Abstract
Burkholderia pseudomallei, the etiologic agent of melioidosis, is responsible for a broad spectrum of illnesses in humans and animals particularly in Southeast Asia and northern Australia, where it is endemic. Burkholderia thailandensis is a nonpathogenic environmental organism closely related to B. pseudomallei. Subtractive hybridization was carried out between these two species to identify genes encoding virulence determinants in B. pseudomallei. Screening of the subtraction library revealed A-T-rich DNA sequences unique to B. pseudomallei, suggesting they may have been acquired by horizontal transfer. One of the subtraction clones, pDD1015, encoded a protein with homology to a glycosyltransferase from Pseudomonas aeruginosa. This gene was insertionally inactivated in wild-type B. pseudomallei to create SR1015. It was determined by enzyme-linked immunosorbent assay and immunoelectron microscopy that the inactivated gene was involved in the production of a major surface polysaccharide. The 50% lethal dose (LD50) for wild-type B. pseudomallei is <10 CFU; the LD50 for SR1015 was determined to be 3.5 × 105 CFU, similar to that of B. thailandensis (6.8 × 105 CFU). DNA sequencing of the region flanking the glycosyltransferase gene revealed open reading frames similar to capsular polysaccharide genes in Haemophilus influenzae, Escherichia coli, and Neisseria meningitidis. In addition, DNA from Burkholderia mallei and Burkholderia stabilis hybridized to a glycosyltransferase fragment probe, and a capsular structure was identified on the surface of B. stabilis via immunoelectron microscopy. Thus, the combination of PCR-based subtractive hybridization, insertional inactivation, and animal virulence studies has facilitated the identification of an important virulence determinant in B. pseudomallei.
Burkholderia pseudomallei, the causative agent of melioidosis, is a gram-negative, facultatively anaerobic, motile bacillus that is commonly found in the soil and stagnant waters in Southeast Asia and northern Australia. Infection by B. pseudomallei is often due to either direct inoculation into wounds and skin abrasions or to inhalation of contaminated material (11, 24, 30). This would explain the prevalence of the disease among rice farmers as well as helicopter pilots in the Vietnam War who developed melioidosis due to inhalation of contaminated dust (24, 47). Melioidosis may present as an acute pneumonia or an acute septicemia, which is the most severe form of the disease. The disease may also manifest as a chronic infection involving long-lasting suppurative abscesses in numerous sites in the body. Infection with B. pseudomallei may even result in a subclinical infection and remain undetected for a number of years. Both the chronic and subclinical forms generally remain undiagnosed until activated by a traumatic event or a decrease in immunocompetence (25).
Both secreted and cell-associated antigens have been identified in B. pseudomallei. Cell-associated antigens include exopolysaccharide (EPS) and lipopolysaccharide (LPS) (5, 8, 51). The EPS produced by B. pseudomallei is an unbranched polymer of repeating tetrasaccharide units with the structure -3)- 2-O-acetyl-β-d-Galp-(1-4)-α-d-Galp-(1-3)-β-d-Galp-(1-5)-β-d- KDOp-(2- (35, 40). The role of EPS in virulence is unknown, but sera from patients with melioidosis have been shown to contain antibodies against EPS (51). The LPS of B. pseudomallei has been reported to contain two types of O-polysaccharide moieties termed type I O-PS and type II O-PS (27, 41). Type II O-PS is an unbranched heteropolymer with repeating d-glucose and l-talose residues with the structure -3)-β-d-glucopyranose-(1-3)-6-deoxy-α-l-talopyranose-(1-, in which approximately 33% of the talose residues contain 2-O-methyl and 4-O-acetyl substituents, while the other l-talose residues contain only 2-O-acetyl substituents. Type II O-PS has been shown to be involved in serum resistance (17). Mutants lacking in type II O-PS were found to be sensitive to the bactericidal activities of 30% normal human serum. Type II O-PS mutants also demonstrated reduced virulence in three animal models of B. pseudomallei infection (17). Type I O-PS is an unbranched homopolymer with the structure -3)-2-O-acetyl-6-deoxy-β-d-manno-heptopyranose-(1-. The role for this polysaccharide in infection was previously undefined.
B. thailandensis is a nonpathogenic soil organism originally isolated in Thailand (6). Based on biochemical, immunological, and genetic data, B. pseudomallei and B. thailandensis are closely related species. However, these two organisms differ in a number of ways and have been classified into two different species (7). The rRNA sequence of B. thailandensis differs from that of B. pseudomallei by 15 nucleotides, and there are significant differences in genomic macrorestriction patterns between these organisms (10). The biochemical profiles of these two species differ in that B. thailandensis can utilize l-arabinose whereas B. pseudomallei does not (7, 62). The most distinct difference between these two species, however, is their relative virulence. The 50% lethal dose (LD50) for B. pseudomallei in the Syrian hamster model of acute melioidosis is <10 organisms, whereas the LD50 for B. thailandensis is approximately 106 organisms (7). It has also been shown that the two species can be differentiated based on their propensity to cause disease in humans. Environmental strains isolated in Thailand that are able to assimilate l-arabinose are not associated with human infection, whereas clinical isolates are not able to utilize l-arabinose (54).
To identify the genetic determinants that confer enhanced virulence in B. pseudomallei, a method combining subtractive hybridization, insertional mutagenesis, and animal virulence studies was developed. The described method should aid in the identification of virulence factors in pathogenic bacteria and provide further insights into microbial diversity and evolution.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. B. pseudomallei, B. thailandensis, B. cepacia, and Escherichia coli were grown at 37°C on Luria-Bertani (LB) broth base (Becton Dickinson) agar plates or in LB broth. B. mallei was grown at 37°C on LB plates or in LB broth supplemented with 4% glycerol and at pH 6.8. For animal studies, B. pseudomallei and B. thailandensis cultures were grown at 37°C in TSBDC medium (6). When appropriate, antibiotics were added at the following concentrations: 50 μg of tetracycline, 100 μg of streptomycin, 100 μg of polymyxin B, 100 μg of trimethoprim, 25 μg of gentamicin, and 25 μg of kanamycin per ml for B. pseudomallei and 100 μg of ampicillin, 25 and 50 μg of kanamycin, 15 μg of tetracycline, and 1.5 mg of trimethoprim per ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| SM10 | Mobilizing strain; transfer genes of RP4 integrated in chromosome; Kmr Sms | 48 |
| SM10λpir | SM10 with a λ prophage carrying the gene encoding the π protein | 37 |
| SURE | e14− (mcrA) Δ(mcrCB-hsdSMR-mrr)171 endA1 supE44 thi-1 gyrA96 relA1 lac recB recJ sbcC umuC::Tn5 uvrC [F′ proAB lacIqZΔM15 Tn10] Kanr Tetr | Stratagene |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | Bethesda Research Laboratories |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| XL10-Gold | TetrΔ (mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte[F′ proAB lacIqZ ΔM15 Tn10 (Tetr) Amy Camr] | Stratagene |
| B. pseudomallei | ||
| 1026b | Clinical isolate; Kmr Gmr Smr Pmr Tcs Tps | 16 |
| SR1015 | 1026b(pSR1015); Smr Tcr | This study |
