Abstract
Bexarotene is an FDA-approved drug for the treatment of cutaneous T-cell lymphoma (CTCL); however, its use provokes or disrupts other retinoid-X-receptor (RXR)-dependent nuclear receptor pathways and thereby incites side effects including hypothyroidism and raised triglycerides. Two novel bexarotene analogs, as well as three unique CD3254 analogs and thirteen novel NEt-TMN analogs, were synthesized and characterized for their ability to induce RXR agonism in comparison to bexarotene (1). Several analogs in all three groups possessed an isochroman ring substitution for the bexarotene aliphatic group. Analogs were modeled for RXR binding affinity, and EC50 as well as IC50 values were established for all analogs in a KMT2A-MLLT3 leukemia cell line. All analogs were assessed for liver-X-receptor (LXR) activity in an LXRE system to gauge the potential for the compounds to provoke raised triglycerides by increasing LXR activity, as well as to drive LXRE-mediated transcription of brain ApoE expression as a marker for potential therapeutic use in neurodegenerative disorders. Preliminary results suggest these compounds display a broad spectrum of off-target activities. However, many of the novel compounds were observed to be more potent than 1. While some RXR agonists cross-signal the retinoic acid receptor (RAR), many of the rexinoids in this work displayed reduced RAR activity. The isochroman group did not appear to substantially reduce RXR activity on its own. The results of this study reveal that modifying potent, selective rexinoids like bexarotene, CD3254, and NEt-TMN can provide rexinoids with increased RXR selectivity, decreased potential for cross-signaling, and improved anti-proliferative characteristics in leukemia models compared to 1.
Keywords: retinoid-x-receptor, retinoid, rexinoid, leukemia, small molecule therapeutic, structure-activity-relationship
1. Introduction
Nuclear receptors (NRs) are ligand-dependent transcription factors that bind DNA sequence-specific motifs in enhancers and promoters to transactivate their target genes [1]. The retinoid X receptors (RXRs) are ligand-activated NRs that have pleiotropic effects including the control of hematopoietic stem cell self-renewal and differentiation. There are three different isoforms of each receptor (α, β, and γ) that are differently expressed in mouse and human tissues [2,3]. The RXRs, often working in concert with other NRs regulate gene transcription through receptor-specific molecular signals. The RXRs are remarkably versatile compared to other NRs, since they partner with many of the NRs to form heterodimers that modulate cell differentiation, migration, proliferation, and metabolic pathways. Several critical NR pathways that are RXR-dependent include those regulated by the retinoic acid receptor (RAR), the vitamin D receptor (VDR), the peroxisome proliferator-activated receptor (PPAR), the thyroid hormone receptor (TR), the farnesoid X receptor (FXR), and the liver-X-receptor (LXR), to cite just a few. All NRs function as transcriptional modulators, most often promoting transcription as a result of the presence of corresponding receptor ligand in addition to any obligate receptor partner. The receptor ligands are often endogenous molecules that bind to a ligand-binding domain (LBD) in the receptor. This subsequently forces the receptor into a new conformation more conducive towards dimerizing with another receptor, recruiting associated co-factors, and finally binding to a high affinity hormone responsive element (HRE) specific to the genes the receptor controls in the DNA. While several HREs have been located proximal to or inside the promoter region of the regulated genes, HREs are increasingly being observed a considerable distance down- or upstream from the regulated genes. The HREs display a shared sequence specificity that includes two repeat hexads enclosing a specific quantity of spacers that separate those inverted, everted, or direct repeats [4]. RAR, TR, and VDR’s HREs comprise half-sites enclosing, respectively, five, four, and three nucleotide spacers [5,6].
Before RXR was well known, TR, RAR, and VDR were believed to assemble into homodimers [7] in order to bind their respective HREs, though heterodimerizing with RXR was later discovered to be the prerequisite for them to bind and activate their HREs [8]. The naturally occurring 9-cis-retinoic acid (9-cis-RA)—a geometric isomer of the all-trans-retinoic acid (ATRA)—was identified as an RXR-specific agonist (a rexinoid) by Zhang and coworkers, who documented that its binding to RXR’s LBD triggers RXR homodimerization and the subsequent association of the homodimer to the RXR responsive elements (RXREs) [9]. In other NR heterodimers where RXR is involved, the LBD of RXR may not necessarily need to possess a rexinoid. For instance, the VDR-RXR heterodimer functions absent a ligand or rexinoid binding to RXR [10]. Conversely, there are examples of RXR heterodimers where a rexinoid bound to RXR enhances that heterodimer’s activity, such as for the LXR-RXR heterodimer [11]. This remarkable versatility—where RXR can partner with many other NRs with and without rexinoids—has led to RXR’s classification as the indispensable, master receptor [12].
A great number of studies reported in the literature concerning RXR partnering with other NRs and comprising many rexinoids have been distilled to yield two primary classifications for RXR heterodimers—permissive and nonpermissive. For purely nonpermissive heterodimers of RXR, only the other NR’s agonist can activate the heterodimer, whereas permissive RXR heterodimers can be activated by either a rexinoid or the partnering NR’s agonists [13]. The RAR-RXR, TR-RXR, and VDR-RXR heterodimers are generally nonpermissive. In the majority, but not every instance, the partnering RXR receptor is “silent” in the TR and VDR heterodimers. The RAR-RXR heterodimer, meanwhile, displays increased activity in the presence of certain rexinoids as well as agonists specific for RAR. The presence of certain rexinoids have been shown to activate RAR-RXR despite the absence of agonists specific for RAR [14]. Thus, the classical idea of purely nonpermissive RXR heterodimers has evolved towards a spectrum of conditions for permissibility, such that some RXR heterodimers, such as RAR-RXR, could be more accurately described as conditionally nonpermissive. The LXR-RXRs, FXR-RXRs, and the PPAR-RXRs, however, are fully permissive.
Potent rexinoids can disrupt the proper functioning of both types of RXR heterodimers, giving rise to pleiotropy by stimulating activity in the permissive RXR heterodimers or by removing RXR from participation in the nonpermissive heterodimers. The tendency to exert pleotropic effects has blocked clinical development of many rexinoids for various therapeutic applications. Rexinoids like 9-cis-RA, for example, arrest activity for VDR-RXR [15,16,17] and TR-RXR [18]. Similarly, the 1,25-dihydroxyvitamin D3 (1,25D) and T3 promote formation of VDR-RXR and TR-RXR, respectively, and thus deplete RXR availability for other RXR-dependent pathways. This effect has been termed cross-receptor squelching and is manifested in the loss of TR function via (1,25D)-VDR-RXR-modulated inhibition [18,19], or similarly, in the loss of VDR activity paralleling T3-TR-RXR-activation [20,21]. However, more than just RXR depletion may account for this crosstalk inhibition. Accordingly, the potency and selectivity are the primary characteristics that must be considered in the development of novel rexinoids to minimize side effects and maximize therapeutic potential. Hence, the approach of slightly modifying the structural features of a parent rexinoid’s structure could impact both characteristics and generate rexinoids with less severe pleiotropy and greater specificity, resulting in specific NR modulators (SNuRMs) [22].
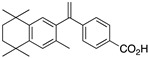
A number of rexinoid SNuRMs can be found in advanced stages of pre-clinical or clinical investigation as therapeutics, especially as preventative or treatment regimens for various cancers where selective RXR versus RAR activation exerts therapeutic effects and avoids RAR toxicities [23] in treating many human cancers. After multiple studies [24,25] that used 9-cis-RA as a starting point to model and design new compounds for RXR-selectivity, 4-[1-(3,5,5,8,8,-pentamethyltetralin-2-yl)ethynyl]benzoic acid (1) [26] was identified as a lead compound in terms of its stability and RXR-selectivity, though there were many additional candidates with equally promising profiles. After Ligand Pharmaceuticals Inc. earned FDA approval for 1 as a treatment for cutaneous T-cell lymphoma, the tradename of “bexarotene”—the more common name by which 1 is known—was assigned. Many studies making very minor modifications of the structure of bexarotene have identified structurally similar analogs of 1 that display similar profiles of activity in RXR-binding and activity—disilabexarotene (2) [27] is a representative example.
Even though bexarotene (1) is an approved treatment for CTCL, there are an increasing number of studies in other human cancers and cancer models such as lung [28], breast [29], and colon cancer [30]. In fact, bexarotene is prescribed in certain cases of non-small cell lung cancer off-label, since a proof-of-concept (POC) clinical trial showed benefits of 1 as a treatment [31,32]. Increasingly, studies are linking suppression of cell-proliferation and apoptosis synergy for combination-chemotherapeutic approaches that target RXR-controlled pathways. There are numerous studies in the literature for bexarotene (1) and analogous synthetic rexinoids exhibiting therapeutic effects for non-insulin-dependent diabetes mellitus murine models likely tied to metabolic impacts from RXR:PPAR activity [33]. While the selective activation of RXR by bexarotene avoids toxicities associated with RAR activity, humans dosed with 1 often suffer hyperlipidemia and hypothyroidism [34], and sometimes cutaneous toxicity, as the most significant side effects. These side effects arise because, much like 9-cis-RA, bexarotene (1) disrupts nonpermissive heterodimers—such as TR-RXR to incite hypothyroidism [35]—and concurrently stimulates permissive heterodimers—such as LXR-RXR to provoke hyperlipidemia [36,37] or cutaneous toxicity [38] via activating RAR at raised dose concentrations. It is difficult to dissociate the potential of a potent synthetic rexinoid to provoke increased activity in RXR’s permissive heterodimers from its selectivity for RXR alone, though many research groups are exploring this area. Developing potent rexinoids that mitigate permissive RXR heterodimer activity is a timely objective since 1 has exhibited potential to stem neurodegenerative progression in models of Alzheimer’s disease [39] (AD) as well as Parkinson’s disease (PD) [40]. A limited POC clinical trial for moderate AD patients treated with 1 or placebo revealed that non-apoE4 genotypes treated with 1 showed a reduction of soluble amyloid beta from cerebrospinal fluid that was statistically significant [41].
