Abstract
Moringa oleifera is a traditional food crop widespread in Asiatic, African, and South American continents. The plant, able to grow in harsh conditions, shows a high nutritional value and medicinal potential evidencing cardioprotective, anti-inflammatory, antioxidant, and antimicrobial properties. The purpose of this study was the phytochemical analysis of M. oleifera and the identification of the antimicrobial compounds by combining a chemical approach with in vitro tests. The metabolite profile of M. oleifera polar and apolar extracts of leaves and seeds were investigated by using Nuclear Magnetic Resonance spectroscopy and Gas Chromatography-Mass Spectrometry. The antimicrobial activity of all of the obtained extract was evaluated against four bacterial pathogens (Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa and Salmonella enterica). The chemical analysis provided a wide set of metabolites that were identified and quantified. Moreover, apolar extracts from seeds showed a significant concentration-dependent antimicrobial activity against S. aureus and S. epidermidis, (4 mg/mL reduced the viability up to 50%) that was associated to the content of specific fatty acids. Our results remarked the advantages of an integrated approach for the identification of plant metabolites and its use in association with biological tests to recognize the compounds responsible for bioactivity without compounds purification.
Keywords: Moringa oleifera, Moringaceae, bioactive metabolites, natural products, NMR, GC-MS, Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa, Salmonella enterica
1. Introduction
Moringa oleifera Lam., also known as miracle, horseradish or drumstick tree, is a plant member of the Moringaceae family able to grow as a short and slender tree. M. oleifera is native to Eastern countries, such as the Himalayas, India, Pakistan, Asia Minor, Africa, and Arabia [1], but today it is also distributed in other countries due to the plant tenacity making possible the cultivation in different habitats (Figure 1). The roots can penetrate deeply into the soil and retain water for long periods, making possible the growth of the plant also in dry and desertic soils and at different rainfall levels [2,3]. The optimal temperature for M. oleifera growth goes from 25 to 40 °C, but it can withstand temperatures ranging from −1 to 3 °C and from 38 to 48 °C, being able to resist to a large variety of environments [4].
Figure 1.
(a) Moringa oleifera tree; (b) Worldwide diffusion of the plant.
The interest in this plant is mainly derived from its traditional uses. In fact, in traditional medicine, M. oleifera parts were used to treat a large variety of conditions, as the plant was believed to possess several properties, such as carminative, anti-inflammatory, laxative, anti-rheumatic activities [1,5].
Today, M. oleifera is used for many purposes such as in human diet and livestock feeding, thanks to the excellent nutritional aspects (high quantities of vitamins, proteins, and amino acids) [2]; in medicine, thanks to the properties mentioned earlier; as fuel wood; for soil conservation (used as green manure) and water purification (the seeds are used for clarification of water) [6].
Both polar and apolar leaf and seed extracts contain several relevant compounds belonging to the classes of fatty acids, alkanes, amino acids, glucosinolates, polyphenols, which make M. oleifera a very interesting plant from a nutritional and a pharmacological point of view [5,7]. Thus, several pharmacological properties have been investigated and attributed to the seed and leaf extracts, e.g., cardiovascular activity [8], anti-inflammatory activity [9], antihypertensive activity [10], radical scavenging and antioxidant activity [11,12], anticancer activity [13,14], hepatoprotective and nutraceutical activity [15,16], anti-allergic activity, antimicrobial [17] and antiviral activity [18].
The aim of this study was to explore the qualitative and quantitative aspects of the metabolite profile of M. oleifera leaves and seeds and to evaluate the antimicrobial activity of their polar and apolar extracts.
The interests in plant metabolite profiling approaches are rapidly growing as they can provide a wide overview of the metabolites present in a plant extract [19]. It is often used to characterize extracts from plant tissues, or to compare extracts from plants grown in different conditions, as it provides a “fingerprint” of the status of the plant.
Here, we performed an untargeted metabolite profiling analysis of leaf and seed extracts using an integrated approach of proton Nuclear Magnetic Resonance (1H-NMR) spectroscopy and Gas Chromatography-Mass Spectrometry (GC-MS), followed by the identification of the main components from the obtained spectra. Then, the extracts were tested against Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa and Salmonella enterica to assess the antimicrobial activity.
2. Results
2.1. NMR Analysis of M. oleifera Polar Extracts
Polar extracts of leaves and seeds of M. oleifera were analyzed in triplicate through 1H NMR spectroscopy obtaining a qualitative and quantitative profile of plant tissues analyzed. Peak by peak analysis of the spectra was performed, with the aid of 2D NMR experiments (COSY, HSQC and HMBC), and by comparison with standard compounds available in the laboratory and reported in the literature.
The main metabolites identified from the NMR spectra are listed in Table 1, where they are divided in chemical structural classes.
Table 1.
1H NMR chemical shifts, assignment, and multiplicity at 600 MHz in D2O of organic compounds detected in the polar extracts of M. oleifera leaves and seeds.
