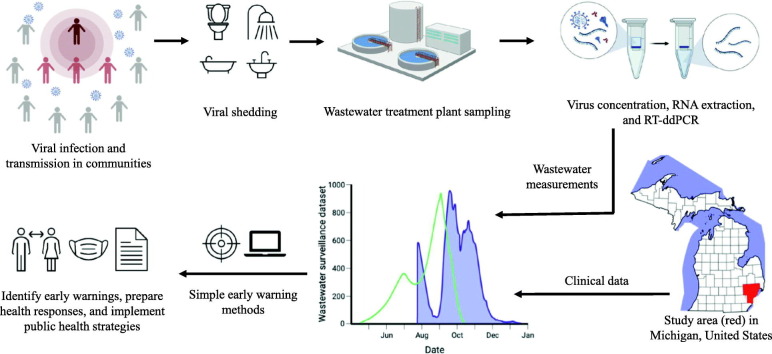
Abstract
Wastewater-based epidemiology (WBE) has drawn great attention since the Coronavirus disease 2019 (COVID-19) pandemic, not only due to its capability to circumvent the limitations of traditional clinical surveillance, but also due to its potential to forewarn fluctuations of disease incidences in communities. One critical application of WBE is to provide “early warnings” for upcoming fluctuations of disease incidences in communities which traditional clinical testing is incapable to achieve. While intricate models have been developed to determine early warnings based on wastewater surveillance data, there is an exigent need for straightforward, rapid, broadly applicable methods for health departments and partner agencies to implement. Our purpose in this study is to develop and evaluate such early-warning methods and clinical-case peak-detection methods based on WBE data to mount an informed public health response. Throughout an extended wastewater surveillance period across Detroit, MI metropolitan area (the entire study period is from September 2020 to May 2022) we designed eight early-warning methods (three real-time and five post-factum). Additionally, we designed three peak-detection methods based on clinical epidemiological data. We demonstrated the utility of these methods for providing early warnings for COVID-19 incidences, with their counterpart accuracies evaluated by hit rates. “Hit rates” were defined as the number of early warning dates (using wastewater surveillance data) that captured defined peaks (using clinical epidemiological data) divided by the total number of early warning dates. Hit rates demonstrated that the accuracy of both real-time and post-factum methods could reach 100 %. Furthermore, the results indicate that the accuracy was influenced by approaches to defining peaks of disease incidence. The proposed methods herein can assist health departments capitalizing on WBE data to assess trends and implement quick public health responses to future epidemics. Besides, this study elucidated critical factors affecting early warnings based on WBE amid the COVID-19 pandemic.
Keywords: Wastewater-based epidemiology, SARS-CoV-2, COVID-19, Early warning methods, Peak-defining methods, Public health response
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been spreading worldwide since its first identification in Wuhan, China, in December 2019. Since SARS-CoV-2 persists in human bodily fluids and excretions, including saliva, sputum, urine, and feces, numerous studies have applied wastewater-based epidemiology (WBE), also known as wastewater surveillance, to monitor COVID-19 infections in various global settings (Ahmed et al., 2020a; Ahmed et al., 2021, Ahmed et al., 2022; Barua et al., 2022; Bivins and Bibby, 2021; Corchis-Scott et al., 2021; Li et al., 2022; Miyani et al., 2020, Miyani et al., 2021; Sherchan et al., 2020; Xiao et al., 2022; Zhao et al., 2022; Zhu et al., 2022). WBE is a comparatively inexpensive and less laborious tool than clinical surveillance for tracking disease incidence and/or prevalence within a large-scale community (Safford et al., 2022; Xagoraraki, 2020; Xagoraraki and O’Brien, 2020). WBE provides a comprehensive and anonymous surveillance of both symptomatic and asymptomatic viral disease, making it an ideal complimentary approach to traditional clinical surveillance testing (Bibby et al., 2021; Safford et al., 2022). A recent study reported that WBE can conserve financial resources without altering surveillance accuracy by replacing some of clinical surveillance programs with WBE-based surveillance (Safford et al., 2022). Perhaps most critically, WBE has the potential to provide early warnings of impending disease outbreaks or surges, if translated effectively to a public health setting (Zhao et al., 2022). “Early warnings” in this context refers to the early detection of relevant pathogen fluctuations within a community, providing a critical window to mount a public health response, prior to lagging parallel trends in clinical cases (Bibby et al., 2021; Olesen et al., 2021). Recent studies have proposed early warning algorithms predicated on intricate statistical models, such as autoregressive time series models (Zhao et al., 2022), artificial neural network models (Zhu et al., 2022), and other advanced statistical and machine learning models (Table S1). These sophisticated models are resource- and time-intensive for health departments to calculate and interpret, particularly in the context of a possible emerging threat. Currently, there are scant studies that have investigated or tested early warning methods for COVID-19, using straightforward, reliable, and rapid approaches for health departments. Some attempts to determine early warnings for COVID-19 clinical cases include: using thresholds for wastewater viral RNA concentrations (Zhu et al., 2021b), calculating Epidemic Volatility Index (EVI) (Kostoulas et al., 2021), implementing statistical thresholds such as mean plus two standard deviations (Bowman et al., 2016; Prabdial-Sing et al., 2021), mean and variance (O’Brien and Clements, 2021), kurtosis and skewness (Harris et al., 2020), assessing the ratio between wastewater viral concentrations and clinical cases (w/c ratio) (D’Aoust et al., 2022; Xiao et al., 2022), estimating the percentage change of wastewater viral concentrations and their relationships to clinical cases (Kumar et al., 2021), etc.
Until now, resources have been spent to generate WBE data that are not fully understood or applied. Wastewater surveillance for pathogens is only beneficial if public health practitioners and partner agencies can apply the results to inform policy decisions and guide actions. Henceforth, we propose three clinical case peak-defining methods (Table 1 ) and eight simple-to-calculate early warning methods (Table 2 ) that can be smoothly implemented by public health departments and partner agencies to provide prompt warnings of impending disease incidences or surges. One of the advantages of these methods is that they encompass both real-time and post-factum analyses. Moreover, these methods combine WBE data with clinical data, though a w/c ratio (D’Aoust et al., 2022; Xiao et al., 2022) using the post-factum methods. Lastly, accuracy of these methods was evaluated via “hit rate”, which is subsequently defined. Results indicate that hit rates for all real-time methods and four post-factum methods could reach 100 % under different circumstances, demonstrating successful discernment of clinical case peaks. Thus, these methods can equip local public health officials with a toolset that integrates wastewater surveillance with traditional clinical surveillance data, to provide early warnings for disease outbreaks or surges, and alert officials and the public when action is needed based on warnings identified by the methods. Besides, we also elucidated the impact of policy changes due to COVID-19, and social events in the Detroit metropolitan area on the wastewater viral concentrations and clinical cases over the past two years. In addition, factors that affect applying WBE for disease surveillance and accuracy of early warning methods are discussed.
Table 1.
Methods of defining peak-ranges of confirmed COVID-19 cases.
| Method | Method description | Early warning level / Cutoff level | Peak range |
|---|---|---|---|
| Method I | Data sequence numerical increase or decrease | Continual increase for ≥14 consecutive data points. Peak range begins at day 0. Peaks at the maximum, and ends symmetrically |
10/6/20–12/24/20, 2/27/21–5/1/21, 7/17/21–10/2/21, 10/29/21–1/22/22, 3/31/22–5/31/22 |
| Method II | Variance / mean | Intersection values > variance/mean threshold | 11/4/20-12/13/20, 3/19/21-5/4/21, 10/18/21-10/25/21, 11/5/21-2/7/22, 5/4/22-5/24/22 |
| Method III | Mean – 0.5standard deviation (SD) | Intersection values > mean-0.5SD threshold | 9/25/20-9/27/20, 10/24/20-1/20/21, 2/8/21-2/21/21, 3/12/21-5/10/21, 5/15/21-5/31/21, 7/29/21-9/6/21, 9/14/21-2/17/22, 4/18/22-5/31/22 |
Table 2.
Early warning methods.
| Type of analysis | Early warning parameter | Early warning level / cutoff level | Abbreviation | Data type |
|---|---|---|---|---|
|
Real-time analysis |
Data sequence numerical increase or decrease | Keep increasing for 3 consecutive data points | OBMN1N2⁎ | N1 N2 gene concentrations |
| Positive percentage change | > 40 % | PPCN1N2 | N1 N2 gene concentrations | |
| Positive percentage change of slope | > 200 % | PPCS200⁎ | N1 N2 gene concentrations | |
|
Post-factum analysis |
Mean + 2 standard deviation | Intersection values higher than mean + 2SD threshold | MSD (B1) ⁎ | N1 N2 gene concentrations and w/c ratio |
| Variance / mean | Intersection values higher than variance/mean threshold | VAM (B2) | ||
| Skewness | Intersection values higher than skewness threshold | SKE (B3) | ||
| Kurtosis | Intersection values higher than kurtosis threshold | KUR (B4) | ||
| 90 percentile | Intersection values higher than 90 percentile threshold | PER90 (B5) ⁎ |
Note: (1) B1, B2, B3, B4, B5 are short representation of each statistical method for visualization purposes. (2) * marked methods were demonstrated in figures in the main text.
