Abstract
Pharma-grade extractive chondroitin sulfate (CS) is widely used for osteoarthritis (OA) treatment. Recently, unsulfated biofermentative chondroitin (BC) proved positive effects in OA in vitro model. This study, based on primary pathological human synoviocytes, aimed to analyze, by a multiplex assay, a panel of OA-related biomarkers in response to short-term treatments with bovine (CSb), pig (CSp) and fish (CSf) chondroitins, in comparison to BC. As expected, all samples had anti-inflammatory properties, however CSb, CSf and especially BC affected more cytokines and chemokines. Based on these results and molecular weight similarity, CSf and BC were selected to further explore the synoviocytes’ response. In fact, Western blot analyses showed CSf and BC were comparable, downregulating OA-related biomarkers such as the proteins mTOR, NF-kB, PTX-3 and COMP-2. Proteomic analyses, performed by applying a nano-LC-MS/MS TMT isobaric labelling-based approach, displayed the modulation of both common and distinct molecules to chondroitin treatments. Thus, CSf and BC modulated the biological mediators involved in the inflammation cascade, matrix degradation/remodeling, glycosaminoglycans’ synthesis and cellular homeostasis. This study helps in shedding light on different molecular mechanisms related to OA disease that may be potentially affected not only by animal-source chondroitin sulfate but also by unsulfated biofermentative chondroitin.
Keywords: extractive chondroitin sulfate, biofermentative unsulfated chondroitin, human primary synoviocytes, intracellular proteomic
1. Introduction
Nowadays, some factors such as longevity and the increase in obesity, have made osteoarthritis (OA) among the most common joint diseases worldwide [1,2,3]. OA causes joint pain and stiffness with progressive cartilage matrix degradation and bone sclerosis, leading to chronic disability that affects the patient’s quality of life [4,5]. The scientific community agrees that synovitis is associated with articular pain and OA advancement [6]. Fibroblast-like synoviocytes represent the major cellular component of synovium that produce many biological mediators (nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), pro-inflammatory interleukins such as IL-1β and IL-6, matrix metalloproteases (MMPs), tissue inhibitor of metalloproteinases (TIMP-1) and cartilage oligomeric matrix protein (COMP)) affecting immune cells’ activation, vascular hyperplasia and inflammation [7,8,9]. It has been reported that collagen and proteoglycan reduction in the synovial fluid increase the inflammatory signaling cascade with a central role played by cytokines [10,11]. In addition, synoviocytes produce hyaluronic acid (HA), through hyaluronic acid synthase (HAS), which is the major substance responsible for the viscoelastic behavior of the synovial fluid. Besides the presence of other proteoglycans and/or glycosaminoglycans (GAGs), HA concentration and molecular weight affect the composition and rheological properties of pathological synovial fluid [12,13]. Currently, OA treatments are based on the use of both non-pharmacological and pharmacological therapies. The first strategy aims at improving patient lifestyle (i.e., losing weight by exercising), while pharmacological approaches are concentrated on the use of analgesics and non-steroidal anti-inflammatory drugs (NSAIDs) [14,15]. Chondroitin sulfate-based treatments have been reported to be endowed with positive effects on OA-suffering patients [16,17,18,19]. Specifically, chondroitin sulfate (CS) is a natural biomacromolecule present in all vertebrates and invertebrates having a role in numerous biological processes such as support for cell growth, wound healing repair and suppression of pro-inflammatory cytokine activities [20,21,22]. It is a linear polysaccharide composed of repeating disaccharide units (D-glucuronic and N-acetyl-D-galactosamine) and sulfate groups at different positions on sugar residues [23]. This biopolymer can be extracted from different animal sources, thus presenting different purity grades [24] and many studies analyzed the potential biological features of different chondroitin sulfation patterns and extractive sources [24,25,26,27]. The recent literature showed that this biopolymer stimulates the chondrocyte’s production of proteoglycans in vitro, inhibits the expression of pro-inflammatory cytokines and counteracts metalloproteases’ activity preventing cartilage damage. The CS action mechanism is not completely revealed, also considering the heterogeneity of the molecule (source, molecular weight (MW), sulfation pattern), and even more BC-specific features have to be investigated since there was a common opinion asserting that sulfation was directly correlated to bioactivity [25]. The presence of CS is predominant in cartilage tissues with anti-inflammatory and chondroprotective effects [28]. Moreover, unsulfated chondroitin is also present at low concentrations in human connective tissues [29]. Although extensive information about the cellular effects of CS is increasingly growing, a wide molecular fingerprint of effects elicited by unsulfated chondroitin is still lacking. For this reason, to deepen our understanding of pharma-grade CS effectiveness and better explore the properties of biofermentative unsulfated chondroitin in counteracting OA degenerative and inflammatory process, experimental tests were performed exploiting a well-established OA in vitro model based on pathological human synoviocytes [30,31]. Specifically, the unsulfated chondroitin (biofermentative chondroitin (BC)) used in the present study has been obtained through a validated fermentative process, followed by extensive purification procedures [32,33]. The introduction of BC in therapeutic applications could represent a potential solution to overcome issues related to ethical and/or religious concerns regarding both the extraction sources and methods [34]. Furthermore, previous studies showed that BC was more effective than CS on an OA in vitro model based on IL-1β-insulted nasal human chondrocytes [35]. More recently, the secretome of BC-stimulated synoviocytes was compared to the fish CS-stimulated ones, reporting a comparable, or even better effect of BC [31]. In addition, the combination of BC with HA was used to assess the potential anti-inflammatory and restorative effects in both pathological chondrocytes and synoviocytes [30]. In the present work, we first investigated the modulation of cytokines, chemokines and growth factors’ profile of primary pathological synoviocytes in response to four different animal-source chondroitins (bovine CSb, pig CSp, fish CSf and biotechnological BC) by multiplex immunoassay. All chondroitins used in this experimental set-up have been analyzed for their purity grade and molecular weight distribution [26]. Based on their higher efficacy in inducing modulation of mediators of inflammation, CSf and BC treatments were selected for an in-depth quantitative proteomic analysis aimed at characterizing the response of osteoarthritic synoviocytes to these two specific chondroitin treatments. In this OA in vitro model, CSf and BC confirmed a similar biological effect as previously observed at the extra-cellular level [31]. Our results provide an overview of proteins modulated by treatments, thus improving our knowledge of pathways affected by different chondroitins potentially related to OA disease.
2. Results
2.1. Bioplex Assay
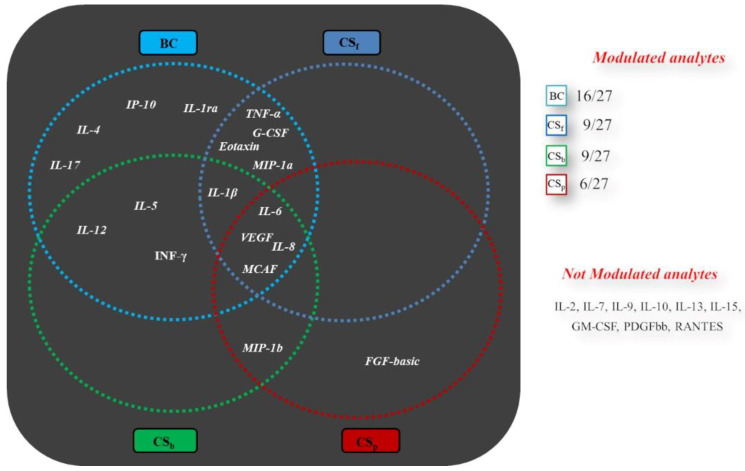
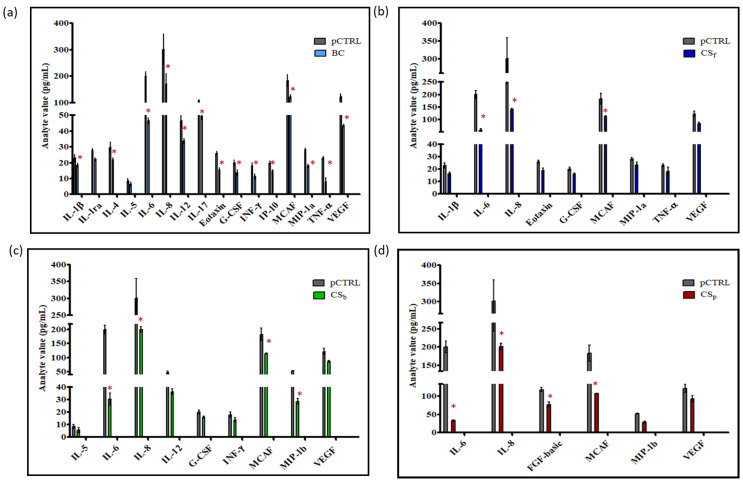
It was previously shown that chondroitin treatments affect the extracellular secretion of important inflammation mediators in pathological synoviocytes [31]. However, the analyses were performed following long incubation times, especially considering the starvation conditions. Here, different chondroitins with respect to origin, sulfation and Mw (Table 1), were assayed for their action after 8 h of contact with cells, and compared to untreated pathological ones, to evaluate if this shorter time-frame could be sufficient to prompt a biological response. The modulated analytes are shown in Figure 1 and the specific measured values for cytokines, chemokines and growth factors in each treatment are reported in Figure 2a–d. In this context, CSp treatment significantly modulated the secretion levels of 6 analytes out of the 27 assayed, while more factors were modulated by CSf and CSb (i.e., 9 out of 27) and BC (16 out of 27). Interestingly, all chondroitins, at the tested concentration, were able to significantly decrease pro-inflammatory biomarkers’ levels, such as IL-6, IL-8 and MCAF (vs. pCTRL, p < 0.05), but BC and CSf treatments also downregulated TNF-α (Figure 2a and Figure 2b, respectively). In addition, BC treatment significantly (p < 0.05) reduced the secreted levels of two growth factors, G-CSF (FC 0.60) and VEGF (FC 0.66); similarly, CSf significantly decreased G-CSF secretion (FC 0.78, p < 0.05) vs. pCTRL (Figure 2b). Overall, these results proved that short-time treatments based on both sulfated and unsulfated chondroitins are also effective in inducing a modulation of different OA-related mediators.