| Compound | Assignment | 1H (ppm) | Multiplicity (J in Hz) | Leaves | Seeds |
|---|---|---|---|---|---|
| Organic acids | |||||
| Citric acid (CI) | α,γ-CH2 | 2.50 ** | d (15.0, 15.0) | x | x |
| α’,γ’-CH2 | 2.68 ** | d | |||
| Fumaric acid (FU) | α-CH | 6.61 * | s | x | |
| Malic acid (MA) | β’-CH2 | 2.43 | dd (15.7, 8.9) | x | x |
| β-CH | 2.78 | dd (15.7, 3.7) | |||
| α-CH | 4.33 * | dd (8.9, 3.7) | |||
| Succinic acid (SU) | α-CH2 | 2.42 ** | s | x | x |
| Acetic acid (AC) | α-CH3 | 1.96 * | s | x | x |
| Amino acids | |||||
| Alanine (Ala) | β-CH3 | 1.51 * | d (7.0) | x | x |
| Asparagine (Asn) | β-CH | 2.84 | dd (17.4, 3.8) | x | x |
| 2.98 * | dd (4.0, 13.0) | ||||
| Isoleucine (Ile) | δ-CH3 | 0.93 | t (7.0) | x | x |
| γ’-CH3 | 1.05 * | d (7.0) | |||
| γ-aminobutyrate (GABA) | β-CH2 | 1.92 | m | x | x |
| α-CH2 | 2.34 | t (7.0) | |||
| γ-CH2 | 3.04 * | t (7.0) | |||
| Leucine (Leu) | δ-CH3 | 1.01 * | d (7.0) | x | x |
| Glutamic acid (Glu) | β, β’-CH | 2.08, 2.16 * | m | x | |
| Phenylalanine (Phe) | CH-4 | 7.35 | t (7.0) | x | x |
| CH-2,6 | 7.45 * | m | |||
| Threonine (Thr) | γ-CH3 | 1.36 * | d (7.0) | x | x |
| Tryptophane (Trp) | CH | 7.74 * | d (7.5) | x | |
| Tyrosine (Tyr) | CH-3,5 | 6.86 ** | d (7.0) | x | x |
| CH-6,8 | 7.10 ** | d (7.0) | |||
| Valine (Val) | γ’-CH3 | 1.03 | d (7.0) | x | x |
| γ-CH3 | 1.08 * | d (7.0) | |||
| Carbohydrates | |||||
| β-Glucose (β-Glc) | CH-1 | 4.60 * | d (8.0) | x | x |
| α-Glucose (α-Glc) | CH-1 | 5.21 * | d (4.0) | x | x |
| Sucrose (Suc) | Glc CH-1 | 5.43 * | d (3.8) | x | x |
| Fru CH-3′ | 4.20 | d (8.5) | |||
| myo-Inositol (Myo) | CH-4 | 3.34 * | t (9.5) | x | x |
| CH-2,5 | 3.58 ** | ||||
| CH-3,6 | 3.66 ** | ||||
| Other compounds | |||||
| Adenosine (Adn) | CH-2 | 8.25 | s | x | x |
| CH-8 | 8.36 * | s | |||
| Caffeic acid (Caf) | α-CH | 6.42 * | d (16.0) | x | |
| Flavonoids (Fla) | CH | 6.53 * | x | ||
| Choline (Cho) | N(CH3)3+ | 3.24 * | s | x | x |
| Ethanolamine (Eta) | β-CH2 | 3.15 * | bt (7.0) | x | |
| Glucomoringin (GMor) | CH3 | 1.19 | d | x | x |
| Glc CH-1 | 4.33 | d (8.0) | |||
| Rha CH-1 | 5.57 * | d (2.0) | |||
| Glucosinolates (GSin) | Rha CH-1 | 5.55 * | d (2.0) | x | |
| Rha CH-1 | 5.53 * | d (2.0) | |||
| Quercetin (Que) | CH | 7.65 * | x | ||
| Trigonelline (Tri) | CH-4 | 8.12 | t | x | x |
| CH-3,5 | 8.88 | t | |||
| CH-1 | 9.17 * | s | |||
* Signal used for quantification. ** Signal partially overlapped.
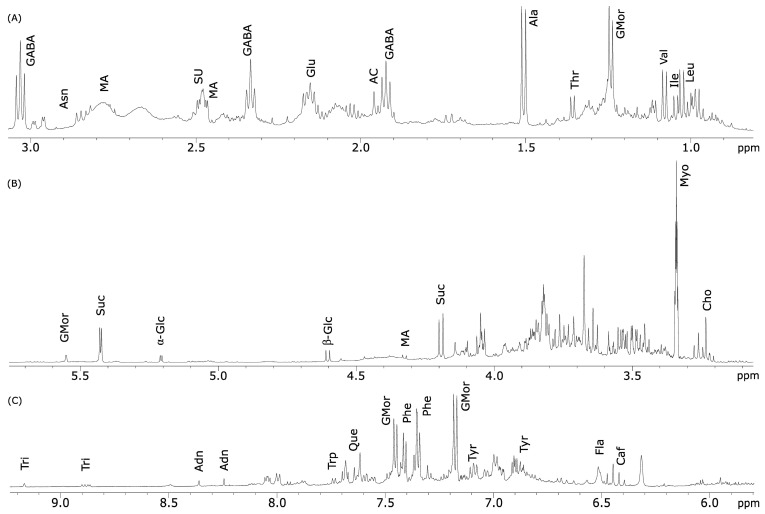
In the high field region of the leaf spectra (Figure 2A), diagnostic signals for the methyl groups of the branched amino acids are shown. Leucine (Leu), isoleucine (Ile) and valine (Val) were identified using the three doublets at δ 1.01 (J = 7.0 Hz), δ 1.05 (J = 7.0 Hz) and δ 1.08 (J = 7.0 Hz), respectively. Threonine (Thr) was also identified by a doublet at δ 1.36 (J = 7.0 Hz), corresponding to the γ-CH3 group, while the β-CH3 group of alanine (Ala) was assigned through a doublet at δ 1.51 (J = 7.0 Hz). This signal was shifted downfield compared to the previous ones because of the proximity to the nitrogen atom. Asparagine (Asn) was identified through the double doublet resonating at δ 2.98 (dd, J = 4.0 and 13.0 Hz), whereas the β-CH and β’-CH groups of glutamic acid (Glu), used for its identification, resonated as multiplets at δ 2.08 and δ 2.16, respectively. The amino acid γ-aminobutyrate (GABA) was identified using the triplet at δ 3.04 (J = 7.0 Hz).
