 1, wild-type LO28; 2, clpE; 3, clpC; 4, clpC clpE. The prfA transcript levels were similar in all of the strains tested. In contrast, the amounts of inlA, inlB, and actA transcripts were similar in the wild type and the clpE mutant but partly reduced in the clpC and clpC clpE mutants.
DISCUSSION
This is the first report which describes a Clp chaperone, the ClpC ATPase, as playing a major role in cell adhesion and invasion of L. monocytogenes. We have demonstrated that the invasion of hepatocytes by a clpC mutant is strongly reduced in the liver during the early phase of infection in the mouse. Hepatocyte invasion is considered a key event during murine listeriosis, and it is known that bacteria accumulate predominantly in the liver, where they replicate until the host mobilizes a protective cellular immune response (6, 33, 35, 40, 45). Several works have clearly shown that L. monocytogenes replicates in hepatocytes rather than in Kupffer cells (6, 20, 33). These results were then confirmed by infecting in vitro hepatocyte and epithelial cell lines, showing dramatic differences visualized by confocal microscopy in the invasive capacity of a clpC mutant. Another important finding was that, in contrast to ClpC, ClpE is not involved in the invasive process during listeriosis. Although ClpC and ClpE are both required for the virulence of L. monocytogenes and for growth in the liver (32, 36, 37), we clearly show that only ClpC promotes in vivo invasion of hepatocytes. Cell adhesion and invasion were strongly reduced for the clpC and clpC clpE mutants, in contrast to the clpE mutant. Although ClpE acts synergistically with ClpC in cell septation, the present data suggest that ClpE may act at a different step during intracellular survival, presumably in the process of survival in macrophages. So, the ClpC ATPase of L. monocytogenes not only contributes to early escape from the phagosomal compartment of macrophages (36) but is also involved at the step of adhesion and invasion, thus explaining the reduced virulence seen in the absence of ClpC.
Several virulence factors, including InlA, InlB, and ActA, are involved in the entry of L. monocytogenes into various cultured cell lines (1, 3, 5, 10, 11, 28). InlB plays an important role in the entry of L. monocytogenes into most cell lines. InlB is a 630-amino-acid surface protein associated with the bacterial surface and is released in culture supernatants. The loose association of InlB at the bacterial surface is mediated by the so-called GW repeats, located in the C-terminal region of InlB, which bind to lipoteichoic acid (5, 23). While the contribution of the released InlB to the entry process is as yet unclear, it is known that InlB plays a role in the process of hepatic infection (9). Recently, a mammalian receptor for InlB invasion, the C1q-binding protein, has been identified in epithelial cells (4). Our invasion assays clearly indicated that clpC is a crucial factor for entry of L. monocytogenes into the murine hepatocyte cell line TIB73. We showed by Western blot analysis that ClpC modulates the expression of the virulence factors InlA, InlB, and ActA, which are known to be directly involved in cell adhesion and invasion. The reduced expression of InlA, InlB, and ActA therefore contributes to the impaired adhesion and invasion capacity of the clpC mutant. The implication of the Clps in the modulation of cell surface proteins has been proposed before. In Yersinia enterocolitica, ClpP modulates the expression of Ail, a 17-kDa cell surface protein that confers the ability to attach to and invade cells in vitro. Ail expression is normally repressed during stationary phase at 28°C in the presence of functional ClpP (34).
Acting as a molecular chaperone means binding to heat-denatured or otherwise damaged proteins and preventing or slowing down their aggregation, triggering proper protein transport and folding through the dissolution of protein aggregates. Our results suggest that ClpC is involved in the proper folding and transport of the invasion factors, like a classical chaperone. We were surprised to find that the ClpC-dependent modulation of these invasion factors occurs at the transcriptional level. Indeed, transcriptional analysis reveals a partial reduction of transcription of inlA, inlB, and actA in the clpC and clpC clpE mutants, in contrast to the clpE mutant. Whether this is due to direct or indirect interactions between chaperones and selected transcriptional factors involved in the expression of these invasive factors remains unclear. It is worth noting that expression of the inlA, inlB, and actA genes is regulated by PrfA and that PrfA expression in the clpC mutant was comparable to that in the wild type. Thus, the expression of these invasion factors is not wholly dependent upon PrfA and it is tempting to speculate about a role for an alternate transcriptional activator.
In conclusion, we show that ClpC, in addition to promoting escape from the phagosome, modulates the expression of invasion factors, thus playing an important role in virulence. This work illustrates the complexity of the molecular mechanisms involving stress proteins and further supports the major role of Clp chaperones in the virulence of intracellular pathogens.
