Abstract
The neisserial fbpABC locus has been proposed to constitute a single transcriptional unit. To confirm this operonic arrangement, transcription assays using reverse transcriptase PCR amplification were conducted with Neisseria meningitidis. The presence of fbpAB and fbpBC transcripts obtained by priming cDNA synthesis with an fbpC-sequence-specific oligonucleotide indicates that fbpABC is organized as a single expression unit. The ratio of fbpA to fbpABC mRNA was approximately between 10- to 20-fold, as determined by real-time quantitative PCR.
The necessity to sequester essential biochemical processes poses a challenge for living cells. Physical compartmentalization created by semipermeable membrane partitions achieves this segregation, but at the predictable expense of imposing a restrictive barrier to the cellular ingress and exit of solutes. Prokaryotes and eukaryotes have surmounted this obstacle by evolving transport systems, termed ATP-binding cassette (ABC) transporters, that couple ATP hydrolysis with substrate translocation across biological membranes (2, 6, 12, 24). These systems exhibit a modular organization comprised of four structural domains that may be expressed as individual polypeptides or may be fused into single multidomain proteins. Two membrane-integral domains span the membrane multiple times and form the passageway through which the solute flux occurs. Two ATP-binding cassettes reside on the cytosolic face of the membrane.
The binding-protein-dependent transporters of gram-negative bacteria represent the best-characterized members of this superfamily. These transporters recruit an auxiliary component, a periplasmic binding protein, that constitutes the major determinant in conferring substrate specificity (2, 6, 7, 12, 20, 26). The genes encoding this transporter complex are arranged as a single transcriptional unit, although apparent exceptions exist in which the periplasmic binding protein genes are unlinked from their cognate permease genes (8, 15, 23).
In the pathogenic neisseria Neisseria gonorrhoeae and Neisseria meningitidis, a gene cluster, termed fbpABC, has been postulated to mediate the delivery of iron across the periplasmic space into the cytoplasm (1, 5). The iron acquisition phenotype of a meningococcal fbpABC mutant supports this proposal for the obligatory participation of fbpABC in neisserial periplasmic iron transport from human transferrin and human lactoferrin (14). This locus displays the signature core components characteristic of binding-protein-dependent ABC transporters. Biochemical studies (5) identify FbpA as the substrate binding protein. The functional assignments of FbpB as the cytoplasmic membrane protein and of FbpC as the ATPase subunit are implied by the deduced amino acid sequence similarity to homologous proteins (1).
It remains unclear whether the genes in this locus are cotranscribed as a single expression unit. Nucleotide sequence (1) and primer extension (9) analyses have mapped a single potential promoter site upstream of fbpA. However, Northern blot hybridization did not disclose the presence of a polycistronic transcript (9). Therefore, this investigation was undertaken to address the proposed operonic organization of fbpABC by conducting transcription assays that exploit the sensitivity of reverse transcriptase (RT)-PCR amplification.
Bacterial strains and growth conditions.
