Abstract
Purpose
To investigate the performance of a combined nasal midturbinate- and oropharyngeal (NAOP) self-swab compared to a deep oropharyngeal (OP) swab by health care workers (HCW) in detecting SARS-CoV-2 in a real-life setting.
Methods
Paired swabs from 1119 participants were included. RT-PCR were used to detect SARS-CoV-2 in both swab samples.
Results
330 participants tested positive. The sensitivity of the combined self-swab and OP swab was 96.9 % and 95.4 % respectively, whereas the Ct-values for self-swabs were significantly lower compared to OP swabs.
Conclusion
The combined NAOP self-swab outperformed the OP swab and thus, the NAOP self-swab may be an alternative sampling method under the given circumstances.
Keywords: Covid-19, SARS-CoV-2, Self-swab, Combined swab, Nasal midturbinate, Oropharyngeal
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has, since its emergence in December 2019, caused an ongoing global pandemic. In Denmark, complimentary community testing for COVID-19 is carried out by “TestCenter Denmark” (TCDK) at local test sites, where both residents and visitors can be tested. The standard routine test is an oropharyngeal (OP) swab performed by health care workers (HCW) and analyzed by RT-PCR targeting the E-gene with E-Sarbeco primers (Corman et al., 2020, Vogels et al., 2020). Self-testing could reduce the need for HCWs, infection risk for individuals, reduction in personal protective equipment and enable a more rapid scalable test capacity. Self-performed swabs show high potential for population testing (Tsang et al., 2021) and this study aims to investigate if the performance of a combined self-swab of nasal midturbinate- and oropharyngeal (NAOP) specimen at least equals OP swabs by HCWs in a community setting.
1. Characteristics of study cohort
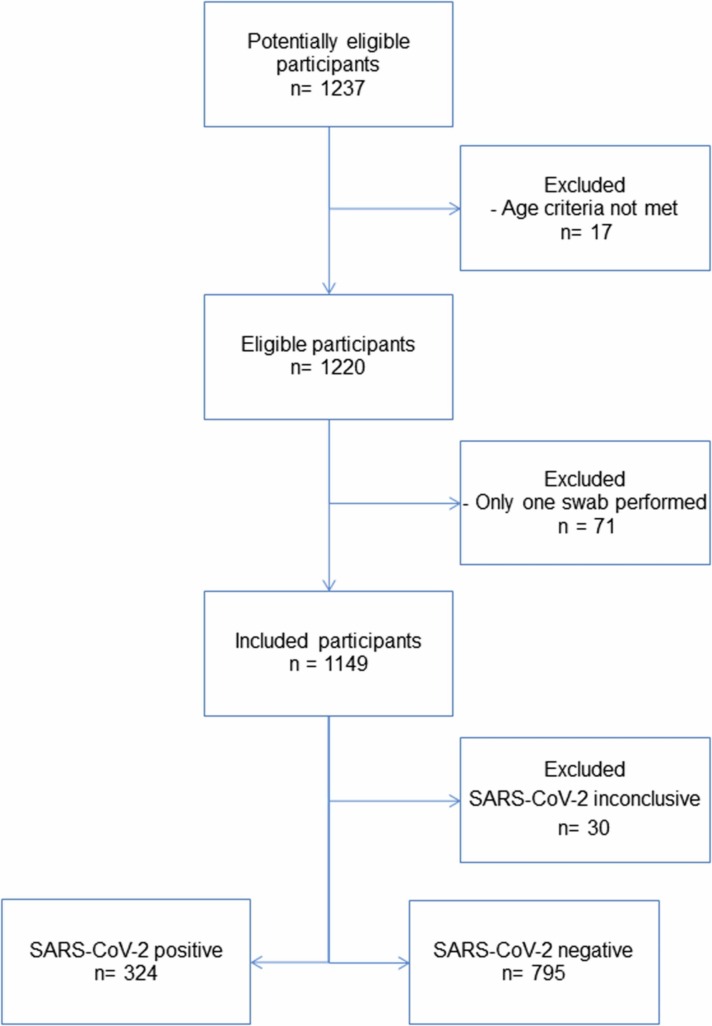
This prevalence study included 1119 participants recruited directly from a TCDK community test site ( Fig. 1). Subjects were approached and, if aged 18 or older, offered to participate in connection with the standard routine OP swab performed by HCWs. Upon agreement, participants were handed a written instruction on how to perform a combined oropharyngeal- and nasal midturbinate (NAOP) swab of both nares using the same flocked swab, and subsequently, break the swab stick and place it in a transport vial (detailed description in Supplement S1). A HCW was present to observe but did not supervise the self-swab. Participants who only performed a single swab were excluded from the study. The cohort included 631 (56.4 %) participants with self-reported symptoms and the age ranged between 18 and 88 (median 40), where 651 (58.2 %) participants were female (Supplementary Table S1).
