Abstract
Staphylococcus aureus pathogenicity is mainly due to the production of a number of secreted and cell surface-associated proteins under the regulation of the agr gene. A region of the agr gene was used to subgroup S. aureus strains according to restriction fragment length polymorphisms. Additionally, strains were subtyped according to the coagulase gene in order to strengthen discriminatory power. Virulence capabilities of agr genotype subgroups were evaluated using an in vitro neutrophil bactericidal assay, which showed that prevalent genotypes were significantly better at evading this primary host defense. Multiplex PCR was then used to detect enterotoxin genes among the genotype subgroups in order to determine possible virulence candidates that enable strains to combat neutrophil killing. The prevalent genotype strains were found to possess higher production capabilities for enterotoxin A than did low-prevalence strains. The significance of enterotoxin A production capabilities in affecting pathogenicity of S. aureus strains was evaluated and found to have a profound effect on neutrophil killing abilities. The use of a large epidemiological database as a tool for subgrouping strains with varying degrees of pathogenicity has allowed the identification of relevant and previously undefined virulence factors that affect a pathogen's capability to overcome host immune defenses.
Staphylococcus aureus is a gram-positive bacterium that has remained a persistent pathogen, causing such infections as endocarditis, meningitis, and toxic shock syndrome in humans. S. aureus also is the leading cause of intramammary infections (mastitis), especially in dairy animals, from whose milk it is frequently isolated (38). Neutrophils are the principle line of defense during the initial stages of mastitis, and the ability of these cells to phagocytize and kill invading bacteria is critically related to the establishment of new intramammary infections (26). Therefore, any bacterially derived component that may compromise neutrophil function would constitute an important virulence factor in the pathogenesis of S. aureus mastitis. Although a number of different virulence factors involved in the pathogenesis of S. aureus mastitis have been identified (38), the differential expression of these factors as it relates to field strain prevalence of S. aureus genotypes has not been investigated. A better understanding of the epidemiology of S. aureus mastitis as it pertains to virulence will provide insight concerning important host-pathogen interactions during the pathogenesis of disease.
Subtyping is an important tool for epidemiologic investigation of bacterial infections. In the past decade, numerous molecular techniques such as multilocus enzyme electrophoresis, phage typing, plasmid DNA restriction patterns, random amplified polymorphic DNA ribotyping, and coagulase genotyping have proved useful in identification and comparison of S. aureus isolates in epidemiological studies (7, 21, 29, 35, 36). However, very few studies have identified S. aureus isolates by the gene polymorphisms among important virulence-related genes. Among the virulence-related genes in S. aureus, we were particularly interested in the accessory gene regulator (agr), which has been shown to regulate the synthesis of many virulence factors during bacterial growth (5, 25). The agr system coordinately down-regulates the production of cell wall-associated proteins and up-regulates secreted proteins at late to stationary growth phase in vitro (16, 24, 25, 27). The agr locus encodes a two-component signal-transducing system consisting of two divergent transcription units driven by promoters P2 and P3 (15). The P3 operon encodes the transcript for RNAIII, the effector of the agr response, while the P2 operon contains transcripts for four open reading frames designated agrA, -B, -C, and -D (6). agrB and -D generate an autoinducing peptide that acts as an activating ligand for agrC. Interestingly, Ji et al. (15, 16) have shown that variations in the gene sequences of agrB and -D result in variation of the autoinducing peptide that, in turn, causes differences in the activation of strains by one another (15, 16). In contrast, mutations of wild-type S. aureus strains resulting in agr deletions reduced persistence of infection, exotoxin synthesis, and binding capabilities and decreased intracellular growth of these strains (3, 11, 37), suggesting that agr itself is an important virulence gene in S. aureus.
The aim of this study was to identify potential S. aureus virulence factors based on their unique expression in predominant field strains isolated from clinical cases of mastitis. In this study, we developed a genotyping method for S. aureus strains based on agr gene polymorphisms. We showed that enterotoxin production capabilities were more pronounced in the prevalent agr genotypes that were more resistant to neutrophil bactericidal activities than were the low-prevalence genotypes. The ability of enterotoxin A to directly modify neutrophil function suggests an important role of these toxins in the pathogenesis of mastitis.
MATERIALS AND METHODS
Bacterial isolates.
The S. aureus strains used in this study included RN6390B, a wild-type agr+ strain (25); RN6911, an agr mutant (25); SA 502A (ATCC 27217); and 255 field isolates. S. aureus field isolates were collected from clinical mastitis bovine milk samples from the Czech Republic (n = 10), France (n = 34), Korea (n = 165), and several locations within the United States (n = 46), (including Indiana, Kentucky, Louisiana, Minnesota, New York, Pennsylvania, Tennessee, Washington, and Wisconsin). The variety of geographical locations from which isolates were collected provided control for regional differences in herd management and herd differences that result in variations in host resistance to disease. All isolates were stored in Trypticase soy broth with 15% glycerol at −70°C until needed. Isolates were cultured on Trypticase soy agar with 5% sheep blood (BiMed, St. Paul, Minn.) for identification based on colony morphology, hemolysis, Gram stain, and acetoin and catalase production. Coagulase production by isolates was determined in a tube test using 0.5 ml of citrate-stabilized rabbit plasma. One colony from an overnight culture on Trypticase soy agar–5% sheep blood was inoculated into a plasma-containing tube and incubated at 37°C. A positive test was determined by clot formation after 1, 4, or 24 h. To differentiate S. aureus from coagulase-positive S. hyicus and S. intermedius, the acetoin test (Voges-Proskauer test) was used as outlined previously (1). All isolates with a questionable acetoin test result were further identified by the API Staph system (bioMerieux Vitek, Hazelwood, Mo.).
