Abstract
Background
In spring of 2022, an outbreak of monkeypox (mpox) spread worldwide. Here, we describe performance characteristics of monkeypox virus (MPXV)-specific and pan-orthopoxvirus qPCR assays for clinical use.
Methods
We validated probe-based qPCR assays targeting MPXV-specific loci F3L and G2R (genes MPXVgp052/OPG065 and MPXVgp002 and gp190/OPG002, respectively) and a pan-orthopoxvirus assay targeting the E9L locus (MPXVgp057/OPG071). Clinical samples and synthetic controls were extracted using the Roche MP96 or Promega Maxwell 48 instrument. qPCR was performed on the AB7500 thermocycler. Synthetic control DNA and high concentration clinical samples were quantified by droplet PCR. Cross-reactivity was evaluated for camelpox and cowpox genomic DNA, vaccinia culture supernatant, and HSV- and VZV-positive clinical specimens. We also tested the performance of the F3L assay using dry swabs, Aptima vaginal and rectal swabs, nasopharyngeal, rectal, and oral swabs, cerebrospinal fluid, plasma, serum, whole blood, breastmilk, urine, saliva, and semen.
Results
The MPXV-F3L assay is reproducible at a limit of detection (LoD) of 65.6 copies/mL of viral DNA in viral transport medium/universal transport medium (VTM/UTM), or 3.3 copies/PCR reaction. No cross-reactivity with herpesviruses or other poxviruses was observed. MPXV-F3L detects MPXV DNA in alternative specimen types, with an LoD ranging between 260-1000 copies/mL, or 5.7-10 copies/PCR reaction. In clinical swab VTM specimens, MPXV-F3L and MPXV-G2R assays outperformed OPXV-E9L by an average of 2.4 and 2.8 Cts, respectively. MPXV-G2R outperformed MPXV-F3L by 0.4 Cts, consistent with presence of two copies of G2R present in labile inverted terminal repeats (ITRs) of MPXV genome.
Conclusions
MPXV is readily detected by qPCR using three clinically validated assays.
Keywords: Monkeypox virus, qPCR, Orthopox, F3L, G2R, Alternative specimen, mpox
1. Introduction
Since its discovery in the 1970s, human cases of monkeypox/mpox have rarely been reported outside of countries in western and central Africa where the causative agent, monkeypox virus (MPXV), is endemic [1]. Mpox is a zoonotic disease, and frequently causes isolated human infections through spillover events [2]. Despite evidence of increased human-to-human transmission and warnings of global spread, the disease was largely neglected until its recent emergence in an outbreak beginning in spring of 2022 [3], [4], [5]. By November 2022, the epidemic had expanded to over 77,000 cases detected in more than 100 countries worldwide. In the United States, at time of writing, more than 28,000 cases have been reported, as well as several deaths [6]. Worldwide, this outbreak of human mpox has disproportionately burdened men who have sex with men [7].
Individuals who contract MPXV may experience fever, lymphadenopathy, and a disseminated papillary rash that can lead to secondary bacterial infections [[8], [9]]. Effects may include significant pain for weeks, as well as a wide range of complications including encephalitis, bronchopneumonia, and vision loss [10]. Immunocompromised individuals, particularly those with AIDS, are at high risk of severe manifestations of MPXV, including death [11]. Prophylactic vaccination with the JYNNEOS vaccinia virus vaccine and treatment with the antiviral tecovirimat (Tpoxx), both originally developed for variola (smallpox), are the major medical countermeasures available for MPXV. Because prevention of MPXV spread and effective treatment of MPXV infection inherently rely on rapid and early detection, development of clinical quantitative PCR (qPCR) assays is critical for patient care and to disrupt networks of transmission. In the context of a developing outbreak of an emerging virus that can be shed in multiple body fluids, evaluation of assay performance in many alternative specimen types is also critical to preparedness efforts [[12], [13]].
MPXV is a member of the orthopoxvirus (OPXV) family of double-stranded DNA viruses. Although they are large (100-300 kb), OPXV genomes are highly conserved across species, making MPXV-specific qPCR assay design a challenge. We evaluated two MPXV-specific qPCR assays that target the F3L and G2R loci, and compared the sensitivity, specificity, and accuracy of these assays to the CDC pan-orthpoxvirus assay OPXV-E9L [14], [15], [16]. We also assessed performance of all three assays in the presence of alphaherpesviruses. We found MPXV-F3L and MPXV-G2R assays to have equivalent analytical sensitivity, with 100% sensitivity at the limit of detection (LoD) of 65.6 copies/mL. OPXV-E9L had a sensitivity of 95% at the same LoD. We demonstrated that the MPXV-F3L assay does not cross react with Camelpox virus (CMLV), Cowpox virus (CPXV) or Vaccinia virus (VACV) and detects MPXV DNA with adequate sensitivity in fourteen different specimen types.
