Abstract
Background
Understanding biological differences between different racial groups of human papillomavirus (HPV)‐associated oropharyngeal squamous cell carcinoma (OPSCC) patients, who have differences in terms of incidence, survival, and tumor morphology, can facilitate accurate prognostic biomarkers, which can help develop personalized treatment strategies.
Methods
This study evaluated whether there were morphologic differences between HPV‐associated tumors from Black and White patients in terms of multinucleation index (MuNI), an image analysis‐derived metric that measures density of multinucleated tumor cells within epithelial regions on hematoxylin–eosin images and previously has been prognostic in HPV‐associated OPSCC patients. In this study, the authors specifically evaluated whether the same MuNI cutoff that was prognostic of overall survival (OS) and disease‐free survival in their previous study, TTR, is valid for Black and White patients, separately. We also evaluated population‐specific cutoffs, TB for Blacks and TW for Whites, for risk stratification.
Results
MuNI was statistically significantly different between Black (mean, 3.88e–4; median, 3.67e–04) and White patients (mean, 3.36e–04; median, 2.99e–04), with p = .0078. Using TTR, MuNI was prognostic of OS in the entire population with hazard ratio (HR) of 1.71 (p = .002; 95% confidence interval [CI], 1.21–2.43) and in White patients with HR of 1.72 (p = .005; 95% CI, 1.18–2.51). Population‐specific cutoff, TW, yielded improved HR of 1.77 (p = .003; 95% CI, 1.21–2.58) for White patients, whereas TB did not improve risk‐stratification in Black patients with HR of 0.6 (p = .3; HR, 0.6; 95% CI, 0.2–1.80).
Conclusions
Histological difference between White and Black patient tumors in terms of multinucleated tumor cells suggests the need for considering population‐specific prognostic biomarkers for personalized risk stratification strategies for HPV‐associated OPSCC patients.
Keywords: Black, HPV, image analysis, MuNI, OPSCC, White
Short abstract
There are substantial differences between different racial groups of human papillomavirus‐related oropharyngeal squamous cell carcinoma (OPSCC) patients, in terms of survival and tumor morphology. In this study, the authors demonstrated a histological difference between tumors from White and Black OPSCC patients using a computational pathology‐based metric called the multinucleation index.
INTRODUCTION
The rising incidence of oropharyngeal squamous cell carcinoma (OPSCC) in the United States has been attributed to human papillomavirus (HPV) infection, with approximately 70% of OPSCCs being HPV‐associated. 1 , 2 The American Joint Committee on Cancer (AJCC) tumor‐node‐metastasis (TNM) staging system remains the standard for clinicians to determine prognosis and to facilitate choice of treatment. It has evolved extensively over the past 50 years and now, with the eighth edition, there are separate TNM staging systems for HPV‐associated and HPV‐independent OPSCC patients. 3 , 4 HPV‐associated OPSCC patients consistently show overexpression of p16 by immunohistochemistry with this surrogate marker becoming the recommended test for all new patients with this cancer type. 5 p16 immunohistochemistry is widely used as a surrogate marker for transcriptionally active HPV for many reasons, including the challenge of performing HPV‐specific testing in large numbers across multiple institutions. p16 is very sensitive for high‐risk HPV and has moderate specificity with a small subset of patients who have p16‐positive, but HPV‐negative, tumors. 6 Along with p16 status, the latest AJCC staging system uses tumor, nodes, and metastases based on the clinical and pathological examination to risk stratify patients. Some classic tumor parameters, such as tumor grade and lymphocytic response, are not prognostic by visual (human) review and have failed to serve as effective prognosticators despite years of attempted application. 7 , 8 One reason for such failures could be the inability of capturing tissue morphology and architecture within the tumor microenvironment (TME), which carries rich prognostic information across myriad different cancer types. 9 Thus, there is a need for accurate, quantitative, prognostic histologic biomarkers to improve patient prognostication and stratification for various treatments.
In several recent studies, population‐specific differences have been examined in terms of several aspects, such as incidence, survival, and tumor morphology in various carcinomas (e.g., head and neck, 10 , 11 , 12 , 13 prostate, 14 breast, 15 , 16 and lung 17 among others). Among head and neck squamous cell carcinoma (HNSCC) patients, White patients greatly outnumber Black patients. 3 , 4 , 18 Studies have shown that a greater percent of White patients have HPV‐associated OPSCC and better overall survival (OS) as compared to Black patients. 19 , 20 , 21 , 22 Among the multiple reasons, one reason might be that the cells in the HPV‐associated tumors had higher CpG methylation, known to turn off gene expression and associated with cancer progression, leading to better response to chemoradiation. 23 , 24 , 25 , 26 , 27 Data from the Surveillance, Epidemiology, and End Results program shows that survival rates for OPSCC are much lower in Black patients. Five‐year survival for Black patients was found to be 39.5% as compared 59% among Whites. 28 Although the Black population tended to present with advanced tumor stage and to have poorer socioeconomic status, O'Neill et al. 11 found that, on average, Black patients have significantly shorter OS than White patients, even when these two groups have similar access to medical care. Although these studies have examined population‐specific differences in survival and tumor biology of patients with OPSCC, 28 , 29 , 30 , 31 whether these findings translate to specifically HPV‐associated OPSCC is still unknown. 32 , 33
Other differences have been found in Black and White HNSCC patients with regard to tumor lymphocytic infiltration and mutational profiles. In a study involving HNSCC patients, inverse correlation was found between lymphocyte infiltration in hematoxylin–eosin (H & E)‐stained tumor tissue sections, local recurrence, and survival; tumor lymphocytic response was also considered an independent predictor of race. 29 White patients were six to seven times less likely to have a host lymphocytic response in comparison to Black patients. 29 The study also revealed that the same groups, Black and White patients, have different tumor mutational profiles, suggesting that differences in survival are inherent to the tumors themselves. For this reason, one expects there to be morphologic differences in TME. 34
For HPV‐associated OPSCC, studies have focused mainly on single‐ or multigene prognostic signatures to be used as biomarkers. 13 Decrease in STAT3 expression and increase in AP1 and nuclear factor–κB have shown to be prognostic for HPV‐associated OPSCC, 13 and analysis of similar expression profiles have led to identification of various gene signatures like two‐gene signature 35 and six‐gene signature 36 among others. However, until now, none of these signatures have been validated as a practical predictor of OS in multiple cohorts, a critical prerequisite step toward translating the signature into a clinical test. 13 Morphometry and architecture of the tissues within TME mirror tumor characteristics and these may be used as potential biomarkers for prediction and prognosis. 13 Lewis et al. previously visually identified tumor cell multinucleation (MN), defined as tumor cells having three or more nuclei in the TME, and found an association with poor survival in HPV‐associated OPSCC patients. 32 However, the recognition and quantification of these morphologic features is time‐intensive and requires human interpretation, leading to interobserver variability and bias. 32
Previously, we presented a computational pathology‐based metric called the multinucleation index (MuNI) to quantify MN density in epithelial (EP) regions. MuNI was validated as being prognostic of OS for 1094 HPV‐associated OPSCC patients. 37 In the present study, we have extended this approach to explore survival outcomes in 744 HPV‐associated OPSCC patients across different racial categories (Black vs. White) collected from five independent institutions. MuNI was used to find differences in Black and White populations for the entire study set, as well as for the individual staging groups, as defined by AJCC's 8th edition.
