Abstract
Fecal microbiota transplantation (FMT) is a promising therapeutic modality for the treatment and prevention of metabolic disease. We previously conducted a double-blind, randomized, placebo-controlled pilot trial of FMT in obese metabolically healthy patients in which we found that FMT enhanced gut bacterial bile acid metabolism and delayed the development of impaired glucose tolerance relative to the placebo control group. Therefore, we conducted a secondary analysis of fecal samples collected from these patients to assess the potential gut microbial species contributing to the effect of FMT to improve metabolic health and increase gut bacterial bile acid metabolism. Fecal samples collected at baseline and after 4 weeks of FMT or placebo treatment underwent shotgun metagenomic analysis. Ultra-high-performance liquid chromatography-mass spectrometry was used to profile fecal bile acids. FMT-enriched bacteria that have been implicated in gut bile acid metabolism included Desulfovibrio fairfieldensis and Clostridium hylemonae. To identify candidate bacteria involved in gut microbial bile acid metabolism, we assessed correlations between bacterial species abundance and bile acid profile, with a focus on bile acid products of gut bacterial metabolism. Bacteroides ovatus and Phocaeicola dorei were positively correlated with unconjugated bile acids. Bifidobacterium adolescentis, Collinsella aerofaciens, and Faecalibacterium prausnitzii were positively correlated with secondary bile acids. Together, these data identify several candidate bacteria that may contribute to the metabolic benefits of FMT and gut bacterial bile acid metabolism that requires further functional validation.
Keywords: bile salt hydrolase (BSH), bile acids, gut microbiota, metagenomics, fecal microbiome transplant (FMT)
1. Introduction
Type 2 diabetes mellitus (T2DM) continues to be a worldwide clinical challenge. The gut microbiota plays an important role in determining host metabolic health and has been associated with T2DM. Studies comparing the composition and function of the fecal microbiota from groups who had T2DM, impaired glucose tolerance, or normal glucose tolerance have reported distinct bacterial compositions. For example, Roseburia and Faecalibacterium prausnitzii are differentially enriched in T2DM [1]. Additionally, decreases in Bacteroidetes and increases in Actinobacteria and Firmicutes are associated with obesity [2,3]. Studies in germ-free mice show that transplantation of the gut microbiota from metabolically healthy vs. metabolically impaired donors transfers these metabolic phenotypes, pointing to a causative role for the gut microbiota in the pathogenesis of metabolic disease [4,5]. Therefore, the gut microbiome is an attractive target for the treatment and prevention of T2DM.
Fecal microbiota transplantation (FMT) is a potential method to target the gut microbiome for T2DM treatment and prevention [6]. FMT has been shown to successfully treat microbiota-related dysfunction, with the treatment of Clostridioides difficile infection being the most notable example of its successful therapeutic use [7,8]. To test the potential utility of FMT for the treatment of metabolic disease, our group previously studied patients with obesity, without metabolic impairment, treated with FMT or placebo. FMT did not induce weight loss but did successfully colonize the gastrointestinal tract of recipients and slowed the development of glucose intolerance compared with placebo, as assessed by mixed meal tolerance testing [9]. The mechanisms for this microbially induced improvement in glucose tolerance are unknown. However, a key mechanism by which the gut microbiota influences host metabolic health is through the production of metabolites, such as short-chain fatty acids (SCFAs) and unconjugated and secondary bile acids. Although SCFA concentrations were not altered by FMT, FMT increased gut bacterial bile acid metabolism compared to placebo resulting in a change in the bile acid profile that mirrored that of the lean donor [10].
Bile acids are a class of bioactive metabolites that signal through bile acid receptors, such as FXR and TGR5, to improve metabolic health. Bile acids are primarily metabolized by the liver and the gut microbiota. Primary bile acids are produced in the liver from cholesterol and are conjugated with taurine or glycine prior to secretion into the gut lumen. Primary bile acids are converted into secondary bile acids, deoxycholic acid (DCA) and lithocholic acid (LCA), by gut bacteria. Bile acids vary in their affinity for bile acid receptors. Therefore, alterations in the bile acid profile can influence metabolic health by altering bile acid receptor signaling. In particular, DCA and LCA are the strongest ligands for TGR5 [11,12,13,14]. Indeed, studies investigating the impact of gut microbiota on metabolic disease often identify gut bacterial bile acid production as a key mechanistic mediator. For example, dietary fiber supplementation has been reported to enhance gut bacterial 6-α-hydroxylation to improve metabolic phenotypes in mice [15]. Furthermore, recent work reports that enhancing gut bacterial bile acid deconjugation through the use of genetically modified microbes improves metabolic parameters in mice [16].