2. Materials and methods
2.1. Ethics and regulatory approvals
Use of excess clinical specimens was approved by the University of Washington Institutional Review Board with a consent waiver (STUDY00010205) or with informed consent (STUDY00000853, STUDY00001055, STUDY00008491).
2.2. Primers and probes
Primers and probes targeting the loci for MPXV-F3L [15], MPXV-G2R [14], and OPXV-E9L [16] were synthesized by ThermoFisher. Sequences used were as follows: F3L forward 5´-CATCTATTATAGCATCAGCATCAGA-3´ and reverse 5´-GATACTCCTCCTCGTTGGTCTAC -3´, probe 5´-FAM/TGTAGGCCGTGTATCAGCATCCATT/BHQ1-3´. G2R forward 5′-GGAAAATGTAAAGACAACGAATACAG-3´ and reverse 5´-GCTATCACATAATCTGGAAGCGTA-3´, probe 5´-FAM/AAGCCGTAATCTATGTTGTCTATCGTGTCC/BHQ1-3´. E9L forward 5´-TCAACTGAAAAGGCCATCTATGA-3´ and reverse 5´-GAGTATAGAGCACTATTTCTAAATCCCA-3´, probe 5´-VIC/CCATGCAATATACGTACAAGATAGTAGCCAAC/QSY-3´.
2.3. Nucleic acid extraction
All samples were handled in a biosafety cabinet prior to viral inactivation. DNA from breastmilk and semen specimens was extracted on the Promega Maxwell 48 with Maxwell RSC Viral Total Nucleic Acid Purification Kit. Breastmilk specimens were spun down for 10 min to separate the fat layer, and the liquid was used for extraction. 220µL of sample were mixed with 200μL extraction mix (181.5μL lysis buffer, 18.2μL proteinase K, and 0.38μL EXOBS internal control). Samples were then incubated for 10 min at room temperature and 10 min at 56°C, then extracted following manufacturer protocols and eluted in 100 μL of Promega elution buffer.
DNA from all other specimen types was extracted on the MagNA Pure 96 with the DNA and Viral NA Small Volume Kit unless otherwise noted. Aptima collection tubes contain a reagent to inactivate MPXV, so 200μL from these tubes was extracted without additional lysis buffer. For lesion, nasopharyngeal, rectal, or oral swabs in viral transport media or universal transport media (VTM/UTM), cerebrospinal fluid (CSF), plasma, serum, whole blood, urine, or saliva, 100μL of sample was added to 100μL of buffer AL (Qiagen). Dry swabs were added to 1 mL VTM/UTM, incubated for 1 min, vortexed for 10 s, and treated as above. All samples were eluted in 100μL Roche elution buffer.
Select specimens were extracted on the MagNA Pure 96 with the DNA and Viral NA Large Volume Kit, with 250μL of sample added to 250μL of buffer AL (Qiagen), then eluted in 50μL Roche elution buffer.
2.4. Qualitative PCR
All F3L and G2R qPCR reactions contained 2.71µL water, 11.95µL No-ROX QuantiTect master mix (Qiagen), 0.55µL ROX QuantiTect master mix (Qiagen), 0.1µL each of 100μM assay-specific forward and reverse primers, 0.05µL of 100μM assay-specific probe, 0.063μL EXOBS internal control primer/probe mix [17], 0.025µL of Uracil-N-Glycosylase (0.025 units, EpiCentre technologies), and 10μL of template DNA. All E9L qPCR reactions contained an additional 0.063µL water in place of EXOBS primer/probe mix. Amplification on the 7500 Real-Time PCR System (Applied Biosystems) consisted of 2 min at 50°C; 15 min at 95°C; and 45 cycles of 1 min at 94°C and 1 min at 60°C [18].
Additional methodological details for ddPCR, accuracy, cross-reactivity and interfering substances, and alternative specimen types are available in Supplemental Methods.