MATERIALS AND METHODS
Patient selection
A total of 1047 OPSCC patients from five independent institutions were identified retrospectively within the period 2009–2020 for the study. If the patient was treated with primary surgery, this slide was from the resection specimen or, if treated with primary (chemo)radiation, then the best representative biopsy slides were selected. OPSCC diagnoses were confirmed by collaborating study pathologists at each of the respective institutions. HPV positivity is determined by p16 immunohistochemistry, a surrogate marker of high‐risk HPV, according to the AJCC guideline. All cases immunoreactive for p16 were read using a ≥ 70% nuclear and cytoplasmic block‐like reaction cutoff as recommended by the College of American Pathologists guidelines and confirmed to be positive by local review. 38
Data set preparation
Patients self‐identified as a certain race as recorded in electronic medical records. The patients who self‐identified as neither Black nor White were excluded from this study. We removed patients with missing follow‐up data or insufficient whole slide image (WSI) quality. Quality was determined by HistoQC, 39 a quality control tool automatically detecting defects on pathology images such as image blurriness, air bubbles, and tissue artifacts. After reviewing the inclusion/exclusion criteria, 744 of the patients were retained for the subsequent feature analysis from Washington University in St. Louis (D1, Black = 3, White = 103), John Hopkins University (D2, Black = 6, White = 114), Cleveland Clinic (D3, Black = 19, White = 299), UH Seidman Cancer Center (D4, Black = 16, White = 17), and Southern California Permanente Medical Group (D5, Black = 10, White = 157). Each patient had a representative H & E slide chosen by the pathologist from the respective institution from the primary oropharyngeal tumor. The representative slides were defined as the one that contained the largest amount of tumor regions. H & E WSIs were acquired at ×40 resolution (0.25 μm/pixel resolution) using a Ventana iScan HT slide scanner. D5 was selected as the training set, and DTR was selected to maintain consistency with the original study. 37 In this manner, we sought to avoid introducing any potential data leakage during validation. DTR was used for cutoff definition and the combination of the remaining four data sets, D1–4, comprised the validation set, DVA. The Black and White patients in DVA comprised the sets, DB and DW, respectively.
Study design
Density of tumor MN events in EP regions (MuNI) was used to compare tumor morphology. MuNI was calculated from H & E WSIs by using two different deep learning models; MMN and MEP. Association of MuNI with survival outcome was measured by comparing OS of high MuNI and low MuNI groups. MuNI stratification was performed based on a cutoff, TTR, calculating the average MuNI value in DTR. Using TTR, association of MuNI with OS was also explored within DB and DW, separately. Additionally, we optimized and evaluated cutoffs for DB and DW separately, denoted as TB and TW, and they were calculated as the average MuNI values of their respective racial categories in DTR. Additionally, MuNI of DB and DW were compared within the various AJCC 8th edition individual stage groups. Model development and cutoff definition were performed on DTR, whereas DVA, DB, and DW were used for validation.
MuNI calculation
The overall schema for inferring tumor MN events is shown in Figure 1. Two deep learning models were used for MuNI calculation: (1) segmentation of epithelial regions, and (2) simultaneous segmentation of tumor MN events and other nuclei. After the models were built, the initial step of MuNI calculation is the extraction of tiles with a size of 2048 × 2048 pixels from tissue regions. The tiles were analyzed with the models to generate their corresponding masks. The total number of multinucleated and total epithelial cells was then counted across the tiles to calculate MuNI for each WSI.
FIGURE 1.
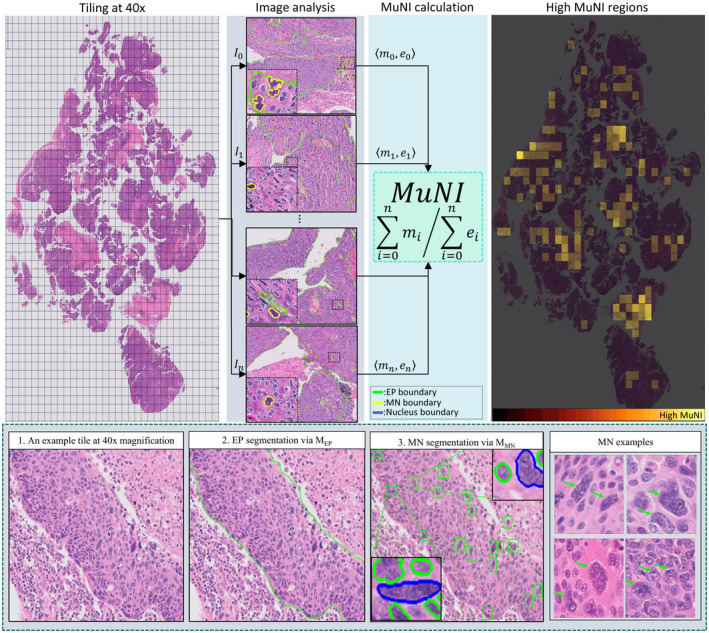
Overall schema of the multinucleation index (MuNI) calculation for a whole slide image (WSI). Epithelial regions and multinucleation (MN) events in every tile were detected using MEP and MMN. After that, the total numbers of epithelial (EP) and MN cells were counted, depicted as m i and e i , respectively, and their ratio was defined as MuNI for the WSI. The last column illustrates MuNI of every tile overlaid as a heatmap on the image. The second row depicts the overall schema of the MN detection from an example image tile. (2) Epithelial regions in the tile were segmented by MEP, shown by green boundaries. (3) MN cells were detected by MMN. Two example MNs automatically detected by MMN are zoomed in for better visualization. The MNs and epithelial cells are shown in blue and green colors, respectively. The last column of the second row illustrates high‐power fields of four images with multinucleated tumor cells, illustrated by green arrows.
Stroma versus epithelium segregation
An in–house‐developed deep learning model was employed for epithelium versus stroma segregator. 37 The model was built using tiles with a size of 512 × 512 pixels at 10× magnification. They were extracted from six WSIs from a publicly available breast cancer data set. 40 An enhanced deep learning model, MEP, based on conditional generative adversarial network were trained ab initio. Output of MEP was a binary map with the same size as the input images, where white and black pixels represented epithelium and other regions, respectively.
MN event detection
Our main collaborating pathologist (JSL) annotated 1002 tumor MN events in 12 WSIs of the training set by locating dots in their centers. The cell boundaries were then identified by using a pretrained nucleus segmentation model. 41 Once the boundaries were obtained, the nuclear segmentation model was fine‐tuned with the annotated tumor multinucleated cells, obtaining MMN, the MN cell segmentation model. To train the model, 256 × 256 images at 40× magnification were fed into the network. Output of MMN is a colored image where multinucleated cells, other segmented cells and background regions, were illustrated via three different colors.
