Abstract
Accurate immunoassays with a good correlation to neutralizing antibodies are required to support SARS-CoV-2 diagnosis, management, vaccine deployment, and epidemiological investigation. We conducted a study to evaluate the performance and correlation of the surrogate virus neutralization test (sVNT) and other commercial immunoassays. We tested 107 sera of COVID-19 confirmed cases from three different time points, 58 confirmed non-COVID-19 sera, and 52 sera collected before the pandemic with two sVNTs, seven chemiluminescent assays, and one fluorescein assay. All assays achieved excellent sensitivity (95%–100%, ≥15 days after onset of illness), specificity (95.5%–100%), and showed moderate to high correlation with GenScript sVNT (r = 0.58 to r = 0.98), except Roche total antibodies (r = 0.48). Vazyme sVNT and Siemens total antibodies showed the highest correlation with GenScript sVNT (r = 0.98 and 0.88, respectively). Median indexes that may be used to estimate sera with the highest ability to inhibit SARS-CoV-2 and ACE-2 receptor attachment (GenScript sVNT inhibition 90%–100%) were 6.9 S/C (Abbott IgG), 161.9 COI (FREND™ IgG), 16.8 AU/ml (Snibe IgG), 40.1 S/CO (Beckman IgG), 281.9 U/ml (Mindray IgG), 712.2 U/ml (Mindray total antibodies), >10 index (Siemens total antibodies), and 95.3% inhibition (Vazyme sVNT). All ten commercial COVID-19 serology assays, with different targeting antigens, demonstrated a reliable performance, supporting the utility of those assays in clinical and research settings. However, further studies using more samples are needed to refine the results of evaluating the performances of these marketed serological assays. Reliable serological assays would be useful for clinicians, researchers and epidemiologists in confirming SARS-CoV-2 infections, observing SARS-CoV-2 transmission, and immune response post infection and vaccination, leading to better management and control of the disease.
Keywords: SARS-CoV-2, Antibodies, Immunoassays, sVNT
SARS-CoV-2; Antibodies; Immunoassays; sVNT.
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has become a global pandemic (WHO 2020). The continuing spread of this virus raised a demand for accurate serological assays to support clinical management (e.g. assessing antibody-mediated therapy) (Lee et al., 2021; Joyner et al., 2021) and diagnosis (for patients presenting later in the disease course) (Watson et al. 2020), vaccine development and monitoring (e.g. measuring immunogenicity and estimating level of protection) (Krajewski et al., 2020), and epidemiological investigation (e.g. tracing the transmission in a community and estimating seroprevalence to quantify the level of herd immunity) (Watson et al. 2020; Peeling et al., 2020; Lisboa Bastos et al., 2020). Numerous products with different platforms have been developed, including lateral flow immunochromatography, enzyme-linked immunosorbent assay (ELISA), chemiluminescent assays, and neutralization assays (Theel et al., 2020; Galipeau et al., 2020).
Amongst these platforms, the neutralizing antibody/neutralization assays are considered the gold standard as these assays measure the function of all binding antibodies, mostly IgG in neutralizing the virus, and therefore reduce or inhibit the attachment of SARS-CoV-2 with the ACE-2 receptor in human cells. Several neutralization assays are currently known, including micro-neutralization, focus forming neutralization, fluorescence-based, pseudovirus neutralization and plaque reduction neutralization assays (Theel et al., 2020; Riepler et al., 2020; Muruato et al., 2020; Bennett et al., 2021). Apart from pseudovirus neutralization assays, these are not easy to perform as they need bio-safety level (BSL) 3 facilities. All assays need highly experienced technicians to perform cell and virus culture and need several days to provide the results (Theel et al., 2020; Galipeau et al., 2020). A breakthrough ELISA based platform, surrogate viral neutralization test (sVNT) was first introduced by researchers from the Duke University and NUS, Singapore, avoiding the use of cell and virus culture and hence, BSL-3 lab facilities. Previous studies have reported that sVNT showed a strong correlation with pseudotype virus neutralisation test (pVNT) and plaque reduction neutralization test (PRNT) (Tan et al., 2020; Meyer et al., 2020). Antibody detection tests use purified proteins of SARS-CoV-2, not viable virus, and can be performed in lower biosafety level laboratories (e.g., BSL-2). However, since not all binding antibodies block infection, these assays do not actually reflect antibody inhibition of SARS-CoV-2 infection (Mardian et al., 2021).
Given that many binding antibodies assays have been widely used in Indonesia prior to the development of the sVNT, the gold standard for serological assays, it is important to evaluate their sensitivity and specificity, their correlation with the sVNT, and the cut-off of these assays that were equivalent to certain inhibition rate of sVNT. The results of this evaluation will provide the rationales whether these assays may replace sVNT in clinical and research settings since most hospitals prefer to run a large number of specimens simultaneously with the automatic and high throughput chemiluminescent assays, which are easy to perform and cheaper. This is useful for hospitals that cannot perform ELISA or run a large number of specimens simultaneously with chemiluminescent assays. In this study, we compared the performance of ten immunoassays that use three different platforms (two sVNTs, seven chemiluminescent assays, and one fluorescein immune based assay), we also measured the correlation between these assays and the cut-offs index from each assay that was equivalent to certain sVNT inhibition rate.
2. Materials and methods
This comparative diagnostic accuracy study was part of the national COVID-19 rapid serology diagnostic validation conducted by the Ministry of Health and has been approved by the National Institute of Health Research and Development (NIHRD) ethics committee. All participants provided written informed consent for the study. It was conducted at the Indonesia Research Partnership on Infectious Diseases (INA-RESPOND) (Karyana et al., 2015) reference laboratory, located at the Tangerang District Hospital during COVID-19 first wave in Indonesia (June–December 2020). We adhered to the principles of the Declaration of Helsinki.
To determine the sample size, we calculated the highest minimum sample size estimation (with unknown pre-specified data from previous research in the early pandemic) to compare two proportions (N1 = N2 = (Zα/2+Zβ)2 ∗ (p1(1 - p1) + p2(1 - p2))/(p1 - p2)2, for a power (β) of 80%, type 1 error (α) of 5%, and the expected sample proportion of 50%) resulted in 192 samples, and for correlation analysis (total N = [(Zα + Zβ)/C]2 + 3, with C = 0.5 ∗ ln [(1 + r)/(1 - r)] and expected correlation coefficient (r) of 0.3) resulted in 170 samples. This study included a total of 217 samples, comprised of 107 COVID-19 samples for sensitivity evaluation and 110 of non-COVID-19 samples (52 samples were collected before the pandemic and 58 samples after the pandemic) for specificity evaluation. The unequal number of samples was due to the difficulty in finding serum samples with sufficient volume for 10 serological assays.