2. Materials and methods
2.1. Sample collection and laboratory analysis
Untreated wastewater samples were collected twice weekly from the Water Resource Recovery Facility (WRRF) of the Great Lakes Water Authority (GLWA) located in Detroit, Michigan, USA, between September 1, 2020, and May 31, 2022. The WRRF receives wastewater via three main interceptors including the Detroit River Interceptor (DRI), the North Interceptor-East Arm (NIEA), and the Oakwood-Northwest-Wayne County Interceptor (ONWI) from its service area that encompasses greater Detroit. Samples were collected from the three interceptors at the point of discharge into the WRRF. Depending on the suspended solids of wastewater, approximately 10 to 50 L of untreated wastewater passed through NanoCeram electropositive cartridge filters at a rate not >11 L/min using a previously described method (Miyani et al., 2021; Zhao et al., 2022). Viruses were eluted within 24 h after sampling, based on a previously described method (Supplementary Materials: Sampling and Virus Elution) (Miyani et al., 2021; Zhao et al., 2022). Bacteriophage Phi6 was used as a proxy virus to evaluate recovery during virus elution and concentration (Kantor et al., 2021; Ye et al., 2016; Zhao et al., 2022). Recoveries obtained ranged from 10.37 % to 58.96 %, with a mean recovery of 24.91 % (±22.89 %) (Zhao et al., 2022). Viral RNA was extracted using Viral RNA QIAGEN kit (QIAGEN, Germantown, MD, USA), following the manufacturer's protocol with the method described previously (Supplementary Materials: RNA Extraction) (Miyani et al., 2021; Zhao et al., 2022). RT-ddPCR was performed on a QX200 AutoDG Droplet Digital PCR system (Bio-Rad, Hercules, CA, USA), using the One-step RT-ddPCR Advanced Kit for Probes (Bio-Rad, Hercules, CA, USA) in a previously described method (Supplementary Materials: RT-ddPCR) (Zhao et al., 2022). The Limit of Blank (LOB) was determined by examining three types of samples using RT-ddPCR, across four consecutive days, including interceptor samples collected before COVID-19 pandemic, nuclease-free water, and negative process control samples from elution and extraction. The samples before COVID-19 pandemic were collected on February 18, 2018, from the ONWI, NIEA, and DRI interceptors at GLWA using the same methods. Limit of Blank (LOB) for N1 gene ddPCR was determined to be 0.09 gc/μL, and the LOB for N2 gene ddPCR was determined to be 0.08 gc/μL (Zhao et al., 2022). Limit of Detection (LOD) of 0.1 gc/μL with 72.92 % confidence for the N1 gene ddPCR and 0.1 gc/μL with 81.25 % confidence for the N2 gene ddPCR were determined (Zhao et al., 2022).
2.2. WBE and clinical data of COVID-19
Throughout our 21-month surveillance of wastewater in the Detroit metropolitan area, the wastewater surveillance data (September 2020 to May 2022) together with clinical data were implemented with eight proposed early-warning methods and three proposed clinical case peak-defining methods. Publicly available clinical data were accessed on August 30, 2022, for the period between September 25, 2020, and May 31, 2022, for the city of Detroit, as well as Wayne, Macomb, and Oakland counties (michigan.gov) shown in Fig. 1a. Clinical data presented as a 7-day moving average (Menkir et al., 2021) was used for further statistical analysis (Fig. 1b). COVID-19 data was only available per city or county within study area. Lastly, each interceptor received wastewater from portions of each city or county, thus, only the total SARS-CoV-2 concentrations could be correlated to the total COVID-19 cases in each jurisdiction.
Fig. 1.
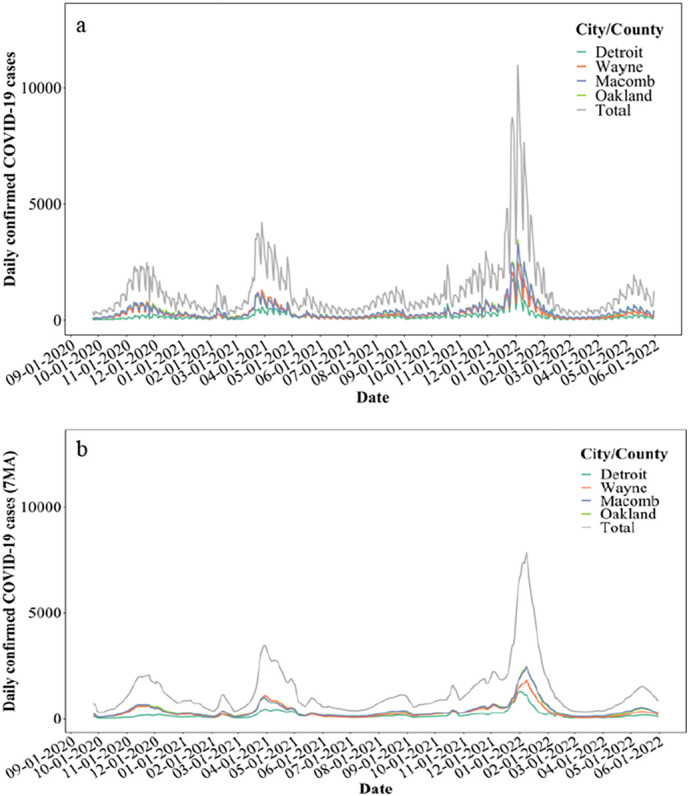
a. COVID-19 cases in the city of Detroit, as well as Wayne, Macomb, and Oakland counties; b. 7-day moving average of COVID-19 cases.
2.3. Data analysis and visualization
Data were tracked and organized with Microsoft Excel (version 16.66.1, Microsoft co. ltd). MATLAB of a 2019b edition (MatLab, 2018) and R version 4.1.3 (Team, 2022) were utilized to perform the early warning analyses, depending primarily on the ggplot2 package for visualization, and the DescTools package for standard deviation, mean, variance, skewness, kurtosis, and quantile for calculation.
Eq. (1). depicts the ratio between the wastewater viral gene concentrations and clinical cases (w/c ratio) (D’Aoust et al., 2022; Xiao et al., 2022), which was first proposed as an indicator of disease incidence based on wastewater surveillance in a recent study (Xiao et al., 2022).
CN1 or N2 gene= N1 or N2 gene concentrations (genomic copies/L, gc/L).
clinical case= daily confirmed COVID-19 cases (7-day moving average)
| (1) |
2.3.1. Methods of defining peaks of COVID-19 cases
Few recent studies have discussed on approaches to defining peaks of clinical cases of COVID-19. O'Brien et al. defined the peak-range of COVID-19 clinical cases to be when the first derivative of cases remains positive for seven consecutive data points (O’Brien and Clements, 2021). Similarly, in this study, we define the peak-range to be when the 7-day moving average of clinical cases continues increasing for over 14 consecutive days (method I, shown in Table 1 and Fig. 2a). The uptick begins at day 0, peaks at the maximum, and ends symmetrically. Method II defines the peak where the intersection values are greater than an established variance/mean threshold, shown in Fig. 2b (Eq. (2)). Similarly, method III uses mean-0.5standard deviation threshold to define the case-peak where the intersection values are greater than the threshold, shown in Fig. 2c (Eq. (3)). Both methods II and III measure the distribution of the clinical cases in the Detroit metropolitan area. The specific “peak ranges” or “surges” determined by these three methods are summarized in Table 1.
| (2) |
Fig. 2.