The bacteria used in this study are listed in Table 1. Neisserial strains were grown on chocolate agar at 35°C in an atmosphere of 5% CO2. A single colony was selected from overnight growth on chocolate agar to inoculate 10 ml of brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) containing a 50 μM concentration of the iron chelator EDDA [ethylenediamine-di(o-hydroxyphenylacetic acid)]. The culture was grown in a shaking incubator at 37°C in the presence of 5% CO2 until mid-logarithmic growth was achieved (equivalent to an optical density at 600 nm [OD600] of 0.685 as measured with a Pye Unicam PU8800 spectrophotometer).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strain | ||
| Neisseria meningitidis B16B6 | Clinical isolate, serogroup B, serotype 2a | C. Frasch |
| Plasmids | ||
| PCR2.1 | TA cloning vector, ampicillin and kanamycin resistance | Invitrogen |
| pCR2FABC | PCR2.1 containing the cloned fbpABC locus from N. meningitidis B16B6 | This study |
| Primers (5′→3′) | ||
| 5′fbpA | CAGCCGTCTGAAAGGAATACACTACACCCG; 5′ oligonucleotide containing the gonococcal uptake sequence (boldface) for the amplification of fbpABC, nucleotide positions 45–70a | 14 |
| 5′fbpAint | GTACAAAATGTCCACACCCGC; 5′ oligonucleotide for the amplification of fbpAB, nucleotide positions 801–821a | This study |
| 3′fbpBint | CAGGCTGATGCGTTTGAGTGC; 3′ oligonucleotide for the amplification of fbpAB, nucleotide positions 1600–1620a | This study |
| 5′fbpBint | CCTTTATCGTCGTCATCCTT; 5′ oligonucleotide for the amplification of fbpBC, nucleotide positions 2321–2340a | This study |
| 3′fbpCint | CGGACGGGAAGGTTGGTATT; 3′ oligonucleotide upstream of the Walker A motif for the amplification of fbpBC, nucleotide positions 2939–2959a | This study |
| 3′fbpCstop | TGCCGCCTTTCAGAGGGTATTTCC; 3′ oligonucleotide reverse primer encompassing the stop codon (boldface) of fbpC, nucleotide positions 3771–3794a | This study |
| 5′HKasdint | GTATCTGATTTCCTGCGCAGCG; 5′ oligonucleotide for the amplification of asd, nucleotide positions 8290–8311b | This study |
| 3′HKasdstop | GCTTACAGGCTGCCCAACACG; 3′ oligonucleotide encompassing the stop codon (boldface) for the amplification of asd, nucleotide positions 8769–8798b | This study |
| 48 | TTCTGCATTCCTTATGCGCATGGATTTC; 3′ oligonucleotide for the amplification of tbpA, nucleotide positions 560–575c | This study |
| 385 | AAATTTGGATCCGAAAGCGAAGATTAG; 5′ oligonucleotide for the amplification of tbpA, nucleotide positions 1940–1965c | This study |
| 560 | CCATGCATCATTTAGGAGGAAATCGATATG; 5′ oligonucleotide containing the Shine-Dalgarno sequence (underlined) and the start codons (boldface) for the amplification of fbpA, nucleotide positions 100–122a | This study |
| 561 | ATCCGATGCATGCTTATTTCATACCGGCTTG; 3′ oligonucleotide upstream of the stem-loop structure (boldface type indicates the stop codon) for the amplification of fbpA, nucleotide positions 1098–1117a | This study |
The positions correspond to the nucleotide sequences of fbpABC (EMBL/GenBank DDBJ Nucleotide Sequence Data Library accession no. U33937) (1).
The positions correspond to the nucleotide sequences of asd (EMBL/GenBank DDBJ Nucleotide Sequence Data Library accession no. AE002557).
The positions correspond to the nucleotide sequences of tbpA (EMBL/GenBank DDBJ Nucleotide Sequence Data Library accession no. Z15129).
DNA isolation and manipulations.
Meningococcal genomic DNA was recovered by standard methods (21). DNA fragments were purified from agarose gels by using either the GeneClean II Purification Matrix kit (BIO 101, Inc., Vista, Calif.) or by passage through NENSORB 20 (NEN, Boston, Mass.) cartridges. DNA sequencing was performed according to the dideoxynucleotide chain-termination method (22) with a PRISM Ready Reaction dye cycle sequencing kit (Applied Biosystems) with fluorescence-labelled synthetic oligonucleotide primers based on the known fbpABC sequence (1). All sequence reactions were run and analyzed on an Applied Biosystems 377XL automated DNA sequencer.
The fbpABC locus from N. meningitidis B16B6 chromosomal DNA was PCR amplified with primer set 5′fbpA and 3′fbpCstop with Pfu DNA polymerase (Stratagene, La Jolla, Calif.). The amplification product was ligated into the TA cloning vector pCR2.1 (Invitrogen, San Diego, Calif.), creating pCR2FABC. This construct was used to generate the standard curves in the quantitative PCR assay.
