Pathways enriched in B-ALL clones with preferential CNS residence include complement component C3 and stem cell, metabolism, and mRNA translation.
Abstract
Central nervous system (CNS) dissemination of B-precursor acute lymphoblastic leukemia (B-ALL) has poor prognosis and remains a therapeutic challenge. Here we performed targeted DNA sequencing as well as transcriptional and proteomic profiling of paired leukemia-infiltrating cells in the bone marrow (BM) and CNS of xenografts. Genes governing mRNA translation were upregulated in CNS leukemia, and subclonal genetic profiling confirmed this in both BM-concordant and BM-discordant CNS mutational populations. CNS leukemia cells were exquisitely sensitive to the translation inhibitor omacetaxine mepesuccinate, which reduced xenograft leptomeningeal disease burden. Proteomics demonstrated greater abundance of secreted proteins in CNS-infiltrating cells, including complement component 3 (C3), and drug targeting of C3 influenced CNS disease in xenografts. CNS-infiltrating cells also exhibited selection for stemness traits and metabolic reprogramming. Overall, our study identifies targeting of mRNA translation as a potential therapeutic approach for B-ALL leptomeningeal disease.
Significance:
Cancer metastases are often driven by distinct subclones with unique biological properties. Here we show that in B-ALL CNS disease, the leptomeningeal environment selects for cells with unique functional dependencies. Pharmacologic inhibition of mRNA translation signaling treats CNS disease and offers a new therapeutic approach for this condition.
This article is highlighted in the In This Issue feature, p. 1
Introduction
Early successes with chemotherapy for B-precursor acute lymphoblastic leukemia (B-ALL) were almost uniformly complicated by relapses in the leptomeninges of the central nervous system (CNS; refs. 1–3). The CNS was deemed a sanctuary site for relapse with sparing of resident leukemic cells due to immune privilege and poor blood–brain barrier penetration of systemic chemotherapies. This led to the implementation of universal CNS-directed therapy with craniospinal irradiation and/or intrathecal chemotherapy—treatments that are associated with long-term effects on neurocognitive function (1). Despite this aggressive treatment strategy, 30% of relapsed B-ALL patients have CNS involvement (4, 5). CNS relapse has a poor prognosis due in part to a lack of effective targeted therapies for CNS leukemia (2, 4–6). Thus, there is a great need for new therapies to treat CNS dissemination in B-ALL.
B-ALL leptomeningeal disease can be recapitulated through xenotransplantation of primary B-ALL patient samples into immunodeficient mice (7–10). Patient-derived xenografts are tractable models for interrogating B-ALL clonal dynamics and therapeutic responses. Like the majority of human patients prior to the introduction of CNS-directed therapies, many mice bearing B-ALL xenografts develop CNS disease (7–10). Human B-ALL samples exhibit intratumoral heterogeneity and are composed of genetically and functionally distinct subclones, though the impact of such diversity on CNS engraftment is unclear (7, 11, 12). Although several prior studies did not find evidence of subclonal selection in the leukemic cells in the CNS (8–10), we recently reported subclonal differences between select pairs of bone marrow (BM) and CNS in xenografts (7). Functional studies have to date largely focused on mechanisms of B-ALL trafficking to the subarachnoid space, a process dependent on signaling through VEGFA (8, 13), IL7 (14), IL15 (15), and ITGA6 (16). However, knowledge of the factors governing B-ALL survival and proliferation within the CNS remains limited.
In this study, we used the B-ALL xenograft model to identify novel requirements for leukemic CNS involvement. Using RNA sequencing (RNA-seq), we identified mRNA translation as a requirement for B-ALL disease in the CNS irrespective of clonal selection and validated this pharmacologically in xenografts with the mRNA translation inhibitor omacetaxine mepesuccinate (OMA). Proteomic analysis suggested increased production of complement component 3 (C3) in CNS xenografts, which we demonstrated to be a key determinant of CNS disease.
Results
Clonal Composition of BM and CNS Blasts in B-ALL Xenografts
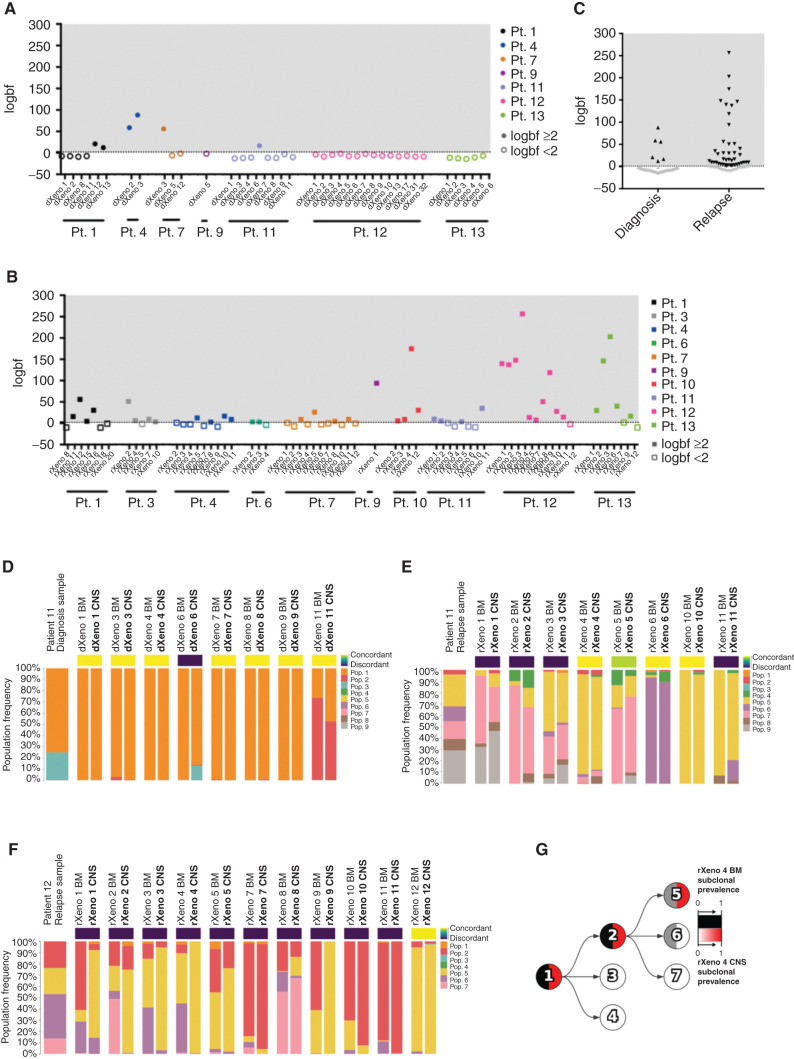
We previously carried out xenografting of paired diagnosis and relapse samples from a cohort of pediatric and adult B-ALL patients (7). The diagnosis and relapse samples of 10 patients (7 diagnosis samples and 10 relapse samples) generated significant CNS disease in xenografts (Supplementary Fig. S1A–S1G; Supplementary Table S1). Leukemic blasts were isolated from dissociated CNS material using Percoll density gradient centrifugation and used for our genomic, transcriptomic, and proteomic studies. CNS cells retained the ability to engraft secondary xenografts in the BM, spleen, and CNS following intrafemoral injection as demonstrated for three primary KMT2A-rearranged samples (Supplementary Fig. S1H–S1J). We used targeted DNA sequencing described in our previous study (7) to gain further insight into the genetic subclonal identity and clonal dynamics in the BM and CNS of primary xenografts of all 10 patient samples (Supplementary Table S2; ref. 7). We used Pairtree (17), the computational tool reported in ref. 10, to quantify the relative prevalence of mutational populations composed of leukemic variants with similar variant allele frequencies in the BM (composed of cells from bilateral femurs and tibias) and CNS of a given recipient mouse for 106 xenografts. Using Bayes factors, we compared a “concordant” model in which leukemic blasts of individual mice shared mutational population frequencies between BM and CNS with a “discordant” model in which the BM and CNS mutational populations could have different frequencies (Fig. 1A–C). We deemed BM and CNS blasts to be genetically discordant if the discordant model was at least 100 times as likely as the concordant model, reflecting the consistently different frequencies of mutational populations between paired BM and CNS samples from individual mice. For diagnosis samples, we found that BM and CNS grafts showed little variation and were almost uniformly concordant, with only 15% demonstrating discordance (6/40 mice; Fig. 1A, C, and D; Supplementary Fig. S2A–S2J). By contrast, 42 of 66 xenografts (64%) generated from relapse samples showed discordance in mutational populations present in BM and CNS of individual mice (Fig. 1B, C, E, and F; Supplementary Fig. S2A–S2J). In the relapse sample of patient 11, as was the case for 7 of 10 patients (14 of 17 samples), no recurrent expansion of any one mutational population was detected in the BM or CNS of multiple mice (Fig. 1E; Supplementary Fig. S2B–S2J). On the contrary, grafts generated from three samples (patient 1 relapse, patient 12 relapse, and patient 13 relapse) exhibited evidence of enrichment for one or more mutational populations in two or more xenografts in BM and/or CNS xenografts (Fig. 1F; Supplementary Fig. S2B and S2J). For example, CNS xenografts derived from the patient 12 relapse sample displayed enrichment for population 5 compared with BM in six xenografts, whereas BM grafts showed greater subclonal diversity with enrichment of populations 6 and 7 (Fig. 1F and G). These data suggest that in many cases, particularly at diagnosis, there is considerable genetic concordance between subclonal diversity of BM and CNS. However, there are also patient samples where genetic discordance can be found between the mutational populations present in the BM and CNS of individual mice, and in some patients, tissue-specific selection for individual subclones occurs; this is more prevalent in relapse samples.