3. Results
3.1. Absolute quantification of synthetic control and high viral load clinical sample
Because MPXV genomic DNA and clinical samples were in short supply in spring 2022, we first validated synthetic DNA purchased from ATCC (VR-3270SD). This template contains primer and probe binding sites for several previously-developed assays, including F3L, G2R, and E9L, and has a copy number of 1 × 108-1 × 109 copies/mL DNA estimated by the manufacturer [14], [15], [16]. Measurements of VR-3270SD lot 70053297 by ddPCR yielded 3.28 × 108 copies/mL DNA for F3L; measurements of VR-3270SD lot 70053666 resulted in equivalent concentrations for all three loci: 1.28 × 108 copies/mL for MPXV-F3L, 1.30 × 108 copies/mL for MPXV-G2R, and 1.29 × 108 copies/mL for OPXV-E9L (Figure S1A-C). In addition, a high viral load remnant clinical specimen (F3L Ct ∼16) used for downstream validation studies was quantified by ddPCR as 5.34 × 108, 8.78 × 108, and 4.78 × 108 copies/mL for F3L, G2R, and E9L respectively. These data are consistent with equal copy numbers of loci present in the ATCC DNA standard and an extra copy of G2R present in MPXV genomes.
3.2. MPXV-F3L and MPXV-G2R assays are highly specific for MPXV DNA
To determine specificity, we first tested 15 HSV-positive, 17 VZV-positive, and 52 known HSV/VZV-negative clinical remnant skin swabs in VTM/UTM using the CDC OPXV-E9L assay and confirmed that they were negative for MPXV. We verified that the F3L assay did not detect MPXV DNA in any of these specimens. The G2R assay similarly did not detect MPXV DNA in the negative or HSV-positive swabs, or in 16 out of 17 VZV-positive swabs; one VZV-positive specimen tested positive for G2R (Ct 39.8) and was negative on repeat (Table 1 ). Overall, the F3L assay had a negative percent agreement of 100% and the G2R assay had a negative percent agreement of 98.8% over 84 samples.
Table 1.
MPXV-F3L, MPXV-G2R, and OPXV-E9L sensitivity and specificity.
| F3L | Known (Contrived) Positive | Known Negative |
|---|---|---|
| Assay Positive | 31 | 0 |
| Assay Negative | 1 | 84 |
| G2R | Known (Contrived) Positive | Known Negative |
|---|---|---|
| Assay Positive | 32 | 1 |
| Assay Negative | 0 | 83 |
| E9L | Known (Contrived) Positive | Known Negative |
|---|---|---|
| Assay Positive | 32 | 0 |
| Assay Negative | 0 | 84 |
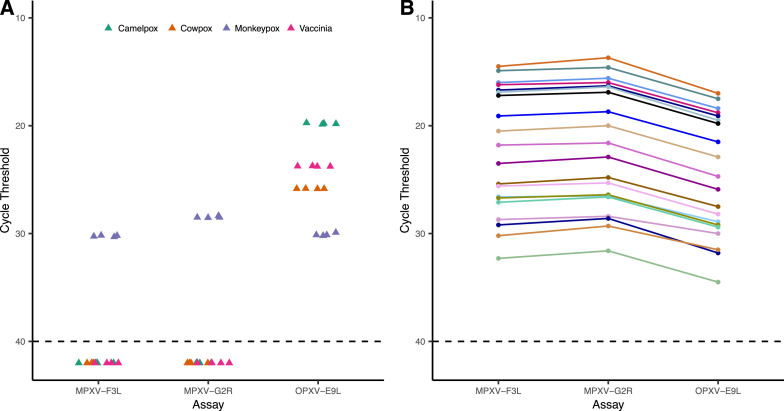
Camelpox virus (CMLV), Cowpox virus (CPXV) and Vaccinia virus (VACV) are the nearest phylogenetic relatives to MPXV based on DNA sequence similarity of conserved OPXV genes[[16], [19]]. Accordingly, we tested VACV viral culture and purified CMLV and CPXV DNAs to assess cross-reactivity with MPXV-F3L and MPXV-G2R. The OPXV-E9L PCR assay served as a positive control for the other OPXV. VACV, CPXV, and CMLV were not detected by either the MPXV-F3L or MPXV-G2R assays, while the OPXV-E9L assay returned average Cts of 23.8, 25.8, and 19.8, respectively (Fig. 1 A).
Fig. 1.