A key pathway in gut bacterial bile acid metabolism is the conversion of conjugated primary bile acids to secondary bile acids through deconjugation followed by 7-α-dehydroxylation. Primary bile acids are first deconjugated by the enzyme bile salt hydrolase (BSH) [17,18]. BSH expression has been identified across all major bacterial divisions and archaeal species in the gut, and elevations in BSH activity improve metabolic outcomes [19]. Furthermore, BSH activity enhances bacterial survival [19]. Therefore, BSH may also be a key determinant of the efficacy by which probiotics and FMT are able to successfully colonize the host [20]. However, the regulation of BSH expression is poorly understood. Unconjugated bile acids are converted to secondary bile acids through 7-α-dehydroxylation, which is a multi-step process that is less widely dispersed throughout the gut microbiome relative to BSH [17,18]. Nevertheless, the gut bacterial species and genes responsible for 7-α-dehydroxylation are still incompletely defined.
Research suggests FMT increases gut microbial diversity and the abundance of beneficial bacteria. Indeed, we found that patients who received FMT show sustained shifts in gut microbiota profiles toward those of the donor, as determined by 16S rRNA gene sequencing. Additionally, bile acid profiles resembled that of the donor [10]. Importantly, this shift in bile acid profile also coincided with a slowing of glycemic impairment compared with placebo [9]. The results from this clinical pilot, which evaluated the effectiveness of FMT in obese metabolically healthy patients, provide an ideal study set to identify gut bacteria involved in gut bacterial bile acid metabolism. Therefore, we assessed the impact of FMT on gut bacterial composition by metagenomics to better understand the dynamic alterations induced by FMT. Furthermore, we assessed the correlation between bacterial abundance determined by metagenomics with bile acid levels, assessed in the same samples, to identify putative bacterial species that may contribute to gut microbial bile acid metabolism.
2. Materials and Methods
2.1. Sample Collection
Secondary analysis was conducted on a single-center, double-blind, randomized, placebo-controlled pilot trial of FMT in obese metabolically normal/healthy patients (body mass index (BMI), 35 kg/m2 or higher without diabetes, metabolic syndrome, or non-alcoholic fatty liver disease). Briefly, patients were randomized 1:1 to receive FMT (via an induction dose of 30 FMT capsules followed by two maintenance doses of 12 capsules at week 4 and week 8) or an identical placebo capsule. A single healthy lean (BMI 17.5 kg/m2) donor was used to generate FMT capsules. A total of 22 patients were enrolled, 11 in each arm, and primarily female [10]. Samples collected at baseline (prior to FMT intervention) and after 4 weeks of intervention were available from 8 placebo and 11 FMT patients. Two patients in the placebo group withdrew from the study, and one placebo sample was unavailable at the 4-week time point. We focused on the 4-week time point because this was the time point at which the most substantial FMT-induced change in the bile acid profile was detected [10].
2.2. Microbiota Analysis
Fecal samples were shipped frozen to the George Washington University Genomics Core for processing. Each sample underwent DNA and RNA extraction in parallel from 250 mg of fecal material using a ZymoBIOMICS DNA/RNA Miniprep Kit (Zymo Research Corporation, Irvine, CA, USA). The resulting DNA was quality controlled using a Thermo Fisher Qubit 3.0 High Sensitivity DNA kit (Life Technologies, Carlsbad, CA, USA) and standardized to 0.2 ng/µL for library construction. Sequencing libraries were prepared, along with a ZymoBIOMICS Microbial Community DNA standard, using a Nextera XT DNA Library Prep kit (Illumina, San Diego, CA, USA) following Illumina’s recommended guidelines. Libraries were quality controlled using a Thermo Fisher Qubit 3.0 High Sensitivity DNA kit and an Agilent 2100 Bioanalyzer High Sensitivity DNA kit (Agilent, Santa Clara, CA, USA) and were subsequently sequenced as paired-end, 2 × 150 bp, using a NextSeq 500 Mid-Output kit (Illumina, San Diego, CA, USA), with a 1% phi X sequencing control spike-in.
2.3. Data Analysis and Statistics
Metagenomic shotgun sequencing read quality was assessed using FastQC and MultiQC [21,22], and host reads were filtered using kneaddata with low quality (Phred scores <28) ends and reads trimmed for downstream analyses. Functional pathways of associated microbes were determined using omePath [23]. Functional associations between metabolites, clinical phenotypes, and microbes were assessed using Tweedievers [24]. Data are presented as the mean ± SEM, and statistical analyses were performed using GraphPad Prism 9.4.1. Data were analyzed by non-parametric Wilcoxon matched-pairs signed rank test. Multiple corrections of statistical tests were applied using the Benjamini and Hochberg false discovery rate (FDR), and differences were considered significant at p ≤ 0.05 unless otherwise noted.