Figure 1.
Differences in clonal composition of BM and CNS B-ALL xenograft engraftment. Mutational population genetic concordance or discordance of CNS blasts as determined using Bayes factors comparing the concordant model, whereby a mouse's BM/CNS samples share mutational population frequencies, with the discordant model, whereby a BM/CNS sample pair can have different mutational population frequencies. We show the logarithm of the Bayes factor (“logbf,” base 10) for the discordant model versus the concordant model for each sample pair in diagnosis xenografts (A; patient 1 n = 6 xenografts; patient 4 n = 2 xenografts; patient 7 n = 3 xenografts; patient 9 n = 1 xenograft; patient 11 n = 8 xenografts; patient 12 n = 14 xenografts; and patient 13 n = 6 xenografts) or relapse xenografts (B; patient 1 n = 7 xenografts; patient 3 n = 5 xenografts; patient 4 n = 9 xenografts; patient 6 n = 3 xenografts; patient 7 n = 11 xenografts; patient 9 n = 1 xenograft; patient 10 n = 4 xenografts; patient 11 n = 8 xenografts; patient 12 n = 11 xenografts; and patient 13 n = 7 xenografts). We used a threshold of logbf e2 to declare discordance for each mouse represented by the dotted line, reflecting that the discordant model was at least 100 times more likely than the concordant model. Filled-in data points have a logbf e2, whereas data points with no fill color have a logbf <2. C, Summary of concordance and discordance calls in diagnosis and relapse patient samples. Black data points have a logbf e2, whereas gray data points have a logbf <2. Mutational population frequencies were computed using Pairtree from patient 11 diagnosis (D; n = 8 xenografts), patient 11 relapse (E; n = 8 xenografts), and patient 12 relapse (F; n = 11 xenografts) samples. (Patient 12 diagnosis in Supplementary Fig. S1I.) G, Evolutionary trajectory of mutational populations for patient 12 is shown in a clone tree determined using Pairtree. Each mutational population is shown as a tree node, with edges indicating evolutionary descent. Each node shows the relative prevalence of mutational population lineages, consisting of mutational populations and their descendants, in BM (black, left half of node) and CNS (red, right half of node) within relapse xenograft 4 from patient 12. Pop., population; Pt., patient. dXeno represents xenografts generated from diagnosis patient samples, and rXeno represents xenografts generated from relapse patient samples.
Leukemia Cells that Disseminate to the CNS Are Transcriptionally Distinct from Cells in the BM Irrespective of Subclonal Composition
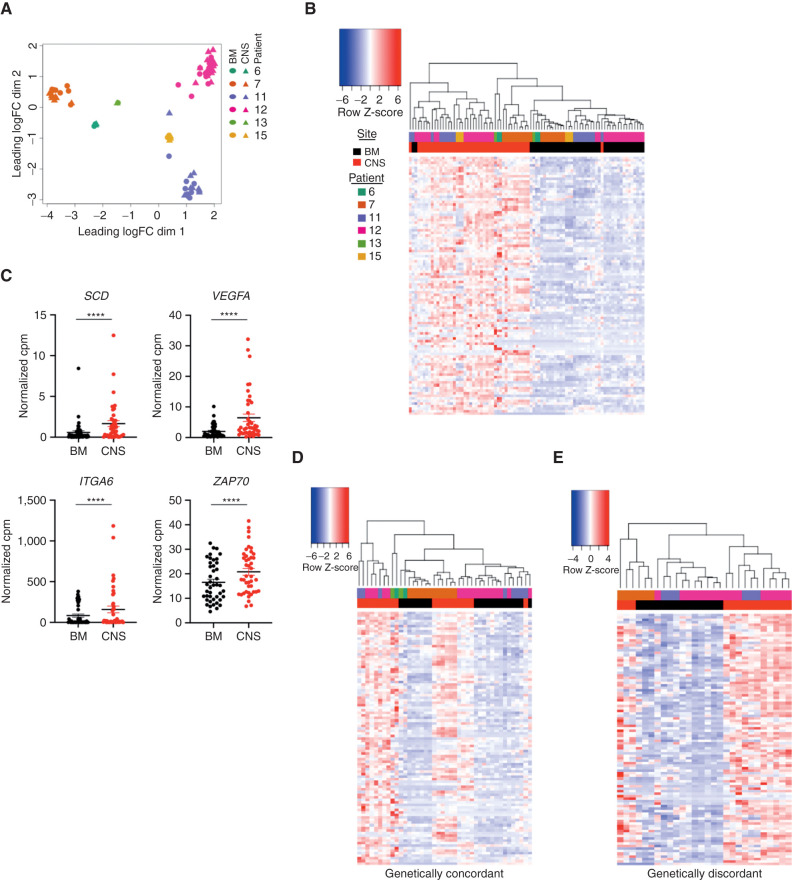
To define the transcriptional landscape of leukemia cells present in the CNS, we performed RNA-seq on 43 pairs of matched BM (cells from bilateral femurs and tibias) and CNS obtained from xenografts from 6 B-ALL patients (patient 6 n = 2 xenografts, patient 7 n = 10, patient 11 n = 9, patient 12 n = 18, patient 13 n = 1, and patient 15 n = 3) with high burden of CNS disease. Xenograft leukemia cells were grouped principally based on the patient from which they were derived (Fig. 2A). However, CNS sites were highly transcriptionally divergent from matched BM sites based on differential gene expression, multidimensional scaling, and unsupervised hierarchical clustering, with 381 CNS-upregulated and 1,064 BM-upregulated genes with greater than twofold change and a false discovery rate (FDR) < 0.05 (Fig. 2B; Supplementary Fig. S3A–S3H). Multiple genes previously associated with CNS dissemination including SCD, VEGFA, ITGA6, and ZAP70 were also upregulated in CNS grafts in our cohort (Fig. 2C; refs. 8, 13, 14, 16, 18, 19). This analysis revealed that CNS blast samples showed a common subset of genes that are significantly upregulated (Fig. 2B and C). Notably, the genes defining the CNS-derived blasts were upregulated in both genetically concordant and discordant CNS blasts (Fig. 2D and E). These data indicate that in B-ALL xenografts, cells in the CNS microenvironment have a distinct transcriptional phenotype compared with those isolated from the BM irrespective of subclonal composition. This may arise from the unique selection pressures in the leptomeningeal microenvironment (20, 21).
Figure 2.
BM and CNS blasts are transcriptionally distinct. A, Multidimensional scaling of RNA-seq gene counts from matched BM (cells from bilateral femurs and tibias; circles) and CNS (triangles) xenografts derived from patient 6 (n = 2 mice), patient 7 (n = 10 mice), patient 11 (n = 9 mice), patient 12 (n = 18 mice), patient 13 (n = 1 mouse), and patient 15 (n = 3 mice). B, Unsupervised hierarchical clustering of matched BM and CNS xenografts based on the normalized gene counts per million (cpm) of the top 100 CNS-upregulated genes by FDR. C, Normalized counts per million for genes indicated in graph title are shown for n = 43 BM and n = 43 CNS xenografts. Line, mean; ****, FDR < 0.001 corrected for multiple hypothesis testing in edgeR. D and E, Expression of the top 100 CNS-upregulated genes from Fig. 1C in the BM and CNS of samples that were also analyzed by targeted sequencing and reported to be genetically concordant (D; n = 21) and discordant (E; n = 28) xenografts from patients 7, 11, 12, and 13, diagnosis and relapse, with samples grouped by unsupervised hierarchical clustering with expression intensity scaled by gene.