Specificity of MPXV-F3L and MPXV-G2R assays. (A) OPXV cross reactivity analysis of MPXV specific assays by testing MXPV, CPXV, CMLV, and VACV samples, with OPXV-E9L used as a positive control. Samples with no detectable MPXV are plotted at Ct of 42. (B). Specificity of all three assays with 20 clinical specimens initially positive by MPXV-F3L, followed by concomitant testing with MPXV-G2R and MPXV-E9L.
3.3. MPXV-F3L, MPXV-G2R, and OPXV-E9L assays are > 96.8% sensitive for MPXV DNA
All three assays were tested for their ability to detect MPXV DNA using contrived positives consisting of a range of dilutions of VR-3270SD spiked into negative skin swabs in VTM (eight replicates at 330,000 copies/mL, 16 at 33,000 copies/mL, and eight at 3,300 copies/mL, based on ddPCR). G2R and E9L assays had 100% positive percent agreement, while a single replicate at 3,300 copies/mL was negative in the F3L assay (G2R Ct = 36.6), resulting in a positive percent agreement of 96.9%. Upon quadruplicate repeat testing, the failed sample had F3L Cts of 32.2, 33.4, 32.9, and 33.0. Overall percent agreement for the three assays were 99.1% for both G2R and F3L, and 100% for E9L.
3.4. MPXV-F3L and MPXV-G2R assays detect MPXV viral DNA
Next, we confirmed that the F3L and G2R assays would accurately identify samples containing MPXV. Due to the short supply of MPXV-positive samples and genomic DNA early in the 2022 MPXV pandemic, we were secured MPXV genomic DNA from BEI Resources, as well as seven MPXV-positive skin swabs in VTM from the WA State Public Health Laboratory (PHL). All known positive samples tested positive for MPXV by both F3L and G2R assays. Because of volume constraints, the PHL samples were diluted at least 1:40 prior to extraction at our lab, resulting in later Ct values for F3L and G2R assays than the Ct values for OPXV-E9L assay used by PHL, and complicating quantitative comparison of the assays. The qualitative interpretation was 100% concordant.
In order to quantitatively compare the ability of the three assays to detect MPXV DNA, we took 20 MPXV-positive clinical swab specimens in VTM (as detected by MPXV-F3L assay) and performed concomitant MPXV-G2R and OPXV-E9L testing on them. In every specimen, the MPXV-specific primers gave lower Ct values with averaging Ct differences of 2.4 (range, 1.3-2.9) and 2.8 (1.6-3.2) lower than OPXV-E9L for MPXV-F3L and MPXV-G2R assays, respectively. In addition, the MPXV-G2R gave a lower Ct value than MPXV-F3L in every specimen, averaging 0.4 Ct difference (0.1-0.9) (Fig. 1B, Supplemental Table 1). This is consistent with two copies of G2R being present in the MPXV genome.
3.5. MPXV-F3L, MPXV-G2R and OPXV-E9L assays are not affected by alphaherpesviruses
We tested if HSV or VZV interfered with the detection of MPXV since co-infections have been reported previously [[7], [20]]. Contrived MPXV-positive controls with no herpesviruses (n=32) resulted in Cts with no significant difference from those MPXV/herpesviruses double positives for all three assays (Supplemental Figure 2, Supplemental Table 2, p>0.05, T test).
3.6. MPXV-F3L, MPXV-G2R, and OPXV-E9L assays have analytic sensitivity of 3.3 copies per PCR reaction
We performed an initial LoD for MPXV-F3L, and MPXV-G2R assays using contrived positive specimens containing VR-3270SD at a range of concentrations (serial dilutions from 330,000-33 copies/mL VTM) tested in quadruplicate. For both assays, all four replicates were detected at 330,000; 33,000; 3,300; and 330 copies/mL, while only 2/4 and 3/4 of replicates amplified at 33 copies/mL with F3L and G2R, respectively.
We next tested twenty additional replicates of contrived positive samples at both 330 copies/mL and 33 copies/mL with F3L and G2R assays. At 330 copies/mL, corresponding to 3.3 copies per PCR reaction, 19/20 were detected for F3L and G2R, with mean Ct values of 34.5 and 36.8 respectively. At 33 copies/mL, corresponding to only 0.3 copies per reaction, 4/20 and 3/20 replicates were detected for F3L and G2R respectively. Therefore, the limit of detection of both F3L and G2R assays is 330 copies/mL using standard extraction methods, or 3.3 copies per reaction.