Figure 2.
1H NMR spectrum of M. oleifera polar leaf extract acquired in D2O at 600 MHz. Spectral regions between (A) 0.5–3.1 ppm vertically expanded (×5); (B) 3.1–5.7 ppm; (C) 5.8–9.2 ppm vertically expanded (×2).
In the low field region of the 1H NMR spectra of leaves (Figure 2C), all of the characteristic groups belonging to aromatic amino acids were found. Going into detail, the multiplet at δ 7.45 and the triplet at at 7.35 were assigned to phenylalanine (Phe), the doublets at δ 6.86 and 7.10 belonged to tyrosine (Tyr) and the doublet at δ 7.74 (J = 7.5 Hz) was indicative of tryptophan (Trp), all of them corresponding to aromatic protons at different positions of the skeleton (see Table 1).
The signals used for the identification of five organic acids were found across all of the spectral regions (Figure 2A–C). Acetic acid (AC) was identified through the singlet at δ 1.96 found in the high field region of the spectra and corresponded to the α-CH3 group. Malic acid (MA) was identified by a double doublet resonating at δ 4.33 (J = 8.9 and 3.7 Hz), while fumaric acid (FU) showed the characteristic singlet at δ 6.61, both of them being associated to a α-CH group. Signals detected and associated to citric acid (CI) and succinic acid (SU), resonating, respectively, at δ 2.50 and 2.60, were overlapped, and the quantification of these two metabolites was not possible.
The region of the spectra between δ 3.35 and δ 4.10 (Figure 2B) was very crowded due to the high number of signals belonging to sugars and sugar alcohols that were not useful for compound identification. The characteristic signal of the anomeric proton (H-1) of α-glucose (α-Glc) resonated at δ 5.21 (d, J = 4.0 Hz), while the anomeric proton of β-glucose (β-Glc) appeared at δ 4.60 (d, J = 8.0 Hz) (Figure 2B and Table 1). Sucrose (Suc) was identified using the anomeric proton signal resonating at δ 5.43 (d, J = 3.8 Hz) due to its glucose moiety. The triplet resonating at δ 3.34 (J = 9.5 Hz) was assigned to myo-inositol (Myo).
Among other compounds, choline (Cho) presence was showed by the characteristic singlet corresponding to its methyl groups, resonating at δ 3.24, and shifted downfield in the spectra because of the presence of nitrogen (Table 1). Glucomoringin (GMor) was identified using the doublet (J = 2.0 Hz) resonating at δ 5.57, corresponding to H-1 of its rhamnose residue.
In the low-field region between 5.7 and 9.2 ppm, caffeic acid (Caf) was identified by a doublet resonating at δ 6.42 (J = 16.0). Signals for flavonoids (Fla) and in particular for quercetin (Que) were found at δ 6.53 and δ 7.65, respectively; both of them were only present in spectra of leaves. Lastly, the nucleoside adenosine (Adn) showed a singlet at δ 8.36 (s, CH-8), while the presence of trigonelline (Tri) was revealed by the characteristic signals resonating at δ 9.17 (s, CH-1) and 8.88 (t, CH-3,5) (Figure 2C and Table 1).
Figure 3 shows the 1H NMR spectrum of the seeds polar extract with compound identification. A close similarity with the NMR spectrum of leaves (Figure 2) was evident, although there were some differences in the metabolite profiles (Table 1).
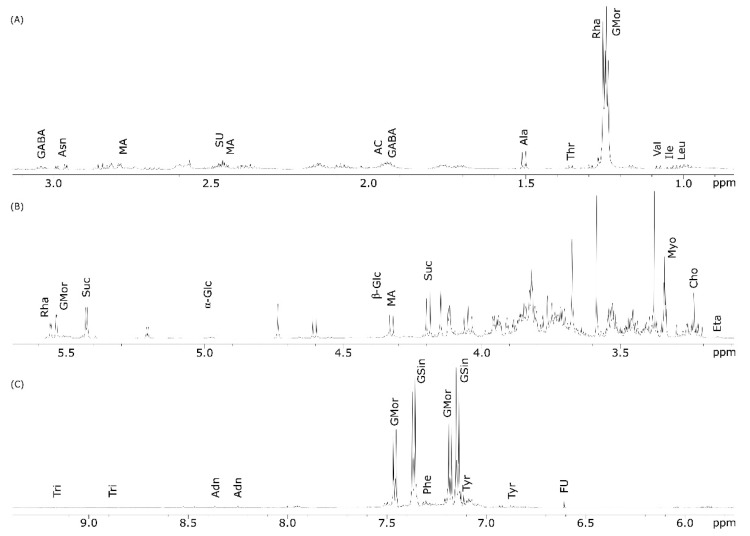
Figure 3.
1H NMR spectrum of M. oleifera polar seed extract run in D2O at 600 MHz. Spectral regions between (A) 0.5–3.1 ppm; (B) 3.1–5.7 ppm; (C) 5.8–9.3 ppm vertically expanded (×3).
Going into detail, signals for flavonoids and, particularly, quercetin were undetectable as well as that for caffeic acid (Figure 3). The amino acids phenylalanine and glutamic acid were not found in the seed extracts, while the signal identifying fumaric acid were additionally present.
The aromatic amino acids were undetectable in the seed extracts. Interestingly, in the aromatic region, additional signals at δ 7.15 and 7.37 (each d, J = 7.0 Hz) appeared; this could be attributed to glucosinolates (GSin) whose presence was confirmed by the signals at δ 5.55 and 5.53 corresponding to H-1 of rhamnose (Figure 3B). Among other compounds, ethanolamine (Eta) was identified using the broad triplet resonating at δ 3.15 (β-CH2, J = 7.0) (Table 1).