Table 1.
Hydrodynamic characterization of chondroitins through Size Exclusion Chromatography coupled to triple array detector (SEC-TDA), molecular weight, polydispersity and purity evaluation.
| Chondroitin | Mw (kDa) | Mw/Mn | Recovery |
|---|---|---|---|
| Chondroitin fish | 32.65 | 1.18 | >96% |
| Chondroitin bovine | 18.0 | 1.7 | >96% |
| Chondroitin pig | 17.7 | 1.24 | 99.3% |
| Chondroitin biofermentative | 31.5 | 1.22 | >96% |
Figure 1.
Schematic representation of biomarkers modulated by all the extractive and biofermentative chondroitins. The analysis was performed by multiplex assay. Fold Change considered vs. pCTRL: ≥1.20 or ≤0.80.
Figure 2.
Multiplex assay. Only the analytes with a 0.8 ≥ fold change ≥ 1.2 (vs. pCTRL) were considered differentially modulated. * p < 0.05 vs. pCTRL. Data were divided as following: (a) pCTRL and BC, (b) pCTRL and CSf, (c) pCTRL and CSb, (d) pCTRL and CSp. pCTRL is the same for all the graphs.
2.2. Western Blot Analyses
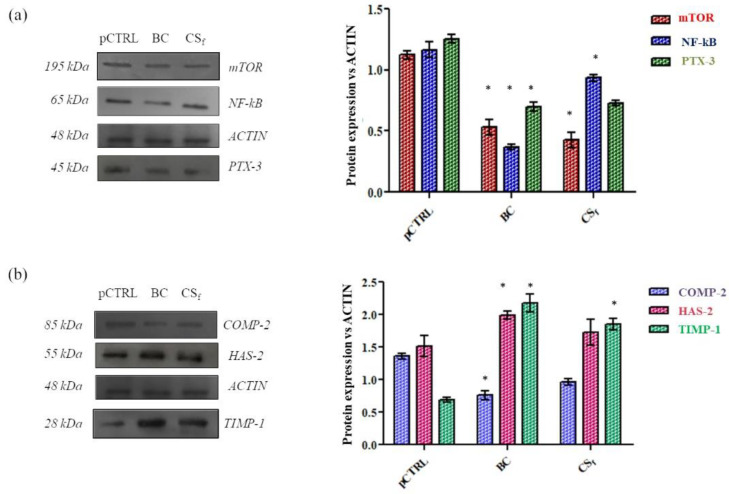
We next analyzed the intra-cellular expression levels of well-recognized OA markers (i.e., COMP-2, NF-kB and PTX-3). Figure 3 displays that, after 24 h of treatment in a culture serum-free medium, BC and CSf modulated the production of all analyzed biomarkers with respect to pCTRL. As expected, based on the source of isolated synoviocytes from OA synovial fluids, we detected basal levels of the proteins mTOR, COMP-2, NF-kB and PTX-3, thus confirming an ongoing inflammatory and degradative process. Both BC and CSf treatments significantly (p < 0.05) downregulated mTOR (by 2.1 and 2.7 fold, respectively) and NF-kB (2.9 and 1.2 fold correspondingly) in comparison to pCTRL. In addition, PTX-3 protein levels were found to be significantly (p < 0.05) reduced, specifically, BC treatment was slightly more effective by 1.8 fold compared to untreated cells while CSf reduced the expression of the same biomarker by about 1.7 fold (Figure 3a). Additionally, in COMP-2 negative modulation, BC also proved more active than CSf with a decrease of 1.8 fold vs. pCTRL, in comparison to the 1.4 fold of the extractive one (Figure 3b). The degradation of the cartilage matrix is related to TIMP-1 expression since it may counteract the activity of metalloproteases. These analyses demonstrated the tested chondroitins, increased TIMP-1 expression, contributing to slow down the degradation process. In fact, TIMP-1 levels were higher (3.14 fold and 2.68 fold) (p < 0.05) in cells treated with BC and CSf in comparison to pCTRL (Figure 3b). Finally, BC induced the increase in HAS-2 protein levels, thus supporting HA biosynthesis. HAS-2 protein expression resulted in being higher by about 1.3 fold for BC treatments, while CSf treatments were similar to the pCTRL outcomes (about 0.90 fold).
Figure 3.
Western blot analysis in CS- and BC-treated OA synoviocytes. ACTIN was used as the loading control. (a) Biomarkers related to inflammatory process; (b) Biomarkers involved in remodeling and/or regeneration of cartilage. Relative changes of treated samples vs. pCTRL are reported as mean + SD. * p < 0.05.
2.3. Proteomic Analysis by High-Resolution nanoLC-MS/MS
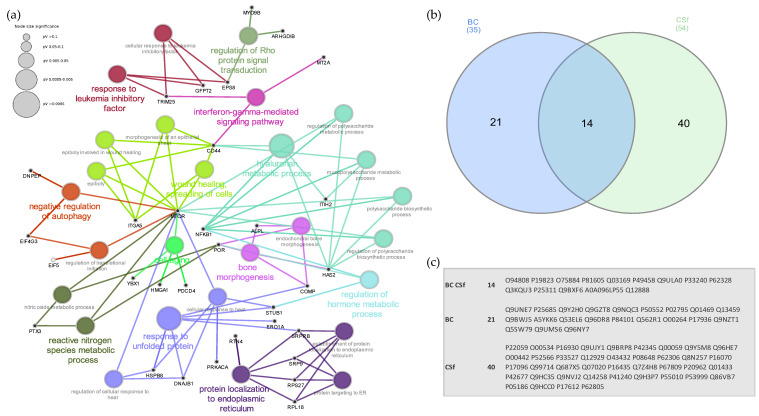
To investigate the effects of BC and CSf treatments on the overall protein profile at the intracellular level, comparative proteomic analyses were carried out by applying a quantitative nanoLC-MS/MS TMT isobaric labelling-based approach on treated and untreated starved human primary-synoviocytes. Using the Proteome Discoverer proteomics software package, we compared protein abundances of CSf-/BC-treated OA synoviocytes with respect to untreated cells. In particular, for each treatment replicate, we required a minimum of two replicates and at least two peptides per protein in at least one out of three conditions for the identification to be considered reliable. According to these criteria, a total of 1558 proteins were identified across the three conditions (Table S1 (Supplementary Materials)). From this list, we then extracted a subset of 75 proteins whose expression levels changed 1.2-fold or more (in any direction) in both (Table 2) or one (Table 3) of the treatment conditions with respect to untreated OA synoviocytes. Bioinformatics enrichment analysis performed for the biological process GO category (Figure 4a) revealed that a significant number of identified proteins are involved in processes related to hyaluronan metabolic process (17.9%), wound healing (14.3%), response to unfolded protein/protein localization to endoplasmic reticulum (10.7%) and bone morphogenesis (7.1%). Proteins related to the reactive nitrogen species metabolic process (7.1%), response to leukemia inhibitory factor (7.1%) and regulation of autophagy (7.1%) were also identified. A small fraction (about 4.9%) of identified proteins was found differentially expressed (0.8 ≥ fold change ≥ 1.2) across the cells treated with either CSf or BC compared to untreated cells (Table 2 and Table 3). We then investigated if there was a correlation of responses to different chondroitin treatments by comparing differentially expressed proteins to different treatments. We found that a specific subset of 14 proteins was commonly differentially expressed in both treatments (Figure 4b). Despite this partial similar response to the different treatments, for some differentially expressed proteins, a distinctive response of OA synoviocytes to BC and CS was also observed thus suggesting the modulation of both common and distinct responses to chondroitin treatments. The heatmap representing the log2 fold-change values of differentially expressed (0.8 ≥ FC ≥ 1.2) in CSf- and/or BC-treated vs. pCTRL synoviocytes identified by high-resolution LC-MS/MS was obtained and is provided as Figure S1 (Supplementary Materials).