A total of 107 serum specimens for the case group were collected from real time polymerase chain reaction (RT-PCR) confirmed COVID-19 patients from several hospitals in Banten province, Indonesia. Sera from the case group were divided into three time point (TP) categories based on onset of illness: (1) 0–7 days, (2) 8–14 days, and (3) ≥15 days. Serum specimens from the control group were 58 sera from suspected COVID-19 patients with negative SARS-CoV-2 RT-PCR twice during acute hospitalization and 52 sera from repository specimens collected <2016 during acute febrile illness requiring hospitalization (AFIRE) and SEPSIS studies (Southeast Asia Infectious Disease Clinical Research 2017; Gasem et al., 2020). Of these 52 sera, 32 convalescent specimens (14–28 days after fever onset) were from patients with confirmed acute infections by other viruses or bacteria (13 influenza A/B, 8 Salmonella typhi, 6 Rickettsia typhi, 5 Escherichia coli, 4 Leptospira spp, 2 chikungunya virus, 2 Streptococcus pneumonia, 1 Klebsiella pneumoniae, 1 Staphylococcus aureus, and 1 Mycobacterium tuberculosis), and 20 sera were from pneumonia patients with unknown pathogens. In addition to the acute infecting pathogens above, 52, 26, 25, 24, and 22 sera also had dengue, chikungunya, Leptospira spp, Salmonella typhi and Rickettsia typhi IgG antibodies, respectively.
These sera were tested using two sVNTs [cPass SARS-CoV-2 Neutralization Antibody Detection Kit (GenScript, Piscataway, NJ, USA) and Anti-SARS-CoV-2 Neutralizing Antibody ELISA Kit (Vazyme, Nanjing, China)], seven automated, high-throughput chemiluminescent assays [ARCHITECT SARS-CoV-2 IgG assay (Abbott, Chicago, IL, USA), Elecsys Anti-SARS-CoV-2 assay (Roche, Basel, Switzerland), MAGLUMI 2019-nCoV IgG (Snibe, Shenzhen, China), Access anti-SARS-CoV-2 IgG (Beckman Coulter, Brea, CA, USA), SARS-CoV-2 S-RBD IgG and total antibodies (Mindray, Shenzhen, China), and ADVIA Centaur SARS-CoV-2 Total (COV2T) (Siemens, Munich, Germany)], and one fluorescein immune based assay FREND™ COVID-19 (NanoEntek, Seoul, Korea) according to the manufacturers’ instructions. Details of products are listed in Table 1. These assays will then be referred to GenScript sVNT, Vazyme sVNT, Abbott IgG, Roche total antibodies, Snibe IgG, Beckman IgG, Mindray IgG, Mindray total antibodies, Siemens total antibodies, and FREND™ IgG.
Table 1.
Details of the evaluated ten commercial immunoassays.
| Product | Manufacturer | Isotype antibodies | Targeted antigens | Cut-off value | Platform |
|---|---|---|---|---|---|
| Abbott ARCHITECT SARS-CoV-2 IgG | Abbott | IgG | Nucleocapsid | 1.4 S/C | Chemiluminescent microparticle immunoassay (CMIA) |
| FRENDTM COVID-19 | NanoEntek, Inc. | IgG | Nucleocapsid | 1 COI | Fluoroimmunoassay (FIA) |
| Elecsys Anti-SARS-CoV-2 | Roche | Total antibodies | Nucleocapsid | 1 COI | Electro-Chemiluminescence Immunoassay (ECLIA) |
| MAGLUMI 2019-nCoV IgG | SNIBE Co., Ltd | IgG | Nucleocapsid and Spike | 1 AU/mL | Chemiluminescence immunoassay (CLIA) |
| Access SARS-CoV-2 IgG | Beckman Coulter, Inc. | IgG | Spike | 1 S/CO | Chemiluminescence immunoassay (CLIA) |
| SARS-CoV-2 S-RBD IgG | Shenzhen Mindray Bio-Medical Electronics Co., Ltd. | IgG | Spike | 6 U/mL | Chemiluminescence immunoassay (CLIA) |
| SARS-CoV-2 Total antibodies | Shenzhen Mindray Bio-Medical Electronics Co., Ltd. | Total antibodies | Spike | 10 U/mL | Chemiluminescence immunoassay (CLIA) |
| ADVIA Centaur SARS-CoV-2 Total (COV2T) | Siemens Healthcare Diagnostics | Total antibodies | Spike | 1 index | Chemiluminescence immunoassay (CLIA) |
| Anti-SARS-CoV-2 Neutralizing Antibody ELISA Kit | Nanjing Vazyme Medical Technology Co., Ltd | Neutralizing antibodies (all isotypes) | Spike | 20 % inhibition rate | surrogate virus neutralisation test (sVNT) ELISA |
| cPass SARS-CoV-2 Neutralization Antibody Detection Kit | GenScript USA Inc. | Neutralizing antibodies (all isotypes) | Spike | 20% inhibition rate | surrogate virus neutralisation test (sVNT) ELISA |
Note: AU (arbitrary unit), U (unit), COI (cut-off index), S/C (signal to cut-off), S/CO (signal to cut-off).
The sensitivity of each assay was calculated based on each time point, whereas the specificity was calculated based on recent non-SARS-CoV-2 infections and repository specimens (collected less than 2016). Sensitivity and specificity were expressed as percentages with 95% confidence interval (CI). The correlation between values of each assay in case sera was calculated using the Pearson correlation test. Based on the results of GenScript inhibition rate, sera from positive COVID-19 were grouped into <20%, 20–49%, >50–79%, 80–89%, and 90–100%. The equivalent index of each assay was estimated using the median value. All statistical analyses were performed using Microsoft Excel (Microsoft Corporation, Washington, US) and all graphs were constructed using GraphPad Prism 9.0 (GraphPad Software, Inc., San Diego, CA).