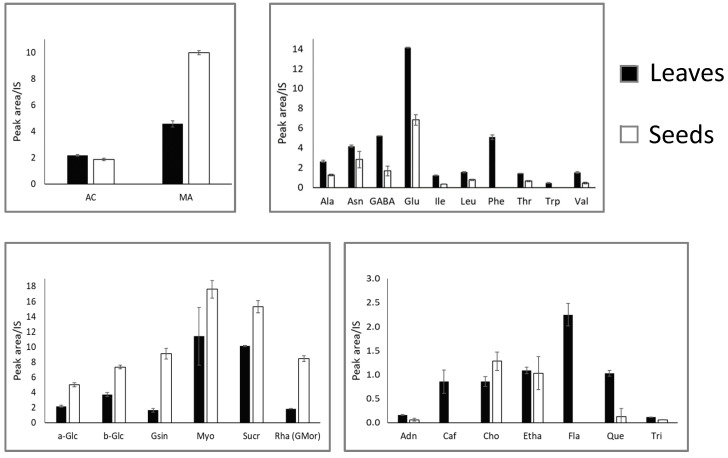
The differences in the quantified metabolites between leaf and seed extracts are shown in Figure 4. Acetic acid and malic acid were the only two organic acids that could be quantified. Acetic acid was slightly more abundant in the leaf extracts. Malic acid quantity was roughly double in seeds compared to the malic acid content of leaves.
Figure 4.
Quantitative data of the metabolites identified in leaves and seeds polar extracts. Top-left, organic acids; top-right, amino acids; bottom-left, sugars; bottom-right, other compounds. Shown data refers to the mean and standard deviation of three replicates.
All of the amino acids were more abundant in leaf extracts than in seed extracts. Glutamic acid was the amino acid present in the larger quantity, and along with phenylalanine they showed the largest difference between the two extracts. The aromatic amino acids phenylalanine and tryptophane were undetectable in seed extracts. (Figure 4).
Conversely, all of the carbohydrates were more abundant in seeds, especially for glucosinolates and glucomoringin, that showed the largest difference among the two extracts. Monosaccharides and disaccharides followed the same trend, but the difference was smaller. Considering the relative and the absolute quantity, carbohydrates were also the most abundant molecules in the leave and seed extracts overall.
Regarding the other compounds, caffeic acid and flavonoids were only detected in leaf extracts. Moreover, the quantity of quercetin was larger in leaf extracts, while ethanolamine and trigonelline showed a comparable quantity in the two extracts. Finally, choline was present in a larger amount in the seed extracts.
2.2. GC-MS Analysis of M. oleifera Apolar Extracts
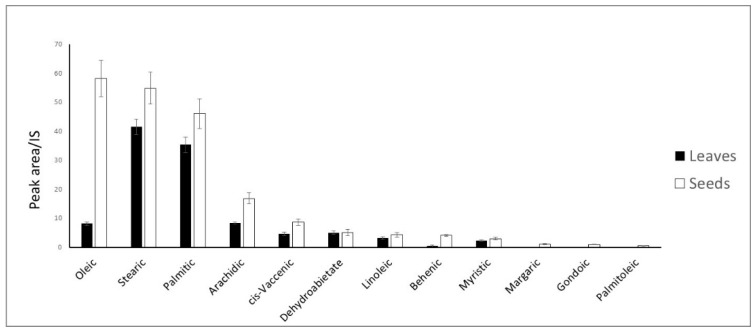
GC-MS spectrometry was used to analyze the apolar extracts from leaves and seeds from a qualitative and quantitative point of view. All of the identified metabolites were present in a larger quantity in apolar seed extract compared to apolar leaf extract (Figure 5).
Figure 5.
Quantitative data of the metabolites identified in leaves and seeds apolar extracts. Shown data refers to the mean and standard deviation of three replicates.
Oleic acid is the one present in the largest amount in the apolar seed extract, followed by stearic acid and palmitic acid. Oleic acid is also the fatty acid that shows the largest concentration difference between leaf and seed apolar extracts. On the contrary, stearic acid was the most abundant fatty acid in leaves, followed by palmitic acid. Arachidic acid was also quite abundant, especially for seed apolar extract.
All of the other detected fatty acids, including cis-vaccenic, linoleic, behenic, myristic, lauric, margaric, gondoic, palmitoleic, and lignoceric acid, were present in minor amounts. Gondoic and palmitoleic acid were only detected in seed extracts.
Oleic acid represents the major metabolite among the unsaturated fatty acids in the seeds (58.18 ± 6.32), while it accounted only for 8.17 ± 0.57 in the leaves. (Figure 5). In addition to fatty acids, two terpenoids were identified as methyl abiet-8-en-18-oate, present only in leaves, and methyl dehydroabietate, detected in both apolar extracts.
2.3. Antimicrobial Activity
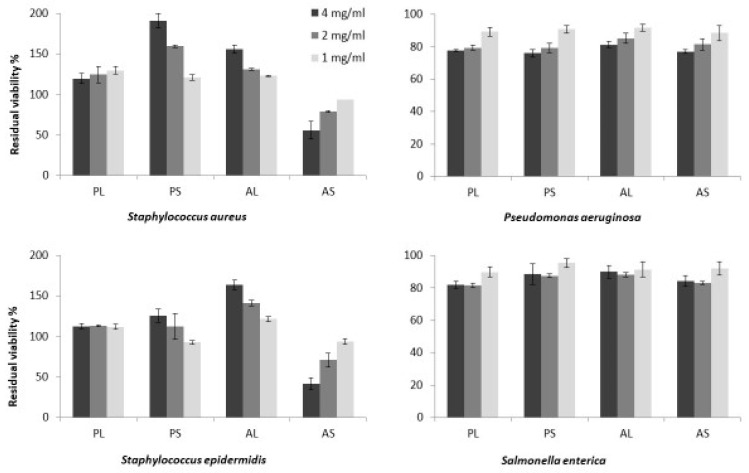
Polar and apolar extracts obtained from the leaves and seeds of M. oleifera were tested for antimicrobial activity against two Gram-positive (Staphylococcus aureus and Staphylococcus epidermidis) and two Gram-negative (Pseudomonas aeruginosa and Salmonella enterica) pathogens (Figure 6).