Table 2.
Proteins differentially expressed (0.8 ≥ FC ≥ 1.2) in both CSf- and BC-treated vs. pCTRL synoviocytes identified by high-resolution LC-MS/MS.
| Accession | Gene Name | Description | CSf vs. pCTRL | BC vs. pCTRL |
|---|---|---|---|---|
| O94808 | GFPT2 | Glutamine—fructose-6-phosphate aminotransferase (isomerizing) 2 | 1.2 | 1.4 |
| Q3KQU3 | MAP7D1 | MAP7 domain-containing protein 1 | 1.2 | 1.4 |
| P62328 | TMSB4X | Thymosin beta-4 | 1.2 | 1.3 |
| Q12888 | TP53BP1 | TP53-binding protein 1 | 1.3 | 1.3 |
| O75884 | RBBP9 | Serine hydrolase RBBP9 | 1.2 | 1.3 |
| Q9ULA0 | DNPEP | Aspartyl aminopeptidase | 1.2 | 1.3 |
| P25311 | AZGP1 | Zinc-alpha-2-glycoprotein | 0.8 | 1.3 |
| P33240 | CSTF2 | Cleavage stimulation factor subunit 2 | 1.2 | 1.2 |
| Q9BXF6 | RAB11FIP5 | Rab11 family-interacting protein 5 | 1.3 | 1.2 |
| Q03169 | TNFAIP2 | Tumor necrosis factor alpha-induced protein 2 | 1.3 | 1.2 |
| P49458 | SRP9 | Signal recognition particle 9 kDa protein | 1.2 | 1.2 |
| A0A096LP55 | UQCRHL | Cytochrome b-c1 complex subunit 6-like, mitochondrial | 1.2 | 1.2 |
| P19823 | ITIH2 | Inter-alpha-trypsin inhibitor heavy chain H2 | 0.6 | 0.8 |
| P81605 | DCD | Dermcidin | 0.5 | 0.7 |
| Q96NY7 | CLIC6 | Chloride intracellular channel protein 6 | 1.2 | 1.3 |
| Q53EL6 | PDCD4 | Programmed cell death protein 4 | 1.2 | 1.3 |
| Q9BWJ5 | SF3B5 | Splicing factor 3B subunit 5 | 1.2 | 1.3 |
| A5YKK6 | CNOT1 | CCR4-NOT transcription complex subunit 1 | 1.2 | 1.3 |
| O00264 | PGRMC1 | Membrane-associated progesterone receptor component 1 | 1.2 | 1.2 |
| P62306 | SNRPF | Small nuclear ribonucleoprotein F | 1.3 | 1.2 |
| P22059 | OSBP | Oxysterol-binding protein 1 | 1.2 | 1.2 |
| Q9BRP8 | PYM1 | Partner of Y14 and mago | 1.3 | 1.2 |
| O43432 | EIF4G3 | Eukaryotic translation initiation factor 4 gamma 3 | 1.2 | 1.2 |
Table 3.
Proteins differentially regulated (0.8 ≥ FC ≥ 1.2) in CSf- or BC-treated vs. pCTRL synoviocytes identified by high-resolution LC-MS/MS.
| Accession | Gene Name | Description | CSf vs. pCTRL | BC vs. pCTRL |
|---|---|---|---|---|
| Q96HE7 | ERO1A | ERO1-like protein alpha | 1.2 | 1.1 |
| Q12929 | EPS8 | Epidermal growth factor receptor kinase substrate 8 | 1.3 | 1.1 |
| Q99714 | HSD17B10 | 3-hydroxyacyl-CoA dehydrogenase type-2 | 1.2 | 1.1 |
| O00442 | RTCA | RNA 3′-terminal phosphate cyclase | 1.3 | 1.1 |
| Q9H3P7 | ACBD3 | Golgi resident protein GCP60 | 1.4 | 1.1 |
| P55010 | EIF5 | Eukaryotic translation initiation factor 5 | 1.2 | 1.1 |
| Q14258 | TRIM25 | E3 ubiquitin/ISG15 ligase TRIM25 | 1.3 | 1.1 |
| Q00059 | TFAM | Transcription factor A, mitochondrial | 1.2 | 1.1 |
| P67809 | YBX1 | Y-box-binding protein 1 | 1.2 | 1.1 |
| Q9HCC0 | MCCC2 | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | 1.2 | 1.1 |
| Q01433 | AMPD2 | AMP deaminase 2 | 1.2 | 1.1 |
| O00534 | VWA5A | von Willebrand factor A domain-containing protein 5A | 1.3 | 1.1 |
| P33527 | ABCC1 | Multidrug resistance-associated protein 1 | 1.2 | 1.1 |
| P17096 | HMGA1 | High mobility group protein HMG-I/HMG-Y | 0.8 | 1.1 |
| P42345 | MTOR | Serine/threonine-protein kinase mTOR | 1.2 | 1.1 |
| P16435 | POR | NADPH--cytochrome P450 reductase | 1.3 | 1.1 |
| Q7Z4H8 | POGLUT3 | Protein O-glucosyltransferase 3 | 1.2 | 1.1 |
| P62805 | H4 | Histone H4 | 1.3 | 1.1 |
| P53999 | SUB1 | Activated RNA polymerase II transcriptional coactivator p15 | 1.2 | 1.0 |
| P17612 | PRKACA | cAMP-dependent protein kinase catalytic subunit alpha | 1.2 | 1.0 |
| Q9UJY1 | HSPB8 | Heat shock protein beta-8 | 1.2 | 1.0 |
| P20962 | PTMS | Parathymosin | 0.8 | 1.0 |
| Q9Y5M8 | SRPRB | Signal recognition particle receptor subunit beta | 1.2 | 1.0 |
| P05186 | ALPL | Alkaline phosphatase, tissue-nonspecific isozyme | 1.2 | 1.0 |
| Q07020 | RPL18 | 60S ribosomal protein L18 | 1.2 | 1.0 |
| P42677 | RPS27 | 40S ribosomal protein S27 | 1.2 | 1.0 |
| Q687X5 | STEAP4 | Metalloreductase STEAP4 | 1.2 | 1.0 |
| Q8N257 | H2BU1 | Histone H2B type 3-B | 1.2 | 1.0 |
| Q86VB7 | CD163 | Scavenger receptor cysteine-rich type 1 protein M130 | 0.7 | 1.0 |
| Q9NVJ2 | ARL8B | ADP-ribosylation factor-like protein 8B | 1.2 | 1.0 |
| P16930 | FAH | Fumarylacetoacetase | 1.2 | 1.0 |
| P08648 | ITGA5 | Integrin alpha-5 | 0.7 | 1.0 |
| P16070 | CD44 | CD44 antigen | 0.8 | 1.0 |
| P52566 | ARHGDIB | Rho GDP-dissociation inhibitor 2 | 0.8 | 0.9 |
| P41240 | CSK | Tyrosine-protein kinase CSK | 0.8 | 0.9 |
| Q9HC35 | EML4 | Echinoderm microtubule-associated protein-like 4 | 0.8 | 0.9 |
| Q96DR8 | MUCL1 | Mucin-like protein 1 | 1.0 | 6.7 |
| Q9GZT8 | NIF3L1 | NIF3-like protein 1 | 1.1 | 1.8 |
| Q9NZT1 | CALML5 | Calmodulin-like protein 5 | 1.1 | 1.7 |
| Q9NQC3 | RTN4 | Reticulon-4 | 1.1 | 1.6 |
| P84101 | SERF2 | Small EDRK-rich factor 2 | 1.0 | 1.3 |
| P02795 | MT2A | Metallothionein-2 | 1.0 | 1.3 |
| P17936 | IGFBP3 | Insulin-like growth factor-binding protein 3 | 1.1 | 1.3 |
| P50552 | VASP | Vasodilator-stimulated phosphoprotein | 1.0 | 1.3 |
| Q9UMS6 | SYNPO2 | Synaptopodin-2 | 1.1 | 1.3 |
| Q5SW79 | CEP170 | Centrosomal protein of 170 kDa | 1.1 | 1.3 |
| Q13459 | MYO9B | Unconventional myosin-IXb | 1.1 | 1.2 |
| Q9UNE7 | STUB1 | E3 ubiquitin-protein ligase CHIP | 1.1 | 1.2 |
| P25685 | DNAJB1 | DnaJ homolog subfamily B member 1 | 0.9 | 1.2 |
| Q9Y2H0 | DLGAP4 | Disks large-associated protein 4 | 1.0 | 1.2 |
| Q01469 | FABP5 | Fatty acid-binding protein 5 | 0.9 | 1.2 |
| Q562R1 | ACTBL2 | Beta-actin-like protein 2 | 0.9 | 0.8 |
Figure 4.