Bacterial DNA lysates.
Bacterial DNA lysates were prepared from 1 ml of an overnight TSB culture. Bacteria were then pelleted and resuspended in 500 μl of 50 mM Tris-HCl buffer (pH 8.3) that contained 50 mM disodium EDTA. Lysis of cells was conducted using 15 U of lysostaphin (Sigma, St. Louis, Mo.) and incubating at 37°C for 30 min. Lysis was completed by adding 1 ml of lysis buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 1% Triton X-100, 0.45% Igepal CA-630, 0.45% Tween 20, and 0.6 μg of proteinase K and incubating for 1 h at 56°C. Proteinase K was inactivated by heating at 95°C for 10 min.
agr genotyping.
In order to amplify the variable region of the agr gene in the S. aureus strains, nested primers were designed using S. aureus sequences available from GenBank and the DNASTAR (Madison, Wis.) software program. The sequences of the outer primers were 5′-ACCAGTTTGCCACGTATCTCA-3′ and 5′-AACCACGACCTTCACCTTTAGTAG-3′. Amplification was conducted in a total volume of 40 μl consisting of 10 μl of cell lysate, 4 μl of 10× buffer, 0.4 μl of deoxyribonucleoside triphosphate (dNTP) (25 mM [each] dATP, dCTP, dGTP, and dTTP), a 2 μM concentration of each primer, 2.4 μl of MgCl2 (25 mM), 2.5 U of Taq DNA polymerase, and 8 μl of nuclease-free water. Reactions were cycled as follows: 95°C for 30 s, 64°C for 60 s, and 72°C for 120 s for 34 cycles.
For the nested primer amplification, 1 μl of the first PCR mixture was added to 39 μl of PCR mixture containing 2 μM concentrations of the second set of primers: 5′-TGCCACGTATCTTCAAA-3′ and 5′-ATAATCATGACGGAACTT-3′. The nested PCR amplification conditions were the same as above with an annealing temperature of 54 instead of 64°C.
Ten microliters of the second PCR mixture was digested at 37°C for 1 h with 2 U of the restriction endonuclease AluI (Promega, Madison, Wis.) according to the manufacturer's instruction. The digested DNA fragments were separated in a 3% agarose gel (Sigma) and visualized in the presence of ethidium bromide under UV light.
Coagulase genotyping.
The coagulase genotyping was performed by a previously described method (1). Ten microliters of DNA lysates was added to a mixture containing a 1 μM concentration of each primer (COAG1, 5′-ATACTCAACCGACGACACCG-3′, and COAG4, 5′-GATTTTGGATGAAGCGGATT-3′), 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl, 1% Triton X-100, a 200 μM concentration of each dNTP, and 1 U of Taq polymerase to a final reaction volume of 40 μl. Each sample was subjected to 40 PCR cycles, consisting of 30 s at 95°C, 2 min at 55°C, and 2 min at 72°C. For the nested-PCR amplification, 1 μl of the first PCR mixture was added to 39 μl of PCR mixture containing a 1 μM concentration of the second set of primers (COAG2, 5′-ACCACAAGGTACTGAATCAACG-3′, and COAG3, 5′-TGCTTTCGATTGTTCGATGC-3′). The nested-PCR amplification was performed using the same conditions as the first PCR. Ten microliters of the second PCR mixture was digested at 37°C for 1 h with 2 U of the restriction endonuclease AluI according to the manufacturer's instruction. The digested DNA fragments were separated in 4% NuSieve GTG agarose gel (FMC BioProducts, Rockland, Maine) and detected in the presence of ethidium bromide under UV illumination.
Detection of enterotoxin genes by multiplex PCR.
Enterotoxin typing was conducted according to the following methodology. In brief, staphylococcal genomic DNA was extracted from lysostaphin-treated cells and processed as described previously (20). The DNA was extracted with phenol-chloroform (1:1, vol/vol) and chloroform and then precipitated with ethanol according to standard techniques (28). Specific primers for staphylococcal enterotoxin A (SEA), SEE, and toxic shock syndrome toxin 1 (TSST-1) were synthesized with a DNA synthesizer (Expedite 8905; Perseptive Co.) as described previously by Johnson et al. (17, 18, 28). The oligonucleotide sequence of each primer is shown in Table 1. The PCR was performed under the following parameters: the reaction mixture consisted of 2.5 μl of 10× reaction buffer without MgCl2 (Promega Corp.); 400 M dNTP; 3 mM MgCl2; 7.5% dimethyl sulfoxide; 50 pmol of primers for sea, seb, sec, sed, and see; 100 pmol of primers for tst; and 100 ng of template DNA; and brought up to a 25-μl final volume with distilled water. Reactions were hot started for 5 min at 95°C and placed on ice, and 1 U of Taq polymerase (Promega Corp.) was added. Each sample was subjected to 30 PCR cycles, consisting of 95°C for 1 min, 2 min at 56°C (for the combination of primer sets for SEA, SEC, and SED) or 50°C (for SEB, SEE, and TSST-1), and 1 min at 72°C. PCR products were separated on a 1.5% agarose gel and visualized under UV illumination.
