Abstract
Cryptosporidiosis is a water- and food-borne zoonotic disease caused by the protozoon parasite of the genus Cryptosporidium. C. hominis and C. parvum are the main two species causing infections in humans and animals. The disease can be transmitted by the fecal–oral route as well as the respiratory route. The infective stage (sporulated oocysts) is resistant to different disinfectants including chlorine. Currently, no effective therapeutic drugs or vaccines are available to treat and control Cryptosporidium infection. To prevent cryptosporidiosis in humans and animals, we need to understand better how the disease is spread and transmitted, and how to interrupt its transmission cycle. This review focuses on understanding cryptosporidiosis, including its infective stage, pathogenesis, life cycle, genomics, epidemiology, previous outbreaks, source of the infection, transmission dynamics, host spectrum, risk factors and high-risk groups, the disease in animals and humans, diagnosis, treatment and control, and the prospect of an effective anti-Cryptosporidium vaccine. It also focuses on the role of the One Health approach in managing cryptosporidiosis at the animal–human–environmental interface. The summarized data in this review will help to tackle future Cryptosporidium infections in humans and animals and reduce the disease occurrence.
Keywords: cryptosporidiosis, one health, poultry, vaccines, epidemiology, waterborne pathogen, foodborne pathogen, outbreaks
1. Introduction
Cryptosporidiosis is an enteric disease caused by a protozoon parasite belonging to the genus Cryptosporidium. It is one of the most prevalent waterborne diseases and the leading cause of waterborne disease outbreaks worldwide [1,2,3]. More than 58 million cases of diarrhea are detected annually in children and are associated with protozoal infections. Specifically, waterborne pathogens such as Cryptosporidium and Giardia were involved in the World Health Organization’s “Neglected Disease Initiative” [4,5,6]. Although Cryptosporidium infections are acute self-limiting gastroenteritis in immunocompetent individuals, chronic and life-threatening diarrheal disease can develop in immunocompromised individuals. Neonates are highly susceptible to infections due to their immature immune system, and they can become infected by ingestion of low doses of the parasite’s oocysts. Annually, diarrheal diseases have caused up to 1.6 million deaths worldwide. One-third of these deaths have been reported in children under 5 years due to contaminated drinking water and poor hygiene [7]. Cryptosporidium causes up to 20% of all cases of diarrhea in children in developing countries and causes fatal complications in HIV-infected persons [8]. Cryptosporidium is also responsible for more than 8 million foodborne illness cases worldwide annually [9]. Cryptosporidiosis primarily affects people who are living in rural and in urban slums, where there is a high probability of disease transmission and spread [10].
The human medical importance of Cryptosporidium was highlighted in 1982, after the CDC report on Cryptosporidium-induced diarrheas in patients infected with Human Immunodeficiency Virus (HIV). The international interest in Cryptosporidium as a public health problem began in 1993 after the largest global waterborne outbreak, when more than 400,000 inhabitants in Milwaukee, Wisconsin, USA were infected with C. hominis due to the consumption of contaminated drinking water [11,12,13]. From 2014 to 2016, the center for disease control (CDC) in the USA reported a doubled increase in the number of Cryptosporidium-associated waterborne infections [14] with an estimated 748,000 annual human cases [15]. In addition, the risk of cryptosporidiosis is increased in developing countries due to poor water and food sanitation [16]. In developing countries, children under five years old are the most affected groups with Cryptosporidium [17]. The oocysts can survive outside the host for several months and retain infectivity, despite adverse environmental conditions such as salinity and the presence of chemicals [18,19,20]. Mixed infections in calves with Cryptosporidium, enterotoxin Escherichia coli (ETEC) as well as the corona- and rotaviruses, are considered the most important reason for the calf diarrhea complex [21]. To date, there are no effective chemotherapeutics for the treatment of cryptosporidiosis [22,23]. Nitazoxanide and halofuginone in humans and animals are the approved drugs against Cryptosporidium infection. However, their application does not guarantee treatment efficacy [24,25,26]. Therefore, the control of cryptosporidiosis should be based mainly on (1) reducing the prevalence of infection, (2) breaking the transmission pathways between animals and humans, and (3) maintaining a good hygienic environment for humans and animals. Information about the route and spread of Cryptosporidium, the magnitude of infections, and the major sub-species prevailing in animals and humans, is important to achieve effective control. This epidemiological information, in addition to the One Health approach, will help to initiate planning for the control of cryptosporidiosis.
2. Life Cycle and Developmental Stages of Cryptosporidium
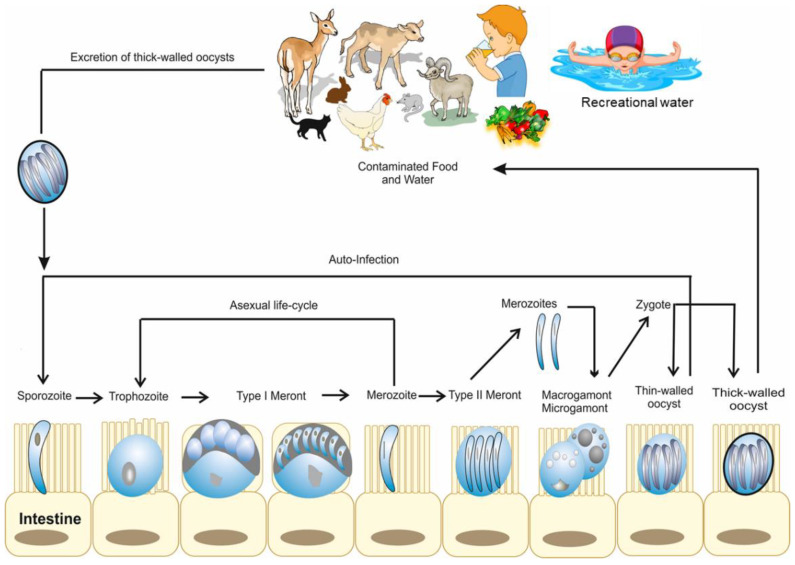
Cryptosporidium belongs to the Coccidia class of the phylum Apicomplexa. Cryptosporidium have some features which differentiate them from all other Coccidia [27], including (1) intracellular and extra-cytoplasmic localization, (2) forming of a “feeder” organ, (3) presence of morphological (thin- or thick-walled) oocysts as well as functional (auto vs. new-infection) types of oocysts, (4) small size of oocysts, (5) missing some morphological characteristics such as sporocysts or micropyles, and (6) the resistance of Cryptosporidium to all the available anti-coccidial drugs [27,28]. Cryptosporidium has a complex monoxenous life cycle, which is divided into two phases: the asexual phase (sporogony and schizogony/merogony) and the sexual (gamogony) phase. They proliferate and differentiate during the invasion of the free-living stages of Cryptosporidium within the parasitophorous vacuole under the brush border of the host cell located outside the cellular cytoplasm [29]. Cryptosporidium parasites can then attach to the cell surface and move along it for a short time using gliding mobility before they start to enter the cell. Cryptosporidium does not completely invade the cells actively, but they provoke the cells to embrace them with a host-cell-derived membrane. Additionally, at the parasite–cell interaction phase, the Cryptosporidium creates an actin-rich disk, a feeder organelle responsible for nutrition intake, as well as a channel into the cytoplasm of the host cell [30]. After Cryptosporidium internalization in the host cells, the sporozoite divides inside the parasitophorous vacuole to approximately 4 µm × 4 µm in diameter as a spherical trophozoite with an excentric cell nucleus. After three asexual divisions (merogony/schizogony), the trophozoite is divided into 5 µm × 5 µm large type-1 meront, which contains eight merozoites. The merozoites and the sporozoites are similar in shape and size; however, the nucleus of the merozoites is located more centrally to the cell compared to the sporozoites. Upon leaving the parasitophorous vacuole, the merozoites begin their asexual development cycle in the epithelial cells and develop Type-I meronts again, then the trophozoite. Otherwise, the merozoites initiate the sexual development cycle through differentiation to type-II meronts. Inside the meront, four merozoites develop by asexual division and after infection of further enterocytes, they are divided into micro- and macro-gametes (gamogony). The immature micro-gamontes are spherical, 5 µm × 4.5 µm in diameter, contain up to 16 peripherally located compact cell nuclei, and are precursors of the developing micro-gametes (Figure 1). They also have stubbed front ends and cell nuclei with no flagella. The mature micro-gametes leave their host cell and fertilize the macrogametes. Macrogametes are spherical, 5 µm × 5 µm in diameter and contain granulated cytoplasm and eccentrically positioned wall-forming bodies. Tandel et al. have suggested the direct development of gametes from type I meronts [31]. The zygote grows by syngamy and then goes through sporogony-a meiosis-like process. The oocysts (thin- or thick-walled) with 4 haploid sporozoites (sporulated oocysts) develop inside the parasitophorous vacuole (Figure 1) [30,32]. Thin-walled oocysts (about 20%) excystate in the host intestinal tract, leading to endogenous autoinfection, and the thick-walled oocysts (about 80%) are extremely resistant to several disinfectants, are excreted with the feces to the environment and can survive outside the host for a long time [33]. The thick-walled oocysts represent the exogenous stage of the Cryptosporidium parasite. Cryptosporidium oocysts are approximately 4µm× 6µm in diameter, spheric to ovoid shape, have a residual body, and four banana-like or comma-shaped sporozoites with a pointed front end and a stubbed hind end, where the nucleus is localized [34,35,36]. The residual bodies are 2.4 µm × 2.5 µm in diameter and consist of a spherical to ovoid membrane-bound globule (1.5 µm × 1.6 µm) and are surrounded by small granules (0.2 µm × 1.2 µm). Cryptosporidium sporozoites are not encapsulated by a sporocyst and the oocyst wall consists of an outer and an inner layer, and a pre-formed junction that extends from one pole of the oocyst to approximately half of the oocyst [34]. Additionally, four sporozoites (5µm× 1µm in diameter) hatch out of the pre-formed joint under the effect of temperature, pH, gall bladder salts, pancreas enzymes, and CO2 of the host gastrointestinal tract. The free sporozoites adhere to the microvilli of the enterocytes and lead to internalization using their proximal end. The sporozoites’ glycoproteins (GP40 and GP900 of 40 kDa and ˃900 kDa) and the circumsporozoite-like glycoprotein (CSL) play an important role in the adhesion and invasion process of the sporozoites to the host cells [32,37]. The host cell surrounds the sporozoites with membrane protrusions and forms a parasitophorous vacuole in the brush border of the enterocyte. Interestingly, the localization of the parasitophorous vacuole by Cryptosporidium spp. is different from that of the other Apicomplexa; thus, Cryptosporidium spp. localization is described as intracellular, but extracytoplasmic [38]. Additionally, the feeder organelle develops at the sporozoite and host cell membrane contact point. They supply the maturing parasite with nutrients and facilitate internalization [39]. The molecular components and mechanisms involved in the Cryptosporidium development cycle have previously been described [30].
Figure 1.
Life cycle and developmental stages of Cryptosporidium in animals and humans [46].
The infectious stage (sporulated oocyst) of Cryptosporidium was reported to be excreted in large numbers in the feces of experimentally infected calves (up to 4 × 107 oocysts per gram of feces) [40], or excreted with the bronchial exudates in the case of respiratory cryptosporidiosis and which immediately contaminated the environment [41]. The sporulated oocysts are very resistant to environmental factors and only a few chemical disinfectants show efficacy against the sporulated oocysts due to their thick wall [42]. Therefore, it is difficult to completely remove the Cryptosporidium oocysts from contaminated drinking water [43]. The thick wall oocysts are sporulated and are infectious when shedding, which can result in immediate infection of new hosts. The infectious dose of Cryptosporidium oocysts for humans is about nine oocysts per Cryptosporidium isolate and about 50 oocysts for calves [44,45]. However, it was reported that 1 to 10 oocysts of Cryptosporidium caused infection for some individuals during the Milwaukee outbreak [42]. Although, one infected host can shed up to 1010 oocysts, which results in a huge infection pressure.
3. Pathogenesis of Cryptosporidium
After ingestion of the thick-walled oocyst with food or water by the host, many signaling molecules are expressed on the sporozoite surface that mediate their attachment and invasion to the host cells. Calcium-dependent protein kinases (CDPKs) were reported to be involved in the regulation of the invasion process of the sporozoite to the host cell [30,47]. Furthermore, Cryptosporidium is embraced by the host cell instead of invading the host cells. Therefore, it stays in an epicellular location and this induces tremendous actin rearrangement in the infected cells [30]. After attachment and invasion of Cryptosporidium, the host–parasite interactions play an important role in pathogenesis [48]. In calves, C. parvum causes acute to chronic catarrhal enteritis that begins in the distal ileum; however, different Cryptosporidium developmental stages were also detected in the duodenum, colon, and part of the cecum. The affected mucosa is hyperemic and edematous and the mesenteric lymph nodes are partially enlarged and edematous [49]. Histologically, mild to moderate villus atrophy associated with occasional villus fusion was observed. The affected crypts are partially dilated and contain neutrophil granulocytes. The lamina propria mucosa also had neutrophil granulocytes and a large mononuclear cell infiltration [50]. In the infected host, epithelial cell degeneration, metaplasia of physiological high prismatic to isoprismatic villus epithelial cells, hyperplastic crypt epithelium, displacement of microvilli in the area of the intracellular parasite stages’ attachment zone, and long microvilli can be seen in the vicinity of the parasite stage [51]. These pathological alterations result in the reduction of the intestinal absorption surface and, consequently, malabsorption. Damage to the intestinal epithelium may also have an impact on the activity of brush border membrane enzymes (glucoamylase, alpha-dextrinase, saccharase, lactase), resulting in a reduction in the small intestine’s carbohydrate digestion ability. As a result, osmotically active particles persist in the intestinal lumen, osmotic diarrhea develops, and water resorption is impeded. Several causes can lead to increased chloride secretion into the gut lumen, including immune response to membrane injury, prostaglandins secreted by enterocytes of intra- and sub-epithelial lymphocytes, and plasma cells and macrophages that enhance blood vessel permeability [50].