3. Results
3.1. Sensitivity
The sensitivity of all assays increased from TP1, TP2, to TP3. In TP1 where specimens were collected in the median day 6 of illness, the sensitivity ranged from 50% (Roche total antibodies) to 73.5% (Vazyme sVNT). In TP2 (median 11 days), the sensitivity increased to 85.7% (Abbott IgG) and 97.1% (Vazyme sVNT). Finally, in TP3 (median 18 days), the sensitivity increased to 95%–100%. The overall sensitivity ranged from 77.6% (Siemens total antibodies) and 90.7% (Vazyme sVNT) (Table 2). The three most sensitive assays (Vazyme sVNT, GenScript sVNT, and Mindray total antibodies) were spike protein-based assay. However, the least sensitive assays were also spike protein-based assay (Siemens total antibodies and Beckman IgG), besides nucleocapsid (NC)-based assay (Roche total antibodies and Abbott IgG) or both spike protein and NC-based assay (Snibe IgG). In terms of antibody isotype target, the three most sensitive assays were targeting IgM and IgG antibodies (Mindray total antibodies) and neutralizing antibodies from all isotypes (Vazyme sVNT and GenScript sVNT). However, the results showed Siemens total antibodies and Roche total antibodies that also target IgM and IgG antibodies are the least sensitive. The sensitivity of each assay per time point is shown in Table 2.
Table 2.
The sensitivity and specificity of each commercial immunoassay.
| Assays | Sensitivity (95% CI) |
Specificity (95% CI) |
|||||
|---|---|---|---|---|---|---|---|
| 1st week (6.1 d) N = 34 |
2nd week (11.2 d) N = 35 |
3rd week (17.8 d) N = 38 |
Overall N = 107 |
<2016 sera N = 52 |
Non-COVID-19 sera N = 58 |
Overall N = 110 |
|
| Abbott IgG | 19 55.9% (46.5–65.3) |
30 85.7% (79.1–92.3) |
38 100% (99.3–100) |
87 81.3% (73.9–88.7) |
52 100% (99,3–100) |
57 98.3% (95.9–100) |
109 99.1% (97.3–100) |
| FRENDTM IgG | 21 61.8% (52.6–71.0) |
33 94.3% (89.9–98.7) |
38 100% (99.3–100) |
92 85.9% (79.3–92.5) |
51 98.1% (95.5–100) |
57 98.3% (95.9–100) |
108 98.2% (95.7–100) |
| Roche total antibodies | 17 50% (41.1–58.9) |
33 94.3% (90.2–98.5) |
37 97.4% (94.6–100) |
87 81.3% (73.9–88.7) |
51 98.1% (95.7–100) |
57 98.3% (95.9–100) |
108 98.2% (95.7–100) |
| Snibe IgG | 17 50% (40.5–59.5) |
31 88.6% (82.6–94.6) |
37 97.4% (94.4–100) |
85 79.4% (71.7–87.1) |
48 92.3% (87.3–97.3) |
57 98.3% (95.9–100) |
105 95.5% (91.6–99.4) |
| Beckman IgG | 18 52.9% (43.4–62.4) |
32 91.4% (86.1–96.7) |
37 97.4% (94.4–100) |
87 81.3% (73.9–88.7) |
52 100% (99.3–100) |
58 100% (99.3–100) |
110 100% (99.3–100) |
| Mindray IgG | 20 58.8% (49.5–68.1) |
32 91.4% (86.1–96.7) |
38 100% (99.3–100) |
90 84.1% (77.2–91.0) |
52 100% (99.3–100) |
58 100% (99.3–100) |
110 100% (99.3–100) |
| Mindray total antibodies | 24 70.6% (61.9–79.2) |
32 91.4% (86.1–96.7) |
38 100% (99.3–100) |
94 87.9% (81.7–94.1) |
52 100% (99.3–100) |
56 96.6% (93.2–99.9) |
108 98.2% (95.7–100) |
| Siemens total antibodies | 16 47.1% (37.6–56.6) |
30 85.7% (79.1–92.3) |
37 97.4% (94.4–100) |
83 77.6% (69.7–85.5) |
51 98.1% (95.5–100) |
58 100% (99.3–100) |
109 99.1% (97.3–100) |
| Vazyme sVNT | 25 73.5% (65.1–81.9) |
34 97.1% (93.9–100) |
38 100% (99.3–100) |
97 90.7% (85.2–96.2) |
51 98.1% (95.5–100) |
56 96.6% (93.2–99.9) |
107 97.3% (94.3–100) |
| GenScript sVNT | 20 58.8% (49.5–68.1) |
33 94.3% (90.2–98.5) |
38 100% (99.3–100) |
91 85.0% (78.2–91.8) |
51 98.1% (95.5–100) |
58 100% (99.3–100) |
109 99.1% (97.3–100) |
3.2. Specificity
The specificity of all assays was excellent, with two assays (Beckman IgG and Mindray IgG) showing specificity of 100%, but Snibe IgG revealed to be the least specific (95.1%). In non-COVID-19 infection specimens, seven different sera were tested positive in six assays (one serum tested positive by both Abbott IgG and FREND™ IgG, two sera by Mindray total antibodies, two sera by Vazyme sVNT, one serum by Roche total antibodies and one serum by Snibe IgG). The non-COVID-19 acute infecting pathogens in these patients were unknown. Snibe IgG showed more cross-reactivity in sera collected before 2016 (four of 52 sera from two confirmed Rickettsiosis patients, an Influenza B patient, and an unknown pathogen patient). GenScript sVNT and Vazyme sVNT assays showed cross-reactivity in the same serum collected before 2016 from a confirmed influenza B patient, whereas Siemens total antibodies showed cross-reactivity in serum from a confirmed leptospirosis patient, Roche total antibodies showed cross-reactivity in serum from a Rickettsiosis patient, and FREND™ IgG showed cross-reactivity in serum from a patient with an unknown pathogen. Details of the characteristics of all false positive sera are listed in Table 3.
Table 3.