TABLE 1.
Nucleotide sequences of primers used in multiplex PCR
| Gene | Primer | 5′-3′ sequence | Location of gene | Product size (bp) |
|---|---|---|---|---|
| sea | SEA-1 | TTGGAAACGGTTAAAACGAA | 490–509 | 121 |
| SEA-2 | GAACCTTCCCATCAAAAACA | 591–610 | ||
| seb | SEB-1 | TCGCATCAAACTGACAAAGG | 634–653 | 477 |
| SEB-2 | GCAGGTACTCTATAAGTGCC | 1091–1110 | ||
| SEC | SEC-1 | GACATAAAAGCTAGGAATTT | 676–695 | 257 |
| SEC-2 | AAATCGGATTAACATTATCC | 913–932 | ||
| sed | SED-1 | TAGATAAAGTTAAAACAAGC | 354–373 | 318 |
| SED-2 | TAACTTACCGTGGACCTTC | 652–671 | ||
| see | SEE-1 | TAGATAAAGTTAAAACAAGC | 491–510 | 169 |
| SEE-2 | TAACTTACCGTGGACCCTTC | 640–659 | ||
| tst | TST-1 | ATGGCTATATACATTCAATT | 251–270 | 350 |
| TST-2 | TTTCCAATAACCACCCGTTT | 581–600 |
Statistical analysis.
Discriminatory powers of both coagulase and agr genotyping were evaluated as described by Hunter (14). Concordance analysis (10) of agr genotype, coagulase genotype, and enterotoxin production capabilities was conducted by using the Minitab (State College, Pa.) statistical program to determine matches and mismatches among isolates within the same and different groups. Pairwise comparisons (number of combinations) were determined with the following formula: Pk,n/k! = n!/k!(n − k)!, where n = number of samples and k = number of combinations chosen. The G test of independence using Yate's correction for continuity (31) was used to evaluate statistical significance.
Bovine blood neutrophil bactericidal assay.
The functional capabilities of bovine neutrophils are a major factor which determines the establishment of new intramammary infections. For this reason, a series of in vitro assays were conducted to assess the relative abilities of certain strains to resist this important host defense mechanism. Bovine neutrophils were isolated from four lactating Holstein cows free of intramammary infection as determined by microbiological analyses of milk samples. Neutrophils were isolated as previously described (2). For the purpose of opsonization, bovine antiserum was collected from cows diagnosed with S. aureus mastitis.
Ten S. aureus isolates were selected from both predominant (n = 5) and rare (n = 5) genotypes to evaluate differences in capabilities of prevalence groups to evade neutrophil killing. Additionally, eight S. aureus isolates were selected from predominant genotypes with enterotoxin genes (n = 4) and without enterotoxin genes (n = 4) to evaluate the effect of enterotoxin genes on neutrophil killing capabilities. Bacteria were prepared by initial culturing in 30 ml of assay medium (RPMI–5% fetal bovine serum–1% l-glutamine) at 37°C for 6 to 12 h. After incubation bacterial concentration was determined via serial dilutions. Bacterial inocula were stored at 4°C, while concentrations were determined and final concentrations were adjusted to 107 CFU/ml in assay medium. The resistance of bacteria to bovine neutrophil bactericidal activities was evaluated by the bactericidal assay as previously described (2). In brief, bacteria were opsonized in 6.25% bovine antiserum for 30 min at 37°C. In a 96-well plate, 100 μl of bacteria (107 CFU/ml) was combined with 100 μl of neutrophils (108/ml) and incubated at 37°C for 1 h. Following incubation, neutrophils were lysed with the addition of 0.2% Saponin (Sigma) followed by the addition of MTT (1 mg/ml; Sigma). Upon color development, the addition of extraction buffer lysed bacterial cells. Plates were read at a wavelength of 595 nm on a microplate reader (Bio-Rad, Hercules, Calif.).
Additional assays were conducted to determine if SEA, the enterotoxin gene type observed most frequently in field isolates, could affect neutrophil bactericidal activity. For experiments involving SEA (Sigma), neutrophils were preincubated with various concentrations of enterotoxin A for 15 min at 37°C in order to determine the direct effect of this enterotoxin on neutrophil killing capabilities. The dessicated SEA contained 10% protein and 90% sodium phosphate buffer by weight; therefore, neutrophils were preincubated with 4 mM sodium phosphate, the highest concentration found when 20 μg SEA was tested, to serve as control. Bacterial strain Newbould 305 was used to determine the effect of SEA on neutrophil bactericidal capability. Bacteria were inoculated as described above and washed three times in phosphate-buffered saline to remove any secreted enterotoxin produced during growth. Bacteria were then resuspended in assay medium and counted as described above. Student's t test was used to compare percentages of bacteria killed for strains within each prevalence or treatment group.
