New advancements in biological image processing, such as object segmentation, tracking1, and machine learning frameworks, have enabled researchers to extract more information and ask additional questions of their image data. Increasingly, these innovations are written in the Python programming language, using its extensive software library (e.g. NumPy2, SciPy3) and its accessibility to researchers at various programming proficiency levels. As the Python software library has grown over the years to address new image processing needs, so too has ImageJ, a Java-based open-source software package and platform widely used for scientific image analysis. ImageJ allows researchers to perform a variety of image processing and analysis tasks such as edge detection, tiled image stitching, object and cell lineage tracking, morphological operations like skeletonization as well as various data projections. All of these operations can be combined to construct complex workflows in the form of macros and scripts. Additionally, ImageJ's functionality has been extended through the use of plugins: new features written in Java and accessible directly from ImageJ, capable of bringing cutting edge technologies to the ImageJ platform. ImageJ supports an active community of software developers who produce and maintain these three extension types, which in recent years include deep learning capabilities4.
Unfortunately, Java-based and Python-based programs do not work together or share data seamlessly. Without the ability to easily exchange data between Python and ImageJ, features must be re-implemented in each respective environment or targeted wrappers built; this is not scalable. Communities across both languages require a bridge enabling seamless feature integration without duplicated effort.
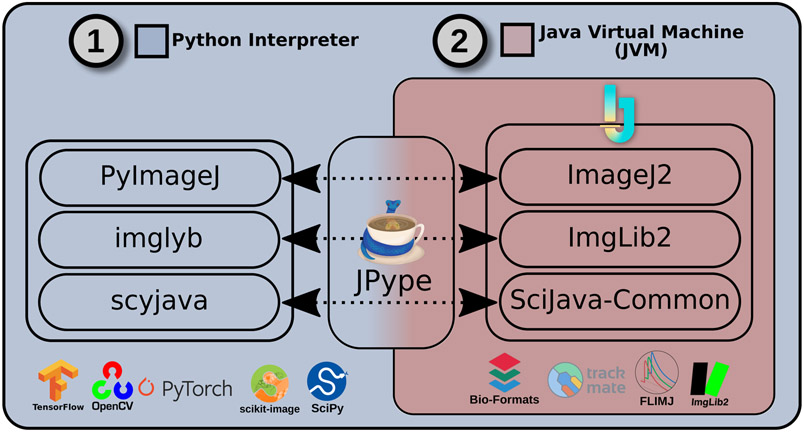
To address this need, we present here PyImageJ, a Python-based package built on ImageJ25 that provides fundamental interoperability between Python and ImageJ-based software including the original ImageJ, ImageJ2, and the Fiji distribution of ImageJ6. With PyImageJ we aim to support both software developers wanting to combine ImageJ and its plugin library with Python-based routines, and bench scientists wanting to do the same within their analysis workflows. PyImageJ is cross-platform, running on Linux, macOS, Windows operating systems and can be installed from PyPI and conda-forge. PyImageJ enables two-way communication between ImageJ and Python by initializing Java as a subprocess of Python, such that any Java-based functionality can be used from Python programs, and new Python-based routines can be written to augment Java programs. The other paradigm, initializing Python as a subprocess of Java, is also useful in some scenarios, and is currently under development. The architecture of ImageJ2 consists of libraries built on two key layers: SciJava (https://scijava.org/), which offers foundational infrastructure and is not image-specific, and ImgLib27, which provides the core image data model (Fig 1). Accordingly, PyImageJ is built on two foundational Python packages: scyjava (https://github.com/scijava/scyjava) and imglyb (https://github.com/imglib/imglyb), which act as Python-based integration layers for the Java-based SciJava and ImgLib2 packages, respectively.
Figure 1.
The software architecture of PyImageJ. Blue: The Python environment and example Python applications. Red: The ImageJ2 software stack with example plugins running in a special Python-integrated JVM. From the Python environment (1) PyImageJ uses JPype (from the scyjava layer) to create the Python-integrated JVM that will run the ImageJ2 software stack. This encapsulated JVM (2) incorporates all the user-requested Java libraries, including ImageJ, ImageJ2, and additional plugins e.g. from Fiji and/or other ImageJ update sites. The top Python layer, PyImageJ, provides access to the ImageJ2 gateway and Python convenience functions. The Python-side imglyb interfaces with the Java-side ImgLib2. Finally, the Python scyjava layer provides the foundational components such as JVM configuration and type conversions.