3. Results
3.1. Microbial Diversity
Patient fecal samples obtained at baseline (prior to FMT intervention) and after 4 weeks of intervention were assessed by shotgun metagenomics. The metagenomic sequencing resulted in an average of 4,015,023 reads per sample, with a minimum of 2,280,276 reads and a maximum of 5,818,393 reads. Of these, 0.031235% were, on average, the host reads, leaving a minimum of 2,279,728 quality microbial reads for microbiome characterization with an average of 4,013,907 quality microbial reads per host individual.
3.2. Impact of FMT on Gut Microbial Composition at 4 Weeks after the Initiation of Intervention
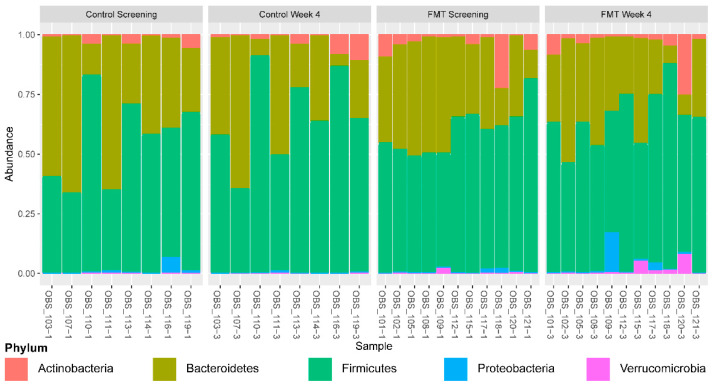
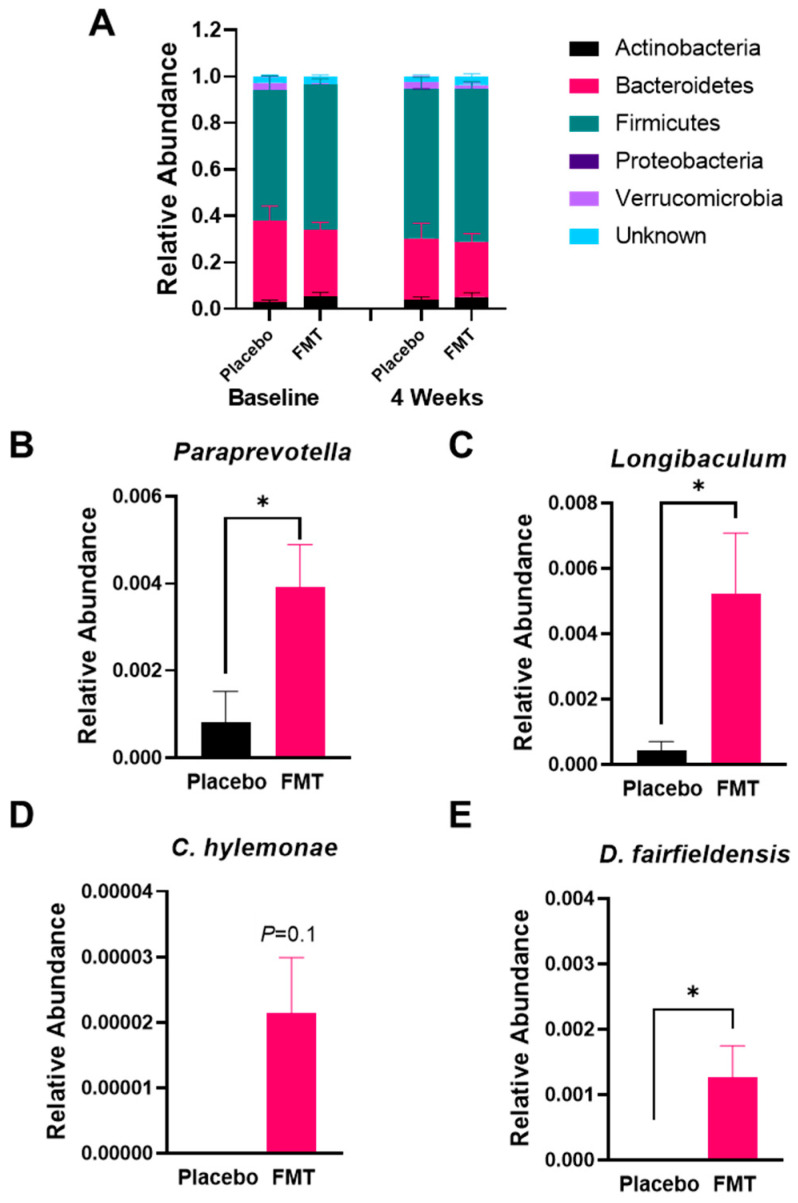
There were no significant baseline clinical differences between FMT and placebo groups [10]. We focused on the fecal sample collected at baseline vs. 4 weeks after the initiation of FMT as this was the time point at which the changes in bile acid levels were most significant [10]. No significant differences in the relative abundance of bacteria were noted at the phyla level (Figure 1 and Figure 2A). At the genus level, FMT-enriched Paraprevotella and Longibaculum (Figure 2B,C, p < 0.05) compared to placebo. On the species level, FMT tended to increase the relative abundance of Clostridium hylemonae, a bacterial species known to convert primary to secondary bile acids (Figure 2D) [17,18]. Finally, FMT increased Desulfovibrio fairfieldensis compared with placebo (Figure 2E, p < 0.05). Of note, Paraprevotella, Longibaculum, Clostridium hylemonae, and Desulfovibrio fairfieldensis did not differ between groups at the baseline.
