Abstract
Several studies have monitored the SARS-CoV-2 variants in Brazil throughout the pandemic. Here, we systematically reviewed and conducted a scientometric analysis of the SARS-CoV-2 genomic surveillance studies using Brazilian samples. A Pubmed database search on October 2022 returned 492 articles, of which 106 were included. Ninety-six different strains were reported, with variant of concern (VOC) gamma (n = 35,398), VOC delta (n = 15,780), and the variant of interest zeta (n = 1983) being the most common. The top three states with the most samples in the published articles were São Paulo, Rio de Janeiro, and Minas Gerais. Whereas the first year of the pandemic presented primary circulation of B.1.1.28 and B.1.1.33 variants, consecutive replacements were observed between them and VOI zeta, VOC gamma, VOC delta, and VOC omicron. VOI mu, VOI lambda, VOC alpha, and VOC beta were also detected but failed to reach significant circulation. Co-infection, re-infection, and vaccine breakthrough reports were found. Article co-citation differed from the co-authorship structure. Despite the limitations, we expect to give an overview of Brazil’s genomic surveillance studies and contribute to future research execution.
Keywords: COVID-19, molecular epidemiology, next-generation sequencing
1. Introduction
The coronavirus disease (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, and was first reported in December 2019. The SARS-CoV-2 virus had been registered in 197 countries by March 2020, when the World Health Organization declared the novel coronavirus outbreak a global pandemic. More than 450 million cases and 6.0 million deaths have been reported worldwide [1]. Although the pandemic remains of concern, SARS-CoV-2 fatalities reduction has been observed mainly due to vaccination programs. Brazil has been heavily affected by COVID-19 [2,3]. Almost 35 million cases and 700,000 deaths have been documented since the first Brazilian case [4].
Viral genetics and evolution have been the main researched topics since the first viral genome was published [5]. Sequencing studies have promptly found viral diversity. Viral nomenclature was standardised in 2020 [6]. In 2021, Greek alphabet letters were introduced [7] to enable clear communication regarding lineages that presented alarming structural mutations: variants of interest (VOIs) and concern (VOCs) [8]. An unprecedented genomic surveillance effort was carried out [9], strengthened by the development, repositioning, and availability of sequencing services (e.g., databases, analysis tools, and other bioinformatics resources) [10,11,12]. A similar trend was observed among research groups previously conducting viral genomic surveillance studies in Brazil [13,14,15]. Here, we systematically reviewed and conducted scientometric analyses using the SARS-CoV-2 genomic surveillance studies characterising Brazilian samples.
2. Materials and Methods
The protocol for this systematic review was registered on PROSPERO [16] (accessible at www.crd.york.ac.uk/PROSPERO/display_record.asp?ID=CRD42021273259 (accessed on 1 December 2022)). Preferred Reporting Items for Systematic reviews and Meta-analysis (PRISMA) guideline was adopted [17]. Study selection was carried out in three phases: identification, screening, and eligibility. Identification was performed on the PubMed database using a structured search argument on 5 October 2022 (Supplementary Material S1). Two independent researchers conducted the screening of the articles. A third researcher solved disagreements. Inclusion criteria were primary articles that address the frequency of SARS-CoV-2 lineages by genotyping and sequencing data using samples from any location of the Brazilian territory. In contrast, exclusion criteria were reviews and primary articles with no SARS-CoV-2 lineage detection or studies with samples from elsewhere.
Two independent researchers conducted metadata extraction. We aimed to obtain the majority of available information since there is no standardisation on reporting genomic surveillance studies. We collected data regarding the: (1) publishing process (author affiliations and State of origin), (2) description of the samples—including either genotyping data or novel genomes used in the posterior analysis in the ‘sample’ category (size, initial and final collection dates, symptomatology, travel history, and nature -human or sewage samples), and (3) genomic surveillance execution (diagnostic gene targets, sequencing platform and metrics, variant calling, and phylogeny method used). Study designs were evaluated with the Joanna Briggs Institute (JBI) Critical Appraisal Tools for Systematic Reviews Checklist for Case Reports or Studies Reporting Prevalence [18]. Statistical descriptive analysis was carried out using software R (Vienna, Austria) (version 4.1.2). The scientometric evaluation was conducted using all included manuscripts in the systematic review. Bibliographic data was downloaded through Europe PMC API using the article digital object identifier (DOI) on 19 October 2022. The association strength was the normalisation method, and clustering was performed using default values on VOSviewer (Leiden, The Netherlands) (version 1.6.17) [19]. The connection between nodes in the maps represents co-authorship (for authors and authors’ affiliation) or citation (for articles). VOSviewer employs a distance-based visualisation of similarities mapping technique to construct a map [19].
3. Results
3.1. Description of the Included Studies
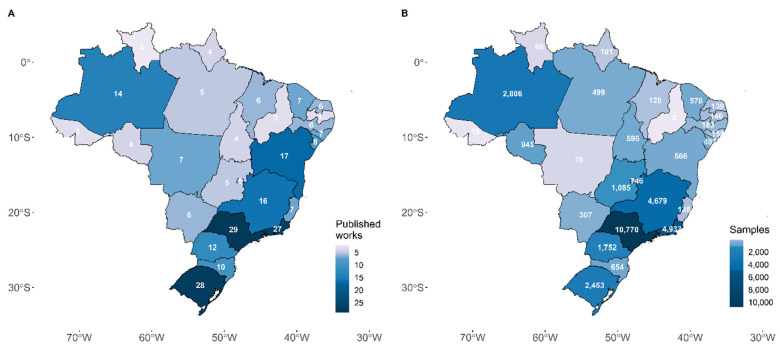
The search led to the screening of 492 articles (Figure S1). Our review included 106 studies, of which 73 were classified as prevalence studies [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91] and 33 were case reports [4,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123] (Table S1). The quality assessment indicated a systematic lack of participant demography and history description in case reports. At the same time, the most critical issue in the prevalence studies was the description of the participants (Figure S2). São Paulo (n = 29, 11.55%), Rio Grande do Sul (n = 28, 11.16%), and Rio de Janeiro (n = 27, 10.76%) were the sampled States with the most significant number of publications (Figure 1A), with Southeastern region being the most represented (n = 20,517, 58.77%). The top three states with the most samples included in published articles were São Paulo (n = 10,770; 30.85%), Rio de Janeiro (n = 4933; 14.13%), and Minas Gerais (n = 4679; 13.4%) (Figure 1B). Most studies included less than 500 samples (n = 92; 86.79%) (Figure S3) sampled from a single state (n = 91; 85.84%) (Figure 2).
Figure 1.
Spatial overview of the genomic surveillance in Brazil. (A) The number of published articles with samples from each Brazilian State. (B) The number of characterised samples from each Brazilian State in the published reports.
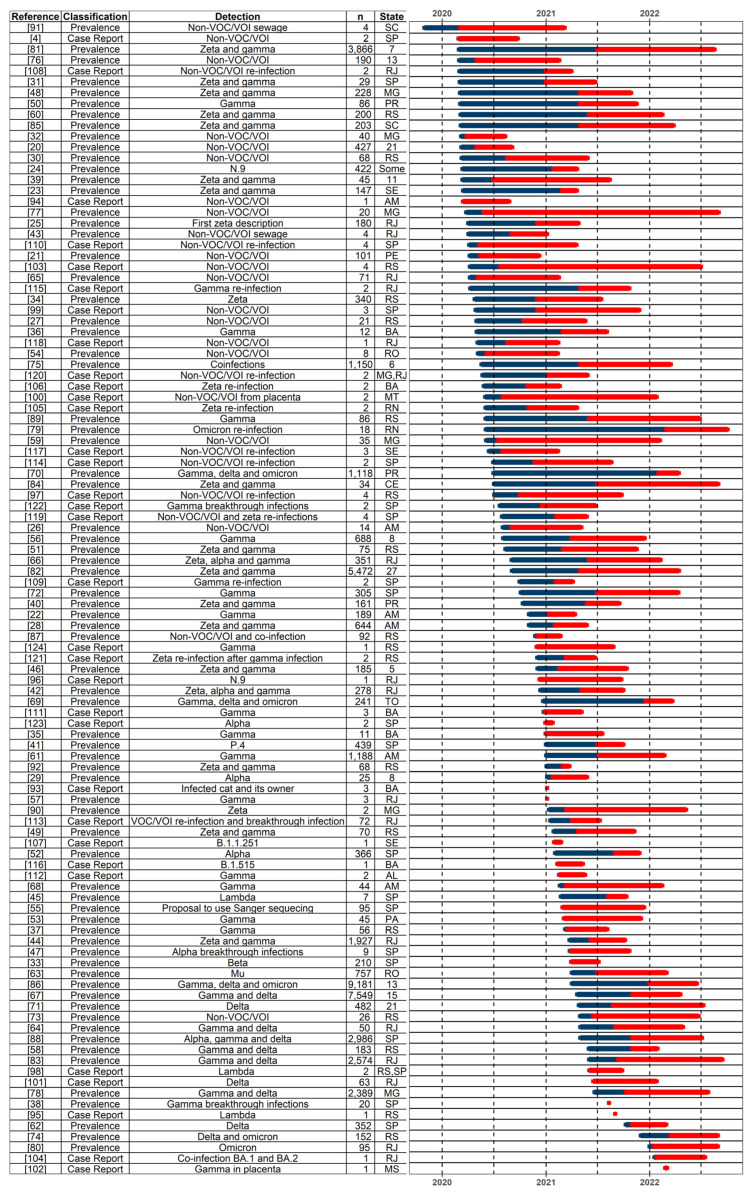
Figure 2.