Detection of enterotoxin protein.
S. aureus strains used in the neutrophil bactericidal assays for toxin evaluation were further studied to determine protein production of enterotoxins under assay conditions. Bacteria were prepared by initial culturing in 30 ml of assay medium (RPMI–5% fetal bovine serum–1% l-glutamine) at 37°C for 6 to 12 h. After incubation, bacterial cultures were centrifuged at 3,500 × g for 5 min at 15°C. Supernatants were then filter sterilized and used in an enzyme immunoassay for the detection of enterotoxins A, B, C, D, and E (RIDASCREEN Set A,B,C,D,E; r-biopharm, Darmstadt, Germany).
RESULTS
agr genotyping.
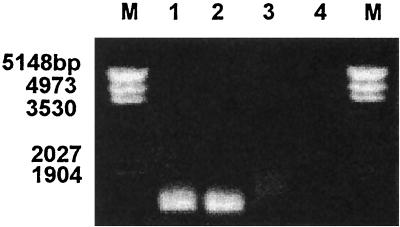
Available agr sequences were analyzed in order to determine which region of the gene had the largest diversity to ensure that a variety of restriction fragment length polymorphism profiles would be attained (Table 2). DNA sequence analysis of several S. aureus strains showed highly conserved regions at the 5′ end of the agrB gene and the 3′ end of the agrC gene (GenBank sequence accession numbers AF001782, AF001783, U85095, and X52543). These conserved sequences were used to design primers to amplify agr gene fragments displaying areas of high divergence found within the stable regions. Specificity of the designed primers was confirmed using Escherichia coli and an agr S. aureus mutant RN6391 (Fig. 1). Sequencing of PCR products from S. aureus ATCC 27217 and S. aureus RN6390 confirmed that the PCR product was the targeted region of the agr locus an expected product of approximately 1,386 bp.
TABLE 2.
agr genotype profiles of S. aureus strains (n = 248)
| Type | agr restriction pattern subgroupsa | % Occurrence |
|---|---|---|
| 1 | 8-480,385,310,257,168,156,104,75 | 47.8 |
| 2 | 4-557,486,181,144 | 17.7 |
| 3 | 4-750,480,385,75 | 9.6 |
| 4 | 6-452,373,188,144,104,75 | 9.6 |
| 5 | 3-680,557,144 | 4.4 |
| 6 | 5-680,557,486,181,144 | 1.6 |
| 7 | 3-750,385,66 | 1.2 |
| 8 | 5-500,257,168,156,75 | 1.2 |
| 9 | 6-620,480,385,188,104,75 | 0.8 |
| 10 | 4-848,257,140,70 | 0.8 |
| 11 | 4-480,257,168,156 | 0.8 |
| 12 | 6-668,405,362,150,104,75 | 0.8 |
| 13 | 3-557,486,144 | 0.8 |
| 14 | 6-747,449,395,333,257,75 | 0.4 |
| 15 | 3-570,480,257 | 0.4 |
| 16 | 3-330,255,168 | 0.4 |
| 17 | 2-643,557 | 0.4 |
| 18 | 5-680,385,181,114,75 | 0.4 |
| 19 | 7-557,480,385,181,168,104,75 | 0.4 |
| 20 | 6-680,480,410,287,104,75 | 0.4 |
First digit indicates number of bands; subsequent values indicate the molecular weight (in thousands) of each band.
FIG. 1.
Nested PCR products of S. aureus ATCC 27217 (lane 1), RN6390 (lane 2), agr mutant RN6391 (lane 4), and E. coli (lane 3).
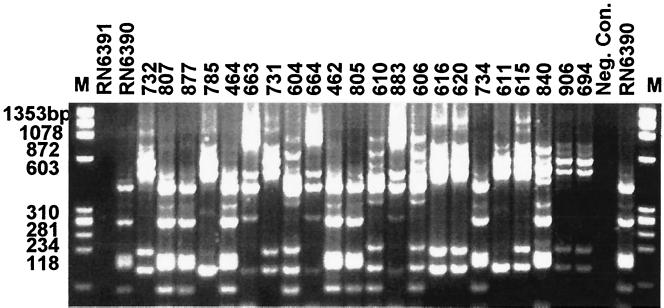
S. aureus strains collected from clinical bovine milk samples from Czech Republic, France, Korea, and several states in the United States were genotyped by agr gene polymorphism. Twenty genotypes have been identified, and an example of an agarose gel electrophoresis is shown in Fig. 2. Genotype profiles were designated by the total number of major bands followed by the estimated molecular weight (in thousands) of each band. The most prevalent type was found to have eight major bands with molecular weights of 480, 385, 310, 257, 168, 156, 104, and 75 and was therefore designated 8:480, 385, 310, 257, 168, 156, 104, 75. The frequencies of all the identified agr genotypes within the tested samples are shown in Table 2. From a total of 255 strains tested, seven samples were nontypeable using agr genotyping methods. agr genotyping methodology had a discriminatory power (D) of 0.7217.
FIG. 2.
Example of restriction fragment profiles of the agr gene for 26 S. aureus strains.
Coagulase genotyping.
