Abstract
A small percentage of natural Escherichia coli isolates (both commensal and pathogenic) have a mutator phenotype related to defects in methyl-directed mismatch repair (MR) genes. We investigated whether there was a direct link between the mutator phenotype and virulence by (i) studying the relationships between mutation rate and virulence in a mouse model of extraintestinal virulence for 88 commensal and extraintestinal pathogenic E. coli isolates and (ii) comparing the virulence in mice of MR-deficient and MR-proficient strains that were otherwise isogenic. The results provide no support for the hypothesis that the mutator phenotype has a direct role in virulence or is associated with increased virulence. Most of the natural mutator strains studied displayed an unusual virulence phenotype with (i) a lack of correspondence between the number of virulence determinants and pathogenicity in mice and (ii) an intermediate level of virulence. On a large evolutionary scale, the mutator phenotype may help parasites to achieve an intermediate rate of virulence which mathematical models predict to be selected for during long-term parasite-host interactions.
Natural bacterial isolates of Escherichia coli and Salmonella enterica have recently been shown to have highly variable mutation rates (15, 18). A small percentage of the natural strains studied were strong mutators (i.e., they have a frequency of mutation 50 to 100 times higher than that of the remaining strains). Most of these strong mutator strains are defective in the methyl-directed mismatch repair (MR) genes (mutS, mutL, mutH, and mutU). The MR system increases the fidelity of replication by correcting DNA biosynthetic errors such as base mismatches and insertion-deletions of up to four bases and inhibits recombination between nonidentical DNA sequences (27).
One of the main issues raised by these data is the possibility that greater genetic variability is associated with pathogenesis. Indeed, the mutator phenotype may enable bacteria to adapt to new niches and to escape immune surveillance by generating numerous mutations (20, 21). In addition, if the mutator phenotype is related to inactivation of the MR system, it may enable bacteria to acquire additional elements such as pathogenicity islands more easily by horizontal transfer (23). MR-defective strains have been identified among commensal and pathogenic (intestinal and extraintestinal) strains (15, 18), but (i) the numbers of strains studied are too small to demonstrate a significantly higher frequency of mutators in pathogenic isolates and (ii) it is difficult to distinguish between commensal and pathogenic strains because enteric E. coli isolated in commensal situations may be the natural reservoir of pathogenic strains (6). Thus, further studies are clearly needed to determine the specific role, if any, of the mutator phenotype in E. coli pathogenesis (30).
In the present study we used two approaches to explore the possible link between mutator phenotype and virulence. First, we studied the relationships between mutation rate and intrinsic virulence for a collection of 88 commensal and extraintestinal pathogenic E. coli isolates. Virulence was assessed by searching for known virulence determinants and by assessing the lethality in a normalized mouse model for extraintestinal virulence (26). Two natural MR-defective isolates from this collection were complemented for the defective MR gene, and their virulence in mice was compared with that of the corresponding isogenic MR-deficient natural isolates.
MATERIALS AND METHODS
Bacterial strains.
Two subsets of E. coli strains were analyzed in this work. First, we studied the virulence in mice of six strong mutator strains: five E. coli strains (TIM28, SA2077, SA1902, M13, and SA1923) from a collection of 504 commensal and pathogenic natural isolates (18) and the ECOR 48 strain from the E. coli reference (ECOR) collection (24). The ECOR collection consists of 72 natural isolates representing the genetic diversity of the species. The rate of mutations to antibiotic resistance of these two collections (the 504 commensal and pathogenic natural isolates and the ECOR collection) have been reported elsewhere (15, 18). The six selected strains correspond to all of the identified strong mutator strains within these two collections. Strong mutators were defined as having a frequency of mutation to rifampin resistance higher than 50 times the median value of the corresponding collection (15, 18), i.e., 7.1 × 10−9 and 7.3 × 10−9, respectively. All six strains are generalized mutators and harbor defects in MR genes identified by complementation with plasmid-borne wild-type MR genes (15, 18) and/or Southern blotting (for mutS mutant strains due to large mutS deletions) (Table 1). M13 and SA1923 strains (mutS-null genotype) were transformed with pBR322 plasmid and pBA40 plasmid, a pBR322 derivative with a wild-type E. coli mutS insert. Both plasmids confer resistance to ampicillin. The origins, frequencies of mutations to rifampin resistance, and genotypes of the strains are shown in Table 1. The E. coli phylogenetic groups (11) to which the strains belong and the presence of virulence determinants were determined as described previously (26). Briefly, the K1 capsular type was determined phenotypically, and PCR was used to detect the remaining virulence determinants (eae, sfa/foc, pap, afa, hly, cnf, aer, and ibe10) (26). The primers used to detect cnf were CNF (5′-CAGTGACCGGATCTCCGTTAT-3′) and CNFREV (5′-CGTGTAATTCTTCTGTACTTCC-3′).