4. Species, Genotypes/Subtypes, and Host Spectrum of Cryptosporidium
Currently, there are more than 40 morphologically and molecular-biologically different Cryptosporidium species [52,53,54,55,56], which infect mammals (Bovidae, Primates, Carnivora, Hares, Equidae, Rabbits, Rhinocerotidae, and Tapiridae), amphibians, birds, and reptiles. Additionally, more than 157 mammalian species were listed as hosts for Cryptosporidium infection [57]. However, Cryptosporidium species including C. hominis, C. bovis, C. parvum, C. ryanae, C. andersoni, C. fayeri, C. canis, C. felis, C. macropodum, C. muris, C. suis, and C. wrairi have been isolated from mammals. C. meleagridis, C. baileyi, and C. galli have been isolated from birds [58], while C. varanii and C. serpentis have been isolated from reptiles and C. fragile has been isolated from amphibians (Table 1) [59]. Additionally, C. rubeyi has been isolated from squirrels, C. scophthalmi from turbot, C. huwi from fish, and C. erinacei from horses and hedgehogs [60]. Human cryptosporidiosis is caused by C. hominis, while C. parvum is considered the zoonotic species of human cryptosporidiosis [61]. Both C. hominis and C. parvum are responsible for more than 90% of human cryptosporidiosis. Although there is host specificity of the Cryptosporidium species, other species such as C. meleagridis, C. baileyi, C. andersoni, C. canis, C. felis, C. bovis, C. suis, C. fayeri, C. scrofarum, C. tyzzeri, C. erinacei, and C. muris have been detected in animal hosts as well as in humans. The aforementioned species and C. parvum have been considered potentially zoonotic species [62,63]. Additionally, humans can also be infected with C. viatorum, C. cuniculus, C. ubiquitum, Chipmunk genotype I, Cryptosporidium horse, and Cryptosporidium mink genotype (Table 1) [64].
Table 1.
Most predominant Cryptosporidium species: major hosts, oocyst sizes and locations [18,58,73,74,75,76].
| Cryptosporidium spp. | Hosts | Sporulated Oocyst Size (µm) | Location |
|---|---|---|---|
| C. hominis | Humans | 4.5 × 5.5 | Small intestine |
| C. parvum | Ruminants, humans, deer | 4.5 × 5.5 | Small intestine |
| C. bovis | Ruminants | 4.2–4.8 × 4.8–5.4 | Small intestine |
| C. andersoni | Ruminants, camel | 5.5 × 7.4 | Abomasum |
| C. ryanae | Ruminants | 3.2 × 3.7 | Small intestine |
| C. xiaoi | Sheep | 3.9 × 3.4 | Small intestine |
| C. ubiquitum | Sheep/wildlife | 5.2 × 4.9 | Small intestine |
| C. meleagridis | Chicken, turkey, humans | 4.5–5.0 × 4.6–5.2 | Intestine |
| C. baileyi | Birds | 6.4 × 6.2 | Cloaca, bursa, respiratory tract |
| C. galli | Birds | 8.0–8.5 × 6.2–6.4 | Proventriculus |
| C. avium | Birds | 5.3–6.9 × 4.3–5.5 | Intestine |
| C. ornithophilusis | Ostrich | 6.13 × 5.15 | Intestine |
| C. proventriculi | Psittaciformes birds | 7.4 × 5.8 | Proventriculus |
| Avian genotype II | Birds | 6.0–6.5 × 4.8–6.6 | Intestine |
| Avian genotype IV | Birds | 8.25 × 6.3 | Intestine |
| Eurasian woodcock genotype | Birds | 8.5 × 6.4 | Intestine |
| C. suis | Pigs, humans | 5.1 × 4.4 | Small intestine |
| C. wrairi | Guinea pigs | 4.0–5.0 × 4.8–5.6 | Small intestine |
| C. cuniculus | Rabbits | 5.9 × 5.4 | Small intestine |
| C. canis | Canids, humans, mink, fox, coyote | 5.0 × 4.7 | Small intestine |
| C. felis | Felids, humans | 4.5 × 5.0 | Small intestine |
| C. saurophilum | Lizards, snakes | 4.2–5.2 × 4.4–5.6 | Intestinal and cloacal mucosa |
| C. serpentis | Snakes, lizards | 4.8–5.6 × 5.6–6.6 | Stomach |
| C. fayeri | Red Kangaroo, marsupials | 4.9 × 4.3 | Intestine |
| C. macropodum | Marsupials | 4.9 × 5.4 | Small intestine |
| C. muris | Rodents, humans | 5.6 × 7.4 | Stomach |
| C. ratti | Rodents | 4.5–5.4 × 4.5–5.0 | Small intestine |
| C. tyzzeri | Mice | 4.6 × 4.2 | Small intestine |
| C. molnari | Fish | 4.7 × 4.5 | Stomach |
| C. scophithalmi | Fish | 3.0–4.7 × 3.7–5.0 | Intestine |
| C. nasorum | Fish | 4.3 × 3.2 | Intestine |
Currently, there are more than 60 reported genotypes of Cryptosporidium that differ in their molecular sequences [56,65]. Cryptosporidium subtypes are distinguished by the number of repeats in each strand. Short, repetitive sequences (R) appear directly after the trinucleotide repeats in some subtypes. In C. parvum, 11 subtype families (IIa- IIk) have been discovered with at least 78 subtypes. Furthermore, in C. hominis, six subtype families have been detected (Ia, Ib, Id, Ie, If, and Ig) with at least 78 subtypes [63,66,67,68]. In C. meleagridis, seven subtype families have been identified (IIIa- IIIg), while six subtype families were identified in C. fayeri (IVa- IVf), and two in C. cuniculus (Va, Vb), Horse genotype (VIa, VIb), and C. tyzzeri (IXa, IXb), whereas one subtype was identified in C. erinacei (XIIIa), Mink genotype (Xa), Ferret genotype (VIIIa), and C. wrairi (VIIa) [63]. Several highly preserved genes, including (1) small subunit rRNA (18S rRNA), (2) Cryptosporidium oocyst wall protein (COWP), (3) heat shock protein (HSP70), and (4) the actin gene, can differentiate between C. parvum and C. hominis. The 18S rRNA gene is crucial because it contains multiple conserved regions within the Cryptosporidium genus. This makes primer development that targets most Cryptosporidium species easier. The amplification of the extracted DNA from the oocysts can be performed using conventional or nested polymerase chain reaction (nPCR). It is difficult to identify the mixed infections of distinct Cryptosporidium genotypes by using PCR with 18S rRNA, COWP, HSP70, and the actin gene. On the other hand, the GP60 gene is advantageous because the species with the highest affinity for the primer (species-specific) will be amplified to a greater extent than the others, allowing the dominant species to be identified alone [69]. Additionally, the 5’ end of the GP60 gene has a highly variable area of microsatellites, which consists of trinucleotide repeats (TCA, TCG, TCT), which all code for the amino acid serine. Amplicon next-generation sequencing (NGS), which can identify low-abundance sequences in mixed infections, has shown that it can identify additional Cryptosporidium gp60 subtypes in various hosts that were not identified by Sanger sequencing [70,71]. This has important implications for tracing the zoonotic transmission of Cryptosporidium, as Sanger sequencing may not detect zoonotic species and subtypes that are present at low abundance and therefore incorrect conclusions regarding zoonotic transmission may be made [72].
5. Epidemiology of Cryptosporidiosis
5.1. Source of Infection and Mode of Transmission
5.1.1. In Humans
The zoonotic transmission of Cryptosporidium can take place via direct contact with an infected person and/or consumption of contaminated drinking water or food and/or inhalation of oocysts from contaminated air with aerosolized droplets or fomites [41,77]. Additionally, synanthropic flies (suborder: Cyclorapha) play a crucial role in the mechanical transmission and spread of infection [78]. There are multiple factors leading to human cryptosporidiosis [61] and the occurrence of outbreaks, such as (1) contaminated drinking water, and unclean recreational/swimming pool water, (2) contaminated foods such as raw fruits and vegetables that were fertilized with contaminated effluent, (3) contact with infected people (hospitals, daycare centers, schools), (4) contact with infected animals (especially calves), and (5) anal sexual contact [42]. Even though cryptosporidiosis is primarily a water-based illness, the risk of foodborne transmission is well known. Food contamination with Cryptosporidium oocysts can occur during food (vegetables, fruits, seafood, and meat) manufacturing, processing, and preparation. The oocysts’ resistance can help them survive various processing procedures, such as chlorine baths and blast freezing [79]. Furthermore, washing fresh fruit may not be enough to eliminate contaminated oocysts, which not only stick to surfaces but can also permeate leafy vegetables through stomatal pores [80,81]. There have been fewer reported foodborne cryptosporidiosis outbreaks than waterborne infections.
5.1.2. In Animals
Calves usually become infected with cryptosporidiosis by ingestion of oocysts from the contaminated environment. There are many possible sources of infection including (1) shedding of infected neighbor animals, (2) contaminated stables, (3) dirty udders and teats of cows, and (4) contaminated water. The subclinical infected adult cattle act as oocysts shedders [82,83], therefore they are considered a potential reservoir for infection. Furthermore, Cryptosporidium infection can be also transmitted by animal handling personnel through dirty shoes and clothes as well as via infected dogs, cats, rodents, wild animals, insects (flies, cockroaches, and beetles), and free-living amoeba [84,85]. Mixed infection of Cryptosporidium together with enterotoxin E. coli, Corona- and Rotaviruses is considered one of the most common causes of neonatal calf diarrhea. The prevalence of bovine cryptosporidiosis ranged between zero and 100% and the prevalence tends to decrease with the increasing age of the animal [86]. There is variation in the tendency of Cryptosporidium species to infect calves in an age-dependent manner. For example, C. parvum is the most prevalent species in calves up to 8 weeks old, while C. bovis is dominant in calves ranging between 2 to 11 months of age [87,88].
5.2. Clinical Signs and High-Risk Groups of Cryptosporidiosis
5.2.1. In Humans
Many risk factors are implicated in the zoonotic transmission of Cryptosporidium infection. These factors include contact with infected animals, age (infection rate is higher in young animals and humans), gender (infection is higher in males compared to females), poverty, overcrowding, season (rise of cases around rainy season), poor water quality, poor hygiene measures, the status of the host immunity, exposure to HIV–infected people [89], and natural disasters (storms, earth erosions, floods) [90]. The high-risk groups of people that can be exposed to Cryptosporidium infection include: (1) children in childcare centers, (2) childcare workers who change children’s diapers, (3) parents or attendees of infected children, (4) the elderly (75 years and older), (5) travelers to/from endemic areas, (6) swimmers who swallow contaminated water, (7) people handling infected animals and birds, (8) people who have been sexually exposed to human feces, (9) people taking care of other people who are infected, (10) people who drink from untreated water such as backpackers, hikers, and campers [91], (11) organ transplant recipients, and (12) other occupational associated groups such as veterinarians, animal handlers (sweepers, vaccinators, debeaking staff), pet owners, and hunters.
The severity of clinical signs in infected humans depends on the age and the immunity of the infected person [92,93]. The incubation period in immunocompetent people is from 5 to 21 days, followed by acute self-limiting diarrhea that lasts 3 to 12 days. The clinical signs range from medium to profuse watery to catarrhal diarrhea, which is often associated with abdominal pain, nausea, vomiting, flatulence, fatigue, and anorexia. Respiratory symptoms such as cough, sneezing, and expectoration may occur after inhalation of oocysts from contaminated air [41,56]. Asymptomatic infection can also occur [94,95]. However, the infection can develop into a chronic and life-threatening disease in immunocompromised persons [95], specifically people suffering a genetic immunological malfunction such as hyper-IgM syndrome, a significant reduction in the number of CD4-lymphocytes such as HIV infection, or those undergoing immunosuppressive therapy after organ transplantation [96]. Cryptosporidium has been isolated from the gallbladder and the respiratory tract of HIV/AIDS patients as well as from patients suffering from severe combined immune deficiencies (SCID), causing cell-mediated immunity deficiency, and extra-intestinal forms (in the ductus pancreaticus a.o. and the respiratory bronchioles) [96]. Differences in clinical symptoms have been noted between C. parvum and C. hominis in children and HIV/AIDS patients, with C. parvum being less virulent than C. hominis [97,98]. In HIV patients, C. parvum infections are mostly associated with vomiting and chronic diarrhea and are more frequent than C. hominis infections [97]. Additionally, Cryptosporidium infection at a young age has been linked to stunted growth and long-term cognitive problems, particularly in children in developing countries [42].
5.2.2. In Livestock Animals
Cryptosporidiosis is more frequent in young calves and the severity of the disease depends on various factors including age, infectious dose, immunity of the host, season, geographical distribution, and mixed infection with other pathogens [99]. The clinical signs vary from asymptomatic to pasty or watery profuse diarrhea, dehydration, and death. Co-infections of C. parvum with enterotoxin E. coli, Coronaviruses, and Rotaviruses can occur within the first three weeks of age and are considered one of the major causes of mortality in calves [1,94]. Neonatal diarrhea with a single or mixed C. parvum infection is characterized by yellowish, profuse diarrhea and is associated with complications such as exsiccosis, metabolic acidosis, and loss of electrolytes [50,100]. Consequently, cryptosporidiosis results in severe economic losses due to morbidity, growth retardation, and treatment costs [101,102]. The prevalence of Cryptosporidium in animals varies according to the geographical area, animal species, rearing forms, and the diagnostic tests. For example, the prevalence reached up to 100% in goats and horses in South America and sheep in Europe. Additionally, the highest prevalence of infection in buffalo was reported in Africa (52.0%) and Asia (50%) compared to other continents. The highest prevalence (more than 50%) in cattle was reported in all the continents except South America. The highest prevalence in pigs was reported in Asia (55.8%) (Table 2).