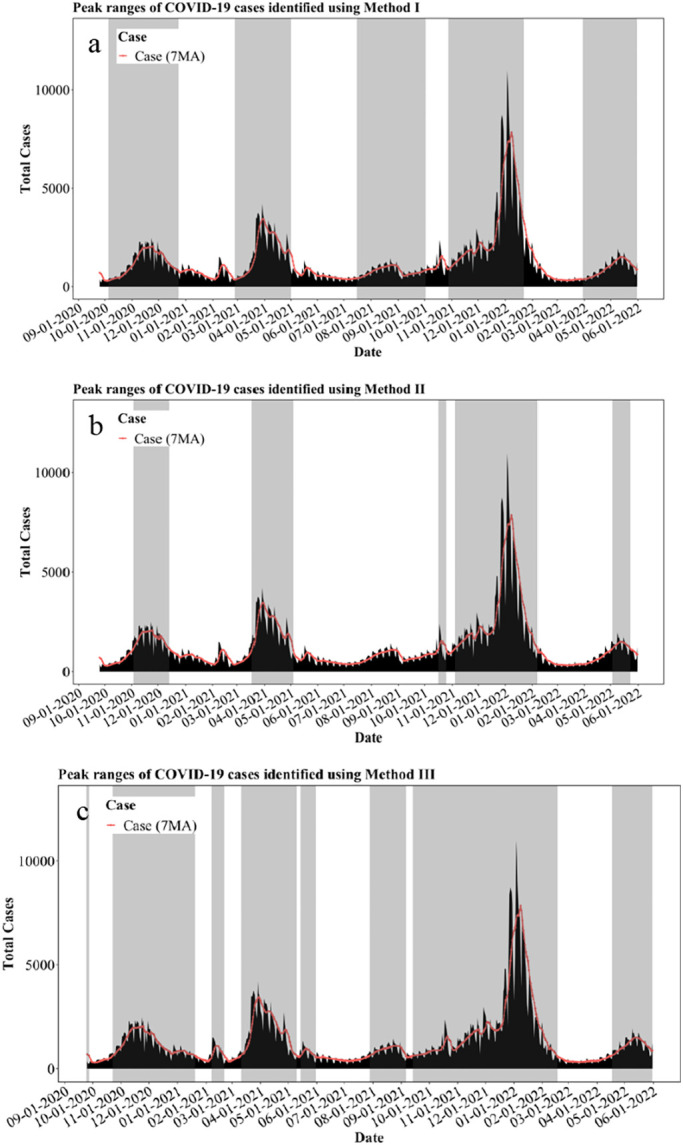
Methods of defining peaks for total COVID-19 cases.
a. Method I defined peaks (gray shaded area) of total COVID-19 cases
b. Method II defined peaks (gray shaded area) of total COVID-19 cases
c. Method III defined peaks (gray shaded area) of total COVID-19 cases.
Vc represents the variance of clinical cases
Mc represents the mean of clinical cases
| (3) |
Sc represents the standard deviation of clinical cases
2.3.2. Real-time early warning methods
As aforementioned, the peak ranges of clinical cases are defined using three methods, namely, methods I, II, and III (Table 1. and Fig. 2). Subsequently, eight methods of early warnings determination (Table 2) were proposed based on literature studies that were elucidated in Section 1. Among them, OBMN1N2, PPCN1N2, and PPCS200 are applied to real-time analysis. OBMN1N2 is applied to real-time N1 and N2 gene concentrations (gc/L) in wastewater, with first early warning dates determined on the final of three consecutively increasing measurements. This method (OBMN1N2) reduces the possibility a “false warning” due to high possible variations of the measured data since OBM requires three consecutive increasing data points to issue a warning. It is also a “non-quantification” method, consistently applicable regardless of the degree of increasing values. The PPCN1N2 method identifies early warnings when the positive percentage change of N1 and N2 gene concentrations (gc/L) are >40 % (Kumar et al., 2021), depicted as Eq. (4):
| (4) |
n indicates the current measurement
n − 1 indicates the previous measurement
Conc.N1 or N2 gene= N1 or N2 gene concentrations (gc/L)
Based on the characteristics of our WBE dataset for the Detroit area, PPCS200 requires the positive percentage change of slope of N1 and N2 gene concentrations (gc/L) to be >200 % to issue early warnings which is depicted in Eq. (6). Percentage change of the slope (Sk) measures the degree of increase between values and can identify values that increased significantly. We have assigned the threshold to be 200 % for the PPCS method, to capture the most meaningful warnings from our WBE data:
| (5) |
| (6) |
n indicates the current measurement or date
n − 1 indicates the previous measurement or date
k indicates the current slope
k − 1 indicates the previous slope
2.3.3. Post-factum early warning methods
Post-factum methods identify early warnings when wastewater surveillance data exceed the thresholds proposed in this study. These methods are designed for post-factum implementations, where both wastewater gene concentration data and clinical data have been reported. An early warning is triggered when the threshold criteria is exceeded. The threshold is computed for an investigation period of interest, after the surges of disease have occurred. For instance, researchers have proposed two standard deviations as an early warning threshold and the time of early warning was determined by the first time the signal exceeded the threshold (Drake and Griffen, 2010). Five statistical thresholds including mean plus two standard deviations (MSD), variance divided by mean (VAM), skewness (SKE), kurtosis (KUR), and 90th-percentile (PER90), were calculated using N1 and N2 gene concentrations (gc/L) and w/c ratio (gc/L/case), to determine early warnings. MSD targets the upper bound limit generated by the two standard deviations away from the mean (Gao et al., 2021; Wang et al., 2017), which is equivalent to a 95 % confidence interval. While PER90 targets the top 10 % of data from the distribution, other studies have applied 70th percentiles, or 80th percentiles to inform early warnings for hand, foot, and mouth disease in China (Gao et al., 2021). VAM is a similar method to MSD, which identifies the variability of the data away from the mean. SKE measures asymmetry of distribution about its mean, and KUR measures the combined weight of a distribution's tails to its center (i.e., whether the plotted shape of the distribution is too sharply “peaked”) (Harris et al., 2020).
2.3.4. Hit rate
Hit rate is introduced to appraise the accuracy of each method – it is defined as the ratio of the number of early warning dates “m” capturing the defined peaks to the total number of early warning dates “n” (see Eq. (7)). The hit rate was calculated in this manner for all early warning methods. As reported by our recent study (Zhao et al., 2022), wastewater signals of N1 and N2 genes preceded the reported clinical cases by up to 5 weeks in the Detroit metropolitan area. Thus, for this study, the number of early warning dates “m” that are said to capture defined peaks must satisfy two criteria: (1) identified early warning dates are located inside the defined peak regions (shaded gray areas in Fig. 2); (2) identified early warning dates are located within a five-week window preceding the defined peak regions.
| (7) |
m= number of early warning dates identified by eight early warning methods, capturing defined peaks, identified by three peak-defining methods
n= total number of early warning dates identified by eight early warning methods
3. Results and discussion
3.1. Wastewater viral concentrations precede disease incidence and can relate to public health policy or community social events
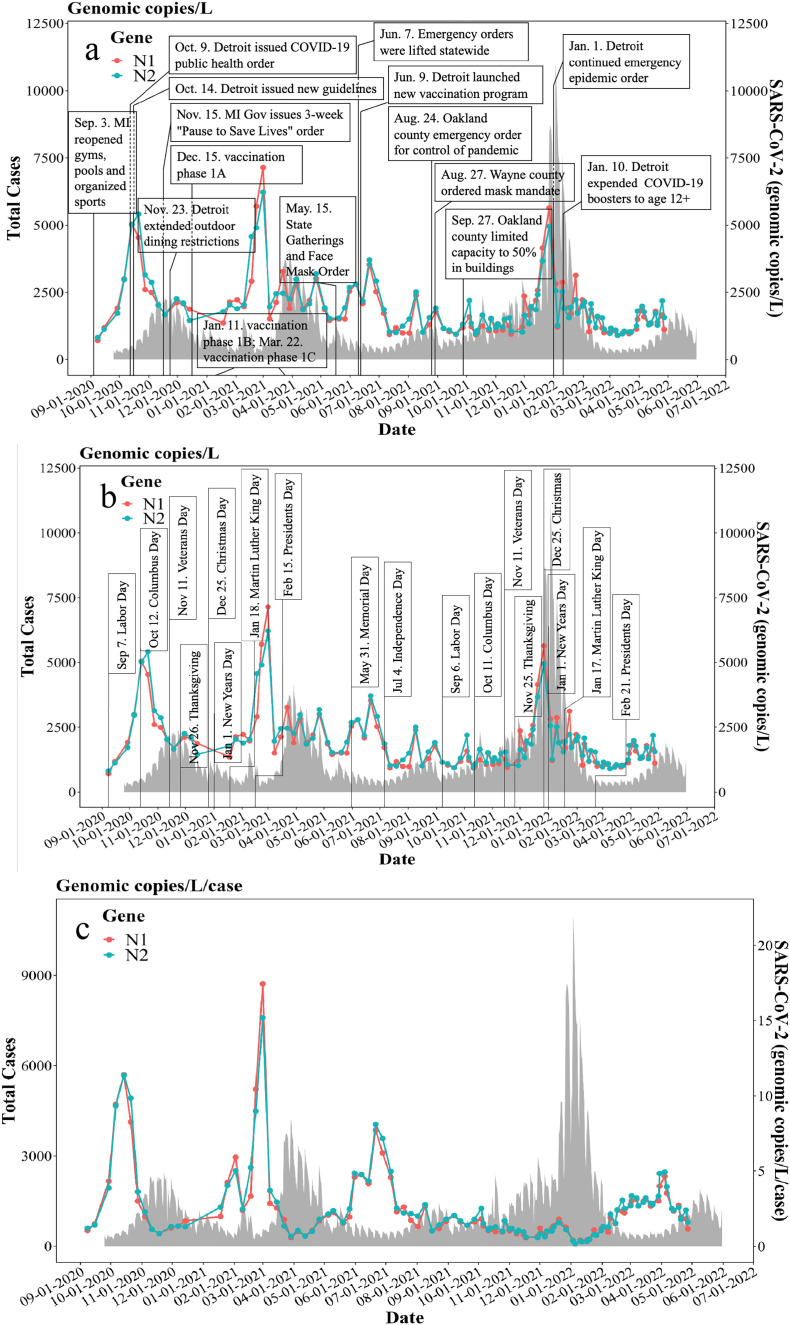
Analysis of our 21-month wastewater surveillance data reveals that the trend of total N1 and N2 gene concentrations preceded and forewarned the trend of total COVID-19 clinical cases (Fig. 3a). Both wastewater viral concentrations and clinical data were compared with calendar dates of major statewide, citywide, and countywide public health policies (Fig. 3a). For instance, both N1 and N2 gene concentrations began to increase shortly after the State of Michigan allowed the reopening of gyms, pools, and permitted organized sports on September 3, 2020. This is suggestive of populations shedding the virus into wastewater after being infected by COVID-19 likely due to unregulated social gatherings (Fig. 3a). Subsequently, both N1 and N2 gene concentrations began to gradually decrease, potentially due to the reduction in SARS-CoV-2 shedding, which persisted up to 24 days (Zhao et al., 2022), as well as due to the implementation of new COVID-19 public health orders and guidelines for Detroit on October 9 and October 14, 2020, respectively (Fig. 3a). Decreasing trends of both wastewater viral concentrations and clinical cases were observed after city of Detroit extended the emergency