Lists of false positives, identified acute infecting pathogens, and results from all assays.
| Sera | Acute infecting pathogen | Ab bott IgG | FRENDTM IgG | Ro che total antibodies | Snibe IgG | Beckman IgG | Mind ray IgG |
Mind ray total antibodies | Sie mens total antibodies | Vazyme sVNT | Gen Script sVNT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 (<2016)1 | R typhi | Neg | Neg | Neg | 3.2 | Neg | Neg | Neg | Neg | Neg | Neg |
| #2 (<2016)2 | Influenza B | Neg | Neg | Neg | 3.1 | Neg | Neg | Neg | Neg | Neg | Neg |
| #3 (<2016)3 | UNK | Neg | Neg | Neg | 3.3 | Neg | Neg | Neg | Neg | Neg | Neg |
| #4 (<2016)3 | R typhi | Neg | Neg | 2.6 | 2.7 | Neg | Neg | Neg | Neg | Neg | Neg |
| #5 (<2016)3 | UNK | Neg | 16.4 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| #6 (<2016)3 | Leptospira spp | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 1.43 | Neg | Neg |
| #7 (<2016)3 | Influenza B | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 41.9 | 47.4 |
| #8 (2020) | UNK | Neg | Neg | Neg | Neg | Neg | Neg | 15.9 | Neg | Neg | Neg |
| #9 (2020) | UNK | Neg | Neg | Neg | 3.2 | Neg | Neg | Neg | Neg | Neg | Neg |
| #10 (2020) | UNK | Neg | Neg | 1.75 | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| #11 (2020) | UNK | Neg | Neg | Neg | Neg | Neg | Neg | 27.9 | Neg | Neg | Neg |
| #12 (2020) | UNK | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 27.8 | Neg |
| #13 (2020) | UNK | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | 20.1 | Neg |
| #14 (2020) | UNK | 2.1 | 46.6 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
There were also 47 sera with IgG antibodies to dengue, 25 with IgG abs to Leptospira spp and 24 with IgG antibodies of murine typhus but remained negative for SARS-CoV-2 antibodies to any assay.
Serum also contained IgG to dengue and chikungunya viruses, and Salmonella typhi.
Serum also contained IgG to dengue and chikungunya viruses, Salmonella typhi and Leptospira spp.
Serum also contained IgG to dengue virus.
3.3. Correlation between assays
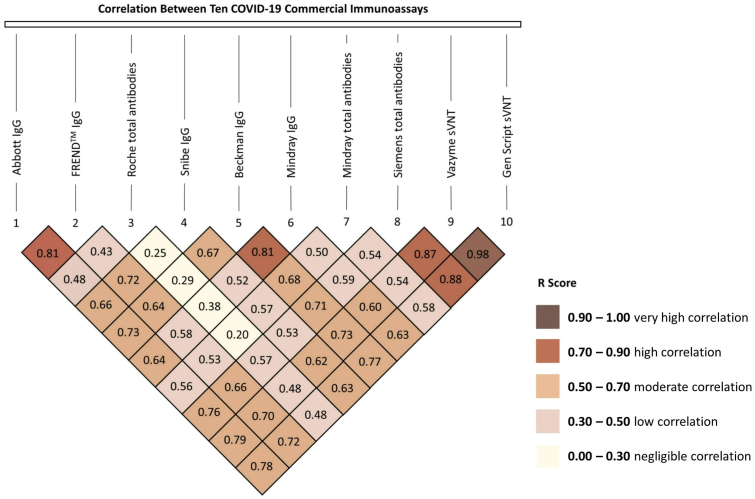
The Pearson correlation between the two sVNTs, GenScript sVNT and Vazyme sVNT was the highest amongst all assays (r = 0.98). A high correlation was also observed between Siemens total antibodies and these two sVNTs (r = 0.88 and r = 0.87, respectively). Among the other spike-based assays, a high correlation was also demonstrated between Beckman IgG and Mindray IgG (r = 0.81), Beckman IgG and Siemens total antibodies (r = 0.71), and Beckman IgG and two sVNTs (r = 0.77 and 0.73, respectively). Despite being a NC-based assay, Abbott IgG had a high correlation with most of the spike-based assays (r = 0.73 to 0.79), except with Mindray IgG and Mindray total antibodies assays (r = 0.64 and 0.56, respectively). Among NC-based assays, the correlation between Abbott IgG and FREND™ IgG was high (r = 0.81), but the correlation with Roche total antibodies was low (r = 0.48). A low correlation was also found between Roche total antibodies and all other assays (r = 0.20 to 0.57). All the values of Pearson's correlation are shown in the inverse pyramid graph (Figure 1).
Figure 1.
Correlation between COVID-19 commercial immunoassays.
3.4. Correlation with GenScript sVNT
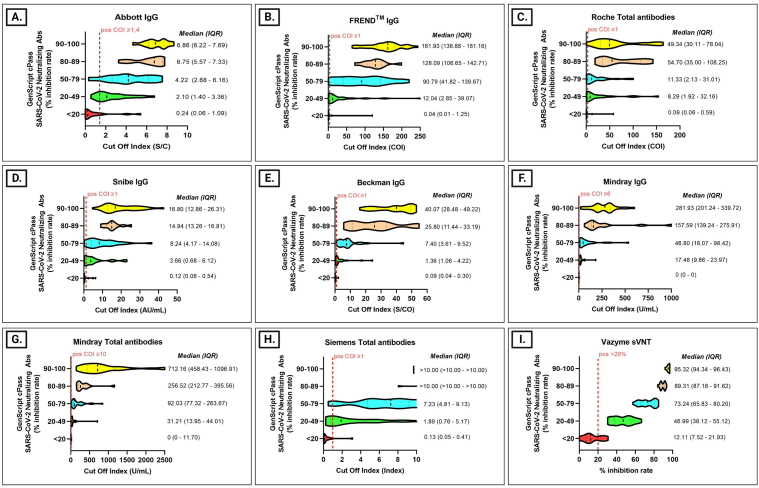
All assays showed moderate to high correlation with GenScript sVNT (r = 0.58 to r = 0.98), except Roche total antibodies (r = 0.48). Vazyme sVNT and Siemens total antibodies showed the highest correlation (r = 0.98 and 0.88, respectively). Using GenScript sVNT results as the reference standard, results from 107 COVID-19 positive sera, 15 were grouped into <20%, 22 into 20%–49%, 28 into 50%–79%, 12 into 80%–89%, and 30 into 90%–100% inhibition rate. In general, the median values of all assays gradually increased from category <20% inhibition to the higher inhibition categories. However, the median value of Roche total antibodies for the 80%–89% inhibition group was higher than the 90%–100% group (54.7 vs 49.3). The median value of Siemens total antibodies had reached the maximum index in the 80%–89% group. In sera that were considered negative by GenScript sVNT (<20%), results from three spike-based assays (Beckman IgG, Mindray IgG, and Siemens total antibodies) were consistent with GenScript sVNT. However, a few positive results were identified from three NC-based assays (Abbott IgG, FREND™ IgG, and Roche total antibodies), one mixed NC and spike-based assay (Snibe IgG), one spike-based assay (Mindray total antibodies), and the other sVNT (Vazyme sVNT). The median index for the highest inhibition group (90%–100%) were 6.9 S/C (Abbott IgG), 161.9 COI (FREND™ IgG), 49.3 COI (Roche total antibodies), 16.8 AU/ml (Snibe IgG), 40.1 S/CO (Beckman IgG), 281.9 U/ml (Mindray IgG), 712.2 U/ml (Mindray total antibodies), >10 index (Siemens total antibodies), and 95.3% inhibition (Vazyme sVNT). Distribution in each GenScript sVNT inhibition rate category was plotted in a violin graph (Figure 2 A-I).