MuNI definition
For calculating MuNI, only the MNs inside epithelial regions were considered, hence the MN figures detected outside epithelium were removed by making use of MEP. MuNI was calculated as the ratio of the number of MN to the total number of EP cells in the following equation, 37
where n is the number of tiles extracted from the WSI and and are the number of detected MNs and epithelial cells in tile i, by making use of MMN and MEP, respectively. To dichotomize the patients into low‐ and high‐risk categories, their MuNIs were thresholded by a cutoff that was determined as the average MuNI values in the training set.
Statistical analysis
Similarity of clinical and pathologic variables between different institutional cohorts was examined using analysis of variance test for continuous measures and χ 2 test for categorical factors (Table 1). MuNIs of the two populations, DW and DB, were compared using Mann–Whitney U test. Additionally, a slightly different approach for comparing the two groups was adopted to alleviate the effect of unbalanced sample sizes: MuNI of DB (i.e., 44 patients) was assessed, whereas for DCA, 50 patients were randomly selected. This step was repeated 200 times, and the histogram of the associated p values was reported. Three cutoffs used for risk stratification, namely TTR, TB, and TW, were calculated as average MuNI in DTR, average MuNI among Whites in DTR, and average MuNI among Blacks in DTR, respectively. Kaplan–Meier (KM) with log‐rank test was performed to compare the survival outcomes among different risk groups. Multivariable Cox proportional hazards model was used to evaluate the independent prognostic ability of MuNI for OS after accounting for age, sex, smoking status, and T stage/N stage categories. OS was defined as the time interval between the date of diagnosis and the date of death and was censored at the date of last follow‐up for still those alive. Disease‐free survival (DFS) is defined as the time from diagnosis to recurrence of tumor or death.
TABLE 1.
Clinical and Pathological Features of all Five Cohorts
| D1, No. (%)a | D2, No. (%)b | D3, No. (%)c | D4, No. (%)d | D5, No. (%)e | ANOVA (two‐sided) p | |
|---|---|---|---|---|---|---|
| No. of patients | 106 | 120 | 318 | 33 | 167 | |
| Age, years | 57.62 ± 9.6 | 57.2 ± 8.3 | 58.73 ± 9.1 | 67.94 ± 8.7 | 56.45 ± 9.3 | .01 |
| Gender | ||||||
| Male | 89 (83.96) | 111 (91.74) | 281 (88.5 | 28 (84.85) | 152 (91.02) | .01 |
| Female | 17 (16.04) | 9 (8.26) | 37 (11.49) | 5 (5.15) | 15 (8.98) | |
| Race | .02 | |||||
| White | 103 (97.17) | 114 (95.00) | 299 (94.02) | 17 (51.52) | 157 (94.01) | |
| Black | 3 (2.83) | 6 (5.00) | 19 (5.98) | 16 (48.48) | 10 (5.99) | |
| Tobacco smoking | .03 | |||||
| Ever | 70 (66.04) | 79 (65.83) | 212 (66.46) | 20 (60.61) | 109 (65.27) | |
| Never | 36 (33.96) | 41 (34.17) | 106 (33.5) | 12 (36.36) | 58 (34.73) | |
| Treatment | <.001 | |||||
| Surgery with adjuvant therapy | 64 (60.38) | 24 (20.00) | 0 (0) | UNK | 95 (56.89) | |
| Surgery alone | 18 (16.98) | 0 (0) | 0 (0) | UNK | 0 (0) | |
| Definitive nonoperative treatment | 24 (22.64) | 96 (80.00) | 318 (100) | UNK | 72 (43.11) | |
| Specimen type | ||||||
| Resection | 82 (77.36) | 25 (20.66) | 0 (0) | UNK | 95 (56.89) | <.001 |
| Biopsy | 24 (22.64) | 96 (79.34) | 318 (100) | UNK | 72 (43.11) | |
| T stage | <.001 | |||||
| T1/T2 | 73 (68.87) | 79 (65.26) | 200 (63.04) | 19 (57.58) | 91 (54.49) | |
| T3/T4 | 33 (31.13) | 41 (34.74) | 118 (37.0) | 14 (42.42) | 76 (45.51) | |
| N stage | <.001 | |||||
| N1/N0 | 77 (72.64) | 84 (70.00) | 205 (64.9) | 7 (21.21) | 123 (73.65) | |
| N2/N3 | 29 (27.36) | 36 (30.00) | 113 (35.1) | 26 (78.79) | 44 (26.35) | |
| Overall stage | <.001 | |||||
| I/II | 87 (82.08) | 92 (76.67) | 237 (74.5) | 20 (60.61) | 125 (74.85) | |
| III/IV | 19 (17.92) | 28 (23.33) | 81 (25.5) | 13 (39.39) | 42 (25.15) | |
| MuNI | (3.62 ± 1.6) × 10−4 | (3.97 ± 1.7) × 10−4 | (3.06 ± 1.7) × 10−4 | (3.72 ± 1.43) × 10−4 | (3.06 ± 1.17) × 10−4 | <.001 |
Note: ± denotes 1 SD below/above the mean.
Abbreviations: ANOVA, analysis of variance; B, Black; MuNI, multinucleation index; SD, standard deviation; UNK, unknown; W, White.
Washington University in St. Louis (B = 3, W = 103).
John Hopkins University (B = 6, W = 114).
Cleveland Clinic (B = 19, W = 299).
UH Seidman Cancer Center (B = 16, W = 17).
Southern California Permanente Medical Group (B = 10, W = 157).
RESULTS
Patient demographics
The median follow‐up duration for the patients was 56.0 months (4.6 years). The median age was 58.2 years. A total of 7% of the patients were Black and 93% were White. Patient demographics for all five cohorts are provided in Table 1. There were statistically significant differences in clinical and pathological features among the patients treated at the five different enrollment locations (Table 1).
Association of MuNI and racial categories with survival outcomes
For DVA, KM survival analysis demonstrated that patients with high‐risk scores defined by MuNI had poorer OS (p = .002, hazard ratio [HR], 1.71; 95% CI, 1.21–2.43) compared to those with lower risk scores (Fig. 2A). Multivariable Cox proportional hazards model revealed that MuNI was prognostic of OS (p = .003; HR, 1.75; 95% CI, 1.21–2.52), controlling age (p = 4e–05; HR, 1.58; 95% CI, 1.30–1.92), sex (p = .05; HR, 0.65; 95% CI, 0.42–1.01), smoking (p = .04; HR, 1.56; 95% CI, 1.02–2.39), O stage (p = .50; HR, 0.87; 95% CI, 0.57–1.32), T stage (p = .03; HR, 1.38; 95% CI, 1.04–1.83), and N stage (p = .13; HR, 1.27; 95% CI, 0.94–1.73) in DVA. Additionally, OS of DB was statistically significantly worse than OS of DW (p = .002; HR, 2.24; 95% CI, 1.1–4.55), as shown in Figure 2B.
FIGURE 2.