ACKNOWLEDGMENTS
We thank Francis Jaubert (Hôpital Necker-Enfants-Malades) for his help with the histological study, J. L. Beretti for excellent technical assistance (Western blot analysis), and The Service Photo at the Pasteur Institute for help with the figures.
This work was supported by INSERM, University of Paris V, and a grant (BMH-4CT 960659) from the EEC.
REFERENCES
- 1.Alvarez-Dominguez C, Vazquez-Boland J A, Carrasco-Marin E, Lopez-Mato P, Leyva-Cobian F. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect Immun. 1997;65:78–88. doi: 10.1128/iai.65.1.78-88.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohne J, Sokolovic Z, Goebel W. Transcriptional regulation of prfA and PrfA-regulated virulence genes in Listeria monocytogenes. Mol Microbiol. 1994;11:1141–1150. doi: 10.1111/j.1365-2958.1994.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 3.Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol. 1997;25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 4.Braun L, Ghebrehiwet B, Cossart P. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 2000;19:1458–1466. doi: 10.1093/emboj/19.7.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun L, Ohayon H, Cossart P. The InlB protein of Listeria monocytogenes is sufficient to promote entry into mammalian cells. Mol Microbiol. 1998;27:1077–1087. doi: 10.1046/j.1365-2958.1998.00750.x. [DOI] [PubMed] [Google Scholar]
- 6.Conlan J W, North R J. Neutrophil-mediated dissolution of infected host cells as a defense strategy against a facultative intracellular bacterium. J Exp Med. 1991;174:741–744. doi: 10.1084/jem.174.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Chastellier C, Berche P. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect Immun. 1994;62:543–553. doi: 10.1128/iai.62.2.543-553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wachter M, Wuenscher M, Chakraborty T. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. Entry of Listeria monocytogenes into hepatocytes requires expression of InlB, a surface protein of the internalin multigene family. Mol Microbiol. 1995;16:251–261. doi: 10.1111/j.1365-2958.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 10.Dramsi S, Dehoux P, Cossart P. Common features of gram-positive bacterial proteins involved in cell recognition. Mol Microbiol. 1993;9:1119–1121. doi: 10.1111/j.1365-2958.1993.tb01241.x. [DOI] [PubMed] [Google Scholar]
- 11.Dramsi S, Kocks C, Forestier C, Cossart P. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotropic activator prfA. Mol Microbiol. 1993;9:931–941. doi: 10.1111/j.1365-2958.1993.tb01223.x. [DOI] [PubMed] [Google Scholar]
- 12.Drevets D A, Sawyer R T, Potter T A, Campbell P A. Listeria monocytogenes infects human endothelial cells by two distinct mechanisms. Infect Immun. 1995;63:4268–4276. doi: 10.1128/iai.63.11.4268-4276.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaillard J L, Berche P, Mounier J, Richard S, Sansonetti P J. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun. 1987;55:2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaillard J L, Berche P, Sansonetti P. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect Immun. 1986;52:50–55. doi: 10.1128/iai.52.1.50-55.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaillard J L, Jaubert F, Berche P. The inlAB locus mediates the entry of Listeria monocytogenes into hepatocytes in vivo. J Exp Med. 1996;183:359–369. doi: 10.1084/jem.183.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol Microbiol. 2000;35:1286–1294. doi: 10.1046/j.1365-2958.2000.01773.x. [DOI] [PubMed] [Google Scholar]
- 17.Gottesman S, Clark W P, Maurizi M R. The ATP-dependent Clp protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate. J Biol Chem. 1990;265:7886–7893. [PubMed] [Google Scholar]
- 18.Gottesman S, Squires C, Pichersky E, Carrington M, Hobbs M, Mattick J S, Dalrymple B, Kuramitsu H, Shiroza T, Foster T, et al. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc Natl Acad Sci USA. 1990;87:3513–3517. doi: 10.1073/pnas.87.9.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray M L, Killinger A H. Listeria monocytogenes and listeric infections. Bacteriol Rev. 1966;30:309–382. doi: 10.1128/br.30.2.309-382.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory S H, Barczynski L K, Wing E J. Effector function of hepatocytes and Kupffer cells in the resolution of systemic bacterial infections. J Leukoc Biol. 1992;51:421–424. doi: 10.1002/jlb.51.4.421. [DOI] [PubMed] [Google Scholar]