(a) Enriched GO network groups using ClueGO/CluePedia-based enrichment. A network view for GO Biological Processes of differentially regulated proteins following BC and CSf treatments. Terms (each represented as node) are functionally grouped based on shared genes (kappa score ≥ 0.4) and are shown with different colors representing the class to which they belong. The specific players (proteins) of each node are highlighted with the respective gene name (in black). The size of the nodes indicates the degree of significance. Within each class, the most significant term (indicated with colored and bold characters) defines the name of the group. Ungrouped terms are not shown. (b) Venn diagram showing the overlapping of differentially expressed proteins after BC (blue) and CSf (green) treatments. Details of the UniProt accession numbers within each subset of the Venn diagram are reported in (c).
3. Discussion
Because resolutive treatments for OA pathology are not currently available, often patients need to use anti-inflammatory drugs or cortisone to reduce pain. Some international Guidelines suggest the use of chondroitin sulfate in oral formulations in the early stage of OA. GAGs-based intra-articular injections of specifically formulated gels may improve functionality through the viscoelastic features, reduce ache and, possibly, in the short term, counteract pathology progression [36,37]. Recently, different scientific approaches are focused on the identification of key biomarkers to evaluate the potential beneficial effects of different treatments. Among these, mass spectrometry approaches play a key role [17,24,38,39]. Indeed, proteomic studies aimed at investigating the response of joint-derived cells to different treatments such as HA, GlcN, extractive CS and, more recently, the recently available high-purity biofermentative unsulfated chondroitin, have been increasing in the last few years. The scientific literature also reports data about the secretome of chondrocyte in in vitro cultures [40,41]. It has been demonstrated that the pro-inflammatory mediators secreted by synovial cells play a key role in the chondrocytes’ activation process by increasing the production of several cytokines and chemokines [42]. For this reason, the response of pathological synoviocytes, at both secretome and proteome levels, following specific GAGs treatments (considering that some of those are injective) is necessary to support the effectiveness of potential OA therapies based on these biopolymers toward the relief of symptoms and movement [6,40]. Recent in vitro and in vivo studies showed a positive effect of CS in human chondrocytes and damaged cartilage [43,44]. The well-accepted scientific hypothesis is the involvement of CS in the inhibition of NF-kB signaling and increasing HA biosynthesis. In this way, CS may reduce the pain through the modulation of the bradykinin (BK) system that is related to the release of IL-6 and IL-8 via p38MAPK and NF-kB activation in synoviocytes and chondrocytes [44]. Most CS used for OA symptomatic management is derived from animal sources such as bovine trachea or porcine (ear and nose) tissues and are namely terrestrial CS. However, fish-derived CS has been also obtained and commercialized for two decades. Specifically, the shark fin-derived product is regularly available, under certification that the protected species are not among the ones sacrificed within the manufacturing process. Terrestrial and marine CS have different biochemical features, presenting diverse molecular weight and sulfation pattern [45,46]. It has been argued that the combination between a specific sequence of disaccharides and the sulfation grade is responsible for a particular biological activity. Accordingly, Pomin et al. 2019 [45] clearly explained that marine CS may be considered a very interesting biomolecule for the presence of sulfation patterns that are not found in terrestrial animals. Marine CS showed anti-inflammatory activities and improved the mechanical performance of cartilage-engineered constructs [46]. Moreover, different studies demonstrated that marine CS has important effects on in vitro chondrocytes’ proliferation and differentiation but the scientific knowledge about its application in OA management is less diffused than the terrestrial CS [45,47]. In addition, very recent studies demonstrated that both marine CS and BC are also related to the differentiation of mesenchymal cells towards the chondrocyte phenotype when added to the culture medium or as a component of chemically modified gelatin-based scaffolds [48,49]. We recently reported that CSf and BC significantly affect the synoviocytes secretome and that BC is able (both alone and coupled to HA) to reduce the inflammatory process in this OA in vitro model [30,31]. Despite the long tradition of using both pharma- and food-grade CS, the specific biological mechanism involved still needs to be fully unraveled. Due to being the polymer dimension and charge density often responsible for interactions with cells and other macromolecules of the extracellular matrix network, the studies aimed at characterizing the bioactivity of the recently available unsulfated BC are of remarkable scientific interest. Recent published data showed that chondroitins (both marine and biofermentative) were able to modulate the secretion of many OA-correlated biomarkers after 48 h of treatment [31]. In the present study, the quantification of these latter (for a total of 27) was performed after a shorter incubation time (8 h) in order to also verify the effectiveness of chondroitin in this condition. The outcomes displayed that the biomarkers strongly involved in the inflammatory cellular response, such as IL-6, IL-8 and MCAF, were significantly reduced by BC and all CS treatments, thus highlighting a generally positive anti-inflammatory effect [50,51]. The analysis of specific inflammatory response-correlated mediators (mTOR, NF-kB, PTX-3 and COMP-2) confirmed the ability of both CSf and BC to affect these proteins [8,52,53,54]. Furthermore, CSf and BC showed similar effectiveness in increasing TIMP-1 levels; this latter is known to contribute to attenuating ECM degradation through the inhibition of metalloproteases action [55]. Another selected biomarker was HAS-2, since this enzyme is involved in HA biosynthesis, which in turn affects the rheological features of synovial fluid, and thus the lubrication and shock absorption ability. HA content and size in synovial fluid may affect joint movement, thus marked upregulation of HAS-2 has to be considered desirable [12,13]. Our study provides new information on the effects of sulfated and unsulfated chondroitins on human OA synoviocytes proteomes by means of a Tandem Mass Tag (TMT)-based high-resolution LC-MS/MS approach. Both treatments affected the expression levels of proteins involved in important biological pathways converging on the mTOR and NF-kB signaling, affecting hyaluronan metabolism and the response to reactive nitrogen species metabolic processes. These findings are coherent with the literature, identifying the PI3K/AKT/mTOR signaling pathway as essential for the metabolism of joint tissues. The mTOR signaling pathway has been related to the development of OA, cartilage degradation, subchondral bone dysfunction and synovial inflammation [56]. Proteins related to the response to leukemia inhibitory factor (LIF) and regulation of autophagy were also differentially expressed following chondroitin treatments. In this respect, both the role of LIF and autophagy are critical among OA progression [56,57]. Furthermore, as previously observed for secretome profiles [31], despite similarities among BC and CSf treatments, for several up- and downregulated proteins a different regulation trend was also observed, suggesting a certain degree of specificity in this specific experimental set-up. From a production point of view, it should be considered that BC is produced by bacterial fermentation, thus overcoming any issues related to the extraction source, such as safety concerns and/or the environmental impact of the manufacturing process. Overall, our results showed that unsulfated chondroitin bioactivity resembles and is comparable to extractive CS, thus increasing our knowledge about biomarkers and treatments that may be supportive in the clinical management of OA-affected patients.
