Abstract
Coronavirus disease 2019 (COVID-19) is especially severe in aged populations1. Vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are highly effective, but vaccine efficacy is partly compromised by the emergence of SARS-CoV-2 variants with enhanced transmissibility2. The emergence of these variants emphasizes the need for further development of anti-SARS-CoV-2 therapies, especially for aged populations. Here we describe the isolation of highly virulent mouse-adapted viruses and use them to test a new therapeutic drug in infected aged animals. Many of the alterations observed in SARS-CoV-2 during mouse adaptation (positions 417, 484, 493, 498 and 501 of the spike protein) also arise in humans in variants of concern2. Their appearance during mouse adaptation indicates that immune pressure is not required for selection. For murine SARS, for which severity is also age dependent, elevated levels of an eicosanoid (prostaglandin D2 (PGD2)) and a phospholipase (phospholipase A2 group 2D (PLA2G2D)) contributed to poor outcomes in aged mice3,4. mRNA expression of PLA2G2D and prostaglandin D2 receptor (PTGDR), and production of PGD2 also increase with ageing and after SARS-CoV-2 infection in dendritic cells derived from human peripheral blood mononuclear cells. Using our mouse-adapted SARS-CoV-2, we show that middle-aged mice lacking expression of PTGDR or PLA2G2D are protected from severe disease. Furthermore, treatment with a PTGDR antagonist, asapiprant, protected aged mice from lethal infection. PTGDR antagonism is one of the first interventions in SARS-CoV-2-infected animals that specifically protects aged animals, suggesting that the PLA2G2D–PGD2/PTGDR pathway is a useful target for therapeutic interventions.
Mice are naturally resistant to infection with SARS-CoV-2, as a result of incompatibility between the viral surface spike (S) glycoprotein and mouse angiotensin-converting enzyme 2 (mACE2)5. To enable murine infection with SARS-CoV-2, human ACE2 (hACE2) has been provided by genetic manipulation of the mouse genome or exogenously using viral vectors. Transgenic expression of hACE2 (refs. 6-9), complete or partial replacement of mACE2 with hACE2 (hACE2 knock-in)10 or administration of viral vectors expressing hACE2 all sensitize mice for infection11,12. Alternatively, the S protein of SARS-CoV-2 has been altered using reverse genetics to enable binding to mACE2 (refs. 13,14). SARS-CoV-2 has been adapted to mice by targeting amino acids at positions 498 and 499 (Q498Y/P499T) or 501 (N501Y). The resulting viruses can infect mice, although the resulting infection is very mild. However, further virus passage through mouse lungs results in increased virulence15. Mice infected with virulent mouse-adapted severe acute respiratory syndrome coronavirus (SARS-CoV) or Middle East respiratory syndrome coronavirus (MERS-CoV) have been similarly developed and are very useful for studies of pathogenesis, including in aged animals.
SARS, MERS and COVID-19 show age-dependent increases in severity1. For example, no individual under the age of 24 years succumbed to SARS during the 2002–2003 epidemic, whereas 50% of those over 65 years died from the infection16. Parallel results were found in C57BL/6 mice infected with SARS. Young C57BL/6 mice were resistant to infection with mouse-adapted SARS but mice became progressively more susceptible beginning at about 5 months of age4. Poor survival correlated with a suboptimal T cell response, which resulted from delayed migration of respiratory dendritic cells to draining lymph nodes. We identified a single prostaglandin, PGD2, that increased in the lungs during ageing and was responsible for impaired migration of dendritic cells4. We also showed that a single phospholipase A2 (PLA2G2D) increased in mouse respiratory dendritic cells during ageing and was responsible for increased levels of PGD2, as well as other prostaglandins in the aged mouse lung3. PGD2 and PLA2G2D seemed to increase during ageing partly in response to inflammaging and concomitant augmented oxidative stress3. Given the similarities between SARS, MERS and SARS-CoV-2 in age-dependent increased disease severity, we postulated that PGD2 and PLA2G2D would have similar roles in aged mice infected with a virulent strain of mouse-adapted SARS-CoV-2.
To generate a virulent mouse-adapted SARS-CoV-2, we inserted a sequence encoding the N501Y substitution into the SARS-CoV-2 genome (rSARS2-N501YP0) using reverse genetics as previously described17. As expected, this alteration rendered mice susceptible to SARS-CoV-2 infection, but BALB/c mice lost only a minimal amount of weight even after administration of 105 plaque-forming units (PFUs) of virus. After 30 passages through mouse lungs, the virus became highly virulent such that 5,000 PFUs caused a lethal disease in young BALB/c mice (Fig. 1a). We sequenced viruses after 10, 20 and 30 passages. Virus from these passages became progressively more lethal (Fig. 1a). Focusing on changes in the S protein, passage 10 virus contained a Q498R substitution, which probably enhanced binding to mACE2 (ref.13; Fig. 2a, b). A similar substitution, Q498H, was identified in other mouse-adapted SARS-CoV-2 strains18-20. By passage 20, an alteration at position 493 (Q493R) was detected. By passage 30, alterations in residues 417 and 484 (K417M and E484K) also arose. E484K does not seem to be required for mouse virulence because it was variably expressed by different isolates that had equivalent lethality (Fig. 2d).
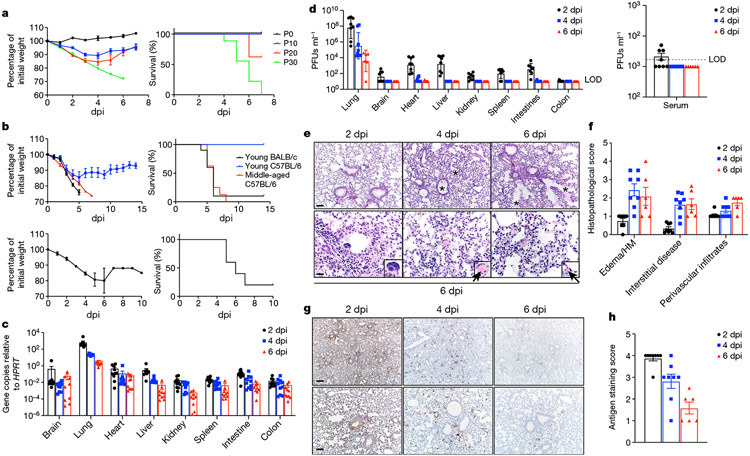
Fig. 1 ∣. Clinical, virological and pathological disease in mice infected with SARS2-N501YMA30.
a, Weight (left) and survival (right) of young BALB/c mice infected with passage (P) 0 (n = 3; 105 PFUs), P10 (n = 8; 9,000 PFUs), P20 (n = 8; 9,000 PFUs) or P30 (n = 9; 5,000 PFUs) SARS-CoV-2. P refers to the number of mouse lung passages. b, Weight (left panels) and survival (right panels) of young BALB/c (n = 10), young C57BL/6 (n = 6) or middle-aged C57BL/6 (n = 8) mice infected with 5,000 PFUs of SARS2-N501YMA30 (upper panel). Weight (left) and survival (right) of middle-aged C57BL/6 (n = 5) mice infected with 5,000 PFUs of BAC-derived SARS2-N501YMA30 (lower panel). Data are representative of two independent experiments. Data in a and b (left) are mean ± s.e.m. c, d, Viral genomic RNA (c) and infectious viral titres (d) at 2, 4 and 6 dpi with 5,000 PFUs of SARS2-N501YMA30 (n = 10 at all time points in c; n = 8 at 2 and 4 dpi, and n = 6 at 6 dpi in d). Serum titres are shown in the right panel of d. LOD, limit of detection. Data in c and d are geometric mean ± geometric s.d. e–h, Lungs from SARS2-N501YMA30-infected mice (n = 8 at 2 and 4 dpi; n = 6 at 6 dpi) were stained with haematoxylin and eosin (H&E) (e) or immunostained for SARS-CoV-2 nucleocapsid (g), and the pathological lesions and staining were quantified (f and h, respectively). e, Infected lungs exhibited airway edema (asterisks, top panels), multinucleated syncytial cells (inset in lower left panel), vascular thrombosis (arrows, insets in lower middle and right insets). Scale bars, 90 μm (top) and 22 μm (bottom); H&E stain. f, Summary scores of lung lesions (n = 8 at 2 and 4 dpi; n = 6 at 6 dpi). HM, hyaline membranes. g, Lungs from SARS2-N501YMA30-infected mice (n = 8 at 2 and 4 dpi; n = 6 at 6 dpi) were immunostained to detect SARS-CoV-2 nucleocapsid protein. Scale bars, 230 μm (top) and 90 μm (bottom). h, Summary scores of nucleocapsid immunostaining of lungs (n = 8 at 2 and 4 dpi; n = 6 at 6 dpi). Data in f and h are mean ± sem.