CNS-Disseminated Blasts Transcriptionally and Phenotypically Resemble Therapy-Resistant Cells
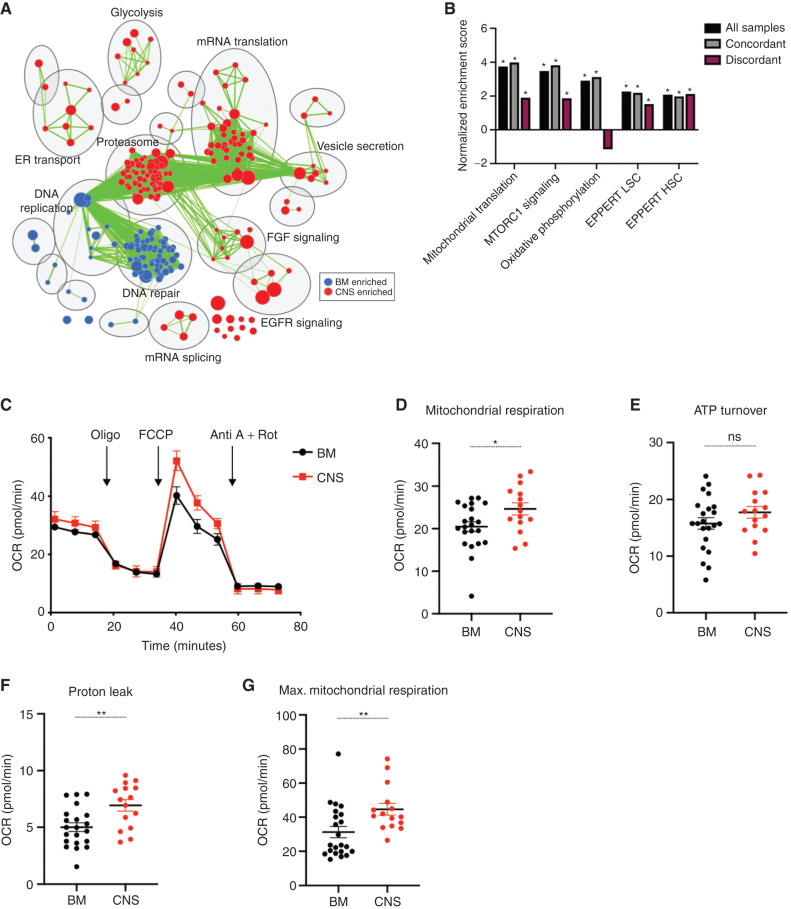
As described in our previous study (10), pathway analysis is more sensitive than simple gene-expression comparisons to find consistent differences between patient samples with widely diverse genetic drivers. Thus, we extended our gene-expression analysis to more broadly investigate the transcriptomic pathways driving the growth of leukemia cells in the CNS across all xenografts. We used gene set enrichment analysis (GSEA; of the data shown in Fig. 2A) for unbiased detection of pathways differentially expressed between blasts in BM and CNS (22). Network analysis grouping differentially expressed gene sets by pathway revealed the top CNS-enriched processes to be the proteasome, mRNA translation initiation and elongation, EGFR signaling, FGF signaling, glycolysis, endoplasmic reticulum (ER) transport, vesicle secretion, and mRNA splicing (Fig. 3A). Relative to matched CNS blasts, BM blasts were enriched for processes including DNA replication and DNA repair (Fig. 3A). Prior studies have shown that treatment-resistant B-ALL xenografts and high-risk diagnosis or relapse B-ALL patient samples express hematopoietic stem cell (HSC) genes and undergo metabolic rewiring that is linked to a stress-tolerant state (7, 23–25). We found similar upregulation of HSC as well as acute myeloid leukemia (AML) stem cell (LSC) gene sets in CNS blasts (Fig. 3B). Furthermore, pathways associated with chemotherapy resistance were upregulated in both concordant and discordant CNS blasts (Fig. 3B). CNS blasts also showed evidence of the metabolic rewiring associated with chemotherapy tolerance and subsequent B-ALL relapse (7), with significant enrichment of gene sets related to mitochondrial translation, oxidative phosphorylation, and mammalian target of rapamycin (mTOR) signaling (Fig. 3B). In keeping with transcriptional upregulation of oxidative phosphorylation and mitochondrial translation gene sets, we performed functional analysis on primary KMT2A-rearranged xenograft cells using flow cytometry and Seahorse XFe analyzer. Evaluation of mitochondrial membrane potential and mitochondrial mass between BM and CNS cells using tetramethylrhodamine ethyl ester (TMRE) and MitoTracker Green demonstrated greater mitochondrial membrane potential per unit mass in CNS blasts than in BM blasts from two of three patient samples (Supplementary Fig. S4A–S4C). Moreover, measuring oxygen consumption rate (OCR) in CNS blasts revealed a remarkable increase in basal mitochondrial respiration, proton leak, and a trend toward greater mitochondrial ATP production (Fig. 3C–F; Supplementary Fig. S4D). We also demonstrated an elevated maximal mitochondrial respiration along with greater mitochondrial spare capacity in CNS blasts over BM blasts (Fig. 3G; Supplementary Fig. S4E). This suggests that CNS blasts have extra mitochondrial reserve capacity available in order to produce more energy or mitochondrial ROS if needed. Overall, CNS blasts have a transcriptional profile and metabolic phenotype that resemble chemotherapy-resistant subclones, and converge on a stress-tolerant state (7, 23–25). Although our study focused on a static analysis of blasts present in the CNS and was not designed to investigate whether these distinct functional properties specifically impact the trafficking, survival, or growth of blasts in the leptomeningeal space, our data argue that improving treatment of CNS disease will require targeting of molecular pathways driving these essential processes.
Figure 3.
CNS xenografts transcriptionally and metabolically resemble therapy-resistant cells. A, Cytoscape map of GSEA-identified differentially enriched gene sets upregulated in CNS and BM xenografts with the top 10 differentially regulated pathways labeled using AutoAnnotate. Node size is proportional to FDRq value of enrichment, and green edges indicate gene overlap between nodes. B, GSEA results showing normalized enrichment scores in CNS versus BM xenograft transcriptomes for gene sets previously associated with B-ALL chemotherapy resistance and clinical relapse in all profiled xenografts (n = 43), concordant xenografts (n = 21), and discordant xenografts (n = 28). *, FDRq score < 0.05. C, Pooled analysis of OCR measured by Seahorse XFe 96 analyzer with additions of oligomycin A (Oligo), carbonyl cyanide-4-phenylhydrazone (FCCP), antimycin A (Anti A), and rotenone (Rot) from BM and CNS KMT2A-rearranged xenografts (n = 3 mice patient 12 and n = 3 mice patient 15, n = 15 replicate wells). Basal respiration (D), ATP-linked respiration (E), proton leak respiration (F), and maximal (Max.) OCR across BM (G; n = 4 mice patient 12 and n = 4 mice patient 15, n = 22 replicate wells for D–G) and CNS (n = 3 mice patient 12 and n = 3 mice patient 15, n = 15 replicate wells for D–G) xenografts. Bars, mean ± SE; P values derived from two-sided unpaired Student t test (D–G), with *, P < 0.05; **, P < 0.01; ns, not significant.
OMA Inhibits mRNA Translation and Impairs CNS Engraftment
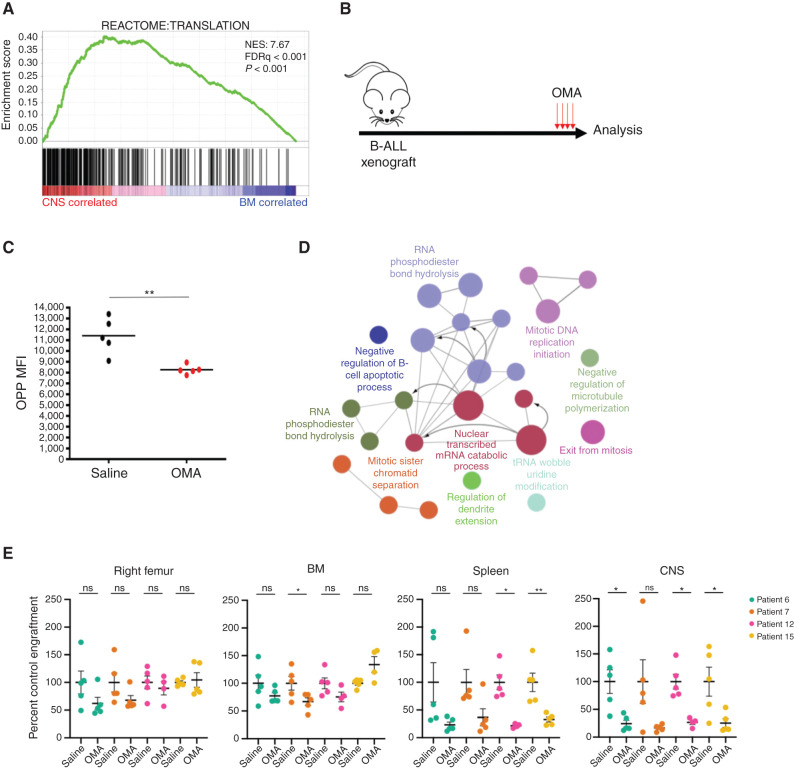
To provide proof of principle that the unique properties we uncovered represent critical dependencies for B-ALL CNS involvement, we focused on mRNA translation as a targetable process that drives CNS disease because mRNA translation and ribosomal biogenesis were the second most highly enriched biological process in CNS blasts (Fig. 4A). Hierarchical clustering and differential expression analysis confirmed upregulation of the leading edge genes from the Reactome “Translation” gene set described in Fig. 4A in the CNS blasts from all six patients (Supplementary Fig. S5A–S5F).
Figure 4.
Blocking mRNA translation as a therapeutic strategy for CNS disease. A, Waveplot from GSEA in Fig. 2A showing the Reactome “Translation” gene set, the third most highly enriched gene set in CNS blasts (positively correlated). Normalized enrichment score (NES) = 7.66; FDRq < 0.001. B, B-ALL xenografts generated by intrafemoral injection of BM blasts (patients 6 and 7) or CNS blasts (patients 12 and 15) were monitored for leukemic engraftment and then treated with 1 mg/kg OMA daily for 4 days or 3 days if significant leukemic morbidity was observed (3 days patient 15; 4 days patients 6, 7, and 12) prior to analysis. C, Translation rate in CNS blasts of saline or OMA-treated patient 12 xenografts measured by O-propargyl-puromycin (OPP) incorporation (n = 5 mice). MFI, mean fluorescence intensity. D, Pathways significantly downregulated in proteomic analysis of CNS blasts in OMA-treated xenografts were identified by ClueGO analysis with Markov Cluster Algorithm (MCL) clustering, displayed as a Cytoscape enrichment map of nodes colored by gene ontology process, with size representing significance and edges showing connection between proteins in different nodes. E, Human B-ALL engraftment in tissues of NSG xenografts treated as in B, with human CD19+CD45+ cell counts normalized to saline-treated controls from the day of sacrifice (n = 5 mice per patient for vehicle; n = 5 mice per patient 6, 7, 12, and 4 mice for patient 15 for OMA). In plots, bars represent mean ± SE; two-sided unpaired t test, with *, P < 0.05; **, P < 0.01; ns, not significant.
