Abstract
Klebsiella pneumoniae is a common cause of septicemia and urinary tract infections. The PCR-supported genomic subtractive hybridization was employed to identify genes specifically present in a virulent strain of K. pneumoniae. Analysis of 25 subtracted DNA clones has revealed 19 distinct nucleotide sequences. Two of the sequences were found to be the genes encoding the transposase of Tn3926 and a capsule polysaccharide exporting enzyme. Three sequences displayed moderate homology with bvgAS, which encodes a two-component signal transduction system in Bordetella pertussis. The rest of the sequences did not exhibit homology with any known genes. The distribution of these novel sequences varied greatly in K. pneumoniae clinical isolates, reflecting the heterogeneous nature of the K. pneumoniae population.
Klebsiella pneumoniae is a common cause of septicemia, pneumonia, and urinary tract infection. As an opportunistic pathogen, K. pneumoniae primarily attacks immunocompromised individuals who are hospitalized and suffer from severe underlying diseases, such as diabetes mellitus or chronic pulmonary obstruction. The virulence factors of K. pneumoniae identified so far include capsular polysaccharides (CPS), lipopolysaccharides, adhesins, and iron acquisition systems. Besides these virulence factors, very little is known about the presence and roles of other gene products that might be participating in the pathogenesis of K. pneumoniae (reviewed in reference 18).
In the last 10 years, the extensive spread of multiple antibiotic-resistant K. pneumoniae strains, especially the extended-spectrum β-lactamase-producing strains, has become a major threat to the ever-increasing number of immunocompromised patients. Therefore, novel targets for drug intervention in the bacterial infections are in urgent demand. Several strategies have been developed in the last few years for the identification of bacterial genes essential for infection. These methods include microarray DNA chips (11), a transposon-based footprinting technique (20), comparative genomics (6), mRNA differential display (1), differential fluorescence detection (25), and in vivo expression technology using the tandem-reporter system (14, 16). The use of these approaches has allowed many different virulence-associated genes to be identified.
Differences in DNA sequence form the basis of the different behavior of bacterial strains. Comparison of the genome sequences of nonpathogenic with pathogenic bacterial strains could yield valuable information. The comparison can be made by a bioinformatic approach if the entire genome sequences of the bacteria are available. Alternatively, the recently developed PCR-based subtractive hybridization method (2, 12, 13) can be used to specifically amplify DNA sequence that are present in the virulent but not the avirulent strain. The genes that are common in both species are subtracted, and the DNA fragments unique to the virulent strain are cloned and characterized. This method requires only a small quantity of genomic DNA and is relatively rapid and simple to perform. We here report the results of using the subtractive hybridization technique in the identification of genomic sequences unique to a highly virulent strain of K. pneumoniae.
The K. pneumoniae strains used in this subtraction study were CG43, a high-virulence strain of the K2 serotype with a 50% lethal dose of 10 CFU for laboratory mice (9), and M5a1, a low-mucoidy, avirulent environmental strain (7). The chromosomal DNA was prepared by incubating the bacteria grown overnight in a buffer containing 50 mM Tris-HCl (pH 8.0) and 0.6% sodium dodecyl sulfate, which was followed by lysozyme digestion, phenol-chloroform extraction, and ethanol precipitation (19). The Bacterial PCR-Select DNA Subtraction Kit (Clontech, La Jolla, Calif.) and a PTC-100 Programmable Thermal Controller (MJ Research, Watertown, Mass.) were used in the DNA subtraction, and the procedure was performed essentially as described by the manufacturer of the kit. After the subtractive hybridization, DNA fragments unique to K. pneumoniae CG43 were amplified by PCR, subcloned into pUC18, and transformed into Escherichia coli XL1-Blue, and the transformants were subjected to β-galactosidase activity selection. Approximately 200 white colonies were obtained, and 30 of them were arbitrarily selected for further analysis. The average size of the subtracted fragments was approximately 400 to 500 bp, as determined by restriction enzyme digestion and agarose gel electrophoresis.