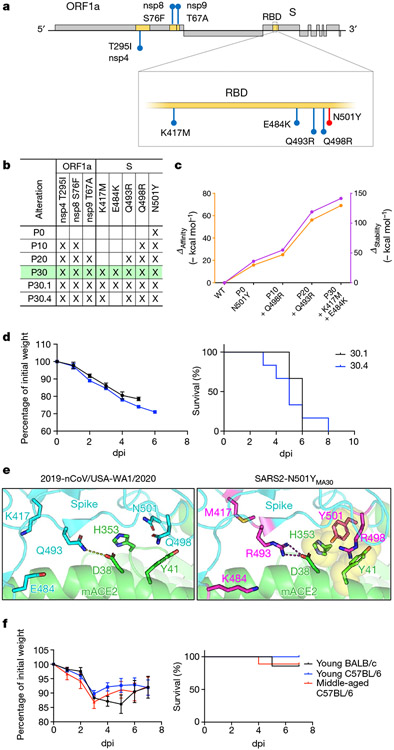
Fig. 2 ∣. Analysis of alterations in SARS2-N501YMA30 that arise during mouse adaptation.
a, Schematic showing the genome of SARS-CoV-2. Initial N501Y alteration engineered into SARS-CoV-2 is shown in red. Viral proteins encoding alterations in SARS2-N501YMA30 are shaded in yellow and the corresponding alterations are highlighted in blue. ORF1a, open reading frame 1a. b, Summary of alterations that emerged in different virus passages. P30 virus is highlighted in green c, In silico modelling of the effects of alterations that emerged in serial mouse passaging on the affinity and stability of the S protein complexes with mACE2. d, Percentage of initial weight (left) and survival (right) of young BALB/c mice infected with viruses from two distinct plaque isolates purified from P30 virus. Virus from plaque 30.1 (black) encodes an extra alteration (E484K) in the S protein compared to virus from plaque 30.4 (blue) (n = 6 for both groups). e, Modelling of the receptor-binding interface between the S protein of 2019-nCoV/USA-WA1/2020 (left) and SARS2-N501YMA30 (right) viruses and mACE2 reveals critical interactions mediated by alterations that emerged through serial passaging. f, Percentage of initial weight (left) and survival (right) of young BALB/c mice and young or old C57BL/6 mice infected with 105 PFUs of B1.351 (n = 7 for young BALB/c; n = 8 for young C57BL/6; n = 9 for middle-aged C57BL/6). Data in d and f (left) are mean ± s.e.m.
In silico modelling of the interaction between mACE2 and the S glycoprotein showed that Y501 in spike allowed π–π interactions and hydrophobic interactions with Y41 and H353 in mACE2, increasing the affinity and stability of the complex (Fig. 2c, e). Two additional spike substitutions (Q498R and Q493R) further increased affinity and stability, mainly driven by R493, which can create a salt bridge with D38 in mACE2 (previously a hydrogen bond between Q493 in spike and D38 in mACE2). Substitutions to other positively charged amino acids in residue 493 of spike have also been described in another mouse-adapted SARS-CoV-2 (ref. 15), suggesting the importance of this salt bridge to D38 in mACE2. The contribution of R498 in spike is probably due to an increase in the buried solvent-accessible surface area of this residue (from 80% to 99%), which improves the overall stability of the molecule and in turn could increase the affinity for mACE2. Residues K417 and E484 in spike do not interact directly with ACE2, and have been reported to have minimal effects on hACE2 binding21. E484K and various alterations in residue 417 have been associated with the variant of concern B.1.351 (E484K + K417N) and P.1 (E484K + K417T). Alterations in positions 417, 484, 493, 498 and 501 were detected in the variant of concern B.1.1.529 (Omicron variant). E484K and K417N have been shown to decrease antibody-mediated neutralization22 and therefore have been proposed to emerge in humans as an evolutionary advantage to evade immune responses. Here we show that E484K and alterations in K417 also occur in the absence of an adaptive immune response, offering an alternative explanation for the evolutionary pressures that drive the appearance of alterations in residues 417 and 484. Previously published models of murine SARS-CoV-2 have also reported substitutions at the same positions as the ones that we observed after 30 mouse lung passages15,18-20,23, demonstrating the advantage of this particular combination of alterations in driving murine lethality.
Alterations in three nonstructural proteins, nsp4, nsp8 and nsp9, were detected at passage 20. T295I in nsp4 and T67A in nsp9 have been described in other models of SARS-CoV-2 mouse adaptation that caused moderate disease but were not lethal in young BALB/c mice15, suggesting that these alterations might be important in driving virulence but are insufficient to cause death. T67A in nsp9 has also been described in a lethal mouse model of SARS (ref. 24), suggesting that this alteration might be important for mouse adaptation not only in SARS-CoV-2 but also in other coronaviruses as well. To assess the importance of Q493R and the nsp alterations in virulence, we infected young BALB/c and middle-aged C57BL/6 mice with the B.1.351 strain, which contains K417N, E484K and N501Y. Infection with a high dose of virus (105 PFUs) resulted in modest weight loss and 20% lethality, indicating that one or more of the alterations detected in nsp4, nsp8, nsp9 and Q493R were required for mouse virulence (Fig. 2f).
Virus after 30 rounds of adaptation in mice was then plaque purified three times, and individual plaques were sequenced. One plaque was chosen for further study and propagated (SARS2-N501YMA30). Of note, SARS2-N501YMA30 grew less well than rSARS2-N501YP0 in human airway epithelial (Calu-3) cells, showing that increased virulence in mice did not result in increased replication in human cells (Extended Data Fig. 1a).
Clinical and pathological characterization
To further assess the virulence of SARS2-N501YMA30, we infected 6–10-week-old BALB/c and C57BL/6 mice and 6–9-month-old C57BL/6 mice intranasally with 5,000 PFUs (Fig. 1b). Administration of 5,000 PFUs of SARS2-N501YMA30 caused a lethal infection in young BALB/c mice and middle-aged C57BL/6 mice but only modest weight loss in young C57BL/6 mice. To confirm that the alterations in SARS2-N501YMA30 were sufficient to explain its virulence, we inserted all of the alterations into a bacterial artificial chromosome (BAC) cDNA clone. Virus rescued from this clone was lethal in 6–9-month-old C57BL/6 mice (Fig. 1b). For most of our subsequent experiments, we infected young BALB/c or middle-aged C57BL/6 mice with SARS2-N501YMA30. After infection of young BALB/c mice with 5,000 PFUs, viral genomic RNA was detected in most organs at all time points, although levels were much higher in the lungs (Fig. 1c). Infectious virus was detected in most organs at 2 days post infection (dpi), but by 4 and 6 dpi, it was detected only in the lungs and, to a lesser extent, the heart (Fig. 1d). Consistent with the presence of virus in organs, we also detected virus in the blood of 4 out of 8 mice at 2 dpi (Fig. 1d, right). To extend these results, we analysed histological samples for evidence of pathological changes and virus antigen (Fig. 1e-h). We detected pathological changes in the lungs and nasal cavity at all times, but not in other organs (Fig. 1e, f and Extended Data Figs. 1b, c and 2). Pathological findings in the lung included perivascular, peribronchial and interstitial infiltration and alveolar edema. We also detected occasional pulmonary vascular thrombi, which are prominent findings in patients with severe COVID-19 (ref.25) and rare multinuclear syncytia (Fig. 1e, insets, bottom panels). In the sinonasal cavity, we detected evidence of infection of the olfactory and respiratory epithelium, with prominent infection of sustentacular cells (Extended Data Fig. 1b, c). Unlike in the case of SARS-CoV-2 infection of K18-hACE2 mice9, there was no pathological or immunohistochemical evidence of infection in the brain (Extended Data Fig. 1d, e).
Immunological responses
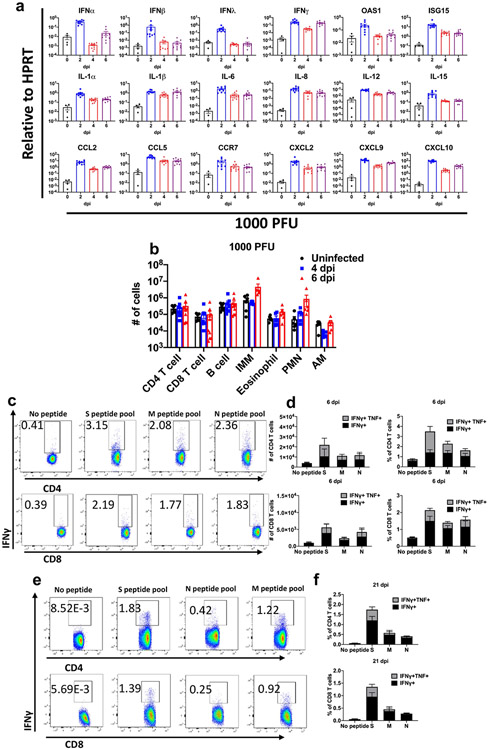
To further characterize the immunological response in the lungs to SARS2-N501YMA30 infection, we measured the levels of several cytokines, chemokines and inflammation-associated molecules in BALB/c lungs at 2, 4 and 6 dpi. Mice were infected with 1,000 PFUs in these assays to ensure that most survived until 6 dpi. Increases in mRNA expression of interferons (IFNs; IFNα, IFNβ, IFNγ and IFNλ), IFN-stimulated gene products (IFN-stimulated gene 15 and 2′-5′-oligoadenylate synthetase 1), cytokines (interleukin (IL)-1α, IL-1β, IL-6, IL-8, IL-12 and IL-15), chemokines (C─C motif chemokine ligand (CCL) 2, CCL5, C-X-C motif chemokine ligand (CXCL) 2, CXCL9 and CXCL10) and C─C motif chemokine receptor (CCR) 7, which is required for cell migration to draining lymph nodes, were observed over the course of the infection, with peak levels of IFNα, IFNβ and IFNλ prominent at 2 dpi. (Extended Data Fig. 3a). Notably, levels of most of these pro-inflammatory molecules remained elevated throughout the infection, as also occurs in the blood of patients with COVID-19 (ref. 26). To determine the character of the cells infiltrating the lungs observed on pathological examination, we immunophenotyped the infiltrating cells at 4 and 6 dpi (Extended Data Fig. 3b). Over time, there was a gradual increase in the numbers of infiltrating CD11b+ cells (macrophages, monocytes and neutrophils; Extended Data Fig. 3b). Virus-specific CD4+ and CD8+ T cells were detected in the lungs at 6 dpi and until at least 21 dpi. These cells expressed IFNγ and tumour necrosis factor (TNF) in response to stimulation with pools of peptides covering the SARS-CoV-2 structural proteins (spike, membrane and nucleocapsid) (Extended Data Fig. 3c-f). The S, N and M proteins all elicited CD4+ and CD8+ T cell responses, with the S protein responsible for the largest responses in both cell types. Lymphopenia and neutrophilia are hallmarks of severe COVID-19 (ref. 27) and were detected in middle-aged mice infected with SARS2-N501YMA30 (Extended Data Fig. 4).