Included studies in the systematic review. Studies are ordered by the beginning of the sampling periods when available. If not, the publication date was used. Non-VOC/VOI detection was omitted when the study reported VOI or VOC. The number of sampled States is shown when there are three or more States. Blue lines represent the sampling period. Red lines represent the time between the end of sampling and the publication date. Dashed lines indicate six months intervals from 1 January 2020. AL—Alagoas, AM—Amazonas, BA—Bahia, CE—Ceará, MG—Minas Gerais, MS—Mato Grosso do Sul, MT—Mato Grosso, PA—Pará, PE—Pernambuco, PR—Paraná, RJ—Rio de Janeiro, RN—Rio Grande do Norte, RO—Rondônia, RS—Rio Grande do Sul, SC—Santa Catarina, SE—Sergipe, SP—São Paulo, TO—Tocantins, VOC—variant of concern, and VOI—variant of interest.
In total, included studies contained 49,409 samples in the included studies (GISAID had 3.7 times more sequences uploaded), of which 571 (1.16%) were present in studies published in 2020, 7655 (15.49%) in 2021, and 41,183 (83.35%) in 2022 (Table S2). Ninety-six different strains were reported, with variant of concern (VOC) gamma (n = 35,398), VOC delta (n = 15,780), and variant of interest (VOI) zeta (n = 1983) being the most common thus far (Table S3). Most studies presented complete information on sample collection date ranges (n = 99, 94.34%), with sampling mostly starting in 2020 (n = 64; 60.38%) and 2021 (n = 33, 31.13%) (Figure 2). The median sampling period was 108.5 days, and the median time between the end of sampling and the publication of 200.5 days.
Several study features were compiled (Table S4). We have observed heterogeneity in methodological features and what authors have decided to report across studies. Clinical data were unavailable in 44 studies (41.51%), whereas outcomes of interest (e.g., death) were almost inexistent. Public laboratories (n = 91; 85.84%) and universities (n = 86, 81.13%) participated in most studies. Two studies were conducted in sewage samples. Pangolin [10] was the leading variant-calling software (n = 75, 70.75%), followed by IQTREE [124] (n = 59, 50.66%) and Nextclade [125] (n = 29, 27.35%). Most studies presented phylogenetic reconstructions (n = 84, 79.24%), with maximum-likelihood models being the most common (n = 79, 74.52%). Short-read platforms (e.g., Illumina MiSeq) account for more than half of the studies. The sequencing metrics were not reported in 56 studies (52.83%). The minimum coverage distribution was concentrated below 3000×, and the minimum genome span was between 75–97% (Figure S3).
3.2. Variant Detection in the Included Studies
The SARS-CoV-2 genomic surveillance in Brazil started in February 2020 in São Paulo, with the first reported patients returning from international destinations [4]. However, a study indicated SARS-CoV-2 mRNA sewage detection in Santa Catarina in November 2019 [90]. Communitarian transmission quickly overcame the importation rate [20,80]. Different SARS-CoV-2 strains descending from clade B.1, which present the structural mutation D614G in the Spike protein, became the most frequent variants detected in the prevalence studies during the first wave in 2020, especially B.1.1.28 and B.1.1.33 [20,21,22,23,24,25,26,27,30,31,32,34,36,39,43,48,50,51,54,56,59,60,65,66,70,72,75,76,78,80,81,83,84,88,90]. One sewage study also found B.1.1.33 circulation in Rio de Janeiro in mid-2020 [43]. The earliest reported re-infection cases were in São Paulo [109] and Rio de Janeiro [107], and co-infections were in Rio Grande do Sul [86], still in 2020.
Cases started to increase again in November 2020. Later that month, the World Health Organization designated VOIs and VOCs. VOI Zeta’s first detection was in Rio de Janeiro, with an inferred emergence date in July 2020 [25]. Zeta was found in several Brazilian States and became the most common variant until February 2021 [23,25,28,31,34,39,40,42,44,46,48,49,51,60,66,80,81,83,84,89,91], being associated with co-infections in Ceará, Rio Grande do Sul, and Bahia [75]. January 2021 was marked by a death surge that peaked in February 2021, coinciding with multiple detections of the recently described VOC gamma [22,28]. That VOC was the most common variant in the first 2021 semester throughout Brazil, associated with increased COVID-19 mortality [22,23,28,31,35,36,37,38,39,40,42,44,46,48,49,50,51,53,56,57,58,60,61,64,66,67,68,69,70,72,77,80,81,82,83,84,85,87,88,91]. In August 2021, VOC delta’s first case was confirmed in Brazil, later spreading all over Brazil [58,62,64,67,69,70,71,74,77,82,85,87]. The gamma replacement was associated with an unexpected substantial decrease in cases and deaths observed up to December 2021.
VOI mu [63], VOI lambda [45,94,97], VOC alpha [29,42,52,66,87], and VOC beta [33] were also detected in Brazil but failed to reach significant circulation. Besides variant detection and prevalence, two crucial issues were explored in the included studies during 2021: re-infections and vaccine breakthrough infections. Re-infections were described in several States as being frequently associated with VOI/VOC infection [96,104,105,108,112,113,114,116,118,119,120]. Vaccine breakthrough studies were less common, with reports found for zeta infections in Rio de Janeiro [112], gamma in Sao Paulo [38,121] and Rio de Janeiro [112], and alpha in São Paulo [47]. Later studies explored clinical outcomes in vaccinated subjects with delta infections [77].
VOC omicron’s first cases were detected at the end of November 2021, with significant circulation reported by December 2021. In 2022, the third and fourth case waves were observed, with daily reported cases reaching over 250,000 in January. Six studies characterised omicron circulation in several States [69,70,74,79,85]. Omicron BA.1/BA.2 co-infection in Rio de Janeiro [103] and BA.1 re-infection in Rio Grande do Norte [78] were reported. A study with samples from Paraná did not indicate changes in lethality during the omicron wave [70]. Further studies will likely be published to uncover further details of the omicron circulation and its sub-variants.
3.3. Scientometric Analysis
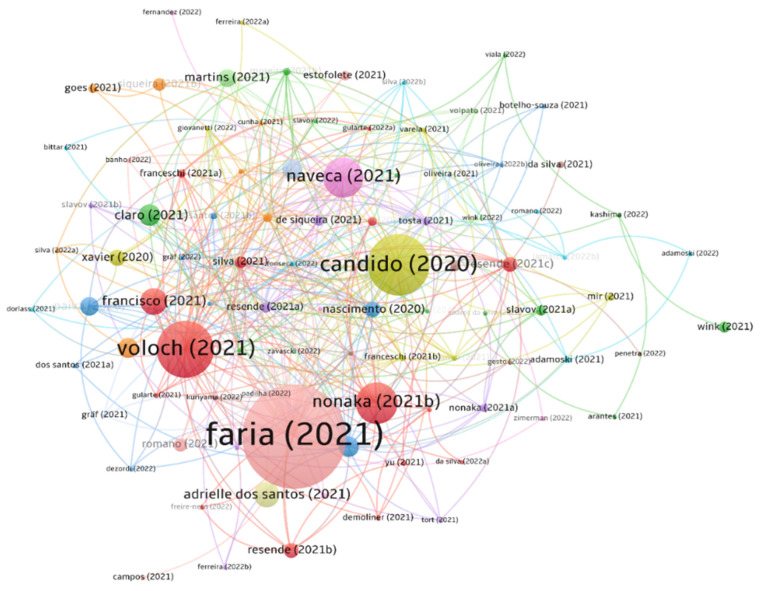
The first scientometric analysis evaluated cross-citations among the included articles (Figure 3). We identified 13 clusters with 338 cross-citations in the network comprised of 95 articles. Eleven manuscripts did not cite another included article. The publications with the highest number of cross-citations were [22] (n = 49), [20] (n = 38), [28] (n = 36), and [25] (n = 26), whereas the highest overall number of citations in the literature until October 2022 was observed in [22] (n = 433), [20] (n = 182), [26] (n = 161), and [105] (n = 95) (Table S5).
Figure 3.
Citation map for articles cross-citations. Connectors indicate article citations. The circle size is weighted by the citation number of each study in the literature up to 25 October 2022. Thirteen clusters were found, and are indicated by different colours. Metadata is available in Table S5 [20,21,22,23,24,25,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,44,45,46,47,48,49,50,51,52,53,54,56,57,58,59,60,61,63,64,65,66,67,68,69,70,72,72,73,74,75,76,77,78,79,80,81,83,84,85,86,87,88,89,90,91,93,94,95,96,97,98,99,100,101,102,104,105,106,107,108,109,110,111,112,114,116,118,119,120,121,122,123].