4. Materials and Methods
4.1. Preparation of CS and BC Solutions
BC was produced in our laboratories through a patented fermentation process using EcK4r3, a specific recombinant strain. The experimental protocols followed were previously reported [32,33]. The obtained BC (MW; 35 ± 3 kDa) had a purity of 95 ± 5% and a low endotoxin content was confirmed through Limulus test (EU/mg < 0.05). The CSs used during this experimental work were provided, in the framework of a collaboration with BioTekNet Scpa, by the IBSA group and were obtained from diverse animal sources, pig (CSp), bovine (CSb) and fish, specifically shark (CSf), with high purity (95 ± 5%), and a very low endotoxin content (0.1 EU/mg). The CSs are manufactured according to ICHQ7 and EudraLex volume 4 part II. Furthermore, the latter were obtained from cartilages collected from species that are not mentioned in the list of protected species of CITIES (the Convention of International Trade in Endangered Species of wild fauna and flora), under the responsibility of the IBSA Chinese subsidiary. BC and CSs were dissolved in Phosphate-Buffered Saline (PBS, pH 7.2; Lonza, Milan, Italy) at a concentration of 16 mg/mL. The pH and osmolality of the obtained solutions were measured to perform experimental tests under physiological conditions (i.e., pH 7.0 ± 0.1 and osmolality 300 mOsm); later they were sterilized by autoclave (1 bar, 121 °C for, 20 min). Lastly, BC- and CS-based gels were diluted 1:5 in a specific culture medium (Dulbecco’s modified Eagle’s medium, Gibco DMEM, without fetal bovine serum Gibco FBS; Fisher Scientific Italia, Milan, Italy) and microfiltered (0.22 µm; Millipore, Milan, Italy).
4.2. OA In Vitro Model and Chondroitin-Based Treatments
As previously reported, human primary articular synoviocytes were isolated from the synovial fluids of OA-affected patients, during knee joint replacement surgical procedures. These operations were performed at the Orthopedics and Traumatology Department of the University Federico II of Naples. All patients gave informed consent, and the procedures were approved by the Internal Ethical Committee. After the isolation, the cell phenotype was confirmed using the protocols previously established in our laboratories [30]. For the cell treatments, we followed the starvation protocol described by Russo et al. 2020 [31] with slight changes. Specifically, the cells were cultured in serum-free medium with or without BC (3.2 mg/mL), and CSs (3.2 mg/mL) for 8 h or 24 h in a standard 24 well culture plate or in a 25 mm2 flask (Falcon DB).
4.3. Bioplex Assay
A 27-plex immunoassay panel based on xMAP technology [58] was used to evaluate the modulation of specific pro-inflammatory and anti-inflammatory cytokines, chemokines and growth factors (Bio-Rad Laboratories s.r.l., Milan, Italy) after the treatments of primary synoviocytes with BC or CSs. After 8 h of treatments, the culture medium for each sample was withdrawn, centrifuged (1500 rpm, 7 min) and used to perform the bioplex assay. The specific panel permitted to evaluate the pro-inflammatory cytokines: IL-1β, IL-1ra, IL-2, IL-5, IL-6, IL-7, IL-9, IL-12, IL-13, IL-15, IL-17, INF-γ, TNF-α; anti-inflammatory cytokines: IL-4, IL-10; growth factors: G-CSF, GM-CSF, FGFbasic, PDGF-bb, VEGF and chemokines: IL-8, MCAF, MIP-1a, MIP-1b, RANTES, Eotaxin, IP-10. The technique was based on the use of magnetic beads labeled with red and infrared fluorophores covered with specific antibodies allowing the simultaneous evaluation of multiple target analytes within a single sample [31]. The binding of each particle with the target analyte detection was performed with a biotinylated antibody and phycoerythrin-conjugated streptavidin. The procedures were carried out following the manufacturer’s instructions using a Bio-Plex array reader (Luminex, Austin), TXAnalyte concentrations (pg/mL) were assessed by a standard curve according to the manufacturer’s protocol. The biological mediators were considered differentially modulated when CSf- or BC-based treatments induced a decrease and/or increase in the biomarker with a ratio 0.8 ≥ fold change ≥ 1.2 in comparison to untreated cells.
4.4. Western Blot Analyses
For Western Blot analyses, 1.0 × 105 cells were seeded in a 25 mm2 flask and treated with BC or CSf for 24 h. Following treatments, cells were harvested and lysed by a Radio-Immunoprecipitation Assay buffer (RIPA buffer 1×) (Cell Signaling Technology, Danvers, MA, USA). Protein concentration for each sample was determined using the Bradford method [59]. Western blotting analyses were performed as previously described [60]. Briefly, aliquots of intracellular proteins (30 µg) were resolved by electrophoresis on 10% SDS-PAGE and transferred to a nitrocellulose membrane (GE, Amersham, Chicago, IL, USA). Equivalent loadings were verified by Ponceau Red (Sigma-Aldrich, St. Louis, MO, USA) staining after transfer. Then, the membrane was blocked by 5% skimmed milk in Tris-buffered saline and 0.05% Tween-20 (TTBS) for 30 min. Primary antibodies to detect mTOR (Cell Signaling Technology, Danvers, MA, USA), COMP-2 (Abcam, Cambridge, UK), NF-kB (Santa Cruz Biotechnology, Dallas, TX, USA), HAS-2 (Santa Cruz Biotechnology, Dallas, TX USA), PTX-3 (Santa Cruz Biotechnology, Dallas, TX, USA) and TIMP-1 (Santa Cruz Biotechnology, Dallas, TX, USA) were diluted 1:500, 1:250, 1:200, 1:200, 1:100, respectively, and incubated overnight at 4 °C. After that, the membrane was extensively washed using TTBS and immunoreactive bands were detected using chemiluminescence suitable horseradish peroxidase-conjugated secondary antibodies diluted 1:5000 (Santa Cruz Biotechnology, Dallas, TX, USA). Following incubation for 2 h at room temperature, signals detection was performed with an ECL system (Merck Millipore, Burlington, MA, USA). Anti-Actin antibody (Santa Cruz Biotechnology, Dallas, TX, USA) was diluted 1:1000 and used to normalize the protein levels of each analyzed biomarker. A semi-quantitative analysis of protein expressions was carried out by using the ImageJ program according to the manufacturer’s protocol.
4.5. Data Analysis
Data are expressed as mean ± standard deviation (SD). The statistical significance of the differences between BC- and CS-treated synoviocytes samples vs. pCTRL was determined using a two-tailed t-test, considering p values < 0.05. As explained in the following, for the proteomic analyses the statistical significance was set to 0.05 (p ≤ 0.05), and the Bonferroni test was used to correct the p-value.
4.6. Sample Preparation for Proteomic Analyses
For proteomic analysis, human primary articular synoviocytes treated and untreated with BC or CSf as described above were lysed in ice-cold lysis buffer (100 mM Triethylammonium bicarbonate TEAB, SDS 1%) and disrupted by two cycles of sonication at a 20% amplitude for 30 sec on ice. Lysates were cleared by centrifugation at 16,000× g for 15 min at 4 °C. Supernatants were transferred into new tubes and treated with 1 Unit of RQ1 DNase (Promega, Milan, Italy) for 1 h at room temperature. Protein concentration was determined by using the Pierce BCA Protein assay kit (Thermo Scientific, Milan, Italy). For each condition, equal amounts of proteins (100 µg in 100 µL of 100 mM TEAB) were reduced with 10 mM Tris-(2-carboxyethyl)-phosphine (TCEP) for 1 h at 55 °C and alkylated with 18 mM iodoacetamide by incubating samples for 30 min at room temperature in the dark. Proteins were then precipitated overnight by adding six volumes of pre-chilled acetone. Following centrifugation at 8000× g for 10 min at 4 °C, protein pellets were resuspended in 100 µL of 100 mM TEAB and digested overnight with MS-grade trypsin (Thermo Scientific, USA) at an enzyme/substrate ratio of 1:40 at 37 °C. The resulting peptide mixtures were chemically labeled with the TMT isobaric tags as previously reported [61,62] using the 128C, 127N and 126 tags for the BC- and CSf-treated and untreated cells, respectively. Briefly, 0.8 mg of TMT reagents in 41 µL of anhydrous acetonitrile were added to each sample. The reaction proceeded for 1 h and then was quenched for 15 min with hydroxylamine to a final concentration of 0.3% (w/w). The two samples were then mixed at equal amounts and diluted in 0.1% TFA/2% CH3CN to a final concentration of 0.5 µg/µL for LC-MS analyses.