Figure 1.
Phylum-level relative abundances across all individual patients. Patient identifiers ending in 1 represent baseline and ending in 3 represent the 4-week time point. Plots are faceted by treatment and time point.
Figure 2.
Impact of FMT on gut microbial composition. (A) Relative abundance of each bacterial phylum. Relative abundance of Paraprevotella (B) and Longibaculum (C), Clostridium hylemonae (D), and Desulfovibrio fairfieldensis (E) in fecal samples from placebo vs. FMT at baseline and after 4 weeks of treatment. Data presented as mean ± SEM. * p < 0.05.
3.3. Impact of FMT on Gene Enrichment and Correlations with Secondary Bile Acid Production
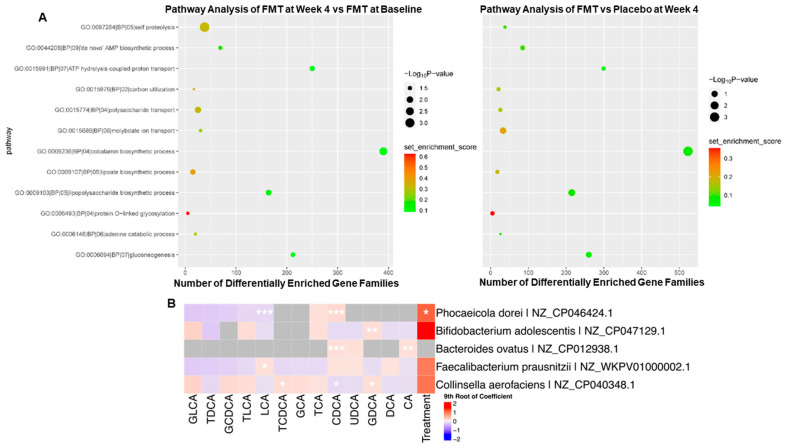
As some of the bacterial species enriched by FMT are implicated in bile acid metabolism, we next determined the effect of FMT on bacterial bile acid metabolic gene copy number. Starting with a broad overview of the impact of FMT on gut microbial gene abundance, we performed pathway analysis using omePath of the metagenomic data (Figure 3A). FMT-enriched genes involved in cell proteolysis pathways. Taking a closer look at bile acid metabolism, FMT did not impact gene abundance for most known gut bacterial bile acid metabolic genes, except for a reduction in BaiB and BaiE. These changes were noted at 4 weeks and not at baseline in the FMT group compared to the placebo. FMT did not impact the gene abundance of other genes involved in bile acid 7-α-dehydroxylation, including BaiCD, BaiA2, BaiF, and BaiH. Further work using metatranscriptomics is warranted to determine the impact of FMT on bacterial bile acid metabolic gene expression.
Figure 3.
Association between bile acids and bacterial species. (A) Assessment of pathways enriched by FMT relative to baseline (left) and relative to placebo control after 4 weeks of treatment (right). (B) Bacterial species and strains that are correlated with gut bacterial-derived bile acids. * p < 0.2, ** p < 0.05, *** p < 0.001.
To identify candidate bacteria involved in gut bacterial bile acid metabolism, we assessed correlations between bacterial species abundance and bile acid profile, with a focus on bile acid products of gut bacterial metabolism, namely unconjugated bile acids, and the secondary bile acids, DCA and LCA. Bile acid levels were measured in the same samples used for metagenomics analysis, as previously described [10]. The impact of FMT on bile acid levels in this sample set has been previously reported [10]. We focused on bacterial species that were positively correlated with bile acid sub-types that are produced, at least in part, through interactions with the gut microbiota with a p-value less than or equal to 0.08. Bacterial species that met these criteria are presented in Figure 3B. Phocaeicola dorei and Bacteroides ovatus were positively correlated with unconjugated chenodeoxycholic acid (CDCA) (p = 7.47 × 10−45 and 2.50 × 10−8). Bifidobacterium adolescentis and Collinsella aerofaciens were positively correlated with the production of DCA (specifically, the glycine-conjugated form, GDCA) (p = 0.0123 and 0.0634). The same strain of B. ovatus was positively correlated with unconjugated cholic acid (CA) (p = 0.0317). Lastly, Faecalibacterium prausnitzii was positively correlated with LCA (p = 0.0634). These data point to a potential role for Phocaeicola dorei and B. ovatus in bile acid deconjugation. Further, these data suggest that Bif. adolescentis, C. aerofaciens, and F. prausnitzii may play a role in the conversion of primary to secondary bile acids, which requires further functional validation.