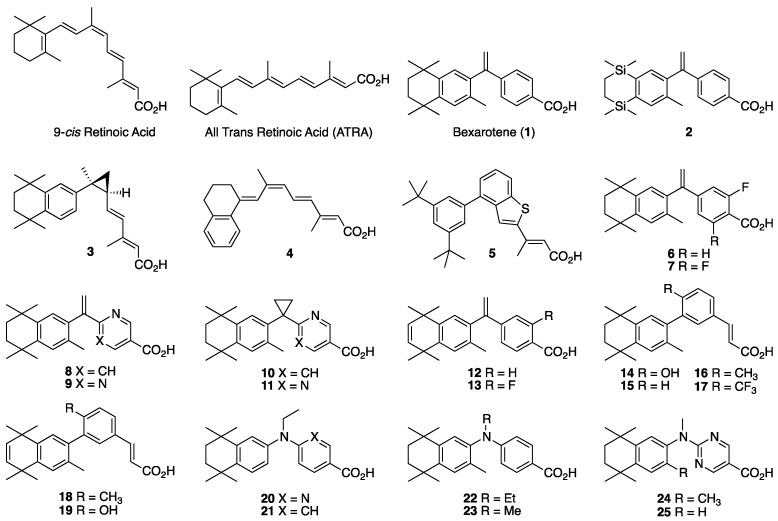
Many groups in this field have successfully used modeling and the structures of potent rexinoids in the literature as the basis to develop new rexinoids that display unique profiles. One well-known rexinoid that is in phase II clinical trials for prostate cancer [42] and pre-clinical trials for PD, AD, and multiple sclerosis is IRX-4204 (3) [43], a chiral rexinoid shown to be more selective and potent for RXR than its enantiomer. The 9cUAB30 (4) [44] is another rexinoid in clinical trials for breast cancer [45,46,47], and studies of its methylated analogs [48,49] have helped elucidate the reasons that 4 does not provoke hyperlipidemia via LXR-RXR agonism. Boehm’s group reported unbranched trienes terminating in carboxylic acids [50], along with analogs incorporating one [51] or fused aryl ring-systems [52]—in the case of the latter, compound 5 [52] is an example. When our group first entered the field, we reported a fluorinated bexarotene analog (6) [53] followed by other halogenated, and even a difluorinated bexarotene analog (7) [54]. The pyridine bexarotene analog (8) [55] and the pyrimidine bexarotene analog (9) [56], as well as LGD100268 (10) [55] and the LGD100268 pyrimidine analog (11) [56] all showed enhanced RXR activity compared to 1 and superior therapeutic effects in mouse models of cancer [28,57]. Installing an unsaturation in bexarotene’s aliphatic ring results in compound 12 [58,59], and the unsaturated-fluorinated bexarotene (13) [56] also activates LXR [60]. CD3254 (14) [61] and CD2915 (15) [62] are two synthetic rexinoids with activity comparable to 1. Our group utilized 14 and 15 to design analogous rexinoids 16–19 [56]. Kakuta and colleagues reported compound 20 (NEt-TMN) [63,64,65,66] as well as its analogous compounds 21 [67,68,69] and 22 [67,68]—all of which showed high potency and selectivity for RXR alongside many other NEt-TMN derivatives that our group reported [70]. Even replacing the ethyl group with a methyl group on the linking nitrogen atom of NEt-TMN leads to potent rexinoids such as 23 [71], 24 [71], and 25 [71] (Figure 1).
Figure 1.
Structures of 9-cis Retinoic Acid, ATRA, Bexarotene (1), and Rexinoids 2–25.
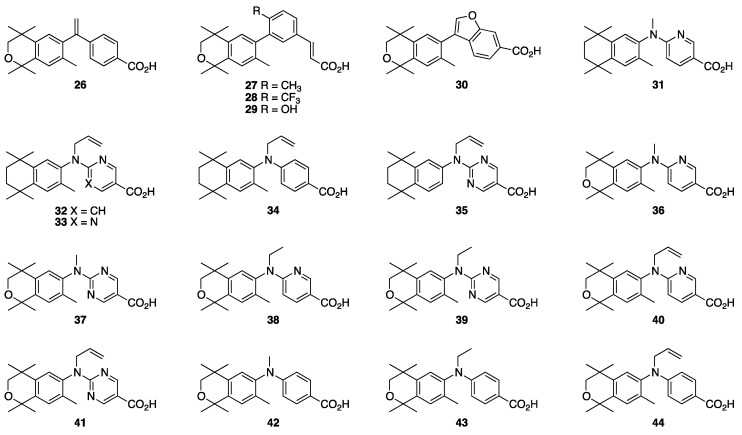
The current study uses many of the above compounds cited above as starting points to investigate how introducing new modifications result in changes to the compounds’ activities. For example, we were interested in substituting an isochroman group for the aliphatic ring system in bexarotene and some of the CD3254 analogs that have been reported, as well as a multiple fused aryl-ring system, and hence we targeted the synthesis of rexinoids 26–30. We were also interested in exploring a pyridine aromatic ring substitution from reported analogs 23–25, so we targeted the synthesis of 31. Due to the potency of NEt-TMN (20) and its analogs, we were curious about the impacts that substituting an allyl group, varying aromatic rings, and adding a methyl group would have on RXR activity for the new rexinoids, so we targeted the synthesis of 32–35. Finally, we were interested in substituting the isochroman group for the aliphatic ring system of NEt-TMN and then varying the N-alkyl chains—including methy, ethyl and allyl—along with different aromatic acid ring systems, and so we targeted 36–44 for synthesis (Figure 2). Interestingly, compound 34 [67] was previously made and disclosed by Kagechika and co-workers, so we were eager to synthesize several possible analogs of it (32, 33, 35, 40, 41, and 44) and compare their activities.
Figure 2.
Structures of Reported Rexinoids 3–27.
While the isochroman group places a fairly polar, hydrogen-bonding oxygen atom in the nonpolar aliphatic ring system for the known parent rexinoids, we hypothesized that it would not disrupt the largely non-polar binding interaction in the RXR LBD. Further, we hypothesized that the isochroman group would make the rexinoids possessing it more resistant to metabolic oxidation. If the isochroman rexinoids were as potent and selective as their non-isochroman counterparts, we also hypothesized that they would possess similar in vitro activities as their parent compounds. Importantly, we wanted to assess these specific hypotheses about the incorporation of an isochroman group across multiple parent compound structures that include bexarotene (1), CD3254 (14), and various NEt-TMN (20) analogs coupled with other potential modifications to assess similarities or differences in the activities of the resulting analogs and their predecessors. Hence, we proceeded with the synthesis and testing of these compounds.
2. Results: Molecular Modeling
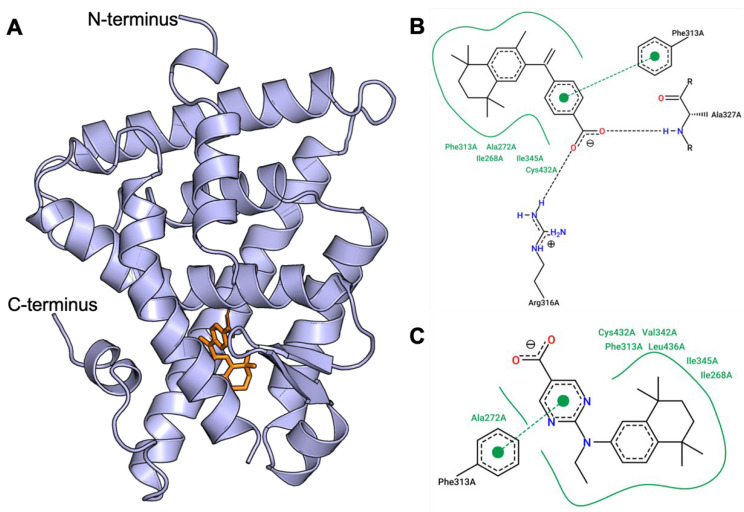
The binding affinity, predicted using AutoDock Vina [72], of human-RXR for each ligand is output as an energy unit (in kcal/mol, Table 1). These predicted ligand-bound RXR complexes were then visually inspected using PyMol (version 2.3, Shrodinger, LLC) (Figure 3). To further analyze and illustrate the interactions between RXR protein residue sidechains with the ligands, PoseView (BioSolvIT [73,74]) was used to generate the detailed two-dimensional depictions (Figure 3B,C). In these two-dimensional depictions, hydrogen bonds are presented as dashed lines between interaction partners, and hydrophobic interactions are depicted as smooth contour lines.
Table 1.
Tabe of Auto-Dock Vina Binding Energies, EC50 (nM) values, 96h IC50 (nM) + 100 nM ATRA values, LXRE Activities (% of Bex), LHS Score (vs. Bex), and RARE Activity (%ATRA at 10 nM).
| Compound | Auto-Dock Vina Scores (kcal/mol) |
EC50 (nM) +/− (SD) |
96 h IC50 (nM) + 100 nM ATRA +/− SD | LXRE Activity (% of Bex) |
LHS Score (vs. Bex) |
RARE Activity (%ATRA at 10 nM) |
|---|---|---|---|---|---|---|
 1 1
|
–12.7 | 17.8 (1.0) | 7.8 (1.1) | 100 | 1 | 37.16 |
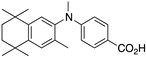 23 23
|
–11.5 | 8.6 (0.2) | 3.7 (1.1) | 99.5 | 0.59 | 24.33 |
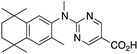 24 24
|
–10.9 | 6.2 (0.1) | 3.9 (1.1) | 105.98 | 0.95 | 32.04 |
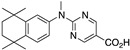 25 25
|
–10.6 | 17.8 (0.2) | 12.9 (1.2) | 80.98 | 0.7 | 14.32 |
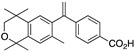 26 26
|
–12.4 | 51.0 (0.1) | 60.0 (1.2) | 213.84 | 2.81 | 10.1 |
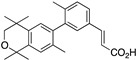 27 27
|
–9.3 | 65.3 (0.1) | 27.6 (1.2) | 253.64 | 1.76 | 19.45 |
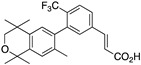 28 28
|
–11.7 | 59.2 (0.1) | 38.4 (1.3) | 208.05 | 1.06 | 15.85 |
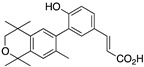 29 29
|
–11.5 | 3.9 (0.5) | 3.7 (1.2) | 221.39 | 1.29 | 54.64 |
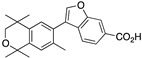 30 30
|
–11.9 | >1000 | >1000 | 161.38 | 4.03 | 7.05 |
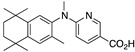 31 31
|
–11.5 | 5.4 (0.1) | 2.1 (1.1) | 114.37 | 1.04 | 48.61 |
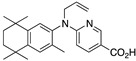 32 32
|
–11.4 | 1.8 (1.1) | 2.0 (1.6) | 92.2 | 0.61 | 61.79 |
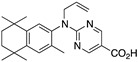 33 33
|
–11.0 | 5.3 (0.2) | 1.3 (1.1) | 103.99 | 0.72 | 39.94 |
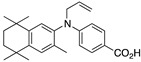 34 34
|
–11.6 | 11.5 (0.2) | 4.1 (1.2) | 77.44 | 0.53 | 18.74 |
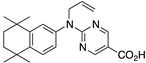 35 35
|
–10.6 | 0.54 (0.11) | 4.68 (1.91) | 94.07 | 0.79 | 28.76 |
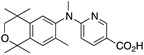 36 36
|
–11.0 | 75.0 (0.1) | 42.3 (1.2) | 51.57 | 0.69 | 6.25 |
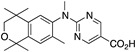 37 37
|
–10.8 | 404.9 (0.1) | >1000 | 62.72 | 1.36 | 4.02 |
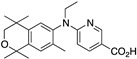 38 38
|
–11.0 | 23.7 (0.2) | 11.6 (1.1) | 79.69 | 0.69 | 12.59 |
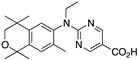 39 39
|
–10.7 | 151.6 (0.1) | >1000 | 61.52 | 0.92 | 4.96 |
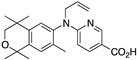 40 40
|
–11.1 | 10.7 (0.2) | 16.6 (1.2) | 73.76 | 0.63 | 11.13 |
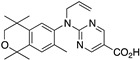 41 41
|
–10.7 | 143.4 (0.3) | 105 (1.6) | 75.66 | 0.71 | 5.16 |
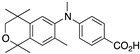 42 42
|
–11.4 | 87.7 (0.1) | 136.7 (1.6) | 61.86 | 0.56 | 7.99 |
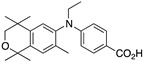 43 43
|
–11.5 | 23.1 (0.1) | 14.8 (1.2) | 71.09 | 0.55 | 9.6 |
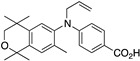 44 44
|
–10.7 | 116.5 (0.1) | ~1000 | 68.04 | 0.68 | 11.13 |
Figure 3.