Figure 6.
Antimicrobial activity of the M. oleifera extracts: PL = polar leaves extract; PS = polar seeds ectract; AL = apolar leaves extract; AS = apolar seeds extract, tested at 4 mg/mL, 2 mg/mL and 1 mg/mL. Bacterial viability was assessed by measuring the optical density at 600 nm and expressed as percentage of residual viability compared to the untreated samples. Showed data refers to the mean and standard deviation of three replicates.
In detail, microbial cultures of all of the tested pathogens were incubated for 24 h in presence and in absence of extracts at 37 °C. Then, the residual viability of treated cultures was evaluated in comparison to the respective untreated samples. Data reported in Figure 6 show that neither polar nor apolar extracts obtained from leaves and seeds significantly affected the microbial growth of the tested Gram-negative bacteria, P. aeruginosa and S. enterica. Whereas, among all, only apolar extracts obtained from seeds (AS) showed a clear antimicrobial activity against the tested Gram-positive pathogens (Figure 6). Moreover, AS induced a reduction in microbial viability in a dose-dependent manner, with an effectiveness of around the 50% at a concentration of 4 mg/mL, on both the tested Staphylococcus species.
3. Discussion
Chemical characterization of M. oleifera was carried out using an integrated approach based on 1H NMR and GC-MS analyses.
Regarding the polar fraction extracted from leaves and seeds, twenty-nine metabolites were identified and quantified (Table 1 and Figure 4). The main metabolites are amino acids, eleven of which were annotated. Generally, they were more abundant in leaves than in seeds. Six carbohydrates were identified and all of them were more abundant in seed than in leaf extracts. Only five organic acids were identified, but signals for succinic and acetic acids were overlapped and, thus, only three of them were quantified. Lastly, among other compounds adenosine, caffeic acid, choline, ethanolamine, quercetin, trigonelline and the general class of the flavonoids were identified and quantified, and all of them were more abundant in leaves except for choline. The chemical composition found by the methods used in this study is similar to the one found by a previous article [20].
The GC-MS analysis resulted in the identification of twelve metabolites, all of them being more present in seeds than in leaves (Figure 5). Oleic acid is the main fatty acid, six times more abundant in seeds than in leaves, and with cis-vaccenic, linoleic, gondoic and palmitoleic acid, it represents the unsaturated fraction of the oil. Conversely, the saturated fraction is composed of stearic, palmitic, arachidic, behenic, myristic and margaric acid.
The antimicrobial assay showed the apolar fraction of the seeds exerts significant antimicrobial activity against the Gram-positive bacteria S. aureus and S. epidermidis, while it was not active against S. enterica. and P. aeruginosa (Figure 6). These results agree with the literature [21,22] reporting on antimicrobial activity for M. oleifera seed oil against S. aureus [21]. Similar activity has been reported for Moringa peregrina oil [22]. Conversely, no antimicrobial effects were found for aqueous extracts from leaves [23]. However, studies have detected antimicrobial activity for polar extracts from leaves and seeds, but at concentrations higher than the ones used in our study [24,25,26]. Moreover, other studies found no antimicrobial effects against S. aureus for seeds petroleum ether extract [27,28].
The different effects between the apolar fractions of seeds and leaves could be due to the remarkable difference in fatty acids content of the two fractions. It is well known that free fatty acids and monoglycerides can exert an antimicrobial effect, especially against Gram-positive bacteria [29,30,31,32] that is dependent on the number of double bonds, since unsaturated fatty acids seem to be more active than saturated, and there is a correlation between the number of double bonds possessed by a fatty acid and its antimicrobial activity [31]. The antimicrobial activity of fatty acids also depends on the length of the carbon chain. The mechanism of action is still under debate, but it could be due to the detergent properties of free fatty acids that can be disruptive for the cellular membrane integrity or for the functionality of the enzymes involved in the electron transport chain.
4. Materials and Methods
4.1. Chemicals and Solvents
n-hexane and methanol were obtained from Delchimica Scientific Laboratories (Naples, Italy). Deuterium oxide (D2O, 99.8 atom% D), used in NMR experiments, was obtained from ARMAR Chemicals (Döttingen, Switzerland). Dimethyl—4—silapentane sodium sulphonate (DSS), used in NMR experiments, was purchased from Merck (Darmstadt, Germany).
4.2. Plant Material
Moringa oleifera dried leaves and seeds were bought from a local shop, in Nablus, Palestine. Leaves and seeds were grinded using a kitchen mixer to obtain a fine powder that was subjected to further extraction procedure and analyses.
4.3. Metabolite Extraction
The metabolite extraction was performed following the method described in de Falco et al. [33]. A total of 4 g of powdered leaves and seeds were extracted with 50 mL of n-hexane, stirring the solution for 1 h. Then, the apolar extracts obtained were filtered, transferred into vials, and dried at room temperature. The remaining pellets were subjected to polar extractions using 50 mL of a methanol/water solution (1:1), stirring the solution for 1 h. The supernatants were then separated by filtration and centrifuged at 3000 rpm for 10 min, RT. The extracts were then dried using a rotary evaporator (30 °C). Both apolar and polar extracts were stored at 4 °C waiting for further analyses. The procedure was performed in triplicate for each plant material analyzed.