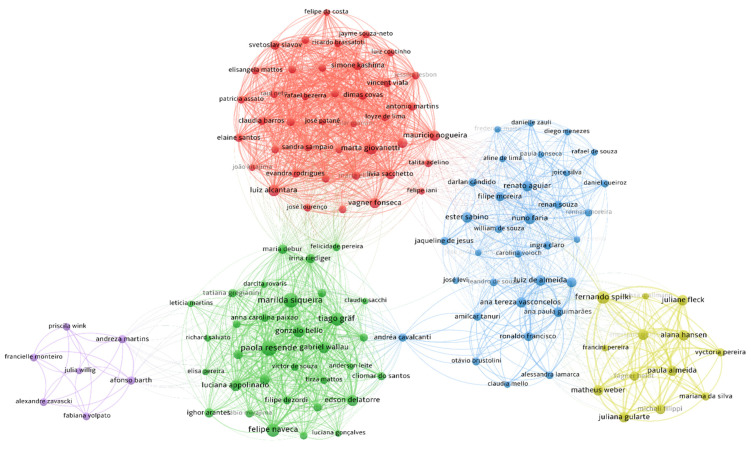
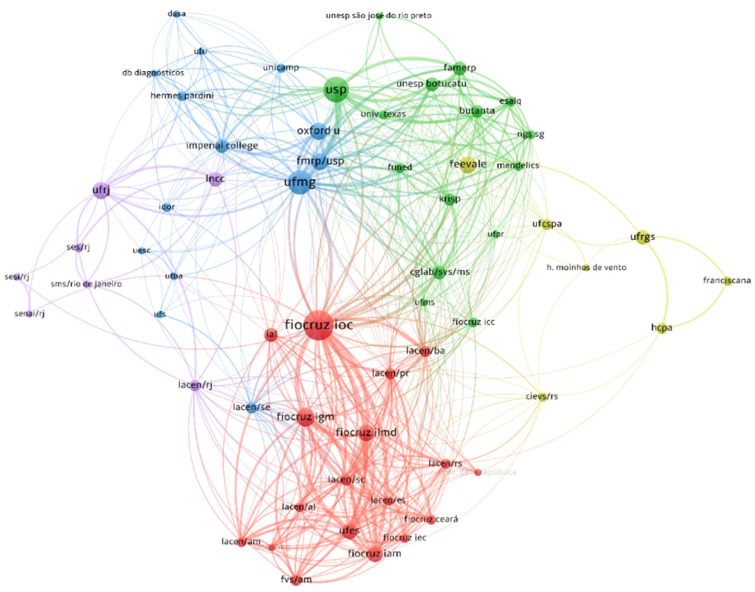
The second scientometric analysis showed authors who published together (Figure 4). The co-authorship network contains 127 authors in five clusters (Table S6). Five groups were also observed in the authors’ affiliation co-authorship network (Figure 5). We also created networks with keywords (Figure S4) and journals (Figure S5).
Figure 4.
Author co-authorship map. Authors with at least four articles were included (n = 127 out of 1190). Connectors indicate co-authorships. Circle sizes were weighted by the number of publications included in the systematic review. Five clusters were found, and are characterised by different colours. Metadata is available in Table S6.
Figure 5.
Organisation co-authorship map. Organisations with at least three articles in the systematic review were included (n = 63 out of 229). Connectors indicate co-authorship. The number of included articles from each organisation weighted circle sizes. Five clusters were found, and are indicated by different colours. Metadata is available in Table S7.
4. Discussion
Genomic surveillance in Brazil has been a prolific field. Here, we were able to review information from 106 publications. We know that the published literature does not fully cover all surveillance conducted, since the shared sequences in GISAID even outnumber the official governmental data in Brazil, a situation only replicated in the USA [126]. Despite the convenience of the pathogen databases, publishing articles on the field is fundamental to reporting the details of the execution process, which is the most reliable way to improve the design of future works.
We identified heterogeneity in sample representation across states. Overrepresentation in the southeast region reflects more extensive infrastructure, human resources, and previous experience executing viral genomic surveillance [13,14,15]. On the other hand, the protagonism of public institutions was homogenous, reinforcing their relevance in generating reliable data to subsidise accurate decision-making in public policy. In the future, stable funding will be required to expand the sequencing capacity and effectiveness in monitoring SARS-CoV-2 and other pathogens of medical interest.
Another interesting trend was that most studies sampled only one state. Broader studies are essential to describe epidemiological diversity under a uniform design and execution. The appropriate report in these works has the potential to reveal local challenges faced during the research process in each State from a comparative perspective. On the other hand, local studies were an essential part of the SARS-CoV-2 monitoring in the country, giving faster, deeper, and more precise responses to the COVID-19 pandemic in different regional scenarios.
Gathering study characteristics may help researchers design future studies. However, we found heterogeneity in the reported items, which made it difficult to conduct proper comparisons between studies. Genomic surveillance still lacks a specific extension of the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement [127]. The closest instrument would be the STROBE—Molecular Epidemiology [128], which does not include relevant information (e.g., sequencing parameters, bioinformatic analysis details).
The scientometric analysis indicated a different structure between co-authorships and article co-citations which could be interpreted as a good indicator of citations happening outside the research groups. However, we still observed articles that did not cite other Brazilian genomic surveillance studies converging in what Clarke and Chalmers once described as “islands in search of continents” [129]: a lack of connection between what is being newly described and what is known in the subject. Whenever possible, original studies could use systematic reviews to contextualise their findings [130].
Our work has limitations. The search argument, although aiming to return the most complete and, at the same time, precise search results, can be too stringent, and we may have missed relevant studies. The area is quickly evolving, and manuscripts have been published with previous variants [131,132] and XAG recombinants while we were updating this systematic review [133,134].
Although the genomic surveillance findings present biological significance, our work is significant from another perspective. Since it is hard to keep track of articles because so many have been published, authors can get a quick field overview, being able to plan new experiments, compare technical aspects, or discuss their results using the existing literature. Therefore, this review reduces initial information overload, accelerating and improving scientific research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14122715/s1. Figure S1: PRISMA flowchart. Figure S2: Quality assessment using the JBI Critical Appraisal Tools in Case Reports (A) and Prevalence Studies (B). Figure S3: Sequencing metrics of the studies included in this review. Figure S4: Keyword co-occurrence map. Figure S5: Journal citation map. Table S1: Dataset with the 106 selected studies. Table S2: Amount of samples in the 106 articles versus sequences included in the GISAID platform from 2020 to 2022. Table S3: Absolute frequencies of all the lineages reported in the 106 selected studies. Table S4: Descriptional panel of nine variables concerning the 106 studies included grouped by study type. Table S5: Studies included in Figure 3. Table S6: Authors included in Figure 4. Table S7: Organizations included in Figure 5. Table S8: Top 30 articles cited in the 106 articles included in the systematic review.
Author Contributions
Conceptualisation and supervision: R.P.d.S.; methodology: D.M., P.L.C.F., J.L.F.d.A. and R.P.d.S.; writing—original draft preparation, D.M. and R.P.d.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Rede Corona-ômica BR MCTI/FINEP affiliated with RedeVírus/MCTI (01.20.0029.000462/20 404096/2020-4; 1227/21 01.22.0074.00); Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (315592/2021-4); Financiadora de Estudos e Projetos—FINEP (0494/20 01.20.0026.00; 1228/21 01.22.0082.00; 1139/20 01.20.0076.00); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES (Finance Code 001); and Fundação de Amparo à Pesquisa do Estado de Minas Gerais—FAPEMIG (APQ-00475-20).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dong E., Du H., Gardner L. An Interactive Web-Based Dashboard to Track COVID-19 in Real Time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castro M.C., Gurzenda S., Turra C.M., Kim S., Andrasfay T., Goldman N. Reduction in Life Expectancy in Brazil after COVID-19. Nat. Med. 2021;27:1629–1635. doi: 10.1038/s41591-021-01437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro M.C., Kim S., Barberia L., Ribeiro A.F., Gurzenda S., Ribeiro K.B., Abbott E., Blossom J., Rache B., Singer B.H. Spatiotemporal Pattern of COVID-19 Spread in Brazil. Science. 2021;372:821–826. doi: 10.1126/science.abh1558. [DOI] [PubMed] [Google Scholar]
- 4.Araujo D.B., Machado R.R.G., Amgarten D.E., Malta F.D.M., de Araujo G.G., Monteiro C.O., Candido E.D., Soares C.P., de Menezes F.G., Pires A.C.C., et al. SARS-CoV-2 Isolation from the First Reported Patients in Brazil and Establishment of a Coordinated Task Network. Mem. Inst. Oswaldo Cruz. 2020;115:e200342. doi: 10.1590/0074-02760200342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konings F., Perkins M.D., Kuhn J.H., Pallen M.J., Alm E.J., Archer B.N., Barakat A., Bedford T., Bhiman J.N., Caly L., et al. SARS-CoV-2 Variants of Interest and Concern Naming Scheme Conducive for Global Discourse. Nat. Microbiol. 2021;6:821–823. doi: 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- 8.WHO Special Edition: Proposed Working Definitions of SARS-CoV-2 Variants of Interest and Variants of Concern. WHO; Geneva, Switzerland: 2021. [Google Scholar]
- 9.Li J., Lai S., Gao G.F., Shi W. The Emergence, Genomic Diversity and Global Spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- 10.O’Toole Á., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021;7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khare S., Gurry C., Freitas L., Schultz M.B., Bach G., Diallo A., Akite N., Ho J., Lee R.T., Yeo W., et al. GISAID’s Role Pandemic Response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faria N.R., Azevedo R.d.S.d.S., Kraemer M.U.G., Souza R., Cunha M.S., Hill S.C., Thézé J., Bonsall M.B., Bowden T.A., Rissanen I., et al. Zika Virus in the Americas: Early Epidemiological and Genetic Findings. Science. 2016;352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adelino T.É.R., Giovanetti M., Fonseca V., Xavier J., de Abreu Á.S., do Nascimento V.A., Demarchi L.H.F., Oliveira M.A.A., da Silva V.L., Cunha G.M., et al. Field and Classroom Initiatives for Portable Sequence-Based Monitoring of Dengue Virus in Brazil. Nat. Commun. 2021;12:2296. doi: 10.1038/s41467-021-22607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naveca F.G., Claro I., Giovanetti M., de Jesus J.G., Xavier J., Iani F.C.D.M., Do Nascimento V.A., de Souza V.C., Silveira P.P., Lourenço J., et al. Genomic, Epidemiological and Digital Surveillance of Chikungunya Virus in the Brazilian Amazon. PLoS Negl. Trop. Dis. 2019;13:e0007065. doi: 10.1371/journal.pntd.0007065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Booth A., Clarke M., Dooley G., Ghersi D., Moher D., Petticrew M., Stewart L. The nuts and bolts of PROSPERO: An international prospective register of systematic reviews. Syst. Rev. 2012;1:2. doi: 10.1186/2046-4053-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.JBI Critical Appraisal Tools. [(accessed on 1 December 2022)]. Available online: https://jbi.global/critical-appraisal-tools.