Table 2.
| Continents | Animal Species | Diagnostic Test * | Prevalence Range |
|---|---|---|---|
| South America | Buffalo | CM, PCR | 9.4–48.2% |
| Cattle | CM, ICT, PCR | 3.0–56.1% | |
| Goat | CM | 4.8–100% | |
| Sheep | CM, PCR | 0.0–25.0% | |
| Pig | CM, PCR | 0.0–2.2% | |
| Horse | CM | 0.0–100% | |
| Calves | CM, ELISA, PCR | 84.2% | |
| North America | Cattle | CM, IFA, PCR | 1.1–78.0% |
| Goat | CM | 20.0–72.5% | |
| Sheep | CM, IFA, PCR | 20.0–77.4% | |
| Pig | CM, IFA | 2.8–19.6% | |
| Horse | CM, IFA, PCR | 0.0–17.0% | |
| Africa | Buffalo | CM, PCR | 1.3–52.0% |
| Cattle | CM, ELISA, PCR | 0.5–86.7% | |
| Goat | CM, ELISA | 0.0–76.5% | |
| Sheep | CM, ELISA, PCR | 1.3–41.8% | |
| Pig | CM, ELISA, IFA, PCR | 13.9–44.9% | |
| Horse | CM, PCR | 0.0–2.9% | |
| Asia | Buffalo | CM, ICT, PCR | 3.6–50.0% |
| Cattle | CM, ICT, IFA, PCR | 1.5–93.0% | |
| Goat | CM, ICT, IFA | 0.0–42.9% | |
| Sheep | CM, ELISA, ICT, PCR | 1.8–66.6% | |
| Pig | CM, IFA, PCR | 0.4–55.8% | |
| Horse | CM, PCR | 2.7–37.0% | |
| Europe | Buffalo | ELISA | 14.7% |
| Cattle | CM, ELISA, ICT, IFA, PCR, QLAT | 0.0–71.7% | |
| Goat | CM, ELISA, IFA | 0.0–93.0% | |
| Sheep | CM, IFA, ELISA | 1.4–100% | |
| Pig | CM, IFA, PCR | 0.1–40.9% | |
| Horse | CM, ELISA, IFA, PCR | 3.4–25.0% | |
| Australia | Buffalo | PCR | 13.1–30.0% |
| Cattle | CM, IFA, PCR | 3.6–73.5% | |
| Goat | PCR | 4.4% | |
| Sheep | PCR | 2.2–81.3% | |
| Pig | CM, PCR | 0.3–22.1% |
* CM, conventional microscopy; IFA, immunofluorescence antibody test; ELISA, enzyme-linked immunosorbent assay; ICT, immunochromatographic test; QLAT, quantitative latex agglutination; and PCR, polymerase chain reaction. The reported prevalence range was summarized from different research articles.
5.2.3. In Poultry
Several Cryptosporidium species can infect birds, including C. meleagridis, C. galli, and C. baileyi [58]. These species have different predilection sites. For example, C. meleagridis and C. baileyi can develop in the small and large intestines as well as the bursa of Fabricius, causing different degrees of enteritis. C. galli was reported to infect finches, chickens, and grosbeaks and infect only the proventriculus, while C. meleagridis was reported to infect turkeys and parrots. C. baileyi is the most common avian Cryptosporidium that can infect chickens, turkeys, cockatiels, quails, ostriches, and ducks [105]. Cryptosporidium species can also multiply in the tissues of the respiratory tract of the infected birds. Additionally, it causes enteritis and renal disease, due to inflammation of Fabricius’ bursa and kidneys [105,106]. There are approximately 11 Cryptosporidium genotypes that have been detected from more than 30 bird species, including avian I–V, duck genotype, goose genotypes I–IV, and the Eurasian Woodcock genotype [76]. However, Cryptosporidium avian genotype III has been associated with chronic vomiting in peach-faced lovebirds (Agapornis roseicollis) [107]. Cryptosporidium species such as C. hominis, C. parvum, and muskrat genotype have also been isolated from Canada geese (Branta canadensis) [108,109,110]. Recently, C. ornithophilusis was isolated from farmed ostrich in the Czech Republic [74], while C. avium was isolated from red-crowned parakeets [75]. The prevalence of Cryptosporidium has been investigated in different species of poultry worldwide. The prevalence ranged between 0.8% in pigeons to 50% in broilers and layers (Table 3). The most detected Cryptosporidium species were C. baileyi, C. meleagridis, C. galli, and C. parvum. In some countries, scientists have also been able to isolate other species such as C. avium from China, C. muris from China and Australia, and C. andersoni from Australia (Table 3).
Table 3.
Prevalence of Cryptosporidium species of birds in different countries.
| Country | Species/Genotype | Host | Prevalence | Reference |
|---|---|---|---|---|
| Brazil | C. meleagridis, C. baileyi | Chicken, turkey, quail | 14.8% | [111] |
| Brazil | C. baileyi, C. parvum, C. meleagridis | Chickens | 12.6% | [112] |
| China | C. baileyi | Chickens | 2.4% | [113] |
| Iraq | C. baileyi, C. parvum, C.galli, C. meliagredis | Broilers, layers | 50% | [114] |
| Iraq | C. parvum and C. baileyi | Wild pigeons | 6.0% | [115] |
| Iran | C. parvum and C. baileyi | Broilers | 8.0% | [116] |
| China | C. parvum and C. baileyi | Wild birds | 8.9% | [117] |
| Bangladesh | C. baileyi, C. meleagridis, C. parvum | Layers, broilers, pigeons | 15.7% | [118] |
| China | C. avium, C. baileyi, C. galli, C. meleagridis | Chickens | 13.7% | [119] |
| Germany | C. parvum, C. baileyi | Turkey, broilers, layers | 7.0% | [58] |
| Spain | C. meleagridis, C. parvum | Wild birds | 8.3% | [120] |
| China | C. baileyi, C. meleagridis | Pigeons | 0.8% | [121] |
| Czech Republic | C. baileyi, C. meleagridis | Red-legged partridge | 22% | [122] |
| China | Avian genotype II, C. baileyi, C. meleagridis | Chickens | 9.9% | [123] |
| China | C. baileyi, C. muris | Ostrich | 10.2% | [123] |
| Vietnam | Avian genotype II | Ostrich | 23.7% | [124] |
| Algeria | C. baileyi, C. meleagridis | Broilers | 9–69.0% | [125] |
| Algeria | C. meleagridis | Turkey | 43.9% | [126] |
| China | C. baileyi, C. meleagridis | Japanese quail | 13.1% | [127] |
| China | C. galli, C. meleagridis, C. baileyi, C. parvum, Avian genotypes I, II, III, V | Pet birds | 8.1% | [128] |
| Brazil | C. baileyi, Avian genotype II, C. galli | Wild birds | 6.6% | [129] |
| China | C. baileyi | Ostrich | 11.7% | [130] |
| China | C. baileyi | Pekin ducks | 16.6% | [131] |
| China | C. baileyi, C. meleagridis | Chickens | 8.9% | [131] |
| USA | C. parvum | Turkey | 6.3% | [132] |
| Brazil | C. baileyi, Avian genotypes I, II, III, C. galli, C. meleagridis, C. parvum | Captive birds | 4.9% | [133] |
| Australia | Avian genotypes I, II, III, C. andersoni, C. baileyi, C. galli, C. muris | Several avian species | 6.3% | [134] |
5.3. Outbreaks of Cryptosporidiosis in Humans
The first waterborne cryptosporidiosis outbreak was reported in 1993 in Milwaukee, Wisconsin (USA), with an estimated 403,000 people affected, 4400 hospitalizations, and more than 100 deaths [11,12,13]. The CDC reported a doubled increase in the number of Cryptosporidium-associated waterborne outbreaks from 2014 to 2017 [14]. Between 2009 and 2017, there were more than 444 reported outbreaks in the USA [2]. The number of outbreaks reported has increased by an average of 13% annually. These outbreaks have resulted in 7465 infected cases with 287 hospitalizations and 1 death. Out of these outbreaks, 156 outbreaks resulted in 4232 cases and 183 hospitalizations and were associated with exposure to Cryptosporidium in pools or waterparks. Among these outbreaks, 14.6% were linked to contact with cattle, and 12.8% were linked to contact with infected persons in childcare settings. Among the 22 foodborne outbreaks, 40.9% were linked to unpasteurized milk and 18.2% were linked to unpasteurized apple cider. However, the mode of transmission was unknown for 14.2% of the outbreaks [2]. Interestingly, salad consumption was incriminated in 35% of cases [9]. Between 2010 and 2020, most of the waterborne outbreaks were caused by C. hominis (72%), while the majority of foodborne outbreaks were caused by C. parvum (96.5%; Table 4) worldwide [135]. Interestingly, most of the reported waterborne outbreaks were linked to swimming pools, whereas most foodborne outbreaks were linked to unpasteurized raw milk and eating salad. During these outbreaks, the most predominant identified C. hominis subtype was IfA12G1 in the USA, IbA10G2 in the UK, Sweden, and Australia, and IbA9G2 in French Guiana and Germany. Over the last 10 years, C. hominis subtype IfA12G1 was responsible for approximately 50% of C. hominis-related waterborne outbreaks in the USA. Furthermore, the most predominant identified C. parvum subtype was IIaA15G2R1 in the USA and UK, and IIaA19G1R1 in Norway (Table 4). The majority (64.3%) of foodborne outbreaks caused by C. parvum were due to IIa and only 35.7% were due to IId subtypes, which are common in livestock, suggesting its important role in foodborne outbreaks [136].
Table 4.
Recent reported outbreaks of human cryptosporidiosis [135].
| Country | Year | No. of Cases | Species/Subtype | Source | No. of Outbreaks | References |
|---|---|---|---|---|---|---|
| USA | 2017 | 41 | C. hominis IfA12G1 and IaA15R3 | Swimming pool | 3 | [137] |
| 2016 | 1373 | C. hominis IbA10G2 and IfA12G1, and C. parvum IIaA17G1R1 and IIaA15G2R1 | Swimming pool and water park | 16 | [137] | |
| 2016 | 10 | C. parvum IIaA15G2R1 and IIaA18G3R1 | Raw cow milk | 2 | [137,138] | |
| 2015 | 55 | C. hominis IfA12G1 | Swimming pool | 1 | [139] | |
| 2015 | 103 | C. parvum IIaA17G2R2 | Raw milk | 1 | [137] | |
| 2014 | 68 | C. hominis IdA17and IfA12G1 | Swimming pool, water slide, and fountain | 4 | [137] | |
| 2014 | 11 | C. parvum IIaA16G3R1 | Unpasteurized goat milk | 1 | [140] | |
| 2013 | 67 | C. hominis IaA28R4 and IfA12G1, and C. parvum (unknown subtype) | Swimming pool, lake, water park, and fountain | 6 | [137] | |
| 2013 | 172 | C. parvum IIaA15G2R1 and C. parvum (unknown subtype) | Drinking water | 3 | [137] | |
| 2013 | 21 | C. parvum IIaA17G2R1 and C. hominis (unknown subtype) | Unknown | 3 | [137] | |
| 2012 | 182 | C. hominis IbA10G2, and C. parvum IIaA16G3R1, IIaA15G2R1, and IIaA16G2R2 | Lake, fountain, water park, and swimming pool | 9 | [137] | |
| 2011 | 44 | C. hominis IaA15R3 and IaA28R4 | Water park and swimming pool | 2 | [137] | |
| 2010 | 162 | C. hominis IaA24R4, IaA28R4, IbA10G2 and IdA15G1 | Splashpad, lake, water park, and swimming pool | 4 | [137] | |
| UK | 2017 | 43 | C. hominis IbA10G2 and IbA12G3 | Swimming pool | 2 | [141] |
| 2016 | 111 | C. hominis IbA10G2 and IdA16, and C. parvum (unknown subtype) | Swimming pool | 10 | [141] | |
| 2015 | 83 | C. hominis IbA10G2 and IaA14R3, and C. parvum IIaA15G2R1 and IIaA26G1R1 | Swimming pool and hydrotherapy pool | 11 | [141] | |
| 2015 | 424 | C. parvum IIdA24G1 | Salad | 1 | [135] | |
| 2014 | 109 | C. hominis IaA14R3, IaA20R3, IbA10G2 and IdA25, and C. parvum IIaA15G2R1 and IIdA17G1 | Swimming pool and hydrotherapy pool | 11 | [141] | |
| 2014 | 12 | C. parvum IIaA15G2R1 | Drinking water | 1 | [141] | |
| 2013 | 94 | C. hominis IbA10G2 and IA14R3 | Swimming pool and paddling pool | 5 | [141] | |
| 2013 | 23 | C. hominis IbA10G2 and IdA18 | Public drinking water supply | 1 | [141] | |
| 2013 | 11 | C. parvum IIaA15G1R1 | Unpasteurization dairy milk | [141] | ||
| 2012 | 176 | C. hominis IbA10G2 and C. hominis (unknown subtype) | Swimming pool and hydrotherapy pool | 10 | [141] | |
| 2012 | 648 | C. parvum IIaA15G2R1 | Pre-cut mixed salad leaves | 1 | [142] | |
| 2011 | 21 | C. hominis IbA10G2 and C. hominis (unknown subtype) | Swimming pool | 1 | [141] | |
| 2010 | 78 | C. hominis (unknown subtype) | Swimming pool | 2 | [143,144] | |
| Sweden | 2019 | 122 | C. parvum IIdA22G1c | Spinach in vegetable juice | 1 | [145] |
| 2011 | 872 + 730 | C. hominis | Public drinking water source | 2 | [146] | |
| 2010 | 27,000 | C. hominis IbA10G2 | Public drinking water source | 1 | [147] | |
| 2010 | 16 + 89 | C. parvum IIdA20G1e and C. parvum IIdA24G1 | Salad garnish on chanterelle sauce | 2 | [148] | |
| French Guiana | 2014 | 12 | C. hominis IbA9G2, IbA10G2, IbA15G1 | Playing and bathing in a river | 1 | [149] |
| Germany | 2013 | 167 | C. hominis IbA9G2 | Playing and bathing in a river | 1 | [150] |
| Ireland | 2012 | 12 | C. parvum IIaA20G3R1 | Public drinking water supply | 1 | [151] |
| Norway | 2018 | 6 | C. parvum IIaA14G1R1 | Apple juice | 1 | [152] |
| 2012 | 145 | C. parvum IIaA19G1R1 | Goat kids and lambs | 1 | [153] | |
| Finland | 2012 | >250 | C. parvum IIdA17G1 | Salad | 5 | [154] |
| South Korea | 2012 | 126 | C. parvum (unknown subtype) | Tap water from the underground water tank | 1 | [155] |
| Australia | 2012 | 18 | C. hominis IbA10G2 | Swimming pool | 1 | [156] |
| Canada | 2010 | 12 | C. hominis (unknown subtype) | Recreational water park | 1 | [3] |
6. Diagnosis of Cryptosporidium
There are several methods used for the detection of Cryptosporidium directly in fecal samples, including microscopy detection of the oocysts either by using flotation or sedimentation techniques to determine the number of oocysts in the stool [33]. The oocyst detection limit using a microscope has been recorded as low as 50,000 to 500,000 oocysts per gram of feces. Direct detection of Cryptosporidium oocysts is usually done by microscopy without any staining and/or by the modified Ziehl–Neelsen stain, where the oocysts are stained purple with a blue background. Fecal smears can be also tested microscopically after staining with the Heine technique or Kinyoun’s Carbol fuchsin staining technique [157]. Additionally, the immunofluorescent antibody-based (IFA) staining techniques using monoclonal antibodies against the oocyst wall antigen are also widely used. These are characterized by high sensitivity and are cheaper compared to other traditional staining methods [73]. In general, the parasitological methods for Cryptosporidium detection do not differentiate between viable and non-viable oocysts.