Fig. 1.
STARD diagram of the study population.
All samples were analyzed according to standard Danish national testing protocol i.e. samples were transported dry to TCDK’s laboratory at Statens Serums Institute (SSI), and analyzed within 48 h. The samples underwent RNA purification on a Beckman Coulter Biomek® i7 automated workstation (Beckman Coulter Life Sciences, Indianapolis, NV, USA) using the Beckman Coulter RNAdvance® Blood kit.RT-PCR for SARS-CoV-2 detectionwas performed using Luna® Universal Probe One-Step RT-qPCR Kit reaction buffer and Luna® WarmStart RT Enzyme mix (New England Biolabs Inc., Ipswich, MA, USA) on a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States). The cycling conditions were reverse transcription at 55⁰C for 10 min, initial denaturation at 95⁰C for 3 min, followed by 45 cycles of denaturation and annealing/extension at 95⁰C for 15 s and 58⁰C at 30 s, respectively.
A sample was considered positive if the Ct-value was between 10 and 38, inconclusive between 38 and 40, and negative if Ct > 40. 299 participants tested positive in both sampling methods, whereas 10 were only positive in the OP swab and 15 in the NAOP swab ( Table 1A). Participants who received an inconclusive result in either sample types, were excluded from the study (Fig. 1).
Table 1.
Comparison of results between the sampling methods A. Contingency table showing the distribution of positives and negatives results between combined NAOP self-swabs and HCW OP swabs. B. Sensitivity of HCW OP swab and combined NAOP self-swab in the entire study population, symptomatic and asymptomatic subgroups. N = number of participants, pos = participants tested positive. The p-value was calculated using McNemar’s exact test.
| A | |||
|---|---|---|---|
| NAOP | |||
| OP | Positive | Negative | Total |
| Positive | 299 | 10 | 309 |
| Negative | 15 | 795 | 810 |
| Total | 314 | 805 | 1119 |
| B | ||||
|---|---|---|---|---|
| Population (N) | Pos | OP sensitivity | NAOP sensitivity | P-value |
| All (1149) | 324 | 95.4 % | 96.9 % | 0.424 |
| Symptomatic participants (642) | 233 | 96.6 % | 97.0 % | 1.000 |
| Asymptomatic participants (507) | 91 | 92.3 % | 96.7 % | 0.344 |
2. Comparison of sensitivities and Ct-values between sampling methods
When assuming all samples testing positive via RT-PCR are true positive, the sensitivity of OP- and NAOP swabs were 95.4 % and 96.9 % respectively (Table 1B), and no significant difference between the performances of the two swabs were found.
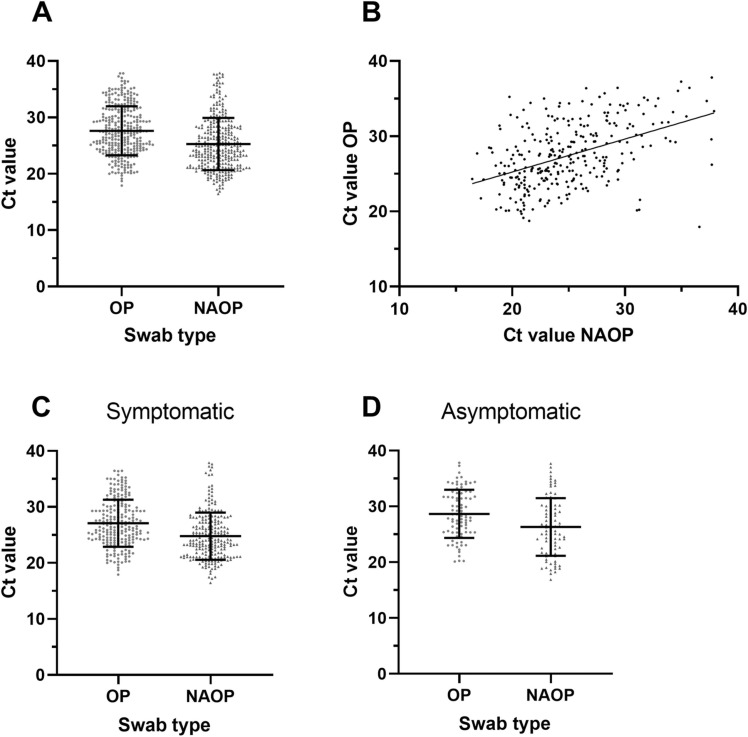
Fig. 2 A shows the Ct-values of all positive swabs, where the mean Ct-value for the NAOP and OP swabs were 25.27 and 27.63, respectively. The difference in Ct-values was statistically significant according to a paired t-test (p < 0.0001). The lower observed Ct-values for the self-collected swabs denote a higher viral load in this sample type and thus, may indicate the presence of virus at both swab sites.