TABLE 1.
Characteristics of the natural mutator E. coli isolates and their derivative strains
| Strain | Origin | Rifra | mut genotype | Phylogenetic groupb | Presence (+) or absence (−) of extraintestinal virulence determinants:
|
Lethality in micec (%) | Virulence phenotyped | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K1 antigen | sfa/foc | pap | afa | hly | cnf | aer | ibe10 | Total no. | |||||||
| TIM28 | Pus | 5.9 × 10−6 | mutL mutant | A | − | − | − | − | − | − | − | − | 0 | 0/20 (0) | Typical |
| IAI77 | UTIe | 2.62 × 10−7 | NDf | B2 | − | + | + | − | + | + | − | − | 4 | 26/50 (52) | Atypical |
| SA2077 | UTI | 5 × 10−7 | mutL mutant | A | − | − | − | + | − | − | − | − | 1 | 11/20 (55) | Atypical |
| ECOR 48 | UTI | 8.8 × 10−7 | mutS mutant | D | + | − | + | − | + | + | − | − | 4 | 3/20 (15) | Atypical |
| IAI68 | UTI | 5 × 10−7 | ND | B2 | − | + | + | − | − | − | + | − | 3 | 26/50 (52) | Atypical |
| SA1902 | UTI | 3.25 × 10−6 | mutS mutant | B2 | + | − | − | − | − | − | + | + | 3 | 1/20 (5) | Atypical |
| M13 | Feces | 6.6 × 10−7 | mutS mutant | B1 | − | − | − | − | − | − | − | − | 0 | 5/20 (25) | Atypical |
| M13/pBR322 | This study | 9 × 10−7 | mutS mutant | 22/52 (42) | |||||||||||
| M13/pBA40 | This study | 3.4 × 10−8 | mutS+ | 27/52 (52) | |||||||||||
| SA1923 | UTI | 4.7 × 10−6 | mutS mutant | D | − | + | + | − | + | + | − | + | 5 | 0/20 (0) | Atypical |
| SA1923/pBR322 | This study | 1.57 × 10−6 | mutS mutant | 0/10 (0) | |||||||||||
| SA1923/pBA40 | This study | 2.73 × 10−8 | mutS+ | 0/10 (0) | |||||||||||
Frequency of mutations to rifampin resistance.
Phylogenetic groups are indicated as given in Herzer et al. (11).
Lethality in mice is indicated as the number of mice killed/number of inoculated mice and as the percentage of inoculated mice that died.
Atypical virulence phenotype is defined as strains with no to one virulence determinant that kill mice or as strains with three to five virulence determinants that are avirulent or kill mice at an intermediate level.
UTI, urinary tract infection.
ND, not determined.
Second, we determined the rate of mutations to rifampin resistance for a collection of 15 commensal and 67 miscellaneous extraintestinal pathogenic strains (26). These 82 strains have been previously studied, and their phylogenetic groups within E. coli, the presence of virulence determinants, and their virulence in mice were determined (26). A selected set of strains from this collection has been reanalyzed for virulence in mice (see below).
Animal model.