AutoDock Vina simulation of bexarotene bound to human RXR protein. (A) Cartoon representation of the human RXR alpha ligand binding domain (PDB:1FBY, blue) and the docked compound bexarotene (orange). N- and C-termini are labeled. (B,C) Two-dimensional depiction of the interactions between protein residue sidechains with bexarotene (B) and compound 26 (C) using PoseView (BioSolvIT [73,74]). In both (B) and (C), hydrogen bonds are presented as dashed lines between interaction partners, and hydrophobic interactions are depicted as smooth contour lines.
The AutoDock Vina docking results showed that the standard compound bexarotene (1), with a score of −12.7 kcal/mol, was the most potent among all compounds. Compound 26 has a comparable score of −12.4 kcal/mol. Additionally, similar interactions with RXR residue sidechains were observed between 1 and 26 such as direct interactions between Phe313 and the aromatic cores of either ligand, as well as hydrophobic residues Ile268, Ile345, and Ala272 bearing common interactions (Figure 3B,C). Several other compounds have similar but slightly lower scores of −11.9 kcal/mol (30), −11.7 kcal/mol (28), −11.6 kcal/mol (34), and −11.5 kcal/mol (23, 29, 31, 32, 42, 43, and 44) (Table 1). Therefore, we can conclude that RXR compounds with a docking score within 10% that of bexarotene could potentially possess comparable, or better, EC50 and IC50 profiles for further study.
3. Results: Chemistry
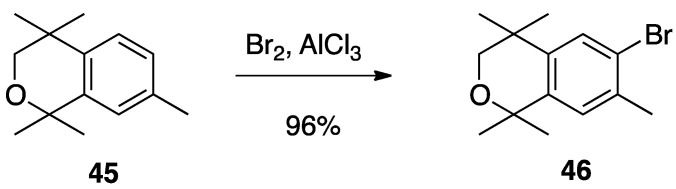
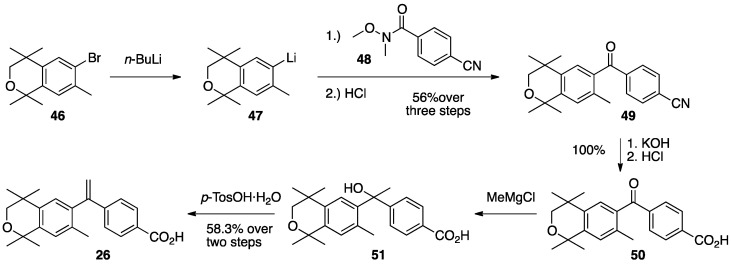
The synthesis of compound 26 begins with the bromination of commercially available 1,1,4,4,7-pentamethylisochroman (45) with bromine in dichloromethane and aluminum chloride catalyst to give 6-bromo-1,1,4,4,7-pentamethylisochroman (46) in 96% yield (Scheme 1) [75].
Scheme 1.
Synthesis of bromo-isochroman 46.
Compound 46 was lithiated by treatment with n-butyllithium, and the resulting aryl lithium reagent (47) was transferred to a solution of the reported amide 48 [59] in THF to provide ketone (49) in 56% purified yield whose nitrile group is then hydrolyzed to a carboxylic acid (50) in quantitative yield. Compound 50, when treated with more than two equivalents of methylmagnesium chloride is converted to alcohol 51 that is then dehydrated to compound 26 in 58.3% over two steps (Scheme 2).
Scheme 2.
Conversion of 46 to lithium-isochroman 47, its reaction with amide-nitrile 48 to give ketone-nitrile 49 that was subsequently hydrolyzed to ketone-acid 50 which was converted to alcohol-acid 51 that was finally dehydrated to isochroman acid 26.
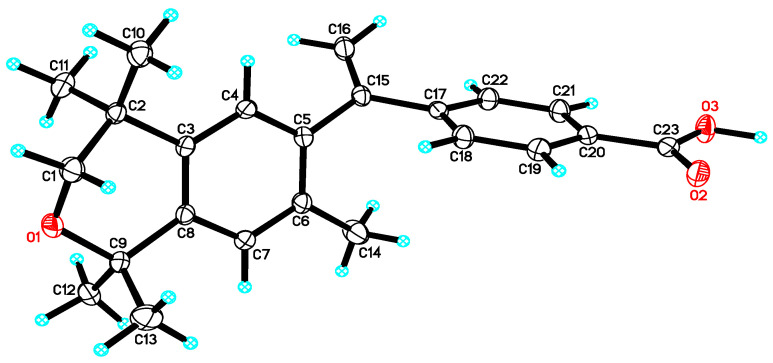
Acid 26 formed single, transparent crystals, so an X-ray diffraction study was conducted to confirm the structure of 26 (Figure 4).
Figure 4.
X-ray crystal structure of a 0.114 × 0.141 × 0.380 mm3 crystal of 26. Thermal ellipsoids are shown at 50% probability level.
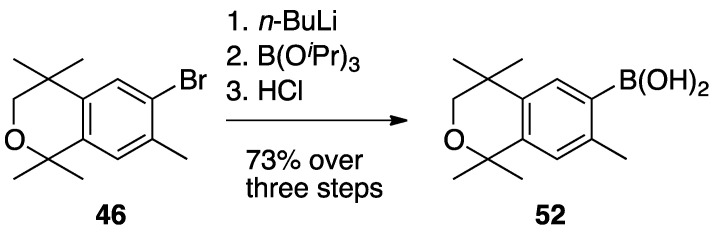
To synthesize compound 27, compound 46 was treated with n-butyllithium and then triisopropylborate to give 52 in 73% yield (Scheme 3).
Scheme 3.
Synthesis of boronic acid 52.
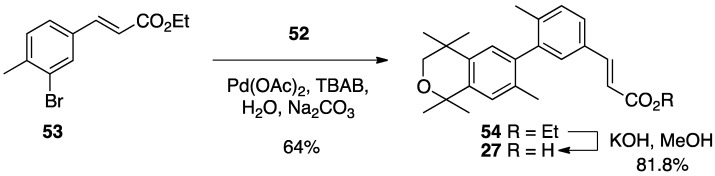
The known ethyl ester 53 [56] was combined with boronic acid 53 along with Pd(II) diacetate, TBAB, and sodium carbonate in water and heated to boiling for 5 min to give 54 in 64% yield, and ethyl ester 54 was saponified with KOH in methanol to give 27 in 81.8% yield after acidification with 1N HCl, filtration, and purification by column chromatography (Scheme 4).
Scheme 4.
Synthesis of biphenyl 54 and its saponification to give acid 27.
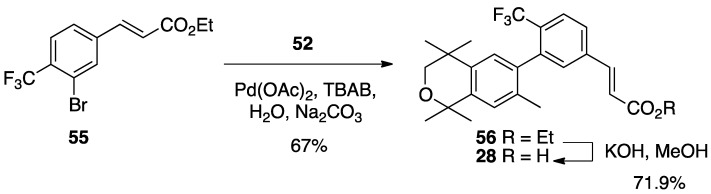
A similar procedure was used to make compounds 28, 29, and 30. For compound 28, the known ethyl ester 55 [56] was combined with boronic acid 52 along with Pd(II) diacetate, TBAB, and sodium carbonate in water and heated to boiling for 5 min to give 56 in 67% yield, and ethyl ester 56 was saponified with KOH in methanol to give 28 in 71.9% yield after acidification with 1N HCl, filtration, and purification by column chromatography (Scheme 5).
Scheme 5.
Synthesis of biphenyl 56 and its saponification to give acid 28.
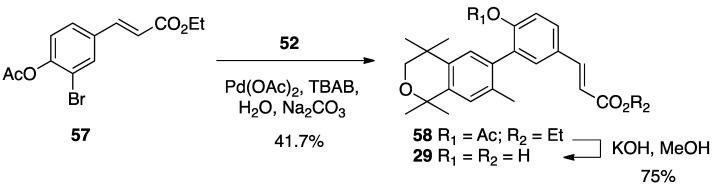
For compound 5, the known ethyl ester 57 [56] was combined with boronic acid 52 along with Pd(II) diacetate, TBAB, and sodium carbonate in water and heated to boiling for 5 min to give 58 in 41.7% yield, and ethyl ester 58 was saponified with KOH in methanol to give 29 in 75% yield after acidification with 1N HCl, filtration, and purification by column chromatography (Scheme 6).
Scheme 6.
Synthesis of biphenyl 58 and its saponification to give acid 29.
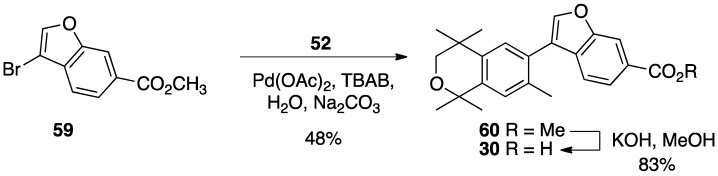
For compound 30, the commercially available methyl ester 59 was combined with boronic acid 52 along with Pd(II) diacetate, TBAB, and sodium carbonate in water and heated to boiling for 5 min to give 60 in 48% yield, and methyl ester 60 was saponified with KOH in methanol to give 30 in 83% yield after acidification with 1N HCl, filtration, and purification by column chromatography (Scheme 7).
Scheme 7.
Synthesis of methyl ester 60 and its saponification to give acid 30.
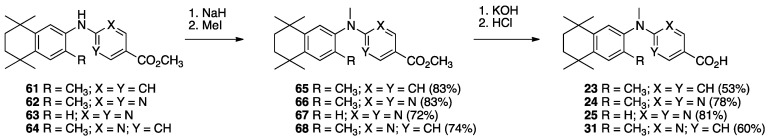
The synthesis of known compounds 23, 24, 25, and novel compound 31 begins with the conversion of known compounds 61–64 [70] to compounds 65–68 by treatment with sodium hydride followed by methyl iodide in DMF with stirring at room temperature in good yields. The methyl esters 65–68 were saponified by treatment with KOH in methanol followed by acidification and purification to give compounds 23–25 and 31, respectively, in moderate to good yields (Scheme 8).
Scheme 8.
Synthesis of methyl esters 65–68 and their saponification to give acids 23–25 and 31.
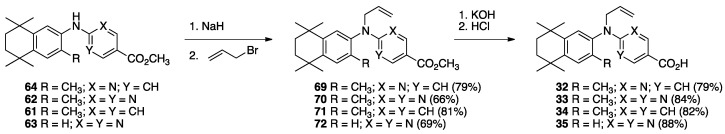
The synthesis of compounds 32–35 begins with the conversion of known compounds [70] 64, 62, 61, and 63 to compounds 69–72 by treatment with sodium hydride followed by allyl bromide in DMF with stirring at room temperature in good yields. The methyl esters 69–72 were saponified by treatment with KOH in methanol followed by acidification and purification to give compounds 32–35 in moderate to good yields (Scheme 9).
Scheme 9.
Synthesis of methyl esters 69–72 and their saponification to give acids 32–35.