The 255 available isolates were subdivided into 40 different coagulase types according to previously designated typing patterns (1). There were four samples that were nontypeable using the coagulase genotyping method. Coagulase typing had a very high D of 0.938. As shown previously, only a few genotypes predominate in each country (32). However, comparisons among countries indicated that predominant types could be distributed in different geographical locations (32).
Detection of enterotoxin genes by multiplex PCR.
The presence of enterotoxin genes was assessed in 211 isolates randomly chosen from the available 255 isolates from various locations. Of the isolates tested, 118 isolates tested negative for enterotoxin genes (56%), and 93 isolates were positive for enterotoxin genes (44%). Of the isolates positive for enterotoxin genes 58 were positive for SEA (62.4%) only, 3 were positive for SEB (3.2%) only, 5 were positive for SEC (5.4%) only, 3 were positive for SED (3.2%) only, and 4 were positive for TSST (4.3%) only. In addition, 20 of the isolates tested positive for more than one enterotoxin gene: 4 isolates were positive for both SEA and SEC (4.3%); 2 isolates were positive for SEA, SEC, and TSST (2.15%); 2 isolates were positive for SEA and TSST (2.15%); 3 isolates tested positive for SEC, SED, and TSST (3.2%); and 9 isolates possessed SEC and TSST (9.7%) genes. Selected strains of S. aureus used in the bactericidal assays also were evaluated for expression of enterotoxin genes. Strains with enterotoxin genes were found to produce the respective enterotoxin protein as evaluated through an enzyme immunoassay. Additionally, strains that tested negative for enterotoxin genes did not test positive for any enterotoxin protein production under assay conditions (data not shown).
Concordance analyses of typing techniques.
All possible pairs of agr genotyping and coagulase genotyping were compared among the 248 samples that were typeable using the two techniques. Pairs of isolates were classified by whether they possessed the same or different agr genotypes and whether they matched or mismatched in coagulase genotypes (Table 3). A total of 30,628 pairwise comparisons for the 248 isolates were possible. For the isolates with the same agr type, 4% had matching and 23% had mismatching coagulase genotypes. For the isolates with different agr types, 2% had matching and 71% had mismatching coagulase genotypes. The overall concordance percentage for coagulase and agr genotypes was 75.2% (simple matching coefficient [S] = 0.752) and had a significance of P < 0.001 (G test of independence, G = 1621.4; df = 1).
TABLE 3.
Concordance analysis of agr gene type and coagulase gene typea
| agr type | No. of isolate pairs (proportion of total comparisons) with coagulase type
|
|
|---|---|---|
| Match | Mismatch | |
| Same | 1,335 (0.04) | 6,998 (0.23) |
| Different | 575 (0.02) | 21,720 (0.71) |
Concordance equals the sum of the same coagulase-match and different coagulase-mismatch entries, expressed as a percentage of the total 30,628 pairwise comparisons. For data presented here, the concordance is 75.2%.
Concordance analysis of genotyping methods and enterotoxin typing.
Of the 211 isolates tested for enterotoxin type, 6 isolates had unidentifiable agr genotypes and 3 isolates had unidentifiable coagulase types. Pairs of isolates were identified by whether they had the same or different enterotoxin type and whether they matched or mismatched in either agr genotype or coagulase genotype (Table 4). Of the total of 21,910 possible pairwise comparisons between enterotoxin type and agr genotype (205 isolates), 10% of the same enterotoxin type had matching agr genotypes and 29% had mismatching agr genotypes. For the isolates with different enterotoxin types, 17% had matching and 44% had mismatching agr genotypes. The overall concordance of enterotoxin type with agr genotype was 54.3% (simple matching coefficient, S = 0.543) and had a significance of P < 0.001 (G test of independence, G = 14.2; df = 1).
TABLE 4.
Concordance analysis of enterotoxin type with agr genotype and coagulase genotypea
| Enterotoxin | No. of isolate pairs (proportion of total comparison) with:
|
|||
|---|---|---|---|---|
|
agr genotype
|
Coagulase genotype
|
|||
| Match | Mismatch | Match | Mismatch | |
| Same | 2,012 (0.10) | 6,038 (0.29) | 636 (0.03) | 7,699 (0.36) |
| Different | 3,518 (0.17) | 9,342 (0.44) | 659 (0.03) | 12,543 (0.58) |
Concordance equals the sum of the same enterotoxin-match and different enterotoxin-mismatch entries, expressed as a percentage of the total pairwise comparisons, i.e., 21,910 for agr genotype and 21,528 for coagulase genotype. Thus, the concordance for agr genotype is 54.3%, and that for coagulase genotype is 61.2%.
In the total of 21,528 possible pairwise comparisons between enterotoxin type and coagulase genotype (208 isolates), 3% of same enterotoxin type had a matching coagulase genotype and 36% had a mismatching coagulase genotypes. For the isolates with different enterotoxin types, 3% had matching and 58% had mismatching coagulase genotypes. The overall concordance of enterotoxin type with coagulase type was 61.2% (simple matching coefficient, S = 0.612) and had a significance of P < 0.001 (G test of independence, G = 64.4; df = 1).
Bovine blood neutrophil bactericidal assay.