4.4. 1H-NMR Analysis
A total of 10 mg of the polar extracts were solubilized in 600 μL of D2O and transferred into a 5 mm NMR tube. Then DSS was added as internal standard at a concentration of 0.2 mg/mL. The NMR spectra were recorded at 298 K on a Varian Unity Inova spectrometer operating at 600 MHz. The 1H-NMR experiments were performed with 128 transients and 16 K complex data point. The recycle time was set to 5 s, and a 45° pulse angle was used. Chemical shifts were referred to DSS signal (Δ 0.00 ppm). All spectra were processed using iNMR program (www.inmr.net, Zimmar Holdings Ltd Company, Westmount, Canada), phased and baseline corrected. In total, 12 spectra were acquired. Quantification was performed by signal integration relative to the internal standard, DSS, as described in Lanzotti et al. [34]. The region of the solvent peaks was excluded from the analysis. Spectral peak assignments of the detected compounds were obtained based on pure standards purchased by Sigma-Aldrich, Saint Louis, MO, USA, and on combined comparison with data reported in the literature and in Human Metabolome Database (HMDB). All spectra were manually phased and baseline corrected.
4.5. GC-MS Analysis
To obtain stable and volatile compounds, apolar extracts were derivatized by using a methanolysis reaction before GC-MS analysis. For this purpose, an aliquot of each apolar extract (0.5 mg) was transferred into a vial and dissolved in 1 mL of methanol + hydrochloric acid 1 N. The vials were vortexed and left at 50 °C overnight, then the reaction mixtures were dried under nitrogen, solubilized in n-hexane and analyzed by GC-MS. According to the method described in Grauso et al. [35]. 1 μL of derivatized samples were injected in a pulsed splitless mode into an Agilent—7820A GC system with 5977E MSD operating in electrospray ionization (EI) mode at 70 eV [35]. The system was equipped with a 30 m × 0.25 mm inner diameter (i.d.) fused—silica capillary column with 0.25 μm HP—5MS stationary phase (Agilent Technologies, Cheadle, UK). The injection temperature was set at 270 °C. Helium was used as carrier gas at a constant flow rate of 1 mL/min. Separation of the apolar extract was achieved using a temperature program of 80 °C for 1 min, then ramped at 10 °C/min to 320 °C and held for 1 min. Both chromatograms and mass spectra were evaluated using the MassHunter Qualitative Analysis B.07.00 (Agilent Technologies, Santa Clara, CA, USA). Mass spectra of all detected compounds were compared with fatty acid methyl esters (FAME) as standard compounds and with spectra obtained by the National Institute of Standard and Technologies library NIST MS search. Data were processed with the AMDIS (Agilent Technologies) software to deconvolute co-eluting peaks. The relative amounts of separated metabolites were calculated from total ion chromatography (TIC) by the computerized integrator and by comparison with internal standard, 1—oleoyl—rac—glycerol, added to the apolar extract as described in Grauso et al. [36].
4.6. Antimicrobial Activity Assay
The study included the following species: Stapyloccocus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa and Salmonella enterica. Isolates were obtained from patients hospitalized at the Medical School of the University of Naples Federico II. Specimens were analyzed using PCR assay as described in Romanelli et al. [37].
The antimicrobial effect of M. oleifera polar and apolar extracts from leaves (PL, AL) and polar and apolar extracts from seeds (PS, AS) were evaluated. Briefly, the wells of a sterile 96-well flat-bottomed polystyrene plate were filled with 200 μL of bacterial culture diluted in Tryptic Soy Broth (TSB) to a final concentration of 1 × 106 colony forming units (CFU) mL−1 in absence and in presence of tested extracts. Each extract was first solubilized in dimethyl sulfoxide (DMSO) and then added to the culture medium and tested at concentrations of 4 mg/mL, 2 mg/mL and 1 mg/mL (DMSO final concentrations ≤ 2.5% v/v). Proper negative controls with only DMSO were included in the experiments. After 24 h incubation at 37 °C, the antimicrobial activity was optically evaluated comparing treated and untreated samples by measuring microbial growth at 600 nm wavelength with a microplate reader.
5. Conclusions
The chemical analysis on polar and apolar extracts of M. oleifera leaves and seeds allowed for the characterization of the metabolite profile, and the further identification and quantification of several primary and secondary metabolites by using a combination of 1H NMR and GC-MS analyses. Moreover, both polar and apolar extracts from leaves and seeds were tested to assess their antimicrobial activity. Only the seeds apolar extracts showed a dose-dependent inhibition against S. aureus and S. epidermidis, and it seems to be due to the remarkably different fatty acid content the two apolar extracts. These findings will be useful for further studies aimed to explore the many other biological potential of this very interesting plant.
Acknowledgments
The research has been developed in the frame of the double degree program between the University of Naples Federico II and the University of Nablus. The NMR and GC-MS analyses were performed at the “Laboratorio di Tecniche Spettroscopiche”, Dipartimento di Farmacia, Università di Napoli Federico II. The assistance of Paolo Luciano is gratefully acknowledged.