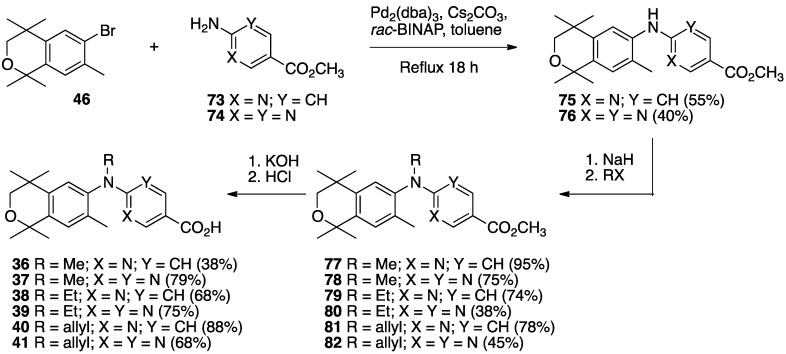
The synthesis of compounds 36–41 begins with the palladium-catalyzed reaction of the known aryl bromide 46 with methyl 6-aminonicotinate (73) or methyl 2-aminopyrimidine-5-carboxylate (74) to give diaryl amines 75 or 76 in moderate yields of 55% and 40%, respectively. The diaryl amines 75 and 76 were treated with sodium hydride followed by methyl iodide, ethyl iodide, or allyl bromide to give methyl esters 77–82 in moderate to good yields which were then saponified by treatment with KOH in methanol followed by acidification and purification to give compounds 36–41 in moderate to good yields (Scheme 10).
Scheme 10.
Synthesis of diaryl amines 75–76, their alkylation to give methyl esters 77–82 and their saponification to give acids 36–41.
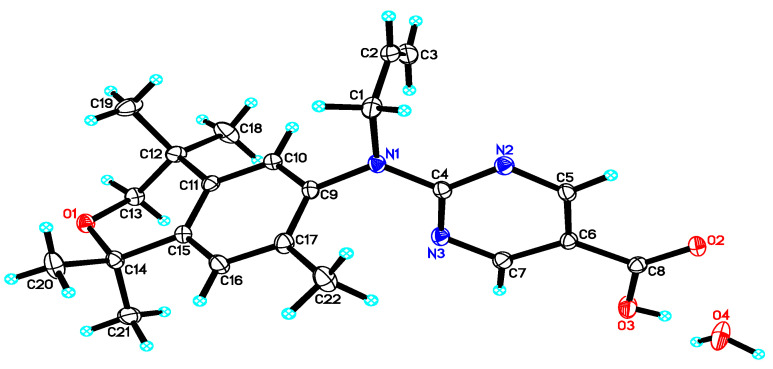
Acid 41 formed single, transparent crystals from d6-DMSO, so an X-ray diffraction study was undertaken to confirm the structure for 41 (Figure 5).
Figure 5.
X-ray crystal structure of a 0.184 × 0.217 × 0.371 mm3 crystal of compound 41 as a monohydrate. Thermal ellipsoids are shown at 50% probability level.
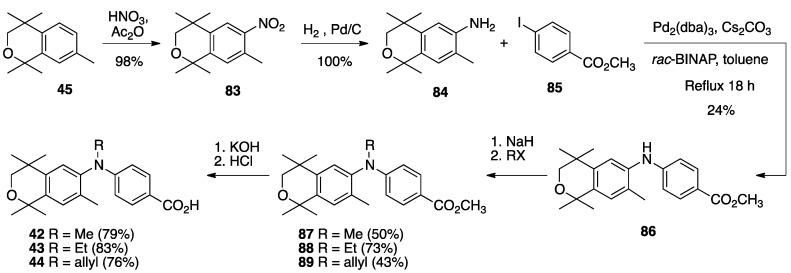
Finally, the synthesis of novel rexinoids 42–44 begins with the nitration of isochroman (45) to give 83 in 98% yield, followed by the hydrogenation of 83 to give aryl amine 84 in quantitative yield. Aryl amine 84 is then combined with 4-iodomethylbenzoate (85) in a palladium-catalyzed reaction to give 86 in 24% yield. Diaryl amine 86 is then treated with sodium hydride followed by methyl iodide, ethyl iodide, or allyl bromide to give methyl esters 87–89 in moderate to good yields and the methyl esters are saponified with potassium hydroxide in methanol followed by acid work-up to give rexinoids 42–44 in good yields after purification (Scheme 11).
Scheme 11.
Synthesis of nitro-isochroman 83, its reduction to aniline 84 and subsequent coupling with 85 to provide 86 and its subsequent alkylation to give methyl esters 87–89 and their saponification to yield acids 42–44.
4. Results and Discussion: Biological Assays
Bexarotene (1) and the isochroman and other analogs of bexarotene, CD3254, and NEt-TMN 26–44 were tested in a KMT2A-MLLT3 cell line to determine RXRα activation EC50 values by both a Luc– and GFP-assay, and then IC50 values for a 96 h cell viability assay were determined both in the presence and absence of 100 nM ATRA and the resulting data is tabulated in Table 1. Compounds 1 and 26–44 were examined for cytotoxicity and mutagenicity in Saccharomyces cerevisiae [76]. None of the compounds were mutagenic or cytotoxic.
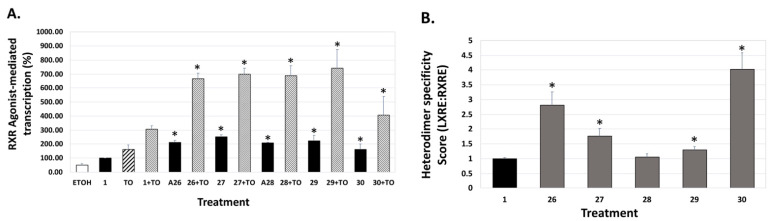
We also evaluated the analogs for their capacity to bind and activate the liver-X-receptor (LXR) using a liver-X-receptor responsive element (LXRE)-based assay, and assessed the effects in the presence vs. absence of an authentic LXR compound (TO901317), as well as with bexarotene alone. LXR is known to regulate inflammatory responses and lipid metabolism in multiple tissues, including the central nervous system, and there is sufficient evidence that strong cholesterol and lipid metabolism in the brain, along with enhanced ApoE expression, is paramount to reducing the risk of human dementias. The “activity assessment” of our novel RXR agonists for their ability to enhance gene expression using a DNA responsive element sequence (LXRE) that is found to occur in the natural human promoter of LXR-RXR controlled genes (such as ApoE) was carried out in U87 glial cells with bexarotene (1) as a comparative control. Using the above assay system, the activation from this natural LXRE was probed with either 100 nM RXR analogs (or bexarotene) alone or in combination with 100 nM of both the RXR agonist and an LXR ligand T0901317 (TO). The employment of both LXR and RXR agonists collectively was anticipated to demonstrate a more vigorous response in LXRE transactivation due to cumulative and/or synergistic effects of dual ligand activation of the RXR-LXR heterodimer. The results demonstrated that in comparison to the parent bexarotene (1) compound alone, separate dosing of the cells with analogs 26, 27, 28, 29, and 30 displayed more LXR/LXRE activity (Figure 6A, p < 0.05).
Figure 6.
RXR agonist potentiation of LXRE-regulated transactivation with or without the T0901317 LXR agonist. (A) U87 glial cells were transfected with an expression vector for human LXRα, an LXRE-luciferase reporter gene with three tandem copies of the LXRE from the human ApoE gene, and a Renilla control plasmid. Cells were transfected for 24 h utilizing a liposome-mediated transfection protocol and then treated with ethanol vehicle, or 100 nM of the indicated compound alone or in combination with 100 nM TO901317 (TO). LXRE-directed activity was compared to compound 1 (Bexarotene), set to 100%. (B) The “Heterodimer Specificity Score” was determined by the LXRE:RXRE ratio with compound 1 set to 1.0. All error bars represent standard deviations; the data are representative of at least three independent experiments with six replicates in each treatment group. * p < 0.05 versus control compound 1.
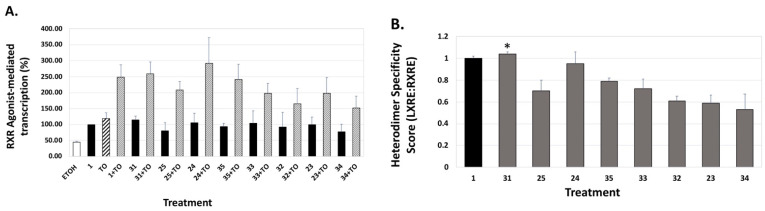
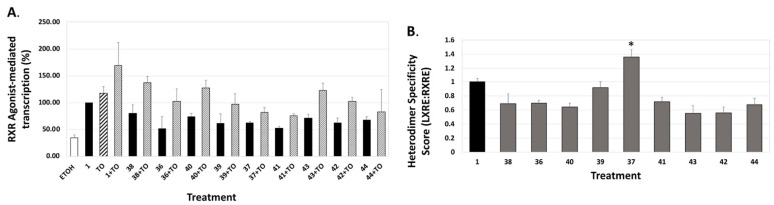
Specifically, the analogs displayed activities ranging from 52% to 254% of the bexarotene control (set to 100%; Table 1). Furthermore, when the LXR synthetic ligand (TO) was used in combination with bexarotene (1) or analogs, similar results were observed (Figure 6A, Figure 7A, and Figure 8A, stippled bars). Although some of the analogs exhibit lower LXR activation when compared to bexarotene (1), it is imperative to evaluate this activity in the background of the RXR-RXR homodimer activity of each analog, and to thus “normalize” the LXR/LXRE heterodimer activation results. The LXRE Heterodimer Specificity (LHS) score (Figure 6B, Figure 7B, and Figure 8B) is thus calculated based on both the RXR homodimer activity, as well as the LXR heterodimer activation. The results of this LHS analysis (Table 1) reveal that many of our novel compounds (e.g., 26, 27, 29, 30, 31, and 37, p < 0.05) display enhanced LXR/LXRE activity via increased heterodimer specificity compared to the parent bexarotene (1).
Figure 7.
RXR agonist potentiation of LXRE-regulated transactivation with or without the T0901317 LXR agonist. (A) U87 glial cells were transfected with an expression vector for human LXRα, an LXRE-luciferase reporter gene with three tandem copies of the LXRE from the human ApoE gene, and a Renilla control plasmid. Cells were transfected for 24 h utilizing a liposome-mediated transfection protocol and then treated with ethanol vehicle, or 100 nM of the indicated compound alone or in combination with 100 nM TO901317 (TO). LXRE-directed activity was compared to compound 1 (Bexarotene), set to 100%. (B) The “Heterodimer Specificity Score” was determined by the LXRE:RXRE ratio with compound 1 set to 1.0. All error bars represent standard deviations; the data are representative of at least three independent experiments with six replicates in each treatment group. * p < 0.05 versus control compound 1.
Figure 8.