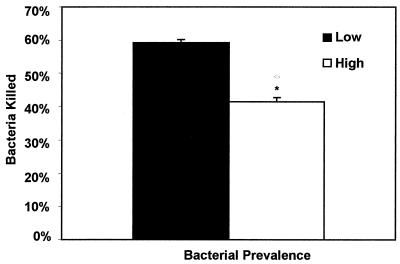
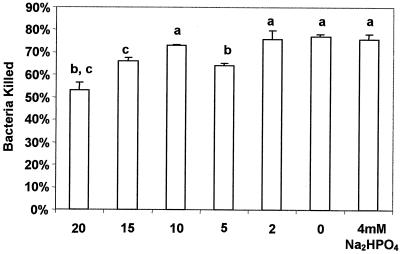
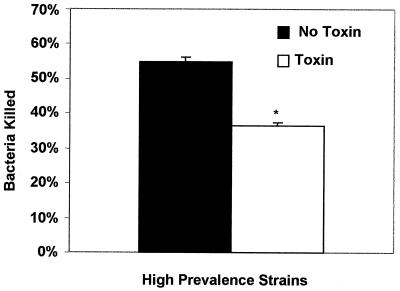
The results of bactericidal assays are shown in Fig. 3 to 5. The mean percentage of killing was 41% (standard error [SE] = 1.4) for the five high prevalence S. aureus strains and 59% (SE = 0.9) for the five low-prevalence strains evaluated. There was a significant difference in bactericidal effects (P = 0.004). Additionally, the mean percentage of killing was 35% (SE = 1.6) for four high-prevalence strains with enterotoxin genes and 54% (SE = 1.2) for strains without enterotoxin genes. A significant difference was found in bactericidal effects (P = 0.005) between those strains with and without enterotoxin genes. Strains with enterotoxin genes were found to produce the respective enterotoxin protein as evaluated through an enzyme immunoassay (data not shown). Additionally, strains that tested negative for enterotoxin genes did not test positive for any enterotoxin protein production under assay conditions. Addition of exogenous SEA to the bactericidal assay resulted in a significant decrease in neutrophil killing capabilities (P < 0.05) at concentrations of 5, 15, and 20 μg.
FIG. 3.
Bovine blood neutrophil bactericidal activity against either high- or low-prevalence S. aureus strains (∗, P < 0.005). Error bars, SE.
FIG. 5.
Bovine blood neutrophil bactericidal activity against S. aureus strain Newbould 305 in the presence of enterotoxin A. Bars labeled with different letters indicate that values differ significantly (P < 0.05). Error bars, SE.
DISCUSSION
Previous work in our laboratory has used the hypervariable region of the coagulase gene to type S. aureus strains (32). The coagulase gene was chosen due to its ubiquitous presence among S. aureus strains, and this gene proved to create a very powerful typing method. In this study, the agr gene also was used to further type strains and provided a means of grouping S. aureus isolates based on a factor that controls virulence-related gene production. The agr gene was used successfully to subtype S. aureus strains, and when the information attained from genotyping of the coagulase gene and the agr gene was combined, the discriminatory power of each method was greatly empowered. The ability to group pathogens based on field prevalence may provide important information pertaining to the coevolution of hosts and pathogens which will influence the genetic diversity of disease-causing microorganisms (13, 22). Selection within the pathogen population will favor mechanisms to avoid host defense and to colonize the host. The pathogens that are most efficient at avoiding the host's defense mechanisms will be the most prevalent type found in the microenvironment of interest. The argument for this phenomenon is supported by the findings in this study in which the abilities of neutrophils to kill different S. aureus genotypes varied with respect to their prevalence in cases of mastitis. We discovered that the most common genotype of S. aureus also was the type against which the neutrophils, and thereby the host's initial defense mechanism, were least efficient. In contrast, the neutrophils were highly efficient against the rarely found genotypes. These results are consistent with our previous reports (2, 32) and suggest that types found in high prevalence have unique characteristics, which in contrast to the rare types, endow them with the superior ability to suppress or resist killing by neutrophils. In the past, in vitro studies of host-pathogen interactions have focused on the use of few selective bacterial strains studied routinely in a laboratory environment (3, 4, 12). However, statistical analysis of a large database of pathogenic strains may provide critical information concerning key virulence-related genes in a prevalent group of disease-causing bacteria that can be evaluated relative to the interaction between pathogen and host in a specific disease model.
Bacterium-host interactions depend on several factors, including the efficiency of the host's defense, growth rate of the bacteria, and production of virulence factors. The neutrophils used in the assays were obtained from the same group of animals, so differences in killing ability cannot be attributed to variations in host defense. In addition, no differences in the growth potential were observed among the strains used in the bactericidal assays. One possible explanation for the variation in killing efficiency may be a consequence of the expression of certain bacterially produced factors. S. aureus has the capacity to synthesize a repertoire of known virulence factors associated with mastitis, including capsular polysaccharide, cell-surface associated proteins, and several hemolytic toxins (33). However, the potential role of enterotoxins in the pathogenesis of S. aureus mastitis is uncertain and has been the topic of several conflicting studies (9, 19). In this study, the use of multiplex PCR technology of the enterotoxin gene was conducted to evaluate the significance of this group of potential virulence factors associated with mastitis-causing S. aureus strains. Use of this technique to evaluate enterotoxin genes eliminated the variations previously observed for enterotoxin detection as a result of various growth conditions influencing the cell density sensing system that controls virulence factor production in S. aureus (16). We found that 44% of S. aureus isolates tested contained enterotoxin genes, and these values are consistent with previous published findings using similar methodologies (20). The significant relationship between enterotoxin types and agr genotypes reported here is supported by previous work in which differences were observed in toxin production due to mutations in the agr gene (11, 25).