epidemic order on January 1, 2022 (Fig. 3a). Xiao et al. reported similar trends of wastewater viral concentrations, as affected by state public health policy in Massachusetts, USA (Xiao et al., 2022). Major public health orders and guidelines can affect wastewater viral concentrations as well as subsequent COVID-19 clinical cases by regulating everyday social gatherings and mitigation efforts (Xiao et al., 2022). Wastewater viral concentrations and clinical data were also compared with public holidays and known large-scale social events in the Detroit metropolitan area (Fig. 3b). Public holidays and social events celebrated in Detroit were seen to be reflected in both the wastewater viral concentration and the clinical data (Fig. 3b). It was observed that both N1 and N2 gene concentrations increased after Labor Day (September 7th, 2020) (Fig. 3b), likely resulting from social gatherings during the holiday, as well as the policy of easing COVID-19 restrictions (September 3rd, 2020) in Michigan (Fig. 3a), leading to potentially high transmissions of COVID-19. Similarly, both N1 and N2 gene concentrations began to increase after Martin Luther King Jr. Day (January 18th, 2021) and peaked shortly after Presidents Day (February 15th, 2021). This increase in gene concentration preceded an increase in COVID-19 cases by 4 to 5 weeks (Zhao et al., 2022). Similar observations can be identified with a steeper increase of wastewater viral concentrations as well as the clinical cases after Veterans Day (November 11th, 2021). Clinical cases surged again in early January of 2022 after the Thanksgiving, Christmas, and New Year's day holidays, when social gatherings might be expected during holiday celebrations (Fig. 3b). Notably, the increase of both N1 and N2 gene concentrations in early September of 2020 and early February of 2021, preceding the increase of clinical cases for 4 to 5 weeks (Zhao et al., 2022), can be related to opening of schools, colleges, and universities in fall and spring semesters (Xiao et al., 2022).
Fig. 3.
a. Major statewide, citywide, and countywide COVID-19 public health policies in the Detroit metropolitan area; b. Major public holidays in Michigan, USA; c. w/c ratio between N1 N2 gene concentrations and 7-day moving average of total COVID-19 cases.
Wastewater surveillance has the ability to monitor virtually most members of a community (with an integrated sewage system), regardless of the presence of disease symptoms or inequity in testing accessibility (Bibby et al., 2021). To quantitively compare wastewater data and clinical cases, a ratio (w/c ratio) between the wastewater viral concentrations and 7-day moving average of daily confirmed COVID-19 clinical cases is adopted from recent studies for this purpose (D’Aoust et al., 2022; Xiao et al., 2022). The w/c ratio could reflect potential undercounting or overcounting of actual clinical cases (D’Aoust et al., 2022; Xiao et al., 2022). Undercounting occurs when people do not seek clinical testing, or when access to testing is restricted due to resource limitations, or when there is an elevated rate of asymptomatic infections. This scenario was evident in the summer of 2021 from June to August where the w/c ratio of both N1 and N2 genes was high, but the confirmed cases were low (Fig. 3c). Furthermore, during the summer, cases were likely to be undercounted due to lack of testing and potentially increasing spreading during summer social activities. Some studies have reported similar issues that can result in undercounting, when the actual number of cases is 12 times larger than reported cases (Lau et al., 2021), and the case-to-report ratio could reach 26 to 32 at the early stage of the pandemic when testing sources were limited (Murhekar et al., 2021). Conversely, overcounting can occur when testing resources are abundant and infected populations get tested multiple times during the entire period of infection. These individuals are counted and reported repeatedly as individual clinical cases, since the shedding duration of SARS-CoV-2 can persist up to 24 days prior to the Omicron surge (Zhao et al., 2022) and throat/nasal swab PCR tests can also remain positive up to nearly 20 days (Xiao et al., 2022). Note that the introduction of w/c ratio also eliminates the noises of N1 and N2 gene concentrations among peaks and accentuate the prominent peaks of N1 and N2 gene w/c ratio preceding major peaks of clinical cases (Fig. 3c). From the inception of the pandemic, the increasing trend of w/c ratio in September and October 2020 provided early warnings of upcoming peaks of clinical cases in late October and November 2020 (Fig. 3c). Similarly, the peaks of w/c ratio in late February 2021 forewarned the impending surge of clinical cases in March 2021. The w/c ratio stayed relatively low and stable before the peak of clinical cases in late December 2021 and early January 2022, which indicated that testing sources were sufficient. Moreover, the State of Michigan (michigan.gov/coronavirus) reported that testing capacity statewide increased to approximately 50,000 test results per day in December 2021, corroborating the accuracy of the low w/c ratio in this period. Because as testing capacity increased throughout November and December of 2021, delayed clinical cases were likely reported simultaneously with newly reported cases but the delayed clinical cases did not contribute to the current wastewater viral concentrations, perhaps resulting from reduced viral shedding, leading to lower w/c ratio. Moreover, the surge of the Omicron variant in November and December 2021 in the Detroit metropolitan area could have contributed to shifting transmission dynamics, leading to a substantial increase in reported clinical cases, and thus, contributing to the relatively low w/c ratio (Auwaerter, 2022; Long et al., 2022; Wiersinga et al., 2020; Zhao et al., 2022).
3.2. Early warnings of COVID-19
Eight early warning methods including both real-time and post-factum methods shown in Table 2 were implemented on N1 and N2 gene concentrations (gc/L) as well as w/c ratio (gc/L/case) to identify early warnings for defined peaks of clinical cases. The real-time methods include OBM, PPC, and PPCS, which were applied to direct measurements of N1 and N2 gene concentrations. The post-factum methods include MSD, VAM, SKE, KUR, and PER, which were applied to N1 and N2 gene concentrations as well as w/c ratio. The accuracy of each method was evaluated by hit rates (Table S3).