Serological methods are considered the best tools for the screening of large numbers of samples, particularly in epidemiological surveys. The serological tests include enzyme-linked immunosorbent assays (ELISA) and enzyme-linked immunoelectron transfer blots (EITB; Western blot). The enzyme immunoassay (EIA) methods have many advantages as they are faster, easy to perform, inexpensive, and more sensitive compared to the immunofluorescence methods [33]. Rapid immunochromatographic (strip) tests can also be used [158,159] as they are used for the detection of the oocyst cell wall proteins using monoclonal antibodies [160].
The molecular diagnosis of Cryptosporidium using nucleic acid detection techniques can differentiate between viable and non-viable oocysts [161]. They can also identify species, genotypes, and subtypes, which is crucial for detecting Cryptosporidium prevalence and transmission routes. [162]. Molecular methods include random amplified polymorphic DNA PCR (RAPD-PCR), single-round and nested PCR, reverse transcription PCR (RT-PCR), arbitrary primed PCR (AP-PCR), single-strand conformation polymorphism (SSCP) analysis, crypto PMA-PCR, real-time PCR followed by restriction fragment length polymorphism (RFLP) analysis, melting curve analysis, microarray, and DNA sequencing [163]. These PCR-based methods are more sensitive than conventional microscopical and serological methods and are considered a gold standard [158]. Molecular techniques are very popular as they are used for the differentiation and genotyping of C. parvum and C. hominis [164]. Molecular diagnosis can detect the target genes of Cryptosporidium such as 18S rRNA, COWP, HSP70, and the actin gene. The 18S rRNA gene-specific PCR is extremely useful for detecting a conserved area in the gene or distinguishing between Cryptosporidium spp. (targeted nucleotide segments with varied nucleotide sequences) [33,165]. Furthermore, restriction enzymes are employed to differentiate species by digesting amplicons into fragments of varying sizes based on the species, causing the products to migrate at different distances on the gel [166]. Gene sequencing can also be used to identify various Cryptosporidium species by using pure DNA that has been amplified using internal primers and tagged with colored nucleotide bases that emit light at different wavelengths [167]. Using the Basic Local Alignment Search Tool, the generated forward and reverse sequences can be assembled into contigs and compared to sequences deposited in the Gene Bank. The dominant species or species with a strong affinity for the primers will be amplified to a greater extent than others, making it difficult to identify the mixed infection using PCR. At gene sequencing, amplification of more than one species manifests itself as multiple peaks in many sites and has difficulty assembling contigs. A combination of various species/genotype-specific primers or cloning of single amplicons produced in the area is required for the successful analysis of mixed infection [168]. Another possibility is to use species-specific primers to undertake GP60 subtype analysis. This GP60 gene is targeted for neutralizing antibodies and is expressed on the apical surface of invading stages (sporozoites and merozoites) [169]. GP60 subtyping can also help in the determination of virulence of different C. parvum and C. hominis subtypes [170].
7. Prevention and Treatment of Cryptosporidium Infection
7.1. One Health Approach for the Control of Cryptosporidiosis
The “One Health” approach is a worldwide strategy that is used to mitigate zoonotic diseases and improve health by preventing infection occurrence at the human–animal–environment interface. Collaboration between all health sectors (veterinarians, occupational health physicians, and public health operators) can help in infection control by enhancing the educational system, status of thinking, legislation, and administrative structures [171]. The One Health approach has been previously proposed to tackle cryptosporidiosis as well as other zoonotic diseases [171,172,173], since there is a critical need for close One-Health-oriented interactions among professionals working in diverse fields such as physicians, veterinarians, diagnosticians, epidemiologists, public health experts, ecologists, economists, social scientists, governments, decision-makers, and pharmaceutical industries. In this review, we propose using the One Health approach as prophylactic prevention for Cryptosporidium infection in humans, animals, and the environment through understanding the disease pathogenesis, life cycle, genomics, epidemiology, previous outbreaks, source and transmission dynamics, host spectrum, risk factors, high-risk groups, disease in animals and humans, diagnosis, treatment and control, and the prospect of effective anti-Cryptosporidium vaccines. The One Health approach includes (1) increasing public health awareness about cryptosporidiosis and its ways of transmission, (2) breaking the parasite’s transmission cycle, (3) epidemiological investigations to identify risk factors, (4) establishing regular surveillance, (5) treating the infected animals to decrease outbreaks in humans, and (6) training the medical and veterinary specialists on the management and diagnosis of the disease and hiring of professional, well-trained personnel.
7.2. Preventive Measures for Cryptosporidium Infection
Due to the absence of effective treatment, the prevention of cryptosporidiosis relies mainly on the elimination and/or reduction of contamination of the environment with infectious oocysts [33]. It is recommended to move animals to clean and dry places and disinfect the contaminated areas, however, this is mostly not applicable on farms with a large number of animals. For humans, continuous disinfection of the contaminated areas will reduce person-to-person transmission in institutional and domestic settings. In general, the infectivity of the oocyst and its survival time will be restored at low temperatures (less than 5 °C) and increased by temperatures higher than 15 °C for 3 months [174]. In general, several physical stresses can affect Cryptosporidium oocysts including irradiation, heat, cold, pressure, and desiccation [19]. The infectivity of C. parvum oocysts at different temperatures is due to the carbohydrate energy reserve of the sporozoites, and the residual bodies including amylopectin (which helps in the excavation process and the host–cell invasion) granules which are used quickly at higher temperatures [33,175]. Increasing the temperature to 64.2 °C or more for 5 min and 72.4 °C for 1 min renders the oocysts non-infectious [176]. Even in the presence of cryoprotectants, C. parvum oocysts can survive at −20 °C for prolonged periods, but not at −70 °C or below [176]. However, ultraviolet (UV) irradiation can render Cryptosporidium oocysts non-infectious [177]. The most effective disinfectants against Cryptosporidium oocysts are those that contain chlorine dioxide, hydrogen peroxide, or ammonia. Although high concentrations and longer exposure to chlorine-, bromine-, and iodine-related compounds can decrease the infectivity of the oocysts, they are limiting their practical applications. Ozone is one of the most effective chemical disinfectants against Cryptosporidium and can be used against Cryptosporidium oocysts in water [33]. It has also been reported that rotifers, which occupy rivers, lakes, seawater, and ponds, and predacious protozoa, can ingest oocysts of C. parvum [178]. Some rotifers were found to discharge oocysts in boluses containing a mixture of other eaten components [179], and therefore they can be used for Cryptosporidium oocyst control in water.
7.3. Treatment of Cryptosporidium Infection
Several active compounds have been tested for their efficacy against Cryptosporidium infections [23]. There are only a few drugs that possess efficacy in vitro [23,180,181,182]. Halofuginone (a bromo-chlorinated quinazoline derivate) is approved for pro- and metaphylactic treatment for animals in Europe. Halofuginone is applied for 7 days at a dose of 100 µg/kg of body weight, starting from the first 24 h after the onset of diarrhea and/or within the first 24–48 h of life as prophylactic. However, symptoms of poisoning include diarrhea, blood in feces, reduction of milk intake, dehydration, exhaustion, and apathy, which can be observed after using a double therapeutic dosage [24]. Furthermore, nitazoxanide (a nitrothiazolylsalicylamide) was approved by the Food and Drug Administration (FDA) for the treatment of cryptosporidiosis in humans ≥ 1 year of age [183]. Nitazoxanide is an oral suspension that is mostly used at a concentration of 100 mg/5 mL for patients ≥ 1 year of age, while tablets at 500 mg for patients ≥ 12 years of age are used for the treatment [91]. Interestingly, approximately 56% of the 71 Cryptosporidium outbreaks were associated with drinking contaminated water [10]. Therefore, control of Cryptosporidium is a major challenge for water treatment professionals. Cryptosporidium oocysts can pass through different types of filters and are not affected by chlorine and chlorine-based disinfectant. Different filtration methods such as direct filtration, conventional filtration, slow-sand filtration, diatomaceous earth filtration, bag filtration cartridge filtration, and membrane filtration are used in the treatment of infected water. The conventional filtration methods using coagulation, flocculation, and sedimentation are capable of the removal of 99% of Cryptosporidium [33]. Sand filtration also uses a biological process to remove Cryptosporidium oocysts from the water supply. UV irradiation can also affect the infectivity of Cryptosporidium oocysts [184,185], suggesting the efficacy of sunlight in the inactivation of oocysts in environmental water reservoirs [73].
7.4. Vaccines Development
Currently, there are no available vaccines to control Cryptosporidium infection in humans and animals [173]. There is a critical need to develop vaccines, particularly for high-risk groups such as young children, malnourished populations, and immunosuppressed persons. It has been reported that vaccinating mother cows against other diarrhea-causing pathogens such as rotavirus, coronavirus, and E. coli may protect against Cryptosporidium infection in calves via colostrum, thus helping the calf to resist the infection during the first weeks of age [186]. To develop an effective vaccine, there is a need to understand the host immune response to infection and the host–parasite interactions [187] as well as understand the innate and adaptive host response [188]. However, the nature of these responses is still unknown and needs further investigation [189,190]. Several trials to produce effective vaccines against cryptosporidiosis have been carried out. It was reported that miRNA plays a crucial role in the protection of the host cell against Cryptosporidium and the regulation of miRNA expression levels in epithelial cells [191], while mannose-binding lectin (MBL) can protect against cryptosporidiosis, especially in children and immunocompromised persons with MBL deficiency [192,193,194]. Additionally, several antigens such as gp15, cp15, and cp23 are being developed as vaccine candidates. The gp15 antigen is substantially conserved between C. parvum and C. hominis, and there is a significant cross-reactivity between both species [195], while cp23 is conserved among C. parvum isolates and found in both the sporozoites and merozoites [187]. Using the cp15 vaccines to immunize pregnant goats protect offspring [196]. The vaccines provide a transient reduction of Cryptosporidium in the stool of vaccinated goats, but they were not fully protected against the infection [197]. Interestingly, the vaccine that contains multiple dominant antigens may enhance protection against the infection. For example, it was reported that cp23 plus cp15 divalent vaccine prolonged the prepatent period and reduced the shedding of the oocyst compared to vaccination with cp23 alone in mice [198]. Furthermore, serum antibodies to both cp23 and gp15 protected diarrhea in immunocompetent persons infected with Cryptosporidium [199,200]. Collectively, the ideal vaccine should (1) provide lifelong immunity in the vaccinated population, (2) protect against species and subtypes of Cryptosporidium to assure cross-protection against the most common species infecting humans, and (3) prevent Cryptosporidium transmission [187,189].
8. Conclusions and Future Perspectives
Cryptosporidium is one of the water- and foodborne pathogens with socioeconomic and public health importance worldwide. The infection is characterized by high morbidity and high mortality. Cryptosporidium infection is ubiquitous and has a high prevalence in animals and humans. Children under 5 years of age and immunocompromised individuals are the most susceptible groups to infections. Cryptosporidiosis in animals has become more common because of environmental contamination in livestock production. Cryptosporidium infection can be transmitted directly via drinking/ingestion of contaminated water or food with sporulated oocysts. Most of the foodborne outbreaks associated with Cryptosporidium are zoonotic. To prevent disease outbreaks, routine surveillance systems and the application of the One Health approach are required. Food safety and water sanitation are required to prevent and/or reduce future outbreaks worldwide. Each of the available diagnostic tools has its limitations in terms of isolation, detection of co-infections with other pathogens, and cost. In developing countries, the true burden of cryptosporidiosis is underestimated and underreported due to the limitation of diagnostic tools, which results in ineffective clinical and public health management of the disease. Therefore, there is a critical need to develop rapid, reliable, and cost-effective diagnostic tests to improve the detection, reporting, and interpretation of results. Cryptosporidium infection prevention and control can be achieved via understanding the sources of the infection (humans and animals), the routes of transmission, the oocyst survival in the environment, and the risk factors. Currently, no effective drugs or vaccines are available to treat and/or prevent infection in animals and humans. There is also a critical need for further studies for the development of effective vaccines. Additionally, more research is needed to develop highly effective disinfection methods for treating Cryptosporidium-contaminated swimming pools and water supplies.