It was interesting to further delineate if the ability of prevalent S. aureus strains to evade host defense mechanisms was related, at least in part, to enterotoxin production capabilities. We showed that strains with enterotoxin genes were significantly (P = 0.005) better at evading this nonspecific cellular defense mechanism of the host compared to strains without these genes. S. aureus enterotoxins have been shown to activate T-cell subsets (34) as well as provide protection against neutrophil apoptosis (23). However, very limited information is available as to the effect of enterotoxins on neutrophil bactericidal activities. In order to establish a direct link between status of the agr gene, enterotoxin genes, and neutrophil killing capabilities, specific S. aureus enterotoxins were studied for the ability to alter bactericidal activities of neutrophils. We assessed the direct effects of SEA on neutrophil bactericidal capabilities since this enterotoxin gene was most commonly observed among the most prevalent agr genotypes. The addition of exogenous SEA was shown to significantly reduce the killing abilities of neutrophils under our assay conditions. Previously, work by Berger et al. (8) observed no effect on SEA on neutrophil bactericidal activity. However, the preincubation of neutrophils with enterotoxin prior to evaluation of killing abilities, as carried out in the present study, may be the deciding factor in the varying results observed. Though there are several putative virulence factors that can account for the effect on neutrophil killing ability observed in Fig. 3, singling out SEA, the most-common enterotoxin gene possessed by the isolates in this study provides a direct correlation between variations in the agr gene sequences, enterotoxin production, and ability to evade neutrophil killing.
The epidemiological study of a large database of disease-causing bacteria can be used to identify uniquely expressed virulence factors that are associated with the ability of pathogens to evade important host defense mechanisms. Using this approach, we showed for the first time that staphylococcus enterotoxins can contribute to the pathogenesis of S. aureus mastitis as suggested by the agr genotype field prevalence data. We also showed that one possible mechanism by which SEA may affect virulence is through its ability to directly hinder neutrophil bactericidal activities. A better understanding of important host-pathogen interactions that contribute to field prevalence of a specific disease may provide greater insight into the development of effective intervention strategies.
FIG. 4.
Bovine blood neutrophil bactericidal activity against high-prevalence S. aureus strain with or without enterotoxin genes. ∗, P = 0.005; error bars, SE.
ACKNOWLEDGMENTS
This research was supported in part by agricultural research funds administered by the Pennsylvania Department of Agriculture (ME445126) and the USDA-BARD Grants Program (US-2648-95).
We appreciate B. Poutrel for providing S. aureus isolates from France and R. P. Novick for providing S. aureus isolates RN6390B and RN6911.
REFERENCES
- 1.Aarestrup F M, Dangler C A, Sordillo L M. Prevalence of coagulase gene polymorphism in Staphylococcus aureus isolates causing bovine mastitis. Can J Vet Res. 1995;59:124–128. [PMC free article] [PubMed] [Google Scholar]
- 2.Aarestrup F M, Scott N L, Sordillo L M. Ability of Staphylococcus aureus coagulase genotypes to resist neutrophil bactericidal activity and phagocytosis. Infect Immun. 1994;62:5679–5682. doi: 10.1128/iai.62.12.5679-5682.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson J C, Williams M R. The contribution of a capsule to survival of staphylococci within bovine neutrophils. J Med Microbiol. 1985;20:317–323. doi: 10.1099/00222615-20-3-317. [DOI] [PubMed] [Google Scholar]
- 5.Balaban N, Novick R P. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3′-end deletion. FEMS Microbiol Lett. 1995;133:155–161. doi: 10.1111/j.1574-6968.1995.tb07877.x. [DOI] [PubMed] [Google Scholar]
- 6.Balban N, Novick R P. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc Natl Acad Sci USA. 1995;92:1619–1623. doi: 10.1073/pnas.92.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumgartner A, Nicolet J, Eggimann M. Plasmid profiles of Staphylococcus aureus causing bovine mastitis. J Appl Bacteriol. 1984;56:159–163. doi: 10.1111/j.1365-2672.1984.tb04708.x. [DOI] [PubMed] [Google Scholar]
- 8.Berger E M, Shibao G A, Brown S N, Repine J E. Variable effect of toxic shock toxins from different sources on neutrophil function in vitro. Inflammation. 1988;12:447–453. doi: 10.1007/BF00919438. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald J R, Hartigan P J, Meaney W J, Smyth C J. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J Appl Microbiol. 2000;88:1028–1037. doi: 10.1046/j.1365-2672.2000.01071.x. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald J R, Meaney W J, Hartigan P J, Smyth C J, Kapur V. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol Infect. 1997;119:261–269. doi: 10.1017/s0950268897007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillaspy A F, Hickmon S G, Skinner R A, Thomas J R, Nelson C L, Smeltzer M S. Role of accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guidry A J, Oliver S P, Squiggins K E, Erbe E F, Dowlen H H, Hambleton C N, Berning L M. Effect of anticapsular antibodies on neutrophil phagocytosis of Staphylococcus aureus. J Dairy Sci. 1991;74:3360–3369. doi: 10.3168/jds.S0022-0302(91)78525-1. [DOI] [PubMed] [Google Scholar]