Animal experiments were performed following the guidelines of the Université de Bretagne. Virulence in mice was tested in this work for the six strong mutator strains from the 504 natural isolates (18) and the ECOR collection (15), for their derivative strains, and for a selected set of strains from the 82-strain collection (26). Pathogen-free females of the outbred white Swiss mice lineage (Rj:Swiss-IOPS Orl) (6 to 8 weeks old, 25 to 30 g) were purchased from the Centre d'Elevage R. Janvier (Le Genest, Saint Isle, France). E. coli strains were cultured in Trypticase soy broth medium. For each strain, at least one series of 10 mice was inoculated with (per mouse) 108 CFU in 0.2 ml of Ringer solution, subcutaneously, on the abdomen, as previously described (26). Strains harboring a plasmid (pBR322 or pBA40) were grown in Trypticase soy broth medium with ampicillin (50 μg/ml). Mice were observed daily for up to 1 week after inoculation. Death and the time to death were recorded for each mouse. Dead animals were dissected, and bacterial populations from the liver, spleen, and kidney of each animal were counted on Trypticase soy plates as previously described (26) for the M13 derivative strains. For mice infected with strains harboring a plasmid, tissues were used to inoculate Trypticase soy plates with or without ampicillin (50 μg/ml) to obtain comparative bacterial counts. In this mouse model, lethality is a clearcut parameter. In a previous study, 84.2% of the strains that killed mice killed 9 or all 10 of the inoculated mice (26). Strains killing 9 or 10 mice were classified as highly virulent, whereas strains killing no mice or only 1 mouse were classified as avirulent. Only 6% of the strains killed two to eight mice (26), and these were classified as intermediate. The mouse virulence phenotype was defined by the number of virulence determinants and the number of mice killed. It has been shown that the number of mice killed is directly proportional to the number of virulence determinants detected in the strain (12, 26). Thus, strains exhibit a typical virulence phenotype if they possess three to five of the tested virulence determinants and are highly virulent or if they possess none or one of the tested virulence determinants and are avirulent. Strains that killed mice but that had none or only one of the virulence determinants tested or that were not highly virulent despite possessing three to five virulence determinants were considered to have an atypical virulence phenotype. Strains with two of the virulence determinants tested were equally likely to be avirulent or highly virulent (26).
Mutation rate.
The frequency of mutagenesis to rifampin resistance (Rifr) was determined in this work for the strains from the collection of 82 commensal and extraintestinal pathogenic strains (26), as previously described (29). A total of 102 to 103 cells from an overnight culture were inoculated on nitrocellulose filters (NC45; Schleicher & Schuell) laid on fresh 869 plates (5 g of NaCl, 10 g of Bacto Tryptone, 5 g of yeast extract, and 15 g of agar per liter). Plates were incubated at 37°C for 24 h. The cells were resuspended in 1 ml of 869 medium and incubated for 1 h at 37°C to allow rifampin resistance to be expressed. Appropriate dilutions were then spread on 869 plates and 869-rifampin (100 μg/ml) plates. The Rifr mutants were scored after incubation for 24 h at 37°C. The frequency of mutation was calculated from four to six independent cultures. The mutation rate was then analyzed both dichotomously (strong mutator versus nonmutator) and as a continuous variable.
Statistical analysis.
The statistical significance of differences between the groups was determined using the χ2 test or the unpaired Student t test.
RESULTS
Characteristics of the six previously identified MR-deficient natural E. coli isolates.
Six previously identified MR-deficient strains (TIM28, SA2077, ECOR48, SA1902, M13, and SA1923) are distributed among the E. coli phylogenetic groups. Strains TIM28, SA2077, and M13 have none or one of the virulence determinants studied, whereas the remaining strains each have more than three virulence determinants (Table 1). The eaeA determinant was not detected in any of the strains studied. The presence of the sfa/foc and/or hly-cnf determinants in strains of the D phylogenetic group (ECOR 48 and SA1923) is striking, since these determinants were previously reported almost exclusively in strains of the B2 phylogenetic group (2, 3, 26).
Lethality in mice of the six previously identified natural MR-deficient E. coli isolates.
We first tested the virulence of MR-deficient natural isolates by assessing their lethality in 10 mice as previously described (26). Three of the six strains (SA2077, ECOR 48, and M13) killed mice, but their virulence level was intermediate, since they each killed only three (ECOR 48) or four (SA2077 and M13) mice. The remaining strains (TIM28, SA1902, and SA1923) did not kill any mice.
Mutation rates of the 82 commensal and pathogenic E. coli strains previously tested for virulence in mice.
Only two strains, IAI77 and IAI68, were strong mutators (Table 1) according the criterion of a frequency of mutations to rifampin resistance higher than 50 times the median value for the collection (5.1 × 10−9). These two strains showed, compared to nonmutator strains, an increase of mutation frequency for several additional genomic targets (nalidixic acid, spectinomycin, streptomycin, phosphomycin resistances, and lacI inactivation) (data not shown). The mutator genes involved in the mutator phenotype of these strains have not been identified (Table 1). The other strains had a wide range of mutation frequencies to rifampin resistance, as previously reported (reference 18 and see below).
Relationships between the virulence phenotype in mice and the mutation rate in the 88 natural E. coli isolates.