To test whether CNS-disseminated B-ALL blasts may be preferentially sensitive to perturbations in protein synthesis, we assessed the effects of in vivo treatment of xenografts with OMA. OMA is an inhibitor of mRNA translation that blocks the ribosome A site and has good blood–brain barrier penetration to the CNS (26). OMA is currently used to treat chronic myelogenous leukemia, is being trialed in AML, and showed activity in a preclinical model of breast cancer metastasis, but was not effective in an early-phase clinical trial against lymphoid leukemias (27–30). Intraperitoneal injection of OMA decreased the rate of mRNA translation in CNS blasts (Fig. 4B and C). Label-free liquid chromatography–mass spectrometry (LC-MS) analysis of CNS blasts from saline- and OMA-treated mice revealed a widespread decrease in protein abundance, with significant downregulation of proteins governing mitosis, negative regulation of B-cell apoptosis, and RNA catabolism (Fig. 4D). In mice with established B-ALL engraftment treated for 4 days (patients 6, 7, and 12) or 3 days (patient 15 due to high pretreatment leukemic morbidity), OMA significantly decreased B-ALL disease burden in the CNS and spleen, with a trend toward a smaller magnitude reduction in BM engraftment that reached significance in xenografts from one patient sample (Fig. 4E; Supplementary Fig. S5G and S5H). Our data strongly suggest that B-ALL CNS disease is critically dependent on mRNA translation.
Posttranscriptional Upregulation of Secretory Machinery and Complement Cascade in CNS Blasts
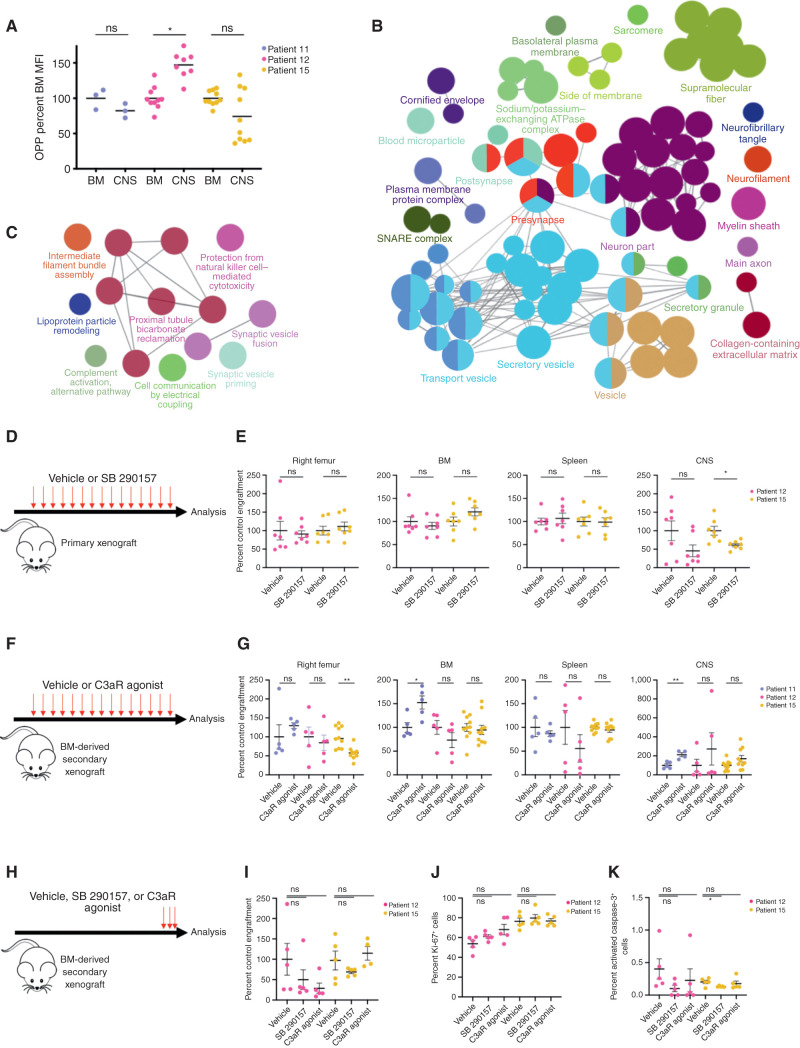
To determine the mechanism by which differences in mRNA translation of CNS blasts might promote B-ALL leptomeningeal disease, we first focused on translation rates. Because increased mRNA translation rates are found in a number of metastatic tumors (31–33), we measured incorporation of the amino acid analogue O-propargyl-puromycin (OPP) in BM and CNS. There was no common trend toward altered CNS translation rate across xenografts from three patients, all of whom had KMT2A-rearranged leukemia (Fig. 5A). We then performed LC-MS on matched BM and CNS derived from these B-ALL patients to identify whole-proteome differences between the sites. Networks of proteins related to synaptic vesicles, secretory granules, neuronal components, and extracellular matrix were identified in CNS, with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis demonstrating protein enrichment of previously described mediators of CNS disease, including VEGF signaling and glycolysis (Fig. 5B; Supplementary Fig. S6A; Supplementary Table S3; refs. 8, 13).
Figure 5.
Posttranscriptional upregulation of complement component 3 promotes leptomeningeal disease in KMT2A-rearranged xenografts. A, Translation rate measured by OPP incorporation in matched BM and CNS blasts from KMT2A-rearranged patient 11, 12, and 15 xenografts represented as OPP mean fluorescence intensity (MFI) normalized to matched BM mean for each sample; n = 3 mice (patient 11 diagnosis), n = 10 mice (patient 12 diagnosis, patient 15 diagnosis). B, Biological processes upregulated as measured by label-free mass spectrometry proteomics of CNS blasts compared with matched BM blasts shown as a Cytoscape enrichment map with Markov Cluster Algorithm (MCL)-clustered, ClueGO-identified nodes colored by gene ontology process, with node size reflecting the number of involved proteins and lines representing proteins shared between nodes. C, Cytoscape map of pathways upregulated in proteins with greater abundance in CNS blasts without differential RNA-seq counts of the corresponding mRNA compared with BM blasts. Circular nodes represent MCL clustering of ClueGO-identified biological processes, with node size reflecting the number of involved proteins and edges representing proteins shared between nodes. D, Primary B-ALL xenografts were treated with 10 mg/kg of the C3a receptor antagonist SB 290157 or DMSO vehicle biweekly from the week after engraftment to endpoint. E, Human B-ALL engraftment in various tissues from D normalized to vehicle; n = 7 mice per group from patient 12 diagnosis and patient 15 diagnosis. F, Secondary B-ALL BM xenografts were treated with vehicle or 10 mg/kg C3a receptor agonist biweekly from the week after engraftment to endpoint. G, Human B-ALL engraftment in various tissues from F normalized to vehicle control; n = 4 (patient 11 CNS), n = 5 (patient 11 BM and patient 12 BM and CNS), or n = 10 (patient 15) mice per group. H, Mice with established secondary B-ALL BM xenografts were treated with vehicle or 10 mg/kg C3a receptor agonist for 3 days immediately prior to endpoint. I, Human B-ALL CNS engraftment from H normalized to vehicle control. Percentage of human B-ALL cells in CNS xenografts from H positive for Ki-67 (J) or activated caspase-3 (K). n = 5 mice per group in I–K. Bars, mean ± SE; two-sided unpaired t test, with *, P < 0.05; **, P < 0.01; ns, not significant.
Because CNS blasts did not consistently differ from BM blasts in their rate of global protein synthesis, we reasoned that CNS-engrafting cells may depend on preferential translation of a specific subset of mRNAs, a phenomenon that occurs in HSCs and embryonic stem cells (34, 35). We searched for pathways in CNS blast–upregulated proteins whose encoding mRNAs were not differentially expressed between BM and CNS sites. The alternative complement signaling cascade was among the most highly upregulated proteomic pathways in non–differentially expressed mRNAs (Fig. 5C). Notably, complement signaling was also the most highly upregulated pathway in CNS blasts among all secreted proteins (Supplementary Fig. S6B) and was enriched across CNS samples from xenografts of all three patients studied regardless of mRNA transcript level (Supplementary Fig. S6A).
Complement Component 3 Promotes Leptomeningeal Disease
To validate that the complement signaling pathway was functionally relevant, we focused on complement C3. Following activation of the lectin, classic, or alternative pathways, cleavage of C3 triggers opsonization, cell lysis, or local inflammation depending on the cleavage product and cellular milieu (36). C3 convertase is a serine protease that cleaves C3 into the inflammatory anaphylotoxin C3a and opsonizing peptide C3b (36). C3 production by metastatic breast and lung carcinoma cells promotes survival in the leptomeningeal space by paracrine signaling through C3a (37). Our proteomic data showed that C3 was more abundant in CNS blasts (Supplementary Table S3). To test the role of C3a signaling in CNS disease, we treated B-ALL primary KMT2A-rearranged xenografts derived from a pediatric (patient 12) and an adult (patient 15) sample with SB 290157, a small-molecule inhibitor of the G protein–coupled C3a receptor shown to be effective in xenograft models of breast and lung carcinoma (ref. 37; Fig. 5D). Mice were treated from the week after cell transplantation until endpoint. SB 290157 decreased leptomeningeal B-ALL engraftment by an average of 50% in CNS xenografts (P = 0.10 patient 12, P = 0.01 patient 15, n = 7 mice DMSO, n = 7 mice SB 290157 per patient) but had no effect on BM or spleen engraftment (Fig. 5E; Supplementary Fig. S6C).