PGD2 signalling and PLA2G2D expression
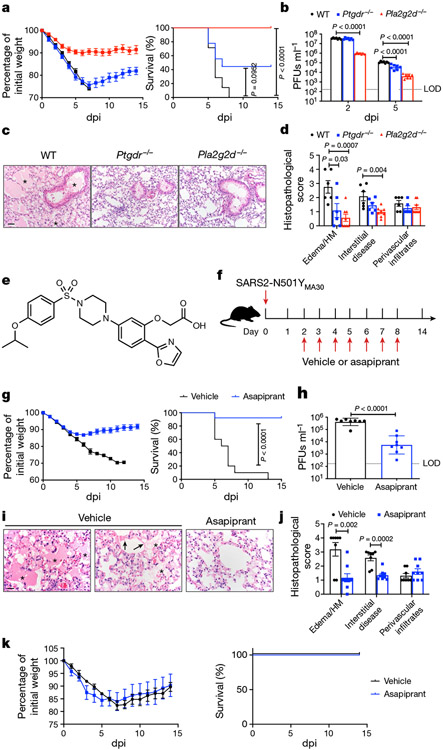
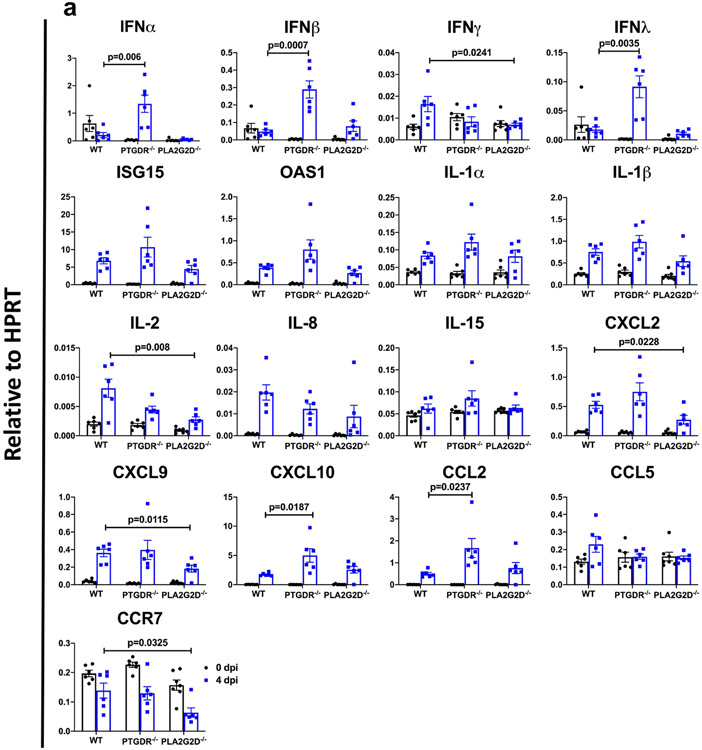
As we previously identified roles for PGD2/PTGDR signalling and PLA2G2D expression by respiratory dendritic cells in enhanced disease in middle-aged mice infected with SARS (refs. 3,4), we next infected 8–12-month-old Pla2g2d−/− and Ptgdr−/− mice with 5,000 PFUs, a lethal dose of SARS2-N501YMA30. Lethality was completely or partially abrogated in the absence of PLA2G2D or PTGDR expression, respectively (Fig. 3a). In both cases, the kinetics of virus clearance was enhanced, probably contributing to diminished disease (Fig. 3b). Further, many of the pathological changes observed in the lungs of infected middle-aged C57BL/6 mice, such as extensive inflammatory infiltration and edema, were diminished in Pla2g2d−/− and Ptgdr−/− mice (Fig. 3c, d). Although the PLA2G2D–PGD2/PTGDR pathway is generally anti-inflammatory, it contributes to the expression of TNF and other pro-inflammatory molecules in some settings28-30. We assessed a possible pro-inflammatory role by measuring levels of several inflammatory mediators in WT and Pla2g2d−/− and Ptgdr−/− mice (Extended Data Fig. 5). In the absence of PTGDR expression, levels of IFNα, IFNβ, IFNλ, CXCL10 and CCL2 all increased, consistent with a predominant anti-inflammatory role for PGD2/PTGDR signalling. By contrast, the absence of PLA2G2D resulted in diminished expression of several molecules, including IL-2, CXCL2, CXCL9 and CCR7. The decreased expression of these molecules is expected to diminish the cellular inflammatory response and possibly decrease a potentially overly robust immune response. These differences in effects on inflammatory molecule expression may occur because the absence of PLA2G2D affects not only PGD2/PTGDR signalling but also the mobilization of omega-3 fatty acids31,32. Together, these results show that PLA2G2D, associated with poor outcomes in mice infected with SARS, also contributes to lethality in mice infected with SARS-CoV-2. The results also demonstrate that these pathological effects were at least partially mediated through the PGD2/PTGDR axis.
Fig. 3 ∣. Absence of PLA2G2D or PTGDR expression, or blocking PGD2/PTGDR signalling, inhibits SARS2-N501YMA30 infection in vivo.
a, Weight (left) and survival (right) of middle-aged WT (n = 8), Ptgdr−/− (n = 9) or Pla2g2d−/− (n = 9) C57BL/6 mice infected with 5,000 PFUs of SARS2-N501YMA30. b, Viral titres in lungs at 2 (n = 7) and 5 dpi (n = 7). P values were determined by a log-rank (Mantel–Cox) test (a) and a two-tailed Student’s t-test (b). P < 0.0001, Pla2g2d−/− versus WT; not significant, Ptgdr−/− versus WT (a). P < 0.0001, Pla2g2d−/− versus WT, 2 dpi; P < 0.0001, Ptgdr−/− versus WT and Pla2g2d−/− versus WT, 5 dpi (b). c, Lungs at 5 dpi from WT mice exhibited edema (marked with asterisks) and cellular infiltration/interstitial thickening whereas these were significantly reduced in lungs from Ptgdr−/− or Pla2g2d−/− mice. H&E stain. Scale bar, 40 μm. d, Summary scores (n = 6 for WT; n = 6 for Ptgdr−/−; n = 8 for Pla2g2d−/−). P values were determined by two-tailed Student’s t-test. P = 0.03, Ptgdr−/− versus WT and P = 0.0007, Pla2g2d−/− versus WT, edema/HM; P = 0.004, Pla2g2d−/− versus WT, interstitial disease. e, Asapiprant structure. f, Experimental design for g and k. Vehicle or asapiprant was administered orally at 2–8 dpi to middle-aged (g–j) or young (k) C57BL/6 mice infected intranasally with SARS2-N501YMA30. g, Weight (left) and survival (right) of vehicle-treated (n = 10) or asapiprant-treated (n = 12) middle-aged mice. The P value was determined by a two-tailed Student’s t-test. P < 0.0001, asapiprant-treated versus vehicle-treated group. h, Lung viral titres of vehicle-treated (n = 8) or asapiprant-treated (n = 8) middle-aged mice at 5 dpi. The P value was determined by a two-tailed Student’s t-test. P < 0.0001, asapiprant-treated versus vehicle-treated group. i, The vehicle-treated group exhibited edema (asterisks) with occasional hyaline membranes (arrows), which were uncommon in asapiprant-treated mice at 5 dpi; H&E stain. Scale bar, 20 μm. j, Summary scores (n = 8 for vehicle-treated mice; n = 9 for asapiprant-treated mice). P values were determined by two-tailed Student’s t-test. P = 0.002, asapiprant-treated versus vehicle-treated group, edema/HM; P = 0.0002, asapiprant-treated versus vehicle-treated group, interstitial disease. HM, hyaline membranes. k, Weight (left) and survival (right) of vehicle-treated (n = 4) or asapiprant-treated (n = 4) young C57BL/6 mice infected with 5,000 PFUs of SARS2-N501YMA30. Data in a, d, g (left), j and k (left) are mean ± s.e.m. Data in b and h are geometric mean ± s.d.