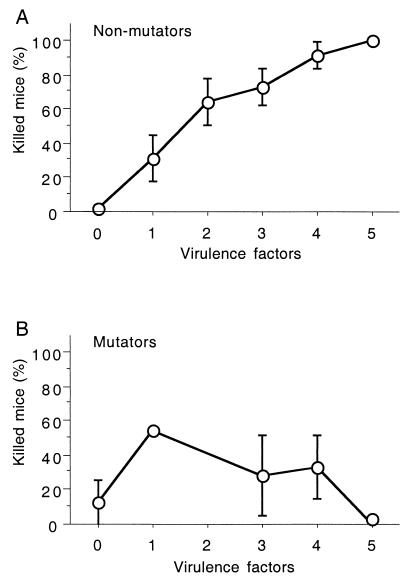
We studied the relationship between virulence phenotype and mutation rate by analyzing both the 82 commensal and pathogenic strains previously tested for virulence in mice and the 6 previously identified MR-deficient natural isolates. These 88 strains consisted of 80 nonmutator strains and 8 strong mutator strains (the 6 previously identified natural MR-deficient isolates and the 2 strong mutators identified in this work from the 82-strain collection [26], IAI68 and IAI77). To avoid any bias in comparing these two collections, we first randomly selected 6 strains among the 82-strain collection and reanalyzed them in the mouse model of virulence. These six strains, belonging to each virulence category (avirulent, intermediate, and highly virulent) were inoculated into series of 10 mice except for a highly virulent strain which had been inoculated to 30 mice. No significant difference was observed in the number of killed mice compared to the published data (26) (data not shown). Second, all of the eight mutator strains were retested to analyze each strain in 20 (TIM28, SA2077, ECOR 48, SA1902, M13, and SA1923) to 50 (IAI77 and IAI68) inoculated mice. A certain level of variability was observed in the number of killed mice (Table 1). However, considering the mean number of killed mice, only one strain, IAI77, which has been tested in a previous study for virulence in mice (26), showed a change in the virulence category (from highly virulent to intermediate) and in the virulence phenotype (from typical to atypical) (Table 1). This strain was later considered to have an intermediate level of virulence and an atypical virulence phenotype. Such a variability can be related to the mutator phenotype, which has been shown to increase loss of functions and fitness during in vitro passages (7). As previously reported (12), for the 80 nonmutator strains of the collection a correlation was found between the number of virulence determinants of the eight studied: K1 antigen; sfa/foc, pap, afa, hly, cnf, aer, and ibe10 genes; and the number of mice killed (Fig. 1A). In contrast, no such correlation was observed for the eight strong mutator strains, indicating unusual behavior of these strains in the mouse model of extraintestinal virulence (Fig. 1B). Of 43 nonmutator strains with none or one of the virulence determinants tested, 38 (88.3%) were avirulent, whereas 19 (82.6%) of 23 nonmutator strains with three to five of the virulence determinants tested were highly virulent. Two of the three strong mutator strains (SA2077 and M13) with none or one of the virulence determinants tested killed mice at an intermediate level of virulence, and all five of five strong mutator strains (IAI77, ECOR48, IAI68, SA1902, and SA1923) with three to five virulence determinants were found to be avirulent or intermediate (Table 2). Overall, 9 of the 66 nonmutator strains (13.6%) and 7 of the 8 strong mutator strains (87.5%) had an atypical virulence phenotype in mice (P < 0.0001). The numbers of nonmutator and mutator strains with atypical virulence phenotypes were also significantly different when the data were stratified by number of virulence determinants (Table 2).
FIG. 1.
Comparison of the percentage of inoculated mice (10 to 50) that died and the number of virulence determinants (K1 antigen and sfa/foc, pap, afa, hly, cnf, aer, and ibe10 genes) per strain for the 80 nonmutator (A) and 8 mutator (B) natural E. coli isolates. The points refer to the mean values for all strains in the group with the indicated number of virulence determinants. The numbers of strains at each point in the curves for virulence determinants 0 to 5 are as follows: 32, 11, 14, 11, 9, and 3 (A) and 2, 1, 0, 2, 2, and 1 (B). Standard error bars are also presented. The absence of the error bar in panel A reflects an absence of variation, whereas in panel B it reflects the presence of only one studied strain.
TABLE 2.