4. Discussion
In the present study, we utilized a fecal sample set from patients receiving FMT or placebo that exhibited alterations in gut bacterial bile acid metabolism to improve our understanding of the gut bacterial species involved in gut bacterial bile acid metabolism and how these pathways are dynamically regulated by FMT. Using metagenomics, we identified an enrichment of Paraprevotella, Longibaculum, Desulfovibrio fairfieldensis, and Clostridium hylemonae in response to FMT. Furthermore, through the assessment of correlations between fecal bile acid levels and bacterial species relative abundances, we identified Bifidobacterium adolescentis, Bacteroides ovatus, Faecalibacterium prausnitzi, and Phocaeicola dorei as potentially contributing to gut bacterial bile acid metabolism. Further work is needed to better understand secondary bile acid metabolism, its roles in metabolic disease, and how it can be manipulated through FMT.
The effect of FMT to enrich Paraprevotella, Longibaculum, C. hylemonae, and D. fairfieldensis may have contributed to the effect of FMT to enhance gut microbial bile acid metabolism and/or slow the development of glucose intolerance. For example, members of the genera Clostridium are the predominant human intestinal species thought to perform 7-α-dehydroxylation of primary bile acids [25]. Additionally, C. hylemonae has been shown to convert CA into DCA in vitro [26]. Furthermore, Paraprevotella abundance was significantly increased after FMT in individuals with functional constipation whose changes in fecal microbiome compositions were measured before and after FMT. This increase in Paraprevotella abundance correlated with improved relief of clinical symptoms measured by three different clinical scales for constipation, suggesting Paraprevotella could improve metabolic dysregulation through gastric motility [27]. The role of Longibaculum in host metabolic health is poorly defined; however, dietary fiber supplementation has been shown to enrich for Longibaculum [28]. D. fairfieldensis is a Gram-negative anaerobic bacillus that has been implicated in bile acid metabolism. Interestingly, D. fairfieldensis bacteremia was found to be associated with choledocholithiasis in a case report [29]. Furthermore, a recent study reports that Desulfovibrionales are enriched in patients with cholelithiasis. Further, the administration of Desulfovibrionales to mice with antibiotic-induced depletion of the gut microbiome increased secondary bile acid production [30]. Desulfovibrionales can reduce taurine into H2S, which has been suggested to facilitate 7-α-dehydroxylation [31]. Together, these data suggest that Desulfovibrionales, and in particular D. fairfieldensis, may play a role in gut bile acid metabolism. Further, these data highlight the potentially important cooperative interactions among bacteria that facilitate gut microbial bile acid metabolism.
In this study, we identified five bacteria that were positively correlated with gut microbiome-derived bile acids. Specifically, Bifidobacterium adolescentis and Collinsella aerofaciens were positively correlated with DCA. Bacteroides ovatus was positively correlated with unconjugated CDCA and CA. Phocaeicola dorei was positively correlated with unconjugated CDCA, and Faecalibacterium prausnitzii was positively correlated with LCA. Thus, our data suggest that Phocaeicola dorei and Bacteroides ovatus may perform bile acid deconjugation. Consistent with this, previous work reports that several Bacteroides strains, including strains of B. ovatus, express BSH [32]. Whether Phocaeicola dorei can perform bile acid deconjugation is unknown and requires further testing. Interestingly, a previous study identified a correlation between Phocaeicola dorei, also named Bacteroides dorei, and the risk of developing type 1 diabetes [33], suggesting a potential metabolic role for this species. The bacteria that were positively correlated with secondary bile acids (Bifidobacterium adolescentis and Faecalibacterium prausnitzii) are known to have BSH functions [34,35]. Faecalibacterium prausnitzii has been connected to anti-inflammatory effects and improvement of intestinal barrier function [36,37]. A role for Collinsella aerofaciens in the production of DCA has not been previously tested. However, Collinsella aerofaciens, previously known as Eubacterium aerofaciens, was found to have NADP-dependent 7-β-hydroxysteriod dehydrogenase activity, which is necessary for the production of hydrophilic secondary bile acids such as ursodeoxycholic acid [38].