- 19.van Eck N.J., Waltman L. Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics. 2010;84:523–538. doi: 10.1007/s11192-009-0146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., Sales F.C.S., et al. Evolution and Epidemic Spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paiva M.H.S., Guedes D.R.D., Docena C., Bezerra M.F., Dezordi F.Z., Machado L.C., Krokovsky L., Helvecio E., da Silva A.F., Vasconcelos L.R.S., et al. Multiple Introductions Followed by Ongoing Community Spread of Sars-Cov-2 at One of the Largest Metropolitan Areas of Northeast Brazil. Viruses. 2020;12:1414. doi: 10.3390/v12121414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A., Sales F.C., Hawryluk I., McCrone J.T., et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dos Santos C.A., Bezerra G.V.B., de Azevedo Marinho A.R.R.A., Alves J.C., Tanajura D.M., Martins-Filho P.R. SARS-CoV-2 Genomic Surveillance in Northeast Brazil: Timing of Emergence of the Brazilian Variant of Concern P1. J. Travel Med. 2021;28:taab066. doi: 10.1093/jtm/taab066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Resende P.C., Gräf T., Paixão A.C.D., Appolinario L., Lopes R.S., Mendonça A.C.D.F., da Rocha A.S.B., Motta F.C., Neto L.G.L., Khouri R., et al. A Potential Sars-Cov-2 Variant of Interest (Voi) Harboring Mutation E484k in the Spike Protein Was Identified within Lineage b.1.1.33 Circulating in Brazil. Viruses. 2021;13:724. doi: 10.3390/v13050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voloch C.M., da Silva Francisco Jr R., de Almeida L.G., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P.D.C., Mariani D., da Costa R.M., Ferreira Jr O.C., et al. Genomic Characterization of a Novel SARS-CoV-2 Lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95:e00119-21. doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolete V.C., Rodrigues P.T., Johansen I.C., Corder R.M., Tonini J., Cardoso M.A., De Jesus J.G., Claro I.M., Faria N.R., Sabino E.C., et al. Interacting Epidemics in Amazonian Brazil: Prior Dengue Infection Associated With Increased Coronavirus Disease 2019 (COVID-19) Risk in a Population-Based Cohort Study. Clin. Infect. Dis. 2021;73:2045–2054. doi: 10.1093/cid/ciab410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franceschi V.B., Caldana G.D., de Menezes Mayer A., Cybis G.B., Neves C.A.M., Ferrareze P.A.G., Demoliner M., de Almeida P.R., Gularte J.S., Hansen A.W., et al. Genomic Epidemiology of SARS-CoV-2 in Esteio, Rio Grande Do Sul, Brazil. BMC Genom. 2021;22:371. doi: 10.1186/s12864-021-07708-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naveca F.G., Nascimento V., de Souza V.C., Corado A.D.L., Nascimento F., Silva G., Costa Á., Duarte D., Pessoa K., Mejía M., et al. COVID-19 in Amazonas, Brazil, Was Driven by the Persistence of Endemic Lineages and P.1 Emergence. Nat. Med. 2021;27:1230–1238. doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- 29.Moreira F.R.R., Bonfim D.M., Zauli D.A.G., Silva J.P., Lima A.B., Malta F.S.V., Ferreira A.C.S., Pardini V.C., Magalhães W.C.S., Queiroz D.C., et al. Epidemic Spread of Sars-Cov-2 Lineage b.1.1.7 in Brazil. Viruses. 2021;13:984. doi: 10.3390/v13060984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mir D., Rego N., Resende P.C., Tort F., Fernández-Calero T., Noya V., Brandes M., Possi T., Arleo M., Reyes N., et al. Recurrent Dissemination of SARS-CoV-2 Through the Uruguayan–Brazilian Border. Front. Microbiol. 2021;12:653986. doi: 10.3389/fmicb.2021.653986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slavov S.N., Giovanetti M., dos Santos Bezerra R., Fonseca V., Santos E.V., Rodrigues E.S., Adelino T., Xavier J., Borges J.S., Evaristo M., et al. Molecular Surveillance of the On-Going SARS-COV-2 Epidemic in Ribeirao Preto City, Brazil. Infect. Genet. Evol. 2021;93:104976. doi: 10.1016/j.meegid.2021.104976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xavier J., Giovanetti M., Adelino T., Fonseca V., Barbosa da Costa A.V., Ribeiro A.A., Felicio K.N., Duarte C.G., Ferreira Silva M.V., Salgado Á., et al. The Ongoing COVID-19 Epidemic in Minas Gerais, Brazil: Insights from Epidemiological Data and SARS-CoV-2 Whole Genome Sequencing. Emerg. Microbes Infect. 2020;9:1824–1834. doi: 10.1080/22221751.2020.1803146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slavov S.N., Patané J.S., Bezerra R.D.S., Giovanetti M., Fonseca V., Martins A.J., Viala V.L., Rodrigues E.S., Santos E.V., Barros C.R., et al. Genomic Monitoring Unveil the Early Detection of the SARS-CoV-2 B.1.351 (Beta) Variant (20H/501Y.V2) in Brazil. J. Med. Virol. 2021;93:6782–6787. doi: 10.1002/jmv.27190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sant’Anna F.H., Muterle Varela A.P., Prichula J., Comerlato J., Comerlato C.B., Roglio V.S., Mendes Pereira G.F., Moreno F., Seixas A., Wendland E.M. Emergence of the Novel SARS-CoV-2 Lineage VUI-NP13L and Massive Spread of P.2 in South Brazil. Emerg. Microbes Infect. 2021;10:1431–1440. doi: 10.1080/22221751.2021.1949948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tosta S., Giovanetti M., Brandão Nardy V., Reboredo de Oliveira da Silva L., Kelly Astete Gómez M., Gomes Lima J., Wanderley Cardoso C., Oliveira Silva T., São Pedro Leal de Souza M., Presta Dia P.H., et al. Short Report: Early Genomic Detection of Sars-Cov-2 p.1 Variant in Northeast Brazil. PLoS Negl. Trop. Dis. 2021;15:e0009591. doi: 10.1371/journal.pntd.0009591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nonaka C.K.V., Gräf T., Barcia C.A.L., Costa V.F., de Oliveira J.L., Passos R.D.H., Bastos I.N., de Santana M.C.B., Santos I.M., de Sousa K.A.F., et al. SARS-CoV-2 variant of concern P.1 (Gamma) infection in young and middle-aged patients admitted to the intensive care units of a single hospital in Salvador, Northeast Brazil, February 2021. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021;111:47–54. doi: 10.1016/j.ijid.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franceschi V.B., Caldana G.D., Perin C., Horn A., Peter C., Cybis G.B., Ferrareze P.A.G., Rotta L.N., Cadegiani F.A., Zimerman R.A., et al. Predominance of the Sars-Cov-2 Lineage p.1 and Its Sublineage p.1.2 in Patients from the Metropolitan Region of Porto Alegre, Southern Brazil in March 2021. Pathogens. 2021;10:988. doi: 10.3390/pathogens10080988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campos K.R., Sacchi C.T., Abbud A., Caterino-De-Araujo A. SARS-CoV-2 Variants in Severely Symptomatic and Deceased Persons Who Had Been Vaccinated against COVID-19 in São Paulo, Brazil. Rev. Panam. Salud Publica/Pan Am. J. Public Health. 2021;45:e126. doi: 10.26633/RPSP.2021.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Resende P.C., Naveca F.G., Lins R.D., Dezordi F.Z., Ferraz M.V.F., Moreira E.G., Coêlho D.F., Motta F.C., Paixão A.C.D., Appolinario L., et al. The Ongoing Evolution of Variants of Concern and Interest of SARS-CoV-2 in Brazil Revealed by Convergent Indels in the Amino (N)-Terminal Domain of the Spike Protein. Virus Evol. 2021;7:veab069. doi: 10.1093/ve/veab069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adamoski D., de Oliveira J.C., Bonatto A.C., Wassem R., Nogueira M.B., Raboni S.M., da Silva Trindade E., de Souza E.M., Gradia D.F. Large-Scale Screening of Asymptomatic Persons for SARS-CoV-2 Variants of Concern and Gamma Takeover, Brazil. Emerg. Infect. Dis. 2021;27:3124–3127. doi: 10.3201/eid2712.211326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bittar C., Possebon F.S., Ullmann L.S., Geraldini D.B., da Costa V.G., de Almeida L.G.P., Paulo P.R., Nascimento-Júnior N.M., Cilli E.M., Artico Banho C., et al. The Emergence of the New, P.4 Lineage of SARS-CoV-2 With Spike L452R Mutation in Brazil. Front. Public Health. 2021;9:745310. doi: 10.3389/fpubh.2021.745310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreira F.R.R., D’Arc M., Mariani D., Herlinger A.L., Schiffler F.B., Rossi Á.D., Leitão I.D.C., Miranda T.D.S., Cosentino M.A.C., Tôrres M.C.D.P., et al. Epidemiological Dynamics of SARS-CoV-2 VOC Gamma in Rio de Janeiro, Brazil. Virus Evol. 2021;7:veab087. doi: 10.1093/ve/veab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prado T., Machado T., Ferreira C., Cristina P., Couto F., Lucia A., Eppinghaus F., Hugo V., Marinho R., Braz S., et al. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191:116810. doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.da Silva Francisco Junior R., Lamarca A.P., de Almeida L.G., Cavalcante L., Machado D.T., Martins Y., Brustolini O., Gerber A.L., de CGuimarães A.P., Gonçalves R.B., et al. Turnover of SARS-CoV-2 Lineages Shaped the Pandemic and Enabled the Emergence of New Variants in the State of Rio de Janeiro, Brazil. Viruses. 2021;13:2013. doi: 10.3390/v13102013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kashima S., Slavov S.N., Giovanetti M., Rodrigues E.S., Patané J.S.L., Viala V.L., Santos E.V., Evaristo M., de Lima L.P.O., Martins A.J., et al. Introduction of SARS-CoV-2 C.37 (WHO VOI Lambda) in the Sao Paulo State, Southeast Brazil. J. Med. Virol. 2022;94:1206–1211. doi: 10.1002/jmv.27389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamarca A.P., de Almeida L.G., Francisco Jr R.D.S., Lima L.F.A., Scortecci K.C., Perez V.P., Brustolini O.J., Sousa E.S.S., Secco D.A., Santos A.M.G., et al. Genomic Surveillance of Sars-Cov-2 Tracks Early Interstate Transmission of p.1 Lineage and Diversification within p.2 Clade in Brazil. PLoS Negl. Trop. Dis. 2021;15:e0009835. doi: 10.1371/journal.pntd.0009835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Souza W.M.D., Muraro P., Souza G.F., Amorim M.R., Sesti-costa R., Mofatto L.S., Forato J., Barbosa P.P., Toledo-teixeira D.A., Bispo-dos-santos K., et al. Infection after Vaccination with Adenovirus-Vectored and Inactivated Vaccines. Viruses. 2021;13:2127. doi: 10.3390/v13112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorlass E.G., Lourenço K.L., Magalhães R.D.M., Sato H., Fiorini A., Peixoto R., Coelho H.P., Telezynski B.L., Scagion G.P., Ometto T., et al. Survey of SARS-CoV-2 genetic diversity in two major Brazilian cities using a fast and affordable Sanger sequencing strategy. Genomics. 2021;113:4109–4115. doi: 10.1016/j.ygeno.2021.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demoliner M., da Silva M.S., Gularte J.S., Hansen A.W., de Almeida P.R., Weber M.N., Heldt F.H., Silveira F., Filippi M., de Abreu Góes Pereira V.M., et al. Predominance of SARS-CoV-2 P.1 (Gamma) lineage inducing the recent COVID-19 wave in southern Brazil and the finding of an additional S: D614A mutation. Infect. Genet. Evol. 2021;96:105134. doi: 10.1016/j.meegid.2021.105134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliveira M.M., Schemberger M.O., Suzukawa A.A., Riediger I.N., do Carmo Debur M., Becker G., Resende P.C., Gräf T., Balsanelli E., de Baura V.A., et al. Re-Emergence of Gamma-like-II and Emergence of Gamma-S:E661D SARS-CoV-2 Lineages in the South of Brazil after the 2021 Outbreak. Virol. J. 2021;18:222. doi: 10.1186/s12985-021-01690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varela A.P.M., Prichula J., Mayer F.Q., Salvato R.S., Sant’Anna F.H., Gregianini T.S., Martins L.G., Seixas A., Veiga A.B.G.D. SARS-CoV-2 introduction and lineage dynamics across three epidemic peaks in Southern Brazil: Massive spread of P.1. Infect. Genet. Evol. 2021;96:105144. doi: 10.1016/j.meegid.2021.105144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slavov S.N., Bezerra R.D.S., Rodrigues E.S., Santos E.V., Borges J.S., de la Roque D.G.L., Patané J.S.L., Lima A.R.J., Ribeiro G., Viala V.L., et al. Genomic monitoring of the SARS-CoV-2 B1.1.7 (WHO VOC Alpha) in the Sao Paulo state, Brazil. Virus Res. 2022;308:198643. doi: 10.1016/j.virusres.2021.198643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Da Silva J.F., Esteves R.J., Siza C., Soares E.P., Ramos T.C., Campelo E.C., da Costa C.F., de Alencar L.C., Cavalcante R.P., Florêncio C.R., et al. Cluster of SARS-CoV-2 Gamma Variant Infections, Parintins, Brazil, March 2021. Emerg. Infect. Dis. 2022;28:262–264. doi: 10.3201/eid2801.211817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Botelho-Souza L.F., Nogueira-Lima F.S., Roca T.P., Naveca F.G., de Oliveria dos Santos A., Maia A.C.S., da Silva C.C., de Melo Mendonça A.L.F., Lugtenburg C.A.B., Azzi C.F.G., et al. SARS-CoV-2 Genomic Surveillance in Rondônia, Brazilian Western Amazon. Sci. Rep. 2021;11:3770. doi: 10.1038/s41598-021-83203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cabral G.B., Ahagon C.M., de Souza Guimarães P.M., Silva Lopez-Lopes G.I., Hussein I.M., Cilli A., de Jesus Alves I., Coelho Bombonatte A.G., do Carmo Sampaio Tavares Timenetsky M., da Silva Santos J.H., et al. Use of Sanger protocols to identify variants of concern, key mutations and track evolution of SARS-CoV-2. J. Virol. Methods. 2022;300:114422. doi: 10.1016/j.jviromet.2021.114422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gräf T., Bello G., Venas T.M.M., Pereira E.C., Paixão A.C.D., Appolinario L.R., Lopes R.S., Mendonça A.C.D.F., Da Rocha A.S.B., Motta F.C., et al. Identification of a Novel SARS-CoV-2 P.1 Sub-Lineage in Brazil Provides New Insights about the Mechanisms of Emergence of Variants of Concern. Virus Evol. 2021;7:veab091. doi: 10.1093/ve/veab091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salles T.S., Cavalcanti A.C., da Costa F.B., Dias V.Z., de Souza L.M., de Meneses M.D.F., da Silva J.A.S., Amaral C.D., Felix J.R., Pereira D.A., et al. Genomic Surveillance of SARS-CoV-2 Spike Gene by Sanger Sequencing. PLoS ONE. 2022;17:e0262170. doi: 10.1371/journal.pone.0262170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gularte J.S., da Silva M.S., Mosena A.C.S., Demoliner M., Hansen A.W., Filippi M., Pereira V.M.A.G., Heldt F.H., Weber M.N., de Almeida P.R., et al. Early introduction, dispersal and evolution of Delta SARS-CoV-2 in Southern Brazil, late predominance of AY.99.2 and AY.101 related lineages. Virus Res. 2022;311:198702. doi: 10.1016/j.virusres.2022.198702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silva A.V.F.G., Menezes D., Moreira F.R.R., Torres O.A., Fonseca P.L.C., Moreira R.G., Alves H.J., Alves V.R., de Resende Amaral T.M., Coelho A.N., et al. Seroprevalence, Prevalence, and Genomic Surveillance: Monitoring the Initial Phases of the SARS-CoV-2 Pandemic in Betim, Brazil. Front. Microbiol. 2022;13:799713. doi: 10.3389/fmicb.2022.799713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wink P.L., Ramalho R., Monteiro F.L., Volpato F.C.Z., Willig J.B., Lovison O.V.A., Zavascki A.P., Barth A.L., Martins A.F. Genomic Surveillance of SARS-CoV-2 Lineages Indicates Early Circulation of P.1 (Gamma) Variant of Concern in Southern Brazil. Microbiol. Spectr. 2022;10:e0151121. doi: 10.1128/spectrum.01511-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naveca F.G., Nascimento V., Souza V., Corado A.D.L., Nascimento F., Silva G., Mejía M.C., Brandão M.J., Costa Á., Duarte D., et al. Spread of Gamma (P.1) Sub-Lineages Carrying Spike Mutations Close to the Furin Cleavage Site and Deletions in the N-Terminal Domain Drives Ongoing Transmission of SARS-CoV-2 in Amazonas, Brazil. Microbiol. Spectr. 2022;10:e02366-21. doi: 10.1128/spectrum.02366-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lima A.R.J., Ribeiro G., Viala V.L., de Lima L.P.O., Martins A.J., Barros C.R.D.S., Marqueze E.C., Bernardino J.D.S.T., Moretti D.B., Rodrigues E.S., et al. SARS-COV-2 Genomic Monitoring in the State of São Paulo Unveils Two Emerging AY.43 Sublineages. J. Med. Virol. 2022;94:3394–3398. doi: 10.1002/jmv.27674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oliveira G.S., Silva-Flannery L., Silva J.F., Siza C., Esteves R.J., Marston B.J., Morgan J., Plucinski M., Roca T.P., Silva A.M.P., et al. Active Surveillance and Early Detection of Community Transmission of SARS-CoV-2 Mu Variant (B.1.621) in the Brazilian Amazon. J. Med. Virol. 2022;94:3410–3415. doi: 10.1002/jmv.27686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuriyama S.N., Farjun B., Henriques-Santos B.M., Cabanelas A., Abrantes J.L., Gesto J., Fidalgo-Neto A.A., Souza T.M.L. SARS-CoV-2 Molecular Epidemiology Can Be Enhanced by Occupational Health: The Experience of Monitoring Variants of Concern in Workplaces in Rio de Janeiro, Brazil. Front. Med. 2022;9:862284. doi: 10.3389/fmed.2022.862284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siqueira J.D., Goes L.R., Alves B.M., Carvalho P.S.D., Cicala C., Arthos J., Viola J.P.B., De Melo A.C., Soares M.A. SARS-CoV-2 Genomic Analyses in Cancer Patients Reveal Elevated Intrahost Genetic Diversity. Virus Evol. 2021;7:veab013. doi: 10.1093/ve/veab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henriques-Santos B.M., Farjun B., Corrêa I.A., de Barros Figueiredo J., Fidalgo-Neto A.A., Kuriyama S.N. SARS-CoV-2 Variant Determination Through SNP Assays in Samples From Industry Workers From Rio de Janeiro, Brazil. Front. Microbiol. 