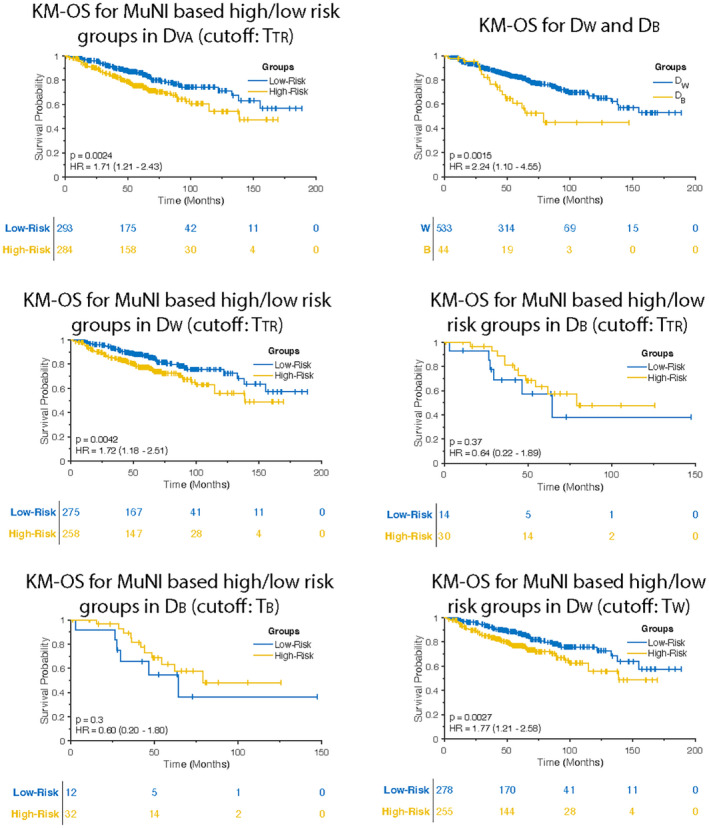
Kaplan–Meier (KM) overall survival (OS) curves in DVA. (A) OS of multinucleation index (MuNI)‐defined high and low‐risk groups using TTR was found statistically significantly different. (B) OS of DB was found statistically significantly shorter than OS of DW. Difference in MuNI could explain the shorter survival of DB patients. Survival differences between the two racial categories could be a factor of histological differences. OS of MuNI‐defined high‐ and low‐risk groups using TTR was statistically significantly different in (C) DW but not in (D) DB. (E) TB yields statistically insignificant separation for DB (p = .3; HR, 0.60; 95% CI, 0.20–1.80). (F) KM OS curve obtained by using TW indicates improved risk stratification for DW.
For DVA, DFS of high‐risk scores defined by MuNI was statistically significantly shorter than DFS of MuNI‐derived low‐risk scores (p = .0003; HR, 1.74; 95% CI, 1.29–2.34) (Fig. 3A). DFS of DB was worse than DFS of DW (p = .034; HR, 1.66; 95% CI, 0.92–2.99) (Fig. 3B).
FIGURE 3.
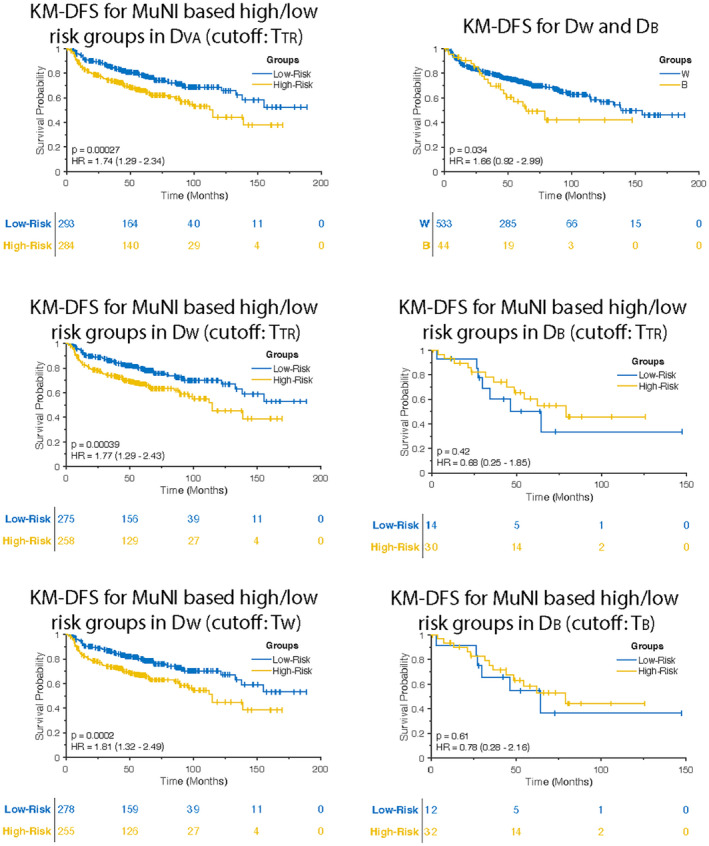
Kaplan–Meier (KM) disease‐free survival (DFS) curves in DVA. (A) DFS of multinucleation index (MuNI)‐defined high‐ and low‐risk groups using TTR was found statistically significantly different. (B) DFS of DB was found statistically significantly shorter than DFS of DW. DFS of MuNI‐defined high‐ and low‐ risk groups using TTR was statistically significantly different in (C) DW but not in (D) DB. (E) KM DFS curve obtained by using TW indicates improved risk stratification for DW. (F) TB yields statistically insignificant separation for DB (p = .61; HR, 0.78; 95% CI, 0.28–2.16).
Population‐specific differences in terms of MuNI
When considering all patients, MuNI was statistically significantly different between DW (mean, 3.88e–04; median, 3.67e–04) and DB (mean, 3.36e–04; median, 2.99e–04) as well (p = .0078) (Fig. 4A). Furthermore, when 50 patients were randomly sampled from DW and compared against the entire DB, 63% and 79% of the times, out of 200 comparisons, p values of the Mann–Whitney U tests were found to be smaller than 0.05 and 0.1, respectively, implying that there is a significant difference in MuNI between DB and DW. A histogram of the p values is shown in Figure 4. Using Mann–Whitney U test, the differences in MuNI between DW and DB patients were also found to be statistically significant for stage I, p = .031 (Fig. 4B). The median, first, and third quartiles of the Black HPV‐associated OPSCC patients were also larger than the corresponding values of the White patients. The p values of the Mann–Whitney U test for comparing the two groups with stages I, II, and III were 0.031, 0.32, and 0.57, respectively (Figs. 4B–D).
FIGURE 4.