Evaluation of RXR agonists to potentiate LXRE-mediated transactivation in the absence and presence of LXR ligand T0901317. (A) U87 glial cells were transfected with an expression vector for human LXRα, an LXRE-luciferase reporter gene with three tandem copies of the LXRE from the human ApoE gene, and a Renilla control plasmid. Cells were transfected for 24 h utilizing a liposome-mediated transfection protocol and then treated with ethanol vehicle, or 100 nM of the indicated compound alone or in combination with 100 nM TO901317 (TO). LXRE-directed activity was compared to compound 1 (Bexarotene), set to 100%. (B) The “Heterodimer Specificity Score” was determined by the LXRE:RXRE ratio with compound 1 set to 1.0. All error bars represent standard deviations; the data are representative of at least three independent experiments with six replicates in each treatment group. * p < 0.05 versus control compound 1.
Finally, even though compound 1 is selective for binding to RXR, it does include some “residual” RAR activity. We assessed the potential of our analogs to induce transcription via the retinoic acid response element (RARE) and retinoic acid receptor. For these studies, we employed human embryonic cells (HEK293) that were transfected with human RARα and dosed with 10 nM of either all-trans retinoic acid (ATRA), the endogenous ligand for RARα, compound 1, or our panel of analogs. The results of the assay revealed that compound 1 displayed an average of 37% of the activity of the ATRA control (set to 100%, Table 1). Analog 32 exhibited the greatest RARE activation at 62% of ATRA, while analog 37 showed the lowest RARE activity at 4%, which is indistinguishable from the ethanol control (Table 1). Importantly, most of our novel analogs (14 out of 18) possess attenuated “cross-over” onto RAR-RARE signaling compared to bexarotene (1).
5. Conclusions
While the substitution of the saturated, pentamethyl-tetrahydronaphthyl ring system of bexarotene, CD3254, or Net-TMN-analog parent compound frameworks with a pentamethyl-isochroman ring system tended to decrease both modeled binding scores as well as RXR EC50 values, there were nevertheless some examples of very potent compounds possessing the pentamethyl-isochroman ring system. Several reported and novel compounds displayed lower or equivalent EC50 values than bexarotene (e.g., reported compounds 23–25 and novel compounds 29, 31–33, 35, and 40), and several reported and novel compounds exhibited lower IC50 values along with 100 nM ATRA (e.g., reported compounds 23 and 24, and novel compounds 29 and 31–35) than bexarotene. It should be noted that compound 29 is an isochroman analog of CD3254 and compound 40 is an isochroman analog of NEt-TMN, so while the isochroman substituted analog tends to reduce potency compared to the parent compounds, rexinoids possessing the isochroman group can still be synthesized that exhibit greater potency than bexarotene. Notably, some of the compounds possessing the greatest RARE activity at 10 nM vs. ATRA also exhibited lower IC50 values along with 100 nM ATRA (e.g., 29 and 31–33). Additionally, the range of LXRE activity vs. bexarotene demonstrated that several compounds exhibit increased LXR activity and specificity vs. bexarotene. In particular, compounds 29 and 31–35 appear to be good candidates for further investigation, and this study suggests that modification of known, potent rexinoids results in novel compounds with unique and promising therapeutic potential meriting further examination. We are actively pursuing additional studies towards these ends.
6. Materials and Methods
Molecular Modeling. The three-dimensional structures of the compounds reported herein were generated using ChemDraw 3D (PerkinElmer Informatics, Waltham, MA, USA), energy minimized, and exported in the Protein Data Bank (PDB) format. The human RXR alpha ligand binding domain structure model was obtained from the PDB (PDB code: 1FBY [77]). The crystallized ligand, 9-cis retinoic acid, was removed from the protein model prior to docking simulations. Further, 9-cis retinoic acid was also used as a positive control in the docking studies presented here. Both the protein and ligand models were prepared using MGLTools (version 1.5.7) [78] and screened virtually using AutoDock Vina [72]. The search space volume (4032 Å3) was determined using MGLTools (center_x = 12.848, center_y = 29.174, center_z = 50.269, size_x = 16, size_y = 14, size_z = 18). The exhaustiveness was set to 8.
Hematopoietic cell culture. Murine bone marrow Kit+ cells were isolated using an Automacs Pro (Miltenyi Biotec, San Diego, CA, USA) according to the manufacturer’s protocol. Kit+ cells were plated in progenitor expansion medium (RPMI 1640 medium, 15% fetal bovine serum, stem factor [50 ng/mL], interleukin 3 [10 ng/mL], Flt3L [25 ng/mL], thrombopoietin [10 ng/mL], L-glutamine [2 mM], sodium pyruvate [1 mM], HEPES buffer [10 mM], penicillin/streptomycin [100 units/mL], and b-mercaptoethanol [50 mM]) overnight and transduced with MSCV-KMT2A-MLLT3 retrovirus by spinfection with 10 μg/mL polybrene and 10 mM HEPES at 2400 rpm, 30 °C for 90 min in an Eppendorf 5810R centrifuge. Cells were transplanted into sublethally irradiated mice and subsequent leukemia harvested 4 to 6 months later. KMT2A-MLLT3 leukemia cells were cultured in vitro using similar media, but without Flt3L or thrombopoietin.
UAS/Gal4 assay. Bone marrow cells from UAS-GFP mice [79] were transduced with retroviruses MSCV-Gal4 (DNA binding domain, DBD)—RXRα (ligand binding domain, LBD)—IRES—mCherry. Gal4 is a yeast transcription factor and the UAS sequence is not recognized by mammalian transcription factors. Cells were treated, and after 48 h, GFP was measured by flow cytometry.
Luciferase detection. HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) [80]. Six hours after transfection, the cells were collected and plated into a 48-well plate in 1% BSA media in biological triplicates and treated with compounds. After 40 h incubation, the cells were harvested and assayed for luciferase (Luc Assay System with Reporter Lysis Buffer, Promega) in a Beckman Coulter LD400 plate reader.
LXRE assay. Human glioblastoma cells (U87) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and used to perform the LXRE-mediated assays. Authentication and validation of these master stocks included both testing for mycoplasma contamination (via Universal Mycoplasma Detection Kit; ATCC, Cat. #30-1012K), as well as short tandem repeat (STR) analysis to confirm cell line identity. For LXRE experiments, cells were seeded at a density of 80,000 cells/well in a 24-well plate and maintained in DMEM (Hyclone) supplemented with 10% fetal bovine serum, 100 µg/mL streptomycin, 100 U/mL penicillin (Invitrogen, Carlsbad, CA, USA) at 37 degrees Celsius, 5% CO2 for 24 h. The cells were transiently transfected in individual wells using Polyethylenimine (PEI) (Polysciences, Inc., Warrington, PA, USA) according to the manufacturer’s protocol. The cells in each well received 250 ng of an LXRE-luciferase reporter gene, 50 ng of CMX-h-LXRα (an expression vector for human LXRα), 50 ng of pSG5-human RXRα (an expression vector for human RXRα), and 20 ng of Renilla control plasmid was used along with 1.25 µL of PEI reagent. After 22–24 h of transfection, the cells were treated with either vehicle control ethanol, reference compound bexarotene (100 nM) alone, or in combination with 100 nM T0901317 (an LXR ligand), or 100 nM of the indicated bexarotene analog either alone or in combination with T0901317, as indicated. All compounds were solubilized in ethanol. After 24 h of treatment, the cells were lysed in 1X passive lysis buffer (Promega, Madison, WI USA) and the amount of reporter gene product (luciferase) was quantified using the Dual-Luciferase Reporter Assay System based on the manufacturer’s protocol (Promega) in a Sirius FB12 luminometer (Berthold Detection Systems, Pforzheim, Germany). Luminescence resulting from the inducible firefly luciferase was divided by luminescence from the constitutively expressed Renilla luciferase in order to normalize for transfection efficacy, cell death, and cellular toxicity from ligand exposure. The data are a compilation of between six to eight independent assays with each treatment group dosed in triplicate for each independent assay. The LXRE-directed transcriptional activation of the reporter gene was measured in comparison to the reference compound bexarotene (1) set to 100%. Error bars on all graphs indicate the standard deviation of the replicate experiments.
RARE assay. Human embryonic kidney cells (HEK293) were plated at 60,000 cells per well in a 24-well plate and maintained as described above. After 22–24 h, the cells were transiently transfected with 250 ng pTK-DR5(X2)-Luc, 25 ng pSG5-human RXRα, and 20 ng of Renilla control plasmid using 1.25 µL polyethylenimine (PEI) per well for 24 h. The sequence of the double DR5 RARE is: 5’-AAAGGTCACCGAAAGGTCACCATCCCGGGAGGTCACCGAAAGGTCACC-3’ (DR5 responsive elements underlined). After 22–24 h of transfection, the cells were treated with ethanol vehicle (0.1%), all-trans-retinoic acid (ATRA, the endogenous ligand for RAR), or the indicated rexinoid analog at a final concentration of 10 nM. After 24 h of treatment, the cells were lysed and the retinoid activity was measured as described above (dual luciferase assay). The activity of compound 1 (or analog) divided by the activity of ATRA (expressed as a percentage) represents the RARE activity. The data (Table 1) are a compilation of between three to four independent assays with each treatment group dosed in triplicate for each independent experiment. The value for the positive control ATRA was set to 100%.
Data analysis. Statistical analysis was performed using Microsoft Excel (Microsoft, Redmond, WA, USA) software. t-tests were performed, as appropriate. All error bars represent the standard deviation. For Figure 6, Figure 7 and Figure 8, data are expressed as means ± SD. Statistical differences between two groups (generally the bexarotene (1) control group versus bexarotene analog group) were determined by a two-sided Student’s t-test. A p-value of less than or equal to 0.05 was considered significant.
Mutagenicity and Toxicity Assay. Mutagenicity and toxicity were assessed in a eukaryotic Saccharomyces cerevisiae model as described previously [76]. Each compound was solubilized in DMSO at increasing concentrations. D7 S. cerevisiae indicator cells were incubated with the compounds for 3 h before plating on selective media or YPD. Growth on plates was used to determine mutagenicity and cytotoxicity. Growth of colonies on the full nutrient YPD plate for each treatment was compared to the DMSO only control. Eleven micrograms per microliter was the highest concentration tested.
X-ray Data Collection, Structure Solution and Refinement. A colorless crystal of indicated dimensions was mounted on a glass fiber and transferred to a Bruker SMART APEX II diffractometer system. The APEX2 (APEX2 Version 2014.11-0, Bruker AXS, Inc.; Madison, WI 2014 USA) program package was used to determine the unit-cell parameters and for data collection (60 sec/frame scan time). The raw frame data were processed using SAINT (SAINT Version 8.34a, Bruker AXS, Inc.; Madison, WI 2013) and SADABS (Sheldrick, G.M. SADABS, Version 2014/5, Bruker AXS, Inc.; Madison, WI 2014 USA) to yield the reflection data file. Subsequent calculations were carried out using the SHELXTL (Sheldrick, G.M. SHELXTL, Version 2014/7, Bruker AXS, Inc.; Madison, WI 2014 USA) program package. For compound 26, the diffraction symmetry was 2/m and the systematic absences were consistent with the monoclinic space group P21/n that was later determined to be correct. The structure for 26 was solved by direct methods and refined on F2 by full-matrix least-squares techniques, and the analytical scattering factors [81] for neutral atoms were used throughout the analysis. Hydrogen atoms in 26, except those associated with C(14), were located from a difference-Fourier map and refined (x,y,z and Uiso), and H(14A), H(14B), and H(14C) were included using a riding model. Least-squares analysis for 26 yielded wR2 = 0.1064 and Goof = 1.025 for 328 variables refined against 3880 data (0.80 Å), R1 = 0.0426 for those 3293 data with I > 2.0σ(I). For compound 41, the diffraction symmetry was 2/m and the systematic absences were consistent with the monoclinic space groups Cc and C2/c, and it was later determined that space group Cc was correct. The structure for 41 was solved by direct methods and refined on F2 by full-matrix least-squares techniques, and again, the analytical scattering factors for neutral atoms were used throughout the analysis. Hydrogen atoms in 41 were located from a difference-Fourier map and refined (x,y,z and Uiso), and there was one water solvent molecule present. Least-squares analysis for 41 yielded wR2 = 0.0792 and Goof = 1.022 for 378 variables refined against 5116 data (0.73 Å), R1 = 0.0322 for those 4861 data with I > 2.0σ(I), and the absolute structure could not be assigned by refinement of the Flack parameter [82]. The wR2 = [Σ[w(Fo2 − Fc2)2]/Σ[w(Fo2)2] ]1/2 and R1 = Σ||Fo| − |Fc||/Σ|Fo|. Goof = S = [Σ[w(Fo2 − Fc2)2]/(n − p)]1/2 where n is the number of reflections and p is the total number of parameters defined.