Fig. 2.
Distributions of Ct-values from the positive samples from both sampling methods A. Ct-values of all positive samples detected by swab type. B. Scatter plot of paired samples positive in both swab types. C. Ct-values of positive samples from the symptomatic subgroup detected by swab type. D. Ct-values of positive samples from the asymptomatic subgroup detected by swab type. Black line at mean with standard deviation (SD) error bars.
Furthermore, a weak correlation between the Ct-values for the NAOP and OP swabs was observed (r = 0.4534, 95 % CI 0.3584–0.5391, p < 0.0001), where increasing Ct-values for the NAOP swabs was only weakly associated with an increased Ct-value for the corresponding OP swab (and vice versa) (Fig. 2B).
3. No heterogeneity for symptomatic and asymptomatic subpopulations
For the groups of symptomatic and asymptomatic participants, no significant difference was observed between the performances of OP- and combined NAOP swabs (Table 1B). However, the NAOP swab showed a tendency towards better performance compared to the OP swabs for the asymptomatic population, with sensitivities of 96.7 % and 92.3 % (p = 0.344), respectively. Similar to the overall findings, the Ct-values of the NAOP swabs were significantly lower compared to the OP swabs in both the symptomatic (p < 0.0001) and asymptomatic (p < 0.0001) population (Fig. 2C and D).
4. Discussion
Self-performed swabs have great potential as they reduce the need for trained HCWs, reduction in personal protective equipment limit exposure to infections for both HCWs and individuals waiting in line for routine testing, and can be used for rapid upscaling. This study showed that the sensitivity of a combined NAOP self-swab was equivalent to an OP swab performed by a HCW. This is in accordance with previous studies and systematic reviews reporting that combined nasal and throat swabs have a percent positive detection equivalent to or higher compared to reference swabs (Lee et al., 2021, Tan et al., 2020, Vlek et al., 2021, Wehrhahn et al., 2020). Furthermore, Tsang et al. found that self-collection of pooled nasal and throat swabs was not associated with any significant impairment of diagnostic accuracy (Tsang et al., 2021).
Although the mean Ct-value of the NAOP self-swab was significantly lower compared to the OP swabs, a higher viral load in the NAOP self-swabs did not equal significantly better sensitivity. All mean Ct-values were < 30 in our study and only a few samples had a Ct-value close to the limit for positive result detection in the PCR analysis. This may explain why no significant difference could be observed between the sensitivities of the sampling methods contrary to the mean Ct-values.
The currently dominating variant, Omicron, has been shown to have a lower viral load in nasopharyngeal samples and improved oral detection compared to the earlier Delta variant (Gert Marais, 2022; Sentis et al., 2022), thus the tissue tropism may vary between variants. Using combined swabs could ensure a high detection rate regardless of changes in viral load between the two tissue sites.
An advantage of our study is the high number of participants, all recruited from a standard community test site. This ensures that the broad general population with both symptomatic and asymptomatic individuals is represented. There was no significant difference in sensitivity between the samplings methods in subgroups. However, a tendency towards higher sensitivity for the NAOP swab in the asymptomatic subgroup, suggests that the combined self-swab has at least the same positive detection rate as the OP swab for asymptomatic subjects.