Next, we tested the effect of a C3a receptor agonist on the secondary transplants of BM from KMT2A-rearranged B-ALL xenografts derived from two pediatric (patients 11 and 12) and one adult (patient 15) patient (Fig. 5F). C3a receptor activation from the time of cell transplantation to endpoint produced a consistent increase in CNS engraftment across recipients, though this only reached statistical significance in xenografts from patient 11 (patient 11 P = 0.004, patient 12 P = 0.37, patient 15 P = 0.081) without impacting BM or spleen engraftment (Fig. 5G; Supplementary Fig. S6D).
To distinguish between the possibility of complement C3a mediating B-ALL dissemination to or invasion of the leptomeninges from C3a-mediated survival within the CNS, we treated mice with established xenografts with SB 290157, C3a receptor agonist, or vehicle immediately prior to endpoint (Fig. 5H). Unlike OMA, treatment of established B-ALL xenografts with SB 290157 or C3a receptor agonist did not appreciably alter disease burden or cell cycling, or induce apoptosis (Fig. 5H–K; Supplementary Fig. S6E). Similarly, dose–response studies of both SB 290157 and the C3a receptor agonist showed no effect on the t(4;11) B-ALL cell line RS4;11 in vitro (Supplementary Fig. S6F). These results suggest that posttranscriptional upregulation of C3 by B-ALL increases CNS disease in vivo by promoting dissemination to rather than survival within the leptomeninges.
Discussion
Here, we have used genomic and proteomic profiling to describe the clonality and biology of B-ALL CNS xenografts and uncovered novel dependencies for CNS involvement in B-ALL. By combining transcriptomic and proteomic profiling in a preclinical xenograft model, we identified altered mRNA translation as a novel regulator and therapeutic vulnerability of CNS disease. Broadly, CNS blasts showed evidence of transcriptional and metabolic rewiring associated with chemotherapy resistance that distinguished them from BM blasts. We also observed recurrent clonal discordance in BM and CNS engraftment in multiple samples from a large patient cohort, with evidence of tissue-specific selection. Despite frequent clonal discordance in populations engrafting the BM and CNS in relapse xenografts, transcriptional and metabolic rewiring was observed in CNS blasts irrespective of clonal selection, suggestive of CNS site–induced changes.
Although the ability to carry out long-term clonal propagation is thought to be a common property among individual B-ALL cells that make up a tumor and the underlying stemness programs are proposed to be plastic in B-ALL, there is clear evidence that B-ALL chemotherapy resistance in vitro and propensity to relapse clinically correlates with a distinct HSC signature expression (7, 24). Additionally, we previously showed activation of mitochondrial translation, oxidative phosphorylation, and mTOR in chemotherapy-resistant, relapse-fated B-ALL (7). Our transcriptomic and metabolic profiling indicated that similar pathways arise in CNS blasts, suggesting that the leptomeningeal environment selects for blasts with enhanced stemness and stress-resistant properties. We hypothesize that these induced cellular properties might relate to their ability to contribute to relapse. The enhanced mitochondrial spare capacity of CNS blasts may reflect an adaptation to the leptomeningeal niche that confers greater physiologic reserve against the stress of chemotherapy (38). Thus, although the CNS is considered a sanctuary site for disease relapse due to immune privilege and poor drug penetration, CNS blasts' inherent or induced transcriptional and metabolic properties may correlate with a greater ability to withstand chemotherapy. This could contribute to their ability to originate relapse disease and warrants further investigation.
The translation of mRNA to synthesize protein is a fundamental cellular process that is a common driver of malignant growth and metastatic spread across multiple cancers (31–33, 39, 40). Indeed, depletion of L-asparagine in the serum and, to a lesser extent in the cerebrospinal fluid (CSF), is a cornerstone of B-ALL therapy that works in part through translational inhibition; translational reprogramming can mediate L-asparaginase resistance in some cancer models (41–43). Many translation inhibitors are thus actively being pursued as antineoplastics, including OMA in tyrosine kinase inhibitor–resistant chronic myeloid leukemia and current clinical trials for AML (30, 40). Intriguingly, KMT2A-rearranged AML shows epigenetic upregulation of ribosomal genes by the KMT2A fusion protein and corresponding sensitivity to OMA in vitro (44). Differential activation of ribosomal machinery was identified as a source of intratumoral heterogeneity in single-cell RNA-seq analysis of primary B-ALL, where it correlated with expression of HSC genes (45). We demonstrate a similar upregulation of stem cell genes, ribosomal components, and translation factors in B-ALL CNS blasts. We found that B-ALL disease in the CNS is exquisitely sensitive to translation inhibition with OMA. OMA rapidly reduced levels of the mitotic machinery and B-cell antiapoptotic proteins within the leukemic xenografts, suggesting that their constant production is required to survive and divide in the leptomeningeal space. Interestingly, OMA prolonged survival in a Bcr–Abl-driven murine model of B-ALL, and although CNS disease burden was not studied, it suggests that all blasts are sensitive to some level of mRNA translation inhibition (46). Nevertheless, this dependency on translation was highly contextual because BM blasts were much less sensitive than CNS blasts in our study. Splenic engraftment was also substantially reduced with OMA, though the mechanisms underlying this and any similarities between spleen- and CNS-engrafting blasts remain outside the scope of the current study.
Complement signaling is a proinflammatory mediator of innate and adaptive immunity and thus can both promote and restrict malignant growth depending on context (47). The C3a receptor is present on choroid plexus epithelial and brain endothelial cells, and C3 signaling has been associated with increased blood–brain barrier permeability (48, 49). C3 drives leptomeningeal metastasis of breast and lung adenocarcinoma by permeabilizing the blood–CSF barrier, helping metastatic cells adapt to the nutrient-poor subarachnoid space (37). Moreover, melanoma patients with leptomeningeal disease have elevated C3 in CSF, and although serum levels of C3 are elevated compared with healthy controls at diagnosis in B-ALL, we are not aware of prior links to CNS disease (50, 51). The increase or decrease in leptomeningeal engraftment of B-ALL xenografts with respective C3a receptor activation or inhibition reveals a novel, C3a-dependent mechanism for B-ALL CNS involvement. The lack of pharmacokinetic data on CNS penetration of these compounds is a limitation of the study. Our data suggest convergent evolution on C3a signaling as a driver of leptomeningeal metastasis across cancers that could have broader clinical implications given the devastating consequences of brain involvement for many cancer types. Intriguingly, targeting C3a signaling was highly contextual. When modulated from engraftment to endpoint, C3a signaling altered B-ALL CNS disease burden in vivo, but administration of C3a receptor agonist or antagonist to mice with established grafts did not alter CNS disease burden, CNS blast cell cycling, or apoptosis. Similarly, targeting the C3a receptor did not affect B-ALL growth in vitro. These results suggest that altered translation of secreted proteins, including C3, may promote CNS disease through non–cell-autonomous mechanisms such as increasing trafficking to or invasion of the leptomeninges. Conversely, proteomic profiling of OMA-treated blasts demonstrated that cells residing within the leptomeningeal space require continuous production of cell-cycle machinery and antiapoptotic factors to survive.
Overall, our findings show promise for translation to the clinic for patients with B-ALL CNS disease. As OMA has been clinically approved as Synribo (Teva Pharmaceuticals), there is a clear pathway for rapid repurposing and clinical testing in B-ALL based on our findings along with the known ability of Synribo to penetrate the CNS following subcutaneous dosing in patients. Although the effects of the drug were most pronounced on extramedullary disease, there was a trend toward a treatment effect on BM-engrafted cells. Clinical study would be required to determine whether relapsed B-ALL patients might benefit most from OMA as a single-agent therapy or as part of combination treatment. Safe and effective doses have not yet been established for pediatric patients, and although young adults with B-ALL may be eligible for trials with adult dosing, early-phase clinical testing would be required prior to Synribo's application to pediatric B-ALL. Targeting CNS disease by inhibiting mRNA translation represents a novel therapeutic strategy for B-ALL that may have potential to improve outcomes for patients with relapsed disease.
Methods
Patient Samples
Primary leukemia cells were obtained upon clinical presentation at diagnosis and relapse from five adult and six pediatric B-ALL patients under protocols approved by the Research Ethics Board of the University Health Network (adult samples) or the St. Jude Institutional Review Board (pediatric samples). All patients or families provided informed written consent. Samples were viably frozen and stored at −150°C prior to retrospective selection based on availability.
Xenograft Generation and CNS Blast Purification
All animal experiments were performed with approval from the University Health Network Animal Resource Centre Review Board. Patient samples were depleted of T cells by cell sorting on a FACSAria III (BD Biosciences) for CD19+CD45dim/− leukemic blasts using the following antibodies: anti-CD19 PE (BD, RRID:AB_2868805), anti-CD3 FITC (BD, RRID:AB_2811220), anti-CD3 APC (Beckman Coulter, clone UCHT1), anti-CD45 APC (BD, RRID:AB_2868745), or anti-CD45 FITC (BD, RRID: AB_400074) as previously described (13). NOD.CB17-PRkdcscidIl2rgtm1Wjl/Szj (NSG) mice (RRID:IMSR_NM-NSG-007) were bred at the University Health Network according to established protocols. Eight- to 12-week-old male or female mice were sublethally irradiated using a Cesium source with 225 cGy 24 hours prior to intrafemoral transplantation of leukemic blasts (7). Mice were grouped by sex for individual experiments. Mice were sacrificed up to 30 weeks after transplant or upon evidence of symptom onset.