An additional LoD study was performed using large volume extraction methods that provided a theoretical 5x concentration of DNA over the standard extraction methods. This study used both synthetic DNA (VR-3270SD, with equal concentration of all three targets) and high-titer clinical specimen diluted in negative clinical specimens. VR-3270SD was first diluted 10-fold in MPXV-negative specimen VTM to final concentrations of 3,300,000-33 copies/mL as well as finer dilutions between 330 and 33 copies/mL, and was tested in quadruplicate (Supplemental Table 4). Linearity was excellent among positive samples (Supplemental Figure 3), for F3L, G2R, and E9L assays with R2 values of 0.9976, 0.9972, and 0.9961 respectively.
For all three assays, 4/4 replicates were detected at 3,300,000-330 and 110 copies/mL dilutions, while only 3/4, 3/4, or 1/4 replicate(s) amplified at 66 copies/mL with F3L, G2R, and E9L assays respectively. Second, 20 replicates each of both VR-3270SD and the high viral load clinical specimen were tested at 330, 110, and 66 copies/mL (copies of F3L for the clinical specimen). For VR-3270SD dilutions, 20/20 replicates amplified at 330 copies/mL with all assays; 19/20, 19/20, and 16/20 replicates amplified at 1.1e2 copies/mL with F3L, G2R, and E9L assays respectively; and 12/20, 11/20, and 13/20 replicates amplified at 66 copies/mL with F3L, G2R, and E9L assays respectively. For clinical specimen dilutions at 66 F3L copies/mL by ddPCR and a theoretical 3.3 copies/rxn, 20/20, 20/20, and 19/20 replicates amplified with F3L, G2R, and E9L assays respectively, with mean Ct values of 35.0, 33.9, and 36.6 respectively (Supplemental Table 5). The LoD for each assay using the higher input volume was 110 copies/mL for the F3L and G2R assays and 330 copies/mL VTM for the OPXV-E9L assay based on the ATCC DNA material and 66 copies/mL VTM for each assay based on ddPCR-determined copy number of a viral positive.
3.7. MPXV DNA is detected in diverse specimen types at a viral load of 1000 copies per mL or lower
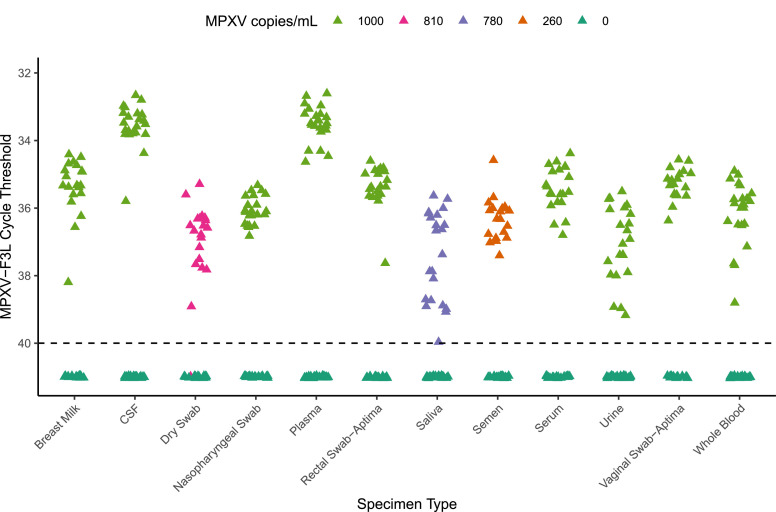
Given the need for testing for MPXV in alternative specimen types, we also validated the F3L assay LoD in multiple other specimen types. Presumptive MPXV-negative cerebrospinal fluid, plasma, serum, urine, breastmilk, whole blood, nasopharyngeal/rectal/oral swabs in VTM/UTM, and vaginal/rectal swabs in Aptima tubes, were spiked to a concentration of 1000 copies/mL of VR-3270SD, followed by extraction and qPCR on both spiked and negative samples. The negative percent agreement for all specimen types was 100%. The positive percent agreement for Aptima vaginal and rectal swabs, nasopharyngeal, cerebrospinal fluid, plasma, breastmilk, and whole blood was 100%. The F3L assay was positive in 22/23 urine specimens and 21/22 serum specimens, for a positive percent agreement of 95.7% and 95.5% respectively (Fig. 2 , Supplemental Table 3). Therefore, we confirmed LoD of F3L for these samples at 1000 copies/mL.
Fig. 2.