To verify if these novel sequences are indeed unique to K. pneumoniae CG43, a Southern blot analysis was performed with each of the subtracted DNA fragments as a probe. Among them, five of the clones were also present in M5a1 and were not investigated further. The nucleotide sequences of the rest of the 25 clones were determined with an ABI-377 Autosequencer. The sequence data were analyzed through the World Wide Web by using the BLAST programs (3), provided by the National Center for Biotechnology Information, and the SeqWeb program of the Genetics Computer Group, provided by the National Health Research Institute, Taiwan, Republic of China.
Analysis of the 25 sequences has revealed a total of 19 distinct sequences. These sequences have been deposited in the EMBL and GenBank sequence databases, and the accession numbers are listed in Table 1. Among them, two sequences were found to match previously identified genes, including genes encoding the transposase of Tn3926 (23) and the capsule polysaccharide export enzyme (4). Three sequences, AJ293850 to AJ293852, displayed moderate similarity to bvgAS (Bordetella virulence genes) of Bordetella pertussis and evgAS, the E. coli homolog of bvgAS (5, 24). The identities between the amino acid sequences predicted from these three clones and BvgAS were 23 to 33%. The rest of the sequences did not exhibit notable homology with any data file in EMBL and GenBank. Further comparison of these 14 sequences with the genome of K. pneumoniae strain MGH78578, elucidated recently by the Genomic Sequencing Center at Washington University (St. Louis, Mo.), has revealed that 11 of the sequences were not present in the database and appeared to be novel. Analysis of the K. pneumoniae MGH78578 contigs containing sequences AJ293847, AJ293848, and AJ293849 did not reveal any known gene that linked closely with these sequences.
TABLE 1.
Characteristics of K. pneumoniae CG43-specific DNA fragments
| Accession no. | Protein sequence identity (%) | Positive bacteremic strains (%) |
|---|---|---|
| AJ293853 | CPS export protein (100) | 76 |
| AJ293846 | Transposase for Tn3926 (93) | 94 |
| AJ293850 | BvgA (33) | 15 |
| AJ293851 | BvgS (25) | 15 |
| AJ293852 | BvgS (23) | 15 |
| AJ293847 | MGH78578 contig 884 | 33 |
| AJ293848 | MGH78578 contig 910 | 46 |
| AJ293849 | MGH78578 contig 915 | 31 |
| AJ276466 | Novel sequence | 46 |
| AJ276464 | Novel sequence | 37 |
| AJ276465 | Novel sequence | 46 |
| AJ276849 | Novel sequence | 24 |
| AJ276850 | Novel sequence | 56 |
| AJ276851 | Novel sequence | 48 |
| AJ276852 | Novel sequence | 70 |
| AJ276853 | Novel sequence | 18 |
| AJ276854 | Novel sequence | 6 |
| AJ276855 | Novel sequence | 32 |
| AJ276857 | Novel sequence | 39 |
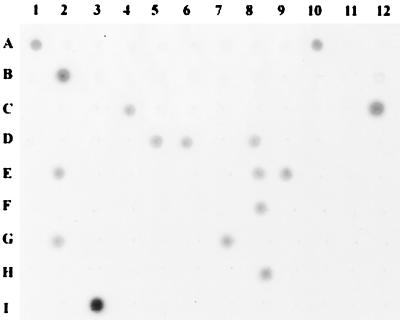
The distribution of a gene in a bacterial population can provide an implication for its physiological role. In order to understand the significance of these novel sequences in pathogenesis, we have investigated the distribution of these sequences in pathogenic K. pneumoniae cells by using dot blot analysis. Ambiguous results were verified by using Southern blot hybridization and PCR. A panel of 96 bacteremic, nonoutbreak strains of K. pneumoniae was chosen on the basis of distinct isolation times and types of primary infection. These clinical isolates, numbered from CG1 to CG129, were recovered from blood specimens of patients obtained during the period of 1987 to 1990 (17). These isolates were identified as K. pneumoniae by results of standard laboratory procedures, such as lack of motility, carbohydrate fermentation patterns, positive Voges-Proskauer test, and negative indole production. Chromosomal DNA was taken from each of the strains, and approximately 3 μg of the purified DNA was spotted onto a Hybond-N+ nylon membrane (Pharmacia Amersham Biotech, Piscataway, N.J.). The DNA blot was alkaline denatured, followed by neutralization and UV cross-linking. Hybridization of the blot was performed with the Gene Images Random Prime Labeling kit (Amersham Pharmacia), and a typical result is shown in Fig. 1.