RNA isolation and RT-PCR.
Total cellular RNA was extracted from mid-logarithmic-phase (OD600 of 0.685) meningococcal cultures by using the RNeasy Midi kit (Qiagen, Inc., Clarita, Calif.) according to the manufacturer's recommendations. To eliminate contaminating genomic DNA, total RNA was subjected to DNase I (amplification grade, Gibco BRL, Life Technologies, Burlington, Canada) treatment as specified by the manufacturer. RNA concentrations were determined by measuring the A260; samples were immediately stored at −70°C. Reverse transcription was performed with the SuperScript II RNase H− RT-PCR kit (Gibco BRL) following the manufacturer's instructions. The indicated gene-specific primers (Table 1) initiated first-strand cDNA synthesis. Thirty-six cycles of PCR amplification were performed with Taq polymerase (Gibco BRL) on a Perkin-Elmer model 480 DNA thermal cycler with denaturation at 94°C for 30 s, primer annealing at 52°C for 30 s, and extension at 72°C for 3 min. Identical aliquots were processed in parallel without the addition of RT, in order to ensure that residual genomic DNA was not serving as the template in the PCR amplification. PCR amplification products were electrophoresed on 1% agarose gels and stained with ethidium bromide. The identity of all RT-PCR amplification fragments was verified by nucleotide sequencing.
QRT-PCR.
For quantitative RT-PCR (QRT-PCR), cDNA synthesis was performed in a 20-μl final volume that included 2 μg of meningococcal total RNA, 100 pmol of random hexamer oligonucleotides (N6) as primers, RT buffer (50 mM Tris [pH 8.3], 75 mM KCl, 1.5 mM MgCl2), 10 mM dithiothreitol, and 1 mM (each) deoxynucleoside triphosphates (dNTPs; dATP, dGTP, dCTP, and dTTP), 100 U of Superscript II RT (Gibco BRL), and 17 U of RNase inhibitor (RNAguard; Amersham Pharmacia Biotech, Inc., Baie d'Urfé, Canada). The RT reaction was performed in an MJ Research minicycler PTC-150 at 22°C for 5 min, followed by incubation at 4°C for 50 min. The samples were heated for 5 min at 95°C to terminate the reaction. Real-time quantitative PCR was performed in 10-μl final volumes in glass capillaries in a LightCycler Instrument (Roche Diagnostics, Laval, Canada) (29). The PCR master mix comprised 1× PCR buffer, 3 mM MgCl2, 1 mg of bovine serum albumin per ml, 0.2 mM dNTPs, 0.5 μM both forward and reverse primers, a 1:3,000 dilution of SYBR Green I (Molecular Probes), and 0.4 U of Platinum Taq (Gibco BRL). Into each capillary tube, 9 μl of PCR master mix and 1 μl of template target DNA (cDNA or pCR2FABC) were loaded. Sealed capillaries were centrifuged prior to placement into the LightCycler carousel. PCR amplification was performed with an initial denaturation at 95°C for 30 s followed by 45 cycles of denaturation at 95°C for 2 s (ramping at 20°C/s), annealing at 52°C for 5 s (ramping at 20°C/s), and elongation at 72°C for 42 s (ramping at 5°C/s). Amplicon specificity was verified by melting curve analyses with the LightCycler software, version 3.39. The identity of the amplicons was also established by confirmation of the expected molecular weight by agarose gel electrophoresis. Optimal conditions for amplification were determined by preliminary experiments with meningococcal genomic DNA. The quantitative PCR experiment was repeated three times, and each experiment produced similar results.
Transcription assays using RT-PCR.