Figure 2.
Violin Plot of nine COVID-19 commercial immunoassays for detecting SARS-CoV-2 antibodies compared to GenScript sVNT. (A). Abbott IgG compared to GenScript sVNT. (B). FREND™ IgG compared to GenScript sVNT. (C). Roche total antibodies compared to GenScript sVNT. (D). Snibe IgG compared to GenScript sVNT. (E). Beckman IgG compared to GenScript sVNT. (F). Mindray IgG compared to GenScript sVNT. (G). Mindray total antibodies compared to GenScript sVNT. (H). Siemens total antibodies compared to GenScript sVNT. (I). Vazyme sVNT compared to GenScript sVNT.
4. Discussion
We evaluated performance characteristics of ten SARS-CoV-2 immunoassays using sera from COVID-19 patients and negative control consisting of sera from confirmed non-COVID-19 patients and before pandemic. In addition to the sensitivity and specificity, we also evaluated the correlation between each assay and proposed equivalent median index corresponding to certain surrogate neutralizing inhibition rates.
The three most sensitive assays were Mindray total antibodies, GenScript sVNT, and Vazyme sVNT. Mindray total antibodies is an assay directed to all antibody isotypes, therefore it outperformed other assays that only detected IgG. For the same reason, GenScript sVNT and Vazyme sVNT measured the function of all antibody isotypes in inhibiting the attachment between spike-receptor binding domain (S-RBD) and ACE2 receptor. Similar results have been reported by Tan et al. which found GenScript sVNT and Vitros OCD anti-SARS-CoV-2 total antibody assay were more sensitive than Abbott IgG, Beckman IgG, Roche total antibodies, and Siemens total antibodies (Tan et al., 2020). Their results were consistent regarding the low sensitivity of Siemens total antibodies despite targeting total antibodies to spike protein. Three assays (Abbott IgG, Roche total antibodies, and FREND™ IgG) targeting the NC showed lower sensitivity than the three most sensitive assays. In addition, the sensitivity of Abbott IgG and FREND™ IgG was also comparable to assays targeting the spike protein (Beckman IgG and Mindray IgG). Our results demonstrated that the sensitivity of the assays was affected by the targeting antibodies rather than the targeting antigen, consistent with previous reports (Qu et al., 2020; Xiang et al., 2020) in contrast to other studies which reported NC-based assays were more sensitive than spike-based assays or vice-versa (Burbelo et al., 2020; Liu et al., 2020; Tang et al., 2020).
Several plausible reasons may explain why the sensitivity in TP1 sera of these assays was higher than previous reports from other studies (Tan et al., 2020; Kweon et al., 2020). First, sera in TP1 were collected mostly from median day 6 (range: 3–7 days) after the onset of illness, the time when sero-conversion begins (Lou et al., 2020). Second, it might correlate with the disease severity of the hospitalized subjects in our study. It has been reported that immune responses in moderate to severe cases appear earlier and stronger than the mild and asymptomatic cases (Edouard et al., 2021; Okba et al., 2020; Yongchen et al., 2020). Another explanation may be the subjective nature of the data, as the accuracy was affected by recalling bias of the patients and the symptoms that were used. Although antibodies would naturally be detected mostly in the second week post infection, the inclusion of TP1 in our study may help in determining which assay was the most sensitive. As in other TPs, the sensitivity among assays was indistinguishable.
All assays showed excellent specificity (above 95%), with Beckman IgG and Mindray IgG reaching 100%. Compared to these two assays, Snibe IgG was less specific (95.1% vs 100%). Although no study has compared Snibe IgG and Beckman IgG performance head-to-head, results from separate reports confirmed that Snibe IgG was less specific (Pere et al., 2020; Oved et al., 2020; Plebani et al., 2020; Van Elslande et al., 2020). Seven false positive results were observed in recent sera from non-COVID-19 infection hospitalized patients. The low positive value in each different assay, suggesting potential interfering factors such as auto-antibodies in chronic inflammatory diseases or other infecting pathogens (Kharlamova et al., 2021). Seven false positive results were also observed in sera collected <2016 suggesting cross-reactivity. Four acute pathogens (Rickettsia typhi, Salmonella typhi, Leptospira spp, and influenza B) identified in five sera (two other sera contained unknown acute pathogen) along with dengue IgG antibodies in all sera. Since these sera may also contain antibodies to other pathogens, cross reactivity to specific pathogen such as other human coronaviruses and zika viruses that have been reported previously (Hicks et al., 2021; Lustig et al., 2021; Lv et al., 2020; Faccini-Martinez et al., 2020) could not be confirmed as we did not test antibodies against these pathogens. However, it was interesting that one serum from a patient with influenza B was positive for SARS-CoV-2 neutralizing antibodies with >40% inhibition both by GenScript sVNT and Vazyme sVNT. Despite the high sequence dissimilarity between two virus families, hemagglutinin (HA) protein reported to cross react with anti-SARS-CoV-2 non-neutralizing antibodies (Lee et al., 2020; Murugavelu et al., 2021). The likelihood of neutralizing antibodies targeting comparable epitopes in influenza viruses and SARS-CoV-2 warrants further research.