- 13.Hafner M S, Nadler S A. Phylogenetic coevolution of parasites and their hosts. Nature. 1988;332:258–259. doi: 10.1038/332258a0. [DOI] [PubMed] [Google Scholar]
- 14.Hunter P R. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji G, Beavis R, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 16.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson W H, Tyler S D. Diagnostic molecular microbiology. Washington, D.C.: American Society for Microbiology; 1993. PCR detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin-1 in Staphylococcus aureus. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.) [Google Scholar]
- 18.Johnson W M, Tyler S D, Ewans E P, Ashton F E, Pollard D R, Rozee K R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson H D, Huda A, Eriksen N H, Jensen N E. Differences between Danish bovine and human Staphylococcus aureus isolates in possession of superantigens. Vet Microbiol. 2000;76:153–162. doi: 10.1016/s0378-1135(00)00232-7. [DOI] [PubMed] [Google Scholar]
- 20.Lee S U, Quesnell M, Lawrence K F, Yoon J W, Park Y H, Davis W C, Falk D, Deobald C F, Bohach G A. Characterization of staphylococcal bovine mastitis isolates using the polymerase chain reaction. J Food Prot. 1998;61:1384–1386. doi: 10.4315/0362-028x-61.10.1384. [DOI] [PubMed] [Google Scholar]
- 21.Matthews R C. PCR fingerprinting microbes by random amplification of polymorphic DNA. J Med Microbiol. 1993;39:161–162. doi: 10.1099/00222615-39-3-161. [DOI] [PubMed] [Google Scholar]
- 22.May R M, Anderson R M. Parasite-host coevolution. Parasitology. 1990;100(Suppl.):89–100. doi: 10.1017/s0031182000073042. [DOI] [PubMed] [Google Scholar]
- 23.Moulding D A, Walter C, Hart C A, Edwards S W. Effects of staphylococcal enterotoxins on human neutrophil functions and apoptosis. Infect Immun. 1999;67:2312–2318. doi: 10.1128/iai.67.5.2312-2318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 25.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth G, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paape M J, Wergin W P, Guidry A J, Pearson R E. Leukocytes-second line of defense against invading mastitis pathogens. J Dairy Sci. 1979;62:135–153. doi: 10.3168/jds.S0022-0302(79)83215-4. [DOI] [PubMed] [Google Scholar]
- 27.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick R P. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Saulnier P, Bourneix C, Prevost G, Andremon A. Random amplified polymorphic DNA assay is less discriminant than pulsed-field gel electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:982–985. doi: 10.1128/jcm.31.4.982-985.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shopsin B, Gomez M, Montgomery S O, Smith D H, Waddington M, Dodge D E, Bost D A, Riehman M, Naidich S, Kreiswirth B N. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokahl R R, Rohlf F J. Analysis of frequencies. New York, N.Y: Freeman; 1995. [Google Scholar]
- 32.Su C, Herblin C, Frieze N, Skardova O, Sordillo L M. Coagulase gene polymorphisms of Staphylococcus aureus isolated from dairy cattle in different geographical areas. Epidemiol Infect. 1999;122:329–336. doi: 10.1017/s0950268899002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutra L, Poutrel B. Virulence factors involved in the pathogenesis of bovine intramammary infections due to S. aureus. J Med Microbiol. 1993;40:79–89. doi: 10.1099/00222615-40-2-79. [DOI] [PubMed] [Google Scholar]
- 34.Tessier P A, Naccache P H, Diener K R, Gladue R P, Neote K S, Clark-Lewis I, McColl S R. Induction of acute inflammation in vivo by staphylococcal superantigens. II. Critical role for chemokines, ICAM-1, and TNF-α. J Immun. 1998;161:1204–1211. [PubMed] [Google Scholar]
- 35.Thomson-Carter F M, Carter P E, Pennington T H. Differentiation of staphylococcal species and strains by ribosomal RNA gene restriction patterns. J Gen Microbiol. 1989;135:2093–2097. doi: 10.1099/00221287-135-7-2093. [DOI] [PubMed] [Google Scholar]
- 36.Wang G, Whittam T S, Berg C M, Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wesson C A, Liou L E, M. T K, Nohach G A, Trumble W R, Bayles K W. Staphylococcus aureus Agr and Sar global regulators influence internalization and induction of apoptosis. Infect Immun. 1998;66:5238–5243. doi: 10.1128/iai.66.11.5238-5243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yancey R J J. Vaccines and diagnostic methods for bovine mastitis: fact and fiction. Adv Vet Med. 1999;41:257–273. doi: 10.1016/s0065-3519(99)80020-2. [DOI] [PubMed] [Google Scholar]