The integrity of the total RNA preparation was assessed by demonstrating the presence of the transcript from the housekeeping gene asd (10) (Fig. 1B, lane h; gene-specific primer 3′HKasdstop for the RT step, primer pair 5′HKasdint and 3′HKasdstop for PCR amplification) and the presence of the iron-regulated transcript tbpA (Fig. 1B, lane d; gene-specific oligonucleotide 48 for the RT step, primers 385 and 48 for the PCR step). The latter result also suggests that the starting RNA preparation is unlikely to be selectively biased against iron-regulated transcripts. The requirement for such a representative mRNA library arises from two considerations. First, the transcription of fbpA is enhanced under iron-limiting conditions (9). Second, given the proposed operonic organization of fbpABC, the expression of the putative polycistronic transcript encompassing this gene cluster would be anticipated to exhibit the same property.
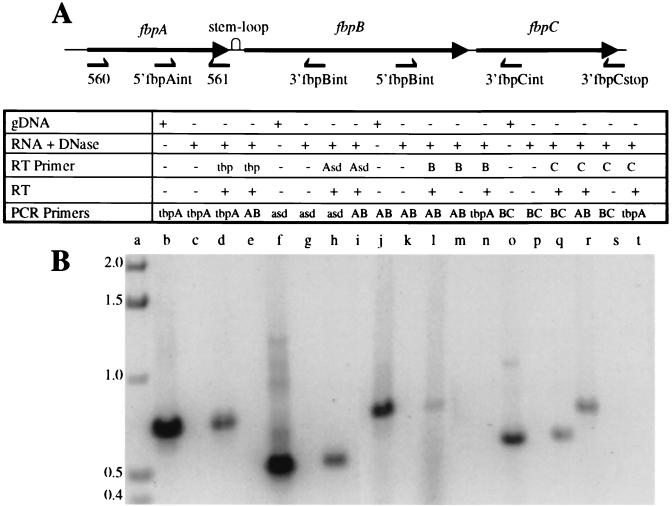
FIG. 1.
(A) The meningococcal fbpABC gene cluster encoding the neisserial iron ABC transporter. The orientations and approximate positions of the gene-specific primers for generating the cDNA and the oligonucleotide primers used in the PCR amplification step of the RT-PCR assays are displayed below the schematic. gDNA, genomic DNA. (B) RT-PCR amplification of total RNA extracted from N. meningitidis B16B6. The PCR amplification products were fractionated in a 1% agarose gel that was stained with ethidium bromide. The ingredients used to generate the amplicon in each lane are depicted in the table above the agarose gel. RT primers are designated as follows: tbp, 48; Asd, 3′HKasdstop; B, 3′fbpBint; and C, 3′fbpCstop. PCR primer pairs are designated as follows: tbpA, 385 and 48; asd, 5′HKasdint and 3′HKasdstop; AB, 5′fbpAint-3′fbpBint; and BC, 5′fbpBint-3′fbpCint. The figure was imaged with a Hewlett-Packard ScanJet HP, edited by using Adobe Photoshop 3.0, and labelled by using Microsoft PowerPoint 97. The sizes of standards in kilobases (lane a) are shown on the left.
The RT-PCR strategy used in the transcription assays was based on the general premise that upstream gene sequences within a given transcript would be readily detected by PCR amplification if these regions formed a continuous message. First-strand synthesis was initiated with gene-specific primers designed to anneal to intragenic sites within fbpB or fbpC and to the region encompassing the fbpC stop codon. Primer pairs were then selected to bracket the intergenic junctions between fbpAB and fbpBC (Fig. 1A). PCR amplification products generated by these oligonucleotides would therefore be contiguous and would be derived from a polycistronic transcript.
Using a cDNA template reverse transcribed from primer 5′fbpBint engineered for sequences situated within fbpB, an amplicon spanning the fbpAB junction was detected (Fig. 1B, lane I; oligonucleotides 5′fbpAint and 3′fbpBint). This result indicates that fbpA and fbpB are cotranscribed.