5. Conclusion
Our study demonstrates that a combined nasal midturbinate and oropharyngeal self-swab has a sensitivity equivalent to or better than a deep oropharyngeal swab performed by a health care worker. The study was conducted in a real-life setting with a large cohort including both symptomatic and asymptomatic participants, and thus suggests NAOP self-swab as an alternative sampling method under the given circumstances.
Ethical statement
This study did not require ethical approval, in accordance with the Danish Act on Research Ethics Review of Health Research Projects of 2018. The planning and execution of this study are in line with the Declaration of Helsinki as revised in 2013.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Sofie Hørlyck: Conceptualization, Methodology, Formal analysis, Project administration, Investigation, Writing – original draft, Visualization. Sofie Holdflod Nielsen: Conceptualization, Methodology, Formal analysis, Project administration, Writing – original draft, Visualization. Tobias Gress: Conceptualization, Methodology, Formal analysis, Project administration, Writing – original draft, Visualization. Uffe Schneider: Conceptualization, Methodology, Writing – original draft, Visualization. Cyril Jean-Marie Martel: Conceptualization, Writing – original draft, Visualization. Nina Steenhaard: Conceptualization. Niels Tobias Gredal: Formal analysis, Project administration. Shila Mortensen: Conceptualization. Arieh S. Cohen: Conceptualization, Writing – original draft, Visualization, Writing – review & editing, Supervision. All authors provided approval of the final version to be published.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank the individuals who volunteered for this study. We thank the collection staff at Test Center Valby and the laboratory staff at TCDK, without whom this study would not have been possible. We thank the following individuals for their invaluable contributions to the design and development of this project: Claus Nielsen, Pia Webster and Ellinor Lindberg Marvin at Statens Serums institut. We would also like to thank the IT Group at TCDK, Statens Serums Institut for assistance with data handling.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jviromet.2022.114667.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
Data will be made available upon request
References
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gert Marais N.-Y.H., Arash Iranzadeh, Deelan Doolabh, Rageema Joseph, Annabel Enoch, Chun-yat Chu, Carolyn Williamson, Adrian Brink, Diana Hardie. Improved oral detection is a characteristic of Omicron infection and has implications for clinical sampling and tissue tropism. J. Clin. Virol. 2022:152. doi: 10.1016/j.jcv.2022.105170. [DOI] [PubMed] [Google Scholar]
- Lee R.A., Herigon J.C., Benedetti A., Pollock N.R., Denkinger C.M. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J. Clin. Microbiol. 2021:59. doi: 10.1128/JCM.02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentis C., Billaud G., Bal A., Frobert E., Bouscambert M., Destras G., Josset L., Lina B., Morfin F., Gaymard A. SARS-CoV-2 omicron variant, lineage BA.1, is associated with lower viral load in nasopharyngeal samples compared to delta variant. Viruses. 2022;14:919. doi: 10.3390/v14050919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.Y., Tey H.L., Lim E.T.H., Toh S.T., Chan Y.H., Tan P.T., Lee S.A., Tan C.X., Koh G.C.H., Tan T.Y., Siau C. The accuracy of healthcare worker versus self collected (2-in-1) oropharyngeal and bilateral mid-turbinate (OPMT) swabs and saliva samples for SARS-CoV-2. PLOS ONE. 2020;15 doi: 10.1371/journal.pone.0244417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang N.N.Y., So H.C., Ng K.Y., Cowling B.J., Leung G.M., Ip D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis. Lancet Infect. Dis. 2021;21:1233–1245. doi: 10.1016/S1473-3099(21)00146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlek A.L.M., Wesselius T.S., Achterberg R., Thijsen S.F.T. Combined throat/nasal swab sampling for SARS-CoV-2 is equivalent to nasopharyngeal sampling. Eur. J. Clin. Microbiol. Amp; Infect. Dis. 2021;40:193–195. doi: 10.1007/s10096-020-03972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Muenker M.C., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O.-E., Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Cold Spring Harbor Laboratory; 2020. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR primer-probe sets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrhahn M.C., Robson J., Brown S., Bursle E., Byrne S., New D., Chong S., Newcombe J.P., Siversten T., Hadlow N. Self-collection: an appropriate alternative during the SARS-CoV-2 pandemic. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available upon request