| SR1016 | 1026b(pSR1016); Smr Tcr | This study |
| DD503 | 1026b derivative; allelic exchange strain; Δ(amrR-oprA)(Kms Gms Sms) rpsL (Smr) | 39 |
| PB401 | DD503 derivative; ΔfliC | Brett et al., unpublished |
| SR1001 | DD503 derivative; ΔfliC wbiE::aacC1 Gmr | This study |
| SLR5 | SR1001 derivative; wcbB::Tn5-OT182 Tcr | This study |
| SLR8 | SR1001 derivative; wzt2::Tn5-OT182 Tcr | This study |
| SLR13 | SR1001 derivative; wcbP::Tn5-OT182 Tcr | This study |
| SLR18 | SR1001 derivative; wcbE::Tn5-OT182 Tcr | This study |
| SLR19 | SR1001 derivative; wcbH::Tn5-OT182 Tcr | This study |
| SR201::Tp | DD503 derivative; wcbC::Tp | This study |
| SR202::Tp | DD503 derivative; wcbA::Tp | This study |
| SR203::Tp | DD503 derivative; yafJ::Tp | This study |
| B. thailandensis E264 | Soil isolate; LPS contains only type II O-PS | 7 |
| B. mallei NCTC 10260 | ||
| B. cepacia complex | ||
| B. cepacia CEP509 (genomovar I) | CFa isolate, Australia | 33 |
| B. multivorans C5393 (formerly B. cepacia genomovar II) | CF isolate, Vancouver, Canada | 33 |
| B. cepacia K56-2 (genomovar III) | CF isolate, Toronto, Canada | 15 |
| B. stabilis (formerly B. cepacia genomovar IV) | ||
| CEP0717 | CF isolate, Calgary, Canada | H. Rabin |
| CEP0467 | CF isolate, Edmonton, Canada | E. Mahenthiralingam |
| J687 | Non-CF isolate, France | 56 |
| CEP0726 | CF isolate, Calgary, Canada | H. Rabin |
| LMG14291 | CF isolate, Belgium | 56 |
| LMG7000 | Blood isolate, Sweden | 56 |
| LMG14294 | CF isolate, Belgium | 33 |
| B. vietnamiensis (also known as B. cepacia genomovar V) LMG10929 | Rice root isolate, Vietnam | 33 |
| Plasmids | ||
| pSKM11 | Positive selection cloning vector; IncP mob; ColE1 ori; Apr Tcs | 38 |
| pPCR | pBluescript II SK(+) derivative; Apr | 3 |
| pZErO-2.1 | Positive selection cloning vector; ColE1; Kmr | Invitrogen |
| pPCR2.1-TOPO | Topoisomerase-mediated cloning vector; Apr Kmr | Invitrogen |
| pDD1015 | Subtractive hybridization product cloned into pZErO-2.1; Kmr | This study |
| pDD1016 | Subtractive hybridization product cloned into pZErO-2.1; Kmr | This study |
| pSR1015 | KpnI-XhoI fragment from pDD1015 cloned into pSKM11 Apr Tcr | This study |
| pSR1016 | KpnI-XhoI fragment from pDD1016 cloned into pSKM11; Apr Tcr | This study |
| pSR1015Bg | 8-kb BglII fragment from SR1015 obtained by self-cloning; Apr Tcr | This study |
| pOT182 | pSUP102(Gm)::Tn5-OT182; Cmr Gmr Apr Tcr | 36 |
| pSLR5B | 8-kb BamHI fragment from SLR5 obtained by self-cloning; Apr | This study |
| pSLR5H | 10-kb HindIII fragment from SLR5 obtained by self-cloning; Apr | This study |
| pSLR13H | 9-kb HindIII fragment from SLR13 obtained by self-cloning; Apr | This study |
| p34E-oriTp | Vector containing self-cloning Tp cassette; dhfrllb-p15A oriV | Brett et al., unpublished |
CF, cystic fibrosis.
Construction and screening of subtractive hybridization libraries.
Subtractive hybridization between B. pseudomallei and B. thailandensis was carried out using a PCR-Select bacterial genome subtraction kit (Clontech) as recommended by the manufacturer except that the hybridization temperature was increased from 63°C to 73°C due to the high G+C content in the genomes of these species. In construction of the subtractive hybridization library, B. pseudomallei genomic DNA was used as the tester and B. thailandensis genomic DNA was used as the driver. The secondary PCR products obtained were cloned into pZErO-2.1 (Invitrogen) and pPCR (Table 1) and were enriched for B. pseudomallei-specific sequences. The subtraction library was screened by sequencing of the tester-specific DNA fragments. The library containing random clones was diluted in sterile phosphate-buffered saline (PBS) to 10−6, and 100 μl was plated on LB plates containing kanamycin (50 μg/ml) and 1 mM isopropylthio-β-d-galactoside (IPTG). Individual colonies were picked and grown overnight at 37°C in LB with kanamycin (50 μg/ml). Plasmid DNA was isolated using a miniprep plasmid isolation kit (Qiagen).
DNA sequencing and analysis.
Automated DNA sequencing was performed by ACGT (Northbrook, Ill.) and the University of Calgary Core DNA Services. The M13 forward primer (dGTAAAACGACGGCCAGT) was used to initiate sequence reactions with the subtractive hybridization clones. DNA flanking the Tn5-OT182 insertions was sequenced using the previously described primers OT182-LT and OT182-RT (16). The DNA flanking the insertion of pSR1015 was sequenced using the pSKM11 primer (38). DNA and protein sequences were analyzed with DNASIS for IBM and with the ORF Finder program at the National Center for Biotechnology Information (NCBI). DNA sequences were analyzed for homology using the BLASTX program through GenBank at NCBI.
Cloning of a subtractive hybridization product and mobilization into wild-type B. pseudomallei.
The DNA insert from pDD1015 was cloned as a KpnI-XhoI fragment into a mobilizable suicide vector, pSKM11 (Table 2). The 373-bp fragment was ligated to pSKM11 digested with the same enzymes to create pSR1015. SM10(pSR1015) was conjugated with B. pseudomallei 1026b using a previously described protocol (16).
TABLE 2.
Recombinant plasmids in the B. pseudomallei-B. thailandensis subtraction library
| Plasmid | Vector | Insert size (bp) | % G+C | Homologuea |
|---|---|---|---|---|
| pDD1000 | pPCR | 326 | 51 | DprA |
| pDD1001 | pPCR | 800 | 44 | None |
| pDD1002 | pPCR | 434 | 50 | GuaA |
| pDD1003 | pPCR | 346 | 51 | None |
| pDD1004 | pPCR | 800 | 44 | None |
| pDD1005 | pPCR | 531 | 46 | Mob protein |
| pDD1006 | pPCR | 353 | 48 | None |
| pDD1007 | pZErO-2.1 | 325 | 51 | None |
| pDD1008 | pZErO-2.1 | 250 | 44 | None |
| pDD1009 | pZErO-2.1 | 350 | 52 | None |
| pDD1012 | pZErO-2.1 | 505 | 47 | None |
| pDD1015 | pZErO-2.1 | 373 | 52 | WbpX |
| pDD1016 | pZErO-2.1 | 259 | 46 | None |
| pDD1017 | pZErO-2.1 | 100 | 50 | None |
| pDD1018 | pZErO-2.1 | 433 | 50 | None |
Animal studies.
The animal model of acute B. pseudomallei infection has been previously described (18). Syrian hamsters (females, 6 to 8 weeks) were injected intraperitoneally with 100 μl of one of a number of serial dilutions of logarithmic-phase cultures in sterile PBS. The five control animals were inoculated with 101 CFU of wild-type B. pseudomallei. The test animals (five per dilution) were inoculated with either 101, 102, or 103 CFU of the mutant strain, SR1015. Blood from two of the test animals was diluted and plated on Ashdown medium with and without the addition of tetracycline (50 μg/ml) to verify the stability of pSR1015 (7). For determination of LD50 for SR1015, hamsters (five per group) were inoculated with 103, 104, 105, and 106 CFU. After 48 h, the LD50 was calculated (42).
Immunoassays.
Immunogold electron microscopy was performed as previously described (17). Samples for Western blot analysis were prepared as previously described (8). Immunoassay was performed with a 1:250 dilution of the primary antibody, polyclonal rabbit antiserum raised to a B. pseudomallei O-PS–flagellin protein conjugate (5, 8). The secondary antibody used was horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG) (Sigma). Enzyme-linked immunosorbent assays (ELISAs) for the presence of EPS were carried out as previously described, using the EPS-specific monoclonal antibody 3015 at a dilution of 1/100 (17, 51, 52). Tn5-OT182 mutants were screened by ELISA according to the same protocol with rabbit polyclonal sera specific for an O-PS–flagellin conjugate containing antibodies to type I O-PS, type II O-PS, and flagella (5).
Southern hybridization.