Virulence in natural nonmutator and strong mutator isolates
| No. of virulence determinants/ straina | No. of strains in each mouse virulence groupb
|
No. of strains (%) with atypical virulence phenotypec
|
||||||
|---|---|---|---|---|---|---|---|---|
| Avirulent
|
Intermediate
|
Highly virulent
|
||||||
| Nonmutator | Mutator | Nonmutator | Mutator | Nonmutator | Mutator | Nonmutator | Mutator | |
| 0–1 | 38 | 1 | 2 | 2 | 3 | 0 | 5 (12) | 2 (66)d |
| 3–5 | 2 | 2 | 2 | 3 | 19 | 0 | 4 (17) | 5 (100)d |
Strains with two virulence determinants were not taken into account since they are equally distributed between the avirulent and highly virulent mouse virulence groups (see Materials and Methods).
The three mouse virulence groups were defined as follows: avirulent group (0 to 1 mouse killed out of 10), intermediate group (2 to 8 mice killed out of 10), and highly virulent group (9 to 10 mice killed out of 10).
Atypical virulence phenotype is defined as strains with none or one virulence determinant that kill mice or strains with three to five virulence determinants that are avirulent or kill mice at an intermediate level.
Significantly different (P = 0.01 and 0.0003 for strains with 0 to 1 and 3 to 5 virulence determinants, respectively).
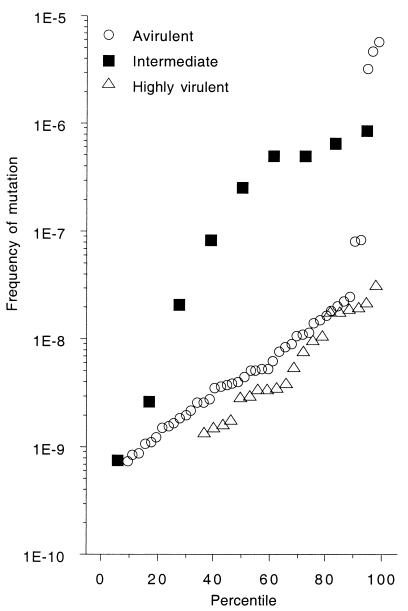
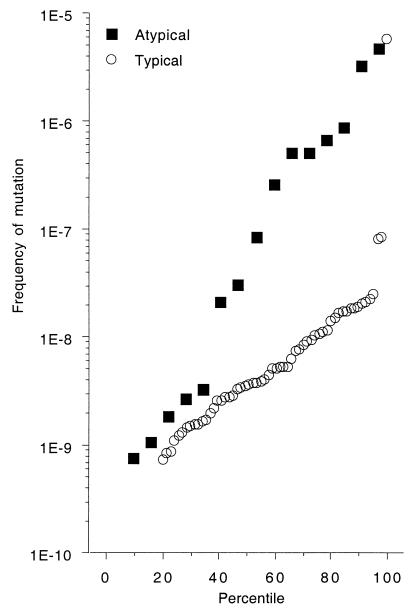
In terms of raw mutation rates without a threshold, no significant difference in mutation rate was observed between highly virulent and avirulent strains. In contrast, strains with an intermediate level of virulence had a significantly higher mutation rate than the other strains (P = 0.0033) (Fig. 2). Similarly, strains with atypical virulence phenotypes had significantly higher mutation rates than strains with typical phenotypes (P < 0.0001) (Fig. 3). Thus, mutator strains had an unusual virulence phenotype involving, in particular, an intermediate level of virulence.
FIG. 2.
Frequency of mutations to rifampin resistance in 88 natural E. coli isolates classified in a mouse model of extraintestinal virulence (26) as highly virulent (9 or 10 mice killed of a total of 10), intermediate (2 to 8 mice killed of a total of 10) or avirulent (0 or 1 mouse killed of a total of 10). The eight strong mutator strains are above the threshold of 2 × 10−7 for the frequency of mutations to rifampin resistance. The three avirulent strains, which exhibit the highest mutation rates of all, encompass one strain (TIM28) devoid of virulence determinants and two strains (SA1923 and SA1902) with numerous virulence determinants.
FIG. 3.
Frequency of mutations to rifampin resistance in 88 natural E. coli isolates classified according to their virulence phenotype in a mouse model of extraintestinal virulence (26) as atypical or typical. See Material and Methods for the definition of these two categories. The eight strong mutator strains are above the threshold of 2 × 10−7 for the frequency of mutations to rifampin.