3.2.1. Early warnings determined by real-time methods
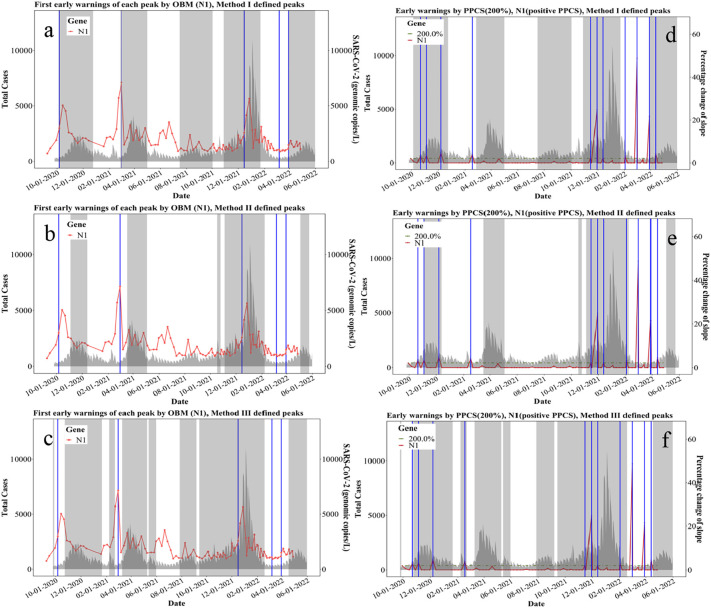
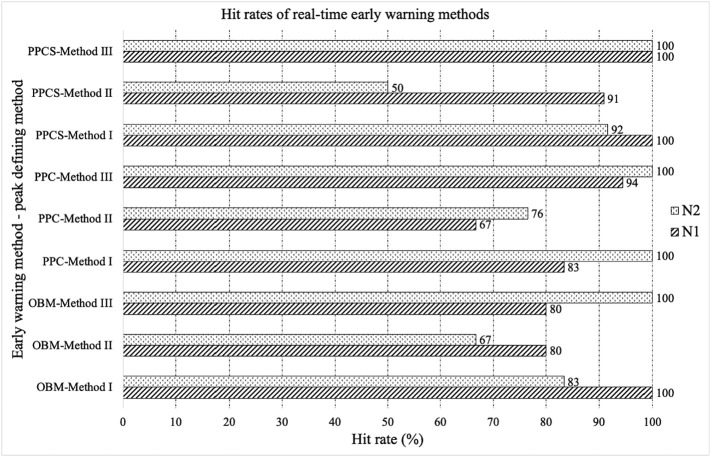
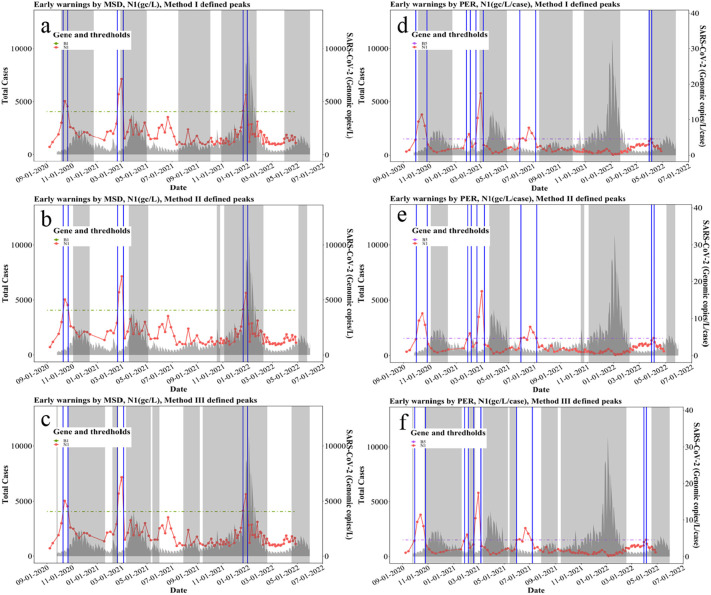
Among real-time methods, OBM method was applied to N1 gene concentration (gc/L) and could reach 100 % hit rate with method I defined peak (Fig. 4a), where all identified early warnings (shown as blue vertical lines in Fig. 4a) are in the defined-peak regions or within a five-week window ahead of the defined-peak regions (shown as gray shaded area in Fig. 4a). In other words, a 100 % hit rate is representative of all identified early warnings successfully forewarning subsequent defined peaks in cases. OBM method could reach 80 % hit rates with both method II and III defined peaks shown in Fig. 4b and c, respectively. Likewise, the application of OBM method to N2 gene concentration (gc/L) results in a 100 % of hit rate with method III defined peaks (Fig. S1c). Specifically, OBM method is based on direct measurements of N1 and N2 gene concentrations and could be immediately applied by health departments after obtaining the data to determine rapid warnings on the upcoming fluctuations of clinical cases. In addition, PPC method could also achieve a 94.44 % hit rate when it is applied to N1 gene concentrations (Fig. S1l) using method III defined peaks, and a 100 % hit rate when it is applied to N2 gene concentrations with both method I (Fig. S1d) and III defined peaks (Fig. S1f). The PPCS method, as applied to N1 gene concentrations, performed the best in terms of higher hit rates where it achieved 90.91 %, 90.91 % and 100 % with method-I, -II, and -III defined peaks, respectively (Fig. 4). PPCS was applied to N2 gene concentrations where it also achieved 91.67 % and 100 % hit rates with method I and III defined peaks, respectively (Table S3, Fig. S1g, S1i). Therefore, PPCS method based on N1 gene concentrations (gc/L) is recommended as a real-time method to capture early warnings with its higher hit rates across three methods for defining clinical peaks. Method III is the recommended as the peak-defining method since it is also more conservative in terms of capturing the most surges and fluctuations of cases. All hit rates developed using real-time methods are summarized in Fig. 6.
Fig. 4.
Real-time early warning methods: OBM and PPCS based on N1 (gc/L):
a. First early warnings of each peak identified by OBM (N1, gc/L) with Method I defined peaks
b. First early warnings of each peak identified by OBM (N1, gc/L) with Method II defined peaks
c. First early warnings of each peak identified by OBM (N1, gc/L) with Method III defined peaks
d. Early warnings identified by PPCS (N1, gc/L) with Method I defined peaks
e. Early warnings identified by PPCS (N1, gc/L) with Method II defined peaks
f. Early warnings identified by PPCS (N1, gc/L) with Method III defined peaks.
Fig. 6.
Hit rates of real-time early warning methods: OBM, PPC, and PPCS, with three peak-defining methods: method I, method II, and method III, based on N1 and N2 gene concentrations (gc/L).
3.2.2. Early warnings determined by post-factum methods
Post-factum methods were applied to N1 and N2 gene concentrations as well as w/c ratio. Selected methods including MSD and PER are illustrated in Fig. 5 . MSD was applied to N1 gene concentrations and reached 100 % hit rates with method I, II, and III defined peaks shown in Fig. 5a, b, and c, respectively, where all identified warnings successfully forewarned the defined peaks. Likewise, PER method was applied to w/c ratio of N1 gene which are illustrated in Fig. 5d, e, and f with method I, II, and III defined peaks, where the hit rates reached 90 %, 70 %, and 100 %, respectively (Table S3). Notably, method II defined less peaks of cases leading to warnings identified by PER between May and July 2021 in vain (Fig. 5e). While method III defined more peaks of cases and covered more data with a wider time range thus leading to higher hit rates (Fig. 5f). From this, we conclude that clinical case peak defining approaches can affect hit rates of early warning methods to forewarn case peaks. Among all post-factum methods applied to N1 and N2 gene concentrations, MSD achieved 100 % hit rates with method I, II, and III defined peaks (Table S3, Fig. 5, Fig. S2). Hence, MSD based on N1 and N2 gene concentrations (gc/L) is recommended as a post-factum method to identify early warnings. In addition, post-factum methods applied to w/c ratio, including MSD (Fig. S3), SKE (Fig. S5), KUR (Fig. S6), and PER (Fig. S2), achieved 100 % hit rates except for VAM (Fig. S4), where KUR completely achieved 100 % hit rates across three methods of defining peaks (Fig. S6) and MSD achieved 100 % hit rates with method-I and -III defined peaks based on both w/c ratio of N1 and N2 genes. Thus, KUR and MSD based on w/c ratio are recommended for as post-factum methods to identify early warnings.
Fig. 5.
Post-factum early warning methods MSD and PER, based on N1 (gc/L) and N1/c (gc/L/case), respectively.
a. Early warnings identified by MSD (N1, gc/L) with Method I defined
b. Early warnings identified by MSD (N1, gc/L) with Method II defined
c. Early warnings identified by MSD (N1, gc/L) with Method III defined
d. Early warnings identified by PER (N1, gc/L/case) with Method I defined peaks
e. Early warnings identified by PER (N1, gc/L/case) with Method II defined peaks
f. Early warnings identified by PER (N1, gc/L/case) with Method III defined peaks.
Worthy of noting, early warning methods achieved generally higher hit rate when they were implemented on N1 and N2 gene concentrations (gc/L) than on the w/c ratio (gc/L/case) dataset. Following this, w/c ratio did not significantly improve the hit rate of early warning methods in our study. Nevertheless, w/c ratio could still be used as an indicator of relationship between actual cases and testing capacity as discussed in Section 3.1 and in recent studies (D’Aoust et al., 2022; Xiao et al., 2022). Among all post-factum methods, MSD method achieved higher hit rates compared with PER, VAM, and SKE methods for both N1 and N2 gene concentrations (gc/L) and w/c ratio (gc/L/case) datasets. In addition, method III could define more peak-ranges and is associated with higher hit rates for both real-time and post-factum methods when compared to method-I and-II.