For Southern hybridization, SstI digests of genomic DNA from B. pseudomallei 1026b and SR1015, B. thailandensis E264, B. mallei NCTC 10260, B. cepacia CEP509 (genomovar I) and K56-2 (genomovar III), B. stabilis LMG14294 and LMG7000, B. vietamiensis LMG10929, and B. multivorans C5393 were transferred to GeneScreen Plus membranes (Du Pont Canada, Lachine, Quebec, Canada), and hybridization was performed at 65°C in 15 ml of 1% sodium dodecyl sulfate (SDS)–10% dextran sulfate–salmon sperm DNA (0.1 mg/ml) according to the manufacturer's recommendations. The 0.4-kb KpnI-XhoI fragment from pDD1015 was used as a probe and labeled with [32P]dCTP using an oligonucleotide labeling kit (Pharmacia Biotech, Inc., Baie d'Urfe, Quebec, Canada).
Tn5-OT182 mutagenesis and screening for type I O-PS mutants.
To screen for mutants deficient in type I O-PS, it was first necessary to create a strain that produced only type I O-PS. This was necessary as the antiserum available was polyclonal antiserum to a flagellin–O-PS conjugate that contains antibodies to flagellin, type I O-PS, and type II O-PS. Therefore, we constructed a strain that was lacking in type II O-PS and flagella, SR1001. Allelic exchange was carried out using two strains that were previously constructed in the laboratory. The donor strain, SM10λpir(pPB611::Gm), has a plasmid containing a copy of the wbiE gene, involved in the synthesis of type II O-PS, which has been mutated by the insertion of a gentamicin resistance (Gm) cassette (17). The recipient strain, PB401, is a B. pseudomallei strain that has a deletion in the fliC gene. SM10λpir(pPB611::Gm) was conjugated to PB401, and transconjugants were selected for by plating on LB containing gentamicin, kanamycin, and polymyxin B. Transconjugants that were Smr, Gmr, and Kms were selected (to select for loss of the vector, pKAS46), and one was designated SR1001. Transposon mutagenesis of SR1001 was performed with Tn5-OT182 according to a previously described protocol (16) except that transconjugants were selected on plates containing gentamicin and tetracycline. Transposon mutants were inoculated into 96-well plates containing 200 μl of LB with gentamicin and tetracycline and grown overnight at 37°C at 250 rpm. A negative growth control well was included for each plate. The wells of a 96-well plate were coated with 10 μl of bacteria and 90 μl of coating buffer (0.05 M carbonate buffer [pH 9.6]), and ELISA was carried out as previously described (5). The primary antibody, polyclonal rabbit antiserum to a B. pseudomallei O-PS–flagellin protein conjugate, was added at a dilution of 1:1,000. The secondary antibody, a goat anti-rabbit IgG-peroxidase conjugate (Sigma), was added at a dilution of 1:1,000. The plates were developed with an HRP color development reagent (Bio-Rad Laboratories) for 30 min. The optical density at 405 nm (OD405) was determined using an ELISA reader. B. pseudomallei 1026b was included as a positive control, and E. coli DH5α was included as a negative control. Transposon mutants that had OD405 readings comparable to the negative control (OD405 = <0.100) were chosen for further analysis.
Construction of allelic exchange mutants.
Allelic exchange was carried out as previously described (17). The allelic exchange vector used in these experiments was pKAS46, an allelic exchange vector based on rpsL for counterselection (49). B. pseudomallei DD503, a double mutant that contains the ΔamrR-oprA and rpsL mutations, was the recipient strain used for all allelic exchange experiments (17, 39). All genes in these experiments were mutated by the insertion of a self-cloning trimethoprim resistance (Tp) cassette from p34EoriTp (P. J. Brett, D. DeShazer, and D. E. Woods, unpublished data). For each allelic exchange experiment, SM10λpir transformed with pKAS46 containing the mutated allele was conjugated to B. pseudomallei DD503 as described above for the construction of SR1001 except that the transconjugants were plated on polymyxin B, kanamycin, and trimethoprim. The Pmr Kmr Tpr transconjugants were subsequently transferred to plates containing streptomycin to select for the loss of pKAS46. Mutant alleles were confirmed by self-cloning and sequencing.
DNA manipulation.
Restriction enzymes and T4 DNA ligase were purchased from Life Technologies (Burlington, Ontario, Canada) and New England Biolabs (Mississauga, Ontario, Canada) and used according to the manufacturer's instructions. DNA fragments used in cloning procedures were excised from agarose gels and purified using a GeneClean II kit (Bio 101, Vista, Calif.) or Qiagen (Mississauga, Ontario, Canada) gel extraction kit. Chromosomal DNA was isolated using a previously described protocol (60). The self-cloning of B. pseudomallei flanking DNA from Tn5-OT182 mutants and from SR1015 was performed as described previously (16).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in the GenBank database under accession number AF228583.
RESULTS
Construction and screening of the B. pseudomallei subtraction library.
Subtractive hybridization was carried out between the virulent B. pseudomallei and the weakly virulent B. thailandensis in order to isolate DNA sequences encoding for virulence determinants unique to B. pseudomallei. The genomic DNA sample from B. pseudomallei containing the sequences of interest was known as the tester DNA, and genomic DNA from B. thailandensis, the reference sample, was called the driver DNA. Tester and driver DNAs were digested and subjected to two rounds of hybridization. The remaining unhybridized sequences were considered tester-specific sequences. To enrich for tester-specific sequences, excess driver DNA was added in the hybridizations. The tester-specific sequences were then amplified by PCR and cloned into pPCR or pZErO-2.1 (Table 1).
Screening of the subtraction library revealed a number of DNA sequences unique to B. pseudomallei. Fifteen distinct plasmid inserts from the library were sequenced (Table 2). The DNA inserts ranged from 100 to 800 bp in length and were found to contain an average G+C content of approximately 44 to 52%, which is considerably lower than the 68% G+C content of the B. pseudomallei chromosome. The DNA sequences were analyzed using the NCBI BLASTX program, and only four of the sequences had homology to predicted proteins present in the GenBank database. One of the plasmid inserts, pDD1000, had homology to DprA, a protein required for chromosomal DNA transformation in Haemophilus influenzae (26). Another insert, pDD1005, had homology to a mobilization protein found in small plasmids (1). The third, pDD1015, was found to share limited homology with WbpX, a glycosyltransferase, from Pseudomonas aeruginosa (45). The fourth, pDD1002, demonstrated homology to GuaA, a GMP synthetase, from Bacillus subtilis (34).
Insertional inactivation of the glycosyltransferase gene in wild-type B. pseudomallei.
The 373-bp DNA insert from pDD1015 was cloned into a mobilizable suicide vector, pSKM11 (Table 1). The resulting plasmid, pSR1015, was mobilized into wild-type B. pseudomallei 1026b to create the mutant strain SR1015. Since the insert from pDD1015 was found to demonstrate homology to a glycosyltransferase from P. aeruginosa, it was postulated that it might encode a protein involved in carbohydrate synthesis.