Comparison of the virulence in mice of MR-deficient and MR-proficient strains.
Two MR-deficient (mutS mutant) strains, one avirulent in mice (SA1923) and the other lethal to mice (M13), were further studied. We used the plasmid-bearing strains (M13/pBR322/pBA40 and SA1923/pBR322/pBA40) and carried out three preliminary experiments. (i) We showed that the lethality of the two strains bearing the plasmid without the mutS insert (pBR322) in 10 mice was similar to that of the natural isolates. (ii) We showed that the strains did not lose the plasmid during growth in the mice by comparing bacterial counts in the organs of killed mice obtained by inoculating Trypticase soy plates with or without ampicillin. (iii) We checked that the strains retained the same phenotype (mutator or nonmutator) after the passage in mice by determining the rifampin mutation rate.
Neither SA1923/pBR322 (mutS mutant) nor SA1923/pBA40 (mutS+) killed mice among the 10 inoculated for each strain (Table 1). Thus, the restoration of a MR-proficient phenotype did not significantly increase virulence.
For the M13 strain, initial experiments showed that the MR-deficient strain killed mice at a low level. We investigated whether the restoration of an MR-proficient phenotype increased or decreased the level of virulence by inoculating two series of 52 mice with 108 CFU of the M13/pBR322 or the M13/pBA40 strain. No significant difference was observed between the two transformed strains as 42 and 52% of the mice were killed by the mutS mutant (M13/pBR322) and wild-type (M13/pBA40) strains, respectively (Table 1). Moreover, the mean bacterial counts determined for the organs of 10 killed mice for each strain were very similar (4 × 107 and 9.2 × 107 CFU/ml for the M13/pBR322 and M13/pBA40 strains, respectively.
In order to determine if the virulence could be differentially increased by experimental infection, three individual colonies per strain were subcultured from the liver of a mouse killed after 1 day of infection with the original M13 strain. The three isolates per strain were tested on three series of 40, 20, and 20 mice. No significant difference in lethality was observed between the two strains since 66 and 46% of the mice were killed by the mutS mutant (M13/pBR322) and wild-type (M13/pBA40) strains, respectively.
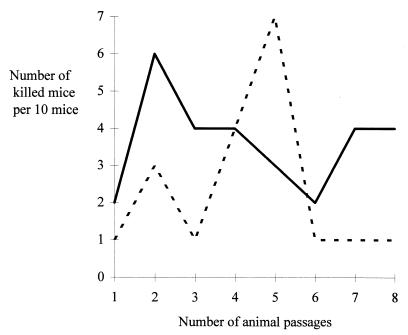
Finally, we investigated the possibility of a differential increase in the virulence of strains over serial animal passages. Ten colonies isolated from mice killed after 1 day of infection were independently subcultured in Trypticase soy broth medium and pooled to obtain 108 CFU for the inoculation of 10 mice. The process was repeated seven times. The number of killed mice was recorded for each passage (Fig. 4). No stable increase in virulence was observed with serial animal passages for either of the strains.
FIG. 4.
Schematic representation of lethality in mice after serial in vivo passages between MR-deficient [M13(pBR322) mutS mutant; dashed line] and MR-proficient [M13(pBA40) mutS wild type; unbroken line] M13 strains.
DISCUSSION
The data presented here suggest that increased mutagenesis does not confer increased virulence and show that the mutator phenotype cannot itself be considered as a virulence factor. Strains with a high mutation rate and several virulence determinants are significantly less virulent than strains with the same number of virulence determinants but with a low mutation rate (Fig. 1). These results are consistent with recent studies, which showed that inactivation of mutS or mutL in S. enterica did not increase virulence as evaluated by the amount of inoculum required to kill 50% of infected mice (4, 10, 32) or the time until the death of the animal (32). However, mutator strains may have an indirect advantage in the presence of antibiotics since it has been shown that the course of infections caused by the mutator strains is unaffected by the presence of a quinolone, whereas with the wild-type strain, antibiotic treatment extends the survival of the mice (D. S. Thaler, personal communication). Indeed, mutator Pseudomonas aeruginosa strains are frequently isolated during chronic lung infections from cystic fibrosis patients who have been repeatedly treated by various antibiotics during several years (ca. 20% of mutators) compared to P. aeruginosa septicemia strains (0% of mutators) (25).