Acknowledgments
This work was also supported by The Rotary Coins for Alzheimer’s Research Trust Fund. We also thank Arizona State University New College Undergraduate Inquiry and Research Experiences (NCUIRE) Program for supporting undergraduate student co-authors. Thanks are given to Felix Grun of the High-Resolution Mass Spectrometry Laboratory at University of California, Irvine (UCI). We thank Gayla Hadwiger and Anh Vu for technical support. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine for the use of the Flow Cytometry Core. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant P30 CA91842.
Abbreviations
| ATRA | all-trans-retinoic acid |
| AD | Alzheimer’s disease |
| 9-cis-RA | 9-cis-retinoic acid |
| CTCL | cutaneous T-cell lymphoma |
| DMF | dimethylformamide |
| DMSO | dimethylsulfoxide |
| DNA | deoxyribonucleic acid |
| FXR | farnesoid-X-receptor |
| HCl | hydrochloric acid |
| HRE | hormone responsive element |
| KOH | potassium hydroxide |
| LBD | ligand binding domain |
| LHS | LXR Heterodimer Specificity |
| LR | lipid risk assessment index |
| LXR | liver-X-receptor |
| LXRE | liver-X-receptor element |
| NaBu | sodium butyrate |
| NR | nuclear receptor |
| POC | proof of concept |
| PPAR | peroxisome proliferator activating receptor |
| RAR | retinoic-acid-receptor |
| RARE | retinoic acid receptor element |
| RXR | retinoid-X-receptor |
| RXRE | retinoid-X-receptor element |
| SNuRMs | specific nuclear receptor modulators |
| SREBP | sterol regulatory element binding protein |
| TR | thyroid hormone receptor |
| VDR | vitamin D receptor |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms232416213/s1.
Author Contributions
Conceptualization, C.E.W., P.W.J., P.A.M., O.d.M. and J.S.W.; methodology, C.E.W., P.W.J., P.A.M., O.d.M., S.R., S.M., W.L., M.-Y.L., Y.N. and J.S.W.; X-ray Diffraction Analysis, J.W.Z.; formal analysis, O.d.M., S.R., M.A.S., M.A.S., G.A.M., I.J.L., D.J.I., T.D.K., S.J.P., A.P., L.T., A.L., I.K., R.P., B.M.S., E.S. and J.S.; writing—original draft preparation, C.E.W., P.W.J., P.A.M., O.d.M., M.-Y.L., Y.N. and J.S.W.; writing—review and editing, C.E.W., P.W.J., P.A.M., O.d.M., M.-Y.L., W.L. and J.S.W.; funding acquisition, C.E.W., P.W.J., P.A.M. and J.S.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
CCDC numbers 2191083 and 2191084 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif, accessed on 15 December 2022.
Conflicts of Interest
The authors declare no conflict of interest. Patent applications covering the technologies described in this work have been applied for on behalf of the Arizona Board of Regents.
Funding Statement
This research was funded by US National Institutes of Health, grant number 1 R15 CA139364-01A2 and 1R15CA249617-01 to C.E.W., P.W.J. and P.A.M., by R01 HL128447 to J.S.W., and by the Siteman Investment Program to J.S.W.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Evans R.M., Mangelsdorf D.J. Nuclear Receptors, RXR, and the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangelsdorf D.J., Umesono K., Evans R.M. The Retinoids. Academic Press; Orlando, FL, USA: 1994. pp. 319–349. [Google Scholar]
- 3.Leid M., Kastner P., Chambon P. Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem. Sci. 1992;17:427–433. doi: 10.1016/0968-0004(92)90014-Z. [DOI] [PubMed] [Google Scholar]
- 4.Olefsky J.M. Nuclear Receptor Minireview Series. J. Biol. Chem. 2001;276:36863–36864. doi: 10.1074/jbc.R100047200. [DOI] [PubMed] [Google Scholar]
- 5.Perlmann T., Rangarajan P.N., Umesono K., Evans R.M. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- 6.Phan T.Q., Jow M.M., Privalsky M.L. DNA recognition by thyroid hormone and retinoic acid receptors: 3,4,5 rule modified. Mol. Cell. Endocrinol. 2010;319:88–98. doi: 10.1016/j.mce.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forman B.M., Yang C.-R., Au M., Casanova J., Ghysdael J., Samuels H.H. A Domain Containing Leucine-Zipper-Like Motifs Mediate Novel in Vivo Interactions between the Thyroid Hormone and Retinoic Acid Receptors. Mol. Endocrinol. 1989;3:1610–1626. doi: 10.1210/mend-3-10-1610. [DOI] [PubMed] [Google Scholar]
- 8.Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X.-K., Lehmann J., Hoffmann B., Dawson M.I., Cameron J., Graupner G., Hermann T., Tran P., Pfahl M. Homodimer formation of retinoid X receptor induced by 9-cis retinoic acid. Nature. 1992;358:587–591. doi: 10.1038/358587a0. [DOI] [PubMed] [Google Scholar]
- 10.Thompson P.D., Remus L.S., Hsieh J.C., Jurutka P.W., Whitfield G.K., Galligan M.A., Dominguez C.E., Haussler C.A., Haussler M.R. Distinct retinoid X receptor activation function-2 residues mediate transactivation in homodimeric and vitamin D receptor heterodimeric contexts. J. Mol. Endocrinol. 2001;27:211–227. doi: 10.1677/jme.0.0270211. [DOI] [PubMed] [Google Scholar]
- 11.Svensson S., Östberg T., Jacobsson M., Norström C., Stefansson K., Hallén D., Johansson I.C., Zachrisson K., Ogg D., Jendeberg L. Crystal structure of the heterodimeric complex of LXRα and RXRβ ligand-binding domains in a fully agonistic conformation. EMBO J. 2003;22:4625–4633. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nahoum V., Pérez E., Germain P., Rodríguez-Barrios F., Manzo F., Kammerer S., Lemaire G., Hirsch O., Royer C.A., Gronemeyer H., et al. Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function. Proc. Natl. Acad. Sci. USA. 2007;104:17323–17328. doi: 10.1073/pnas.0705356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forman B.M., Umesono K., Chen J., Evans R.M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 14.Lala D.S., Mukherjee R., Schulman I.G., Koch S.S.C., Dardashti L.J., Nadzan A.M., Croston G.E., Evans R.M., Heyman R.A. Activation of specific RXR heterodimers by an antagonist of RXR homodimers. Nature. 1996;383:450–453. doi: 10.1038/383450a0. [DOI] [PubMed] [Google Scholar]
- 15.Lemon B.D., Freedman L.P. Selective effects of ligands on vitamin D3 receptor- and retinoid X receptor-mediated gene activation in vivo. Mol. Cell. Biol. 1996;16:1006–1016. doi: 10.1128/MCB.16.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald P.N., Dowd D.R., Nakajima S., Galligan M.A., Reeder M.C., Haussler C.A., Ozato K., Haussler M.R. Retinoid X receptors stimulate and 9-cis retinoic acid inhibits 1,25-dihydroxyvitamin D3-activated expression of the rat osteocalcin gene. Mol. Cell. Biol. 1993;13:5907–5917. doi: 10.1128/MCB.13.9.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson P.D., Jurutka P.W., Haussler C.A., Whitfield G.K., Haussler M.R. Heterodimeric DNA Binding by the Vitamin D Receptor and Retinoid X Receptors Is Enhanced by 1,25-Dihydroxyvitamin D3 and Inhibited by 9-cis-Retinoic Acid: Evidence for Allosteric Receptor Interactions. J. Biol. Chem. 1998;273:8483–8491. doi: 10.1074/jbc.273.14.8483. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann J.M., Zhang X.K., Graupner G., Lee M.O., Hermann T., Hoffmann B., Pfahl M. Formation of retinoid X receptor homodimers leads to repression of T3 response: Hormonal cross talk by ligand-induced squelching. Mol. Cell. Biol. 1993;13:7698–7707. doi: 10.1128/MCB.13.12.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yen P.M., Liu Y., Sugawara A., Chin W.W. Vitamin D receptors repress basal transcription and exert dominant negative activity on triiodothyronine-mediated transcriptional activity. J. Biol. Chem. 1996;271:10910–10916. doi: 10.1074/jbc.271.18.10910. [DOI] [PubMed] [Google Scholar]
- 20.Raval-Pandya M., Freedman L.P., Li H., Christakos S. Thyroid hormone receptor does not heterodimerize with the vitamin D receptor but represses vitamin D receptor-mediated transactivation. Mol. Endocrinol. 1998;12:1367–1379. doi: 10.1210/mend.12.9.0165. [DOI] [PubMed] [Google Scholar]
- 21.Thompson P.D., Hsieh J.C., Whitfield G.K., Haussler C.A., Jurutka P.W., Galligan M.A., Tillman J.B., Spindler S.R., Haussler M.R. Vitamin D receptor displays DNA binding and transactivation as a heterodimer with the retinoid X receptor, but not with the thyroid hormone receptor. J. Cell. Biochem. 1999;75:462–480. doi: 10.1002/(SICI)1097-4644(19991201)75:3<462::AID-JCB11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 22.Altucci L., Leibowitz M.D., Ogilvie K.M., de Lera A.R., Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat. Rev. Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann J.M., Jong L., Fanjul A., Cameron J.F., Lu X.P., Haefner P., Dawson M.I., Pfahl M. Retinoids selective for retinoid X receptor response pathways. Science. 1992;258:1944–1946. doi: 10.1126/science.1335166. [DOI] [PubMed] [Google Scholar]
- 24.Jong L., Lehmann J.M., Hobbs P.D., Harlev E., Huffman J.C., Pfahl M., Dawson M.I. Conformational effects on retinoid receptor selectivity. 1. Effect of 9-double bond geometry on retinoid X receptor activity. J. Med. Chem. 1993;36:2605–2613. doi: 10.1021/jm00070a003. [DOI] [PubMed] [Google Scholar]
- 25.Dawson M.I., Jong L., Hobbs P.D., Cameron J.F., Chao W.-R., Pfahl M., Lee M.-O., Shroot B., Pfahl M. Conformational Effects on Retinoid Receptor Selectivity. 2. Effects of Retinoid Bridging Group on Retinoid X Receptor Activity and Selectivity. J. Med. Chem. 1995;38:3368–3383. doi: 10.1021/jm00017a021. [DOI] [PubMed] [Google Scholar]