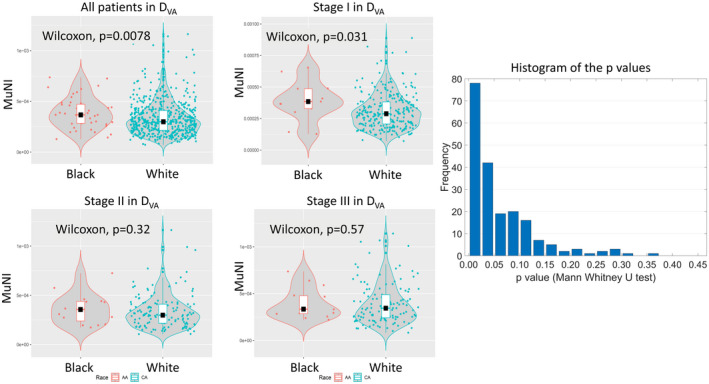
Comparison of Black and White patients. Violin plots show multinucleation index measurements for Black and White human papillomavirus–associated oropharyngeal squamous cell carcinoma patients. (A) All patients in DVA, AJCC 8th edition (B) stage I alone, (C) stage II alone, and (D) stage III alone. (E) The histogram of the p values obtained after applying Mann–Whitney U testing 200 times illustrates the statistical differences between randomly selected subsets of DW and the entire DB. The box plots inside the violin plots demonstrate the median, first, and third quartiles of distributions.
Evaluating population‐specific cutoffs for MuNI‐based risk stratification
Using TTR, KM survival analysis demonstrated that patients with high‐risk scores defined by MuNI had poorer OS (p = .004; HR, 1.72; 95% CI, 1.18–2.51) compared to those with lower risk scores in DW (n = 533) (Fig. 2C). On the other hand, the same cutoff yielded statistically insignificant p and HR values for the DB patients (n = 44; p = .37; HR, 0.64; 95% CI, 0.22–1.89) (Fig. 2D). Additionally, TW yielded improved HR value (p = .003; HR, 1.77; 95% CI, 1.21–2.58) in DW (Fig. 2E), whereas TB did not improve risk‐stratification in DB, generating p and HR values that were not statistically significant (p = .3; HR, 0.6; 95% CI, 0.2–1.8) (Fig. 2F).
DFS of patients with high‐risk scores defined by TTR had poorer (p = .0004; HR, 1.77; 95% CI, 1.29–2.43) compared to those with lower risk scores in DW (Fig. 3C). The same cutoff yielded p and HR values of 0.42 and 0.68 (95% CI, 0.25–1‐85), respectively, in DB (Fig. 3D). Similar to OS, TW yielded improved HR value (p = .0002; HR, 1.81; 95% CI, 1.32–2.49) in DW (Fig. 3E) in predicting DFS, whereas TB did not improve risk‐stratification in DB (p = .61; HR, 0.78; 95% CI, 0.28–2.16) (Fig. 3F).
DISCUSSION
In studies investigating OPSCC patients with different races (Black vs. White), differences in survival, genetic, and environmental factors have been identified that may explain poorer survival and increased likelihood of disease recurrence in Black versus White patients. 42 For example, higher degrees of chromosomal instability have been observed among Black patients with HNSCC. 43 , 44 In a study, tumors of Black patients with HNSCC were found to be 1.4 times more likely to show loss of the CDKN2A gene as compared to White patients. 34 The same study revealed that Black patients were also 57% less likely to have a loss of the SCYA3 gene as compared to White patients. This gene is known as macrophage inflammatory protein‐1 alpha (MIP‐1α), playing an important role in lymph node metastasis in HNSCC. On the other hand, several studies have focused on histomorphometric differences between different populations under the assumption that genetic and epigenetic alterations within tumor are reflected as distinctive phenotypic variations in tumor morphology. Bhargava et al. 45 found significant morphological differences in intratumoral stromal regions of surgically excised prostate specimens between Black and White men with prostate cancer. Based on these findings, they demonstrated that a population‐specific stromal signature was more prognostic of recurrence post‐surgery in Black men compared to a population‐agnostic model. A related study in endometrial carcinomas 46 revealed that patterns of arrangement of tumor infiltrating lymphocytes (TILs) in the stroma were differentially prognostic in Black and White women, indicating spatial TIL patterns being more strongly associated with likelihood of recurrence in Black women compared to White women. These studies provide further quantitative evidence of morphologic differences, as captured by computational pathology‐derived image biomarkers, between different populations. The present study is one example of such assessment by quantifying multinucleated tumor cells (MNs) within the tumor microenvironment.
In a review for using p16 as a surrogate for HPV positivity, concerning 28 studies (a total of 31 analyses) from a period of 2014 to 2016, the median sensitivity and specificity was found to be 95.4% and 87.3%, respectively. 6 Studies using high risk HPV mRNA‐based testing such as RTPCR and RNA in situ hybridization have shown higher correlations with p16 positivity in the United States, where there is a high attributable fraction of high‐risk HPV‐positive OPSCC. The studies indicate that patients with HPV‐associated tumors can be identified with p16 immunohistochemistry, however, a subset of patients who are HPV‐negative but are classified as HPV‐positive does exist. 6
Although the mechanism of neoplastic MN is not known, its association with tumor progression has been shown in several other studies. 47 , 48 Wilms tumor (nephroblastoma) of the kidney and central nervous system gliomas have used anaplasia and MN as prognostic factors. 49 , 50 Lewis et al. 32 previously identified anaplasia and MN as novel prognostic features in HPV‐associated OPSCC patients. In our previous study, we presented a tissue‐nondestructive, reproducible, and cost‐efficient artificial intelligence‐enabled biomarker, termed MuNI, that reflects the frequency of MNs among epithelial cells. It was validated as being prognostic of major clinical outcome, OS and DFS. 37 Similarly, in the present study, we identified a strong association of the predictions of the MuNI with OS and DFS, independent from other clinical factors despite the significant differences between the enrollment locations (Table 1). The previous study did not selectively focus on key population subgroups such as Black and White. Given the potential molecular and morphologic heterogeneity across different populations, in this study, we specifically focused on a systematic analysis of differences in MuNI across Black and White patients. As described in previous related studies on prostate and uterine carcinomas, 45 , 46 the goal was to unearth morphological differences between different key populations to pave the way for developing population‐specific prognostic tools in OPSCC patients. We demonstrated that MuNIs of Black patients were statistically significantly higher than those of White patients overall (p = .0078). The presence of high frequency of MN events in the Black patients and their poorer survival suggest that the difference is not only related to a lower socioeconomic status, which has been suggested, 11 but also possibly because of a more aggressive disease phenotype that is manifested in the morphology. Interestingly, we also found significant differences between the two populations while considering AJCC 8th edition stage I (p = .031) (Fig. 4).
Although the model was prognostic for DVA (Fig. 2A) as well as DW (Figs. 2C,E), it was not prognostic for DB (Figs. 2D,F). However, these findings should not be construed as suggesting that MuNI is not prognostic in Black patients, but rather the fact that thresholds employed for risk stratification were derived from a primarily White population. The threshold defined for MuNI‐derived risk stratification, TTR, was calculated as the average of MuNIs in DTR, comprising a 95% White versus 5% Black population. Black patients constitute a small fraction of the population because incident‐rate of HPV‐associated OPSCC is much higher for White patients than their Black counterparts. 18 The lack of significant p and HR values for the DB patients might be explained by unbalanced sample sizes in the study population. The model is biased toward majority of the population (DW). Even though these findings suggest using different cutoffs for different populations the sample size of Black patients in this study is too small to calculate a reliable cutoff, which is one limitation of the study. With a larger number of Black patients, population‐tailored prognostic models could be developed. A larger data set of Black patients from multiple institutions will help us to explore (1) whether MuNI is associated independently with survival differences between Black and White patients, and (2) whether population‐specific treatment management guidelines based on a reliable model more specific toward Black population can be defined. Another limitation of the study is that the study used self‐declared race information of patients as opposed to genetic ancestry. Because White patients might have different genetic origins, such as European, Middle Eastern, and North African, 13 , 50 there might exist substantial amounts of tumor phenotypical heterogeneity in this population as a consequence of diverse genetic ancestry. Additionally, the previous study demonstrated that patients treated with primary chemoradiation (biopsy specimens only) could be risk‐stratified to almost the same extent as surgically treated patients by MuNI. However, impact of any intrinsic differences between biopsy and resection on MuNI‐based differences across Black and White patients has not been addressed in this study due to the limited sample size.