3.3. Factors affecting early warnings based on WBE and other uncertainties
Few studies have reported on straightforward, easily applied, and rapid early warning methods based on WBE for COVID-19 or other diseases (Bowman et al., 2016; Harris et al., 2020; Kostoulas et al., 2021; O’Brien and Clements, 2021; Prabdial-Sing et al., 2021; Zhu et al., 2021b). The early warning methods developed in the present study including both real-time and post-factum methods, effectively provide early warnings for defined peaks of COVID-19 cases in the Detroit metropolitan area. These methods can potentially be applied to other geographic regions and pathogens; however, further analysis will be necessary. None of the methods designed in this study is perfectly accurate when capturing early warnings of diseases, as numerous complex factors can affect WBE or associated reporting of clinical cases, including physiological, health system, laboratory-based and logistic factors (summarized in Table S2) (Bibby et al., 2021; Kumar et al., 2021; Zhu et al., 2022). According to the authors of a recent WBE study conducted in Boston, Massachusetts, USA, WBE-based early warning methods should not be used in isolation, but rather in conjunction with other methods, given the complexity of factors and various unknowns (Xiao et al., 2022). Despite this recommendation, WBE-based early warning methods, could be essential in providing prompt warnings to impending epidemics, thus aiding health departments to mobilize and craft policy. Critical factors and uncertainties related to early warning methods are elucidated in the following sections.
3.3.1. Physiological factors
Shifting viral shedding cycles and dynamics may affect the accuracy of the relationship between WBE and case incidence, and therefore may affect the early warning potential of WBE (Bibby et al., 2021). Viral shedding dynamics can vary among individuals, variants, and so forth (Bibby et al., 2021; Zhao et al., 2022). Shedding of SARS-CoV-2 began for up to 7 days prior to symptoms onset during the initial stage of the COVID-19 pandemic, but this duration declined to maximum of 3 days during the Omicron surge (Auwaerter, 2022; Cheng et al., 2020; Long et al., 2022; Wiersinga et al., 2020; Zhao et al., 2022). Shedding duration of SARS-CoV-2 shortened from a maximum of 24 days to 10 days during the Omicron surge (Lamers et al., 2022; Zhao et al., 2022). All of the aforementioned dynamics would affect the measured relationship between wastewater viral concentrations and clinical cases, such as the w/c ratio.
Another physiological factor is the poorly understood role of asymptomatic disease transmitters, which lead to an undercounting of true infection cases in communities that employ traditional clinical testing methods (Bibby et al., 2021). Clinical case numbers are not an ideal measure of disease prevalence in a given community, particularly when asymptomatic infections dominate. In this setting, early warning methods based on WBE may not be able to effectively provide early warnings of cases as it will be difficult to establish case-number as reference or baseline. In addition, peak defining approaches can be significantly affected by underestimations, leading to inaccuracy in early warning methods.
The proportion of infected populations who shed detectable levels of virus in their stool and viral load distribution throughout a day, are both significant factors since they have a direct impact on wastewater viral concentrations (Jones et al., 2020). Some studies estimate 48 % to 67 % of infected individuals shed SARS-CoV-2 in their stool (Ahmed et al., 2021). It was also demonstrated that 40 % of infected populations shed SARS-CoV-2 virus RNA in their stool (Kirby et al., 2021). Viral load in wastewater throughout a given day is not evenly distributed, and therefore, some studies have suggested that optimization of sampling strategy coupled with a standardization based on toilet flushing frequency and wastewater travel time could improve the accurate detection of viral signals (Zhu et al., 2021a; Zhu et al., 2021b).
3.3.2. Health system factors
Health system factors encompass a wide range of variables, including the delay of clinical data reporting (Torres et al., 2021; Zhao et al., 2022), prolonged duration of clinical data processing (Contreras et al., 2020), under-reporting of the true number of cases (Kronbichler et al., 2020; Salath et al., 2020), and other inconsistencies in reporting. These factors have the potential to have a prodigious effect on the accuracy of peak-defining methods, including those outlined in this study. These factors also contribute to the disparities of clinical case-numbers, which affect clinical cases used as the reference or benchmark for early warning methods, such as the w/c ratio (Fig. 3c). For instance, at the beginning of the COVID-19 pandemic, several clinical studies reported a delay of nearly 7 days from onset of symptoms to clinical testing, resulting in delayed reporting of clinical cases which already contributed to the wastewater viral concentrations (Huang et al., 2020; Zhao et al., 2022). In some cases, jurisdictions or countries accumulate delays of between 4 and 18 days (Català et al., 2021). Disparities in clinical data can furthermore be influenced by testing hesitancy or eagerness (Jimenez et al., 2021), the turnaround time for screening of cases (Larremore et al., 2021), the availability and accessibility of diagnostic tests (Olesen et al., 2021), and so forth. A shortage of testing supplies and lack of testing in some resource-constrained countries has caused a pileup of clinical samples awaiting results and delays in reporting cases, therefore, inevitably leading to inaccurate representation of actual cases (Torres et al., 2021), which could affect methods of defining peaks thus affecting the accuracy of early warning methods based on WBE.
3.3.3. Laboratory analysis and other uncertainties
Detectable viral signals in wastewater are critical to achieve early warning methods based on WBE. Therefore, potential decay of virus during wastewater transport in the municipal wastewater collection system, sampling, and transportation of samples to laboratories may alter the accuracy of early warning methods (Ahmed et al., 2020b; Bibby et al., 2021). Some recent studies pointed out that sample transportation and laboratory processing including sample concentration, nucleic acid extraction, and PCR-based nucleic acid quantification, may take from one to three days (Bibby et al., 2021; Zhao et al., 2022), but under some scenarios this time range could be easily exceeded, leading to inevitable decay of viral signals in any of the processes. Moreover, recovery efficiency could also vary among different sampling and analytical methods (Ahmed et al., 2020c), introducing more uncertainties in the results.
Recent studies have investigated additional factors that can influence early warning potential of WBE, including sample site geographic distribution, fair sample representation of demographics/communities, uneven mixing of wastewater during sampling and laboratory analysis, dilution of viral RNA during rainfall events, and climate variability (Ahmed et al., 2020b; Bibby et al., 2021; Butler et al., 1995; Kumar et al., 2021; Zhao et al., 2022; Zhu et al., 2022). In addition, the persistence of SARS-CoV-2 in wastewater may be affected by environmental factors. These include temperature, organic matter, and microorganisms, which contribute to more uncertainties of measuring wastewater viral concentrations, ultimately affecting accuracy of early warning methods (Xiao et al., 2022). Overall, the accuracy of reported clinical data and WBE workflows are the primary influencers of effective early warning methods.
3.3.4. Discussions, limitations, and future directions
Real-time methods focus on rapid early warnings using current or recent wastewater measurements for predicting future surging of cases. Real-time methods can be implemented rapidly upon obtaining the wastewater measurements. For instance, the OBM method only requires three consecutive increasing measurements to issue an early warning. The early warning signals determined by real-time methods precede the upcoming increase in cases. Thus, these methods can trigger alerts in real time before surging COVID-19 cases that are subsequently reported by health agencies. Real-time methods can be an effective decision-making tool for public health officials during an on-going epidemic.
Post-factum methods focus on after-the-fact analysis of wastewater and clinical data over an entire period and provide analysis of warnings for surges of cases after they have occurred. Post-factum methods can be used as complementary methodologies to real-time methods. Post-factum methods, such as the MSD method, concentrate on providing an overall picture of early warnings for an entire period of investigations which can identify all early warnings for an entire time (Bowman et al., 2016; Prabdial-Sing et al., 2021). Post-factum methods can be useful to health agencies to plan and design health measures for the next potential epidemic.
Admittedly, this study has various limitations. First, eight early warning methods and three peak-defining methods were successfully applied to WBE data collected from the Detroit metropolitan area, but these methods have not been validated in other regions. More in-depth investigations are warranted to develop and apply early warning methods based on this study. Secondly, the w/c ratio was adopted from a study that assumed that viral shedding did not change significantly over the course of the entire pandemic (Xiao et al., 2022). However, viral shedding patterns and dynamics changed during the study period for the Detroit area where the dominant variant changed from Alpha to Beta, Gamma, Delta, and Omicron variants and so forth (Xiao et al., 2022; Zhao et al., 2022). For example, the lag time of wastewater surveillance preceding the clinical testing declined from five weeks to two weeks during the Omicron surge owning predominately to the changing viral shedding dynamics (Zhao et al., 2022), potentially affecting the performance of early warning methods. Third, the w/c ratio did not capture the third major peak of clinical cases in late December of 2021 and the beginning of January 2022, which perhaps was a consequence of changing shedding dynamics associated with Omicron, as well as the rapidly increasing testing capacity in December 2021 in the State of Michigan leading redundant counting of clinical cases, which we discussed thoroughly in 3.1. Finally, the approaches of defining peaks of COVID-19 cases were potentially specific to our sampling demographic and geographic sampling distribution in the Detroit metropolitan area in this study. Exploration of other methods for defining peaks of clinical cases for other regions seems warranted.