- 21.Hanawa T, Yamamoto T, Kamiya S. Listeria monocytogenes can grow in macrophages without the aid of proteins induced by environmental stresses. Infect Immun. 1995;63:4595–4599. doi: 10.1128/iai.63.12.4595-4599.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 23.Jonquieres R, Bierne H, Fiedler F, Gounon P, Cossart P. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol Microbiol. 1999;34:902–914. doi: 10.1046/j.1365-2958.1999.01652.x. [DOI] [PubMed] [Google Scholar]
- 24.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 25.Leimeister-Wachter M, Domann E, Chakraborty T. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J Bacteriol. 1992;174:947–952. doi: 10.1128/jb.174.3.947-952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackaness G B. Cellular resistance to infection. J Exp Med. 1962;116:381–406. [PubMed] [Google Scholar]
- 27.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 28.Mengaud J, Ohayon H, Gounon P, Mège R M, Cossart P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 29.Milohanic E, Pron B, Berche P, Gaillard J L. Identification of new loci involved in adhesion of Listeria monocytogenes to eukaryotic cells. Microbiology. 2000;146:731–739. doi: 10.1099/00221287-146-3-731. [DOI] [PubMed] [Google Scholar]
- 30.Mounier J, Ryter A, Coquis-Rondon M, Sansonetti P J. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect Immun. 1990;58:1048–1058. doi: 10.1128/iai.58.4.1048-1058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nair S, Derre I, Msadek T, Gaillot O, Berche P. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol Microbiol. 2000;35:800–811. doi: 10.1046/j.1365-2958.2000.01752.x. [DOI] [PubMed] [Google Scholar]
- 32.Nair S, Frehel C, Nguyen L, Escuyer V, Berche P. ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol Microbiol. 1999;31:185–196. doi: 10.1046/j.1365-2958.1999.01159.x. [DOI] [PubMed] [Google Scholar]
- 33.North R J. Cellular mediators of anti-Listeria immunity as an enlarged population of short lived, replicating T cells. Kinetics of their production. J Exp Med. 1973;138:342–355. doi: 10.1084/jem.138.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pederson K J, Carlson S, Pierson D E. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol Microbiol. 1997;26:99–107. doi: 10.1046/j.1365-2958.1997.5551916.x. [DOI] [PubMed] [Google Scholar]
- 35.Rosen H, Gordon S, North R J. Exacerbation of murine listeriosis by a monoclonal antibody specific for the type 3 complement receptor of myelomonocytic cells. Absence of monocytes at infective foci allows Listeria to multiply in nonphagocytic cells. J Exp Med. 1989;170:27–37. doi: 10.1084/jem.170.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouquette C, de Chastellier C, Nair S, Berche P. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol Microbiol. 1998;27:1235–1245. doi: 10.1046/j.1365-2958.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- 37.Rouquette C, Ripio M T, Pellegrini E, Bolla J M, Tascon R I, Vazquez-Boland J A, Berche P. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol Microbiol. 1996;21:977–987. doi: 10.1046/j.1365-2958.1996.641432.x. [DOI] [PubMed] [Google Scholar]
- 38.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 39.Sheehan B, Kocks C, Dramsi S, Gouin E, Klarsfeld A D, Mengaud J, Cossart P. Molecular and genetic determinants of the Listeria monocytogenes infectious process. Curr Top Microbiol Immunol. 1994;192:187–216. doi: 10.1007/978-3-642-78624-2_9. [DOI] [PubMed] [Google Scholar]
- 40.Siddique I H, McKenzie B E, Sapp W J, Rich P. Light and electron microscopic study of the livers of pregnant mice infected with Listeria monocytogenes. Am J Vet Res. 1978;39:887–892. [PubMed] [Google Scholar]
- 41.Sokolovic Z, Fuchs A, Goebel W. Synthesis of species-specific stress proteins by virulent strains of Listeria monocytogenes. Infect Immun. 1990;58:3582–3587. doi: 10.1128/iai.58.11.3582-3587.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokolovic Z, Riedel J, Wuenscher M, Goebel W. Surface-associated, PrfA-regulated proteins of Listeria monocytogenes synthesized under stress conditions. Mol Microbiol. 1993;8:219–227. doi: 10.1111/j.1365-2958.1993.tb01566.x. [DOI] [PubMed] [Google Scholar]
- 43.Squires C, Squires C L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tilney L G, Portnoy D A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood S, Maroushek N, Czuprynski C J. Multiplication of Listeria monocytogenes in a murine hepatocyte cell line. Infect Immun. 1993;61:3068–3072. doi: 10.1128/iai.61.7.3068-3072.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]