Human cells were purified from the injected femur (right femur), BM (noninjected left femur, left and right tibia), and spleen as previously described (7). Secondary xenografts refer to the second passage of B-ALL cells in a xenograft, and tertiary xenografts refer to the third passage of B-ALL cells in a xenograft. The CNS comprising the brain, spinal cord, and meninges was dissected into phosphate-buffered saline (PBS) with 5% fetal bovine serum (FBS) and then mechanically dissociated by crushing through a 70-µm filter. Leukemic blasts were purified from dissociated CNS tissue using a discontinuous 70%/37%/30% Percoll density gradient with leukemic cells found by aspiration of the 37%/70% interface after centrifugation.
Human engraftment was analyzed using human-specific antibodies for CD45 (V500, BD clone H130; FITC, BD clone H130), CD19 (BD, RRID:AB_2868805), CD34 (APC-Cy7, BD, clone 581), CD3 (APC, BD, Clone UCHT1), CD33 (PE-Cy7, BD, RRID: AB_399961; APC, BD RRID: AB_2868824), and CD44 (PE, BD, RRID: AB_394000; FITC, BD, RRID: AB_400360) on a BD Biosciences LSRII or Celesta Cytometer. Viable BM and spleen cells were counted using a Vicell Cell Counter (Beckman Coulter), and viable CNS cells were counted using trypan blue staining and a hemocytometer. Human cells from the BM used for DNA and RNA isolation were depleted of murine cells using the Miltenyi Mouse Cell Depletion Kits (Miltenyi Biotec) or by cell sorting samples (<20% engraftment) to a purity of >90% as determined by flow cytometry. CNS cells of >90% human cells were selected for RNA isolation and >60% for DNA isolation without further purification due to limited cell numbers. Serial sections from formalin-fixed, paraffin-embedded tissues from select xenografts were stained with hematoxylin/eosin or human CD45 (Agilent clone 2B11 + PD7/26, M0701) after antigen retrieval for confirmation of human engraftment.
RNA-seq and Pathway Analysis
Total RNA was extracted from 1–2 × 106 human purified primary xenograft BM or CNS (>85% human engraftment) using TRIzol and ethanol precipitation as previously described (52). Libraries were prepared using TruSeq Stranded Total RNA Library Ribo-Zero Gold Prep Kits (Illumina) and were sequenced at The Centre for Applied Genomics (TCAG; SickKids, Toronto, Canada) with a HiSeq 4000 Sequencer (Illumina) or HiSeq 2500 Sequencer (patient 15 samples; Illumina) with an average of 3.192 × 107 reads per sample and an average 84.3% uniquely mapping reads. Reads were trimmed with Cutadapt and subjected to FastQC quality control analysis. RNA-seq reads were aligned against GRCh38 using STAR 2.4.2b with default parameters (53). Genes were annotated with gencode v23. Counts were obtained using HTSeq v0.7.2 with the gene_name set as the ID attribute (54). Differentially expressed genes were identified using edgeR (version 3.16.5; ref. 55). Pathway analysis was performed using preranked GSEA (version 2.0) run in classic mode with a custom group of gene sets (Bader Lab, http://download.baderlab.org/EM_Genesets/, December 1, 2019 version; ref. 22). Cytoscape v3.6.1 was used to visualize differentially enriched gene sets that were grouped using a Markov Cluster Algorithm (MCL) and annotated using AutoAnnotate v1.2 plugin (56–58).
Metabolic Profiling
Fluorescent dye staining for mitochondrial content was performed on primary xenograft cells by incubation at 37°C with 1 mmol/L MitoTracker Green (M7514) and 1 µmol/L TMRE (T668) according to the manufacturer's instructions (Thermo Fisher) and then analyzed using a BD LSRII Cytometer (BD Biosciences). All respiratory profiles were performed using the XF96 extracellular flux analyzer (Seahorse Bioscience). Prior to respiration analysis, dead cells were removed from BM and CNS xenografts with a magnetic dead cell–depletion kit (Miltenyi Biotec). A total of 5 × 105 BM and CNS cells in 180 µL of XF media (Seahorse Bioscience), supplemented with 11 mmol/L glucose, 2 mmol/L glutamine, and 1 mmol/L pyruvate (pH 7.4), were seeded onto XFe96 plates (102416-100, Agilent Technologies) and incubated at 37°C, in CO2 free incubator, for 1 hour. OCR and extracellular acidification rate were then evaluated using an XF96 extracellular flux analyzer (Seahorse Bioscience). To measure ATP-linked OCR, maximal respiration, and mitochondrial-dependent basal OCR and proton leak, 1 µmol/L oligomycin A, 0.5 µmol/L carbonyl cyanide-4-phenylhydrazone (FCCP), and 1 µmol/L antimycin A and 1 µmol/L rotenone were added, respectively.
Targeted DNA Sequencing and Analysis
Human engrafted cells from BM and CNS tissue (purified for human cells as above) were subjected to targeted sequencing of their patient-identified variants as previously described (7). Briefly, DNA libraries were prepared using 250 to 500 ng of DNA and the NEXTflex DNA-SEQ Library Prep Kit (BiooScientific) with NEXTflex-96 DNA Barcodes (BiooScientific) and sequenced on a HiSeq 2500 sequencer. Mutational clustering to determine population frequencies and temporal ordering of mutations to infer their evolutionary relationships was performed using Pairtree as previously described (refs. 7, 17; Supplementary Methods). To determine genetic concordance of mutational populations between CNS blasts and BM blasts of individual xenografts, we compared a concordant model of the data with a discordant model of the data using Bayes factors (see Supplementary Methods).
Clone trees were built using the Pairtree algorithm, which is published at https://github.com/morrislab/pairtree. Code for running Pairtree on the data associated with this study, and for computing the probability of discordance between paired tissue samples using Pairtree output, is available at https://github.com/morrislab/cns-ball-discordance/.
Xenograft Drug Assays
CNS cells from primary mice (patients 12 and 15) or BM cells from tertiary mice (patients 6 and 7) were transplanted intrafemorally into sublethally irradiated NSG mice and monitored for signs of CNS disease (domed heads, poor eye grooming). After confirmation of leukemic engraftment by flow cytometry of peripheral blood, or at the earliest sign of CNS disease, mice were randomized to receive saline vehicle or 1 mg/kg OMA (Sigma-Aldrich) by daily intraperitoneal injection for 4 days—patients 6, 7, and 12 (patient 6 relapse, patient 7 diagnosis, and patient 12 diagnosis)—or 3 days—patient 15 prior to sacrifice. Mice with patient 12 and patient 15 xenografts were symptomatic. The number of days of treatment was determined based on the physical condition of control mice.
For C3a receptor inhibition assays, NSG mice were sublethally irradiated and transplanted intrafemorally with T-cell–depleted primary patient B-ALL cells from diagnosis sample (n = 2, patient 12 or 15) as described above and subsequently treated with 1% DMSO in PBS or 10 mg/kg SB 290157 (EMD Millipore) dissolved in DMSO and resuspended in PBS at 1% v/v by intraperitoneal injection twice weekly until they displayed evidence of CNS disease.
C3a receptor agonist assays were performed by transplanting sublethally irradiated NSG mice intrafemorally with 1.5 × 105 BM cells from three primary B-ALL xenografts (patient 11 diagnosis, patient 12 diagnosis, and patient 15 diagnosis), and treating intraperitoneally twice weekly with the C3a receptor agonist CAS 944997-60-8 (VDM Bio) dissolved in 5% DMSO, 45% PEG-200, and 50% 0.9% saline vehicle until they displayed evidence of CNS disease (patient 15) or until there was evidence of substantial peripheral blood engraftment detected by flow cytometry (patients 11 and 12).
For C3a receptor inhibitor or agonist treatment of established xenografts based on peripheral blood flow cytometry, NSG mice were sublethally irradiated and transplanted intrafemorally with 7.5 × 105 primary B-ALL BM xenograft cells from patient 12 or 15. Eight weeks after engraftment, mice were administered 10 mg/kg SB 290157 (EMD Millipore), C3a receptor agonist CAS 944997-60-8 (VDM Bio), or vehicle solvent 5% DMSO, 45% PEG-200, and 50% 0.9% saline for three daily doses prior to analysis 36 hours posttreatment.
Group sizes were determined by balancing sample size for statistical analysis and experimental feasibility for xenograft models through our experience with the model systems. Animals were randomized to their experimental groups after leukemic transplantation and were euthanized at prespecified endpoints of hunching, poor self-care, or domed heads in OMA and SB 291057 trials. Researchers were not blinded to treatment groups. Mice treated with the C3a receptor agonist or vehicle were sacrificed after confirming leukemic engraftment with >30% human peripheral blood leukocytes, a threshold that predates the onset of CNS symptoms in our model. Investigators were not blinded to study groups. One mouse randomized to be treated with OMA died of unknown causes prior to treatment, in keeping with spontaneous death in the immunodeficient xenograft B-ALL model, and thus was excluded from analysis. One CNS sample from the patient 11 C3a receptor agonist group was lost due to technical failure and could not be included in the final analysis.