4.7. High-Resolution nanoLC−Tandem Mass Spectrometry
Aliquots of TMT-labeled samples (2.5 µg) were analyzed in triplicate by high-resolution nanoLC−Tandem Mass Spectrometry using a Q-Exactive Orbitrap mass spectrometer equipped with an EASY-Spray nano-electrospray ion source (Thermo Fisher Scientific, Germany) and coupled to a Thermo Scientific Dionex UltiMate 3000RSLC nanosystem (Thermo Fisher Scientific) as previously reported [63]. The solvent composition was 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). Peptides were loaded on a trapping PepMap™ 100 μ Cartridge Column C18 (300 μm × 0.5 cm, 5 μm, 100 Å) and desalted with solvent A for 3 min at a flow rate of 10 μL/min. After trapping, the eluted peptides were separated on an EASY-Spray analytical column (50 cm × 75 μm ID PepMap RSLC C18, 3 μm, 100 Angstrom), heated at 35 °C, at a flow rate of 300 nL/min applying the following gradient: 5% B for 3 min; from 5% to 27.5% B in 222 min; from 27.5% to 40% B in 10 min; from 40% to 95% B in 1 min. Washing (95% B for 4 min) and re-equilibration (5% B for 24 min) steps were always included at the end of the gradient. Eluting peptides were analyzed on the Q-Exactive mass spectrometer operating in positive polarity mode with a capillary temperature of 280 °C and a potential of 1.9 kV applied to the capillary probe. Full MS survey scan resolution was set to 70,000 with an automatic gain control (AGC) target value of 3 × 106 for a scan range of 375–1500 m/z and maximum ion injection time (IT) of 60 ms. The mass (m/z) 445.12003 was used as lock mass. A data-dependent top 12 method was operated, during which high-energy collisional dissociation (HCD) spectra were obtained at a 35000 MS2 resolution with an AGC target of 1 × 105 for a scan range of 200–2000 m/z, maximum IT of 120 ms, 1.6 m/z isolation width and normalized collisional energy (NCE) of 32. Precursor ions targeted for HCD were dynamically excluded for 30 s. Full scans and Orbitrap MS/MS scans were acquired in profile mode, whereas ion trap mass spectra were acquired in centroid mode. Charge state recognition was enabled by excluding unassigned and 1, 7, 8, >8 charged states. All data were acquired with the Xcalibur 3.1 software (Thermo-Fisher Scientific).
4.8. Protein Identification and Quantitation
For data processing, the acquired raw files were analyzed with the Thermo Scientific Proteome Discoverer 2.4 software (Thermo Fisher Scientific) using the SEQUEST HT search engine. The HCD MS/MS spectra were searched against the Homo sapiens database (release 2019_11, 20,380 entries) assuming trypsin (Full) as the digestion enzyme and two allowed numbers of missed cleavage sites. Mass tolerances were set to 10 ppm and 0.02 Da for precursor and fragment ions, respectively. Oxidation of methionine (+15.995 Da) was set as a dynamic modification. Carbamidomethylation of cysteine (+57.021 Da) and the TMT label on lysines and the N-terminus (229.1629) were set as static modifications. False discovery rates (FDRs) for peptide spectral matches (PSMs) were calculated and filtered using the Percolator node in Proteome Discoverer that was run with the following settings: Maximum Delta Cn 0.05, a strict target FDR of 0.01, a relaxed target FDR of 0.05 and validation based on q-value. Protein identifications were accepted when the protein FDR was below 1% and when present in at least two out of three replicate injections with at least two peptides. Fold Change (FC) thresholds of proteins identified in BC and CSf-treated vs. untreated cells were set at ±1.2.
4.9. Bioinformatic Analyses
The clustered heatmap of the proteins differentially expressed in at least one out of two treatments was generated by using the CIMminer freely available web-server tool (http://discover.nci.nih.gov/cimminer/ accessed on 5 December 2021) with unsupervised clustering set on raws only with the following parameters: average linkage, Euclidean distance, and quantile binning. The ClueGO v2.5.8 + CluePedia v1.5.4, a Cytoscape v3.8.2 plug-in, was used to visualize the non-redundant biological process GO terms of proteins up- and downregulated following BC or CSf treatments in functionally organized networks reflecting the relationships between the biological terms based on the similarity of their linked gene/proteins [40]. Biomarkers related to inflammation and selected for Western blot analyses (i.e., mTOR, COMP-2, NF-kB, HAS-2, TIMP-1 and PTX-3) were included in the input list to verify the occurrence of specific connections between target analytes and differentially expressed proteins. For the enrichment of biological terms and groups, the two-sided (Enrichment/Depletion) tests based on the hypergeometric distribution was used. The statistical significance was set to 0.05 (p ≤ 0.05), and the Bonferroni step down adjustment was used to correct the p-value for the terms and the groups created by ClueGO. To diminish the redundancy of the terms shared by similar associated proteins, the network specificity was set to Medium, and the GO Term grouping option was selected, which allows the maintenance of the most representative parent or child term. The parameters were: kappa score threshold set to 0.4; Leading Group term based on: Highest Significance; % gene for Group Merge: 50; % Terms for Group Merge: 50.
5. Conclusions
This study emphasizes the importance of robust in vitro models, resembling OA pathology, to compare chondroitin of different origins, with an extensive molecular fingerprint, based on multiplex assay and quantitative proteomic approaches, to shed light on the peculiar biological features of these intriguing macromolecules. Considering that chondroitin-based treatments have more and more relevance in the management of osteoarthritis, which reports an increasing number of patients every year, a better understanding of the biochemical characteristics of the animal source and biotechnological chondroitins, also in relation to the purity, molecular weight and sulfation pattern, may help in assessing their therapeutic use, either contemporary to other pharmacological treatments or as a main drug to counteract joint-disease progression and patient pain.
Acknowledgments
The authors would like to acknowledge Elisabetta Cassese for her technical support in the SEC-TDA analyses for the diverse chondroitins. Moreover, the authors acknowledge Carlo Ruosi, professor in orthopaedics at the University Federico II of Naples, for providing the biological material during surgery.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232415865/s1.
Author Contributions
Conceptualization, A.C. and C.S.; methodology, V.V., A.S. and R.R.; software, V.V. and R.R.; validation, V.V. and R.R.; formal analysis, V.V., R.R. and M.V.; investigation, V.V., R.R., M.V. and A.A.; data curation; V.V., A.S., R.R., A.C., D.C. and C.S.; writing—original draft preparation, V.V., R.R., A.C. and C.S.; writing-review and editing, D.C., A.C. and C.S.; visualization, V.V. and R.R.; supervision, D.C., P.V.P., A.C. and C.S.; project administration, C.S. and A.C.; funding acquisition, C.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study did not involve humans or animals. The primary cells were derived and used according to a registered protocol approved by the University’s Internal Ethical Committee for these types of in vitro experiments (AOU-SUN reg. no. 0003711/2015).
Informed Consent Statement
Not applicable. This manuscript is not based on clinical studies, thus patients were not involved.