Author Contributions
Conceptualization, Y.A.H.; validation and data curation, Y.A.H., H.M.H.; formal analysis, investigation, Y.A.H.; writing—original draft preparation, Y.A.H.; writing—review and editing, Y.A.H., H.M.H.; visualization, Y.A.H. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
There is no funding for this study.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Helmy Y.A., El-Adawy H., Abdelwhab E.M. A Comprehensive Review of Common Bacterial, Parasitic and Viral Zoonoses at the Human-Animal Interface in Egypt. Pathogens. 2017;6:33. doi: 10.3390/pathogens6030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gharpure R., Perez A., Miller A.D., Wikswo M.E., Silver R., Hlavsa M.C. Cryptosporidiosis Outbreaks—United States, 2009–2017. Mmwr. Morb. Mortal Wkly Rep. 2019;68:568–572. doi: 10.15585/mmwr.mm6825a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins J., Hague H., Hudgin G., Ross L., Moore D. International Perspectives: An Outbreak of Cryptosporidium at a Recreational Water Park in Niagara Region, Canada. J. Environ. Heath. 2013;75:28–33. [PubMed] [Google Scholar]
- 4.Savioli L., Smith H., Thompson A. Giardia and Cryptosporidium join the ‘Neglected Diseases Initiative’. Trends Parasitol. 2006;22:203–208. doi: 10.1016/j.pt.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Helmy Y.A., Klotz C., Wilking H., Krucken J., Nockler K., Von Samson-Himmelstjerna G., Zessin K.H., Aebischer T. Epidemiology of Giardia duodenalis infection in ruminant livestock and children in the Ismailia province of Egypt: Insights by genetic characterization. Parasites Vectors. 2014;7:321. doi: 10.1186/1756-3305-7-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helmy Y.A., Spierling N.G., Schmidt S., Rosenfeld U.M., Reil D., Imholt C., Jacob J., Ulrich R.G., Aebischer T., Klotz C. Occurrence and distribution of Giardia species in wild rodents in Germany. Parasites Vectors. 2018;11:213. doi: 10.1186/s13071-018-2802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dadonaite B., Ritchie H., Roser M. Diarrheal Diseases. In Our World in Data. [(accessed on 26 January 2021)]. Available online: https://ourworldindata.org/diarrheal-diseases.
- 8.Mosier D.A., Oberst R.D. Cryptosporidiosis. A global challenge. Ann. N. Y. Acad. Sci. 2000;916:102–111. doi: 10.1111/j.1749-6632.2000.tb05279.x. [DOI] [PubMed] [Google Scholar]
- 9.Ryan U., Hijjawi N., Xiao L. Foodborne cryptosporidiosis. Int. J. Parasitol. 2018;48:1–12. doi: 10.1016/j.ijpara.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Putignani L., Menichella D. Global distribution, public health and clinical impact of the protozoan pathogen Cryptosporidium. Interdiscip. Perspect. Infect. Dis. 2010;2010:753512. doi: 10.1155/2010/753512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacKenzie W.R., Hoxie N.J., Proctor M.E., Gradus M.S., Blair K.A., Peterson D.E. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 12.Peng M.M., Xiao L., Freeman A.R., Arrowood M.J., Escalante A.A., Weltman A.C., Ong C.S., Mac Kenzie W.R., Lal A.A., Beard C.B. Genetic polymorphism among Cryptosporidium parvum isolates: Evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson R.C., Palmer C.S., O’Handley R. The public health and clinical significance of Giardia and Cryptosporidium in domestic animals. Vet. J. 2008;177:18–25. doi: 10.1016/j.tvjl.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CDC Crypto Outbreaks Linked to Swimming Have Doubled since 2014. [(accessed on 26 January 2021)];2017 Available online: https://www.cdc.gov/media/releases/2017/p0518-cryptosporidium-outbreaks.html.
- 15.Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.-A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States–Major pathogens. Emerg. Inf. Dis. 2011;17:7. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DuPont H.L. Persistent diarrhea: A clinical review. JAMA Netw. 2016;315:2712–2723. doi: 10.1001/jama.2016.7833. [DOI] [PubMed] [Google Scholar]
- 17.Khalil I.A., Troeger C., Rao P.C., Blacker B.F., Brown A., Brewer T.G., Colombara D.V., De Hostos E.L., Engmann C., Guerrant R.L., et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: A meta-analyses study. Lancet Glob. Health. 2018;6:e758–e768. doi: 10.1016/S2214-109X(18)30283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunnotel O., Lowery C.J., Moore J.E., Dooley J.S., Xiao L., Millar B.C., Rooney P.J., Snelling W.J. Cryptosporidium. Lett. Appl. Microbiol. 2006;43:7–16. doi: 10.1111/j.1472-765X.2006.01936.x. [DOI] [PubMed] [Google Scholar]
- 19.Fayer R., Trout J.M., Jenkins M.C. Infectivity of Cryptosporidium parvum oocysts stored in water at environmental temperatures. J. Parasitol. 1998;84:1165–1169. doi: 10.2307/3284666. [DOI] [PubMed] [Google Scholar]
- 20.Smith H.V., Caccio S.M., Cook N., Nichols R.A., Tait A. Cryptosporidium and Giardia as foodborne zoonoses. Vet. Parasitol. 2007;149:29–40. doi: 10.1016/j.vetpar.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Gulliksen S.M., Jor E., Lie K.I., Hamnes I.S., Loken T., Akerstedt J., Osteras O. Enteropathogens and risk factors for diarrhea in Norwegian dairy calves. J. Dairy Sci. 2009;92:5057–5066. doi: 10.3168/jds.2009-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahiduzzaman M., Daugschies A. Therapy and prevention of cryptosporidiosis in animals. Vet. Parasitol. 2012;188:203–214. doi: 10.1016/j.vetpar.2012.03.052. [DOI] [PubMed] [Google Scholar]
- 23.Stockdale H.D., Spencer J.A., Blagburn B.L. Prophylaxis and Chemotherapy. In: Fayer R., Xiao L., editors. Cryptosporidium and Cryptosporidiosis. CRC Press; Boca Raton, FL, USA: 2008. pp. 255–287. [Google Scholar]
- 24.Silverlas C., Bjorkman C., Egenvall A. Systematic review and meta-analyses of the effects of halofuginone against calf cryptosporidiosis. Prevent. Vet. Med. 2009;91:73–84. doi: 10.1016/j.prevetmed.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Brainard J., Hammer C.C., Hunter P.R., Katzer F., Hurle G., Tyler K. Efficacy of halofuginone products to prevent or treat cryptosporidiosis in bovine calves: A systematic review and meta-analyses. Parasitology. 2021;148:408–419. doi: 10.1017/S0031182020002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sparks H., Nair G., Castellanos-Gonzalez A., White A.C., Jr. Treatment of Cryptosporidium: What We Know, Gaps, and the Way Forward. Curr. Trop. Med. Rep. 2015;2:181–187. doi: 10.1007/s40475-015-0056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hijjawi N.S., Meloni B.P., Ryan U.M., Olson M.E., Thompson R.C. Successful in vitro cultivation of Cryptosporidium andersoni: Evidence for the existence of novel extracellular stages in the life cycle and implications for the classification of Cryptosporidium. Int. J. Parasitol. 2002;32:1719–1726. doi: 10.1016/S0020-7519(02)00199-6. [DOI] [PubMed] [Google Scholar]
- 28.Smith H.V., Corcoran G.D. New drugs and treatment for cryptosporidiosis. Curr. Opin. Infect. Dis. 2004;17:557–564. doi: 10.1097/00001432-200412000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Leitch G.J., He Q. Cryptosporidiosis-an overview. J. Biomed. Res. 2012;25:1–16. doi: 10.1016/S1674-8301(11)60001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lendner M., Daugschies A. Cryptosporidium infections: Molecular advances. Parasitology. 2014;141:1511–1532. doi: 10.1017/S0031182014000237. [DOI] [PubMed] [Google Scholar]
- 31.Tandel J., English E.D., Sateriale A., Gullicksrud J.A., Beiting D.P., Sullivan M.C., Pinkston B., Striepen B. Life cycle progression and sexual development of the apicomplexan parasite Cryptosporidium parvum. Nat. Microbiol. 2019;4:2226–2236. doi: 10.1038/s41564-019-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Hara S.P., Chen X.M. The cell biology of Cryptosporidium infection. Microbes Infect. 2011;13:721–730. doi: 10.1016/j.micinf.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fayer R., Xiao L. Cryptosporidium and Cryptosporidiosis. 2nd ed. CRC Press; Boca Raton, FL, USA: Taylor & Francis Group; London, UK: 2007. p. 580. [Google Scholar]
- 34.Upton S.J., Current W.L. The species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) infecting mammals. J. Parasitol. 1985;71:625–629. doi: 10.2307/3281435. [DOI] [PubMed] [Google Scholar]
- 35.Current W.L., Reese N.C. A comparison of endogenous development of three isolates of Cryptosporidium in suckling mice. J. Protozool. 1986;33:98–108. doi: 10.1111/j.1550-7408.1986.tb05567.x. [DOI] [PubMed] [Google Scholar]
- 36.Fayer R., Ungar B.L. Cryptosporidium spp. and cryptosporidiosis. Microbiol. Rev. 1986;50:458–483. doi: 10.1128/mr.50.4.458-483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith H.V., Nichols R.A., Grimason A.M. Cryptosporidium excystation and invasion: Getting to the guts of the matter. Trends Parasitol. 2005;21:133–142. doi: 10.1016/j.pt.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 38.O’Hara S.P., Yu J.R., Lin J.J. A novel Cryptosporidium parvum antigen, CP2, preferentially associates with membranous structures. Parasitol. Res. 2004;92:317–327. doi: 10.1007/s00436-003-1057-5. [DOI] [PubMed] [Google Scholar]
- 39.Zapata F., Perkins M.E., Riojas Y.A., Wu T.W., Le Blancq S.M. The Cryptosporidium parvum ABC protein family. Mol. Biochem. Parasitol. 2002;120:157–161. doi: 10.1016/S0166-6851(01)00445-5. [DOI] [PubMed] [Google Scholar]
- 40.Fayer R., Gasbarre L., Pasquali P., Canals A., Almeria S., Zarlenga D. Cryptosporidium parvum infection in bovine neonates: Dynamic clinical, parasitic and immunologic patterns. Int. J. Parasitol. 1998;28:49–56. doi: 10.1016/S0020-7519(97)00170-7. [DOI] [PubMed] [Google Scholar]
- 41.Sponseller J.K., Griffiths J.K., Tzipori S. The evolution of respiratory Cryptosporidiosis: Evidence for transmission by inhalation. Clin. Microbiol. Rev. 2014;27:575–586. doi: 10.1128/CMR.00115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dillingham R.A., Lima A.A., Guerrant R.L. Cryptosporidiosis: Epidemiology and impact. Microbes Infect. 2002;4:1059–1066. doi: 10.1016/S1286-4579(02)01630-1. [DOI] [PubMed] [Google Scholar]
- 43.Nascimento M.F., Ginoris Y.P., Brandão C.C.S. Cryptosporidium Oocysts Removal by Upflow Direct Filtration: Pilot Scale Assessment. Water. 2020;12:1328. doi: 10.3390/w12051328. [DOI] [Google Scholar]
- 44.Okhuysen P.C., Chappell C.L., Crabb J.H., Sterling C.R., DuPont H.L. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 1999;180:1275–1281. doi: 10.1086/315033. [DOI] [PubMed] [Google Scholar]
- 45.Moore D.A., Atwill E.R., Kirk J.H., Brahmbhatt D., Alonso L.H., Hou L., Singer M.D., Miller T.D. Prophylactic use of decoquinate for infections with Cryptosporidium parvum in experimentally challenged neonatal calves. J. Am. Vet. Med. Assoc. 2003;15:839–845. doi: 10.2460/javma.2003.223.839. [DOI] [PubMed] [Google Scholar]
- 46.Helmy Y.A. Ph.D. Dissertation. Freie Universität; Berlin, Germany: 2014. Epidemiological investigations on the public health significance of Cryptosporidium parasites in livestock and people in the Ismailia Canal Zone of Egypt.77p [Google Scholar]
- 47.Etzold M., Lendner M., Daugschies A., Dyachenko V. CDPKs of Cryptosporidium parvum-stage-specific expression in vitro. Parasitol. Res. 2014;113:2525–2533. doi: 10.1007/s00436-014-3902-0. [DOI] [PubMed] [Google Scholar]
- 48.Tzipori S., Ward H. Cryptosporidiosis: Biology, pathogenesis and disease. Microbes Infect. 2002;4:1047–1058. doi: 10.1016/S1286-4579(02)01629-5. [DOI] [PubMed] [Google Scholar]
- 49.Fayer R., Speer C.A., Dubey J.P. General Biology of Cryptosporidium. In: Dubey J.P., Speer C.A., Fayer R., editors. Cryptosporidiosis of Man and Animals. CRC Press; Boca Raton, FL, USA: 1990. pp. 1–29. [Google Scholar]
- 50.Tzipori S., Smith M., Halpin C., Angus K.W., Sherwood D., Campbell I. Experimental cryptosporidiosis in calves: Clinical manifestations and pathological findings. Vet. Record. 1983;112:116–120. doi: 10.1136/vr.112.6.116. [DOI] [PubMed] [Google Scholar]
- 51.Heine J., Pohlenz J.F., Moon H.W., Woode G.N. Enteric lesions and diarrhea in gnotobiotic calves monoinfected with Cryptosporidium species. J. Infect. Dis. 1984;150:768–775. doi: 10.1093/infdis/150.5.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fayer R., Santin M. Cryptosporidium xiaoi n. sp. (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries) Vet. Parasitol. 2009;164:192–200. doi: 10.1016/j.vetpar.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 53.Elwin K., Hadfield S.J., Robinson G., Crouch N.D., Chalmers R.M. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int. J. Parasitol. 2012;42:675–682. doi: 10.1016/j.ijpara.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 54.Lebbad M., Beser J., Insulander M., Karlsson L., Mattsson J.G., Svenungsson B., Axen C. Unusual cryptosporidiosis cases in Swedish patients: Extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. Parasitology. 2013;140:1735–1740. doi: 10.1017/S003118201300084X. [DOI] [PubMed] [Google Scholar]
- 55.Adamu H., Petros B., Zhang G., Kassa H., Amer S., Ye J., Feng Y., Xiao L. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Neg. Trop. Dis. 2014;8:e2831. doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed S.A., Karanis P. Cryptosporidium and Cryptosporidiosis: The Perspective from the Gulf Countries. Int. J. Environ. Res. Public Heath. 2020;17:6824. doi: 10.3390/ijerph17186824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fayer R. General Biology. In: Fayer R., Xiao L., editors. Cryptosporidium and Cryptosporidiosis. CRC Press; Boca Raton, FL, USA: 2008. pp. 1–42. [Google Scholar]
- 58.Helmy Y.A., Krucken J., Abdelwhab E.M., von Samson-Himmelstjerna G., Hafez H.M. Molecular diagnosis and characterization of Cryptosporidium spp. in turkeys and chickens in Germany reveals evidence for previously undetected parasite species. PLoS ONE. 2017;12:e0177150. doi: 10.1371/journal.pone.0177150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mamedova S., Karanis P. Cryptosporidium spp. Infections in Livestock and Wild Animals in Azerbaijan Territory. Environ. Sci. Proceed. 2020;2:44. doi: 10.2166/wh.2021.050. [DOI] [PubMed] [Google Scholar]
- 60.Zahedi A., Paparini A., Jian F., Robertson I., Ryan U. Public health significance of zoonotic Cryptosporidium species in wildlife: Critical insights into better drinking water management. Int. J. Parasitol. Parasites Wildl. 2016;5:88–109. doi: 10.1016/j.ijppaw.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lendner M., Etzold M., Daugschies A. Cryptosporidiosis—An update. Berl. Und Munch. Tierarztl. Wochenschr. 2011;124:473–484. [PubMed] [Google Scholar]
- 62.Helmy Y.A., Krucken J., Nockler K., von Samson-Himmelstjerna G., Zessin K.H. Molecular epidemiology of Cryptosporidium in livestock animals and humans in the Ismailia province of Egypt. Vet. Parasitol. 2013;193:15–24. doi: 10.1016/j.vetpar.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 63.Ryan U., Fayer R., Xiao L. Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology. 2014;141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- 64.CDC Cryptosporidiosis. [(accessed on 1 August 2020)]; Available online: https://www.cdc.gov/dpdx/cryptosporidiosis/index.html.