FIG. 1.
Distribution of bvgS-like gene in K. pneumoniae clinical isolates. Cell lysates of 95 different K. pneumoniae blood isolates were spotted on a nitrocellulose membrane, and the presence of the bvgS homolog was detected by using labeled AJ293852 DNA as a probe. A1, K. pneumoniae CG43; A2, M5a1; A3, E. coli XL-1Blue; A4 to I2, different K. pneumoniae clinical isolates; I3, purified AJ293852 DNA.
As expected, all the novel sequences were found to be present in K. pneumoniae CG43 and absent in M5a1. The distribution of these sequences in the clinical isolates of K. pneumoniae is somewhat random, ranging from 6 to 94%. The results suggest that the pathogenic K. pneumoniae population is highly heterogeneous. Except for CG43 and CG13, none of the other bacterial strains possess all these sequences. On the other hand, none of the sequences are present in all the clinical isolates. Therefore, these DNA sequences may serve as supplementary genetic markers for epidemiological typing of K. pneumoniae strains.
In general, genes identified with subtractive hybridization are not essential for the growth of the bacteria in regular enriched media. Rather, they are likely to play a role in selective circumstances, such as colonization and multiplication in the host. This notion is reflected in this study by the identification of genes responsible for CPS biosynthesis. In vitro, CPS is not essential for the growth of K. pneumoniae in regular culture media, since CPS-deficient mutants can be readily isolated (9). However, CPS has been shown to be a major virulence factor in K. pneumoniae, presumably functioning as a barrier against antibacterial reactions, such as phagocytosis by polymorphonucleated cells in the host (10, 21, 22). The identification of several DNA segments of the capsular biosynthesis gene cluster may explain why strain CG43 is so much more mucoid and virulent than M5a1. Thus, it will be interesting to know whether some of these newly identified genes might be responsible for different degrees of virulence. Further analysis of these DNA sequences will provide important information on the pathogenesis of K. pneumoniae.
The B. pertussis bvgAS gene encodes a two-component signal transduction system that plays an important role in pathogenesis. It has been shown that when B. pertussis invades the host, BvgS is responsible for sensing the external signals and switches on the transcriptional activator BvgA by phosphorylation. The activated BvgA then regulates the expression of a number of virulence factors, such as pertussis toxin and adenylate cyclase toxin (8, 15). On the basis of their sequence similarity, the regulatory mechanism exerted by the K. pneumoniae bvgAS homolog identified in this study may be very similar to that of bvgAS in B. pertussis. The presence of the bvgAS-like genes in only about 15% of the blood isolates of K. pneumoniae (Fig. 1) suggests that it is not essential for the survival of the bacterium in blood circulation. Gene disruption experiments investigating the physiological role of this novel two-component system are ongoing.
Nucleotide sequence accession numbers. Novel sequences have been deposited to the EMBL and GenBank databases under accession no. AJ276464 to AJ276466, AJ276849 to AJ276855, AJ276857, and AJ293846 to AJ293853.
Acknowledgments
This work was supported in part by grants from the National Science Council of the Republic of China to H.L.P. (NSC-86-2316-B182-007) and to H.Y.C. (NSC-86-2314-B182-080).