Antagonizing PGD2/PTGDR signalling
Although several drugs with inhibitory activity against PLA2 molecules have been described33, none is specific for murine PLA2G2D. Next, we attempted to block PGD2/PTGDR signalling using indomethacin, a nonsteroidal anti-inflammatory drug (NSAID) inhibitor of cyclooxygenase 1 and 2. In addition, because prostaglandins are also involved in thrombus formation, NSAIDs, such as aspirin, might have a role in preventing their formation by inhibiting platelet-derived prostaglandin synthesis. However, clinical studies show that therapy with NSAIDs, including aspirin, are generally ineffective in ameliorating severe COVID-19 (refs. 34-38). Consistent with these results, indomethacin treatment of infected mice resulted in no changes in morbidity and mortality (Extended Data Fig. 6a, b) probably because the production of all downstream lipid mediators is inhibited. However, asapiprant ([2-(oxazol-2-yl)-5-(4-{4-[(propan-2-yl)oxy]phenylsulfonyl} piperazine-1-yl)phenoxy] acetic acid) (also known as BGE-175), a potent and specific antagonist of human PGD2/PTGDR signalling with a mean inhibition constant Ki of 0.44 nM, has been described previously39 (Fig. 3e). Asapiprant affinity for PTGDR was 300- to >15,000-fold higher than for any other prostanoid receptor tested39. We administered asapiprant to middle-aged C57BL/6 mice by once daily oral gavage (30 mg kg−1), starting at 2 dpi with 5,000 PFUs of SARS2-N501YMA30 and for 6 days thereafter (Fig. 3f). We chose to start treatment at 2 dpi because this time is relevant for possible use in symptomatic patients. In middle-aged mice infected with a lethal dose of SARS2-N501YMA30, asapiprant reduced mortality from 100% to <10%, with concomitant effects on weight loss (Fig. 3g). As expected on the basis of the studies of Pla2g2d−/− and Ptgdr−/− mice described above, drug treatment resulted in faster kinetics of virus clearance (Fig. 3h) and diminished pathological changes in the lungs (Fig. 3i, j). Asapiprant also enhanced migration of respiratory dendritic cells from lungs infected with SARS2-N501YMA30 to the draining mediastinal lymph nodes (Extended Data Fig. 6c) and reversed neutrophilia in the lungs and to a lesser extent in the blood (Extended Data Fig. 7). By contrast, treatment with the drug did not reduce the weight loss observed in young C57BL/6 mice infected with SARS2-N501YMA30 (Fig. 3k). The effects of asapiprant were not limited to SARS-CoV-2 infection because drug treatment 2 days after infection with SARS also prevented lethality (Extended Data Fig. 6e). The effects were specific for signalling through PTGDR because treatment with an antagonist of CRTH2 signalling (fevipiprant) did not improve outcomes (Extended Data Fig. 6d).
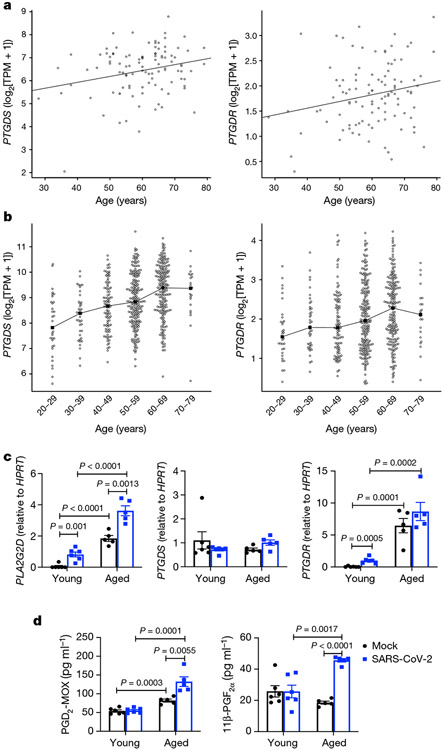
To understand the role of PLA2G2D expression and PGD2/PTGDR signalling in the clinical setting, we first analysed the literature for studies of age-dependent changes in lung levels of PGD2 synthases (PGD2 synthase (PTGDS) and haematopoietic prostaglandin D synthase), the enzymes required for conversion of prostaglandin H2 to PGD2, Ptgdr and Pla2g2d mRNA transcripts. Using RNA-sequencing data from 103 healthy lungs in a study of fibrosis40 (Gene Expression Omnibus (GEO) accession GSE150910) and 578 lungs from the Genotype-Tissue Expression (GTEx) portal41, we found that levels of PTGDS and PTGDR increased with ageing (Fig. 4a, b). Next, we obtained peripheral blood mononuclear cells from young and aged human individuals and cultured them as described in the Methods to obtain dendritic cells. After 14 days, we infected these cells with SARS-CoV-2. SARS-CoV-2 causes an abortive infection of human dendritic cells42. We then measured mRNA expression levels of PLA2G2D, PTGDR and PTGDS and cell supernatant levels of PGD2-methoxime (PDG2-MOX), a stabilized version of PGD2, and 11β-prostaglandin F2α (11β-PGF2α), a PGD2 enzymatic product. Expression of PLA2G2D and PTGDR increased with ageing and that of PLA2G2D further increased after SARS-CoV-2 infection (Fig. 4c). PGD2 increased with ageing and SARS-CoV-2 infection further enhanced expression by dendritic cells from aged individuals (Fig. 4d). 11β-PGF2α increased in dendritic cells from aged people in response to infection (Fig. 4d).
Fig. 4 ∣. Expression of PTGDS, PTGDR, PLA2G2D, PGD2, and 11β-PGF2α increases in human samples with age.
a, b, Expression of PGD2 genes (PTGDS (left panels) and PTGDR (right panels)) versus age in 103 healthy human lungs from GEO series GSE150910 (a) and 578 lungs from the GTEx project (b). The filled squares in b indicate the median expression for each age group. Expression levels of PTGDS and PTGDR were significantly associated with age in both studies (two-tailed Student’s t-test P values P = 0.0108 and 0.0305, respectively, in a; analysis of variance F-test P values P < 0.0001 and 0.0028, respectively, in b). TPM, transcripts per million. c, Dendritic cells derived from peripheral blood mononuclear cells from 6 young (24–39 years, mean 29 years) and 5 aged (56–73 years, mean 62 years) donors were mock infected or infected with SARS-CoV-2. Cells and supernatants were collected at 5 dpi. RNA isolated from cells was assessed for transcript levels of the indicated genes. d, Levels of the indicated prostaglandins in cell supernatants measured by enzyme-linked immunosorbent assay. Data in c, d are mean ± s.e.m. P values determined by two-tailed Student’s t-test (c, d).
Discussion
We describe a mouse-adapted SARS-CoV-2, isolated from mouse lungs after 30 serial passages. Unlike previously described mouse-adapted viruses, this strain is highly virulent in young and old mice, and infects only the lungs and not the brain. Further, it contains a set of alterations that are also observed in the B.1.351, P.1 and B.1.1.529 strains (K417N/T, E484K, Q493R, Q498R and N501Y) although K417N/T is replaced by K417M in SARS2-N501YMA30. All five amino acids are located in the receptor-binding domain, and E484 and N501 are hotspots for recognition by neutralizing antibodies22. Our results show that the selection of K417M (and by extension, presumably K417N) and E484K does not result solely from evasion of the antibody response because they arose in mice in the absence of any immune pressure. Infection of mice with B.1.351 resulted in mild disease, indicating that changes outside the S protein were critical for mouse virulence. T295I, located in the transmembrane domain of nsp4, and Q493K in the receptor-binding domain of S were identified in another variant of mouse-adapted SARS-CoV-2 (ref. 15) so alterations at these sites are prime candidates for conferring virulence in mice.
Our results identify a key role for PGD2/PTGDR signalling in enhanced COVID-19 disease severity during ageing. We show that delivery of a PTGDR antagonist at 2 dpi, a time comparable to when drug might be used in patients, converted a lethal to a sublethal infection. Asapiprant has been used in previous studies of allergic rhinitis and asthma. In a rat model of asthma, oral asapiprant reduced inflammatory cell numbers in bronchoalveolar lavage fluid and reduced the increase in airway hyperresponsiveness39. Asapiprant has been evaluated in 18 clinical studies (phases 1, 2 and 3) (2,000 patients and 200 healthy volunteers) conducted in the USA, the UK, France and Japan for the treatment of allergic rhinitis, alone or in combination with cetirizine, an anti-histamine43. Once-daily dosing of asapiprant for up to 4 weeks was considered safe and well tolerated. Decreased airway resistance after PGD2 challenge and decreased rhinitis symptom scores after allergen exposure were demonstrated in these studies, consistent with on-target pharmacodynamics43. Asapiprant is now being used in randomized, placebo-controlled clinical trials of hospitalized individuals with COVID-19 who are at risk of respiratory failure (https://clinicaltrials.gov/ct2/show/NCT04705597). Together, our results show that infection of mice with SARS2-N501YMA30 recapitulates many of the findings observed in patients with severe COVID-19 and also identify PGD2/PTGDR signalling and PLA2G2D as useful targets for therapy in aged individuals.