The advantages of this study include the application of metagenomics to the analysis of the gut microbiome of individuals receiving FMT or placebo control. Additionally, the results from this secondary analysis are from individuals with obesity such that bacteria identified from this specific population can better inform FMT for the treatment of obesity and metabolic disease. Limitations of this study include the small sample size. While bile acid gene abundance was studied, metatranscriptomics analysis is needed to assess the impact of FMT on gene expression. Further work is needed to functionally validate bacteria identified as potentially contributing to the effects of FMT on gut bacterial bile acid metabolism. Together, these data demonstrate that FMT can alter the composition of bile acids and bacterial communities in the gut microbiome.
Acknowledgments
We thank Castle Raley and the George Washington Genomics Core for sample processing and sequencing for metagenomics.
Author Contributions
Investigation: J.-M.B., T.D., C.L., Z.M., J.R.M., K.A.C., A.R., B.H.M., J.R.A. and B.P.C. Conceptualization, Resources: J.R.A. and B.P.C. Methodology: J.-M.B., T.D., C.L., Z.M., J.R.M., K.A.C., A.R., B.H.M., J.R.A. and B.P.C. Formal Analysis: J.-M.B., T.D., C.L., Z.M., J.R.M., K.A.C., A.R. and B.P.C. Writing—Original Draft Preparation: J.-M.B. Writing—Review and Editing: J.-M.B., T.D., C.L., Z.M., J.R.M., C.C.T., K.A.C., A.R., B.H.M., J.R.A. and B.P.C. Supervision, Funding Acquisition: K.A.C., A.R. and B.P.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Institutional Review Board at the Brigham and Women’s Hospital, and all patients provided written informed consent before participation (NCT02741518). In addition, Food and Drug Administration approval via an investigational new drug application (16936, 2016) was obtained. All authors had access to the study data and approved the final manuscript.
Informed Consent Statement
All patients provided written informed consent before participation.
Data Availability Statement
Sequence data generated by this project are deposited in NCBI Sequence Read Archive (SRA) and associated with BioProject PRJNA904790.
Conflicts of Interest
J.-M.B., C.L., Z.M., J.R.M., K.A.C., A.R. and B.P.C. have no relevant conflict of interest to declare. J.R.A. consults for and has research support from Finch Therapeutics Group, Janssen, Pfizer, Abbvie, Iterative Sopes, Seres Therapeutics, Ferring, Merck, Bristol Myer Squibb and has research support from Pfizer and Merck. T.D. has research support from AMPEL BioSolutions. B.H.M. has received consultancy fees from Finch Therapeutics Group and Ferring Pharmaceuticals. C.C.T. consults for Apollo Endosurgery, Boston Scientific, Medtronic, Enterasense Ltd., EnVision Endoscopy, Fractyl, Fujifilm, GI Dynamics, GI Windows, Lumendi, Olympus, USGI Medical, Xenter, Endoquest Robotics. He has received Research Support from Apollo Endosurgery, Boston Scientific, ERBE, Fujifilm, GI Dynamics, Lumedi Olympus, USGI Medical. He serves on Advisory Boards for Fractyl, Fujifilm, USGI Medical, Xenter and Endoquest Robotics. He is a founder, board member and receives ownership interest from Enterasense Ltd., EnVision Endoscopy and GI Windows, He is on a speakers bureau for Boston Scientific, Fujifilm and Olympus. He receives royalty payments from GI Windows, EndoSim and Enterasense Ltd.
Funding Statement
Research reported in this publication was supported by the National Center for Complementary and Integrative Health of the National Institutes of Health under Award number R21AT010956 to B.P.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Metabolomics studies were performed at the MRC-NIHR National Phenome Centre at Imperial College London; this center receives financial support from the Medical Research Council (MRC) and National Institute of Health Research (NIHR) (grant number MC_PC_12025). B.H.M. is the recipient of an NIHR Academic Clinical Lectureship (CL-2019-21-002) and was formerly in receipt of an MRC Clinical Research Training Fellowship (MR/R000875/1). The Division of Digestive Diseases and MRC-NIHR National Phenome Centre at Imperial College London receive financial and infrastructure support from the NIHR Imperial Biomedical Research Centre (BRC) based at Imperial College Healthcare NHS Trust and Imperial College London.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karlsson F.H., Tremaroli V., Nookaew I., Bergström G., Behre C.J., Fagerberg B., Nielsen J., Bäckhed F. Gut Metagenome in European Women with Normal, Impaired and Diabetic Glucose Control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., et al. A Core Gut Microbiome in Obese and Lean Twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Human Gut Microbes Associated with Obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh P.J., Backhed F., Fulton L., Gordon J.I. Marked Alterations in the Distal Gut Microbiome Linked to Diet-Induced Obesity. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marotz C.A., Zarrinpar A. Treating Obesity and Metabolic Syndrome with Fecal Microbiota Transplantation. Yale J. Biol. Med. 2016;89:383–388. [PMC free article] [PubMed] [Google Scholar]