Despite these limitations, this is the first study to assess population‐specific differences by quantifying tumor histology from routine H & E tissue slides, specifically frequency of tumor MN events within epithelial regions. The findings of this exploratory study provide some biological insight into differences in tumor morphology between Black and White patients in HPV‐associated OPSCC. The findings, along with the latest staging criteria, could pave the way for developing more effective and more personalized treatment strategies by creating population‐specific prognostic models.
AUTHOR CONTRIBUTIONS
Can F. Koyuncu: Conceptualization, formal analysis, methodology, software, visualization, and writing–original draft. Reetoja Nag: Formal analysis, methodology, software, visualization, and writing–original draft. James S. Lewis Jr: Conceptualization, data curation, formal analysis, supervision, and visualization. Anant Madabhushi: Conceptualization, data curation, formal analysis, funding acquisition, resources, supervision, and visualization. Vidya Sankar Viswanathan: Data curation. Vlad C. Sandulache: Data curation. Quintin Pan: Data curation. Deborah J. Chute: Data curation. Wade L. Thorstad: Data curation. Farhoud Faraji: Data curation. Justin A. Bishop: Data curation. Mitra Mehrad: Data curation. Patricia D. Castro: Data curation. Andrew G. Sikora: Data curation. Lester D.R. Thompson: Data curation. Rebecca D. Chernock: Data curation. Krystle A. Lang Kuhs: Data curation. Jay K. Wasman: Data curation. Jingqin R. Luo: Data curation. David J. Adelstein: Data curation. Shlomo A. Koyfman: Data curation. Pingfu Fu: Formal analysis. All authors have made substantial contributions to investigation, project administration, validation, and writing–review and editing.
CONFLICTS OF INTEREST
Rebecca D. Chernock reports funding from Caris Life Sciences and Merck. Deborah J. Chute reports book royalties. Germán Corredor reports a grant from the US Department of Defense. Farhoud Faraji reports funding from the National Institute on Deafness and Other Communication Disorders and Stand Up To Cancer. James S. Lewis Jr reports a grant from National Institutes of Health and payment as an expert witness from Shook, Hardy, and Bacon. Jingqin R. Luo reports a grant from National Institutes of Health. Anant Madabhushi reports grants from AstraZeneca, Boehringer Ingelheim, and Bristol‐Myers Squibb; consulting fees from Aiforia, Caris, and SimBioSys; participation as a fiduciary officer with Picture Health; and stock options with Elucid Bioimaging. Andrew G. Sikora reports consulting fees from F. Hoffmann‐La Roche. Lester D.R. Thompson reports funding from Elsevier. Jun Xu reports a grant from the National Natural Science Foundation of China. Kailin Yang reports grants from the Conquer Cancer Foundation, the National Cancer Institute, and RSNA Research and Education Foundation; and travel funding from Case Comprehensive Cancer Center and Case Western Reserve University. The other authors made no disclosures.
FUNDING INFORMATION
Research reported in this publication was supported by the National Cancer Institute under award numbers R01CA268207‐01A1, R01CA249992‐01A1, R01CA202752‐01A1, R01CA208236‐01A1, R01CA216579‐01A1, R01CA220581‐01A1, R01CA257612‐01A1, 1U01CA239055‐01, 1U01CA248226‐01, 1U54CA254566‐01, National Heart, Lung and Blood Institute 1R01HL15127701A1, R01HL15807101A1, National Institute of Biomedical Imaging and Bioengineering 1R43EB028736‐01, National Center for Research Resources under award number 1 C06 RR12463‐01, VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Servicethe Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH‐19‐1‐0668), the Prostate Cancer Research Program (W81XWH‐20‐1‐0851), the Lung Cancer Research Program (W81XWH‐18‐1‐0440, W81XWH‐20‐1‐0595), the Peer Reviewed Cancer Research Program (W81XWH‐18‐1‐0404, W81XWH‐21‐1‐0345, W81XWH‐21‐1‐0160, and W81XWH‐22‐1‐0236), the Kidney Precision Medicine Project (KPMP) Glue Grant and sponsored research agreements from Bristol Myers‐Squibb, Boehringer‐Ingelheim, Eli‐Lilly and Astrazeneca. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
ACKNOWLEDGMENTS
This study was reviewed and approved by the institutional review boards of Washington University in St. Louis, Missouri, USA; Johns Hopkins University in Baltimore, Maryland, USA; the Cleveland Clinic in Cleveland, Ohio, USA; Case Western Reserve University in Cleveland, Ohio, USA; Cleveland University Hospital Seidman Cancer Center in Cleveland, Ohio, USA; the Southern California Permanente Medical Group in Los Angeles, California, USA; Vanderbilt University Medical Center in Nashville, Tennessee, USA; and the Michael E. DeBakey VA Medical Center in Houston, Texas, USA. This study was supported by the National Cancer Institute (R01CA249992‐01A1, R01CA202752‐01A1, R01CA208236‐01A1, R01CA216579‐01A1, R01CA220581‐01A1, R01CA257612‐01A1, 1U01CA239055‐01, 1U01CA248226‐01, and 1U54CA254566‐01), National Heart, Lung and Blood Institute (1R01HL15127701A1 and R01HL15807101A1), National Institute of Biomedical Imaging and Bioengineering (1R43EB028736‐01), National Center for Research Resources (1 C06 RR12463‐01), a VA Merit Review Award (IBX004121A) from the US Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH‐19‐1‐0668), the Prostate Cancer Research Program (W81XWH‐15‐1‐0558 and W81XWH‐20‐1‐0851), the Lung Cancer Research Program (W81XWH‐18‐1‐0440 and W81XWH‐20‐1‐0595), the Peer Reviewed Cancer Research Program (W81XWH‐18‐1‐0404 and W81XWH‐21‐1‐0345), the Kidney Precision Medicine Project (KPMP) Glue Grant, and sponsored research agreements from Bristol Myers‐Squibb, Boehringer‐Ingelheim, and AstraZeneca. Vlad C. Sandulache is supported by Career Development Award (IK2 CX001953) from the US Department of Veterans Affairs Clinical Sciences R&D Service. Farhoud Faraji is supported by Stand Up to Cancer (308268‐00001) Precision Therapy for Fanconi Anemia and HPV‐related Head & Neck Cancer and National Institutes of Health (T32DC000028). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the US Department of Veterans Affairs, the Department of Defense, or the US Government.