Author Contributions
Conceptualization, R.C. and V.L.; methodology, A.A., B.d.F., A.R. and L.G.; validation, L.G., R.C. and V.L.; investigation, A.A., B.d.F., M.A., A.R. and L.G.; data curation, A.A., B.d.F., M.A. and A.R.; writing—original draft preparation, A.A. and V.L.; writing—review and editing, M.S., R.C. and V.L.; visualization, A.A., B.d.F. and A.R.; supervision, R.C. and V.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of M. oleifera extracts are available from the authors.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Anwar F., Latif S., Ashraf M., Gilani A.H. Moringa oleifera: A food plant with multiple medicinal uses. Phytother. Res. 2007;21:17–25. doi: 10.1002/ptr.2023. [DOI] [PubMed] [Google Scholar]
- 2.Rani E.A., Arumugam T. Moringa oleifera (Lam)-A nutritional powerhouse. J. Crop Weed. 2017;13:238–246. [Google Scholar]
- 3.Chaudhary K., Chaurasia S. Neutraceutical Properties of Moringa oleifera: A Review. Eur. J. Pharm. Med. Res. 2017;4:646–655. [Google Scholar]
- 4.Palada M.C. Moringa (Moringa oleifera Lam.): A versatile tree crop with horticultural potential in the subtropical United States. HortScience. 1996;31:794–797. doi: 10.21273/HORTSCI.31.5.794. [DOI] [Google Scholar]
- 5.Anzano A., Ammar M., Papaianni M., Grauso L., Sabbah M., Capparelli R., Lanzotti V. Moringa oleifera Lam.: A Phytochemical and Pharmacological Overview. Horticulturae. 2021;7:409. doi: 10.3390/horticulturae7100409. [DOI] [Google Scholar]
- 6.Abdulsalam S., Gital A.A., Misau I.M., Suleiman M.S. Water clarification using Moringa oleifera seed coagulant: Maiduguri raw water as a case study. J. Food Agric. Environ. 2007;5:302–306. [Google Scholar]
- 7.Shakour ZT A., Radwa H., Elshamy A.I., El Gendy AE N.G., Wessjohann L.A., Farag M.A. Dissection of Moringa oleifera leaf metabolome in context of its different extracts, origin and in relationship to its biological effects as analysed using molecular networking and chemometrics. Food Chem. 2023;399:133948. doi: 10.1016/j.foodchem.2022.133948. [DOI] [PubMed] [Google Scholar]
- 8.Panda S., Kar A., Sharma P., Sharma A. Cardioprotective potential of N,α-l-rhamnopyranosyl vincosamide, an indole alkaloid, isolated from the leaves of Moringa oleifera in isoproterenol induced cardiotoxic rats: In Vivo and in vitro studies. Bioorganic Med. Chem. Lett. 2013;23:959–962. doi: 10.1016/j.bmcl.2012.12.060. [DOI] [PubMed] [Google Scholar]
- 9.Kooltheat N., Pankla Sranujit R., Chumark P., Potup P., Laytragoon-Lewin N., Usuwanthim K. An ethyl acetate fraction of Moringa oleifera Lam. inhibits human macrophage cytokine production induced by cigarette smoke. Nutrients. 2014;6:697–710. doi: 10.3390/nu6020697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dangi S.Y., Jolly C.I., Narayanan S. Antihypertensive activity of the total alkaloids from the leaves of Moringa oleifera. Pharm. Biol. 2002;40:144–148. doi: 10.1076/phbi.40.2.144.5847. [DOI] [Google Scholar]
- 11.Araújo L.C.C., Aguiar J.S., Napoleão T.H., Mota F.V.B., Barros A.L.S., Moura M.C., Coriolano M.C., Coelho L.C.B.B., Silva T.G., Paiva P.M.G. Evaluation of cytotoxic and anti-inflammatory activities of extracts and lectins from Moringa oleifera seeds. PLoS ONE. 2013;8:e81973. doi: 10.1371/journal.pone.0081973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y., Chen G., Guo M. Correlations between phytochemical fingerprints of Moringa oleifera leaf extracts and their antioxidant activities revealed by chemometric analysis. Phytochem. Anal. 2021;32:698–709. doi: 10.1002/pca.3016. [DOI] [PubMed] [Google Scholar]
- 13.Khalafalla M.M., Abdellatef E., Dafalla H.M., Nassrallah A.A., Aboul-Enein K.M., Lightfoot D.A., El-Deeb F.E., El-Shemy H.A. Active principle from Moringa oleifera Lam leaves effective against two leukemias and a hepatocarcinoma. Afr. J. Biotechnol. 2010;9:8467–8471. doi: 10.5897/AJB10.996. [DOI] [Google Scholar]
- 14.Tiloke C., Anand K., Gengan R.M., Chuturgoon A.A. Moringa oleifera and their phytonanoparticles: Potential antiproliferative agents against cancer. Biomed. Pharmacother. 2018;108:457–466. doi: 10.1016/j.biopha.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 15.Almatrafi M.M., Vergara-Jimenez M., Murillo A.G., Norris G.H., Blesso C.N., Fernandez M.L. Moringa leaves prevent hepatic lipid accumulation and inflammation in guinea pigs by reducing the expression of genes involved in lipid metabolism. Int. J. Mol. Sci. 2017;18:1330. doi: 10.3390/ijms18071330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter N., Siddhuraju P., Becker K. Evaluation of nutritional quality of moringa (Moringa oleifera Lam.) leaves as an alternative protein source for Nile tilapia (Oreochromis niloticus L.) Aquaculture. 2003;217:599–611. doi: 10.1016/S0044-8486(02)00497-0. [DOI] [Google Scholar]
- 17.Abd Rani N.Z., Kumolosasi E., Jasamai M., Jamal J.A., Lam K.W., Husain K. In Vitro anti-allergic activity of Moringa oleifera Lam. extracts and their isolated compounds. BMC Complement. Altern. Med. 2019;19:1–16. doi: 10.1186/s12906-019-2776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong Y., Riaz Rajoka M.S., Zhang M.X., He Z. Isolation and identification of two new compounds from the seeds of Moringa oleifera and their antiviral and anti-inflammatory activities. Nat. Prod. Res. 2020;36:974–983. doi: 10.1080/14786419.2020.1851218. [DOI] [PubMed] [Google Scholar]