- 7.Kassam Z., Lee C.H., Yuan Y., Hunt R.H. Fecal Microbiota Transplantation for Clostridium Difficile Infection: Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2013;108:500–508. doi: 10.1038/ajg.2013.59. [DOI] [PubMed] [Google Scholar]
- 8.Hourigan S.K., Ahn M., Gibson K.M., Pérez-Losada M., Felix G., Weidner M., Leibowitz I., Niederhuber J.E., Sears C.L., Crandall K.A., et al. Fecal Transplant in Children with Clostridioides Difficile Gives Sustained Reduction in Antimicrobial Resistance and Potential Pathogen Burden. Open Forum Infect. Dis. 2019;6:ofz379. doi: 10.1093/ofid/ofz379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allegretti J.R., Kassam Z., Hurtado J., Marchesi J.R., Mullish B.H., Chiang A., Thompson C.C., Cummings B.P. Impact of Fecal Microbiota Transplantation with Capsules on the Prevention of Metabolic Syndrome among Patients with Obesity. Hormones. 2021;20:209–211. doi: 10.1007/s42000-020-00265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allegretti J.R., Kassam Z., Mullish B.H., Chiang A., Carrellas M., Hurtado J., Marchesi J.R., McDonald J.A.K., Pechlivanis A., Barker G.F., et al. Effects of Fecal Microbiota Transplantation with Oral Capsules in Obese Patients. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2020;18:855–863.e2. doi: 10.1016/j.cgh.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton J.P., Xie G., Raufman J.-P., Hogan S., Griffin T.L., Packard C.A., Chatfield D.A., Hagey L.R., Steinbach J.H., Hofmann A.F. Human Cecal Bile Acids: Concentration and Spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G256–G263. doi: 10.1152/ajpgi.00027.2007. [DOI] [PubMed] [Google Scholar]
- 12.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., et al. A G Protein-Coupled Receptor Responsive to Bile Acids. J. Biol. Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 13.Makishima M., Okamoto A.Y., Repa J.J., Tu H., Learned R.M., Luk A., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B. Identification of a Nuclear Receptor for Bile Acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 14.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Nakamura T., Itadani H., Tanaka K. Identification of Membrane-Type Receptor for Bile Acids (M-BAR) Biochem. Biophys. Res. Commun. 2002;298:714–719. doi: 10.1016/S0006-291X(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 15.Makki K., Brolin H., Petersen N., Henricsson M., Christensen D.P., Khan M.T., Wahlström A., Bergh P.-O., Tremaroli V., Schoonjans K., et al. 6α-Hydroxylated Bile Acids Mediate TGR5 Signalling to Improve Glucose Metabolism upon Dietary Fiber Supplementation in Mice. Gut. 2022 doi: 10.1136/gutjnl-2021-326541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell B.J., Brown S.D., Siguenza N., Mai I., Saran A.R., Lingaraju A., Maissy E.S., Dantas Machado A.C., Pinto A.F.M., Sanchez C., et al. Intestinal Transgene Delivery with Native E. coli Chassis Allows Persistent Physiological Changes. Cell. 2022;185:3263–3277.e15. doi: 10.1016/j.cell.2022.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridlon J.M., Kang D.-J., Hylemon P.B. Bile Salt Biotransformations by Human Intestinal Bacteria. J. Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Ridlon J.M., Kang D.J., Hylemon P.B., Bajaj J.S. Bile Acids and the Gut Microbiome. Curr. Opin. Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones B.V., Begley M., Hill C., Gahan C.G.M., Marchesi J.R. Functional and Comparative Metagenomic Analysis of Bile Salt Hydrolase Activity in the Human Gut Microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullish B.H., McDonald J.A.K., Pechlivanis A., Allegretti J.R., Kao D., Barker G.F., Kapila D., Petrof E.O., Joyce S.A., Gahan C.G.M., et al. Microbial Bile Salt Hydrolases Mediate the Efficacy of Faecal Microbiota Transplant in the Treatment of Recurrent Clostridioides Difficile Infection. Gut. 2019;68:1791–1800. doi: 10.1136/gutjnl-2018-317842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews S. Babraham Bioinformatics—FastQC a Quality Control Tool for High Throughput Sequence Data. [(accessed on 17 October 2022)]. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 22.