Sensitivity of MPXV-F3L With Diverse Specimens. MPXV-F3L Ct values of contrived MPXV positive breastmilk, CSF, dry swabs, nasopharyngeal swabs (NP), plasma, rectal swabs, saliva, semen, urine, vaginal swabs, and whole blood samples (n=17-24). Samples with no detectable MPXV are plotted at Ct of 41.
Once MPXV-positive specimens were available, and specifically for specimen types in which DNA degradation might be a concern (specifically dry swabs, semen, and saliva), we performed our validation using diluted MPXV clinical specimen rather than synthetic DNA. The high viral load clinical specimen was spiked into semen at 260 copies/mL, and into saliva and oral and rectal swabs at 780 copies/mL. Dry swabs were spiked with 810 copies per swab. Negative percent agreement of the F3L assay of semen, saliva, rectal, oral, and dry swabs was 100%. Positive percent agreement was 100% for semen, saliva, and rectal swabs, and 95% (19/20) for oral and dry swabs. Therefore, the F3L LoDs were confirmed in semen at 260 copies/mL (5.7 copies per PCR reaction), 780 copies/mL (7.8 copies per PCR reaction) for saliva, oral, and rectal swabs, and 810 copies/swab (8.1 copies per PCR reaction) for dry swabs.
4. Discussion
Prior work on MPXV diagnostics have determined performance characteristics for each of these assays individually MPXV-F3L, MPXV-G2R, and OPXV-E9L individually with either synthetic constructs or purified MPXV virions, but have not compared their performance to each other [[14], [15], [16], [21], [22]]. Our work strengthens and advances this work via a systematic comparison of all three assays in tandem with both synthetic constructs and viral specimens using a rigorous LoD validation. In addition, we assess the MPXV-F3L assay performance in the presence of different potential viral interferents and validated MPXV detection in 14 diverse specimen types at an LoD of 1000 copies/mL or lower. Through extracting from up to 250 μL of specimen, we have demonstrated the LoD for MPXV-F3L, MPXV-G2R, and OPXV-E9L assays to be as low as 66 copies/mL, or 3.3 copies per PCR reaction. Our analytical sensitivity compares favorably with a recent evaluation of the PKamp Monkeypox Virus RT-PCR assay, which also uses the F3L gene as its target [21].
Although overall both MPXV-specific assays had very similar performance characteristics, F3L had a slightly lower Ct on average on synthetic positive control material and was present in a core genomic region, leading us to use it as our primary clinical assay for high-volume testing of swabs in VTM [18]. In clinical samples, G2R detection was associated with slightly lower Ct values than F3L, likely due to the presence of two copies of the gene in the MPXV ITR regions. However, ITR regions of poxviruses are often subject to genomic rearrangements, especially as they spread to new hosts [[23], [24]]. Reports of rare deletions in the G2R target that affected detection of MPXV DNA led the CDC to issue an advisory on September 2nd, 2022, urging caution in the interpretation of negative results returned from the G2R assay if clinical suspicion for MPXV was high [25]. To date, no reports of deletions in F3L in MPXV have occurred.
Limitations of our work include the lack of housekeeping gene specimen adequacy controls within the qPCR reaction. Although specimen adequacy controls have shown association with MPXV viral loads in MPXV-positive specimens [26], there has not been a rigorous evaluation of the value added by specimen adequacy controls, though they are now requested in the FDA assay authorization template. We also did not determine the absolute lower limit of detection for all alternative specimen types, due to limited matrices and the adequacy of confirmatory LoDs of <1000 copies/mL for each specimen type.
Altogether, we find very similar performance characteristics for the MPXV-F3L, MPXV-G2R, and OPXV-E9L assays for the detection of MPXV. Each assay was highly sensitive for MPXV DNA, with an LoD around 3 copies per reaction, near the limit of stochasticity. The F3L assay is specific for MPXV DNA, and does not cross-react with herpesviruses or other orthopoxviruses. This diagnostic will be an important defense for reducing transmission of MPXV during the ongoing outbreak.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: ALG reports contract testing from Abbott, Cepheid, Novavax, Pfizer, Janssen and Hologic and research support from Gilead and Merck, outside of the described work. Dr Kachikis reported serving as a research consultant for Pfizer and GlaxoSmithKline on maternal immunization-related projects in 2020 and as an unpaid consultant for GlaxoSmithKline in 2022 outside the submitted work. Dr Kachikis reported receiving grant support from Merck and Pfizer outside the submitted work.