Overall, numerous prospects extended from this study could inspire applications of WBE data and development of early warning methods of WBE for public health benefits. The eight early warning methods described here are straightforward and easily applied, and could forewarn defined peaks with high hit rates, especially the real-time methods OBM and PPCS. The real-time methods require merely the direct measurements of N1 and N2 gene concentrations as well as simple statistical calculations, which are easily applied tools for public health departments to apply on WBE datasets to determine early warnings rapidly. Three methods of defining peaks are easily applied as well. The early warning methods and peak-defining methods proposed in this study attempt to provide rapid and straightforward approaches to determine early warnings for health departments, partner agencies, and the public, instead of applying intricate and sophisticated models. Additionally, this study demonstrates the impact of public policies on wastewater viral concentrations and subsequent clinical cases in Detroit metropolitan area for approximately two years. Combining wastewater data with clinical cases for the Detroit area, such as application of w/c ratio, could allow health departments to understand the actual infections and testing conditions in communities. However, more studies are warranted to establish a standard framework for defining peaks of clinical cases, apply and develop early warning methods that are easily applied by health departments, to use early warnings in a timely manner. This study highlights the impact of public health policy on measured wastewater viral concentrations and clinical COVID-19 cases in Detroit. Future research should integrate public policy at a granular level.
4. Conclusions
This study introduced eight (three real-time and five post-factum) early-warning methods based on wastewater surveillance data and three peak-defining methods based on clinical data that can be easily implemented by public health departments and partner agencies to warn of viral disease fluctuations. Hit rates were calculated to evaluate the efficacy of early warning methods in predicting clinical case surges. Applying these methods to a 21-month WBE data set in the Detroit metropolitan area in Michigan amid the COVID-19 pandemic, we conclude that wastewater viral signals preceded the reported clinical cases. Both viral signal and clinical cases corresponded to social events and reflected implementation of public health policies. The early-warning methods based on WBE were proven to be efficient during the study period, as evinced by hit rates. Hit rates for early warning methods were affected by the method for defining peaks in clinical cases. Method III for defining peaks (peak defined as clinical data values higher than mean – 0.5 standard deviation of all values) identified most peaks in clinical cases and was associated with higher hit rates across all WBE based early-warning methods. Among all real-time methods, PPCS method (positive percentage change of slope >200 %) achieved higher hit rates. Among all post-factum methods, KUR (values greater than kurtosis) and MSD (values greater than mean + 2 standard deviation) methods achieved higher hit rates.
CRediT authorship contribution statement
Liang Zhao: Methodology, Data acquisition, Data curation, Formal analysis, Investigation, Visualization, Software, Writing – original draft, Writing - review & editing. Yangyang Zou: Methodology, Investigation, Software, Writing - review & editing. Randy E. David: Writing - review & editing. Scott Withington: Writing - review & editing. Stacey McFarlane: Writing - review & editing. Russell A. Faust: Writing - review & editing. John Norton: Funding acquisition, Writing - review & editing. Irene Xagoraraki: Conceptualization, Funding acquisition, Methodology, Investigation, Project administration, Resources, Supervision, Writing - review & editing.
Funding
This study was funded by the Michigan Department of Health and Human Services (MDHHS) and the Great Lakes Water Authority (GLWA).
Declaration of competing interest
All authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank the Michigan Department of Health and Human Services (MDHHS) and the Great Lakes Water Authority (GLWA) for funding this research.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.161152.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Simpson S.L., Bertsch P.M., Ehret J., Hosegood I., Metcalfe S.S., Smith W.J.M., Thomas K.V., Tynan J., Mueller J.F. Wastewater surveillance demonstrates high predictive value for COVID-19 infection on board repatriation flights to Australia. Environ. Int. 2022;158 doi: 10.1016/j.envint.2021.106938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwaerter M.D.P. Coronavirus COVID-19 (SARS-CoV-2). The Johns Hopkins University. 2022. https://www.hopkinsguides.com/hopkins/view/Johns_Hopkins_ABX_Guide/540747/all/Coronavirus_COVID_19__SARS_CoV_2_
- Barua V.B., Juel M.A.I., Blackwood A.D., Clerkin T., Ciesielski M., Sorinolu A.J., Holcomb D.A., Young I., Kimble G., Sypolt S., Engel L.S., Noble R.T., Munir M. Tracking the temporal variation of COVID-19 surges through wastewater-based epidemiology during the peak of the pandemic: a six-month long study in Charlotte, North Carolina. Sci. Total Environ. 2022;814 doi: 10.1016/j.scitotenv.2021.152503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Bivins A., Wu Z., North D. Making waves: plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021;202(July) doi: 10.1016/j.watres.2021.117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Bibby K. Wastewater surveillance during mass COVID-19 vaccination on a college campus. Environ. Sci. Technol. Lett. 2021;8(9):792–798. doi: 10.1021/acs.estlett.1c00519. [DOI] [PubMed] [Google Scholar]
- Bowman L.R., Tejeda G.S., Coelho G.E., Sulaiman L.H., Gill B.S., McCall P.J., Olliaro P.L., Ranzinger S.R., Quang L.C., Ramm R.S., Kroeger A., Petzold M.G. Alarm variables for dengue outbreaks: a multi-centre study in Asia and Latin America. PLoS ONE. 2016;11(6) doi: 10.1371/journal.pone.0157971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D., Friedler E., Gatt K. Characterising the quantity and quality of domestic wastewater inflows. Water Sci. Technol. 1995;31(7):13–24. doi: 10.1016/0273-1223(95)00318-H. [DOI] [Google Scholar]
- Català M., Pino D., Marchena M., Palacios P., Urdiales T., Cardona P.-J., Alonso S., López-Codina D., Prats C., Alvarez-Lacalle E. Robust estimation of diagnostic rate and real incidence of COVID-19 for European policymakers. PLoS ONE. 2021;16(1) doi: 10.1371/journal.pone.0243701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.-Y., Jian S.-W., Liu D.-P., Ng T.-C., Huang W.-T., Lin H.-H., Team T.C.-19. O.I. Contact tracing assessment of COVID-19 transmission dynamics in taiwan and risk at different exposure periods before and after symptom onset. JAMA Internal Medicine. 2020;180(9):1156–1163. doi: 10.1001/jamainternmed.2020.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras S., Biron-Lattes J.P., Villavicencio H.A., Medina-Ortiz D., Llanovarced-Kawles N., Olivera-Nappa Á. Statistically-based methodology for revealing real contagion trends and correcting delay-induced errors in the assessment of COVID-19 pandemic. Chaos Solitons Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corchis-Scott R., Geng Q., Seth R., Ray R., Beg M., Biswas N., Charron L., Drouillard K.D., D’Souza R., Heath D.D., Houser C., Lawal F., McGinlay J., Menard S.L., Porter L.A., Rawlings D., Scholl M.L., Siu K.W.M., Tong Y., McKay R.M.L.… Averting an outbreak of SARS-CoV-2 in a university residence hall through wastewater surveillance. Microbiol. Spectrum. 2021;9(2) doi: 10.1128/Spectrum.00792-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust P.M., Tian X., Towhid S.T., Xiao A., Mercier E., Hegazy N., Jia J.-J., Wan S., Kabir M.P., Fang W., Fuzzen M., Hasing M., Yang M.I., Sun J., Plaza-Diaz J., Zhang Z., Cowan A., Eid W., Stephenson S., Delatolla R.… Wastewater to clinical case (WC) ratio of COVID-19 identifies insufficient clinical testing, onset of new variants of concern and population immunity in urban communities. Sci. Total Environ. 2022;853 doi: 10.1016/j.scitotenv.2022.158547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J.M., Griffen B.D. Early warning signals of extinction in deteriorating environments. Nature. 2010;467(7314):456–459. doi: 10.1038/nature09389. [DOI] [PubMed] [Google Scholar]
- Gao Q., Liu Z., Xiang J., Tong M., Zhang Y., Wang S., Zhang Y., Lu L., Jiang B., Bi P. Forecast and early warning of hand, foot, and mouth disease based on meteorological factors: evidence from a multicity study of 11 meteorological geographical divisions in mainland China. Environ. Res. 2021;192 doi: 10.1016/j.envres.2020.110301. [DOI] [PubMed] [Google Scholar]