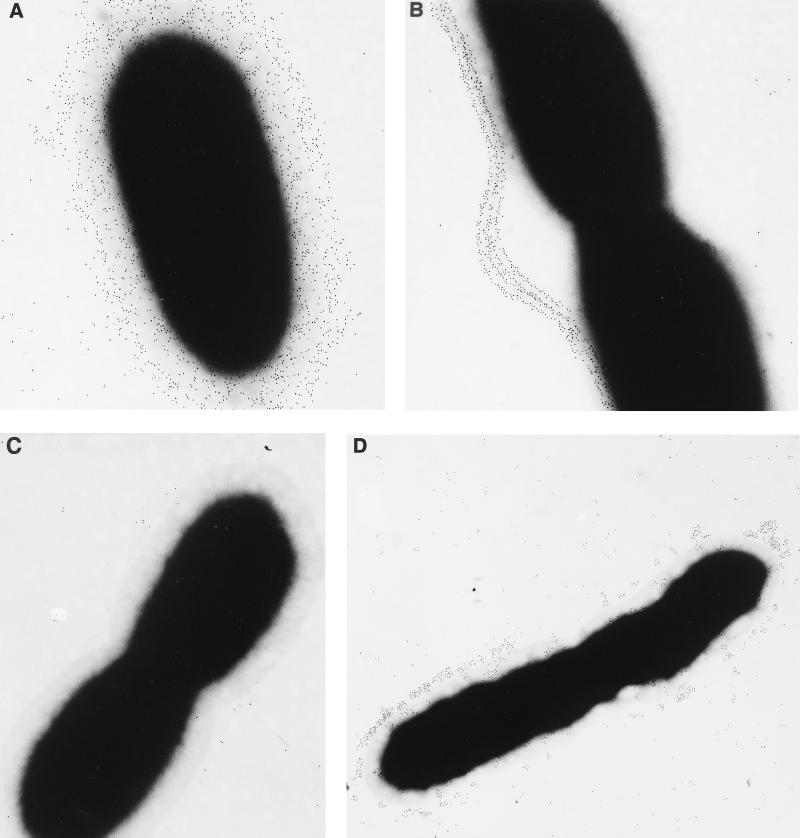
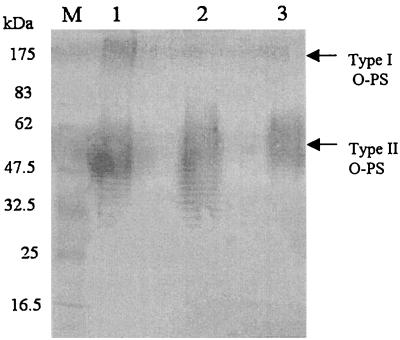
To define the phenotype of SR1015, an ELISA was performed with the EPS-specific monoclonal antibody 3015, and B. pseudomallei 1026b and SR1015 were both found to contain EPS (data not shown) (52). SR1015 was also shown to contain type II O-PS and to be serum resistant (data not shown). Immunogold electron microscopy studies using rabbit polyclonal sera specific for a type I O-PS–flagellin conjugate was performed on the parent strain, 1026b, and SR1015 (Fig. 1). B. pseudomallei 1026b reacted with antibodies to both flagellin and type I O-PS, as was evident by the distribution of gold particles around the bacterial surface and extending out along the flagella (Fig. 1A). The distribution of the gold particles around the outer surface of the bacteria corresponds to the type I O-PS structure, which is known to extend out beyond the type II O-PS. Unlike B. pseudomallei 1026b, SR1015 reacted only with the antibodies to flagellin, as the gold particles were found associated only with the flagella (Fig. 1B). B. thailandensis, the negative control, did not react with the antibodies either to flagellin or to type I O-PS (Fig. 1C). Western blot analysis of proteinase K-digested whole cells from B. pseudomallei 1026b, B. thailandensis E264, and B. pseudomallei SR1015 using rabbit polyclonal sera raised to O-PS–flagellin protein conjugate confirmed the lack of type I O-PS in SR1015 (Fig. 2). Type I and type II O-PS were stained in B. pseudomallei 1026b, while only type II O-PS was stained in the lanes corresponding to B. pseudomallei SR1015 and B. thailandensis. These results indicated that we had identified and insertionally inactivated a gene involved in the synthesis of the type I O-PS of B. pseudomallei.
FIG. 1.
Immunogold electron microscopy of B. pseudomallei 1026b (A) and SR1015 (B), B. thailandensis E264 (C), and B. stabilis LMG7000 (D). Bacteria were reacted with polyclonal rabbit antiserum directed against an O-PS–flagellin protein conjugate absorbed with B. thailandensis E264 to remove the antibodies directed against type II O-PS, washed, and reacted with a goat anti-rabbit IgG-gold (5 nm) conjugate. Original magnification, ×30,000.
FIG. 2.
Western blot analysis of LPS isolated from B. pseudomallei 1026b and SR1015 and B. thailandensis E264. Bacteria were reacted with proteinase K, subjected to SDS-polyacrylamide gel electrophoresis, electroblotted, and reacted with polyclonal rabbit antiserum raised to an O-PS–flagellin protein conjugate from B. pseudomallei. Lane M, prestained protein molecular weight standards (New England Biolabs); lane 1, B. pseudomallei 1026b; lane 2, B. pseudomallei SR1015; lane 3, B. thailandensis E264. The apparent molecular masses of the prestained proteins are indicated.
SR1015 is avirulent in the animal model of infection.
SR1015 was tested for virulence in the Syrian hamster model of acute septicemic melioidosis. The LD50 for SR1015 after 48 h was 3.5 × 105 CFU, while the LD50 of the parent strain, 1026b, was <10 CFU. The LD50 for SR1015 was similar to that for the weakly virulent B. thailandensis (6.8 × 105 CFU). This demonstrates that SR1015 is severely attenuated for virulence in this animal model of melioidosis and that type I O-PS is a major virulence determinant of B. pseudomallei.
Cloning and sequencing of the genetic loci required for type I O-PS production and export.
Two methods were used to clone the genes involved in the production and export of type I O-PS. The DNA flanking the insertion of pSR1015 was cloned from SR1015 and sequenced. We also used transposon mutagenesis to clone the genes involved in production of the polysaccharide; this was done to obtain any unlinked genes that may be involved in polysaccharide production. Approximately 1,300 transposon mutants were screened for loss of type I O-PS by ELISA. Six mutants were identified, and the DNA flanking the transposon insertion was cloned and sequenced. The Tn5-OT182 mutants SLR5, SLR8, SLR13, SLR18, and SLR19 mapped to the same region of the chromosome (Fig. 3). Sequence analysis of the cloned fragments revealed the presence of 20 potential open reading frames involved in the synthesis and export of type I O-PS (Fig. 3). The open reading frames that predicted proteins involved in polysaccharide biosynthesis were found to demonstrate homology to proteins involved in the synthesis of a polysaccharide structure composed primarily of mannose (Table 3). The other reading frames in the locus predicted proteins involved in the transport of capsular polysaccharides in a variety of bacteria, particularly those that produce group 2 and group 3 capsular polysaccharides (Table 3 and reference 59). The genes responsible for the production of type I O-PS was found to be similar to other loci encoding for capsular polysaccharides in that they are divergently transcribed (Fig. 3 and reference 44). The gene cluster involved in the production of this polysaccharide is also similar to group 3 capsule gene clusters in that there are no genes encoding KpsF and KpsU, which are present in group 2 capsule gene clusters (59). However, the organization of the B. pseudomallei type I O-PS gene cluster differs in that it does not contain two export regions flanking a single biosynthetic region as seen in other group 3 capsule polysaccharide clusters (12). The biosynthetic genes identified thus far are not organized into one continuous transcriptional unit; instead, wcbB, manC, and wcbP are separated from the rest of the biosynthetic genes. Another interesting feature is that kpsC is usually found next to kpsS in other group 2 and 3 clusters, unlike the case for wcbA and wcbO in B. pseudomallei (Fig. 3 and reference 59). The promoter sequences of the transcriptional regions of the type I O-PS cluster have yet to be identified. The overall G+C content of this region is about 58%, lower than the G+C content of the rest of the chromosome (68%). The low G+C content in these clusters suggests that polysaccharide genes have a common origin and may have been transferred horizontally between species (21).
FIG. 3.
Organization of the chromosomal region containing the genes responsible for the synthesis and export of type I O-PS in B. pseudomallei. The upper part shows the locations of the genes. The direction of transcription is represented by arrows, and gene names are indicated. The locations of Tn5-OT182 insertions are represented by triangles. Mutants constructed by allelic exchange are shown. The straight line indicates insertion of the Tp cassette into the gene of interest. The horizontal line below the genetic map represents B. pseudomallei chromosomal DNA; the locations of relevant restriction endonuclease recognition sites (Bg, BglII; E, EcoRI; H, HindIII; K, KpnI; B, BamHI) are shown.
TABLE 3.