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahnavard A., Mann B., Giri A., Chatterjee R., Crandall K.A. Metabolite, Protein, and Tissue Dysfunction Associated with COVID-19 Disease Severity. Sci. Rep. 2022;12:12204. doi: 10.1038/s41598-022-16396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallick H., Chatterjee S., Chowdhury S., Chatterjee S., Rahnavard A., Hicks S.C. Differential Expression of Single-Cell RNA-Seq Data Using Tweedie Models. Stat. Med. 2022;41:3492–3510. doi: 10.1002/sim.9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitahara M., Takamine F., Imamura T., Benno Y. Assignment of Eubacterium sp. VPI 12708 and Related Strains with High Bile Acid 7α-Dehydroxylating Activity to Clostridium Scindens and Proposal of Clostridium hylemonae sp. Nov., Isolated from Human Faeces. Pt 3Int. J. Syst. Evol. Microbiol. 2000;50:971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- 26.Ridlon J.M., Kang D.-J., Hylemon P.B. Isolation and Characterization of a Bile Acid Inducible 7α-Dehydroxylating Operon in Clostridium hylemonae TN271. Anaerobe. 2010;16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Li N., Chen Q., Qin H. Fecal Microbiota Transplantation Modulates the Gut Flora Favoring Patients with Functional Constipation. Front. Microbiol. 2021;12:700718. doi: 10.3389/fmicb.2021.700718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massot-Cladera M., Azagra-Boronat I., Franch À., Castell M., Rodríguez-Lagunas M.J., Pérez-Cano F.J. Gut Health-Promoting Benefits of a Dietary Supplement of Vitamins with Inulin and Acacia Fibers in Rats. Nutrients. 2020;12:2196. doi: 10.3390/nu12082196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pimentel J.D., Chan R.C. Desulfovibrio Fairfieldensis Bacteremia Associated with Choledocholithiasis and Endoscopic Retrograde Cholangiopancreatography. J. Clin. Microbiol. 2007;45:2747–2750. doi: 10.1128/JCM.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu H., Shao W., Liu Q., Liu N., Wang Q., Xu J., Zhang X., Weng Z., Lu Q., Jiao L., et al. Gut Microbiota Promotes Cholesterol Gallstone Formation by Modulating Bile Acid Composition and Biliary Cholesterol Secretion. Nat. Commun. 2022;13:252. doi: 10.1038/s41467-021-27758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Eldere J., Celis P., De Pauw G., Lesaffre E., Eyssen H. Tauroconjugation of Cholic Acid Stimulates 7α-Dehydroxylation by Fecal Bacteria. Appl. Environ. Microbiol. 1996;62:656–661. doi: 10.1128/aem.62.2.656-661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon S., Yu J., McDowell A., Kim S.H., You H.J., Ko G. Bile Salt Hydrolase-Mediated Inhibitory Effect of Bacteroides Ovatus on Growth of Clostridium Difficile. J. Microbiol. 2017;55:892–899. doi: 10.1007/s12275-017-7340-4. [DOI] [PubMed] [Google Scholar]
- 33.Davis-Richardson A.G., Ardissone A.N., Dias R., Simell V., Leonard M.T., Kemppainen K.M., Drew J.C., Schatz D., Atkinson M.A., Kolaczkowski B., et al. Bacteroides Dorei Dominates Gut Microbiome Prior to Autoimmunity in Finnish Children at High Risk for Type 1 Diabetes. Front. Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grill J.P., Manginot-Dürr C., Schneider F., Ballongue J. Bifidobacteria and Probiotic Effects: Action of Bifidobacterium Species on Conjugated Bile Salts. Curr. Microbiol. 1995;31:23–27. doi: 10.1007/BF00294629. [DOI] [PubMed] [Google Scholar]
- 35.Kim G.-B., Brochet M., Lee B.H. Cloning and Characterization of a Bile Salt Hydrolase (Bsh) from Bifidobacterium adolescentis. Biotechnol. Lett. 2005;27:817–822. doi: 10.1007/s10529-005-6717-3. [DOI] [PubMed] [Google Scholar]
- 36.Miquel S., Leclerc M., Martin R., Chain F., Lenoir M., Raguideau S., Hudault S., Bridonneau C., Northen T., Bowen B., et al. Identification of Metabolic Signatures Linked to Anti-Inflammatory Effects of Faecalibacterium prausnitzii. mBio. 2015;6:e00300-15. doi: 10.1128/mBio.00300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quévrain E., Maubert M.A., Michon C., Chain F., Marquant R., Tailhades J., Miquel S., Carlier L., Bermúdez-Humarán L.G., Pigneur B., et al. Identification of an Anti-Inflammatory Protein from Faecalibacterium prausnitzii, a Commensal Bacterium Deficient in Crohn’s Disease. Gut. 2016;65:415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirano S., Masuda N. Characterization of NADP-Dependent 7β-Hydroxysteroid Dehydrogenases from Peptostreptococcus Productus and Eubacterium Aerofaciens. Appl. Environ. Microbiol. 1982;43:1057–1063. doi: 10.1128/aem.43.5.1057-1063.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data generated by this project are deposited in NCBI Sequence Read Archive (SRA) and associated with BioProject PRJNA904790.