- 65.Nichols R.A., Connelly L., Sullivan C.B., Smith H.V. Identification of Cryptosporidium species and genotypes in Scottish raw and drinking waters during a one-year monitoring period. Appl. Environ. Microbiol. 2010;76:5977–5986. doi: 10.1128/AEM.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valenzuela O., Gonzalez-Diaz M., Garibay-Escobar A., Burgara-Estrella A., Cano M., Durazo M., Bernal R.M., Hernandez J., Xiao L. Molecular Characterization of Cryptosporidium spp. in Children from Mexico. PLoS ONE. 2014;9:e96128. doi: 10.1371/journal.pone.0096128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jex A.R., Gasser R.B. Genetic richness and diversity in Cryptosporidium hominis and C. parvum reveals major knowledge gaps and a need for the application of “next generation” technologies-research review. Biotechnol. Adv. 2010;28:17–26. doi: 10.1016/j.biotechadv.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Xiao L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 69.Silverlås C. Ph.D. Thesis. Swedish University of Agricultural Sciences; Uppsala, Sweden: 2010. Cryptosporidium Infection in Dairy Cattle. [Google Scholar]
- 70.Zahedi A., Gofton A.W., Greay T., Monis P., Oskam C., Ball A., Bath A., Watkinson A., Robertson I., Ryan U. Profiling the diversity of Cryptosporidium species and genotypes in wastewater treatment plants in Australia using next generation sequencing. Sci. Total Environ. 2018;644:635–648. doi: 10.1016/j.scitotenv.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 71.Zahedi A., Gofton A.W., Jian F., Paparini A., Oskam C., Ball A., Robertson I., Ryan U. Next Generation Sequencing uncovers within-host differences in the genetic diversity of Cryptosporidium gp60 subtypes. Int. J. Parasitol. 2017;47:601–607. doi: 10.1016/j.ijpara.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 72.Ryan U., Zahedi A., Feng Y., Xiao L. An update on zoonotic Cryptosporidium species and genotypes in humans. Animal. 2021;11:3307. doi: 10.3390/ani11113307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caccio S.M., Widmer G. Cryptosporidium: Parasite and Disease. Springer; Berlin/Heidelberg, Germany: 2014. [Google Scholar]
- 74.Holubová N., Tůmová L., Sak B., Hejzlarová A., Konečný R., McEvoy J., Kváč M. Description of Cryptosporidium ornithophilus n. sp. (Apicomplexa: Cryptosporidiidae) in farmed ostriches. Parasites Vectors. 2020;13:340. doi: 10.1186/s13071-020-04191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holubová N., Sak B., Horčičková M., Hlásková L., Květoňová D., Menchaca S., McEvoy J., Kváč M. Cryptosporidium avium n. sp.(Apicomplexa: Cryptosporidiidae) in birds. Parasitol. Res. 2016;115:2243–2251. doi: 10.1007/s00436-016-4967-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ryan U. Cryptosporidium in birds, fish and amphibians. Exp. Parasitol. 2010;124:113–120. doi: 10.1016/j.exppara.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 77.Karanis P. Cryptosporidium: Waterborne and foodborne transmission and worldwide outbreaks; Proceedings of the Euro-Mediterranean Conference for Environmental Integration; Sousse, Tunisia. 22–25 November 2017; pp. 41–44. [Google Scholar]
- 78.Graczyk T.K., Fayer R., Cranfield M.R., Mhangami-Ruwende B., Knight R., Trout J.M., Bixler H. Filth flies are transport hosts of Cryptosporidium parvum. Emerg. Infect. Dis. 1999;5:726–727. doi: 10.3201/eid0505.990520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duhain G.L., Minnaar A., Buys E.M. Effect of chlorine, blanching, freezing, and microwave heating on Cryptosporidium parvum viability inoculated on green peppers. J. Food Prot. 2012;75:936–941. doi: 10.4315/0362-028X.JFP-11-367. [DOI] [PubMed] [Google Scholar]
- 80.Macarisin D., Bauchan G., Fayer R. Spinacia oleracea L. leaf stomata harboring Cryptosporidium parvum oocysts: A potential threat to food safety. Appl. Environ. Microbiol. 2010;76:555–559. doi: 10.1128/AEM.02118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Macarisin D., Santin M., Bauchan G., Fayer R. Infectivity of Cryptosporidium parvum oocysts after storage of experimentally contaminated apples. J. Food Prot. 2010;73:1824–1829. doi: 10.4315/0362-028X-73.10.1824. [DOI] [PubMed] [Google Scholar]
- 82.Lorenzo Lorenzo M.J., Ares-Mazas E., de Maturana I.V.M. Detection of oocysts and IgG antibodies to Cryptosporidium parvum in asymptomatic adult cattle. Vet. Parasitol. 1993;47:9–15. doi: 10.1016/0304-4017(93)90171-I. [DOI] [PubMed] [Google Scholar]
- 83.Villacorta I., Ares-Mazas E., Lorenzo M.J. Cryptosporidium parvum in cattle, sheep and pigs in Galicia (N.W. Spain) Vet. Parasitol. 1991;38:249–252. doi: 10.1016/0304-4017(91)90134-H. [DOI] [PubMed] [Google Scholar]
- 84.Scheid P.L., Schwarzenberger R. Free-living amoebae as vectors of cryptosporidia. Parasitol. Res. 2011;109:499–504. doi: 10.1007/s00436-011-2287-6. [DOI] [PubMed] [Google Scholar]
- 85.Pumipuntu N., Piratae S. Cryptosporidiosis: A zoonotic disease concern. Vet. World. 2018;11:681–686. doi: 10.14202/vetworld.2018.681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Santin M., Trout J.M., Fayer R. A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet. Parasitol. 2008;155:15–23. doi: 10.1016/j.vetpar.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 87.Fayer R., Santin M., Trout J.M. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet. Parasitol. 2007;145:260–266. doi: 10.1016/j.vetpar.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 88.Santin M., Trout J.M. Livestock. In: Fayer R., Xiao L., editors. Cryptosporidium and Cryptosporidiosis. CRC Press; Boca Raton, FL, USA: 2008. pp. 451–483. [Google Scholar]
- 89.Korpe P.S., Valencia C., Haque R., Mahfuz M., McGrath M., Houpt E., Kosek M., McCormick B.J.J., Penataro Yori P., Babji S., et al. Epidemiology and Risk Factors for Cryptosporidiosis in Children From 8 Low-income Sites: Results From the MAL-ED Study. Clin. Infect. Dis. 2018;67:1660–1669. doi: 10.1093/cid/ciy355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lal A., Fearnley E., Wilford E. Local weather, flooding history and childhood diarrhoea caused by the parasite Cryptosporidium spp.: A systematic review and meta-analysis. Sci. Total Environ. 2019;674:300–306. doi: 10.1016/j.scitotenv.2019.02.365. [DOI] [PubMed] [Google Scholar]
- 91.CDC Parasites-Cryptosporidium (also known as “Crypto”) [(accessed on 1 August 2020)]; Available online: https://www.cdc.gov/parasites/crypto/infection-sources.html.