Data Availability Statement
All data critical for reader understanding and outcomes’ discussion are reported within the manuscript or as part of the Supplementary Materials. Additional raw data are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The research was initially supported by PON03PE00060_3; successively by MISE-Incube. Vassallo received a 1 year research fellowship from the Invitalia-funded project Altergon CdS000463.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Saito T., Tanaka S. Molecular mechanisms underlying osteoarthritis development: Notch and NF-κB. Arthritis Res. Ther. 2017;19:94. doi: 10.1186/s13075-017-1296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace I.J., Worthington S., Felson D.T., Jurmain R.D., Wren K.T., Maijanen H., Woods R.J., Lieberman D.E. Knee osteoarthritis has doubled in prevalence since the mid-20th century. Proc. Natl. Acad. Sci. USA. 2017;114:9332–9336. doi: 10.1073/pnas.1703856114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plotnikoff R., Karunamuni N., Lytvyak E., Penfold C., Schopflocher D., Imayama I., Johnson S.T., Raine K. Osteoarthritis prevalence and modifiable factors: A population study. BMC Public Health. 2015;15:1195. doi: 10.1186/s12889-015-2529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glyn-Jones S., Palmer A.J., Agricola R., Price A.J., Vincent T.L., Weinans H., Carr A.J. Osteoarthritis. Lancet. 2015;386:376–387. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y.J., Chang W.A., Wu L.Y., Huang C.F., Chen C.H., Kuo P.L. Identification of Novel Genes in Osteoarthritic Fibroblast-Like Synoviocytes Using Next-Generation Sequencing and Bioinformatics Approaches. Int. J. Med. Sci. 2019;16:1057–1071. doi: 10.7150/ijms.35611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathiessen A., Conaghan P.G. Synovitis in osteoarthritis: Current understanding with therapeutic implications. Arthritis Res. Ther. 2017;19:18. doi: 10.1186/s13075-017-1229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartok B., Firestein G.S. Fibroblast-like synoviocytes: Key effector cells in rheumatoid arthritis. Immunol. Rev. 2010;1:233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calamia V., Fernàndez-Puente P., Mateos J., Lourido L., Rocha B., Montell E., Vergés J., Ruiz-Romero C., Blanco F.J. Pharmacoproteomic Study of Three Different Chondroitin Sulfate Compounds on Intracellular and Extracellular Human Choncrocyte Proteomes. Mol. Cell. Proteom. 2012;11:013417. doi: 10.1074/mcp.M111.013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C.T., Lin Y.T., Chinag B.L., Lin Y.H., Hou S.M. High molecular weight hyaluronic acid down-regulates the gene expression of osteoarthritis-associated cytokines and enzymes in fibroblast-like synoviocytes from patients with early osteoarthritis. Osteoarthr. Cartil. 2006;14:1237–1247. doi: 10.1016/j.joca.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Haidar O., Nathanael O.N., Staunton C., Bavan S., O’Brien1 F., Zouggari S., Sharif U., Mobasheri A., Kumagai K., Barrett-Jolley R. Pro-inflammatory Cytokines Drive Reregulation of Potassium Channel Expression in Primary Synovial Fibroblasts. Front. Physiol. 2020;11:226. doi: 10.3389/fphys.2020.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papalia R., Vadalà G., Torre G., Perna M., Saccone L., Denaro V. The cytokinome in osteoarthritis, a new paradigm in diagnosis and prognosis of cartilage disease. J. Biol. Regul. Homeost. Agents. 2016;30:77–83. [PubMed] [Google Scholar]
- 12.Firestein G.S. Etiology and pathogenesis of rheumatoid arthritis. In: Firestein G.S., Budd R.C., Harris T., McInnes I.B., Ruddy S., Sergent J.S., editors. Kelly’s Textbook of Rheumatology. Volume 8. Saunders Elsevier; Philadelphia, PA, USA: 2009. pp. 1035–1086. [Google Scholar]
- 13.Saari H., Konttinen Y.T., Friman C., Sorsa T. Differential effects of reactive oxygen species on native synovial fluid and purified human umbilical cord hyaluronate. Inflammation. 1993;17:403–415. doi: 10.1007/BF00916581. [DOI] [PubMed] [Google Scholar]
- 14.Zhu X., Sang L., Wu D., Rong J., Jiang L. Effectiveness and safety of glucosamine and chondroitin for the treatment of osteoarthritis: A meta-analysis of randomized controlled trials. J. Orthop. Surg. Res. 2018;13:17. doi: 10.1186/s13018-018-0871-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majeed M.H., Sherazi S.A.A., Bacon D., Bajwa Z.H. Pharmacological treatment of pain in osteoarthritis: A descriptive review. Curr. Rheumatol. Rep. 2018;20:88. doi: 10.1007/s11926-018-0794-5. [DOI] [PubMed] [Google Scholar]
- 16.David-Raoudi M., Deschrevel B., Leclercq S., Galéra P., Boumediene K., Pujol J.P. Chondroitin sulfate increases hyaluronan production by human synoviocytes through differential regulation of hyaluronan synthases: Role of p38 and Akt. Arthritis Rheum. 2009;60:760–770. doi: 10.1002/art.24302. [DOI] [PubMed] [Google Scholar]
- 17.Henrotin Y., Marty M., Mobasheri M. What is the current status of chondroitin sulfate and glucosamine for the treatment of knee osteoarthritis? Maturitas. 2014;78:184–187. doi: 10.1016/j.maturitas.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Bruyere O., Cooper C., Pelletier J.P. An algorithm recommendation for the management of knee osteoarthritis in Europe and internationally: A report from a task force of the European Society for Clinical and Economic Aspects of Osteoporosis and Osteoarthritis (ESCEO) Semin. Arthritis Rheum. 2014;44:253–263. doi: 10.1016/j.semarthrit.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Reginster J.Y., Veronese N. Highly purified chondroitin sulfate: A literature review on clinical efficacy and pharmacoeconomic aspects in osteoarthritis treatment. Aging Clin. Exp. Res. 2021;33:37–47. doi: 10.1007/s40520-020-01643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stellavato A., Restaino O.F., Vassallo V., Finamore R., Ruosi C., Cassese E., De Rosa M., Schiraldi C. Comparative Analyses of Pharmaceuticals or Food Supplements Containing Chondroitin Sulfate: Are Their Bioactivities Equivalent? Adv. Ther. 2019;36:3221–3237. doi: 10.1007/s12325-019-01064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stellavato A., Restaino O.F., Vassallo V., Cassese E., Finamore R., Ruosi C., Schiraldi C. Chondroitin Sulfate in USA Dietary Supplements in Comparison to Pharma Grade Products: Analytical Fingerprint and Potential Anti-Inflammatory Effect on Human Osteoartritic Chondrocytes and Synoviocytes. Pharmaceutics. 2021;13:737. doi: 10.3390/pharmaceutics13050737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volpi N. Quality of different chondroitin sulfate preparations in relation to their therapeutic activity. Review. J. Pharm. Pharmacol. 2009;61:1271–1280. doi: 10.1211/jpp.61.10.0002. [DOI] [PubMed] [Google Scholar]
- 23.Hatano S., Watanabe H. Regulation of Macrophage and Dendritic Cell Function by Chondroitin Sulfate in Innate to Antigen-Specific Adaptive Immunity. Front. Immunol. 2020;11:232. doi: 10.3389/fimmu.2020.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volpi N. Chondroitin Sulfate Safety and Quality. Molecules. 2019;24:1447. doi: 10.3390/molecules24081447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Restaino O.F., De Rosa M., Cimini D., Schiraldi C. Manufacturing chondroitin sulfate: From animal source extraction to biotechnological production. In: Pomin V.H., editor. Chondroitin Sulfate: Structure, Uses and Health Implications. Nova Science; New York, NY, USA: 2013. pp. 41–56. [Google Scholar]
- 26.Restaino O.F., Finamore R., Diana P., Marseglia M., Vitiello M., Casillo A., Bedini E., Parrilli M., Corsaro M.M., Trifuoggi M., et al. A multi-analytical approach to better assess the keratin sulfate contamination in animal origin chondroitin sulfate. Anal. Chim. Acta. 2017;15:59–70. doi: 10.1016/j.aca.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Volpi N., Maccari F. Two analytical approaches to the evaluation of chondroitin sulfate in European food supplements. Sep. Sci. 2008;1:3–8. [Google Scholar]
- 28.Wang J., Guleria S., Koffas M.A.G., Yan Y. Microbial production of value-added nutraceuticals. Curr. Opin. Biotechnol. 2016;37:97–104. doi: 10.1016/j.copbio.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Laurent C., Hellström S. Extracellular matrix components reflect the dynamics of a healing tympanic membrane perforation—A histochemical study. Int. J. Biochem. Cell Biol. 1997;29:221–229. doi: 10.1016/S1357-2725(96)00135-5. [DOI] [PubMed] [Google Scholar]
- 30.Vassallo V., Stellavato A., Cimini D., Pirozzi A.V.A., Alfano A., Cammarota M., Balato G., D’Addona A., Ruosi C., Schiraldi C. Unsulfated biotechnological chondroitin by itself as well as in combination with high molecular weight hyaluronan improves the inflammation profile in osteoarthritis in vitro model. J. Cell. Biochem. 2021 doi: 10.1002/jcb.29907. ahead of print . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo R., Vassallo V., Stellavato A., Valletta M.A., Cimini D., Pedone P.V., Schiraldi C., Chambery A. Differential secretome profiling of human osteoarthritic synoviocytes treated with biotechnological unsulfated and marine sulfated chondroitins. Int. J. Mol. Sci. 2020;21:3746. doi: 10.3390/ijms21113746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiraldi C., Cimini D., De Rosa M. Production of chondroitin sulfate and chondroitin. Appl. Microbiol. Biotechnol. 2010;87:1209–1220. doi: 10.1007/s00253-010-2677-1. [DOI] [PubMed] [Google Scholar]