Similarly, the presence of the predicted PCR fragments straddling the fbpAB (Fig. 1B, lane r; primers 5′fbpAint and 3′fbpBint) and fbpBC (Fig. 1B, lane q; primers 5′fbpBint and 3′fbpCint) intergenic regions, when cDNA generated from the fbpC-specific oligonucleotide 3′fbpCstop was used as template, indicates that fbpA, fbpB, and fbpC are cotranscribed. Thus, the aggregate RT-PCR data illustrate that fbpABC is organized as a single polycistronic transcriptional unit.
The results from a series of control experiments conducted concurrently with each of the four sets of RT-PCR assays confirmed the substrate quality and guaranteed the specificity of each component of the RT-PCRs. First, signals for the specific PCR products were lost when DNase I-treated total RNA served as the template (Fig. 1B, lanes c, g, k, and p), ensuring that residual genomic DNA had not contaminated the starting RNA preparations. Second, the presence of the expected amplicon when genomic DNA acted as the template (Fig. 1B, lanes b, f, j, and o) demonstrated the fidelity of the PCR primers. Third, the absence of a PCR product when reciprocal PCRs were primed with a heterologous oligonucleotide pair (Fig. 1B, lanes e, i, n, and t) verified the authenticity of the cDNA template generated by the gene-specific reverse primer. Last, the inability to PCR amplify the desired product when RT was omitted (Fig. 1B, lanes m and s) (data not shown) validated the compulsory requirement of this enzyme for the initiation of cDNA synthesis.
Transcript quantity.
Real-time PCR studies were used to determine the relative abundance of fbpA-, fbpAB-, and fbpBC-bearing transcripts.
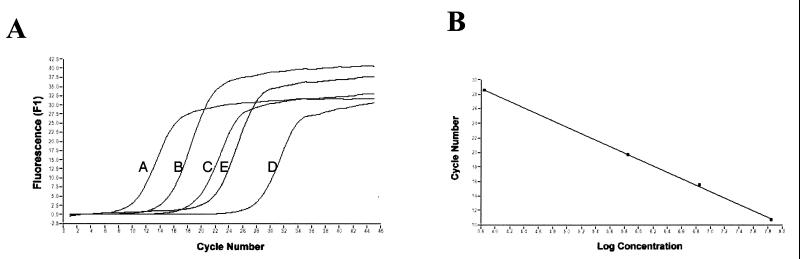
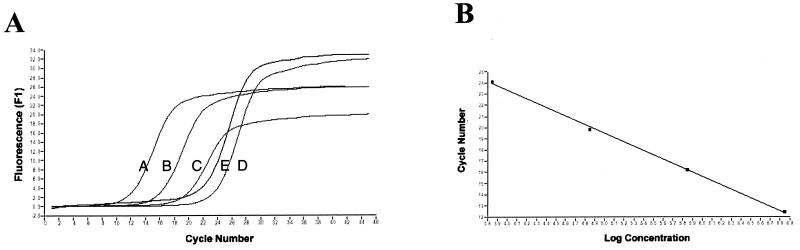
The test samples were cDNA primed with random hexamers, and 10-fold serial dilutions of pCR2FABC DNA were employed to generate the standard curves. Kinetic curves are shown for four concentrations of DNA (Fig. 2A and 3A). For each DNA template concentration, a single PCR product of the expected size for primer pairs 560 and 561 and 5′fbpAint and 3′fbpBint was detected by gel electrophoresis, and amplicon fidelity was confirmed by melting curve analysis (data not shown). Each kinetic curve was defined by a cycle threshold value (Ct) which marks the fractional cycle number during the logarithmic phase at which the fluorescence of a given sample becomes significantly different from the baseline signal. The Ct value also represents the crossover point between the kinetic curve and an arbitrary fluorescence level, which for all of the experiments presented is 1.5. Ct values are inversely proportional to the log of the initial template concentration and thus are used to calculate transcript copy number. The target message in the unknown sample is quantified by measuring Ct and by using the calibration curve performed during the same experiment to determine the starting target message quantity. As depicted in the calibration curves for each primer pair (Fig. 2B and 3B), the kinetic PCR assay exhibited a dynamic range of at least 4 orders of magnitude.