Although a strain with numerous virulence determinants was still avirulent for the mice after MR complementation and no increase in virulence was observed in a mutator virulent strain, even after serial in vivo passages, our experiments correspond to ca. 10 to 300 generations in the animal, a very brief time scale with respect to the evolutionary fate of mutator strains. In nature and on a larger evolutionary scale, the greater genomic variability of MR-deficient strains may enable avirulent commensal strains to become pathogenic (30) by acquiring virulence determinants (9), deleting stretches of DNA (19), or adapting existing functions to this end (22, 28). In addition, pathogenic bacterial cells can evade the host immune system by lipopolysaccharide antigenic variation (22). Most of these gene modifications are associated with the presence of repeated sequences and involve microsatellite instability or homeologous recombination, both of which are increased by MR defects (27).
The principal characteristic of mutator strains, compared to nonmutator strains, that we observed was their atypical behavior in the mouse model with respect to their virulence determinants. Five of the seven natural mutator strains with an atypical virulence phenotype killed mice at an intermediate level (Table 1). The low number of mice killed by the M13 and SA2077 isolates, which are devoid of the virulence determinants tested, may be indicative of strains in the process of becoming virulent. Similarly, the detection of numerous virulence determinants in D strains such as SA1923 and ECOR 48, some of which were considered specific to the B2 phylogenetic group (sfa/foc and hly-cnf), may indicate acquisition by horizontal transfer from B2 group strains as has previously been suggested (2, 3). However, these determinants may be inefficient or suboptimal because they are not yet in an appropriate genetic background. Alternatively, higher levels of mutagenesis may attenuate the virulence of a strain. This may explain why the SA1902, IAI77, and IAI68 mutator strains, which belong to the B2 phylogenetic group and which possess numerous virulence determinants, are not highly virulent in mice. It may also be an alternative explanation for the absence or low level of virulence of the SA1923 and ECOR 48 strains. Mutagenesis may affect the virulence genes themselves or the “fine-tuning” of the interaction between genetic background and virulence genes (7). Serial-passage experiments select strains with high levels of virulence (5), but conditions may exist in nature in which there is an advantage for bacteria to be commensal or slightly virulent and to avoid debilitating or killing their hosts (17). Indeed, recent mathematical models showed that maximum transmission is obtained by parasites with intermediate rates of growth or virulence (1, 16). Mutator phenotype may facilitate conversion to this intermediate status by increasing the rate at which rare adaptive mutations are generated. Physiological conditions for the development of virulence are far more complex than our simple model since bacteria have opportunities to acquire genes by horizontal transfer from other bacteria. The development of more sophisticated models using mixed infections or numerous in vivo passages will be useful.
This work should also be considered in light of the finding that some cancers, such as human hereditary nonpolyposis colorectal cancer (HNPCC), result from defects in the human counterparts of the bacterial MR genes and that such defects create a mutator phenotype (14). The mutator phenotype is not itself oncogenic, in the same way that MR genes are not themselves virulence genes, but instead it increases the probability of mutations in other genes that would otherwise occur at low frequency (13). Interestingly, it has been shown that patients with HNPCC have better prognoses than those with sporadic colorectal carcinoma and that they have a lower probability of metastasis (8, 31). This situation may be considered to be equivalent to the intermediate level of virulence in the mouse model of most of the E. coli mutator strains and may account for the genetic burden of the mutator phenotype.
ACKNOWLEDGMENTS
We thank Christine Amorin for skillful technical assistance. We are grateful to Miroslav Radman and Jacques Elion for constant encouragement and to Patricia Escobar-Páramo, Pilar Francino, and Justin Courcelle for the editing of the manuscript. We acknowledge an anonymous reviewer for his help in improving the manuscript.
Grant support was provided by the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires.