Methods
Generation of SARS-CoV-2 encoding N501Y alteration in the S protein and recombinant SARS2-N501YMA30
p-BAC SARS-CoV-2 carrying the sequence of the isolate Wuhan-Hu-1 (WT-SARS-CoV-2 BAC) was constructed on the basis of a previously described method44. The only difference between the Wuhan-Hu-1 and 2019n-CoV/USA-WA1/2019 strains is one amino acid change in ORF8 and 2 silent alterations (in ORF1a and ORF1b). Two silent alterations were introduced into the BAC clone to enable differentiation from wild-type SARS-CoV-2 (A20085G and G26840C). p-BAC SARS-CoV-2 with a N501Y substitution in spike (N501Y-SARS-CoV-2 BAC) or mouse-adapted alterations were generated using a lambda red recombination system with I-SceI homing endonuclease as previously described17. To generate N501Y-SARS-CoV-2 BAC, forward and reverse primers with overlapping SARS-CoV-2 spike sequence with the N501Y alteration followed by sequence complementary to the target plasmid (pEP-KanS) were synthesized (Invitrogen). A PCR fragment with overlapping ends carrying SARS-CoV-2 sequence flanking the kanamycin-resistance marker was amplified with the primers described using pEP-KanS as template. The PCR fragment was purified using a gel purification kit (Invitrogen). Purified PCR fragment was electroporated into GS1783 strains of Escherichia coli carrying p-BAC SARS-CoV-2. Successful recombinants were selected with kanamycin resistance. The kanamycin marker was removed by arabinose induction of I-SceI cleavage followed by homologous recombination of the overlapping ends. Successful recombinants were selected by replica plating for the loss of kanamycin resistance. Recombinants were purified and sequenced to confirm the introduction of alteration into the BAC clone. The primer sequences used to introduce the N501Y alteration are as follows: forward, 5′-CCTTGTAATGGTGTTGAAGGTTTTAATTGTTACTTTCCTTTACAATCATAT
GGTTTCCAACCCACTTATGGTGTTGGTTAGGATGACGACGATAAGTAG-3′; reverse, 5′-CATGTAGAAGTTCAAAAGAAAGTACTACTACTCTGTATGGTTGGTAACCAACA
CCATTAGTGGGTTGGAAACCATCAACCAATTAACCAATTCTGATTAG-3′. For generation of SARS2-N501YMA30, four substitutions (E484K, Q493R, Q498R and N501Y) in the S protein (4S) were introduced in a single round of recombination, followed by the K417M substitution in the S protein, the nsp4 (T295I) substitution and transcriptional regulatory sequence (TRS) alteration of the M protein (A26058G). The nsp8 (S76F) and nsp9 (T67A) alterations were introduced using CRISPR–Cas9 editing as described in the following section. The primer sequences used for the 4S alterations are as follows: forward, 5′-CGGTAGCACACCTTGTAATGGTGTTAAAGGTTTTAATTGTTACTTTCCTTTA
CGATCATATGGTTTCCGACCCACTTATGAGGATGACGACGATAAGTAG-3′; reverse, 5′-GTATGGTTGGTAACCAACACCATAAGTGGGTCGGAAACCATATGATCGTAAA
GGAAAGTAACAATTAAAACCTTTCAACCAATTAACCAATTCTGATTAG-3′. The primer sequences used for the K417M alteration are as follows: forward, 5′-GAGGTGATGAAGTCAGACAAATCGCTCCAGGGCAAACTGGAATGATTGCT
GATTATAATTATAAAAGGATGACGACGATAAGTAG-3′; reverse, 5′-CAGCCTGTAAAATCATCTGGTAATTTATAATTATAATCAGCAATCATTCCA
GTTTGCCCTGGAGCGATCAACCAATTAACCAATTCTGATTAG-3′. The primer sequences used for the nsp 4 alteration are as follows: forward, 5′-GACATATCAGCATCTATAGTAGCTGGTGGTATTGTAGCTATCGTAGTA
ATATGCCTTGCCTACTATTTTATGAGAGGATGACGACGATAAGTAG-3′; reverse, 5′-CACCAAAAGCTCTTCTAAACCTCATAAAATAGTAGGCAAGGCATATTACT
ACGATAGCTACAATACCCAACCAATTAACCAATTCTGATTAG-3′. The primer sequences used for the TRS alteration of the M protein are as follows: forward, 5′-CTGGTCTAAACGAACTAAATATTATATTAGTTTTTCTGTTTGGAGCTTTAA
TTTTAGCCATGGCAGAAGGATGACGACGATAAGTAG-3′; reverse, 5′-CAACGGTAATAGTACCGTTGGAATCTGCCATGGCTAAAATTAAAGCT
CCAAACAGAAAAACTAATATAATCAACCAATTAACCAATTCTGATTAG-3′. Base changes are shown in italics; sequences complementary to pEP-KanS are shown in bold.
Next, the nsp8 (S76F) and nsp9 (T67A) alterations of SARS2-N501YMA30 were introduced using CRISPR–Cas technology to generate a BAC cDNA clone with a full set of SARS2-N501YMA30 alterations. The alteration-encoding PCR product was linked and amplified from two PCR fragments with either the nsp8 or the nsp9 alteration by two-step PCR. Guide RNAs targeting the desired cleavage site were designed using online resources (https://chopchop.cbu.uib.no). DNA oligonucleotide sequences were generated with the EnGen sgRNA template oligonucleotide designer (NEB; https://sgrna.neb.com/#!/sgrna) to include essential elements for guide RNA transcription and RNA scaffold for Cas9 recognition. DNA oligonucleotides were chemically synthesized and transcribed with an EnGen sgRNA synthesis kit (NEB) according to the manufacturer’s protocol. sgRNAs were purified using Monarch kits for RNA cleanup (NEB). CRISPR–Cas9 cleavage was performed according to the manufacturer’s protocol (NEB). Cleaved BAC products were recovered by gel purification using the PureLink Quick Gel Extraction and PCR Purification Combo Kit (Invitrogen). Purified BAC products were incubated with alteration-encoding PCR products for in vitro recombination with Gibson assembly (NEB) according to the manufacturer’s protocol. Ligation products were transformed into DH10β cells. Bacterial colonies derived from the ligation products were cultured for DNA extraction to isolate mutant BACs. Introduction of alterations was verified by Sanger sequencing. Oligonucleotide sequences for sgRNAs are as follows: sgRNA1, 5′-TTCTAATACGACTCACTATAGTTTAGATATATGAATTCACAGTTTTAGA
GCTAGA-3′; sgRNA2, 5′-TTCTAATACGACTCACTATAGTCTGTAACAAACCTACAAGGGTTTT
AGAGCTAGA-3′. Target DNA sequences are underlined. Primer sequences for amplifying PCR product encoding the nsp8 alteration are as follows: forward, 5′-ACAGGAGTTTAGATATATGAATTCACAGGGAC-3′; reverse, 5′-GTCCTCAAATCTAGCCTGTTTA-3′. Primer sequences for amplifying PCR product encoding the nsp9 alteration are as follows: forward, 5′-TAAACAGGCTAGATTTGAGGAC-3′; reverse, 5′-GTGTGTCTGTAACAAACCTACAAGGTGGTTCCAGTTCTGCATAGAT-3′. Base changes are indicated in bold.
A 2 μg quantity of mutant BACs was transfected into Vero E6 cells (ATCC) with Lipofectamine 3000 (Invitrogen) in a 6-well plate according to the manufacturer’s protocol. Cells were monitored daily for cytopathic effects (CPEs). Cultures were collected when the CPE was >50% by freezing at −80 °C. rSARS2-N501YP0 and rSARS2-N501YMA30 were further passaged in Calu-3 cells in DMEM supplemented with 20% fetal bovine serum (FBS). Virus titres were determined by plaque assay.
Mice, cells and virus
BALB/c and C57BL/6 mice of 6–10 weeks or 6 months of age were obtained from Charles River Laboratories. Mice were maintained in the Animal Care Unit at the University of Iowa under standard conditions of dark/light cycle, ambient temperature and humidity. Mice were randomly assigned to different groups, with numbers per group sufficient to obtain statistical significance. 2019-nCoV/USA-WA1/2020 (accession number MN985325.1) and 20H/501Y.V2 (B.1.351, BEI catalogue number NR-54009) were obtained from BEI and passaged in Calu-3 2B4 cells45. Calu-3 2B4 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 20% FBS. Vero E6 cells (ATCC CRL-1586) were grown in DMEM supplemented with 10% FBS. All viruses were sequenced after propagation and found to match the input strain.
Serial in vivo passaging of virus in mice
Serial blind passage of rSARS2-N501YP0 through mouse lungs was performed in 8–10-week-old BALB/c mice. In brief, two mice were each inoculated with 50 μl of the virus intranasally at each passage. At 2/3 dpi, mice were euthanized, and lungs were pooled, homogenized in PBS and used to infect naive mice. After 30 passages, virus was obtained and plaque purified three times using Vero E6 cells. Several isolates were obtained and a single one was chosen for further study (SARS2-N501YMA30). SARS2-N501YMA30 was further propagated in Calu-3 2B4 cells.
Mouse infection
Mice were anaesthetized with ketamine–xylazine and infected intranasally with the indicated amount of virus in a total volume of 50 μl DMEM. Animal weight and health were monitored daily. All experiments with SARS-CoV-2 were performed in a biosafety level 3 (BSL3) laboratory at the University of Iowa. All animal studies were approved by the University of Iowa Animal Care and Use Committee and meet stipulations of the Guide for the Care and Use of Laboratory Animals.
Virus titre by plaque assay
At the indicated times, mice were euthanized and transcardially perfused with PBS. Organs were collected and homogenized before clarification by centrifugation and titring. Virus or tissue homogenate supernatants were serially diluted in DMEM. Twelve-well plates of VeroE6 cells were inoculated at 37 °C in 5% CO2 for 1 h and gently rocked every 15 min. After removing the inocula, plates were overlaid with 0.6% agarose containing 2% FBS. After 3 days, overlays were removed, and plaques visualized by staining with 0.1% crystal violet. Viral titres were quantified as PFUs per ml tissue.
Collection of whole blood/serum
Mice were anaesthetized by intraperitoneal injection of ketamine–xylazine. Blood was collected through retro-orbital bleed with a capillary tube (Fisher Scientific). Blood was allowed to clot at room temperature for 30 min. Serum was clarified by centrifugation and transferred to a new tube for storage at −80 °C. For collection of whole blood, heparinized capillary tubes were used (Fisher Scientific).