Acknowledgements
The authors thank our study participants for their time and effort to donate specimens to the University of Washington Medical Center. AK is supported by NIH K23 AI153390-01.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105373.
Appendix. Supplementary materials
References
- 1.Ladnyj ID, Ziegler P, Kima E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull. World Health Organ. 1972;46:593–597. [PMC free article] [PubMed] [Google Scholar]
- 2.Berthet N, Descorps-Declère S, Besombes C, Curaudeau M, Nkili Meyong AA, Selekon B, Labouba I, Gonofio EC, Ouilibona RS, Simo Tchetgna HD, Feher M, Fontanet A, Kazanji M, Manuguerra J-C, Hassanin A, Gessain A, Nakoune E. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. 1. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-92315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomassen HA, Fuller T, Asefi-Najafabady S, Shiplacoff JAG, Mulembakani PM, Blumberg S, Johnston SC, Kisalu NK, Kinkela TL, Fair JN, Wolfe ND, Shongo RL, LeBreton M, Meyer H, Wright LL, Muyembe J-J, Buermann W, Okitolonda E, Hensley LE, Lloyd-Smith JO, Smith TB, Rimoin AW. Pathogen-host associations and predicted range shifts of human monkeypox in response to climate change in central Africa. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0066071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva NIO, de Oliveira JS, Kroon EG, Trindade G de S, Drumond BP. Here, there, and everywhere: the wide host range and geographic distribution of zoonotic orthopoxviruses. 1. Viruses. 2021;13 doi: 10.3390/v13010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nolen LD, Osadebe L, Katomba J, Likofata J, Mukadi D, Monroe B, Doty J, Hughes CM, Kabamba J, Malekani J, Bomponda PL, Lokota JI, Balilo MP, Likafi T, Lushima RS, Ilunga BK, Nkawa F, Pukuta E, Karhemere S, Tamfum J-JM, Nguete B, Wemakoy EO, McCollum AM, Reynolds MG. Extended human-to-human transmission during a monkeypox outbreak in the Democratic Republic of the Congo. Emerg. Infect. Dis. 2016;22:1014–1021. doi: 10.3201/eid2206.150579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC . Cent Dis Control Prev; 2022. Monkeypox in the U.S.https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html Retrieved 17 August 2022. [Google Scholar]
- 7.Thornhill JP, Barkati S, Walmsley S, Rockstroh J, Antinori A, Harrison LB, Palich R, Nori A, Reeves I, Habibi MS, Apea V, Boesecke C, Vandekerckhove L, Yakubovsky M, Sendagorta E, Blanco JL, Florence E, Moschese D, Maltez FM, Goorhuis A, Pourcher V, Migaud P, Noe S, Pintado C, Maggi F, Hansen A-BE, Hoffmann C, Lezama JI, Mussini C, Cattelan A, Makofane K, Tan D, Nozza S, Nemeth J, Klein MB, Orkin CM. Monkeypox virus infection in humans across 16 countries — April–June 2022. N. Engl. J. Med. 2022;0 doi: 10.1056/NEJMoa2207323. null. [DOI] [PubMed] [Google Scholar]
- 8.McCollum AM, Damon IK. Human monkeypox. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2014;58:260–267. doi: 10.1093/cid/cit703. [DOI] [PubMed] [Google Scholar]
- 9.Ježek Z, Szczeniowski M, Paluku KM, Mutombo M. Human monkeypox: clinical features of 282 patients. J. Infect. Dis. 1987;156:293–298. doi: 10.1093/infdis/156.2.293. [DOI] [PubMed] [Google Scholar]
- 10.Monkeypox. https://www.who.int/news-room/fact-sheets/detail/monkeypox. Retrieved 18 August 2022.