FIG. 2.
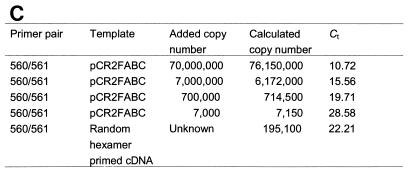
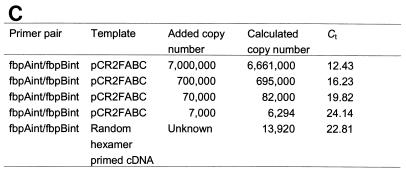
(A) Kinetic PCR curves for primer pair 560 and 561. Relative fluorescence output is plotted versus PCR cycle number. Kinetic curves are for 107 (A), 106 (B), 105 (C), and 103 (D) pCR2FABC DNA template copy numbers and for random-hexamer-primed cDNA (E). (B) Calibration curve for primer pair 560 and 561. The Ct value from each kinetic curve is plotted versus the log of the initial DNA concentration. (C) Copy number measured by real-time QRT-PCR. The copy number of standard pCR2FABC DNA added to the reactions is calculated from the moles of standard pCR2FABC added multiplied by Avogadro's number (6 × 1023). Given the Ct of each sample, the initial copy number is calculated from the calibration curve conducted during the same experiment. Each assay was performed in triplicate, and representative data from one such experiment are shown. The figure was imaged with a Hewlett-Packard ScanJet HP, edited by using Adobe Photoshop 3.0, and labelled by using Microsoft PowerPoint 97.
FIG. 3.
(A) Kinetic PCR curves for primer pair 5′fbpAint and 3′fbpBint. Relative fluorescence output is plotted versus PCR cycle number. Kinetic curves are for 106 (A), 105 (B), 104 (C), and 103 (D) pCR2FABC DNA template copy numbers and for random hexamer-primed cDNA (E). (B) Calibration curve for primer pair 3′fbpBint. The Ct value from each kinetic curve is plotted versus the log of the initial DNA concentration. (C) Copy number measured by real-time QRT-PCR. The copy number of standard pCR2FABC DNA added to the reactions is calculated from moles of standard pCR2FABC added multiplied by Avogadro's number (6 × 1023). Given the Ct of each sample, the initial copy number is calculated from the calibration curve conducted during the same experiment. Each assay was performed in triplicate, and representative data from one such experiment are shown. The figure was imaged with a Hewlett-Packard ScanJet HP, edited by using Adobe Photoshop 3.0, and labelled by using Microsoft PowerPoint 97.
These experiments revealed that fbpA mRNA was expressed at a 10- to 20-fold-higher level than the fbpAB transcript (Fig. 2 and 3). Similar ratios were observed when the level of fbpA transcript was compared to that of the fbpBC-expressing transcript (data not shown). These results indicate a preferential accumulation of fbpA transcript relative to full-length fbpABC mRNA.
The evidence provided in this study unequivocally shows that the meningococcal fbpABC locus is transcribed as a single contiguous message, and, therefore, this gene cluster is organized as a polycistronic operon. A prior report employing RT-PCR amplification was unable to detect either fbpC or fbpBC transcripts (25). The reasons for the discordant results are unclear, but differences in the primer design and in the RT-PCR amplification protocol represent two potential explanations.
The fbpAB and fbpBC transcripts detected in this study are likely translationally active, because transcription and translation are coupled processes in prokaryotes. Implicit in this observation is a functional role for both FbpB and FbpC in neisserial periplasmic iron transport from human transferrin and human lactoferrin. However, the mandatory participation of FbpC remains uncertain, because a previous investigation showed that a gonococcal fbpC mutant is unimpaired in the ability to access iron from human transferrin and human lactoferrin for growth (25).