Present addresses: Vlad C. Sandulache, ENT Section, Operative Care Line, Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX, USA; Bobby R. Alford Department of Otolaryngology Head and Neck Surgery, Baylor College of Medicine, Houston, Texas, USA; and Center for Translational Research on Inflammatory Diseases, Michael E. DeBakey Veterans Affairs Medical Center, Houston, Texas, USA.
Can Koyuncu, Reetoja Nag, Cheng Lu, Germán Corredor, Vidya Sankar Viswanathan, Zelin Zhang, and Anant Madabhushi, Biomedical Engineering Department, Emory University, Atlanta, GA, USA.
The first two authors contributed equally to this article.
REFERENCES
- 1. Denoix PF. [Note on the possible role of the International Union against Cancer in nomenclature, classification, analytical index, bibliography and documentation]. Acta Unio Int Contra Cancrum 1952;8(Special No):92–96, 136, 139. [PubMed] [Google Scholar]
- 2. Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of human papillomavirus‐positive head and neck squamous cell carcinoma. J Clin Oncol. 2015;33(29):3235‐3242. doi: 10.1200/JCO.2015.61.6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Sullivan B, Huang SH, Su J, et al. Development and validation of a staging system for HPV‐related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON‐S): a multicentre cohort study. Lancet Oncol. 2016;17(4):440‐451. doi: 10.1016/S1470-2045(15)00560-4 [DOI] [PubMed] [Google Scholar]
- 4. Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24‐35. doi: 10.1056/NEJMoa0912217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Satgunaseelan L, Virk S, Lum T, Gao K, Clark J, Gupta R. The role of p16 expression in oral squamous cell carcinoma. Pathology. 2015;47:S53. doi: 10.1097/01.PAT.0000461456.42149.fc [DOI] [PubMed] [Google Scholar]
- 6. Jouhi L, Hagström J, Atula T, Mäkitie A. Is p16 an adequate surrogate for human papillomavirus status determination? Curr Opin Otolaryngol Head Neck Surg. 2017;25(2):108‐112. doi: 10.1097/MOO.0000000000000341 [DOI] [PubMed] [Google Scholar]
- 7. Amin MB, Greene FL, Edge SB, et al. The Eighth Edition AJCC Cancer Staging Manual: continuing to build a bridge from a population‐based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67(2):93‐99. doi: 10.3322/caac.21388 [DOI] [PubMed] [Google Scholar]
- 8. Pollaers K, Hinton‐Bayre A, Friedland PL, Farah CS. AJCC 8th Edition oral cavity squamous cell carcinoma staging—is it an improvement on the AJCC 7th Edition? Oral Oncol. 2018;82:23‐28. doi: 10.1016/j.oraloncology.2018.04.018 [DOI] [PubMed] [Google Scholar]
- 9. Lu C, Lewis JS, Dupont WD, Plummer WD, Janowczyk A, Madabhushi A. An oral cavity squamous cell carcinoma quantitative histomorphometric‐based image classifier of nuclear morphology can risk stratify patients for disease‐specific survival. Mod Pathol. 2017;30(12):1655‐1665. doi: 10.1038/modpathol.2017.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brandwein‐Gensler M, Teixeira MS, Lewis CM, et al. Oral squamous cell carcinoma: histologic risk assessment, but not margin status, is strongly predictive of local disease‐free and overall survival. Am J Surg Pathol. 2005;29(2):167‐178. doi: 10.1097/01.pas.0000149687.90710.21 [DOI] [PubMed] [Google Scholar]
- 11. O'Neill WQ, Wasman J, Thuener J, et al. African Americans With p16+ and p16‐ oropharyngeal squamous cell carcinomas have distinctly poor treatment outcomes independent of medical care access. JCO Oncol Pract. 2021;17(5):e695‐e702. doi: 10.1200/OP.20.01105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richardson LM, Hill JN, Smith BM, et al. Patient prioritization of comorbid chronic conditions in the veteran population: implications for patient‐centered care. SAGE Open Med. 2016;4:205031211668094. doi: 10.1177/2050312116680945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koyuncu C, Lu C, Zhang Z, et al. Tumor cell multinucleation is more frequent in African‐American oropharyngeal squamous cell carcinoma patients than Caucasian‐American ones‐implications for outcome differences. Mod Pathol. 2020;33(3):1208‐1209. [Google Scholar]
- 14. Bhargava HK, Leo P, Elliott R, et al. Computer‐extracted stromal features of African‐Americans versus Caucasians from H&E slides and impact on prognosis of biochemical recurrence. J Clin Oncol. 2018;36(suppl 15):12075. doi: 10.1200/JCO.2018.36.15_suppl.12075 [DOI] [Google Scholar]
- 15. Abdou Y, Attwood K, Cheng TYD, et al. Racial differences in CD8+ T cell infiltration in breast tumors from Black and White women. Breast Cancer Res. 2020;22(1):62. doi: 10.1186/s13058-020-01297-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoskins KF, Calip GS. Racial/ethnic differences in the 21‐gene recurrence score assay among women with breast cancer‐reply. JAMA Oncol. 2021;7(8):1248‐1249. doi: 10.1001/jamaoncol.2021.1959 [DOI] [PubMed] [Google Scholar]
- 17. Ellis L, Canchola AJ, Spiegel D, Ladabaum U, Haile R, Gomez SL. Racial and ethnic disparities in cancer survival: the contribution of tumor, sociodemographic, institutional, and neighborhood characteristics. J Clin Oncol. 2018;36(1):25‐33. doi: 10.1200/JCO.2017.74.2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo T, Rettig E, Fakhry C. Understanding the impact of survival and human papillomavirus tumor status on timing of recurrence in oropharyngeal squamous cell carcinoma. Oral Oncol. 2016;52:97‐103. doi: 10.1016/j.oraloncology.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chernock RD, Zhang Q, El‐Mofty SK, Thorstad WL, Lewis JS. Human papillomavirus‐related squamous cell carcinoma of the oropharynx: a comparative study in Whites and African Americans. Arch Otolaryngol Head Neck Surg. 2011;137(2):163‐169. doi: 10.1001/archoto.2010.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Worsham MJ, Stephen JK, Chen KM, et al. Improved survival with HPV among African Americans with oropharyngeal cancer. Clin Cancer Res. 2013;19(9):2486‐2492. doi: 10.1158/1078-0432.CCR-12-3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stephen JK, Chen KM, Shah V, et al. Human papillomavirus outcomes in an access‐to‐care laryngeal cancer cohort. Otolaryngol Head Neck Surg. 2012;146(5):730‐738. doi: 10.1177/0194599811434482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ragin C, Liu JC, Jones G, et al. Prevalence of HPV infection in racial–ethnic subgroups of head and neck cancer patients. Carcinogenesis. 2017;38(2):218‐229. doi: 10.1093/carcin/bgw203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Richards KL, Zhang B, Baggerly KA, et al. Genome‐wide hypomethylation in head and neck cancer is more pronounced in HPV‐negative tumors and is associated with genomic instability. PLoS One. 2009;4(3):e4941. doi: 10.1371/journal.pone.0004941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sartor MA, Dolinoy DC, Jones TR, et al. Genome‐wide methylation and expression differences in HPV(+) and HPV(−) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6(6):777‐787. doi: 10.4161/epi.6.6.16216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus‐positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261‐269. doi: 10.1093/jnci/djn011 [DOI] [PubMed] [Google Scholar]