- 11.Miller MJ. Severe Monkeypox in Hospitalized patients — United States, August 10–October 10, 2022. MMWR Morb. Mortal. Wkly Rep. 2022;71 doi: 10.15585/mmwr.mm7144e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perchetti GA, Nalla AK, Huang M-L, Zhu H, Wei Y, Stensland L, Loprieno MA, KR Jerome, Greninger AL. Validation of SARS-CoV-2 detection across multiple specimen types. J. Clin. Virol. Off. Publ. Pan. Am. Soc. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veintimilla C, Catalán P, Alonso R, de Viedma DG, Pérez-Lago L, Palomo M, Cobos A, Aldamiz-Echevarria T, Muñoz P. The relevance of multiple clinical specimens in the diagnosis of monkeypox virus, Spain, June 2022. Euro Surveill Bull. Eur. Sur. Mal. Transm. Eur. Commun. Dis. Bull. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.33.2200598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Zhao H, Wilkins K, Hughes C, Damon IK. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J. Virol. Methods. 2010;169:223–227. doi: 10.1016/j.jviromet.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maksyutov RA, Gavrilova EV, Shchelkunov SN. Species-specific differentiation of variola, monkeypox, and varicella-zoster viruses by multiplex real-time PCR assay. J. Virol. Methods. 2016;236:215–220. doi: 10.1016/j.jviromet.2016.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Olson VA, Laue T, Laker MT, Damon IK. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. Off. Publ. Pan. Am. Soc. Clin. Virol. 2006;36:194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allele-specific PCR for determination of IL28B genotype. J. Clin. Microbiol. 2022 doi: 10.1128/JCM.02084-12. https://journals.asm.org/doi/10.1128/JCM.02084-12?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed |. Retrieved 4 October. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieberman NAP, Mathias PC, Bradley BT, Greninger AL. 2022. Clinical performance and trends during the first two months of monkeypox Virus PCR testing at two United States reference labs. preprint. Infectious Diseases (except HIV/AIDS). [DOI] [PMC free article] [PubMed]
- 19.Emerson GL, Li Y, Frace MA, Olsen-Rasmussen MA, Khristova ML, Govil D, Sammons SA, Regnery RL, Karem KL, Damon IK, Carroll DS. The phylogenetics and ecology of the orthopoxviruses endemic to North America. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes CM, Liu L, Davidson WB, Radford KW, Wilkins K, Monroe B, Metcalfe MG, Likafi T, Lushima RS, Kabamba J, Nguete B, Malekani J, Pukuta E, Karhemere S, Muyembe Tamfum J-J, Okitolonda Wemakoy E, Reynolds MG, Schmid DS, McCollum AM. A tale of two viruses: coinfections of monkeypox and varicella zoster virus in the Democratic Republic of Congo. Am. J. Trop. Med. Hyg. 2021;104:604–611. doi: 10.4269/ajtmh.20-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mondolfi AP, Guerra S, Muñoz M, Luna N, Hernandez MM, Patino LH, Reidy J, Banu R, Shrestha P, Liggayu B, Umeaku A, Chen F, Cao L, Patel A, Hanna A, Li S, Look A, Pagani N, Albrecht R, Pearl R, Garcia-Sastre A, Bogunovic D, Palacios G, Bonnier L, Cera F, Lopez H, Calderon Y, Eiting E, Mullen K, Shin SJ, Lugo LA, Urbina AE, Starks C, Koo T, Uychiat P, Look A, van Bakel H, Gonzalez-Reiche A, Betancourt AF, Reich D, Cordon-Cardo C, Simon V, Sordillo EM, Ramírez JD. Evaluation and validation of an RT-PCR assay for specific detection of monkeypox virus (MPXV) J. Med. Virol. 2022 doi: 10.1002/jmv.28247. [DOI] [PubMed] [Google Scholar]
- 22.Uhteg K, Mostafa HH. Validation and implementation of an orthopoxvirus qualitative real-time PCR for the diagnosis of monkeypox in the clinical laboratory. J. Clin. Virol. Off. Publ. Pan. Am. Soc. Clin. Virol. 2022;158 doi: 10.1016/j.jcv.2022.105327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, Geballe AP, Malik HS. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. 2012;150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sereewit J, Lieberman NAP, Xie H, Bakhash SAKM, Nunley BE, Chung B, Mills MG, Roychoudhury P, Greninger AL. ORF-interrupting mutations in Monkeypox virus genomes from Washington and Ohio, 2022. 11. Viruses. 2022;14 doi: 10.3390/v14112393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.2022. Lab Alert: MPXV TNF Receptor Gene Deletion May Lead to False Negative Results with Some MPXV Specific LDTs. https://www.cdc.gov/locs/2022/09-02-2022-lab-alert-MPXV_TNF_Receptor_Gene_Deletion_May_Lead_False_Negative_Results_Some_MPXV_Specific_LDTs.html. Retrieved 21 October 2022.
- 26.Mostafa HH. Importance of internal controls to monitor adequate specimen collection: The case of orthopoxvirus real-time PCR. J. Clin. Virol. Off. Publ. Pan. Am. Soc. Clin. Virol. 2022;156 doi: 10.1016/j.jcv.2022.105294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.