There are several possible explanations for this observation, but no version supplies an immediately patent answer. First, functional disruption of fbpC may have unmasked the presence of an unidentified subsidiary ABC transporter involved in neisserial periplasmic iron transport. Such an explanation is unlikely, since in an antecedent study, an fbpABC mutant, which might also be anticipated to exhibit an iron acquisition phenotype, similar to that of the fbpC mutant, is incompetent in iron utilization (14). Second, iron transport in the fbpC mutant may have been restored by the presence of another chromosomal wild-type copy of fbpABC. The absence of other gonococcal gene loci displaying significant sequence homology to fbpABC in an analysis of the assembled contigs deposited in the ongoing gonococcal and meningococcal genome projects renders this explanation unlikely. Third, iron transport in the fbpC mutant may have been rescued by complementation with a heterologous ATPase subunit. Such functional exchange has occurred only in the context in which the heterologous complementing ATPase component is significantly overexpressed with respect to its respective cognate integral membrane protein (11, 28). Because this requirement has not been directly satisfied in the defined fbpC mutant, this explanation also appears unlikely to apply.
The enhanced amount of the fbpA-bearing transcript compared to the full-length fbpABC mRNA has several significant implications. First, this result supplies a molecular correlate for the observation that FbpA is synthesized in excess of the permease components FbpB and FbpC (1, 3). A cardinal feature of bacterial binding-protein-dependent importers is the preferential production of the periplasmic binding protein constituent (27). This characteristic is functionally relevant because the efficiency of the transport process is critically dependent upon the preservation of such a stoichiometry (16).
Second, this result suggests that segmental differences in transcript stability may account for the differential expression of individual genes in the fbpABC operon. Such differential rates of transcript decay underlie the preferential accumulation of the periplasmic binding protein MalE in the E. coli maltose transporter malEFGK (7). The increased stability of the malE transcript is a consequence of a stem-loop structure located in the malEF intergenic region (18, 19). Many stem-loop structures serve as barriers to 3′→5′ exonucleases (13, 19) by impeding the processive action of these enzymes, thereby increasing the chemical longevity of upstream mRNA. The sequence comprising the neisserial fbpAB intercistronic junction exhibits the potential to adopt a similar conformation (1), raising the intriguing speculation that this secondary structure represents the structural determinant of fbpA transcript stability.
In summary, we have established that the meningococcal fbpABC locus exhibits an operonic organization. The genetic (4), structural, and immunological (17) conservation of fbpABC in the pathogenic Neisseria spp. suggests that the results from this investigation apply to N. gonorrhoeae.
Acknowledgments
This work was supported by a grant (MT-15111) from the Medical Research Council of Canada. V.D. is the recipient of a summer studentship from the Alberta Heritage Foundation for Medical Research.
We thank R. Chalus for excellent technical assistance with the use of the LightCycler Instrument.
REFERENCES
- 1.Adhikari P, Berish S A, Nowalk A J, Veraldi K L, Morse S A, Mietzner T A. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J Bacteriol. 1996;178:2145–2149. doi: 10.1128/jb.178.7.2145-2149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames G F-L. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 3.Berish S A, Chen C-Y, Mietzner T A, Morse S A. Expression of a functional neisserial fbp gene in Escherichia coli. Mol Microbiol. 1992;6:2607–2615. doi: 10.1111/j.1365-2958.1992.tb01438.x. [DOI] [PubMed] [Google Scholar]
- 4.Berish S A, Kapczunski D, Morse S A. Nucleotide sequence of the Fbp gene from Neisseria meningitidis. Nucleic Acids Res. 1990;18:4596. doi: 10.1093/nar/18.15.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C-Y, Berish S A, Morse S A, Mietzner T A. The ferric iron-binding protein of pathogenic Neisseria spp. functions as a periplasmic transport protein in iron acquisition from human transferrin. Mol Microbiol. 1993;10:311–318. doi: 10.1111/j.1365-2958.1993.tb01957.x. [DOI] [PubMed] [Google Scholar]