Histology and immunohistochemistry
Mice were anaesthetized by intraperitoneal injection of ketamine–xylazine and perfused transcardially with PBS. Tissues were fixed in zinc formalin. For routine histology, tissue sections (≈4 μm each) were stained with H&E. Tissues were evaluated for the presence of edema or hyaline membranes (0–4) using distribution-based ordinal scoring: 0, none; 1, <25%; 2, 26–50%; 3, 51–75%; 4, >75% of tissue fields. Perivascular lymphoid inflammation was evaluated by severity-based ordinal scoring: 0, absent; 1, minor (solitary to loose infiltration of cells); 2, moderate (small to medium aggregates); 3, severe (robust aggregates that are circumferential around vessels and can extend into adjacent parenchyma). Interstitial scores were evaluated using a modified H score for the presence of interstitial disease: 0, none; 1, minor (increased cellularity in septa); 2, moderate (cellular infiltrates with some thickened septa and uncommon inflammatory cells extending into lumen); 3, severe (cellular infiltrates in thickened septa and filling in air-space lumens with atelectasis and/or evidence of diffuse alveolar damage (edema or hyaline membranes)). For each score, this was multiplied by the percentage of lung affected, and then summed for each lung and divided by 100 to yield a final score of 0–3. For SARS-CoV-2 antigen detection, slides were incubated with blocking reagent (10% normal goat serum for 30 min) followed by rabbit monoclonal antibody against SARS-CoV-2 N protein (1:20,000 dilution for 60 min, number 40143-R019, Sino Biological), and then incubated with rabbit Envision (Dako) and diaminobenzidine (Dako) as chromogen. Lung tissue sections were evaluated by a boarded veterinary pathologist familiar with the model using the post-examination method of masking46. IHC was ordinally scored on the percentage distribution of staining in the tissues: 0, absent; 1, 0–25%; 2, 26–50%; 3, 51–75%; 4, >75% of tissue.
Lung cell preparation and antibodies for flow cytometric analysis
Animals were anaesthetized with ketamine–xylazine and perfused transcardially with 10 ml PBS. Lungs were removed, minced and digested in HBSS buffer consisting of 2% fetal calf serum, 25 mM HEPES, 1 mg ml−1 collagenase D (Roche) and 0.1 mg ml−1 DNase (Roche) at 37 °C for 30 min. Single-cell suspensions were prepared by passage through a 70-μm cell strainer. Cells were enumerated with a Scepter 2.0 cell counter (MilliporeSigma). Whole blood was treated with ACK lysis buffer for 1 min. Cells were washed and pelleted. Cells were then washed and blocked with 1 μg anti-CD16/anti-CD32 (2.4G2) at 4 °C for 20 min and surface stained with the following antibodies at 4 °C for 30 min: V450 anti-mouse CD45 (clone 30-F11; catalogue number 560501); APC anti-mouse CD220 (clone RA3-6B2, catalogue number 553092); APC/cyanine 7 anti-mouse CD3e (clone 145-2C11, catalogue number 100330); APC/cyanine 7 anti-mouse CD11c (clone HL3; catalogue number 561241); FITC anti-mouse Ly6G (clone 1A8; catalogue number 127606); PE anti-mouse CD11b (clone M1/70; catalogue number 101208); PE/cyanine 7 anti-mouse CD8 (clone 53-6.7; catalogue number 100722); PerCP/cyanine 5.5 anti-mouse CD4 (clone RM4.5; catalogue number 550954); FITC anti-mouse Ly6G (1A8; catalogue number 127606); PerCP/cyanine 5.5 anti-mouse Ly6C (clone HK1.4; catalogue number 128012); CD64 (clone X54-5/7.1; catalogue number 139304). All antibodies were used at 1:200 dilution. Cells were washed, fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences). The gating strategy is shown in Supplementary Fig. 1.
For intracellular cytokine staining (ICS), lymphocytes were cultured in 96-well dishes at 37 °C for 5–6 h in the presence of 2 μM peptide pool and brefeldin A (BD Biosciences). Cells were then labelled for cell-surface markers, fixed/permeabilized with Cytofix/Cytoperm Solution (BD Biosciences) and labelled with anti-IFNγ and anti-TNF (1:100 dilution). All flow cytometry data were acquired using a BD FACSVerse and analysed with FlowJo software.
RNA isolation and RT–qPCR
Total RNA was extracted from tissues using TRIzol (Invitrogen) or a Direct-zol RNA Miniprep kit (Zymo Research) according to the manufacturer’s protocol. Following DNase treatment, 200 ng of total RNA was used as a template for first-strand cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The resulting cDNA was subjected to amplification of selected genes by real-time quantitative PCR using Power SYBR Green PCR Master Mix (Applied Biosystems). Average values from duplicates of each gene were used to calculate the relative abundance of transcripts normalized to HPRT and presented as 2−ΔCT. The primers used for cytokine and chemokines were reported previously47. For detection of viral genomes, the following primers were used to amplify the genomic RNA for the N protein: 2019-nCoV_N1-F: 5′-GACCCCAAAATCAGCGAAAT-3′; 2019-nCoV_N1-R: 5′-TCTGGTTACTGCCAGTTGAATCTG-3′. The following primers were used to amplify the subgenomic RNA for the E protein: F, 5′-CGATCTCTTGTAGATCTGTTCTC-3′; R, 5′-ATATTGCAGCAGTACGCACACA-3′.
Viral whole-genome sequencing
Samples were treated with TRIzol reagent for viral inactivation and RNA extraction. Sequencing libraries were prepared using the sequencing protocol (v2) and primer pools (v3) from the ARTIC network (https://doi.org/10.17504/protocols.io.bdp7i5rn). Reads were sequenced on a MinION sequencer from Oxford Nanopore Technologies and consensus sequences were generated using Medaka following the ARTIC network nCoV-2019 bioinformatics protocol v1.1.0 (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html). A total of 249,944 average reads ± 82,509 (standard deviation) was obtained per sample. After quality filtering and alignment, the average sequencing depth was >200× per amplicon for each sample. The consensus sequence of P10 and P20 virus from mouse lung homogenates contained areas with low sequencing depth; to accurately cover these gaps, we amplified the missing regions using individual PCR reactions and performed Sanger sequencing on the amplicons.
In silico structural modelling and analysis
A homology model for mACE2 (NP_001123985.1) was generated using YASARA and docked into the crystal structure of complexed hACE2 and spike RBD (PDB: 6M0J). Alteration calculations were performed on the mACE2–spike complex in Bioluminate (Schrodinger Release 2020-4) by first using the Protein Preparation Wizard and then the Residue Scanning module. Stability and affinity calculations were performed optimizing for the affinity, and backbone minimization was used with a cutoff of 5 Å. Figures were generated in PyMOL. Software used was installed and configured by SBGrid (ref. 48).
Treatment with indomethacin, asapiprant and fevipiprant.
Male and female 7–8-month-old C57BL/6 mice were infected with SARS2-N501YMA30 (5,000 PFUs). For indomethacin treatment, mice were treated daily from days 0–4 or 2–6 after infection with 0.2 ml PBS or 10 mg kg−1 indomethacin diluted in 0.2 ml PBS. For asapiprant treatment, mice were treated daily at 2–8 dpi with 0.2 ml vehicle (0.5% carboxymethyl-cellulose sodium (NaCMC), Sigma) or 30 mg kg−1 asapiprant (BioAge). For fevipiprant treatment, mice were treated with 5 mg kg−1 fevipiprant (Medkoo) diluted in 0.2 ml vehicle twice daily. Drugs and vehicle/PBS were administered through oral gavage.
Generation of human dendritic cells.
Human peripheral blood cells were obtained from anonymous donors at the DeGowin Blood Center at the University of Iowa. Consent forms were approved by the University of Iowa Institutional Review Board. To obtain monocytes, peripheral blood mononuclear cells were isolated by density gradient centrifugation through Ficoll-Paque PLUS density gradient medium (Cytiva) and cultured at a seeding density of 1 × 106 cells ml−1 in RP-10 medium (RPMI-1640 medium (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals)), 2 mM l-glutamine and 100 ng ml−1 granulocyte macrophage colony-stimulating factor (PeproTech) + 50 ng ml−1 IL-4 (PeproTech)) at 37 °C for 96 h. Plates were then washed with Hanks’ balanced salt solution lacking divalent cations (Invitrogen) to remove non-adherent cells. Adherent cells were then trypsinized, pelleted and cultured for 10 days.
Statistics and reproducibilty
Differences in mean values between groups were analysed by analysis of variance and Student’s t-tests and differences in survival were analysed by log-rank (Mantel–Cox) tests using Microsoft Excel and GraphPad Prism 8. All results are expressed as mean ± s.e.m. and were corrected for multiple comparisons. P < 0.05 was considered statistically significant.
The data in Figs. 1, 2d, f and 3 were pooled from two independent experiments, except for those in Fig. 3k, which shows representative data from two independent experiments. The data in Fig. 3c, i are representative of two independent experiments with similar results.
Extended Data
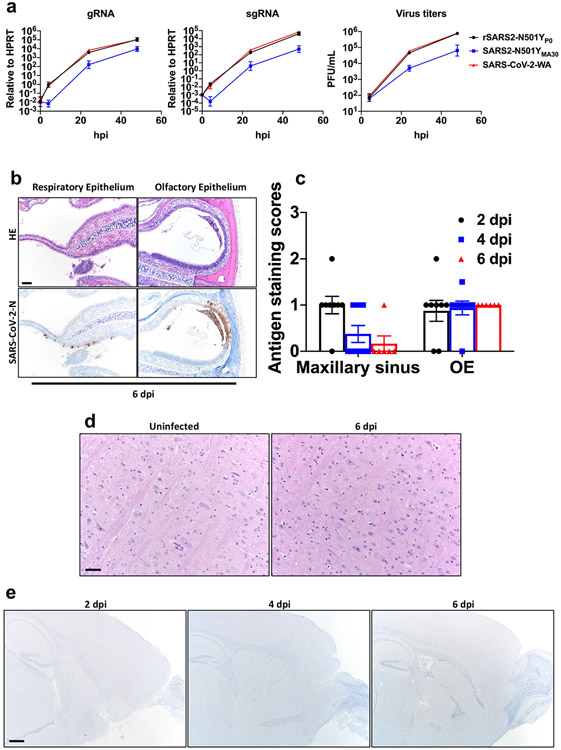
Extended Data Fig. 1 ∣. Infection of human respiratory cells and distribution of SARS2-N501YMA30 antigen in sinonasal cavity and brain.
a, Quantification of genomic RNA (gRNA), subgenomic RNA (sgRNA) and virus titers in Calu-3 cells at the indicated times after infection with 0.01 MOI of the indicated viruses. Data in a are geometric mean ± geometric SD and are representative of two independent experiments. b, Sinonasal cavity from young BALB/c mice infected with 5000 PFU of SARS2-N501YMA30, H&E stain (top panels). Regions of respiratory epithelium and olfactory epithelium exhibited uncommon regional scattered (bottom-left panel) to localized SARS-CoV-2 nucleocapsid immunostaining, (bottom panels). Scale bar 90 μm. OE: olfactory epithelium. c, Summary scores of nucleocapsid staining, as described in Methods (n = 8 at 2, 4 dpi; n = 6 at 6 dpi). Data in c are mean ± s.e.m. d, Brains from uninfected or infected young BALB/c mice at 6 dpi lacked overt lesions, H&E stain. Scale bar 45 μm. e, Brains from uninfected mice or young BALB/c mice infected with 5000 PFU at 2,4, and 6 dpi revealed no SARS-CoV-2 nucleocapsid immunostaining. Scale bar 460 μm. Data were pooled from two independent experiments. Data in b,d are representative of two independent experiments with similar results.