REFERENCES
- 1.Abu Kwaik Y, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants N S, Fradkov A, Diatchenko L, Hill J E, Siebert P D, Lukyanov S A, Sverdlov E D, Berg D E. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:13108–13113. doi: 10.1073/pnas.95.22.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local aligment search tools. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 1995;177:1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aricó B, Scarlato V, Monack D M, Falkow S, Rappuoli R. Structural and genetic analysis of the bvg locus in Bordetella species. Mol Microbiol. 1991;5:2481–2492. doi: 10.1111/j.1365-2958.1991.tb02093.x. [DOI] [PubMed] [Google Scholar]
- 6.Arigoni F, Talabot F, Peitsch M, Edgerton M D, Meldrum E, Allet E, Fish R, Jamotte T, Ourchod M L, Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16:851–857. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 7.Bender H. Cyclodextrin-Glucanotransferase von Klebsiella pneumoniae. 1. Synthese, Reinigung und Eigenschaften des Enzyms von Klebsiella pneumoniae M5a1. Arch Microbiol. 1977;111:271–282. doi: 10.1007/BF00549366. [DOI] [PubMed] [Google Scholar]
- 8.Boucher P E, Stibitz S. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J Bacteriol. 1995;177:6486–6491. doi: 10.1128/jb.177.22.6486-6491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang H Y, Deng W L, Lee J H, Fu T F, Peng H L. Virulence and outer membrane properties of a Klebsiella pneumoniae galU mutant. Microb Pathog. 1996;20:255–261. doi: 10.1006/mpat.1996.0024. [DOI] [PubMed] [Google Scholar]
- 10.Cryz S J, Furer E, Germanier R. Protection against fatal Klebsiella pneumoniae burn wound sepsis by passive transfer of anticapsular polysaccharide. Infect Immun. 1984;45:139–142. doi: 10.1128/iai.45.1.139-142.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Saizieuu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–50. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 12.Duguid J R, Dinauer M C. Library subtraction of in vitro cDNA libraries to identify differentially expressed genes in scrapie infection. Nucleic Acids Res. 1990;18:2789–2792. doi: 10.1093/nar/18.9.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedrick S M, Cohen D J, Neilson E A, Davis M M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 14.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karimova G, Bellalou J, Ullmann A. Phosphorylation-dependent binding of BvgA to the upstream region of the cyaA gene of Bordetella pertussis. Mol Microbiol. 1996;20:489–496. doi: 10.1046/j.1365-2958.1996.5231057.x. [DOI] [PubMed] [Google Scholar]
- 16.Mahan M J, Tobias J W, Slauch J M, Collier P C, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng H L, Wang P Y, Wu C L, Chiu C T, Chang H Y. Molecular epidemiology of Klebsiella pneumoniae. Chin J Microbiol Immunol. 1991;24:264–271. [PubMed] [Google Scholar]
- 18.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simoons-Smit A M, Verweij-van Vught A M, MacLaren D M. The role of K antigen as virulence factors in Klebsiella. J Med Microbiol. 1986;21:133–137. doi: 10.1099/00222615-21-2-133. [DOI] [PubMed] [Google Scholar]
- 22.Tomás J M, Benedí V J, Ciurana B, Jofre J. Role of capsule and O antigen in resistance of Klebsiella pneumoniae to serum bactericidal activity. Infect Immun. 1986;54:85–89. doi: 10.1128/iai.54.1.85-89.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner A K, Grinsted J. DNA sequence of the transposase gene of the class II transposon, Tn3926. Nucleic Acids Res. 1989;17:1757. doi: 10.1093/nar/17.4.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Utsumi R, Katayama S, Taniguchi M, Horie T, Ikeda M, Igaki S, Nakagawa H, Miwa A, Tanabe H, Noda M. Newly identified genes involved in the signal transduction of Escherichia coli K-12. Gene. 1994;140:73–77. doi: 10.1016/0378-1119(94)90733-1. [DOI] [PubMed] [Google Scholar]
- 25.Valdivia R H, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:67–78. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]