In all drug treatments, CNS engraftment in drug treatment groups was normalized to the average of simultaneously vehicle-treated mice from the same patient samples by expressing drug-treated engraftment as a percentage of the control mean allowing engraftment comparison across experiment replicates and patient samples [percent control engraftment = ({{number of CD19+CD45+ cells extracted from tissue of treated mouse}/{average number of CD19+CD45+ cells extracted from tissue in vehicle controls from same experiment}} × 100)].
Translation Assay, Protein Quantification, and Pathway Analysis
Nascent protein formation was measured through incorporation of OPP according to the manufacturer's instructions (Invitrogen) by 1 × 105 freshly isolated, unsorted BM or CNS blasts incubated for 30 minutes at 37°C in Dulbecco's modified Eagle medium with 5% FBS and quantified on a BD LSRII Cytometer (BD Biosciences) from xenograft patient samples with greater than 90% human BM and CNS engraftment. A total of 1 × 105 cells from BM and CNS taken from primary xenografts (patient 11 diagnosis n = 4, patient 12 diagnosis n = 3, patient 15 diagnosis n = 5) or secondary xenografts (saline-treated CNS patient 12 diagnosis and patient 15 diagnosis n = 5, and OMA-treated patient 12 diagnosis n = 4, patient 15 diagnosis n = 5) were prepared for mass spectrometry as previously described (Supplementary Table S2; ref. 59). Label-free peptide quantitation yielded protein intensities that were analyzed using MaxQuant and significantly differentially abundant peptides were identified using label-free quantification–adjusted iBAQ spectral counts in edgeR (OMA treatment) or Proteome Discoverer 2.2 (BM vs. CNS blasts; Thermo Fisher Scientific; ref. 60). Functional enrichment analysis of differentially abundant peptides was performed using STRING version 10.5 and visualized with Cytoscape v3.6.1 with MCL clustering (ClusterMaker2 version 1.3), with annotation by ClueGO 2.5.3 (61–63).
Cell-Cycle and Apoptosis Analysis
Purified CNS blasts were stained with anti-human CD45 (APC, BD clone 2D1, RRID: AB_400555), fixed with Cytofix/Cytoperm (BD Biosciences, RRID: AB_2869008), and then stained with anti–Ki-67 (FITC, BD clone B56, RRID: AB_396302) and anti–active caspase-3 (PE, BD clone C92–605, RRID: AB_393906). Prior to analysis on a BD Celesta Flow Cytometer (BD Biosciences), cells were stained with Hoechst 33342 Dye (Thermo Fisher). Cells negative for Ki-67 with 2n DNA content per Hoechst staining were considered in phase G0, Ki-67+ and 2n were G1, Ki-67+ and 2 to 4n DNA content were S–G2–M.
Leukemia In Vitro Culture
RS4;11 cells (RRID: CVCL_0093) were obtained from the American Type Culture Collection and cultured in RPMI-1640 media supplemented with 10% FBS. Confirmation testing was not performed. Cells were cultured in media containing a dilution series of DMSO vehicle, SB 290157 (EMD Millipore), or C3a receptor antagonist (VDM Bio) for 72 hours prior to quantification of live cells per well based on exclusion of SYTOX Blue viability dye (Thermo Fisher) as per the manufacturer's instructions on a BD LSRII cytometer (BD Biosciences).
Graphs and Statistical Analysis
Prism version 8 (GraphPad) was used for statistical analyses unless otherwise described. Plots were made using Prism, gplots, or ggplot2 in the R programming environment (64, 65).
Data and Materials Availability
Targeted DNA-sequencing data are available in ref. 7. RNA-seq data have been submitted to the European Genome-phenome Archive with the following data accession numbers: EGA study number EGAS00001005647 and EGA dataset EGAD00001008183. LC-MS data have been submitted to the ProteomeXchange Consortium with data set identifier PXD022411.
Authors' Disclosures
E. Waanders reports grants from Dutch Cancer Society during the conduct of the study. C.G. Mullighan reports personal fees from Illumina during the conduct of the study, as well as grants from Pfizer and AbbVie, and other support from Amgen outside the submitted work. J.E. Dick reports grants from Joseph M. West Family Memorial Fund Research Award, Dutch Cancer Society (KUN2012-5366), Novo Nordisk Foundation (Novo Nordisk Foundation Center for Stem Cell Biology, DanStem; grant number NNF17CC0027852), Princess Margaret Cancer Centre Foundation, Ontario Institute for Cancer Research through funding provided by the Government of Ontario, Canadian Institutes for Health Research (Foundation: 154293, operating grant number 130412, operating grant number 89932), Canadian Cancer Society (grant number 703212), Terry Fox Research Institute (Program Project Grant), and Canada Research Chair during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Clinical Characteristics of B-ALL Patient Samples.
B-ALL Xenografts Used in Current Study.
Differentially abundant proteins as quantified by label-free liquid chromatography mass spectrometry in KMT2A-r B-ALL xenografts.
Supplementary Figures S1-S6
Acknowledgments
The authors thank the patients and their families who contributed to this study. They thank the University Health Network/SickKids Flow Cytometry Facility and Robert Lopez and Mario D'Souza from the University Health Network Animal Resource Centre. The authors thank Jenny Ho for experimental assistance and Ethan Kulman for assistance with figure formatting. R.J. Vanner is the recipient of the Joseph M. West Family Memorial Fund Research Award, E. Waanders was awarded Dutch Cancer Society KUN2012-5366, and B. Porse was supported through a center grant from the Novo Nordisk Foundation (Novo Nordisk Foundation Center for Stem Cell Biology, DanStem; grant number NNF17CC0027852). C.G. Mullighan was supported by grants from the National Cancer Institute (NCI): R35 CA297695 and P50 CA021765. J.E. Dick was supported by funds from the Princess Margaret Cancer Centre Foundation, Ontario Institute for Cancer Research through funding provided by the Government of Ontario, Canadian Institutes for Health Research (Foundation: 154293, operating grant number 130412, operating grant number 89932), Canadian Cancer Society (grant number 703212), Terry Fox Research Institute Program Project Grant, and a Canada Research Chair.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors' Contributions
R.J. Vanner: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. S.M. Dobson: Conceptualization, formal analysis, investigation, methodology, writing–review and editing. O.I. Gan: Investigation, writing–review and editing. J. McLeod: Investigation. E.M. Schoof: Data curation, investigation, writing–review and editing. I. Grandal: Investigation. J.A. Wintersinger: Data curation, formal analysis, investigation, writing–review and editing. L. Garcia-Prat: Investigation, writing–review and editing. M. Hosseini: Formal analysis, investigation, writing–review and editing. S.Z. Xie: Formal analysis. L. Jin: Investigation. N. Mbong: Investigation. V. Voisin: Investigation, methodology. M. Chan-Seng-Yue: Formal analysis. J.A. Kennedy: Data curation, writing–review and editing. E. Waanders: Data curation. Q. Morris: Supervision. B. Porse: Supervision. S.M. Chan: Supervision. C.J. Guidos: Supervision. J.S. Danska: Supervision, writing–review and editing. M.D. Minden: Supervision. C.G. Mullighan: Conceptualization, supervision, methodology, writing–review and editing. J.E. Dick: Conceptualization, resources, supervision, funding acquisition, methodology, writing–review and editing.