Genes involved in the production and export of type I O-PS in B. pseudomallei and homologous proteins located in the nonredundant sequence database
| Gene | Size (bp) | Homologue, strain | Putative function | % Identity | % Similarity |
|---|---|---|---|---|---|
| manC | 1,427 | ManC, Escherichia coli | Mannose-1-phosphate guanyltransferase | 55 | 70 |
| ManC, Salmonella enterica serovar Typhimurium | 54 | 68 | |||
| ManC, Klebsiella pneumoniae | 48 | 65 | |||
| wcbA | 2,015 | KpsC, E. coli | Capsule polysaccharide export protein | 37 | 52 |
| LipA, Neisseria meningitidis | 39 | 55 | |||
| PhyA, Pseudomonas multocida | 35 | 51 | |||
| wcbB | 1,097 | WbpX, P. aeruginosa | Glycosyltransferase/mannosyltransferase | 33 | 49 |
| ManB, Aquifex aeolicus | 26 | 44 | |||
| MtfA, Archaeoglobus fulgidis | 30 | 47 | |||
| wcbC | 1,163 | KpsD, E. coli | Capsule export periplasmic protein | 26 | 39 |
| CtrA, N. meningitidis | 40 | 58 | |||
| BexD, Haemophilus influenzae | 37 | 58 | |||
| ′manB | 203 | XanA, Xanthomonas campestris | Phosphomannomutase | 35 | 48 |
| ManB, K. pneumoniae | 29 | 49 | |||
| Rfk9, E. coli | 35 | 53 | |||
| wcbD | 1,148 | BexC, H. influenzae | Capsule export inner membrane protein | 40 | 62 |
| CtrB, N. meningitidis | 38 | 60 | |||
| KpsE, E. coli | 26 | 48 | |||
| wzm2 | 410 | CtrC, N. meningitidis | Capsule export inner membrane protein | 53 | 73 |
| BexB, H. influenzae | 28 | 50 | |||
| KpsM, E. coli | 28 | 50 | |||
| wzt2 | 746 | BexA, H. influenzae | ATP-binding protein | 59 | 75 |
| CtrD, N. meningitidis | 57 | 72 | |||
| KpsT, E. coli | 46 | 66 | |||
| wcbE | 1,523 | MtfB, A. aeolicus | Mannosyltransferase/glycosyltransferase | 28 | 42 |
| WbpX, P. aeruginosa | 38 | 55 | |||
| ManB, Synechocystis spp. | 30 | 48 | |||
| wcbF | 1,379 | Putative, Homo sapiens | Heparan-sulfate 6-sulfotransferase | 24 | 39 |
| Putative Arabidopsis thaliana | En/Spm transposon protein | 37 | 52 | ||
| wcbG | 941 | SyfB, Helicobacter pylori | Phenylalanyl-tRNA synthetase | 29 | 46 |
| wcbH | 1,796 | MtfA, Archaeoglobus spp. | Mannosyltransferase/glycosyltransferase | 23 | 40 |
| Putative, Synechocystis spp. | 25 | 42 | |||
| Putative, Streptomyces coelicolor | 28 | 41 | |||
| wcbI | 1,187 | NifQ, Enterobacter agglomerans | Nitrogen fixation protein | 28 | 41 |
| wcbJ | 842 | Rbd1, Methanobacterium thermoautotrophicum | dTDP-4-dehydrorhamnose reductase | 23 | 39 |
| RmlD, Mycobacterium tuberculosis | 24 | 40 | |||
| wcbK | 1,013 | Gm4D, E. coli | GDP-mannose dehydratase | 29 | 48 |
| Gm4D, Yersinia pseudotuberculosis | 32 | 49 | |||
| Gm4D, Vibrio cholerae | 30 | 48 | |||
| wcbL | 1,040 | Putative, Campylobacter jejuni | Sugar kinase | 40 | 56 |
| Rv0115, M. tuberculosis | Lincomycin production | 39 | 52 | ||
| LmbP, Synechocystis spp. | 24 | 36 | |||
| gmhA | 593 | LpcA, H. pylori | Phosphoheptose isomerase | 50 | 68 |
| GmhA, Methanococcus jannaschii | 54 | 72 | |||
| LpcA, H. influenzae | 45 | 60 | |||
| wcbM | 692 | RmlA2, M. tuberculosis | Mannose-1-phosphate guanyltransferase | 32 | 46 |
| Putative, C. jejuni | Sugar-phosphate nucleotidyltransferase | 41 | 58 | ||
| wcbN | 353 | YaeD, E. coli | Hypothetical intergenic protein | 40 | 62 |
| YaeD, H. influenzae | Hypothetical intergenic protein | 40 | 58 | ||
| wcbO | 731 | KpsS, E. coli | Capsule polysaccharide export protein | 34 | 46 |
| PhyB, Pasteurella multocida | 29 | 45 | |||
| LipB, N. meningitidis | 27 | 44 | |||
| wcbP | 1,950 | YooP, M. tuberculosis | Oxidoreductase | 37 | 52 |
| HetN, Anabaena spp. | 34 | 52 |
The genes involved in the production of the type I O-PS have been named according to the bacterial polysaccharide gene nomenclature scheme (43). The gene products associated with the type I O-PS cluster and their homologues are listed in Table 3. Mutations constructed in a number of these genes have confirmed their role in the production of type I O-PS. One gene that is required for the production of the polysaccharide is wcbA (Fig. 3; Table 3). The wcbA gene and wcbO predict proteins that demonstrate homology to the KpsC and KpsS proteins of E. coli and the LipA and LipB proteins of Neisseria meningitidis, respectively (Table 3). These proteins are involved in the processing and export of capsular polysaccharide in these organisms (22, 44). To confirm the role of the wcbA gene in capsule production, an allelic exchange mutant was constructed by the insertion of a Tp cassette (Fig. 3). The resulting strain, SR202::Tp, did not produce polysaccharide and demonstrated attenuated virulence in the hamster model, similar to SR1015 (data not shown).
The wcbC, wcbD, wzm2, and wzt2 genes encode proteins that demonstrate homology to proteins involved in the transport of capsular polysaccharides (Table 3) (20, 28, 46). The wcbC gene predicts a protein that shares homology with KpsD, a periplasmic protein involved in capsule polysaccharide export in E. coli (21, 28, 61). An isogenic mutant was constructed by the insertion of a Tp cassette into the wcbC gene (Fig. 3). The resulting strain, SR201::Tp, was still virulent in the hamster model, and type I O-PS was detected on Western blots (data not shown). This is in contrast to the phenotype observed with E. coli kpsD mutants (61). The gene products encoded by wzm2 and wzt2 are homologous to the KpsM and KpsT proteins of E. coli, CtrC and CtrD of N. meningitidis, and BexA and BexB of H. influenzae (Table 3). These proteins are ATP-binding cassette (ABC) transporters that comprise an inner membrane polysaccharide export system (50). The wzm2 and wzt2 gene products of B. pseudomallei likely comprise an ABC transporter system that is involved in the transport of the type I O-polysaccharide across the cytoplasmic membrane. The termination codon of the wzm2 gene overlaps the initiation codon of the wzt2 gene, suggesting that these two genes are translationally coupled. The kpsM and kpsT genes of E. coli are organized into a single transcriptional unit, and both genes are translationally coupled (44). These genes have been designated wzm2 and wzt2 since wzm and wzt have previously been identified and are associated with the type II O-PS gene cluster (17). A hydrophobicity plot of the predicted wzm2 gene product revealed a hydrophobic protein with multiple-membrane spanning domains, like KpsM, that may act as an integral membrane protein for the export of polysaccharide (29). Analysis of the primary amino acid sequence of the predicted Wzt2 protein from B. pseudomallei has shown that this protein contains a conserved ATP-binding motif, including an A site (GGNGAGKST) and a B site (DCFLIDE) (57). The wzt2 gene was found to be necessary for the production of type I O-PS in B. pseudomallei. In SLR18, the insertion of Tn5-OT182 in the wzt2 gene resulted in a loss of type I O-PS.
The wcbB, wcbE, and wcbH genes encode for proteins that demonstrate homology to different mannosyltranferases or glycosyltransferases from a variety of bacterial species (Table 3). Since type I O-PS is a homopolymer of mannoheptopyranosyl residues, it is likely that these genes are involved in the biosynthesis of this polysaccharide. The wcbB gene encodes for a protein with homology to a glycosyltransferase, WbpX, from P. aeruginosa as well as to mannosyltransferases from a variety of bacteria (45). The function of glycosyltransferases is to catalyze the sequential transfer of sugar residues from nucleotide precursors to the membrane-bound acceptor, undecaprenol phosphate-P-GlcpNAc (58). The wcbB gene product is likely involved in the transfer of mannose residues in the synthesis of type I O-PS. This gene was determined to be required for the synthesis of type I O-PS based on two lines of evidence: the insertional inactivation of this gene using pSKM11 rendered the mutant strain, SR1015, negative for type I O-PS production; and a transposon mutant, SLR5 (Fig. 3), lacked type I O-PS due to the insertion of Tn5-OT182 in the wcbB gene. The wcbE and wcbH genes both predict proteins with homology to mannosyltransferases and are both required for the production of type I O-PS. This is supported by the fact that the insertion of Tn5-OT182 in both the wcbH and wcbE genes (SLR19 and SLR18, respectively) resulted in mutant strains lacking type I O-PS (Fig. 3). Furthermore, an internal fragment of the wcbE gene was cloned into pSKM11 and used to insertionally inactivate this gene in B. pseudomallei. The resulting strain, SR1016, was found to lack type I O-PS and demonstrated attenuated virulence in the animal model (data not shown).