- 33.Cimini D., De Rosa M., Carlino E., Ruggiero A., Schiraldi C. Homologous overexpression of RfaH in E. coli K4 improves the production of chondroitin-like capsular polysaccharide. Microb. Cell Fact. 2013;12:46. doi: 10.1186/1475-2859-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agiba A.M. Nutraceutical formulations containing glucosamine and chondroitin sulphate in the treatment of osteoarthritis: Emphasis on clinical efficacy and formulation challenges. Int. J. Curr. Pharm. Res. 2017;2:1–7. doi: 10.22159/ijcpr.2017v9i2.17380. [DOI] [Google Scholar]
- 35.Stellavato A., Tirino V., de Novellis F., Della Vecchia A., Cinquegrani F., De Rosa M., Papaccio G.P., Schiraldi C. Biotechnological Chondroitin a Novel Glycosamminoglycan With Remarkable Biological Function on Human Primary Chondrocytes. J. Cell. Biochem. 2016;117:2158–2169. doi: 10.1002/jcb.25556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Gatta A., Stellavato A., Vassallo V., Di Meo C., Toro G., Iolascon G., Schiraldi C. Hyaluronan and Derivatives: An In vitro Multilevel Assessment of Their Potential in Visco supplementation. Polymers. 2021;13:3208. doi: 10.3390/polym13193208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severino V., Farina A., Chambery A. Analysis of secreted proteins. Methods Mol. Biol. 2013;1002:37–60. doi: 10.1007/978-1-62703-360-2_4. [DOI] [PubMed] [Google Scholar]
- 38.Lourido L., Calamia V., Mateos J., Fernandez-Puente P., Fernandez-Tajes J., Blanco F.J., Ruiz-Romero C. Quantitative proteomic profiling of human articular cartilage degradation in osteoarthritis. J. Proteome Res. 2014;13:6096–6106. doi: 10.1021/pr501024p. [DOI] [PubMed] [Google Scholar]
- 39.McInnes I.B., Schett G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 40.López-Reyes A., Medina-Luna D., Santamaría-Olmedo M., Martínez-Flores K., Zamudio-Cuevas Y., Fernández-Torres J., Martínez-Nava G.A., Olivos-Meza A., Camacho-Rea C., Fernández-Moreno M., et al. Soluble inflammatory mediators of synoviocytes stimulated by monosodium urate crystals induce the production of oxidative stress, pain, and inflammation mediators in chondrocytes: Secretome of synoviocytes induces chondrocyte damage. Clin. Rheumatol. 2021;40:3265–3271. doi: 10.1007/s10067-021-05676-w. [DOI] [PubMed] [Google Scholar]
- 41.Zheng L., Zhang Z., Sheng P., Mobasheri A. The role of metabolism in chondrocyte dysfunction and the progression of osteoarthritis. Ageing Res. Rev. 2021;66:101249. doi: 10.1016/j.arr.2020.101249. [DOI] [PubMed] [Google Scholar]
- 42.Imagawa K., de Andrés M.C., Hashimoto K., Pitt D., Itoi E., Goldring M.B., Roach H.I., Oreffo R.O. The epigenetic effect of glucosamine and a nuclear factor-kappa B (NFkB) inhibitor on primary human chondrocytes—Implications for osteoarthritis. Biochem. Biophys. Res. Commun. 2011;405:362–367. doi: 10.1016/j.bbrc.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jawed H., Anjum S., Awan S.I., Simjee S.U. Anti-arthritic effect of GN1, a novel synthetic analog of glucosamine, in the collagen-induced arthritis model in rats. Inflamm. Res. 2011;60:1113–1120. doi: 10.1007/s00011-011-0375-9. [DOI] [PubMed] [Google Scholar]
- 44.Simental-Mendía M., Sánchez-García A., Vilchez-Cavazos F., Acosta-Olivo C.A., Peña-Martínez V.M., Simental-Mendía L.E. Effect of glucosamine and chondroitin sulfate in symptomatic knee osteoarthritis: A systematic review and meta-analysis of randomized placebo-controlled trials. Rheumatol. Int. 2018;38:1413–1428. doi: 10.1007/s00296-018-4077-2. [DOI] [PubMed] [Google Scholar]
- 45.Pomin V.H., Vignovich W.P., Gonzales A.V., Vasconcelos A.A., Mulloy B. Galactosaminoglycans: Medical Applications and Drawbacks. Molecules. 2019;24:2803. doi: 10.3390/molecules24152803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vázquez J.A., Fraguas J., Novoa-Carvallal R., Reis R.L., Antelo L.T., Pérez-Martín R.I., Valcarcel J. Isolation and Chemical Characterization of Chondroitin Sulfate from Cartilage By-Products of Blackmouth Catshark (Galeus melastomus) Mar. Drugs. 2018;16:344. doi: 10.3390/md16100344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valcarcel J., Novoa-Carballal R., Pérez-Martín R.I., Reis R.L., Vázquez J.A. Glycosaminoglycans from marine sources as therapeutic agents. Biotechnol. Adv. 2017;35:711–725. doi: 10.1016/j.biotechadv.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Alessio N., Stellavato A., Aprile D., Cimini D., Vassallo V., Di Bernardo G., Galderisi U., Schiraldi C. Timely Supplementation of Hydrogels Containing Sulfated or Unsulfated Chondroitin and Hyaluronic Acid Affects Mesenchymal Stromal Cells Commitment Toward Chondrogenic Differentiation. Front. Cell Dev. Biol. 2021;9:641529. doi: 10.3389/fcell.2021.641529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vassallo V., Tsianaka A., Alessio N., Grübel J., Cammarota M., Tovar G.E.M., Southan A., Schiraldi C. Evaluation of novel biomaterials for cartilage regeneration based on gelatin methacryloyl interpenetrated with extractive chondroitin sulfate or unsulfated biotechnological chondroitin. J. Biomed. Mater. Res. A. 2022 doi: 10.1002/jbm.a.37364. ahead of print . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doss F., Menard J., Hauschild M., Kreutzer H.J., Mittlmeier T., Müller-Steinhardt M., Müller B. Elevated IL-6 levels in the synovial fluid of osteoarthritis patients stem from plasma cells. Scand. J. Rheumatol. 2007;36:136–139. doi: 10.1080/03009740701250785. [DOI] [PubMed] [Google Scholar]
- 51.Matsushima K., Oppenheim J.J. Interleukin 8 and MCAF: Novel inflammatory cytokines inducible by IL 1 and TNF. Cytokine. 1989;1:2–13. doi: 10.1016/1043-4666(89)91043-0. [DOI] [PubMed] [Google Scholar]
- 52.Tak P.P., Firestein G.S. NF-kappaB: A key role in inflammatory diseases. Clin. Investig. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Padeh S., Farzam N., Chayen G., Gerstein M., Berkun Y. Pentraxin 3 is a marker of early joint inflammation in patients with juvenile idiopathic arthritis. Immunol. Res. 2013;56:444–450. doi: 10.1007/s12026-013-8417-8. [DOI] [PubMed] [Google Scholar]
- 54.Sun K., Luo J., Guo J., Yao X., Jing X., Guo F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: A narrative review. Osteoarthr. Cartil. 2020;28:400–409. doi: 10.1016/j.joca.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 55.Arpino V., Brock M., Gill S.E. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 2015;44:247–254. doi: 10.1016/j.matbio.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Jiang Y., Xiao Q., Hu Z., Pu B., Shu J., Yang Q., Lao H., Hao J. Tissue levels of leukemia inhibitory factor vary by osteoarthritis grade. Orthopedics. 2014;37:e460–e464. doi: 10.3928/01477447-20140430-57. [DOI] [PubMed] [Google Scholar]
- 57.Ran D., Zheng-Zhao L. The Role of Autophagy in Osteoarthritis. Front. Cell Dev. 2020;8:608388. doi: 10.3389/fcell.2020.608388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miceli M., Dell’Aversana C., Russo R., Rega C., Cupelli L., Ruvo M., Altucci L., Chambery A. Secretome profiling of cytokines and growth factors reveals that neuro-glial differentiation is associated with the down-regulation of Chemokine Ligand 2 (MCP-1/CCL2) in amniotic fluid derived-mesenchymal progenitor cells. Proteomics. 2016;16:674–688. doi: 10.1002/pmic.201500223. [DOI] [PubMed] [Google Scholar]
- 59.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 60.Stellavato A., Abate L., Vassallo V., Donniacuo M., Rinaldi B., Schiraldi C. An in vitro study to assess the effect of hyaluronan-based gels on muscle-derived cells: Highlighting a new perspective in regenerative medicine. PLoS ONE. 2020;15:e0236164. doi: 10.1371/journal.pone.0236164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russo R., Matrone N., Belli V., Ciardiello D., Valletta M., Esposito S., Pedone P.V., Ciardiello F., Troiani T., Chambery A. Macrophage Migration Inhibitory Factor Is a Molecular Determinant of the Anti-EGFR Monoclonal Antibody Cetuximab Resistance in Human Colorectal Cancer Cells. Cancers. 2019;11:1430. doi: 10.3390/cancers11101430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bindea G., Galon J., Mlecnik B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cimini D., Russo R., D’Ambrosio S., Dello Iacono I., Rega C., Carlino E., Argenzio O., Russo L., D’Abrosca B., Chambery A., et al. Physiological characterization and quantitative proteomic analyses of metabolically engineered E. coli K4 strains with improved pathways for capsular polysaccharide biosynthesis. Biotechnol. Bioeng. 2018;115:1801–1814. doi: 10.1002/bit.26597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data critical for reader understanding and outcomes’ discussion are reported within the manuscript or as part of the Supplementary Materials. Additional raw data are available from the corresponding author upon request.