References
- 1. Pui CH, Thiel E. Central nervous system disease in hematologic malignancies: historical perspective and practical applications. Semin Oncol 2009;36:S2–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaudichon J, Jakobczyk H, Debaize L, Cousin E, Galibert MD, Troadec MBet al. Mechanisms of extramedullary relapse in acute lymphoblastic leukemia: reconciling biological concepts and clinical issues. Blood Rev 2019;36:40–56. [DOI] [PubMed] [Google Scholar]
- 3. Pui CH, Howard SC. Current management and challenges of malignant disease in the CNS in paediatric leukaemia. Lancet Oncol 2008;9:257–68. [DOI] [PubMed] [Google Scholar]
- 4. Fielding AK, Richards SM, Chopra R, Lazarus HM, Litzow MR, Buck Get al. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood 2007;109:944–50. [DOI] [PubMed] [Google Scholar]
- 5. Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med 2015;373:1541–52. [DOI] [PubMed] [Google Scholar]
- 6. Frishman-Levy L, Izraeli S. Advances in understanding the pathogenesis of CNS acute lymphoblastic leukaemia and potential for therapy. Br J Haematol 2017;176:157–67. [DOI] [PubMed] [Google Scholar]
- 7. Dobson SM, García-Prat L, Vanner RJ, Wintersinger J, Waanders E, Gu Zet al. Relapse-fated latent diagnosis subclones in acute B lineage leukemia are drug tolerant and possess distinct metabolic programs. Cancer Discov 2020;10:568–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kato I, Nishinaka Y, Nakamura M, Akarca AU, Niwa A, Ozawa Het al. Hypoxic adaptation of leukemic cells infiltrating the CNS affords a therapeutic strategy targeting VEGFA. Blood 2017;129:3126–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Williams MTS, Yousafzai YM, Elder A, Rehe K, Bomken S, Frishman-Levy Let al. The ability to cross the blood-cerebrospinal fluid barrier is a generic property of acute lymphoblastic leukemia blasts. Blood 2016;127:1998–2006. [DOI] [PubMed] [Google Scholar]
- 10. Elder A, Bomken S, Wilson I, Blair HJ, Cockell S, Ponthan Fet al. Abundant and equipotent founder cells establish and maintain acute lymphoblastic leukaemia. Leukemia 2017;122:2577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SMet al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 2011;469:356–61. [DOI] [PubMed] [Google Scholar]
- 12. Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LAet al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 2011;469:362–7. [DOI] [PubMed] [Google Scholar]
- 13. Münch V, Trentin L, Herzig J, Demir S, Seyfried F, Kraus JMet al. Central nervous system involvement in acute lymphoblastic leukemia is mediated by vascular endothelial growth factor. Blood 2017;130:643–54. [DOI] [PubMed] [Google Scholar]
- 14. Alsadeq A, Lenk L, Vadakumchery A, Cousins A, Vokuhl C, Khadour Aet al. IL7R is associated with CNS infiltration and relapse in pediatric B-cell precursor acute lymphoblastic leukemia. Blood 2018;132:1614–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cario G, Izraeli S, Teichert A, Rhein P, Skokowa J, Moricke Aet al. High interleukin-15 expression characterizes childhood acute lymphoblastic leukemia with involvement of the CNS. J Clin Oncol 2007;25:4813–20. [DOI] [PubMed] [Google Scholar]
- 16. Yao H, Price TT, Cantelli G, Ngo B, Warner MJ, Olivere Let al. Leukaemia hijacks a neural mechanism to invade the central nervous system. Nature 2018;560:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wintersinger JA, Dobson SM, Stein LD, Dick JE, Morris QD. Reconstructing complex cancer evolutionary histories from multiple bulk DNA samples using Pairtree. BioRxiv 2020.11.06.372219 [Preprint]. 2021. Available from: 10.1101/2020.11.06.372219. [DOI] [PMC free article] [PubMed]
- 18. Alsadeq A, Fedders H, Vokuhl C, Belau NM, Zimmermann M, Wirbelauer Tet al. The role of ZAP70 kinase in acute lymphoblastic leukemia infiltration into the central nervous system. Haematologica 2017;102:346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Savino AM, Fernandes SI, Olivares O, Zemlyansky A, Cousins A, Markert EKet al. Metabolic adaptation of acute lymphoblastic leukemia to the central nervous system microenvironment depends on stearoyl-CoA desaturase. Nat Cancer 2020;1:998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science 2007;316:600–4. [DOI] [PubMed] [Google Scholar]
- 21. Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JSet al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell 2008;13:483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MAet al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ebinger S, Özdemir EZ, Ziegenhain C, Tiedt S, Alves CC, Grunert Met al. Characterization of rare, dormant, and therapy-resistant cells in acute lymphoblastic leukemia. Cancer Cell 2016;30:849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mullighan CG, Su X, Zhang J, Radtke I, Phillips LAA, Miller CBet al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009;360:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morris V, Marion W, Hughes T, Sousa P, Sensharma P, Pikman Yet al. Single-cell analysis reveals mechanisms of plasticity in leukemia initiating cells. BioRxiv2020.04.29.066332 [Preprint].2021. Available from: https://doi.org/10.1101/2020.04.29.066332.
- 26. Savaraj N, Feun LG, Lu K, Leavens M, Moser R, Fields WSet al. Central nervous system (CNS) penetration of homoharringtonine (HHT). J Neurooncol 1987;5:77–81. [DOI] [PubMed] [Google Scholar]
- 27. Tan CT, Luks E, Bacha DM, Steinherz P, Steinherz L, Mondora A. Phase I trial of homoharringtonine in children with refractory leukemia. Cancer Treat Rep 1987;71:1245–8. [PubMed] [Google Scholar]
- 28. Ebright RY, Lee S, Wittner BS, Niederhoffer KL, Nicholson BT, Bardia Aet al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 2020;367:1468–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nicolini FE, Lipton JH, Kantarjian H, Wetzler M, Akard LP, Baccarani Met al. Subcutaneous omacetaxine mepesuccinate in patients with chronic phase (CP) or accelerated phase (AP) chronic myeloid leukemia (CML) resistant/intolerant to two or three approved tyrosine-kinase inhibitors (TKIs). J Clin Oncol 30,2012(suppl; abstr 6513). [Google Scholar]
- 30. Lam SSY, Ho ESK, He BL, Wong WW, Cher CY, Ng NKLet al. Homoharringtonine (omacetaxine mepesuccinate) as an adjunct for FLT3-ITD acute myeloid leukemia. Sci Transl Med 2016;8:359ra129. [DOI] [PubMed] [Google Scholar]
- 31. Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden Jet al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 2015;34:2032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher Aet al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012;485:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wurth L, Papasaikas P, Olmeda D, Bley N, Calvo GT, Guerrero Set al. UNR/CSDE1 drives a post-transcriptional program to promote melanoma invasion and metastasis. Cancer Cell 2016;30:694–707. [DOI] [PubMed] [Google Scholar]
- 34. Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SKet al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 2018;173:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MNet al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 2017;67:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ricklin D, Reis ES, Mastellos DC, Gros P, Lambris JD. Complement component C3 - the “Swiss Army Knife” of innate immunity and host defense. Immunol Rev 2016;274:33–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boire A, Zou Y, Shieh J, Macalinao DG, Pentsova E, Massagué J. Complement component 3 adapts the cerebrospinal fluid for leptomeningeal metastasis. Cell 2017;168:1101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Porporato PE, Filigheddu N, Pedro JMBS, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res 2018;28:265–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer 2017;17:332. [DOI] [PubMed] [Google Scholar]
- 40. Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov 2015;14:261–78. [DOI] [PubMed] [Google Scholar]
- 41. Pathria G, Lee JS, Hasnis E, Tandoc K, Scott DA, Verma Set al. Translational reprogramming marks adaptation to asparagine restriction in cancer. Nat Cell Biol 2019;21:1590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rizzari C, Lanvers-Kaminsky C, Valsecchi MG, Ballerini A, Matteo C, Gerss Jet al. Asparagine levels in the cerebrospinal fluid of children with acute lymphoblastic leukemia treated with pegylated-asparaginase in the induction phase of the AIEOP-BFM ALL 2009 study. Haematologica 2019;104:1812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marini BL, Perissinotti AJ, Bixby DL, Brown J, Burke PW. Catalyzing improvements in ALL therapy with asparaginase. Blood Rev 2017;31:328–38. [DOI] [PubMed] [Google Scholar]
- 44. Lenard A, Xie HM, Pastuer T, Shank T, Libbrecht C, Kingsley Met al. Epigenetic regulation of protein translation in KMT2A-rearranged AML. Exp Hematol 2020;85:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caron M, St-Onge P, Sontag T, Wang YC, Richer C, Ragoussis Iet al. Single-cell analysis of childhood leukemia reveals a link between developmental states and ribosomal protein expression as a source of intra-individual heterogeneity. Sci Rep 2020;10:8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y, Hu Y, Michaels S, Segal D, Brown D, Li S. Inhibitory effects of omacetaxine on leukemic stem cells and BCR-ABL-induced chronic myeloid leukemia and acute lymphoblastic leukemia in mice. Leukemia 2019;23:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roumenina LT, Daugan MV, Petitprez F, Sautès-Fridman C, Fridman WH. Context-dependent roles of complement in cancer. Nat Rev Cancer 2019;19:698–715. [DOI] [PubMed] [Google Scholar]
- 48. Propson NE, Roy ER, Litvinchuk A, Köhl J, Zheng H. Endothelial C3a receptor mediates vascular inflammation and blood-brain barrier permeability during aging. J Clin Invest 2021;131:e140966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jacob A, Alexander JJ. Complement and blood–brain barrier integrity. Mol Immunol 2014;61:149–52. [DOI] [PubMed] [Google Scholar]
- 50. Smalley I, Law V, Wyatt C, Evernden B, Fang B, Koomen JMet al. Proteomic analysis of CSF from patients with leptomeningeal melanoma metastases identifies signatures associated with disease progression and therapeutic resistance. Clin Cancer Res 2020;26:1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cavalcante Mde S, de Souza Cavalcante M, Torres-Romero JC, Lobo MDP, Mendes FB, Bezerra LPet al. A panel of glycoproteins as candidate biomarkers for early diagnosis and treatment evaluation of B-cell acute lymphoblastic leukemia. Biomarker Res 2016;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rio DC, Ares M, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010;2010:pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- 53. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha Set al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2014;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage Det al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kucera M, Isserlin R, Arkhangorodsky A, Bader GD. AutoAnnotate: a Cytoscape app for summarizing networks with semantic annotations. F1000Res 2016;5:1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oesper L, Merico D, Isserlin R, Bader GD. WordCloud: a Cytoscape plugin to create a visual semantic summary of networks. Source Code Biol Med 2011;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lechman ER, Gentner B, Ng SWK, Schoof EM, van Galen P, Kennedy JAet al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell 2016;29:214–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- 61. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas Jet al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su Get al. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 2011;12:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky Aet al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wilkinson L. ggplot2: elegant graphics for data analysis by WICKHAM, H. Biometrics 2011;67:678–9. [Google Scholar]
- 65. Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw Aet al. gplots: various R programming tools for plotting data. R package version2009;2:1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical Characteristics of B-ALL Patient Samples.
B-ALL Xenografts Used in Current Study.
Differentially abundant proteins as quantified by label-free liquid chromatography mass spectrometry in KMT2A-r B-ALL xenografts.
Supplementary Figures S1-S6