- 19.Sumner L.W., Mendes P., Dixon R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry. 2003;62:817–836. doi: 10.1016/S0031-9422(02)00708-2. [DOI] [PubMed] [Google Scholar]
- 20.Mahmud I., Kamal C., Arezue B. Tissue-specific metabolic profile study of Moringa oleifera L. using nuclear magnetic resonance spectroscopy. Plant Tissue Cult. Biotechnol. 2014;24:77–86. doi: 10.3329/ptcb.v24i1.19214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinesha B.L., Nidoni U., Ramachandra C.T., Naik N., Sankalpa K.B. Effect of extraction methods on physicochemical, nutritional, antinutritional, antioxidant and antimicrobial activity of Moringa (Moringa oleifera Lam.) seed kernel oil. J. Appl. Nat. Sci. 2018;10:287–295. doi: 10.31018/jans.v10i1.1619. [DOI] [Google Scholar]
- 22.Lalas S., Gortzi O., Athanasiadis V., Tsaknis J., Chinou I. Determination of Antimicrobial Activity and Resistance to Oxidation of Moringa peregrina Seed Oil. Molecules. 2012;17:2330–2334. doi: 10.3390/molecules17032330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyo B., Masika P.J., Muchenje V. Antimicrobial activities of Moringa oleifera Lam leaf Extracts. Afr. J. Biotechnol. 2011;11:2797–2802. doi: 10.5897/AJB10.686. [DOI] [Google Scholar]
- 24.Jabeen R., Shahid M., Jamil A., Ashraf M. Microscopic evaluation of the antimicrobial activity of seed extracts of Moringa oleifera. Pak. J. Bot. 2008;40:1349–1358. [Google Scholar]
- 25.Oluduro A.O. Evaluation of Antimicrobial properties and nutritional potentials of Moringa oleifera Lam. leaf in South-Western Nigeria. Malays. J. Microbiol. 2012;8:59–67. doi: 10.21161/mjm.02912. [DOI] [Google Scholar]
- 26.Ndhlala A.R., Mulaudzi R., Ncube B., Abdelgadir H.A., Du Plooy C.P., Van Staden J. Antioxidant, antimicrobial and phytochemical variations in thirteen Moringa oleifera Lam. cultivars. Molecules. 2014;19:10480–10494. doi: 10.3390/molecules190710480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saadabi A.M., Abu Zaid I.E. An in vitro antimicrobial activity of Moringa oleifera L. against different groups of microorganisms. Aust. J. Basic Appl. Sci. 2011;5:129–134. [Google Scholar]
- 28.Ruttarattanamongkol K., Petrasch A. Antimicrobial activities of Moringa oleifera seed and seed oil residue and oxidative stability of its cold pressed oil compared with extra virgin olive oil. Songklanakarin J. Sci. Technol. 2015;37:587–594. [Google Scholar]
- 29.Kodicek E., Worden A.N. The effect of unsaturated fatty acids on Lactobacillus helveticus and other Gram-positive microorganisms. Biochem. J. 1945;39:78–85. doi: 10.1042/bj0390078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galbraith H., Miller T.B., Paton A.M., Thompson J.K. Antibacterial activity of long chain fatty acids and the reversal with calcium, magnesium, ergocalciferol and cholesterol. J. Appl. Bacteriol. 1971;34:803–813. doi: 10.1111/j.1365-2672.1971.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 31.Desbois A.P., Smith V.J. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010;85:1629–1642. doi: 10.1007/s00253-009-2355-3. [DOI] [PubMed] [Google Scholar]
- 32.Churchward C.P., Alany R.G., Snyder L.A.S. Alternative antimicrobials: The properties of fatty acids and monoglycerides. Crit. Rev. Microbiol. 2018;44:561–570. doi: 10.1080/1040841X.2018.1467875. [DOI] [PubMed] [Google Scholar]
- 33.de Falco B., Grauso L., Fiore A., Bonanomi G., Lanzotti V. Metabolomics and chemometrics of seven aromatic plants: Carob, eucalyptus, laurel, mint, myrtle, rosemary and strawberry tree. Phytochem. Anal. 2022;33:696–709. doi: 10.1002/pca.3121. [DOI] [PubMed] [Google Scholar]
- 34.Lanzotti V., Anzano A., Grauso L., Zotti M., Sacco A., Senatore M., Moreno M., Diano M., Parente M., Esposito S., et al. NMR metabolomics and chemometrics of lettuce, Lactuca sativa L., under different foliar organic fertilization treatments. Plants. 2022;11:2164. doi: 10.3390/plants11162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grauso L., Emrick S., Bonanomi G., Lanzotti V. Metabolomics of the alimurgic plants Taraxacum officinale, Papaver rhoeas and Urtica dioica by combined NMR and GC–MS analysis. Phytochem. Anal. 2019;30:535–546. doi: 10.1002/pca.2845. [DOI] [PubMed] [Google Scholar]
- 36.Grauso L., Zotti M., Sun W., de Falco B., Lanzotti V., Bonanomi G. Spectroscopic and multivariate data-based method to assess the metabolomic fingerprint of Mediterranean plants. Phytochem. Anal. 2019;30:572–581. doi: 10.1002/pca.2862. [DOI] [PubMed] [Google Scholar]
- 37.Romanelli A., Moggio L., Montella R.C., Campiglia P., Iannaccone M., Capuano F., Pedone C., Capparelli R. Peptides from Royal Jelly: Studies on the antimicrobial activity of jelleins, jelleins analogs and synergy with temporins. J. Pept. Sci. 2011;17:348–352. doi: 10.1002/psc.1316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.