Extended Data Fig. 2 ∣. Histological analysis of extrapulmonary tissue.
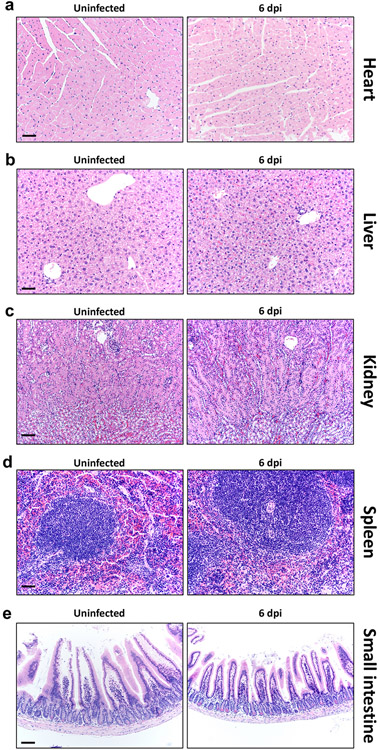
Mice infected with 5000 PFU of SARS2-N501YMA30 were sacrificed at 6 dpi with 5000 PFU of SARS2-N501YMA30 and tissues were prepared for histological examination. Tissues from uninfected mice were analyzed in parallel. a–e, heart (a), liver (b), kidney (c), spleen (d), and small intestine (e) were studied. No overt, group-specific lesions were observed. Scale bars, 45 μm (a, b, d) and 90 μm (c, e), H&E stain. Two sections of each organ from 6 mice per group were evaluated. Representative images from two independent experiments are shown.
Extended Data Fig. 3 ∣. Inflammatory mediators and immune effector cells in infected lungs.
Mice were infected with 1000 PFU SARS2-N501YMA30 to ensure survival until 6 dpi. a, Cytokine and chemokine transcripts were measured by qRT–PCR after isolation of RNA from the lungs of uninfected (0 dpi) and infected young BALB/c mice. Each lung was collected from an individual mouse. Mock (0 dpi), n = 4; 2, 4, and 6 dpi, n = 10. b, Quantification of immune cells (as gated in Supplementary Fig. 1) in the lungs (n = 6 for uninfected group; n = 9 for CD4, CD8 and B cells; n = 6 for IMM, eosinophils, PMNs and AM, for both 4 and 6 dpi) each lung was collected from an individual mouse). IMM: inflammatory monocytes/macrophages; PMN: neutrophils; AM: alveolar macrophages. c, e, Representative FACS plots of IFNγ+ CD4 and CD8 T cells after stimulation with indicated peptide pools in the lungs of young BALB/c mice at 6 (c) and 21 (e) dpi. d, f, Summary data for IFNγ and TNF expression are shown (n = 5, 6 dpi; n = 4, 21 dpi). Data in a, b, d are mean ± s.e.m. a and b are pooled data from two independent experiments. Data in c-f are from one of two independent experiments.
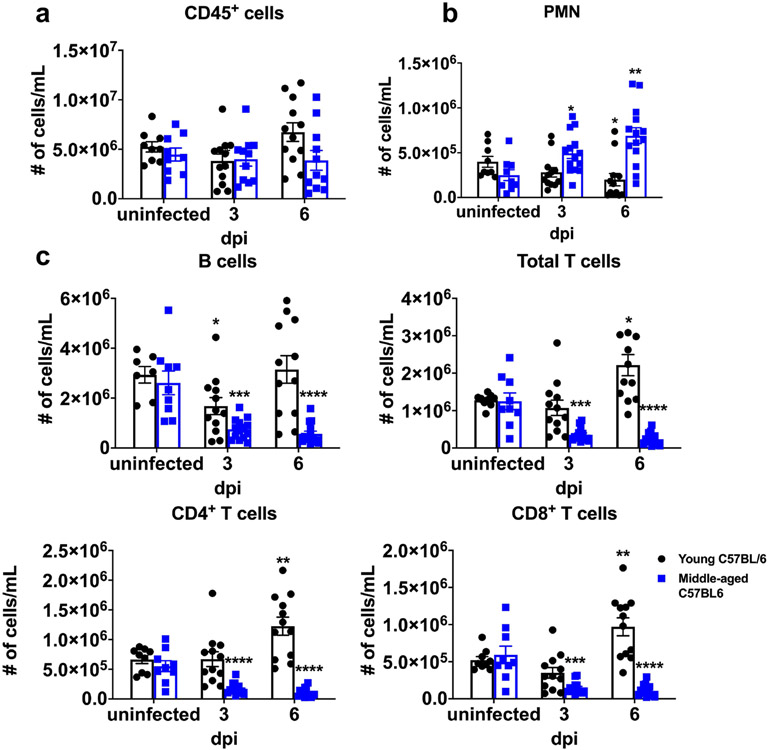
Extended Data Fig. 4 ∣. Lymphopenia in SARS2-N501YMA30-infected middle-aged mice.
Numbers of immune cells in the blood of young and middle-aged C57BL/6 mice at 3 and 6 days after infection with 5000 PFU of SARS2-N501YMA30 (n = 9 for uninfected controls in both groups; n = 14 for middle-aged mice at 3 and 6 dpi; n = 12 for young mice at 3 and 6 dpi). a. CD45+ cells; b. PMN: Polymorphonuclear cells. c. B and T cells. P values determined by two-tailed Student’s t test. P values indicated are determined from the corresponding time point compared to uninfected control of the same group. P = 0.0483 (6 dpi vs uninfected, young C57BL/6; PMN); P = 0.0117 (3 dpi vs uninfected, middle-aged C57BL/6, PMN); P = 0.0016 (6 dpi vs uninfected, middle-aged C57BL/6, PMN); P = 0.0264 (3 dpi vs uninfected, young C57BL/6; B cells); P = 0.0001 (3 dpi vs uninfected, 6 dpi vs uninfected middle-aged C57BL/6, B cells); P = 0.0109 (6 dpi vs uninfected, young C57BL/6; Total T cells); P = 0.0001 (3 dpi vs uninfected, middle-aged C57BL/6, Total T cells); P < 0.0001 (6 dpi vs uninfected, middle-aged C57BL/6, Total T cells); P = 0.0072 (6 dpi vs uninfected, young C57BL/6; CD4+ T cells); P < 0.0001 (3 dpi vs uninfected, 6 dpi vs uninfected, middle-aged C57BL/6, CD4+ T cells); P = 0.0065 (6 dpi vs uninfected, young C57BL/6; CD8+ T cells); P = 0.0001 (3 dpi vs uninfected, middle-aged C57BL/6, CD8+ T cells); P < 0.0001 (6 dpi vs uninfected, middle-aged C57BL/6, CD8+ T cells). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Data in a are mean ± s.e.m. Data are pooled from two independent experiments.
Extended Data Fig. 5 ∣. Cytokine and chemokine expression profile in middle-aged WT, PTGDR−/− or PLA2G2D−/− C57BL/6 mice after SARS2-N501YMA30 infection.
a, Middle-aged WT, PTGDR−/− and PLA2G2D−/− C57BL/6 mice were infected with 5000 PFU SARS2-N501YMA30. Cytokine and chemokine transcripts were detected by qRT–PCR from the lungs of uninfected (0 dpi) and infected mice at 4 dpi. Each lung was collected from an individual mouse. n = 6 for all groups at 0 and 4 dpi except for PLA2G2D−/− mice at 0 dpi where n = 7. Data are mean ± s.e.m. Data are pooled from two independent experiments.
Extended Data Fig. 6 ∣. Effects of asapiprant, indomethacin and fevipiprant in aged mice infected with SARS-CoV-2 or SARS-CoV.
a, Schematic showing experimental design for results shown in panel b (mouse image with BioRender. com). b, Percentage of initial weight and survival of middle-aged C57BL/6 mice infected with 5000 PFU of SARS2-N501YMA30 after indomethacin treatment (n = 5 in all groups except in the group when indomethacin was given at 0 dpi, where n = 4). c, Middle-aged C57BL/6 mice were instilled with CFSE intranasally 6 h before infection with 5000 PFU of SARS2-N501YMA30. One group of mice received vehicle or asapiprant at 1 and 2 dpi (1+2) while another group of mice received drug treatment only at 2 dpi (2). Mice were euthanized at 3 dpi and lung draining lymph node (DLN; mediastinal) were harvested and analyzed with flow cytometry. rDC migration from the lung to DLN was measured as the frequency of CFSE+CD11c+ cells in the DLN (n = 4). d, Percentage of initial weight and survival of middle-aged C57BL/6 mice infected with 5000 PFU of SARS2-N501YMA30 with fevipiprant (CRTH2 inhibitor) treatment (two times a day; days 2 to 8; 5 mg/kg) (n = 4, vehicle; n = 5, fevipiprant). e, Percentage of initial weight and survival of middle-aged C57BL/6 mice infected with 104 PFU of SARS-CoV, with vehicle or asapiprant treatment from day 2 to 8 (n = 5, untreated; n = 12, vehicle and asapiprant). P values determined by two-tailed Student’s t test in a and log-rank (Mental-Cox) in e. Data are mean ± s.e.m. and are representative of at least two independent experiments.