Another gene required for the production of type I O-PS is wcbP. This gene predicts a protein that shares homology to the YooP protein of Mycobacterium tuberculosis (Table 3). The YooP protein has been characterized as a putative oxidoreductase based on sequence comparisons (13). The function of the predicted wcbP gene product in B. pseudomallei is unclear; however, the insertion of Tn5-OT182 into this gene in the mutant strain SLR13 rendered the organism negative for the production of type I O-PS.
The yafJ gene encodes a protein of 278 amino acids that demonstrates homology to the YafJ protein from E. coli (Table 3). The YafJ protein is a putative amidotransferase in these organisms (2). The yggB gene encodes a protein of 235 amino acids with homology to the YggB protein of E. coli (Table 3). The function of this protein is unclear, but it has been defined as a hypothetical 30.9-kDa protein in an intergenic region (2). The G+C contents of these genes are 65.7% for yafJ and 65.4% for yggB, which is higher than the rest of the polysaccharide cluster and consistent with the G+C content of the B. pseudomallei chromosome. Southern blot analysis using these genes as probes has demonstrated their presence in B. thailandensis (data not shown); therefore, it is unlikely that these genes are required for the production of type I O-PS (Fig. 3). An allelic exchange mutant containing a Tp cassette in the yafJ gene was constructed. The resulting strain, SR203::Tp, was found to be virulent in hamsters (data not shown).
The type I O-PS is also present in B. mallei and the B. cepacia complex but not in B. thailandensis.
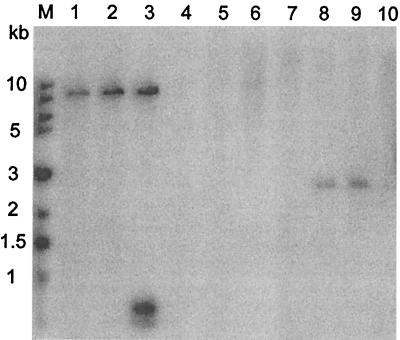
Southern blot analysis using a probe containing the A-T-rich glycosyltransferase fragment from pDD1015 confirmed that the fragment was present in B. pseudomallei 1026b and SR1015 but not in B. thailandensis (Fig. 4). B. mallei and the B. cepacia complex were also tested for the presence of this fragment. It was found that the probe hybridized to an SstI fragment in B. mallei and the B. stabilis (formerly genomovar IV) strains LMG7000 and LMG14294. The B. cepacia complex has recently been divided into two genomovars (B. cepacia genomovar I and genomovar III) and three species (B. multivorans, B. vietnamiensis, and B. stabilis) (55, 56). None of the strains tested from B. cepacia genomovars I and III, B. multivorans, or B. vietnamiensis were found to contain this DNA fragment. Southern blot analysis was carried out on five other B. stabilis strains: CEP0717, CEP0467, J687, CEP0726, and LMG14291. All of the B. stabilis strains tested hybridized to the probe from pDD1015 (data not shown). The presence of type I O-PS was confirmed in the B. stabilis strain LMG7000 by immunoelectron microscopy. As seen in Fig. 1D, B. stabilis LMG7000 showed reactivity to the type I O-PS antibodies, but lacked a uniform distribution of the polysaccharide on the cell surface, and therefore appears to produce less of this polysaccharide than B. pseudomallei 1026b.
FIG. 4.
Southern hybridization analysis of genomic DNA from Burkholderia spp. digested with SstI. A 0.4-kb KpnI-XhoI fragment from pDD1015 was used as a probe. Lane 1, B. mallei NCTC 10260; lane 2, B. pseudomallei 1026b; lane 3, B. pseudomallei SR1015; lane 4, B. thailandensis E264; lane 5, B. vietnamiensis LMG10929; lane 6, B. cepacia CEP509 (genomovar I); lane 7, B. cepacia K56-2 (genomovar III); lane 8, B. stabilis LMG14294; lane 9, B. stabilis LMG7000; lane 10, B. multivorans C5393.
Further Southern blot experiments were carried out to confirm the absence of the type I O-polysaccharide in B. thailandensis. B. thailandensis was hybridized with probes corresponding to a number of genes involved in the synthesis of the type I O-PS. The following genes were demonstrated by Southern hybridization to be present in B. pseudomallei but absent in B. thailandensis: wcbA, wcbC, wcbD, wzm2, wzt2, wcbE, wcbF, wcbH, wcbK, gmhA, and wcbO (data not shown).
DISCUSSION
Although melioidosis is less common outside of Southeast Asia and northern Australia, it may be underdiagnosed in other regions, and it poses a concern due to increased travel and military involvement in regions where the disease is endemic (14). Recently, our attention has been focused on the identification of genetic determinants that contribute to the pathogenesis of B. pseudomallei infections. To obtain virulence determinants unique to B. pseudomallei, we used subtractive hybridization between this organism and a related nonpathogenic organism, B. thailandensis.
Analysis of the subtractive hybridization library revealed that B. pseudomallei contains a number of DNA sequences that are not found in B. thailandensis (Table 2). One of the subtraction clones, pDD1015, demonstrated weak homology to a glycosyltransferase, WbpX, from P. aeruginosa (45). The insert from pDD1015 was cloned into a mobilizable suicide vector, pSKM11, for insertional inactivation of the glycosyltransferase gene in wild-type B. pseudomallei. The resulting strain, SR1015, was markedly less virulent than the parent strain in an animal model. This demonstrated that B. pseudomallei contains DNA sequences encoding for virulence determinants that are not found in B. thailandensis and that the glycosyltransferase gene may encode an important virulence determinant in B. pseudomallei. Using antibodies to type I O-PS, we determined that SR1015 harbored a mutation in a glycosyltransferase gene involved in the production of type I O-PS.
Sequence analysis of the DNA flanking the glycosyltransferase gene revealed the presence of at least 20 open reading frames involved in the synthesis and export of type I O-PS (Fig. 3; Table 3). The genes identified encode for proteins that are similar to proteins involved in the biosynthesis and export of capsular polysaccharides, particularly those involved in the production of group 3 capsular polysaccharides. Group 3 capsules include the E. coli K10 capsule and may also include the H. influenzae group b capsule and the capsule produced by N. meningitidis serogroup B (59). Group 3 capsules are always coexpressed with O serogroups, are not thermoregulated, are transported by an ABC-2 exporter system, and do not contain the kpsU and kpsF genes, and usually the gene clusters map near the serA locus (59). Thus far, no serA locus that is associated with the type I O-PS cluster has been identified, but this polysaccharide is coexpressed with O antigen and lacks the kpsU and kpsF genes, and genes encoding for a putative ABC-2 transporter have been identified. The genes involved in the production of group 3 capsules are organized into regions and are divergently transcribed. Regions 1 and 3 are generally conserved and contain genes involved in export of the polysaccharide. These regions flank region 2, which contains the biosynthetic genes and is not conserved between serotypes (44). The genetic organization of the type I O-PS is also similar to that of other capsule gene clusters in that the genes are organized into more than one transcriptional unit and appear to be divergently transcribed. However, the organization of the B. pseudomallei type I O-PS cluster differs in that the biosynthetic genes identified thus far are not organized into one biosynthetic region. yafJ and yggB are likely not involved in the production of type I O-PS since they have a high G+C content (62 to 65%), they are present in B. thailandensis, and a mutation in yafJ (SR203::Tp) did not reduce virulence in hamsters (data not shown). We are also currently constructing a mutant in the polyketide synthase gene that lies downstream of the wcbP gene in order to define this end of the type I O-PS cluster.