- 92.Chen X.M., Keithly J.S., Paya C.V., LaRusso N.F. Cryptosporidiosis. N. Engl. J. Med. 2002;346:1723–1731. doi: 10.1056/NEJMra013170. [DOI] [PubMed] [Google Scholar]
- 93.Tzipori S. Cryptosporidiosis in perspective. Adv. Parasitol. 1988;27:63–129. doi: 10.1016/S0065-308X(08)60353-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Current W.L., Garcia L.S. Cryptosporidiosis. Clin. Microbiol. Rev. 1991;4:325–358. doi: 10.1128/CMR.4.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Farthing M.J. Clinical aspects of Human Cryptosporidiosis. In: Tetry F., editor. Cryptosporidiosis and Microsporidiosis. Karger; Basel, Switzerland: 2000. pp. 50–74. [DOI] [PubMed] [Google Scholar]
- 96.Hunter P.R., Nichols G. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin. Microbiol. Rev. 2002;15:145–154. doi: 10.1128/CMR.15.1.145-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cama V.A., Ross J.M., Crawford S., Kawai V., Chavez-Valdez R., Vargas D., Vivar A., Ticona E., Navincopa M., Williamson J., et al. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J. Infect. Dis. 2007;196:684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- 98.Hunter P.R., Hughes S., Woodhouse S., Raj N., Syed Q., Chalmers R.M., Verlander N.Q., Goodacre J. Health sequelae of human cryptosporidiosis in immunocompetent patients. Clin. Infect. Dis. 2004;39:504–510. doi: 10.1086/422649. [DOI] [PubMed] [Google Scholar]
- 99.Hatam-Nahavandi K., Ahmadpour E., Carmena D., Spotin A., Bangoura B., Xiao L. Cryptosporidium infections in terrestrial ungulates with focus on livestock: A systematic review and meta-analysis. Parasites Vectors. 2019;12:453. doi: 10.1186/s13071-019-3704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaske M., Smolka K., Andresen U. Neonatal diarrhea in the calf—Etiology and path physiology. Der. Prakt. Tierarzt. 2008;89:852–859. [Google Scholar]
- 101.De Graaf D.C., Vanopdenbosch E., Ortega-Mora L.M., Abbassi H., Peeters J.E. A review of the importance of cryptosporidiosis in farm animals. Int. J. Parasitol. 1999;29:1269–1287. doi: 10.1016/S0020-7519(99)00076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McDonald V. Host cell-mediated responses to infection with Cryptosporidium. Parasit. Immunol. 2000;22:597–604. doi: 10.1046/j.1365-3024.2000.00343.x. [DOI] [PubMed] [Google Scholar]
- 103.Bertoni E., Barragán A.A., Bok M., Vega C., Martínez M., Gil J.F., Cimino R.O., Parreño V. Assessment of Influential Factors for Scours Associated with Cryptosporidium sp., Rotavirus and Coronavirus in Calves from Argentinean Dairy Farms. Animals. 2021;11:2652. doi: 10.3390/ani11092652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lombardelli J.A., Tomazic M.L., Schnittger L., Tiranti K.I. Prevalence of Cryptosporidium parvum in dairy calves and GP60 subtyping of diarrheic calves in central Argentina. Parasitol. Res. 2019;118:2079–2086. doi: 10.1007/s00436-019-06366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiao L., Fayer R., Ryan U., Upton S.J. Cryptosporidium taxonomy: Recent advances and implications for public health. Clin. Microbiol. Rev. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lindsay D.S., Blagburn B.L. Cryptosporidiosis Man and Animals. CRC Press; Boca Raton, FL, USA: 1990. Cryptosporidiosis in birds; pp. 133–148. [Google Scholar]
- 107.Makino I., Abe N., Reavill D.R. Cryptosporidium avian genotype III as a possible causative agent of chronic vomiting in peach-faced lovebirds (Agapornis roseicollis) Avian Dis. 2010;54:1102–1107. doi: 10.1637/9227-123009-Case.1. [DOI] [PubMed] [Google Scholar]
- 108.Jellison K.L., Distel D.L., Hemond H.F., Schauer D.B. Phylogenetic analysis of the hypervariable region of the 18S rRNA gene of Cryptosporidium oocysts in feces of Canada geese (Branta canadensis): Evidence for five novel genotypes. App. Environ. Microbiol. 2004;70:452–458. doi: 10.1128/AEM.70.1.452-458.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jellison K.L., Lynch A.E., Ziemann J.M. Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ. Sci. Technol. 2009;43:4267–4272. doi: 10.1021/es900081m. [DOI] [PubMed] [Google Scholar]
- 110.Zhou L., Kassa H., Tischler M.L., Xiao L. Host-adapted Cryptosporidium spp. in Canada geese (Branta canadensis) Appl. Environ. Microbiol. 2004;70:4211–4215. doi: 10.1128/AEM.70.7.4211-4215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.da Cunha M.J.R., Cury M.C., Santín M. Molecular characterization of Cryptosporidium spp. in poultry from Brazil. Res. Vet. Sci. 2018;118:331–335. doi: 10.1016/j.rvsc.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 112.Santana B.N., Kurahara B., Nakamura A.A., da Silva Camargo V., Ferrari E.D., da Silva G.S., Nagata W.B., Meireles M.V. Detection and characterization of Cryptosporidium species and genotypes in three chicken production systems in Brazil using different molecular diagnosis protocols. Prev. Vet. Med. 2018;151:73–78. doi: 10.1016/j.prevetmed.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 113.Cao S., Xu M., Jiang Y., Liu H., Yuan Z., Sun L., Cao J., Shen Y. Prevalence and Genetic Characterization of Cryptosporidium, Giardia and Enterocytozoon in Chickens From Ezhou, Hubei, China. Fron. Vet. Sci. 2020;7:30. doi: 10.3389/fvets.2020.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.AL-Ghezey M.R., Al-Zubaidi M.T. Molecular detection of Cryptosporidium parasite in chickens (Broiler and layer) in Thi-Qar province. Iraqi J. Vet. Sci. 2020;34:441–445. [Google Scholar]
- 115.Altamimi M.K., Al-Zubaidi M.T. Cryptosporidium Spp.: Conventional and molecular and molecular detection in wild pigeons (COLUMBA LIVIA) in Babylon province, Iraq. Plan. Arch. 2020;20:1544–1548. [Google Scholar]
- 116.Shahbazi P., Aligolzadeh A., Khordadmehr M., Farhang H.H., Katiraee F. Molecular study and genotyping of Cryptosporidium baileyi and Cryptosporidium parvum from free-range and commercial broiler chickens in Guilan province, Iran. Comp. Immunol. Microbiol. Infect. Dis. 2020;69:101411. doi: 10.1016/j.cimid.2019.101411. [DOI] [PubMed] [Google Scholar]
- 117.Jian Y., Zhang X., Li X., Schou C., Charalambidou I., Ma L., Karanis P. Occurrence of Cryptosporidium and Giardia in wild birds from Qinghai Lake on the Qinghai-Tibetan Plateau, China. Parasitol. Res. 2021;120:615–628. doi: 10.1007/s00436-020-06993-w. [DOI] [PubMed] [Google Scholar]
- 118.Kabir M.H.B., Han Y., Lee S.-H., Nugraha A.B., Recuenco F., Murakoshi F., Xuan X., Kato K. Prevalence and molecular characterization of Cryptosporidium species in poultry in Bangladesh. One Health. 2020;9:100122. doi: 10.1016/j.onehlt.2020.100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liao C., Wang T., Koehler A.V., Fan Y., Hu M., Gasser R.B. Molecular investigation of Cryptosporidium in farmed chickens in Hubei Province, China, identifies ‘zoonotic’subtypes of C. meleagridis. Parasit Vectors. 2018;11:484. doi: 10.1186/s13071-018-3056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reboredo-Fernandez A., Ares-Mazas E., Caccio S.M., Gomez-Couso H. Occurrence of Giardia and Cryptosporidium in wild birds in Galicia (Northwest Spain) Parasitology. 2015;142:917–925. doi: 10.1017/S0031182015000049. [DOI] [PubMed] [Google Scholar]
- 121.Li J., Lin X., Zhang L., Qi N., Liao S., Lv M., Wu C., Sun M. Molecular characterization of Cryptosporidium spp. in domestic pigeons (Columba livia domestica) in Guangdong Province, Southern China. Parasitol. Res. 2015;114:2237–2241. doi: 10.1007/s00436-015-4415-1. [DOI] [PubMed] [Google Scholar]
- 122.Máca O., Pavlásek I. First finding of spontaneous infections with Cryptosporidium baileyi and C. meleagridis in the red-legged partridge Alectoris rufa from an aviary in the Czech Republic. Vet. Parasitol. 2015;209:164–168. doi: 10.1016/j.vetpar.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 123.Wang L., Xue X., Li J., Zhou Q., Yu Y., Du A. Cryptosporidiosis in broiler chickens in Zhejiang Province, China: Molecular characterization of oocysts detected in fecal samples. Parasite. 2014;21:36. doi: 10.1051/parasite/2014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Qi M., Huang L., Wang R., Xiao L., Xu L., Li J., Zhang L. Natural infection of Cryptosporidium muris in ostriches (Struthio camelus) Vet. Parasitol. 2014;205:518–522. doi: 10.1016/j.vetpar.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 125.Laatamna A.E., Holubova N., Sak B., Kvac M. Cryptosporidium meleagridis and C. baileyi (Apicomplexa) in domestic and wild birds in Algeria. Folia Parasitol. 2017;64:2017.018. doi: 10.14411/fp.2017.018. [DOI] [PubMed] [Google Scholar]
- 126.Baroudi D., Khelef D., Goucem R., Adjou K.T., Adamu H., Zhang H., Xiao L. Common occurrence of zoonotic pathogen Cryptosporidium meleagridis in broiler chickens and turkeys in Algeria. Vet. Parasitol. 2013;196:334–340. doi: 10.1016/j.vetpar.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 127.Wang R., Wang F., Zhao J., Qi M., Ning C., Zhang L., Xiao L. Cryptosporidium spp. in quails (Coturnix coturnix japonica) in Henan, China: Molecular characterization and public health significance. Vet. Parasitol. 2012;187:534–537. doi: 10.1016/j.vetpar.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 128.Qi M., Wang R., Ning C., Li X., Zhang L., Jian F., Sun Y., Xiao L. Cryptosporidium spp. in pet birds: Genetic diversity and potential public health significance. Exp. Parasitol. 2011;128:336–340. doi: 10.1016/j.exppara.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 129.da Paixão Sevá A., Funada M.R., Richtzenhain L., Guimarães M.B., de Oliveira Souza S., Allegretti L., Sinhorini J.A., Duarte V.V., Soares R.M. Genotyping of Cryptosporidium spp. from free-living wild birds from Brazil. Vet. Parasitol. 2011;175:27–32. doi: 10.1016/j.vetpar.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 130.Wang R., Qi M., Jingjing Z., Sun D., Ning C., Zhao J., Zhang L., Xiao L. Prevalence of Cryptosporidium baileyi in ostriches (Struthio camelus) in Zhengzhou, China. Vet. Parasitol. 2011;175:151–154. doi: 10.1016/j.vetpar.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 131.Wang R., Jian F., Sun Y., Hu Q., Zhu J., Wang F., Ning C., Zhang L., Xiao L. Large-scale survey of Cryptosporidium spp. in chickens and Pekin ducks (Anas platyrhynchos) in Henan, China: Prevalence and molecular characterization. Avian Pathol. 2010;39:447–451. doi: 10.1080/03079457.2010.518314. [DOI] [PubMed] [Google Scholar]
- 132.McEvoy J., Giddings C. Cryptosporidium in commercially produced turkeys on-farm and postslaughter. Lett. App. Microbiol. 2009;48:302–306. doi: 10.1111/j.1472-765X.2008.02516.x. [DOI] [PubMed] [Google Scholar]
- 133.Nakamura A.A., Simões D.C., Antunes R.G., da Silva D.C., Meireles M.V. Molecular characterization of Cryptosporidium spp. from fecal samples of birds kept in captivity in Brazil. Vet. Parasitol. 2009;166:47–51. doi: 10.1016/j.vetpar.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 134.Ng J., Pavlasek I., Ryan U. Identification of novel Cryptosporidium genotypes from avian hosts. Appl. Environ. Microbiol. 2006;72:7548–7553. doi: 10.1128/AEM.01352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zahedi A., Ryan U. Cryptosporidium—An update with an emphasis on foodborne and waterborne transmission. Res. Vet. Sci. 2020;132:500–512. doi: 10.1016/j.rvsc.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 136.Feng Y., Li N., Roellig D.M., Kelley A., Liu G., Amer S., Tang K., Zhang L., Xiao L. Comparative genomic analysis of the IId subtype family of Cryptosporidium parvum. Int. J. Parasitol. 2017;47:281–290. doi: 10.1016/j.ijpara.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.CDC . National Outbreak Reporting System Dashboard. U.S. Department of Health and Human Services, CDC; Atlanta, Georgia: [Google Scholar]
- 138.Doman B., Selvage W.D., Ferrreira J., Roelling D.M., Kahler A. Cryptosporidium Outbreak Associated with the Consumption of Unpasteurized Cow Milk, New Mexico. [(accessed on 1 August 2020)]. Available online: https://cste.confex.com/cste/2017/webprogram/Paper8610.html.