- 26.Boehm M.F., Zhang L., Badea B.A., White S.K., Mais D.E., Berger E., Suto C.M., Goldman M.E., Heyman R.A. Synthesis and Structure-Activity Relationships of Novel Retinoid X Receptor-Selective Retinoids. J. Med. Chem. 1994;37:2930–2941. doi: 10.1021/jm00044a014. [DOI] [PubMed] [Google Scholar]
- 27.Daiss J.O., Burschka C., Mills J.S., Montana J.G., Showell G.A., Fleming I., Gaudon C., Ivanova D., Gronemeyer H., Tacke R. Synthesis, Crystal Structure Analysis, and Pharmacological Characterization of Disila-bexarotene, a Disila-Analogue of the RXR-Selective Retinoid Agonist Bexarotene. Organometallics. 2005;24:3192–3199. doi: 10.1021/om040143k. [DOI] [Google Scholar]
- 28.Zhang D., Leal A.S., Carapellucci S., Shahani P.H., Bhogal J.S., Ibrahim S., Raban S., Jurutka P.W., Marshall P.A., Sporn M.B., et al. Testing Novel Pyrimidinyl Rexinoids: A New Paradigm for Evaluating Rexinoids for Cancer Prevention. Cancer Prev. Res. 2019;12:211–224. doi: 10.1158/1940-6207.CAPR-18-0317. [DOI] [PubMed] [Google Scholar]
- 29.Yen W.-C., Prudente R.Y., Lamph W.W. Synergistic effect of a retinoid X receptor-selective ligand bexarotene (LGD1069, Targretin) and paclitaxel (Taxol) in mammary carcinoma. Breast Cancer Res. Treat. 2004;88:141–148. doi: 10.1007/s10549-004-1426-5. [DOI] [PubMed] [Google Scholar]
- 30.Cesario R.M., Stone J., Yen W.-C., Bissonnette R.P., Lamph W.W. Differentiation and growth inhibition mediated via the RXR:PPARγ heterodimer in colon cancer. Cancer Lett. 2006;240:225–233. doi: 10.1016/j.canlet.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Yen W.-C., Corpuz M.R., Prudente R.Y., Cooke T.A., Bissonnette R.P., Negro-Vilar A., Lamph W.W. A Selective Retinoid X Receptor Agonist Bexarotene (Targretin) Prevents and Overcomes Acquired Paclitaxel (Taxol) Resistance in Human Non–Small Cell Lung Cancer. Clin. Cancer Res. 2004;10:8656–8664. doi: 10.1158/1078-0432.CCR-04-0979. [DOI] [PubMed] [Google Scholar]
- 32.Dragnev K.H., Petty W.J., Shah S.J., Lewis L.D., Black C.C., Memoli V., Nugent W.C., Hermann T., Negro-Vilar A., Rigas J.R., et al. A Proof-of-Principle Clinical Trial of Bexarotene in Patients with Non–Small Cell Lung Cancer. Clin. Cancer Res. 2007;13:1794–1800. doi: 10.1158/1078-0432.CCR-06-1836. [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee R., Davies P.J.A., Crombie D.L., Bischoff E.D., Cesario R.M., Jow L., Hamann L.G., Boehm M.F., Mondon C.E., Nadzan A.M., et al. Sensitization of diabetic and obese mice to insulin by retinoid X receptor agonists. Nature. 1997;386:407–410. doi: 10.1038/386407a0. [DOI] [PubMed] [Google Scholar]
- 34.Sherman S.I., Gopal J., Haugen B.R., Chiu A.C., Whaley K., Nowlakha P., Duvic M. Central Hypothyroidism Associated with Retinoid X Receptor–Selective Ligands. N. Engl. J. Med. 1999;340:1075–1079. doi: 10.1056/NEJM199904083401404. [DOI] [PubMed] [Google Scholar]
- 35.Li D., Li T., Wang F., Tian H., Samuels H.H. Functional Evidence for Retinoid X Receptor (RXR) as a Nonsilent Partner in the Thyroid Hormone Receptor/RXR Heterodimer. Mol. Cell. Biol. 2002;22:5782–5792. doi: 10.1128/MCB.22.16.5782-5792.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Field F.J., Born E., Mathur S.N. LXR/RXR ligand activation enhances basolateral efflux of beta-sitosterol in CaCo-2 cells. J. Lipid Res. 2004;45:905–913. doi: 10.1194/jlr.M300473-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Murthy S., Born E., Mathur S.N., Field F.J. LXR/RXR activation enhances basolateral efflux of cholesterol in CaCo-2 cells. J. Lipid Res. 2002;43:1054–1064. doi: 10.1194/jlr.M100358-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Thacher S.M., Standeven A.M., Athanikar J., Kopper S., Castilleja O., Escobar M., Beard R.L., Chandraratna R.A.S. Receptor Specificity of Retinoid-Induced Epidermal Hyperplasia: Effect of RXR-Selective Agonists and Correlation with Topical Irritation. J. Pharmacol. Exp. Ther. 1997;282:528–534. [PubMed] [Google Scholar]
- 39.Cramer P.E., Cirrito J.R., Wesson D.W., Lee C.Y.D., Karlo J.C., Zinn A.E., Casali B.T., Restivo J.L., Goebel W.D., James M.J., et al. ApoE-Directed Therapeutics Rapidly Clear β-Amyloid and Reverse Deficits in AD Mouse Models. Science. 2012;335:1503. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFarland K., Spalding T.A., Hubbard D., Ma J.-N., Olsson R., Burstein E.S. Low Dose Bexarotene Treatment Rescues Dopamine Neurons and Restores Behavioral Function in Models of Parkinson’s Disease. ACS Chem. Neurosci. 2013;4:1430–1438. doi: 10.1021/cn400100f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cummings J.L., Zhong K., Kinney J.W., Heaney C., Moll-Tudla J., Joshi A., Pontecorvo M., Devous M., Tang A., Bena J. Double-blind, placebo-controlled, proof-of-concept trial of bexarotene Xin moderate Alzheimer’s disease. Alzheimers Res. Ther. 2016;8:4. doi: 10.1186/s13195-016-0173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabbinavar F.F., Zomorodian N., Rettig M., Khan F., Greenwald D.R., Davidson S.J., DiCarlo B.A., Patel R., Pandit L., Chandraratna R., et al. An open-label phase II clinical trial of the RXR agonist IRX4204 in taxane-resistant, castration-resistant metastatic prostate cancer (CRPC) J. Clin. Oncol. 2014;32:5073. doi: 10.1200/jco.2014.32.15_suppl.5073. [DOI] [Google Scholar]
- 43.Vuligonda V., Thacher S.M., Chandraratna R.A. Enantioselective syntheses of potent retinoid X receptor ligands: Differential biological activities of individual antipodes. J. Med. Chem. 2001;44:2298–2303. doi: 10.1021/jm0100584. [DOI] [PubMed] [Google Scholar]
- 44.Muccio D.D., Brouillette W.J., Breitman T.R., Taimi M., Emanuel P.D., Zhang X.-k., Chen G.-q., Sani B.P., Venepally P., Reddy L., et al. Conformationally Defined Retinoic Acid Analogues. 4. Potential New Agents for Acute Promyelocytic and Juvenile Myelomonocytic Leukemias. J. Med. Chem. 1998;41:1679–1687. doi: 10.1021/jm970635h. [DOI] [PubMed] [Google Scholar]
- 45.Atigadda V.R., Vines K.K., Grubbs C.J., Hill D.L., Beenken S.L., Bland K.I., Brouillette W.J., Muccio D.D. Conformationally Defined Retinoic Acid Analogues. 5. Large-Scale Synthesis and Mammary Cancer Chemopreventive Activity for (2E,4E,6Z,8E)-8-(3‘,4‘-Dihydro-1‘(2‘H)-naphthalen-1‘-ylidene)-3,7-dimethyl-2,4,6-octatrienoic Acid (9cUAB30) J. Med. Chem. 2003;46:3766–3769. doi: 10.1021/jm030095q. [DOI] [PubMed] [Google Scholar]
- 46.Kolesar J.M., Hoel R., Pomplun M., Havighurst T., Stublaski J., Wollmer B., Krontiras H., Brouillette W., Muccio D., Kim K., et al. A Pilot, First-in-Human, Pharmacokinetic Study of 9cUAB30 in Healthy Volunteers. Cancer Prev. Res. 2010;3:1565–1570. doi: 10.1158/1940-6207.CAPR-10-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen N.J., Wylie R.C., Phipps S.M., Love W.K., Andrews L.G., Tollefsbol T.O. The low-toxicity 9-cis UAB30 novel retinoid down-regulates the DNA methyltransferases and has anti-telomerase activity in human breast cancer cells. Int. J. Oncol. 2007;30:641–650. doi: 10.3892/ijo.30.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atigadda V.R., Xia G., Desphande A., Boerma L.J., Lobo-Ruppert S., Grubbs C.J., Smith C.D., Brouillette W.J., Muccio D.D. Methyl substitution of a rexinoid agonist improves potency and reveals site of lipid toxicity. J. Med. Chem. 2014;57:5370–5380. doi: 10.1021/jm5004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desphande A., Xia G., Boerma L.J., Vines K.K., Atigadda V.R., Lobo-Ruppert S., Grubbs C.J., Moeinpour F.L., Smith C.D., Christov K., et al. Methyl-substituted conformationally constrained rexinoid agonists for the retinoid X receptors demonstrate improved efficacy for cancer therapy and prevention. Bioorganic Med. Chem. 2014;22:178–185. doi: 10.1016/j.bmc.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michellys P.Y., Ardecky R.J., Chen J.H., Crombie D.L., Etgen G.J., Faul M.M., Faulkner A.L., Grese T.A., Heyman R.A., Karanewsky D.S., et al. Novel (2E,4E,6Z)-7-(2-alkoxy-3,5-dialkylbenzene)-3-methylocta-2,4,6-trienoic acid retinoid X receptor modulators are active in models of type 2 diabetes. J. Med. Chem. 2003;46:2683–2696. doi: 10.1021/jm020340q. [DOI] [PubMed] [Google Scholar]
- 51.Michellys P.Y., Ardecky R.J., Chen J.H., D’Arrigo J., Grese T.A., Karanewsky D.S., Leibowitz M.D., Liu S., Mais D.A., Mapes C.M., et al. Design, synthesis, and structure-activity relationship studies of novel 6,7-locked-[7-(2-alkoxy-3,5-dialkylbenzene)-3-methylocta]-2,4,6-trienoic acids. J. Med. Chem. 2003;46:4087–4103. doi: 10.1021/jm020401k. [DOI] [PubMed] [Google Scholar]
- 52.Michellys P.Y., D’Arrigo J., Grese T.A., Karanewsky D.S., Leibowitz M.D., Mais D.A., Mapes C.M., Reifel-Miller A., Rungta D., Boehm M.F. Design, synthesis and structure-activity relationship of novel RXR-selective modulators. Bioorganic Med. Chem. Lett. 2004;14:1593–1598. doi: 10.1016/j.bmcl.2003.12.089. [DOI] [PubMed] [Google Scholar]