The polysaccharide with the structure -3)-2-O-acetyl-6-deoxy-β-d-manno-heptopyranose-(1- was originally isolated and characterized as an O-PS component of LPS in B. pseudomallei and was designated type I O-PS (41). However, our results suggest that this polysaccharide is a capsule rather than an O-PS moiety. The genes involved in the production of this capsule demonstrate strong homology to the genes involved in the production of capsular polysaccharides in many organisms, including N. meningitidis, H. influenzae, and E. coli. In addition, the export genes associated with this cluster are not associated with the previously characterized O-PS gene cluster (17). Western blot analysis of proteinase K cell extracts (Fig. 2) and silver staining (data not shown) have shown that this polysaccharide has a high molecular mass (200 kDa) and lacks the banding pattern seen with O-PS moieties. Studies by our laboratory have indicated that mutants in the production of the core oligosaccharide of the LPS are still capable of producing this polysaccharide (9). Based on the above criteria and the genetic similarity to group 3 capsules, we propose that this polysaccharide is a group 3 capsule.
Capsule production has been correlated with virulence in many bacteria, particularly those causing serious invasive infections of humans (4). Our studies have demonstrated that this capsule is critical for the virulence of B. pseudomallei. However, its specific role in infection has yet to be elucidated. A number of functions have been suggested for polysaccharide capsules: prevention of desiccation for transmission and survival, adherence for colonization, resistance to complement-mediated phagocytosis and complement-mediated killing, and resistance to specific host immunity due to a poor antibody response to the capsule (44). Preliminary studies have shown that type I O-PS is not involved in serum resistance. SR1015 was tested for resistance to killing by 30% normal human serum and was found to be resistant to killing (data not shown). Studies to define the role of the capsule in infection are under way.
Genomic DNAs from B. mallei NCTC 10260 and seven strains of B. stabilis were shown to hybridize to the glycosyltransferase probe from pDD1015. Immunoelectron microscopic analysis demonstrated that B. stabilis LMG7000 contained this capsule (Fig. 1). Interestingly, B. stabilis LMG7000 was noted to produce less of this polysaccharide than B. pseudomallei 1026b and lacked a uniform distribution of the polysaccharide on the cell surface. The importance of the capsule in infection by B. stabilis has yet to be elucidated. The results of our study demonstrating the presence of this capsule in B. stabilis corresponds with its recent classification as a novel species (56). This capsule may be an additional tool to aid in the identification of B. stabilis strains.
Virulence genes of a number of pathogenic bacteria are located on pathogenicity islands (PAIs), regions on the bacterial chromosome that are present in the genome of pathogenic strains but rarely present in those of nonpathogenic strains. The PAIs may range in size from about 30 kb to 200 kb and often differ in G+C content from the remaining bacterial genome; the PAIs are often associated with the carriage of many virulence genes. These genetic units are often flanked by direct repeats and may be associated with tRNA genes or insertion sequence (IS) elements at their boundaries. They may also be associated with the presence of mobility genes, such as IS elements, integrases, transposases, and origins of plasmid replication. These DNA regions are considered to be unstable in that they may be subject to deletion with high frequency or undergo duplications and amplifications (23). A number of PAIs have been described for both gram-positive and gram-negative bacteria, and the application of subtraction hybridization has been used to successfully identify such genetic elements (23, 32). The subtractive hybridization that was carried out between B. pseudomallei and B. thailandensis led to the identification of a number of sequences that were found to be A-T rich compared to the rest of the B. pseudomallei chromosome. This, combined with the fact that insertional mutagenesis of the glycosyltransferase gene identified by this method resulted in an avirulent strain, suggests that we may have identified DNA sequences from a putative PAI and that the capsular polysaccharide gene cluster may be located on this island. It is possible that B. pseudomallei, B. mallei, and B. stabilis acquired DNA encoding for capsule as well as other potential, yet unidentified virulence factors by horizontal transfer recently in evolution. B. pseudomallei is known to contain IS elements that are present in B. cepacia but not in B. thailandensis (31). However, IS elements have not yet been identified in association with the capsule gene cluster. Further studies are under way to determine whether a PAI exists in these organisms and whether the capsule gene cluster is located on such a genetic element.
The identification of bacterial virulence genes has traditionally relied on empirical predictions of putative virulence determinants and inactivation of the genes encoding for these putative virulence determinants by any number of methods, followed by comparisons of virulence between mutant and wild-type infection models (19). Tools such as in vivo expression technology and differential fluorescence technology have been developed to facilitate the identification of expressed sequences under a given set of circumstances within a test host; however, these approaches do not necessarily lead to the identification of virulence determinants (53). The method for identification of virulence genes described herein should be applicable to a broad range of pathogenic bacteria. The combination of PCR-based subtractive hybridization, insertional mutagenesis, and an animal infection model provides for the efficient detection of virulence genes. While we have applied the method to the pathogen B. pseudomallei in our current studies, it could be applied to any species and for which only a few prerequisites are in place. These prerequisites include related virulent and avirulent strains, suitable suicide vectors for insertional inactivation, and an infection model for differentiation of virulent and avirulent strains. The described method should lead to the identification of relevant virulence determinants for a number of bacterial species and further the understanding of molecular pathogenesis.
ACKNOWLEDGMENTS
This work was supported by Department of Defense contract DAMD 17-98-C-8003, the Medical Research Council of Canada, and the Canadian Bacterial Diseases Network of Centers of Excellence. D.D. was a recipient of an Alberta Heritage Foundation for Medical Research (AHFMR) Fellowship award.
We thank Trish Darling and Amie White for excellent technical assistance. We thank Ivo Steinmetz for the EPS-specific monoclonal antibody 3015 and Malcolm Perry for many helpful discussions. We also thank E. Mahenthiralingam and P. Vandamme for sharing data prior to publication.