- 139.Fill M.-M.A., Lloyd J., Chakraverty T., Sweat D., Manners J., Garman K., Hlavsa M.C., Roellig D.M., Dunn J.R., Schaffner W. Cryptosporidiosis outbreak associated with a single hotel. J. Environ. Health. 2017;79:16–23. [PubMed] [Google Scholar]
- 140.Rosenthal M., Pedersen R., Leibsle S., Hill V., Carter K., Roellig D.M. Cryptosporidiosis Associated with Consumption of Unpasteurized Goat Milk—Idaho, 2014. Morb. Mortal. Wkly. Rep. 2015;64:194. [PMC free article] [PubMed] [Google Scholar]
- 141.Chalmers R.M., Robinson G., Elwin K., Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasites Vectors. 2019;12:1–13. doi: 10.1186/s13071-019-3354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McKerr C., Adak G.K., Nichols G., Gorton R., Chalmers R.M., Kafatos G., Cosford P., Charlett A., Reacher M., Pollock K.G. An outbreak of Cryptosporidium parvum across England & Scotland associated with consumption of fresh pre-cut salad leaves, May 2012. PLoS ONE. 2015;10:e0125955. doi: 10.1371/journal.pone.0125955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chalmers R.M. Waterborne outbreaks of cryptosporidiosis. Ann. Dell’istituto Super. Di Sanita. 2012;48:429–446. doi: 10.4415/ANN_12_04_10. [DOI] [PubMed] [Google Scholar]
- 144.McCann R., Jones R., Snow J., Cleary P., Burgess S., Bothra V., Chalmers R. An outbreak of cryptosporidiosis at a swimming club–can rapid field epidemiology limit the spread of illness? Epidemiol. Infect. 2014;142:51–55. doi: 10.1017/S0950268813001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Whitworth J. Food Safety News. [(accessed on 1 August 2020)]. Available online: https://www.foodsafetynews.com/2020/03/five-foodborne-outbreaks-added-to-cryptosporidium-rise-in-sweden/
- 146.Rehn M., Wallensten A., Widerström M., Lilja M., Grunewald M., Stenmark S., Kark M., Lindh J. Post-infection symptoms following two large waterborne outbreaks of Cryptosporidium hominis in Northern Sweden, 2010–2011. BMC Public Health. 2015;15:529. doi: 10.1186/s12889-015-1871-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Widerström M., Schönning C., Lilja M., Lebbad M., Ljung T., Allestam G., Ferm M., Björkholm B., Hansen A., Hiltula J. Large outbreak of Cryptosporidium hominis infection transmitted through the public water supply, Sweden. Emerg. Infec. Dis. 2014;20:581. doi: 10.3201/eid2004.121415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gherasim A., Lebbad M., Insulander M., Decraene V., Kling A., Hjertqvist M., Wallensten A. Two geographically separated food-borne outbreaks in Sweden linked by an unusual Cryptosporidium parvum subtype, October 2010. Eurosurveillance. 2012;17:20318. doi: 10.2807/ese.17.46.20318-en. [DOI] [PubMed] [Google Scholar]
- 149.Mosnier E., Martin N., Razakandrainibe R., Dalle F., Roux G., Buteux A., Favennec L., Brousse P., Guarmit B., Blanchet D. Cryptosporidiosis outbreak in immunocompetent children from a remote area of French Guiana. Am. J Trop. Med. Hyg. 2018;98:1727–1732. doi: 10.4269/ajtmh.17-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gertler M., Dürr M., Renner P., Poppert S., Askar M., Breidenbach J., Frank C., Preußel K., Schielke A., Werber D. Outbreak of Cryptosporidium hominis following river flooding in the city of Halle (Saale), Germany, August 2013. BMC Infect. Dis. 2015;15:88. doi: 10.1186/s12879-015-0807-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mahon M., Doyle S. Waterborne outbreak of cryptosporidiosis in the South East of Ireland: Weighing up the evidence. Irish J. Med. Sci. 2017;186:989–994. doi: 10.1007/s11845-016-1552-1. [DOI] [PubMed] [Google Scholar]
- 152.Robertson L., Temesgen T.T., Tysnes K., Eikås J. An apple a day: An outbreak of cryptosporidiosis in Norway associated with self-pressed apple juice. Epidemiol. Infect. 2019;147:e139. doi: 10.1017/S0950268819000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lange H., Johansen Ø., Vold L., Robertson L., Anthonisen I., Nygard K. Second outbreak of infection with a rare Cryptosporidium parvum genotype in schoolchildren associated with contact with lambs/goat kids at a holiday farm in Norway. Epidemiol. Infect. 2014;142:2105–2113. doi: 10.1017/S0950268813003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Åberg R., Sjöman M., Hemminki K., Pirnes A., Räsänen S., Kalanti A., Pohjanvirta T., Caccio S., Pihlajasaari A., Toikkanen S. Cryptosporidium parvum caused a large outbreak linked to frisée salad in Finland, 2012. Zoonoses Public Health. 2015;62:618–624. doi: 10.1111/zph.12190. [DOI] [PubMed] [Google Scholar]
- 155.Moon S., Kwak W., Lee S., Kim W., Oh J., Youn S.-K. Epidemiological characteristics of the first water-borne outbreak of cryptosporidiosis in Seoul, Korea. J. Korean Med. Sci. 2013;28:983. doi: 10.3346/jkms.2013.28.7.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ng-Hublin J., Hargrave D., Combs B., Ryan U. Investigation of a swimming pool-associated cryptosporidiosis outbreak in the Kimberley region of Western Australia. Epidemiol. Infect. 2015;143:1037–1041. doi: 10.1017/S095026881400106X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chartier C., Rieux A., Delafosse A., Lehebel A., Paraud C. Detection of Cryptosporidium oocysts in fresh calf faeces: Characteristics of two simple tests and evaluation of a semi-quantitative approach. Vet. J. 2013;198:148–152. doi: 10.1016/j.tvjl.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 158.Helmy Y.A., Krucken J., Nockler K., von Samson-Himmelstjerna G., Zessin K.H. Comparison between two commercially available serological tests and polymerase chain reaction in the diagnosis of Cryptosporidium in animals and diarrhoeic children. Parasitol. Res. 2014;113:211–216. doi: 10.1007/s00436-013-3645-3. [DOI] [PubMed] [Google Scholar]
- 159.El-Missiry A., Abd El-Hameed L., Saad G., El-Badry A., Helmy Y., Shehata M. Molecular genetic characterization of human Cryptosporidium isolates and their respective demographic, environmental and clinical manifestations in Egyptian diarrheic patients. Parasitol. United J. 2019;12:187–196. doi: 10.21608/puj.2019.15158.1050. [DOI] [Google Scholar]
- 160.Papini R., Cardini G. Evaluation of a rapid Cryptosporidium/Giardia immunochromatographic test for diagnosis of giardiasis in dogs. Revue Méd. Vét. 2006;157:490–493. [Google Scholar]
- 161.Brescia C.C., Griffin S.M., Ware M.W., Varughese E.A., Egorov A.I., Villegas E.N. Cryptosporidium propidium monoazide-PCR, a molecular biology-based technique for genotyping of viable Cryptosporidium oocysts. Appl. Environ. Microbiol. 2009;75:6856–6863. doi: 10.1128/AEM.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Caccio S.M., Thompson R.C., McLauchlin J., Smith H.V. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 2005;21:430–437. doi: 10.1016/j.pt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 163.Egyed Z., Sreter T., Szell Z., Beszteri B., Dobos-Kovacs M., Marialigeti K., Cornelissen A.W., Varga I. Polyphasic typing of Cryptosporidium baileyi: A suggested model for characterization of cryptosporidia. J. Parasitol. 2002;88:237–243. doi: 10.1645/0022-3395(2002)088[0237:PTOCBA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 164.Morgan U.M., Constantine C.C., O’Donoghue P., Meloni B.P., O’Brien P.A., Thompson R.C. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am. J. Trop. Med. Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 165.Helmy Y.A., Von Samson-Himmelstjerna G., Nockler K., Zessin K.H. Frequencies and spatial distributions of Cryptosporidium in livestock animals and children in the Ismailia province of Egypt. Epidemiol. Infect. 2015;143:1208–1218. doi: 10.1017/S0950268814001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ghaffari S., Kalantari N., Hart C.A. A Multi-Locus Study for Detection of Cryptosporidium Species Isolated from Calves Population, Liverpool; UK. Int. J. Mol. Cell Med. 2014;3:35–42. [PMC free article] [PubMed] [Google Scholar]
- 167.Morrison L.J., Mallon M.E., Smith H.V., MacLeod A., Xiao L., Tait A. The population structure of the Cryptosporidium parvum population in Scotland: A complex picture. Infect. Gen. Evol. 2008;8:121–129. doi: 10.1016/j.meegid.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xiao L., Fayer R. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int. J. Parasitol. 2008;38:1239–1255. doi: 10.1016/j.ijpara.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 169.Cevallos A.M., Zhang X., Waldor M.K., Jaison S., Zhou X., Tzipori S., Neutra M.R., Ward H.D. Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect. Immun. 2000;68:4108–4116. doi: 10.1128/IAI.68.7.4108-4116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li N., Xiao L., Cama V.A., Ortega Y., Gilman R.H., Guo M., Feng Y. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg. Infect. Dis. 2013;19:1573–1582. doi: 10.3201/eid1910.121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fawzy M., Helmy Y.A. The one health approach is necessary for the control of Rift Valley fever infections in Egypt: A comprehensive review. Viruses. 2019;11:139. doi: 10.3390/v11020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control. J. Clin. Med. 2020;9:1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Innes E.A., Chalmers R.M., Wells B., Pawlowic M.C. A one health approach to tackle cryptosporidiosis. Trends Parasitol. 2020;36:290–303. doi: 10.1016/j.pt.2019.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.King B.J., Keegan A.R., Monis P.T., Saint C.P. Environmental temperature controls Cryptosporidium oocyst metabolic rate and associated retention of infectivity. App. Environ. Microbiol. 2005;71:3848–3857. doi: 10.1128/AEM.71.7.3848-3857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jenkins M., Trout J.M., Higgins J., Dorsch M., Veal D., Fayer R. Comparison of tests for viable and infectious Cryptosporidium parvum oocysts. Parasitol. Res. 2003;89:1–5. doi: 10.1007/s00436-002-0720-6. [DOI] [PubMed] [Google Scholar]
- 176.Fayer R. Effect of high temperature on infectivity of Cryptosporidium parvum oocysts in water. Appl. Environ. Microbiol. 1994;60:2732–2735. doi: 10.1128/aem.60.8.2732-2735.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sivaganesan M., Sivaganesan S. Effect of lot variability on ultraviolet radiation inactivation kinetics of Cryptosporidium parvum oocysts. Environ. Sci. Technol. 2005;39:4166–4171. doi: 10.1021/es0489083. [DOI] [PubMed] [Google Scholar]
- 178.Stott R., May E., Ramirez E., Warren A. Predation of Cryptosporidium oocysts by protozoa and rotifers: Implications for water quality and public health. Water Sci. Technol. 2003;47:77–83. doi: 10.2166/wst.2003.0166. [DOI] [PubMed] [Google Scholar]
- 179.Fayer R., Trout J.M., Walsh E., Cole R. Rotifers Ingest Oocysts of Cryptosporidium parvum. J. Eukar. Microbiol. 2000;47:161–163. doi: 10.1111/j.1550-7408.2000.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 180.Mead J.R. Cryptosporidiosis and the challenges of chemotherapy. Drug Resis. Updates. 2002;5:47–57. doi: 10.1016/S1368-7646(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 181.Schneider A., Wendt S., Lübbert C., Trawinski H. Current pharmacotherapy of cryptosporidiosis: An update of the state-of-the-art. Expert Opin. Pharmacother. 2021;22:2337–2342. doi: 10.1080/14656566.2021.1957097. [DOI] [PubMed] [Google Scholar]
- 182.Van Voorhis W.C., Hulverson M.A., Choi R., Huang W., Arnold S.L.M., Schaefer D.A., Betzer D.P., Vidadala R.S.R., Lee S., Whitman G.R., et al. One health therapeutics: Target-Based drug development for cryptosporidiosis and other apicomplexa diseases. Vet. Parasitol. 2021;289:109336. doi: 10.1016/j.vetpar.2020.109336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cabada M.M., White A.C., Jr. Treatment of cryptosporidiosis: Do we know what we think we know? Curr. Opin. Infect. Dis. 2010;23:494–499. doi: 10.1097/QCO.0b013e32833de052. [DOI] [PubMed] [Google Scholar]
- 184.Johnson A.M., Linden K., Ciociola K.M., De Leon R., Widmer G., Rochelle P.A. UV inactivation of Cryptosporidium hominis as measured in cell culture. Appl. Environ. Microbiol. 2005;71:2800–2802. doi: 10.1128/AEM.71.5.2800-2802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rochelle P.A., Fallar D., Marshall M.M., Montelone B.A., Upton S.J., Woods K. Irreversible UV inactivation of Cryptosporidium spp. despite the presence of UV repair genes. J. Euk. Microbiol. 2004;51:553–562. doi: 10.1111/j.1550-7408.2004.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 186.Innes E.A., Bartley P.M., Rocchi M., Benavidas-Silvan J., Burrells A., Hotchkiss E., Chianini F., Canton G., Katzer F. Developing vaccines to control protozoan parasites in ruminants: Dead or alive? Vet. Parasitol. 2011;180:155–163. doi: 10.1016/j.vetpar.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 187.Mead J.R. Prospects for immunotherapy and vaccines against Cryptosporidium. Human Vac. Immun. 2014;10:1505–1513. doi: 10.4161/hv.28485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ryan U., Zahedi A., Paparini A. Cryptosporidium in humans and animals–A one health approach to prophylaxis. Parasit. Immunol. 2016;38:535–547. doi: 10.1111/pim.12350. [DOI] [PubMed] [Google Scholar]
- 189.Ludington J.G., Ward H.D. Systemic and mucosal immune responses to cryptosporidium–Vaccine development. Curr. Trop. Med. Rep. 2015;2:171–180. doi: 10.1007/s40475-015-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Borad A., Ward H. Human immune responses in cryptosporidiosis. Future Microbiol. 2010;5:507–519. doi: 10.2217/fmb.09.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ming Z., Zhou R., Chen X.-M. Regulation of host epithelial responses to Cryptosporidium infection by microRNAs. Parasit. Immunol. 2017;39:e12408. doi: 10.1111/pim.12408. [DOI] [PubMed] [Google Scholar]
- 192.Kirkpatrick B., Huston C., Wagner D., Noel F., Rouzier P., Pape J., Bois G., Larsson C., Alston W., Tenney K. Serum mannose-binding lectin deficiency is associated with cryptosporidiosis in young Haitian children. Clin. Infect. Dis. 2006;43:289–294. doi: 10.1086/505396. [DOI] [PubMed] [Google Scholar]
- 193.Kelly P., Jack D.L., Naeem A., Mandanda B., Pollok R.C., Klein N.J., Turner M.W., Farthing M.J. Mannose-binding lectin is a component of innate mucosal defense against Cryptosporidium parvum in AIDS. Gastroenterology. 2000;119:1236–1242. doi: 10.1053/gast.2000.19573. [DOI] [PubMed] [Google Scholar]
- 194.Carmolli M., Duggal P., Haque R., Lindow J., Mondal D., Petri W.A., Jr., Mourningstar P., Larsson C.J., Sreenivasan M., Khan S. Deficient serum mannose-binding lectin levels and MBL2 polymorphisms increase the risk of single and recurrent Cryptosporidium infections in young children. J. Infect. Dis. 2009;200:1540–1547. doi: 10.1086/606013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lucio-Forster A., Griffiths J.K., Cama V.A., Xiao L., Bowman D.D. Minimal zoonotic risk of cryptosporidiosis from pet dogs and cats. Trends Parasitol. 2010;26:174–179. doi: 10.1016/j.pt.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 196.Sagodira S., Buzoni-Gatel D., Iochmann S., Naciri M., Bout D. Protection of kids against Cryptosporidium parvum infection after immunization of dams with CP15-DNA. Vaccine. 1999;17:2346–2355. doi: 10.1016/S0264-410X(99)00041-9. [DOI] [PubMed] [Google Scholar]
- 197.Roche J.K., Rojo A.L., Costa L.B., Smeltz R., Manque P., Woehlbier U., Bartelt L., Galen J., Buck G., Guerrant R.L. Intranasal vaccination in mice with an attenuated Salmonella enterica Serovar 908htr A expressing Cp15 of Cryptosporidium: Impact of malnutrition with preservation of cytokine secretion. Vaccine. 2013;31:912–918. doi: 10.1016/j.vaccine.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Liu K., Zai D., Zhang D., Wei Q., Han G., Gao H., Huang B. Divalent Cp15–23 vaccine enhances immune responses and protection against Cryptosporidium parvum infection. Parasit. Immunol. 2010;32:335–344. doi: 10.1111/j.1365-3024.2009.01191.x. [DOI] [PubMed] [Google Scholar]
- 199.Riggs M.W. Recent advances in cryptosporidiosis: The immune response. Microb. Inf. 2002;4:1067–1080. doi: 10.1016/S1286-4579(02)01631-3. [DOI] [PubMed] [Google Scholar]
- 200.Frost F.J., Tollestrup K., Craun G.F., Fairley C.K., Sinclair M.I., Kunde T.R. Protective immunity associated with a strong serological response to a Cryptosporidium-specific antigen group, in HIV-infected individuals. J. Inf. Dis. 2005;192:618–621. doi: 10.1086/431681. [DOI] [PubMed] [Google Scholar]