- 53.Wagner C.E., Jurutka P.W., Marshall P.A., Groy T.L., van der Vaart A., Ziller J.W., Furmick J.K., Graeber M.E., Matro E., Miguel B.V., et al. Modeling, Synthesis and Biological Evaluation of Potential Retinoid X Receptor (RXR) Selective Agonists: Novel Analogues of 4-[1-(3,5,5,8,8-Pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic Acid (Bexarotene) J. Med. Chem. 2009;52:5950–5966. doi: 10.1021/jm900496b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furmick J.K., Kaneko I., Walsh A.N., Yang J., Bhogal J.S., Gray G.M., Baso J.C., Browder D.O., Prentice J.L.S., Montano L.A., et al. Modeling, Synthesis and Biological Evaluation of Potential Retinoid X Receptor-Selective Agonists: Novel Halogenated Analogues of 4-[1-(3,5,5,8,8-Pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic Acid (Bexarotene) ChemMedChem. 2012;7:1551–1566. doi: 10.1002/cmdc.201290042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boehm M.F., Zhang L., Zhi L., McClurg M.R., Berger E., Wagoner M., Mais D.E., Suto C.M., Davies P.J.A., Heyman R.A., et al. Design and Synthesis of Potent Retinoid X Receptor Selective Ligands That Induce Apoptosis in Leukemia Cells. J. Med. Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- 56.Jurutka P.W., Kaneko I., Yang J., Bhogal J.S., Swierski J.C., Tabacaru C.R., Montano L.A., Huynh C.C., Jama R.A., Mahelona R.D., et al. Modeling, Synthesis, and Biological Evaluation of Potential Retinoid X Receptor (RXR) Selective Agonists: Novel Analogues of 4-[1-(3,5,5,8,8-Pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic Acid (Bexarotene) and (E)-3-(3-(1,2,3,4-tetrahydro-1,1,4,4,6-pentamethylnaphthalen-7-yl)-4-hydroxyphenyl)acrylic Acid (CD3254) J. Med. Chem. 2013;56:8432–8454. doi: 10.1021/jm4008517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liby K., Rendi M., Suh N., Royce D.B., Risingsong R., Williams C.R., Lamph W., Labrie F., Krajewski S., Xu X., et al. The Combination of the Rexinoid, LG100268, and a Selective Estrogen Receptor Modulator, Either Arzoxifene or Acolbifene, Synergizes in the Prevention and Treatment of Mammary Tumors in an Estrogen Receptor–Negative Model of Breast Cancer. Clin. Cancer Res. 2006;12:5902–5909. doi: 10.1158/1078-0432.CCR-06-1119. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L., Badea B.A., Enyeart D., Berger E.M., Mais D.E., Boehm M.F. Syntheses of isotopically labeled 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethenyl]benzoic acid (LGD1069), a potent retinoid x receptor-selective ligand. J. Label. Comp. Radiopharm. 1995;36:701–712. doi: 10.1002/jlcr.2580360712. [DOI] [Google Scholar]
- 59.Faul M.M., Ratz A.M., Sullivan K.A., Trankle W.G., Winneroski L.L. Synthesis of Novel Retinoid X Receptor-Selective Retinoids. J. Org. Chem. 2001;66:5772–5782. doi: 10.1021/jo0103064. [DOI] [PubMed] [Google Scholar]
- 60.Marshall P.A., Jurutka P.W., Wagner C.E., van der Vaart A., Kaneko I., Chavez P.I., Ma N., Bhogal J.S., Shahani P., Swierski J.C., et al. Analysis of differential secondary effects of novel rexinoids: Select rexinoid X receptor ligands demonstrate differentiated side effect profiles. Pharm. Res. Perspect. 2015;3:e00122. doi: 10.1002/prp2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santín E.P., Germain P., Quillard F., Khanwalkar H., Rodríguez-Barrios F., Gronemeyer H., de Lera Á.R., Bourguet W. Modulating Retinoid X Receptor with a Series of (E)-3-[4-Hydroxy-3-(3-alkoxy-5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)phenyl]acrylic Acids and Their 4-Alkoxy Isomers. J. Med. Chem. 2009;52:3150–3158. doi: 10.1021/jm900096q. [DOI] [PubMed] [Google Scholar]
- 62.Gianni M., Ponzanelli I., Mologni L., Reichert U., Rambaldi A., Terao M., Garattini E. Retinoid-dependent growth inhibition, differentiation and apoptosis in acute promyelocytic leukemia cells. Expression and activation of caspases. Cell Death Differ. 2000;7:447–460. doi: 10.1038/sj.cdd.4400673. [DOI] [PubMed] [Google Scholar]
- 63.Fujii S., Ohsawa F., Yamada S., Shinozaki R., Fukai R., Makishima M., Enomoto S., Tai A., Kakuta H. Modification at the acidic domain of RXR agonists has little effect on permissive RXR-heterodimer activation. Bioorg. Med. Chem. Lett. 2010;20:5139–5142. doi: 10.1016/j.bmcl.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 64.Ohsawa F., Morishita K.-I., Yamada S., Makishima M., Kakuta H. Modification at the Lipophilic Domain of RXR Agonists Differentially Influences Activation of RXR Heterodimers. ACS Med. Chem. Lett. 2010;1:521–525. doi: 10.1021/ml100184k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kakuta H., Yakushiji N., Shinozaki R., Ohsawa F., Yamada S., Ohta Y., Kawata K., Nakayama M., Hagaya M., Fujiwara C., et al. RXR Partial Agonist CBt-PMN Exerts Therapeutic Effects on Type 2 Diabetes without the Side Effects of RXR Full Agonists. ACS Med. Chem. Lett. 2012;3:427–432. doi: 10.1021/ml300055n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohsawa F., Yamada S., Yakushiji N., Shinozaki R., Nakayama M., Kawata K., Hagaya M., Kobayashi T., Kohara K., Furusawa Y., et al. Mechanism of Retinoid X Receptor Partial Agonistic Action of 1-(3,5,5,8,8-Pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)-1H-benzotriazole-5-carboxylic Acid and Structural Development To Increase Potency. J. Med. Chem. 2013;56:1865–1877. doi: 10.1021/jm400033f. [DOI] [PubMed] [Google Scholar]
- 67.Kagechika H., Koichi S., Sugioka T., Sotome T., Nakayama Y., Doi K. Retinoid Activity Regulators. WO9845242A1. European Patent. 1998 October 15;
- 68.Ohta K., Tsuji M., Kawachi E., Fukasawa H., Hashimoto Y., Shudo K., Kagechika H. Potent Retinoid Synergists with a Diphenylamine Skeleton. Biol. Pharm. Bull. 1998;21:544–546. doi: 10.1248/bpb.21.544. [DOI] [PubMed] [Google Scholar]
- 69.Ohta K., Kawachi E., Fukasawa H., Shudo K., Kagechika H. Diphenylamine-based retinoid antagonists: Regulation of RAR and RXR function depending on the N-substituent. Bioorg. Med. Chem. 2011;19:2501–2507. doi: 10.1016/j.bmc.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 70.Heck M.C., Wagner C.E., Shahani P.H., MacNeill M., Grozic A., Darwaiz T., Shimabuku M., Deans D.G., Robinson N.M., Salama S.H., et al. Modeling, Synthesis, and Biological Evaluation of Potential Retinoid X Receptor (RXR)-Selective Agonists: Analogues of 4-[1-(3,5,5,8,8-Pentamethyl-5,6,7,8-tetrahydro-2-naphthyl)ethynyl]benzoic Acid (Bexarotene) and 6-(Ethyl(5,5,8,8-tetrahydronaphthalen-2-yl)amino)nicotinic Acid (NEt-TMN) J. Med. Chem. 2016;59:8924–8940. doi: 10.1021/acs.jmedchem.6b00812. [DOI] [PubMed] [Google Scholar]
- 71.Ohta K., Kawachi E., Inoue N., Fukasawa H., Hashimoto Y., Itai A., Kagechika H. Retinoidal pyrimidinecarboxylic acids. Unexpected diaza-substituent effects in retinobenzoic acids. Chem. Pharm. Bull. 2000;48:1504–1513. doi: 10.1248/cpb.48.1504. [DOI] [PubMed] [Google Scholar]
- 72.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stierand K., Rarey M. Drawing the PDB: Protein−Ligand Complexes in Two Dimensions. ACS Med. Chem. Lett. 2010;1:540–545. doi: 10.1021/ml100164p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stierand K., Rarey M. From Modeling to Medicinal Chemistry: Automatic Generation of Two-Dimensional Complex Diagrams. ChemMedChem. 2007;2:853–860. doi: 10.1002/cmdc.200700010. [DOI] [PubMed] [Google Scholar]
- 75.Tachdjian C., Guo J., Boudjelal M., Al-Shamma H.A., Giachino A.F., Jakubowicz-Jaillardon K., Chen Q., Zapf J.W., Pfahl M. Preparation of Substituted Isochroman Compounds for the Treatment of Metabolic Disorders, Cancer and Other Diseases. CN1774246A. Patent. 2004 May 17;
- 76.Jurutka P.W., di Martino O., Reshi S., Mallick S., Sabir Z.L., Staniszewski L.J.P., Warda A., Maiorella E.L., Minasian A., Davidson J., et al. Modeling, Synthesis, and Biological Evaluation of Potential Retinoid-X-Receptor (RXR) Selective Agonists: Analogs of 4-[1-(3,5,5,8,8-Pentamethyl-5,6,7,8-tetrahyro-2-naphthyl)ethynyl]benzoic Acid (Bexarotene) and 6-(Ethyl(4-isobutoxy-3-isopropylphenyl)amino)nicotinic Acid (NEt-4IB) Int. J. Mol. Sci. 2021;22:1237. doi: 10.3390/ijms222212371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Egea P.F., Mitschler A., Rochel N., Ruff M., Chambon P., Moras D. Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harris R., Olson A.J., Goodsell D.S. Automated prediction of ligand-binding sites in proteins. Proteins Struct. Funct. Bioinf. 2008;70:1506–1517. doi: 10.1002/prot.21645. [DOI] [PubMed] [Google Scholar]
- 79.Di Martino O., Niu H., Hadwiger G., Kuusanmaki H., Ferris M.A., Vu A., Beales J., Wagner C., Menéndez-Gutiérrez M.P., Ricote M., et al. Endogenous and combination retinoids are active in myelomonocytic leukemias. Haematologica. 2021;106:1008–1021. doi: 10.3324/haematol.2020.264432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Di Martino O., Ferris M.A., Hadwiger G., Sarkar S., Vu A., Menéndez-Gutiérrez M.P., Ricote M., Welch J.S. RXRA DT448/9PP generates a dominant active variant capable of inducing maturation in acute myeloid leukemia cells. Haematologica. 2021;107:417–426. doi: 10.3324/haematol.2021.278603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilson A.J.C., Hahn T., Wilson A.J.C., Shmueli U., Crystallography I.U.O. International Tables for Crystallography, Volume C: Mathematical, Physical and Chemical Tables. Springer; Dordrecht, The Netherland: 1992. [Google Scholar]
- 82.Parsons S., Flack H.D., Wagner T. Use of intensity quotients and differences in absolute structure refinement. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2013;69:249–259. doi: 10.1107/S2052519213010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CCDC numbers 2191083 and 2191084 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif, accessed on 15 December 2022.