2022;12:757783. doi: 10.3389/fmicb.2021.757783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silva J.P., de Lima A.B., Alvim L.B., Malta F.S., Mendonça C.P., Fonseca P.L., Moreira F.R., Queiroz D.C., Ferreira J.G., Ferreira A.C., et al. Delta Variant of SARS-CoV-2 Replacement in Brazil: A National Epidemiologic Surveillance Program. Viruses. 2022;14:847. doi: 10.3390/v14050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zimerman R.A., Ferrareze P.A.G., Cadegiani F.A., Wambier C.G., do Nascimento Fonseca D., De Souza A.R., Goren A., Rotta L.N., Ren Z., Thompson C.E. Comparative Genomics and Characterization of SARS-CoV-2 P.1 (Gamma) Variant of Concern From Amazonas, Brazil. Front. Med. 2022;9:806611. doi: 10.3389/fmed.2022.806611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Souza U.J.B., Dos Santos R.N., de Melo F.L., Belmok A., Galvão J.D., de Rezende T.C.V., Cardoso F.D.P., Carvalho R.F., da Silva Oliveira M., Ribeiro Junior J.C., et al. Genomic Epidemiology of SARS-CoV-2 in Tocantins State and the Diffusion of P.1.7 and AY.99.2 Lineages in Brazil. Viruses. 2022;14:659. doi: 10.3390/v14040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adamoski D., Baura V.A.D., Rodrigues A.C., Royer C.A., Aoki M.N., Tschá M.K., Bonatto A.C., Wassem R., Nogueira M.B., Raboni S.M., et al. SARS-CoV-2 Delta and Omicron Variants Surge in Curitiba, Southern Brazil, and Its Impact on Overall COVID-19 Lethality. Viruses. 2022;14:809. doi: 10.3390/v14040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arantes I., Gomes Naveca F., Gräf T., COVID-19 Fiocruz Genomic Surveillance Network. Miyajima F., Faoro H., Luz Wallau G., Delatorre E., Reis Appolinario L., Cavalcante Pereira E., et al. Emergence and Spread of the SARS-CoV-2 Variant of Concern Delta across Different Brazilian Regions. Microbiol. Spectr. 2022;10:e0264121. doi: 10.1128/spectrum.02641-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Banho C.A., Sacchetto L., Campos G.R.F., Bittar C., Possebon F.S., Ullmann L.S., Marques B.D.C., da Silva G.C.D., Moraes M.M., Parra M.C.P., et al. Impact of SARS-CoV-2 Gamma Lineage Introduction and COVID-19 Vaccination on the Epidemiological Landscape of a Brazilian City. Commun. Med. 2022;2:41. doi: 10.1038/s43856-022-00108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.da Silva M.S., Gularte J.S., Demoliner M., Hansen A.W., Heldt F.H., Filippi M., Luckmann C.B., Pereira V.M.D.A.G., de Almeida Vaucher R., dos Santos Barboza V., et al. Brief Dispersion of a Putative, B.1.1.28-Derived SARS-CoV-2 Lineage Harboring Additional N234P and E471Q Spike Protein Mutations in Individuals Crossing the Argentina-Brazil Border. Travel Med. Infect. Dis. 2022;49:102390. doi: 10.1016/j.tmaid.2022.102390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.da Silva M.S., Gularte J.S., Filippi M., Demoliner M., Girardi V., Mosena A.C.S., Pereira V.M.D.A.G., Hansen A.W., Weber M.N., de Almeida P.R., et al. Genomic and Epidemiologic Surveillance of SARS-CoV-2 in Southern Brazil and Identification of a New Omicron-L452R Sublineage. Virus Res. 2022;321:198907. doi: 10.1016/j.virusres.2022.198907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dezordi F.Z., Resende P.C., Naveca F.G., Do Nascimento V.A., de Souza V.C., Paixão A.C.D., Appolinario L., Lopes R.S., da Fonseca Mendonça A.C., da Rocha A.S.B., et al. Unusual SARS-CoV-2 Intrahost Diversity Reveals Lineage Superinfection. Microb. Genomics. 2022;8:000751. doi: 10.1099/mgen.0.000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Resende P.C., Delatorre E., Gräf T., Mir D., Motta F.C., Appolinario L.R., Paixão A.C.D.D., Mendonça A.C.D.F., Ogrzewalska M., Caetano B., et al. Evolutionary Dynamics and Dissemination Pattern of the SARS-CoV-2 Lineage B.1.1.33 During the Early Pandemic Phase in Brazil. Front. Microbiol. 2021;11:615280. doi: 10.3389/fmicb.2020.615280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fonseca P.L.C., Moreira F.R.R., De Souza R.M., Guimarães N.R., Carvalho N.O., Adelino T.E.R., Alves H.J., Alvim L.B., Candido D.S., Coelho H.P., et al. Tracking the Turnover of SARS-CoV-2 VOCs Gamma to Delta in a Brazilian State (Minas Gerais) with a High-Vaccination Status. Virus Evol. 2022;8:veac064. doi: 10.1093/ve/veac064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freire-neto F.P., Id D.G.T., Cunha D.C.S., Morais C., Tavares C.P.M., Gurgel G.P., Medeiros S.D.N., Santos C., Sales A.D.O., Id S.M.B.J. SARS-CoV-2 Reinfections with BA. 1 (Omicron) Variant among Fully Vaccinated Individuals in Northeastern Brazil. PLoS Negl. Trop. Dis. 2022;16:e0010337. doi: 10.1371/journal.pntd.0010337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gesto J.S.M., Cabanelas A., Farjun B., dos Santos M.C., Fidalgo-Neto A.A., Kuriyama S.N., Souza T.M.L. Implemented Occupational Health Surveillance Limits the Spread of SARS-CoV-2 Omicron at the Workplace. Front. Med. 2022;9:910176. doi: 10.3389/fmed.2022.910176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giovanetti M., Slavov S.N., Fonseca V., Wilkinson E., Tegally H., Patané J.S.L., Viala V.L., San E.J., Rodrigues E.S., Santos E.V., et al. Genomic Epidemiology of the SARS-CoV-2 Epidemic in Brazil. Nat. Microbiol. 2022;7:1490–1500. doi: 10.1038/s41564-022-01191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gräf T., Bello G., Naveca F.G., Gomes M., Cardoso V.L.O., da Silva A.F., Dezordi F.Z., dos Santos M.C., de Oliveira Santos K.C., Batista É.L.R., et al. Phylogenetic-Based Inference Reveals Distinct Transmission Dynamics of SARS-CoV-2 Lineages Gamma and P.2 in Brazil. iScience. 2022;25:104156. doi: 10.1016/j.isci.2022.104156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamarca1 A.P., De Almeida1 G.P., Da Silva R., Junior1 F., Cavalcante1 L., Brustolini1 O., Gerber1 A.L., Paula De C Guimarães1 A., Henrique De Oliveira2 T., Ramos É., et al. Phylodynamic Analysis of SARS-CoV-2 Spread in Rio de Janeiro, Brazil, Highlights How Metropolitan Areas Act as Dispersal Hubs for New Variants. Microb. Genom. 2022;8:mgen000859. doi: 10.1099/mgen.0.000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.da Silva Oliveira F.A., de Holanda M.V., Lima L.B., Dantas M.B., Duarte I.O., de Castro L.G.Z., de Oliveira L.L.B., Paier C.R.K., Moreira-Nunes C.D.F.A., Lima N.C.B., et al. Genomic Surveillance: Circulating Lineages and Genomic Variation of SARS-CoV-2 in Early Pandemic in Ceará State, Northeast Brazil. Virus Res. 2022;321:198908. doi: 10.1016/j.virusres.2022.198908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Padilha D.A., Filho V.B., Moreira R.S., Soratto T.A.T., Maia G.A., Christoff A.P., Barazzetti F.H., Schörner M.A., Ferrari F.L., Martins C.L., et al. Emergence of Two Distinct SARS-CoV-2 Gamma Variants and the Rapid Spread of P.1-like-II SARS-CoV-2 during the Second Wave of COVID-19 in Santa Catarina, Southern Brazil. Viruses. 2022;14:695. doi: 10.3390/v14040695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Romano C.M., de Oliveira C.M., da Silva L.S., Levi J.E. Early Emergence and Dispersal of Delta SARS-CoV-2 Lineage AY.99.2 in Brazil. Front. Med. 2022;9:930380. doi: 10.3389/fmed.2022.930380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Francisco R.D.S., Benites L.F., Jr., Lamarca A.P., de Almeida L.G.P., Hansen A.W., Gularte J.S., Demoliner M., Gerber A.L., de C Guimarães A.P., Antunes A.K.E., et al. Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 2021;296:198345. doi: 10.1016/j.virusres.2021.198345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Viala V.L., Slavov S.N., de Lima L.P.O., Lima A.R.J., Ribeiro G., Martins A.J., Petry B., Banho C.A., Dos Santos Barros C.R., Moncau C.T., et al. The Divergent Pattern of SARS-CoV-2 Variant Predominance and Transmission Dynamics in the Brazilian Island of Ilhabela. Viruses. 