REFERENCES
- 1.Antia R, Lipsitch M. Mathematical models of parasite responses to host immune defences. Parasitology. 1997;115:S155–S167. doi: 10.1017/s003118209700200x. [DOI] [PubMed] [Google Scholar]
- 2.Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998;177:642–650. doi: 10.1086/514217. [DOI] [PubMed] [Google Scholar]
- 3.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1988;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campoy S, Pérez de Rozas A M, Barbé J, Badiola I. Virulence and mutation rates of Salmonella typhimurium strains with increased mutagenic strength in a mouse model. FEMS Microbiol Lett. 2000;187:145–150. doi: 10.1111/j.1574-6968.2000.tb09151.x. [DOI] [PubMed] [Google Scholar]
- 5.Ebert D. Experimental evolution of parasites. Science. 1998;282:1432–1435. doi: 10.1126/science.282.5393.1432. [DOI] [PubMed] [Google Scholar]
- 6.Eisenstein B I. Enterobacteriaceae. In: Mandell G L, Douglas R G, Bennet J E, editors. Principles and practice of infectious disease. 3rd ed. New York, N.Y: Churchill Livingstone; 1990. pp. 1658–1673. [Google Scholar]
- 7.Funchain P, Yeung A, Stewart J L, Lin R, Slupska M M, Miller J H. The consequence of growth of a mutator strain of Escherichia coli as measured by loss of functions among multiple gene targets and loss of fitness. Genetics. 2000;154:959–970. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gryfe R, Kim H, Hsieh E T, Aronson M D, Holowaty E J, Bull S B, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 9.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 10.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 11.Herzer P J, Inouye S, Inouye M, Whittman T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson J R, Kuskowski M. Clonal origin, virulence factors, and virulence. Infect Immun. 2000;68:424–425. doi: 10.1128/iai.68.1.424-425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinzler K W, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 14.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, Guan X Y, Zhang J, Metzer P S, Yu J W, Kao F T, Chen D J, Cerosaletti K M, Fournier R E K, Todd S, Lewis T, Leach R J, Naylor S L, Weissenbach J, Mecklin J P, Järvinen H, Petersen G M, Hamilton S R, Green J, Jass J, Watson P, Lynch H T, Trent J M, de la Chapelle A, Kinzler K W, Vogelstein B. Mutations of mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 15.LeClerc J E, Baouguang L, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 16.Lenski R E, May R M. The evolution of virulence in parasites and pathogens: reconciliation between two competing hypotheses. J Theor Biol. 1994;169:253–265. doi: 10.1006/jtbi.1994.1146. [DOI] [PubMed] [Google Scholar]
- 17.Lipsitch M, Moxon E R. Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 1997;5:31–37. doi: 10.1016/S0966-842X(97)81772-6. [DOI] [PubMed] [Google Scholar]
- 18.Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- 19.Maurelli A T, Fernandez R E, Bloch C A, Rod C K, Fasano A. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metzgar D, Wills C. Evidence for the adaptive evolution of mutation rates. Cell. 2000;101:581–584. doi: 10.1016/s0092-8674(00)80869-7. [DOI] [PubMed] [Google Scholar]
- 21.Moxon E R, Thaler D S. The tinkerer's evolving tool-box. Nature. 1997;387:659–662. doi: 10.1038/42607. [DOI] [PubMed] [Google Scholar]
- 22.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 23.Mühldorfer I, Hacker J. Genetic aspects of Escherichia coli virulence. Microb Pathog. 1994;16:171–181. doi: 10.1006/mpat.1994.1018. [DOI] [PubMed] [Google Scholar]
- 24.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural population. J Bacteriol. 1984;157:690–692. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver A, Cantŏn R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 26.Picard B, Sevali Garcia J, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radman M, Matic I, Halliday J A, Taddei F. Editing DNA replication and recombination by mismatch repair: from bacterial genetics to mechanisms of predisposition to cancer in humans. Philos Trans R Soc Lond B. 1995;347:97–103. doi: 10.1098/rstb.1995.0015. [DOI] [PubMed] [Google Scholar]
- 28.Sokurenko E V, Chesnokova V, Dykhuizen D E, Ofek I, Wu I, X R, Krogfelt K A, Struve C, Schembri M A, Hasty D L. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taddei F, Matic I, Radman M. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc Natl Acad Sci USA. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taddei F, Matic I, Godelle B, Radman M. To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol. 1997;5:427–429. doi: 10.1016/S0966-842X(97)01157-8. [DOI] [PubMed] [Google Scholar]
- 31.Watson P, Liu K M, Rodriguez-Bigas M A, Smyrk T, Lemon S, Shashidharan M, Franklin B, Karr B, Thorson A, Lynch H T. Colorectal carcinoma survival among hereditary nonpolyposis colorectal carcinoma family members. Cancer. 1998;83:259–266. [PubMed] [Google Scholar]
- 32.Zahrt T C, Buchmeir N, Maloy S. Effect of mutS and recD mutations on Salmonella virulence. Infect Immun. 1999;67:6168–6172. doi: 10.1128/iai.67.11.6168-6172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]