The first layer, scyjava, utilizes the jgo project (https://github.com/scijava/jgo) to retrieve the ImageJ2 Java libraries, and JPype (https://jpype.readthedocs.io/en/latest/) to create a special Python-integrated Java environment which includes them. This design enables scyjava to transparently download and cache Java libraries packaged from remote online repositories, start the Java environment as a subprocess with those libraries included, wrap Java classes as dynamically generated Python classes with all the same functions, and convert common data structures such as lists, sets, and dictionaries/maps between Java and Python. Notably, the scyjava package is potentially useful for any in-process integration of Java-based libraries into Python programs and can be used independently of PyImageJ.
Exchanging image data between Python and ImageJ is accomplished through the imglyb layer, which provides zero-copy access to NumPy arrays and metadata-rich xarray data through shared memory. Using shared memory to store image data not only reduces memory use and processing time, but also enables the user to see the results of ImageJ processing immediately on Python-based images. ImageJ images that have been converted into an appropriate Python type (i.e. NumPy or xarray) can be accessed by Python-based image processing tools such as napari8, a fast and interactive multi-dimensional image viewer, or CellProfiler9, a workflow tool for reproducibly scaling analyses to large batches of data. Improved performance has already been shown with the RunImageJScript CellProfiler plugin which, for example, enables a user to apply models from Trainable Weka Segmentation10 (an ImageJ plugin) as one step in their feature classification workflow. PyImageJ offers the user interactive access to the full ImageJ2 Application Programming Interface (API), including all of ImageJ2’s functionality and plugins, as well as the original ImageJ API, accessible via ImageJ2’s backwards compatibility legacy layer. PyImageJ also supports a headless mode without graphical user interface (GUI) elements, enabling workflows on systems with no computer monitor (e.g. a remote server)—all ImageJ2 commands are available in headless mode, although functions of the original ImageJ are limited by its underlying dependence on GUI elements.
In summary, PyImageJ gives users access to the best of both Python and ImageJ, by fundamentally integrating the two software ecosystems. PyImageJ supports robust data interoperability between both Python and ImageJ, enabling users to create workflows that incorporate both Python and ImageJ elements.
Acknowledgements
This package was only made possible through the work of jgo and SciJava plugin frameworks. We are grateful for the contributions of Phillip Hanslovsky, the architect of imglyb and co-author of jgo. The authors also thank several individuals for various contributions and suggestions including Ellen TA Dobson, Jan Eglinger, Sam Griffin, Robert Haase, Yang Liu, Hadrien Mary, and Leon Yang. We also thank the PyImageJ user community for their great input and feedback.
This work has been supported by the National Institutes of Health [P41GM135019 to A.E.C., B.A.C., and K.W.E., T32CA009206 to M.A.P., T32GM008349 to M.A.P.]; Chan Zuckerberg Initiative funding to B.A.C., C.T.R. and K.W.E.; National Science Foundation 1429045 to K.W.E.; and additional internal funding from the Laboratory for Optical and Computational Instrumentation (LOCI) and the Morgridge Institute for Research.
Footnotes
Conflicts of interest: none declared.
Data Availability
All data used for PyImageJ usecases are available at https://github.com/imagej/pyimagej/tree/master/doc/sample-data.
Code Availability Statement
The source code, documentation, tutorials and use cases for PyImageJ, which is made available under the open-source Apache software license, can be found online at https://github.com/imagej/pyimagej.
References
- 1.Tinevez J-Y et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Harris CR et al. Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virtanen P et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gómez-de-Mariscal E et al. DeepImageJ: A user-friendly environment to run deep learning models in ImageJ. Nat. Methods 18, 1192–1195 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Rueden CT et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pietzsch T, Preibisch S, Tomančák P & Saalfeld S ImgLib2—generic image processing in Java. Bioinformatics 28, 3009–3011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sofroniew N et al. napari/napari: 0.4.15rc1. (Zenodo, 2022). doi: 10.5281/zenodo.6325333. [DOI] [Google Scholar]
- 9.Stirling DR et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinformatics 22, 433 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arganda-Carreras I et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used for PyImageJ usecases are available at https://github.com/imagej/pyimagej/tree/master/doc/sample-data.
The source code, documentation, tutorials and use cases for PyImageJ, which is made available under the open-source Apache software license, can be found online at https://github.com/imagej/pyimagej.