A very high correlation (r = 0.98) between the two sVNT assays (Genescript sVNT and Vazyme sVNT), and the cross-reactivity to the same sera indicated the similarity of these two assays in their S-RBD and ACE-2 receptor design. A slightly better sensitivity but also slightly less specificity of Vazyme sVNT compared to GenScript sVNT might be associated with inter-assay variability that may have occurred as we used a manual ELISA machine, particularly in sera with low level neutralizing antibodies. A high correlation between GenScript sVNT with Siemens total antibodies and Beckman IgG (r = 0.88 and r = 0.77, respectively) was conceivable as both assays were spike-based. However, the correlation with NC-based assays, Abbott IgG and FREND™ IgG were also high (r = 0.78 and 0.72), suggesting that circulating NC antibodies in each positive sera in our study were comparable to the circulating antibodies against the spike protein. Having accurate spike-based and NC-based targeted serology assays would assist distinguish spike-based vaccine response from natural infection and research anti-NC dynamic response in inactivated viral vaccines (Dorschug et al., 2021; Azak et al., 2021). Since the correlation between GenScript sVNT and PRNT was high (Tan et al., 2020), the correlation between assays with GenScript sVNT might be expected to reflect their correlation with PRNT as well. The low correlation between Roche total antibodies and GenScript sVNT (r = 0.48) than other assays with GenScript sVNT (r = 0.58 to 0.88) in our study supported previous reports that showed Roche total antibodies has a lower correlation with neutralizing antibody titers than other assays such as Abbott IgG and Euroimmune (r = 0.29 vs 0.47 and 0.46) (Tang et al., 2020). Similarly, Padoan et al. and Therrien et al. reported the correlation between neutralizing antibody titers and Roche total antibodies was 0.214 while Abbott IgG was 0.69 (Padoan et al., 2020), and Roche total antibodies was 0.54 while Abbott IgG was 0.64, Siemens total antibodies was 0.71 and Beckman IgG was 0.73–0.77 (Therrien et al., 2021).
To assist centers that cannot perform any kind of neutralizing assays, we proposed to use index values that are equivalent to a certain inhibition rate of GenScript sVNT. We chose 20% inhibition as the first group as this is the positive cut-off of GenScript sVNT. All assays showed a median index less than their negative cut-off. We chose 50%, 80% and 90% inhibition in the following groups as they are commonly used in analyzing the PRNT results (Theel et al., 2020; Galipeau et al., 2020). Additionally, we would like to see if there was any difference between the 80%–89%, and 90%–100% groups. The median index of all assays gradually increased, consistent with the increasing inhibition rate, except Roche total antibodies and Siemens total antibodies in the two highest categories. Roche total antibodies's median index in the 90%–100% inhibition category was lower than the 80%–89% category and Siemens total antibodies' similar median index for both groups suggested these two assays could not distinguish the difference in these two groups. Median indexes from assay with a good correlation with GenScript sVNT in the inhibition group of GenScript sVNT 80%–89% and 90%–100% may be proposed as a proxy to high titer neutralizing antibodies in convalescent plasma. It was consistent with the suggested high titer by the FDA, which is equivalent to GenScript's sVNT inhibition ≥80% (FDA 2021).
The study has several strengths. First, we look at the sensitivity and specificity of ten commercial assays using ten immunoassays, with different platforms, antibodies and antigen targets available in Indonesia around the time of the first COVID-19 wave (June–December 2020) prior to vaccine deployment. Second, we measured the correlation coefficient between each assay and the cut-off index from each assay that was equivalent to a certain sVNT inhibition rate. Third, we separate the sensitivity analysis into three-time points ((1) 0–7 days, (2) 8–14 days, and (3) ≥15 days). Although antibodies would naturally be detected mainly in the second-week post infection, including TP1 in our study may help determine the most sensitive assay. Fourth, we also included repository samples from various infectious disease cases before COVID-19 pandemic for specificity analysis.
The main limitation of our evaluation was the absence of viral neutralizing tests as the gold standard since our laboratory was not equipped with BSL-3 facilities. Also, we may need more positive specimens used to estimate the assays' indexes that are equivalent to GenScript sVNT inhibition rates. However, the current results are consistent with other findings and may provide an estimation for hospitals that do not have the competence to conduct neutralization assays. As our negative sera were only partly tested for pathogens, we were not able to determine which pathogens caused the cross-reactivity that occurred in 14 of 110 sera. Though, from the available data, they might provide novel information regarding cross-reactivity with pathogens such as Leptospira spp, Rickettsia typhi, Salmonella typhi, and influenza B.
In conclusion, we characterised the performance of ten commercial immunoassays with three different platforms. All of them showed excellent sensitivity and specificity, though a few of them performed significantly better than others. Our study also reveals further research that should be done, including testing on well-characterized negative sera for better understanding about cross-reactivity, in silico analysis to identify similarities in epitopes between SARS-CoV-2 with the potential pathogens, testing with larger numbers of positive sera to provide a better correlation picture amongst assays and robust equivalent indexes to the inhibition rate. Commercial immunoassays should be evaluated regularly due to their importance in the clinical and scientific landscape in guiding public health officers, especially after vaccine deployment.
Declarations
Author contribution statement
Dewi Lokida; Herman Kosasih: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Muhammad Karyana; Retna Indah Sugiyono; Irmansyah: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yan Mardian: Analyzed and interpreted the data; Wrote the paper.
Dona Arlinda: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Nurhayati Lukman: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Gustiani Salim; Deni Pepy Butar Butar; Adhella Menur Naysilla: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the National Institute of Health Research and Development, Ministry of Health, Jakarta, Indonesia [2071.053.003.051].
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no competing interests.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to thank Aly Diana and Antonius Arditya Pradana for their technical assistance with the manuscript.