- 6.Doige C A, Ames G F-L. ATP-dependent transport systems in bacteria and humans: relevance to cystic fibrosis and multidrug resistance. Annu Rev Microbiol. 1993;47:291–319. doi: 10.1146/annurev.mi.47.100193.001451. [DOI] [PubMed] [Google Scholar]
- 7.Ehrmann M, Ehrle R, Hofmann E, Boos W, Schlosser A. The ABC maltose transporter. Mol Microbiol. 1998;29:685–694. doi: 10.1046/j.1365-2958.1998.00915.x. [DOI] [PubMed] [Google Scholar]
- 8.Fleischmann R D, Adams M D, White O, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 9.Forng R-Y, Ekechukwu C R, Subbarao S, Morse S A, Genco C A. Promoter mapping and transcriptional regulation of the iron-regulated Neisseria gonorrhoeae fbpA gene. J Bacteriol. 1997;179:3047–3052. doi: 10.1128/jb.179.9.3047-3052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatten L A, Schweizer H P, Averill N, Wang L, Schryvers A B. Cloning and characterization of the Neisseria meningitidis asd gene. Gene. 1993;129:123–128. doi: 10.1016/0378-1119(93)90707-a. [DOI] [PubMed] [Google Scholar]
- 11.Hekstra D, Tommassen J. Functional exchangeability of the ABC proteins of the periplasmic binding protein-dependent transport systems Ugp and Mal of Escherichia coli. J Bacteriol. 1993;175:6546–6552. doi: 10.1128/jb.175.20.6546-6552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 13.Hiles I D, Gallagher M P, Jamieson D J, Higgins C F. Molecular characterization of the oligopeptide permease of Salmonella typhimurium. J Mol Biol. 1987;195:125–142. doi: 10.1016/0022-2836(87)90332-9. [DOI] [PubMed] [Google Scholar]
- 14.Khun H H, Kirby S D, Lee B C. A Neisseria meningitidis fbpABC mutant is incapable of using nonheme iron for growth. Infect Immun. 1998;66:2330–2336. doi: 10.1128/iai.66.5.2330-2336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linton K J, Higgins C F. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 16.Mademidis A, Koster W. Transport activity of FhuA, FhuC, FhuD, and FhuB derivatives in a system free of polar effects, and stoichiometry of components involved in ferrichrome uptake. Mol Gen Genet. 1998;258:156–165. doi: 10.1007/s004380050718. [DOI] [PubMed] [Google Scholar]
- 17.Mietzner T A, Barnes R C, Jeanlouis Y A, Shafer W M, Morse S A. Distribution of an antigenically related iron-regulated protein among the Neisseria spp. Infect Immun. 1986;51:60–68. doi: 10.1128/iai.51.1.60-68.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newbury S F, Smith N H, Higgins C F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987;51:1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- 19.Newbury S F, Smith N H, Robinson E C, Hiles I D, Higgins C F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987;48:297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- 20.Quiocho F A, Ledvina P S. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of a common theme. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saurin W, Dassa E. In search of Mycoplasma genitalium lost substrate-binding proteins: sequence divergence could be the result of a broader substrate specificity. Mol Microbiol. 1996;22:389–390. [PubMed] [Google Scholar]
- 24.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 25.Sebastian S, Genco C A. FbpC is not essential for iron acquisition in Neisseria gonorrhoeae. Infect Immun. 1999;67:3141–3145. doi: 10.1128/iai.67.6.3141-3145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam R, Saier M H., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres A G, Payne S M. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 28.Wilken S, Schmees G, Schneider E. A putative helical domain in the Malk subunit of the ATP-binding-cassette transport system for maltose of Salmonella typhimurium (MalFGK2) is crucial for the interaction with MalF and MalG. A study using the LacK protein of Agrobacterium radiobacter as a tool. Mol Microbiol. 1996;22:655–666. doi: 10.1046/j.1365-2958.1996.d01-1724.x. [DOI] [PubMed] [Google Scholar]
- 29.Wittwer C T, Ririe K M, Andrew R V, David D A, Gundry R A, Balis U J. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]