2022;14:1481. doi: 10.3390/v14071481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zavascki A.P., Vieceli T., Wink P.L., Volpato F.C.Z., Monteiro F.L., Willig J.B., Ferreira C.F., Arns B., Magalhães Costa G.O., de Souza Niches M., et al. Evaluation of Clinical Course of Gamma (P.1) Variant of Concern versus Lineages in Hospitalised Patients with COVID-19 in a Reference Center in Brazil. Am. J. Trop. Med. Hyg. 2022;107:245–251. doi: 10.4269/ajtmh.21-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miguita L., Martins-Chaves R.R., Geddes V.E.V., da Rocha Mendes S., dos Santos Costa S.F., Fonseca P.L.C., Menezes D., de Souza R.M., Queiroz D.C., Alves H.J., et al. Biosafety in Dental Health Care During the COVID-19 Pandemic: A Longitudinal Study. Front. Oral Health. 2022;3:871107. doi: 10.3389/froh.2022.871107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fongaro G., Stoco P.H., Souza D.S.M., Grisard E.C., Magri M.E., Rogovski P., Schörner M.A., Barazzetti F.H., Christoff A.P., de Oliveira L.F.V., et al. The presence of SARS-CoV-2 RNA in human sewage in Santa Catarina, Brazil, November 2019. Sci. Total Environ. 2021;778:146198. doi: 10.1016/j.scitotenv.2021.146198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martins A.F., Zavascki A.P., Wink P.L., Volpato F.C.Z., Monteiro F.L., Rosset C., De-Paris F., Ramos Á.K., Barth A.L. Detection of SARS-CoV-2 Lineage P.1 in Patients from a Region with Exponentially Increasing Hospitalisation Rate, February 2021, Rio Grande Do Sul, Southern Brazil. Eurosurveillance. 2021 February 20;26:2100276. doi: 10.2807/1560-7917.ES.2021.26.12.2100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carlos R.S.A., Mariano A.P.M., Maciel B.M., Gadelha S.R., de Melo Silva M., Belitardo E.M.M.A., Rocha D.J.P.G., de Almeida J.P.P., Pacheco L.G.C., Aguiar E.R.G.R., et al. First Genome Sequencing of SARS-CoV-2 Recovered from an Infected Cat and Its Owner in Latin America. Transbound. Emerg. Dis. 2021;68:3070–3074. doi: 10.1111/tbed.13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nascimento V.A.D., Corado A.D.L.G., Nascimento F.O.D., Costa Á.K.A.D., Duarte D.C.G., Luz S.L.B., Gonçalves L.M.F., Jesus M.S.D., Costa C.F.D., Delatorre E., et al. Genomic and Phylogenetic Characterisation of an Imported Case of Sars-Cov-2 in Amazonas State, Brazil. Mem. Inst. Oswaldo Cruz. 2020;115:e200310. doi: 10.1590/0074-02760200310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wink P.L., Volpato F.C.Z., Monteiro F.L., Willig J.B., Zavascki A.P., Barth A.L., Martins A.F. First Identification of SARS-CoV-2 Lambda (C.37) Variant in Southern Brazil. Infect. Control Hosp. Epidemiol. 2021;2021:1–2. doi: 10.1017/ice.2021.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tort L.F.L., Ribeiro I.P., Menezes L.S.R., Dos Santos A.A.C., Santos M.P., Damasceno L., Silva P.C.R., de Siqueira M.A.M.T., Brasil P., Bonaldo M.C. SARS-CoV-2 Variant N.9 Identified in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. 2021;116:e210166. doi: 10.1590/0074-02760210166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gularte J.S., da Silva M.S., Demoliner M., Hansen A.W., Heldt F.H., Silveira F., Filippi M., Pereira V.M.D.A.G., da Silva F.P., Mallmann L., et al. Reinfection Cases by Closely Related SARS-CoV-2 Lineages in Southern Brazil. Braz. J. Microbiol. 2021;52:1881–1885. doi: 10.1007/s42770-021-00621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arantes I.G., Salvato R.S., Gregianini T.S., Martins L.G., Barth A.L., Martins A.F., Paixão A.C.D., Appolinario L., Lopes R.S., da Fonseca Mendonça A.C., et al. Multiple Introductions of SARS-CoV-2 C.37 Lambda Lineage in the Southern Brazilian Region. J. Travel Med. 2021;28:taab153. doi: 10.1093/jtm/taab153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cunha M.D.P., Vilela A.P.P., Molina C.V., Acuna S.M., Muxel S.M., Barroso V.D.M., Baroni S., Gomes de Oliveira L., Angelo Y.D.S., Peron J.P.S., et al. Atypical Prolonged Viral Shedding With Intra-Host SARS-CoV-2 Evolution in a Mildly Affected Symptomatic Patient. Front. Med. 2021;8:760170. doi: 10.3389/fmed.2021.760170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ferreira M.D.F.C., Pavon J.A.R., Napoleão A.C.B., Figueiredo G.M.D.P., Florêncio P.C.B., dos Santos Arantes R.B., Rizzo P.S., Carmo M.A.M.V., Nakazato L., Dutra V., et al. Clinical and Genomic Data of Sars-Cov-2 Detected in Maternal–Fetal Interface during the First Wave of Infection in Brazil. Microbes Infect. 2022;24:104949. doi: 10.1016/j.micinf.2022.104949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lamarca A.P., de Almeida L.G.P., da Silva Francisco R., Cavalcante L., Jr., Machado D.T., Brustolini O., Gerber A.L., Guimarães A.P.C., Policarpo C., Oliveira G.D.S., et al. Genomic Surveillance Tracks the First Community Outbreak of the SARS-CoV-2 Delta (B.1.617.2) Variant in Brazil. J. Virol. 2022;96:e0122821. doi: 10.1128/JVI.01228-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fernandez Z., Lichs G.G.C., Zubieta C.S., Machado A.B., Ferreira M.A., Valente N., Keren T., Arantes I., Nacife V., Pereira E.C., et al. Case Report: SARS-CoV-2 Gamma Isolation From Placenta of a Miscarriage in Midwest, Brazil. Front. Med. 2022;9:839389. doi: 10.3389/fmed.2022.839389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gularte J.S., da Silva M.S., Filippi M., Demoliner M., Schallenberger K., Hansen A.W., de Abreu Góes Pereira V.M., Heldt F.H., Girardi V., Weber M.N., et al. Viral Isolation Allows Characterisation of Early Samples of SARS-CoV-2 Lineage B1.1.33 with Unique Mutations (S: H655Y and T63N) Circulating in Southern Brazil in 2020. Braz. J. Microbiol. 2022;53:1313–1319. doi: 10.1007/s42770-022-00789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Oliveira C.M., Romano C.M., Sussuchi L., Cota B.D.C.V., Levi J.E. SARS-CoV-2 BA.1 and BA.2 Co-infection Detected by Genomic Surveillance in Brazil, January 2022. Arch. Virol. 2022;167:2271–2273. doi: 10.1007/s00705-022-05532-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Resende P.C., Bezerra J.F., Vasconcelos R.H.T., Arantes I., Appolinario L., Mendoncą A.C., Paixao A.C., Duarte A.C., Silva T., Rocha A.S., et al. Severe Acute Respiratory Syndrome Coronavirus 2 P.2 Lineage Associated with Re-infection Case, Brazil, June-October 2020. Emerg. Infect. Dis. 2021;27:1789–1794. doi: 10.3201/eid2707.210401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nonaka C.K.V., Franco M.M., Gräf T., De Lorenzo Barcia C.A., De Ávila Mendonça R.N., De Sousa K.A.F., Costa Neiva L.M., Fosenca V., Mendes A.V.A., De Aguiar R.S., et al. Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil. Emerg. Infect. Dis. 2021;27:1522–1524. doi: 10.3201/eid2705.210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dos Santos C.A., Bezerra G.V.B., De Azevedo Marinho A.R.R.A., Sena L.O.C., Alves J.C., De Souza M.S.F., De Oliveira Góes M.A., Teixeira D.C.P., Silva P.C.R., De Siqueira M.A.M.T., et al. First Report of SARS-CoV-2 B.1.1.251 Lineage in Brazil. J. Travel Med. 2021;28:taab033. doi: 10.1093/jtm/taab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fintelman-Rodrigues N., Da Silva A.P.D., Dos Santos M.C., Saraiva F.B., Ferreira M.A., Gesto J., Rodrigues D.A.S., Vale A.M., De Azevedo I.G., Soares V.C., et al. Genetic Evidence and Host Immune Response in Persons Reinfected with SARS-CoV-2, Brazil. Emerg. Infect. Dis. 2021;27:1446–1453. doi: 10.3201/eid2705.204912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Romano C.M., Felix A.C., de Paula A.V., de Jesus J.G., Andrade P.S., Cândido D., de Oliveira F.M., Ribeiro A.C., da Silva F.C., Inemami M., et al. Sars-Cov-2 Reinfection Caused by the p.1 Lineage in Araraquara City, Sao Paulo State, Brazil. Rev. Inst. Med. Trop. 2021;63:e36. doi: 10.1590/s1678-9946202163036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Amorim M.R., Souza W.M., Barros A.C.G., Toledo-Teixeira D.A., Bispo-Dos-Santos K., Simeoni C.L., Parise P.L., Vieira A., Forato J., Claro I.M., et al. Respiratory Viral Shedding in Healthcare Workers Reinfected with SARS-CoV-2, Brazil, 2020. Emerg. Infect. Dis. 2021;27:1737–1740. doi: 10.3201/eid2706.210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.de Siqueira I.C., Camelier A.A., Maciel E.A.P., Nonaka C.K.V., Neves M.C.L.C., Macêdo Y.S.F., de Sousa K.A.F., Araujo V.C., Paste A.A., de Freitas Souza B.S., et al. Early Detection of P.1 Variant of SARS-CoV-2 in a Cluster of Cases in Salvador, Brazil. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021;108:252–255. doi: 10.1016/j.ijid.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.da Silva J.C., Félix V.B., Leão S.A.B.F., Trindade-Filho E.M., Scorza F.A. New Brazilian Variant of the SARS-CoV-2 (P1/Gamma) of COVID-19 in Alagoas State. Braz. J. Infect. Dis. Off. Publ. Braz. Soc. Infect. Dis. 2021;25:101588. doi: 10.1016/j.bjid.2021.101588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goes L.R., Siqueira J.D., Garrido M.M., Alves B.M., Pereira A.C.P.M., Cicala C., Arthos J., Viola J.P.B., Soares M.A. New Infections by SARS-CoV-2 Variants of Concern after Natural Infections and Post-Vaccination in Rio de Janeiro, Brazil. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2021;94:104998. doi: 10.1016/j.meegid.2021.104998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Camargo C.H., Gonçalves C.R., Pagnoca E.V.R.G., Campos K.R., Montanha J.O.M., Flores M.N.P., Soares M.M.C.N., Binhardi F.M.T., Ferreira P.M., Yu A.L.F., et al. SARS-CoV-2 Re-infection in a Healthcare Professional in Inner Sao Paulo during the First Wave of COVID-19 in Brazil. Diagn. Microbiol. Infect. Dis. 2021;101:115516. doi: 10.1016/j.diagmicrobio.2021.115516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Penetra S.L.S., da Silva M.F.B., Resende P., Pina-Costa A., Santos H.F.P., Guaraldo L., Calvet G.A., Ogrzewalska M., Arantes I., Zukeram K., et al. Post-Acute COVID-19 Syndrome after Reinfection and Vaccine Breakthrough by the SARS-CoV-2 Gamma Variant in Brazil. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2022;114:58–61. doi: 10.1016/j.ijid.2021.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pereira F., Tosta S., Lima M.M., Reboredo de Oliveira da Silva L., Nardy V.B., Gómez M.K.A., Lima J.G., Fonseca V., de Oliveira T., Lourenço J., et al. Genomic Surveillance Activities Unveil the Introduction of the SARS-CoV-2 B. 1.525 Variant of Interest in Brazil: Case Report. J. Med. Virol. 2021;93:5523–5526. doi: 10.1002/jmv.27086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dos Santos L.A., de Góis Filho P.G., Silva A.M.F., Santos J.V.G., Santos D.S., Aquino M.M., de Jesus R.M., Almeida M.L.D., da Silva J.S., Altmann D.M., et al. Recurrent COVID-19 Including Evidence of Reinfection and Enhanced Severity in Thirty Brazilian Healthcare Workers. J. Infect. 2021;82:399–406. doi: 10.1016/j.jinf.2021.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Siqueira J.D., Goes L.R., Alves B.M., da Silva A.C.P., de Carvalho P.S., Cicala C., Arthos J., Viola J.P.B., Soares M.A. Distinguishing SARS-CoV-2 Bonafide Re-Infection from Pre-Existing Minor Variant Reactivation. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2021;90:104772. doi: 10.1016/j.meegid.2021.104772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu A.L.F., Liphaus B.L., Ferreira P.M., Tanamachi A.T., Masuda E.T., Trevisan C.M., Lucas P.C.D.C., Bugno A., Carvalhanas T.R.M.P. Sars-Cov-2 Reinfection: Report of Two Cases in Southeast Brazil. Rev. Inst. Med. Trop. 2021;63:e50. doi: 10.1590/s1678-9946202163050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fonseca V., de Jesus R., Adelino T., Reis A.B., de Souza B.B., Ribeiro A.A., Guimarães N.R., Livorati M.T.F.P., de Lima Neto D.F., Kato R.B., et al. Genomic Evidence of SARS-CoV-2 Reinfection Case with the Emerging B.1.2 Variant in Brazil. J. Infect. 2021;83:237–279. doi: 10.1016/j.jinf.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Silva M.S.D., Demoliner M., Hansen A.W., Gularte J.S., Silveira F., Heldt F.H., Filippi M., Pereira V.M.D.A.G., Silva F.P.D., Mallmann L., et al. Early Detection of SARS-CoV-2 P.1 Variant in Southern Brazil and Reinfection of the Same Patient by P.2. Rev. Inst. Med. Trop. 2021;63:e58. doi: 10.1590/s1678-9946202163058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Estofolete C.F., Banho C.A., Campos G.R.F., Marques B.C., Sacchetto L., Ullmann L.S., Possebon F.S., Machado L.F., Syrio J.D., Araújo Junior J.P., et al. Case Study of Two Post Vaccination SARS-CoV-2 Infections with P1 Variants in Coronavac Vaccinees in Brazil. Viruses. 2021;13:1237. doi: 10.3390/v13071237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Claro I.M., da Silva Sales F.C., Ramundo M.S., Candido D.S., Silva C.A.M., de Jesus J.G., Manuli E.R., de Oliveira C.M., Scarpelli L., Campana G., et al. Local Transmission of SARS-CoV-2 Lineage B.1.1.7, Brazil, December 2020. Emerg. Infect. Dis. 2021;27:970–972. doi: 10.3201/eid2703.210038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Volpato F.C.Z., Wink P.L., Monteiro F.L., Willig J.B., Vieceli T., Martins A.F., Zavascki A.P., Barth A.L. Early Detection of the SARS-CoV-2 P.1 Variant in Rio Grande Do Sul, Brazil: A Case Report. Infect. Control Hosp. Epidemiol. 2021;1:1–2. doi: 10.1017/ice.2021.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aksamentov I., Roemer C., Hodcroft E., Neher R. Nextclade: Clade Assignment, Mutation Calling and Quality Control for Viral Genomes. J. Open Source Softw. 2021;6:3773. doi: 10.21105/joss.03773. [DOI] [Google Scholar]
- 126.Chen Z., Azman A.S., Chen X., Zou J., Tian Y., Sun R., Xu X., Wu Y., Lu W., Ge S., et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat. Gen. 2022;54:499–507. doi: 10.1038/s41588-022-01033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.von Elm E., Altman D.G., Egger M., Pocock S.J., Gøtzsche P.C., Vandenbroucke J.P., STROBE Initiative The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. J. Clin. Epidemiol. 2008;61:344–349. doi: 10.1016/j.jclinepi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 128.Gallo V., Egger M., McCormack V., Farmer P.B., Ioannidis J.P., Kirsch-Volders M., Matullo G., Phillips D.H., Schoket B., Stromberg U., et al. STrengthening the Reporting of OBservational studies in Epidemiology--Molecular Epidemiology STROBE-ME: An extension of the STROBE statement. J. Clin. Epidemiol. 2021;64:1350–1363. doi: 10.1016/j.jclinepi.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 129.Clarke M., Chalmers I. Discussion sections in reports of controlled trials published in general medical journals: Islands in search of continents? JAMA. 1998;280:280–282. doi: 10.1001/jama.280.3.280. [DOI] [PubMed] [Google Scholar]
- 130.Draborg E., Andreasen J., Nørgaard B., Juhl C.B., Yost J., Brunnhuber K., Robinson K.A., Lund H. Systematic reviews are rarely used to contextualise new results-a systematic review and meta-analysis of meta-research studies. Syst. Rev. 2022;11:189. doi: 10.1186/s13643-022-02062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pinheiro J.R., Dos Reis E.C., Farias J.P., Fogaça M.M.C., da Silva P.S., Santana I.V.R., Rocha A.L.S., Vidal P.O., Simões R.D.C., Luiz W.B., et al. Impact of Early Pandemic SARS-CoV-2 Lineages Replacement with the Variant of Concern P.1 (Gamma) in Western Bahia, Brazil. Viruses. 2022;14:2314. doi: 10.3390/v14102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alves H.A., de Araújo J.L.F., Fonseca P.L.C., Moreira F.R.R., Bonfim D.M., Queiroz D.C., Miguita L., de Souza R.M., Geddes V.E.V., Costa W.C., et al. Monitoring the Establishment of VOC Gamma in Minas Gerais, Brazil: A Retrospective Epidemiological and Genomic Surveillance Study. Viruses. 2022;14:2424. doi: 10.3390/v14122747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Queiroz D.C., Carobin N.V., de Araújo e Santos L.C.G., Fonseca P.L.C., Braga-Paz I.L., Dias R.C., Ferreira J.G.G., Freitas T.R., Menezes D., Nolasco S.C.V.M., et al. SARS-CoV-2 Omicron BA.1, BA.2, and XAG identification during routine surveillance on a university campus in Belo Horizonte, Brazil, 2022. Braz. J. Microbiol. 2022;53:2009–2014. doi: 10.1007/s42770-022-00848-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Silva T.S., Salvato R.S., Gregianini T.S., Gomes I.A., Pereira E.C., de Oliveira E., de Menezes A.L., Barcellos R.B., Godinho F.M., Riediger I., et al. Molecular characterisation of a new SARS-CoV-2 recombinant cluster XAG identified in Brazil. Front. Med. 2022;9:1008600. doi: 10.3389/fmed.2022.1008600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.