References
- Azak E., Karadenizli A., Uzuner H., Karakaya N., Canturk N.Z., Hulagu S. 'Comparison of an inactivated Covid 19 vaccine-induced antibody response with concurrent natural Covid19 infection. Int. J. Infect. Dis. 2021;113:58–64. doi: 10.1016/j.ijid.2021.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.S., Postnikova E.N., Liang J., Gross R., Mazur S., Dixit S., Lukin V.V., Kocher G., Yu S., Georgia-Clark S., Gerhardt D., Cai Y., Marron L., Holbrook M.R. bioRxiv; 2021. 'Scalable, Micro-neutralization Assay for Qualitative Assessment of SARS-CoV-2 (COVID-19) Virus-Neutralizing Antibodies in Human Clinical Samples. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Cohen J.I. 'Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschug A., Frickmann H., Schwanbeck J., Yilmaz E., Mese K., Hahn A., Gross U., Zautner A.E. Diagnostics; Basel): 2021. 'Comparative Assessment of Sera from Individuals after S-Gene RNA-Based SARS-CoV-2 Vaccination with Spike-Protein-Based and Nucleocapsid-Based Serological Assays; p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edouard S., Colson P., Melenotte C., Di Pinto F., Thomas L., La Scola B., Million M., Tissot-Dupont H., Gautret P., Stein A., Brouqui P., Parola P., Lagier J.C., Raoult D., Drancourt M. Evaluating the serological status of COVID-19 patients using an indirect immunofluorescent assay, France. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:361–371. doi: 10.1007/s10096-020-04104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccini-Martinez A.A., Rivero R., Garay E., Garcia A., Mattar S., Botero Y., Galeano K., Miranda J., Martinez C., Guzman C., Arrieta G., Contreras H., Kerguelen H., Moscote M., Brango E., Contreras V. 'Serological cross-reactivity using a SARS-CoV-2 ELISA test in acute Zika virus infection, Colombia. Int. J. Infect. Dis. 2020;101:191–193. doi: 10.1016/j.ijid.2020.09.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . Convalescent Plasma EUA Letter of Authorization; 2021. [Google Scholar]
- Galipeau Y., Greig M., Liu G., Driedger M., Langlois M.A. 'Humoral responses and serological assays in SARS-CoV-2 infections. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasem M.H., Kosasih H., Tjitra E., Alisjahbana B., Karyana M., Lokida D., Neal A., Liang C.J., Aman A.T., Arif M., Sudarmono P., Suharto, Merati T.P., Lisdawati V., Siswanto, Siddiqui S., Lane H.C., Ina-Respond for 'An observational prospective cohort study of the epidemiology of hospitalized patients with acute febrile illness in Indonesia. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0007927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J., Klumpp-Thomas C., Kalish H., Shunmugavel A., Mehalko J., Denson J.P., Snead K.R., Drew M., Corbett K.S., Graham B.S., Hall M.D., Memoli M.J., Esposito D., Sadtler K. 'Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal betacoronaviruses. J. Clin. Immunol. 2021;41:906–913. doi: 10.1007/s10875-021-00997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner M.J., Carter R.E., Senefeld J.W., Klassen S.A., Mills J.R., Johnson P.W., Theel E.S., Wiggins C.C., Bruno K.A., Klompas A.M., Lesser E.R., Kunze K.L., Sexton M.A., Diaz Soto J.C., Baker S.E., Shepherd J.R.A., van Helmond N., Verdun N.C., Marks P., van Buskirk C.M., Winters J.L., Stubbs J.R., Rea R.F., Hodge D.O., Herasevich V., Whelan E.R., Clayburn A.J., Larson K.F., Ripoll J.G., Andersen K.J., Buras M.R., Vogt M.N.P., Dennis J.J., Regimbal R.J., Bauer P.R., Blair J.E., Paneth N.S., Fairweather D., Wright R.S., Casadevall A. Convalescent plasma antibody levels and the risk of death from covid-19. N. Engl. J. Med. 2021;384:1015–1027. doi: 10.1056/NEJMoa2031893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyana M., Kosasih H., Samaan G., Tjitra E., Aman A.T., Alisjahbana B., Fatmawati, Gasem M.H., Arif M., Sudarmono P., Suharto, Merati T.P., Lane C., Siswanto, Siddiqui S. 'INA-RESPOND: a multi-centre clinical research network in Indonesia. Health Res. Pol. Syst. 2015;13:34. doi: 10.1186/s12961-015-0024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharlamova N., Dunn N., Bedri S.K., Jerling S., Almgren M., Faustini F., Gunnarsson I., Ronnelid J., Pullerits R., Gjertsson I., Lundberg K., Manberg A., Pin E., Nilsson P., Hober S., Fink K., Fogdell-Hahn A. 'False positive results in SARS-CoV-2 serological tests for samples from patients with chronic inflammatory diseases. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.666114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski R., Golebiowska J., Makuch S., Mazur G., Agrawal S. Update on serologic testing in COVID-19. Clin. Chim. Acta. 2020;510:746–750. doi: 10.1016/j.cca.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon O.J., Lim Y.K., Kim H.R., Kim M.C., Choi S.H., Chung J.W., Lee M.K. 'Antibody kinetics and serologic profiles of SARS-CoV-2 infection using two serologic assays. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Pinho M.P., Buckley P.R., Woodhouse I.B., Ogg G., Simmons A., Napolitani G., Koohy H. 'Potential CD8+ T cell cross-reactivity against SARS-CoV-2 conferred by other coronavirus strains. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.579480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.T., Girardin R.C., Dupuis A.P., Kulas K.E., Payne A.F., Wong S.J., Arinsburg S., Nguyen F.T., Mendu D.R., Firpo-Betancourt A., Jhang J., Wajnberg A., Krammer F., Cordon-Cardo C., Amler S., Montecalvo M., Hutton B., Taylor J., McDonough K.A. Neutralizing antibody responses in COVID-19 convalescent sera. J. Infect. Dis. 2021;223:47–55. doi: 10.1093/infdis/jiaa673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Ahmad Khan F. 'Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. 'Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou B., Li T.D., Zheng S.F., Su Y.Y., Li Z.Y., Liu W., Yu F., Ge S.X., Zou Q.D., Yuan Q., Lin S., Hong C.M., Yao X.Y., Zhang X.J., Wu D.H., Zhou G.L., Hou W.H., Li T.T., Zhang Y.L., Zhang S.Y., Fan J., Zhang J., Xia N.S., Chen Y. 'Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig Y., Keler S., Kolodny R., Ben-Tal N., Atias-Varon D., Shlush E., Gerlic M., Munitz A., Doolman R., Asraf K., Shlush L.I., Vivante A. 'Potential antigenic cross-reactivity between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and dengue viruses. Clin. Infect. Dis. 2021;73:e2444–e2449. doi: 10.1093/cid/ciaa1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H., Wu N.C., Tsang O.T., Yuan M., Perera Rapm, Leung W.S., So R.T.Y., Chan J.M.C., Yip G.K., Chik T.S.H., Wang Y., Choi C.Y.C., Lin Y., Ng W.W., Zhao J., Poon L.L.M., Peiris J.S.M., Wilson I.A., Mok C.K.P. 'Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardian Y., Kosasih H., Karyana M., Neal A., Lau C.Y. 