- 26. Thompson LDR, Burchette R, Iganej S, Bhattasali O. Oropharyngeal squamous cell carcinoma in 390 patients: analysis of clinical and histological criteria which significantly impact outcome. Head Neck Pathol. 2020;14(3):666‐688. doi: 10.1007/s12105-019-01096-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shield KD, Ferlay J, Jemal A, et al. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012: lip, oral cavity, and pharyngeal cancers. CA Cancer J Clin. 2017;67(1):51‐64. doi: 10.3322/caac.21384 [DOI] [PubMed] [Google Scholar]
- 28. Osazuwa‐Peters N, Massa ST, Christopher KM, Walker RJ, Varvares MA. Race and sex disparities in long‐term survival of oral and oropharyngeal cancer in the United States. J Cancer Res Clin Oncol. 2016;142(2):521‐528. doi: 10.1007/s00432-015-2061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shavers VL, Harlan LC, Winn D, Davis WW. Racial/ethnic patterns of care for cancers of the oral cavity, pharynx, larynx, sinuses, and salivary glands. Cancer Metastasis Rev. 2003;22(1):25‐38. doi: 10.1023/A:1022255800411 [DOI] [PubMed] [Google Scholar]
- 30. Nichols AC, Bhattacharyya N. Racial differences in stage and survival in head and neck squamous cell carcinoma. Laryngoscope. 2007;117(5):770‐775. doi: 10.1097/MLG.0b013e318033c800 [DOI] [PubMed] [Google Scholar]
- 31. Yu AJ, Choi JS, Swanson MS, et al. Association of race/ethnicity, stage, and survival in oral cavity squamous cell carcinoma: a SEER study. OTO Open. 2019;3(4):2473974X1989112. doi: 10.1177/2473974X19891126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewis JS, Scantlebury JB, Luo J, Thorstad WL. Tumor cell anaplasia and multinucleation are predictors of disease recurrence in oropharyngeal squamous cell carcinoma, including among just the human papillomavirus‐related cancers. Am J Surg Pathol. 2012;36(7):1036‐1046. doi: 10.1097/PAS.0b013e3182583678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheth S, Farquhar DR, Lenze NR, et al. Decreased overall survival in black patients with HPV‐associated oropharyngeal cancer. Am J Otolaryngol. 2021;42(1):102780. doi: 10.1016/j.amjoto.2020.102780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Worsham MJ, Stephen JK, Lu M, et al. Disparate molecular, histopathology, and clinical factors in head and neck squamous cell carcinoma racial groups. Otolaryngol Head Neck Surg. 2012;147(2):281‐288. doi: 10.1177/0194599812440681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu X, Li M, Hu J, et al. Expression profile analysis identifies a two‐gene signature for prediction of head and neck squamous cell carcinoma patient survival. J Cancer Res Ther. 2018;14(7):1525‐1534. doi: 10.4103/jcrt.JCRT_557_18 [DOI] [PubMed] [Google Scholar]
- 36. Wang J, Chen X, Tian Y, et al. Six‐gene signature for predicting survival in patients with head and neck squamous cell carcinoma. Aging (Albany NY). 2020;12(1):767‐783. doi: 10.18632/aging.102655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koyuncu CF, Lu C, Bera K, et al. Computerized tumor multinucleation index (MuNI) is prognostic in p16+ oropharyngeal carcinoma. J Clin Invest. 2021;131(8):e145488. doi: 10.1172/JCI145488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis JS, Beadle B, Bishop JA, et al. Human papillomavirus testing in head and neck carcinomas: guideline from the College of American Pathologists. Arch Pathol Lab Med. 2018;142(5):559‐597. doi: 10.5858/arpa.2017-0286-CP [DOI] [PubMed] [Google Scholar]
- 39. Janowczyk A, Zuo R, Gilmore H, Feldman M, Madabhushi A. HistoQC: an open‐source quality control tool for digital pathology slides. JCO Clin Cancer Inform. 2019;3:1‐7. doi: 10.1200/CCI.18.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aresta G, Araújo T, Kwok S, et al. BACH: grand challenge on breast cancer histology images. Med Image Anal. 2019;56:122‐139. doi: 10.1016/j.media.2019.05.010 [DOI] [PubMed] [Google Scholar]
- 41. Mahmood F, Borders D, Chen RJ, et al. Deep adversarial training for multi‐organ nuclei segmentation in histopathology images. IEEE Trans Med Imaging. 2020;39(11):3257‐3267. doi: 10.1109/TMI.2019.2927182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wiencke JK. Impact of race/ethnicity on molecular pathways in human cancer. Nat Rev Cancer. 2004;4(1):79‐84. doi: 10.1038/nrc1257 [DOI] [PubMed] [Google Scholar]
- 43. Yuan J, Hu Z, Mahal BA, et al. Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell. 2018;34(4):549‐560.e9. doi: 10.1016/j.ccell.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reed AL, Califano J, Cairns P, et al. High frequency of p16 (CDKN2/MTS‐1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56(16):3630‐3633. [PubMed] [Google Scholar]
- 45. Bhargava HK, Leo P, Elliott R, et al. Computationally derived image signature of stromal morphology is prognostic of prostate cancer recurrence following prostatectomy in African American patients. Clin Cancer Res. 2020;26(8):1915‐1923. doi: 10.1158/1078-0432.CCR-19-2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Azarianpour Esfahani S, Fu P, Mahdi H, Madabhushi A. Computational features of TIL architecture are differentially prognostic of uterine cancer between African and Caucasian American women. J Clin Oncol. 2021;39(suppl 15):5585. doi: 10.1200/JCO.2021.39.15_suppl.5585 [DOI] [Google Scholar]
- 47. Ariizumi T, Ogose A, Kawashima H, Hotta T, Umezu H, Endo N. Multinucleation followed by an acytokinetic cell division in myxofibrosarcoma with giant cell proliferation. J Exp Clin Cancer Res. 2009;28(1):44. doi: 10.1186/1756-9966-28-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17(2):157‐162. doi: 10.1016/j.gde.2007.02.011 [DOI] [PubMed] [Google Scholar]
- 49. Molony P, Werner R, Martin C, et al. Tumor cell anaplasia and multinucleation as prognosticators in oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2020;14(3):606‐615. doi: 10.1007/s12105-019-01081-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Popov SD, Sebire NJ, Vujanic GM. Wilms' tumour—histology and differential diagnosis. In: van den Heuvel‐Eibrink MM, ed. Wilms Tumor. Codon Publications; 2016. Accessed September 1, 2021. http://www.ncbi.nlm.nih.gov/books/NBK373364/ [PubMed] [Google Scholar]