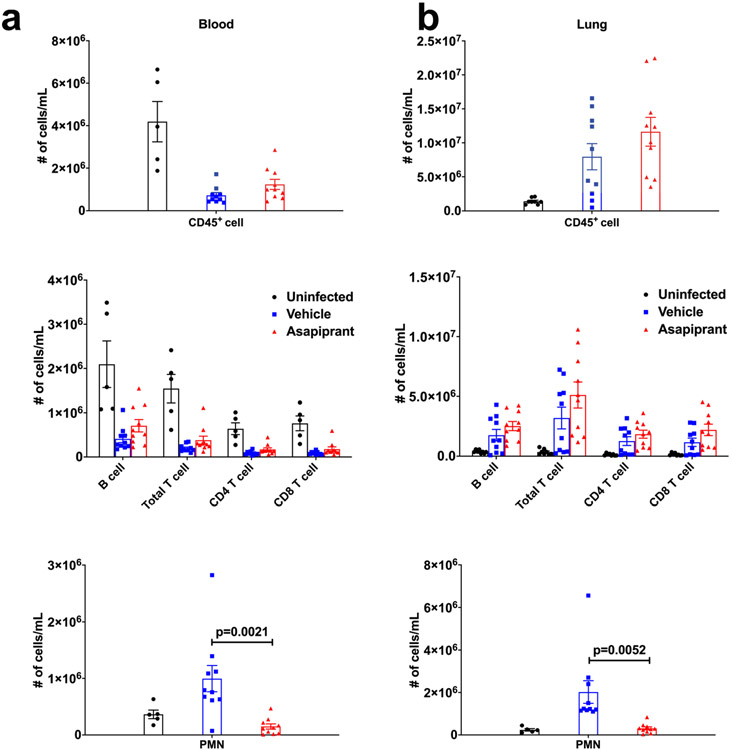
Extended Data Fig. 7 ∣. Asapiprant reversed neutrophilia but not lymphopenia in infected mice.
Numbers of immune cells in the blood (a) and lung (b) of middle-aged C57BL/6 mice at 6 days after infection with 5000 PFU of SARS2-N501YMA30 with vehicle or asapiprant treatment starting at 2 dpi (n = 5 for uninfected controls; n = 10 vehicle- and asapiprant-treated mice). PMN: Polymorphonuclear neutrophils. P values determined by two-tailed Student’s t test. Data are mean ± s.e.m. Data are pooled from two independent experiments.
Supplementary Material
Acknowledgements
This work is supported in part by grants from the National Institutes of Health USA (NIH; P01 AI060699 (S.P. and P.B.M.) and R01 AI129269 (S.P.)) and BIOAGE Labs (S.P.). The Pathology Core is partially supported by the Center for Gene Therapy for Cystic Fibrosis (NIH grant P30 DK-54759) and the Cystic Fibrosis Foundation. P.B.M. is supported by the Roy J. Carver Charitable Trust. L.-Y.R.W. is supported by Mechanism of Parasitism Training Grant (T32 AI007511). We thank M. Gelb (University of Washington) for Pla2g2d−/− mice.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-022-04630-3.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Competing interests K.W., K.K., F.A., J.R. and K.F. are employees of BIOAGE Labs. All other authors have no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-022-04630-3.
Data availability
The data supporting the findings of this study are documented within the paper and are available from the corresponding authors upon request. Human datasets were obtained from the GEO accession GSE150910 and the GTEx portal (https://www.gtexportal.org/home/) for analysis. The sequence of the mouse-adapted virus was deposited in GISAID (https://www.gisaid.org/) (accession ID EPI_ISL_1666328). Source data are provided with this paper.
References
- 1.Channappanavar R & Perlman S Age-related susceptibility to coronavirus infections: role of impaired and dysregulated host immunity. J. Clin. Invest 130, 6204–6213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plante JA et al. The variant gambit: COVID-19’s next move. Cell Host Microbe 29, 508–515 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijay R et al. Critical role of phospholipase A2 group IID in age-related susceptibility to severe acute respiratory syndrome-CoV infection. J. Exp. Med 212, 1851–1868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao J, Zhao J, Legge K & Perlman S Age-related increases in PGD2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J. Clin. Invest 121, 4921–4930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan Y, Shang J, Graham R, Baric RS & Li F Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol 94, e00127–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao L et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583, 830–833 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Jiang RD et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182, 50–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler ES et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol 21, 1327–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng J et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun SH et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28, 124–133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassan AO et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182, 744–753 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 182, 734–743 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinnon KH III et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu H et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369, 1603–1607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leist SR et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peiris JS, Guan Y & Yuen KY Severe acute respiratory syndrome. Nat. Med 10, S88–S97 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehr AR Bacterial artificial chromosome-based lambda red recombination with the I-SceI homing endonuclease for genetic alteration of MERS-CoV. Methods Mol. Biol 2099, 53–68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang K et al. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine 67, 103381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J et al. Mouse-adapted SARS-CoV-2 replicates efficiently in the upper and lower respiratory tract of BALB/c and C57BL/6J mice. Protein Cell 11, 776–782 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y et al. SARS-CoV-2 rapidly adapts in aged BALB/c mice and induces typical pneumonia. J. Virol 95, e02477–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starr TN et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell 184, 2362–2371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun H et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat. Commun 12, 5654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts A et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3, e5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ackermann M et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med 383, 120–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galani IE et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol 22, 32–40 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Zhang X et al. Viral and host factors related to the clinical outcome of COVID-19. Nature 583, 437–440 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Hirano Y et al. Synergistic effect of PGD2 via prostanoid DP receptor on TNF-α-induced production of MCP-1 and IL-8 in human monocytic THP-1 cells. Eur. J. Pharmacol 560, 81–88 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Hirano Y et al. Prostanoid DP receptor antagonists suppress symptomatic asthma-like manifestation by distinct actions from a glucocorticoid in rats. Eur. J. Pharmacol 666, 233–241 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Ullah MA, Rittchen S, Li J, Hasnain SZ & Phipps S DP1 prostanoid receptor activation increases the severity of an acute lower respiratory viral infection in mice via TNF-α-induced immunopathology. Mucosal Immunol. 14, 963–972 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miki Y et al. Dual roles of group IID phospholipase A2 in inflammation and cancer. J. Biol. Chem 291, 15588–15601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miki Y et al. Lymphoid tissue phospholipase A2 group IID resolves contact hypersensitivity by driving antiinflammatory lipid mediators. J. Exp. Med 210, 1217–1234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oslund RC, Cermak N & Gelb MH Highly specific and broadly potent inhibitors of mammalian secreted phospholipases A2. J. Med. Chem 51, 4708–4714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drake TM et al. Non-steroidal anti-inflammatory drug use and outcomes of COVID-19 in the ISARIC Clinical Characterisation Protocol UK cohort: a matched, prospective cohort study. Lancet Rheumatol. 3, e498–e506 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.RECOVERY Collaborative Group. Aspirin in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet 10.1016/S0140-6736(21)01825-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kragholm K, Torp-Pedersen C & Fosbol E Non-steroidal anti-inflammatory drug use in COVID-19. Lancet Rheumatol. 3, e465–e466 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santoro F et al. Antiplatelet therapy and outcome in COVID-19: the Health Outcome Predictive Evaluation Registry. Heart 10.1136/heartjnl-2021-319552 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voruganti D, Bassareo PP, Calcaterra G & Mehta JL Does aspirin save lives in patients with COVID-19? Heart 10.1136/heartjnl-2021-320255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi G et al. Effect of the potent and selective DP1 receptor antagonist, asapiprant (S-555739), in animal models of allergic rhinitis and allergic asthma. Eur. J. Pharmacol 765, 15–23 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Furusawa H et al. Chronic hypersensitivity pneumonitis, an interstitial lung disease with distinct molecular signatures. Am. J. Respir. Crit. Care Med 202, 1430–1444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carithers LJ & Moore HM The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank 13, 307–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng J et al. Severe acute respiratory syndrome coronavirus 2-induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J. Infect. Dis 223, 785–795 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marone G et al. Prostaglandin D2 receptor antagonists in allergic disorders: safety, efficacy, and future perspectives. Expert Opin. Investig. Drugs 28, 73–84 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Almazan F et al. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. MBio 4, e00650–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshikawa T et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS ONE 5e8729 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyerholz DK & Beck AP Principles and approaches for reproducible scoring of tissue stains in research. Lab. Invest 98, 844–855 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Li K et al. Middle East respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J. Infect. Dis 213, 712–722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morin A et al. Collaboration gets the most out of software. Elife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are documented within the paper and are available from the corresponding authors upon request. Human datasets were obtained from the GEO accession GSE150910 and the GTEx portal (https://www.gtexportal.org/home/) for analysis. The sequence of the mouse-adapted virus was deposited in GISAID (https://www.gisaid.org/) (accession ID EPI_ISL_1666328). Source data are provided with this paper.