- Harris M.J., Hay S.I., Drake J.M. Early warning signals of malaria resurgence in Kericho, Kenya. Biol. Lett. 2020;16(3):20190713. doi: 10.1098/rsbl.2019.0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Cao B.… Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez M.E., Rivera-Núñez Z., Crabtree B.F., Hill D., Pellerano M.B., Devance D., Macenat M., Lima D., Martinez Alcaraz E., Ferrante J.M., Barrett E.S., Blaser M.J., Panettieri R.A., Jr., Hudson S., v. Black and latinx community perspectives on COVID-19 mitigation behaviors, testing, and vaccines. JAMA Network Open. 2021;4(7) doi: 10.1001/jamanetworkopen.2021.17074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol. 2021;55(6):3514–3519. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Kirby A.E., Walters M.S., Jennings W.C., Fugitt R., LaCross N., Mattioli M., Marsh Z.A., Roberts V.A., Mercante J.W., Yoder J., Hill V.R. Using wastewater surveillance data to support the COVID-19 response — United States, 2020–2021. MMWR Morb. Mortal. Wkly Rep. 2021;70(36):1242–1244. doi: 10.15585/mmwr.mm7036a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostoulas P., Meletis E., Pateras K., Eusebi P., Kostoulas T., Furuya-Kanamori L., Speybroeck N., Denwood M., Doi S.A.R., Althaus C.L., Kirkeby C., Rohani P., Dhand N.K., Peñalvo J.L., Thabane L., BenMiled S., Sharifi H., Walter S.D. The epidemic volatility index, a novel early warning tool for identifying new waves in an epidemic. Sci. Rep. 2021;11(1):23775. doi: 10.1038/s41598-021-02622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronbichler A., Kresse D., Yoon S., Lee K.H., Effenberger M., Shin J.il. Asymptomatic patients as a source of COVID-19 infections: a systematic review and meta-analysis. International Journal of Infectious Diseases. 2020;98:180–186. doi: 10.1016/j.ijid.2020.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Joshi M., Patel A.K., Joshi C.G. Unravelling the early warning capability of wastewater surveillance for COVID-19: a temporal study on SARS-CoV-2 RNA detection and need for the escalation. Environ. Res. 2021;196(December 2020) doi: 10.1016/j.envres.2021.110946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Mykytyn A.Z., Breugem T.I., Groen N., Knoops K., Schipper D., van Acker R., van den Doel P.B., Bestebroer T., Koopman C.D., Reusken C., Muraro M.J., GeurtsvanKessel C.H., van Royen M.E., Peters P.J., Zhang J., Haagmans B.L. BioRxiv; 2022. SARS-CoV-2 Omicron Efficiently Infects Human Airway, but not Alveolar Epithelium. 2022.01.19.476898. [DOI] [Google Scholar]
- Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A., Tambe M., Mina M.J., Parker R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021;7(1) doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H., Khosrawipour T., Kocbach P., Ichii H., Bania J., Khosrawipour V. Evaluating the massive underreporting and undertesting of COVID-19 cases in multiple global epicenters. Pulmonology. 2021;27(2):110–115. doi: 10.1016/j.pulmoe.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Miyani B., Zhao L., Spooner M., Gentry Z., Zou Y., Rhodes G., Li H., Kaye A., Norton J., Xagoraraki I. Surveillance of SARS-CoV-2 in nine neighborhood sewersheds in Detroit Tri-County area, United States: assessing per capita SARS-CoV-2 estimations and COVID-19 incidence. Sci. Total Environ. 2022;851 doi: 10.1016/j.scitotenv.2022.158350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long B., Carius B.M., Chavez S., Liang S.Y., Brady W.J., Koyfman A., Gottlieb M. Clinical update on COVID-19 for the emergency clinician: presentation and evaluation. Am. J. Emerg. Med. 2022;54:46–57. doi: 10.1016/j.ajem.2022.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MatLab P. MathWorks Inc; Natick MA USA: 2018. 9.7. 0.1190202 (R2019b) [Google Scholar]
- Menkir T.F., Chin T., Hay J.A., Surface E.D., de Salazar P.M., Buckee C.O., Watts A., Khan K., Sherbo R., Yan A.W.C., Mina M.J., Lipsitch M., Niehus R. Estimating internationally imported cases during the early COVID-19 pandemic. Nat. Commun. 2021;12(1):311. doi: 10.1038/s41467-020-20219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyani B., Fonoll X., Norton J., Mehrotra A., Xagoraraki I. SARS-CoV-2 in Detroit wastewater. J. Environ. Eng. 2020;146(11):06020004. [Google Scholar]
- Miyani B., Zhao L., Spooner M., Buch S., Gentry Z., Mehrotra A., Xagoraraki I.… Early warnings of COVID-19 second wave in Detroit. J. Environ. Eng. 2021;147(8):06021004. [Google Scholar]
- Murhekar M.V., Bhatnagar T., Selvaraju S., Saravanakumar V., Thangaraj J.W.V., Shah N., Kumar M.S., Rade K., Sabarinathan R., Asthana S., Balachandar R., Bangar S.D., Bansal A.K., Bhat J., Chopra V., Das D., Deb A.K., Devi K.R., Dwivedi G.R., Zaman K.… SARS-CoV-2 antibody seroprevalence in India, August–September, 2020: findings from the second nationwide household serosurvey. Lancet Glob. Health. 2021;9(3):e257–e266. doi: 10.1016/S2214-109X(20)30544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien D.A., Clements C.F. Early warning signal reliability varies with COVID-19 waves. Biol. Lett. 2021;17(12) doi: 10.1098/rsbl.2021.0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen S., Imakaev M., Duvallet C. Making waves: defining the lead time of wastewater-based epidemiology for COVID-19. Water Res. 2021;117433 doi: 10.1016/j.watres.2021.117433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabdial-Sing N., Motaze V., Manamela J., McCarthy K., Suchard M. Establishment of outbreak thresholds for hepatitis a in South Africa using laboratory surveillance, 2017–2020. Viruses. 2021;13(12):2470. doi: 10.3390/v13122470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford H.R., Karen S., Heather B.N. Wastewater analysis can be a powerful public health tool—if it's done sensibly. Proc. Natl. Acad. Sci. 2022;119(6) doi: 10.1073/pnas.2119600119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salath M., Althaus C.L., Neher R., Stringhini S., Hodcroft E., Fellay J., Zwahlen M., Senti G., Battegay M., Wilder-Smith A., Eckerle I., Egger M., Low N. COVID-19 epidemic in Switzerland: on the importance of testing, contact tracing and isolation. Swiss Med. Wkly. 2020 doi: 10.4414/smw.2020.20225. [DOI] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.C. 2022. R: A Language and Environment for Statistical Computing. R Version 4.1. 3. [Google Scholar]
- Torres I., Sippy R., Sacoto F. Assessing critical gaps in COVID-19 testing capacity: the case of delayed results in Ecuador. BMC Public Health. 2021;21(1):637. doi: 10.1186/s12889-021-10715-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Jiang Y., Michael E., Zhao G. How to select a proper early warning threshold to detect infectious disease outbreaks based on the China infectious disease automated alert and response system (CIDARS) BMC Public Health. 2017;17(1):570. doi: 10.1186/s12889-017-4488-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Xagoraraki I. Can we predict viral outbreaks using wastewater surveillance? J. Environ. Eng. 2020;146(11):01820003. [Google Scholar]
- Xagoraraki I., O’Brien E. Women in Water Quality. Springer; Cham: 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [Google Scholar]
- Xiao A., Wu F., Bushman M., Zhang J., Imakaev M., Chai P.R., Duvallet C., Endo N., Erickson T.B., Armas F., Arnold B., Chen H., Chandra F., Ghaeli N., Gu X., Hanage W.P., Lee W.L., Matus M., McElroy K.A., Alm E.J.… Metrics to relate COVID-19 wastewater data to clinical testing dynamics. Water Res. 2022;212 doi: 10.1016/j.watres.2022.118070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhao L., Zou Y., Li Y., Miyani B., Spooner M., Gentry Z., Jacobi S., David R.E., Withington S., McFarland S., Faust R., Sheets J., Kaye A., Broz J., Gosine A., Mobley P., Busch A.W.U., Norton J., Xagoraraki I. Five-week warning of COVID-19 peaks prior to the omicron surge in Detroit, Michigan using wastewater surveillance. Sci. Total Environ. 2022;157040 doi: 10.1016/j.scitotenv.2022.157040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Oishi W., Maruo C., Saito M., Chen R., Kitajima M., Sano D. Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks. Sci. Total Environ. 2021;767 doi: 10.1016/j.scitotenv.2021.145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Oishi W., Saito M., Kitajima M., Sano D. Early warning of COVID-19 in Tokyo via wastewater-based epidemiology: how feasible it really is? J. Water Environ. Technol. 2021;19(3):170–183. doi: 10.2965/jwet.21-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Oishi W., Maruo C., Bandara S., Lin M., Saito M., Kitajima M., Sano D. COVID-19 case prediction via wastewater surveillance in a low-prevalence urban community: a modeling approach. J. Water Health. 2022;20(2):459–470. doi: 10.2166/wh.2022.183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.