'Review of current COVID-19 diagnostics and opportunities for further development. Front. Med. 2021;8 doi: 10.3389/fmed.2021.615099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Reimerink J., Torriani G., Brouwer F., Godeke G.J., Yerly S., Hoogerwerf M., Vuilleumier N., Kaiser L., Eckerle I., Reusken C. 'Validation and clinical evaluation of a SARS-CoV-2 surrogate virus neutralisation test (sVNT) Emerg. Microb. Infect. 2020;9:2394–2403. doi: 10.1080/22221751.2020.1835448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruato A.E., Fontes-Garfias C.R., Ren P., Garcia-Blanco M.A., Menachery V.D., Xie X., Shi P.Y. 'A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020;11:4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugavelu P., Perween R., Shrivastava T., Singh V., Ahmad Parray H., Singh S., Chiranjivi A.K., Thiruvengadam R., Singh S., Yadav N., Jakhar K., Sonar S., Mani S., Bhattacharyya S., Sharma C., Vishwakarma P., Khatri R., Kumar Panchal A., Das S., Ahmed S., Samal S., Kshetrapal P., Bhatnagar S., Luthra K., Kumar R. 'Non-neutralizing SARS CoV-2 directed polyclonal antibodies demonstrate cross-reactivity with the HA glycans of influenza virus. Int. Immunopharm. 2021;99 doi: 10.1016/j.intimp.2021.108020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken Cbem, Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. 'Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved K., Olmer L., Shemer-Avni Y., Wolf T., Supino-Rosin L., Prajgrod G., Shenhar Y., Payorsky I., Cohen Y., Kohn Y., Indenbaum V., Lazar R., Geylis V., Oikawa M.T., Shinar E., Stoyanov E., Keinan-Boker L., Bassal R., Reicher S., Yishai R., Bar-Chaim A., Doolman R., Reiter Y., Mendelson E., Livneh Z., Freedman L.S., Lustig Y. 'Multi-center nationwide comparison of seven serology assays reveals a SARS-CoV-2 non-responding seronegative subpopulation. EClinicalMedicine. 2020;29 doi: 10.1016/j.eclinm.2020.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padoan A., Bonfante F., Pagliari M., Bortolami A., Negrini D., Zuin S., Bozzato D., Cosma C., Sciacovelli L., Plebani M. Analytical and clinical performances of five immunoassays for the detection of SARS-CoV-2 antibodies in comparison with neutralization activity. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Wedderburn C.J., Garcia P.J., Boeras D., Fongwen N., Nkengasong J., Sall A., Tanuri A., Heymann D.L. 'Serology testing in the COVID-19 pandemic response. Lancet Infect. Dis. 2020;20:e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pere H., Wack M., Vedie B., Demory Guinet N., Kassis Chikani N., Janot L., Belec L., Veyer D. 'Sequential SARS-CoV-2 IgG assays as confirmatory strategy to confirm equivocal results: Hospital-wide antibody screening in 3,569 staff health care workers in Paris. J. Clin. Virol. 2020;132 doi: 10.1016/j.jcv.2020.104617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plebani M., Padoan A., Negrini D., Carpinteri B., Sciacovelli L. Diagnostic performances and thresholds: the key to harmonization in serological SARS-CoV-2 assays? Clin. Chim. Acta. 2020;509:1–7. doi: 10.1016/j.cca.2020.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X., Zhu Q., Liu L. 'Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71:2255–2258. doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riepler L., Rossler A., Falch A., Volland A., Borena W., von Laer D., Kimpel J. Vol. 9. Vaccines; Basel): 2020. (Comparison of Four SARS-CoV-2 Neutralization Assays). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southeast Asia Infectious Disease Clinical Research, Network 'Causes and outcomes of sepsis in southeast Asia: a multinational multicentre cross-sectional study. Lancet Global Health. 2017;5:e157–e167. doi: 10.1016/S2214-109X(17)30007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I., Tiu C., Hu Z., Chen V.C., Young B.E., Sia W.R., Tan Y.J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.F. 'A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Tan S.S., Saw S., Chew K.L., Huak C.Y., Khoo C., Pajarillaga A., Wang W., Tambyah P., Ong L., Jureen R., Sethi S.K. 'Head-to-head evaluation on diagnostic accuracies of six SARS-CoV-2 serological assays. Pathology. 2020;52:770–777. doi: 10.1016/j.pathol.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.S., Case J.B., Franks C.E., Chen R.E., Anderson N.W., Henderson J.P., Diamond M.S., Gronowski A.M., Farnsworth C.W. 'Association between SARS-CoV-2 neutralizing antibodies and commercial serological assays. Clin. Chem. 2020;66:1538–1547. doi: 10.1093/clinchem/hvaa211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W., Farnsworth C.W. 'Clinical performance of two SARS-CoV-2 serologic assays. Clin. Chem. 2020;66:1055–1062. doi: 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theel E.S., Couturier M.R., Filkins L., Palavecino E., Mitchell S., Campbell S., Pentella M., Butler-Wu S., Jerke K., Dharmarha V., McNult P., Schuetz A.N. 'Application, verification, and implementation of SARS-CoV-2 serologic assays with emergency use authorization. J. Clin. Microbiol. 2020;59 doi: 10.1128/JCM.02148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien C., Serhir B., Belanger-Collard M., Skrzypczak J., Shank D.K., Renaud C., Girouard J., Loungnarath V., Carrier M., Brochu G., Tourangeau F., Gilfix B., Piche A., Bazin R., Guerin R., Lavoie M., Martel-Laferriere V., Fortin C., Benoit A., Marcoux D., Gauthier N., Laumaea A.M., Gasser R., Finzi A., Roger M. 'Multicenter evaluation of the clinical performance and the neutralizing antibody activity prediction properties of 10 high-throughput serological assays used in clinical laboratories. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.02511-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J., Decru B., Jonckheere S., Van Wijngaerden E., Houben E., Vandecandelaere P., Indevuyst C., Depypere M., Desmet S., Andre E., Van Ranst M., Lagrou K., Vermeersch P. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin. Microbiol. Infect. 2020;26:1557 e1–57 e7. doi: 10.1016/j.cmi.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J., Richter A., Deeks J. 'Testing for SARS-CoV-2 antibodies. BMJ. 2020;370:m3325. doi: 10.1136/bmj.m3325. [DOI] [PubMed] [Google Scholar]
- WHO . 2020. WHO Director-General's Opening Remarks at the media Briefing on COVID-19 - 11 March 2020. [Google Scholar]
- Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., Zhou Q., Ye H., Ma Y., Li H., Wei X., Cai P., Ma W.L. 'Antibody detection and dynamic characteristics in patients with coronavirus disease 2019. Clin. Infect. Dis. 2020;71:1930–1934. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J., Chen Y., Gu B. 'Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg. Microb. Infect. 2020;9:833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.