REFERENCES
- 1.Antoine R, Locht C. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from Gram-positive organisms. Mol Microbiol. 1992;6:1785–1799. doi: 10.1111/j.1365-2958.1992.tb01351.x. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Borovkov A Y, Rivkin M I. Xcm-I containing vector for direct cloning of PCR products. BioTechniques. 1997;22:812–814. doi: 10.2144/97225bm04. [DOI] [PubMed] [Google Scholar]
- 4.Boulnois G J, Roberts I S. Genetics of capsular polysaccharide production in bacteria. Curr Top Microbiol Immunol. 1990;150:1–18. doi: 10.1007/978-3-642-74694-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Brett P J, Woods D E. Structural and immunologic characterization of Burkholderia pseudomallei O-polysaccharide–flagellin protein conjugates. Infect Immun. 1996;64:2824–2828. doi: 10.1128/iai.64.7.2824-2828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brett P J, DeShazer D, Woods D E. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol Infect. 1997;118:137–148. doi: 10.1017/s095026889600739x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brett P J, DeShazer D, Woods D E. Burkholderia thailandensis sp. nov., description of a Burkholderia pseudomallei-like species. Int J Syst Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 8.Bryan L E, Wong S, Woods D E, Dance D A, Chaowagul W. Passive protection of diabetic rats with antisera specific for the polysaccharide portion of the lipopolysaccharide isolated from Pseudomonas pseudomallei. Can J Infect Dis. 1994;5:170–178. doi: 10.1155/1994/856850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burtnick M N, Woods D E. Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob Agents Chemother. 1999;43:2648–2656. doi: 10.1128/aac.43.11.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaiyaroj S C, Kotrnon K, Koonpaew S, Anantagool N, White N J, Sirisinha S. Differences in genomic macrorestriction patterns of arabinose-positive (Burkholderia thailandensis) and arabinose-negative Burkholderia pseudomallei. Microbiol Immunol. 1999;43:625–630. doi: 10.1111/j.1348-0421.1999.tb02449.x. [DOI] [PubMed] [Google Scholar]
- 11.Chaowagul W, White N J, Dance D A, Wattanagoon Y, Naigowit P, Davis T M, Looareesuwan S, Pitakwatchara N. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J Infect Dis. 1989;159:890–899. doi: 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- 12.Clarke B R, Pearce R, Roberts I S. Genetic organization of the Escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J Bacteriol. 1999;181:2279–2285. doi: 10.1128/jb.181.7.2279-2285.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 14.Dance D A B. Melioidosis. Rev Med Microbiol. 1990;1:143–150. [Google Scholar]
- 15.Darling P, Chan M, Cox A D, Sokol P A. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect Immun. 1998;66:874–877. doi: 10.1128/iai.66.2.874-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeShazer D, Brett P J, Carylon R, Woods D E. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J Bacteriol. 1997;179:2116–2125. doi: 10.1128/jb.179.7.2116-2125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeShazer D, Brett P J, Woods D E. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol. 1998;30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
- 18.DeShazer D, Woods D E. Animal models of melioidosis. In: Zak O, Sande M, editors. Handbook of animal models of infection. London, England: Academic Press; 1999. pp. 199–203. [Google Scholar]
- 19.Falkow S. Molecular Koch's postulates. Rev Infect Dis. 1988;10:S274–S278. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 20.Frosch M, Edwards U, Bousset K, Krausse B, Weisgerber C. Evidence for a common molecular origin of capsular gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 21.Frosch M, Müller D, Bousset K, Müller A. Conserved outer membrane protein of Neisseria meningitidis involved in capsule expression. Infect Immun. 1992;60:798–803. doi: 10.1128/iai.60.3.798-803.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frosch M, Müller A. Phospholipid substitution of capsular polysaccharide and mechanisms of capsule formation in Neisseria meningitidis. Mol Microbiol. 1993;8:483–493. doi: 10.1111/j.1365-2958.1993.tb01592.x. [DOI] [PubMed] [Google Scholar]
- 23.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 24.Howe C, Sampath A, Spotnitz M. The pseudomallei group: a review. J Infect Dis. 1971;124:598–606. doi: 10.1093/infdis/124.6.598. [DOI] [PubMed] [Google Scholar]
- 25.Ip M, Osterberg L G, Chau P Y, Raffin T A. Pulmonary melioidosis. Chest. 1995;108:1420–1424. doi: 10.1378/chest.108.5.1420. [DOI] [PubMed] [Google Scholar]
- 26.Karudapuram S, Zhao X, Barcak G J. DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid transformation. J Bacteriol. 1995;177:3235–3240. doi: 10.1128/jb.177.11.3235-3240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knirel Y A, Paramonov N A, Shashkov A S, Kochetkov N K, Yarullin R G, Farber S M, Efremenko V I. Structure of the polysaccharide chains of Pseudomonas pseudomallei lipopolysaccharides. Carbohydr Res. 1992;233:185–193. doi: 10.1016/s0008-6215(00)90930-3. [DOI] [PubMed] [Google Scholar]
- 28.Kroll S, Loynds B, Brophy L, Moxon E. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol. 1990;4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Leelarasamee A, Bovornkitti S. Melioidosis: review and update. Rev Infect Dis. 1989;11:413–425. doi: 10.1093/clinids/11.3.413. [DOI] [PubMed] [Google Scholar]
- 31.Mack K, Titball R W. The detection of insertion sequences within the human pathogen Burkholderia pseudomallei which have been identified previously in Burkholderia cepacia. FEMS Microbiol Lett. 1998;162:69–74. doi: 10.1111/j.1574-6968.1998.tb12980.x. [DOI] [PubMed] [Google Scholar]
- 32.Mahairas G G, Sabo P J, Hickey M J, Singh D V, Stover C K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1284. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahenthiralingam E, Coenye T, Chung J W, Speert D P, Govan J R W, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantsala P, Zalkin H. Cloning and sequence of Bacillus subtilis purA and guaA, involved in the conversion of IMP to AMP and GMP. J Bacteriol. 1992;174:1883–1890. doi: 10.1128/jb.174.6.1883-1890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masoud H, Ho M, Schollaardt T, Perry M B. Characterization of the capsular polysaccharide of Burkholderia pseudomallei 304b. J Bacteriol. 1997;179:5663–5669. doi: 10.1128/jb.179.18.5663-5669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merriman T R, Lamont I L. Construction and use of a self-cloning promoter probe vector for Gram-negative bacteria. Gene. 1993;126:17–23. doi: 10.1016/0378-1119(93)90585-q. [DOI] [PubMed] [Google Scholar]
- 37.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mongkolsuk S, Rabibhadana S, Vattanaviboon P, Loprasert S. Generalized and mobilizable positive-selective cloning vectors. Gene. 1994;143:145–146. doi: 10.1016/0378-1119(94)90620-3. [DOI] [PubMed] [Google Scholar]
- 39.Moore R A, DeShazer D, Reckseidler S, Weissman A, Woods D E. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nimtz M, Wray V, Domke T, Brenneke B, Haussler S, Steinmetz I. Structure of an acidic exopolysaccharide of Burkholderia pseudomallei. Eur J Biochem. 1997;250:608–616. doi: 10.1111/j.1432-1033.1997.0608a.x. [DOI] [PubMed] [Google Scholar]
- 41.Perry M B, MacLean L L, Schollaardt T, Bryan L E, Ho M. Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect Immun. 1995;63:3348–3352. doi: 10.1128/iai.63.9.3348-3352.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 43.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;498:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 44.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 45.Rocchetta H L, Burrows L L, Pacan J C, Lam J S. Three rhamnosyltransferases responsible for assembly of the A-band d-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol Microbiol. 1998;28:1103–1119. doi: 10.1046/j.1365-2958.1998.00871.x. [DOI] [PubMed] [Google Scholar]
- 46.Rosenow C, Esumeh F, Roberts I S, Jann K. Characterization and localization of the KpsE protein from Escherichia coli K5, which is involved in polysaccharide export. J Bacteriol. 1995;177:1137–1143. doi: 10.1128/jb.177.5.1137-1143.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanford J P. Pseudomonas species (including melioidosis and glanders) In: Mandell G L, Douglas R G Jr, Bennett J E, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone; 1995. pp. 2003–2009. [Google Scholar]
- 48.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 49.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 50.Smith A N, Boulnois G J, Roberts I S. Molecular analysis of the Escherichia coli K5 kps locus: identification and characterization of an inner-membrane capsular polysaccharide transport system. Mol Microbiol. 1990;4:1863–1869. doi: 10.1111/j.1365-2958.1990.tb02035.x. [DOI] [PubMed] [Google Scholar]
- 51.Steinmetz I, Rohde M, Brenneke B. Purification and characterization of an exopolysaccharide of Burkholderia (Pseudomonas) pseudomallei. Infect Immun. 1995;63:3959–3965. doi: 10.1128/iai.63.10.3959-3965.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinmetz I, Reganzerowski A, Brenneke B, Haussler S, Simpson A, White N J. Rapid identification of Burkholderia pseudomallei by latex agglutination based on an exopolysaccharide-specific monoclonal antibody. J Clin Microbiol. 1999;37:225–228. doi: 10.1128/jcm.37.1.225-228.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strauss E J, Falkow S. Microbial pathogenesis: genomics and beyond. Science. 1997;276:707–712. doi: 10.1126/science.276.5313.707. [DOI] [PubMed] [Google Scholar]
- 54.Trakulsomboon S, Dance D A B, Smith M D, White N J, Pitt T L. Ribotype differences between clinical and environmental isolates of Burkholderia pseudomallei. J Med Microbiol. 1997;46:565–570. doi: 10.1099/00222615-46-7-565. [DOI] [PubMed] [Google Scholar]
- 55.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 56.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, De Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 59.Whitfield C, Roberts I S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 60.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]
- 61.Wunder D E, Aaronson W, Hayes S F, Bliss J M, Silver R P. Nucleotide sequence and mutational analysis of the gene encoding KpsD, a periplasmic protein involved in transport of polysialic acid in Escherichia coli K1. J Bacteriol. 1994;176:4025–4033. doi: 10.1128/jb.176.13.4025-4033.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wuthiekanun V, Smith M D, Dance D A, Walsh A L, Pitt T L, White N J. Biochemical characteristics of clinical and environmental isolates of Burkholderia pseudomallei. J Med Microbiol. 1996;45:408–412. doi: 10.1099/00222615-45-6-408. [DOI] [PubMed] [Google Scholar]