Abstract
The adherence mechanism of Aeromonas caviae Sch3N to HEp-2 cells was initially investigated through four mini-Tn5 mutants that showed a 10-fold decrease in adherence. These mutants lost motility, flagella, and their lipopolysaccharide (LPS) O antigen (O-Ag). Three genes, flmB-neuA-flmD, were found to be interrupted by the transposon insertions; additionally, two other genes, one lying upstream (flmA) and one downstream (neuB), were found to be clustered in the same operon. While the flmA and flmB genes were present in all mesophilic Aeromonas spp. (A. hydrophila, A. caviae, A. veronii bv. veronii, and A. veronii bv. sobria) tested, this was not the case for the neuA-flmD-neuB genes. Construction and characterization of flmB insertion mutants in five other mesophilic Aeromonas strains revealed the loss of motility, flagella, and adherence but did not alter the LPS composition of these strains. Taking the above findings into consideration, we conclude (i) that flagella and possibly the LPS O-Ag are involved in the adherence of the mesophilic Aeromonas to human epithelial cells; (ii) flmA and flmB are genes widely distributed in the mesophilic Aeromonas and are involved in flagella assembly, and thus adherence; and (iii) in A. caviae Sch3N the flmA and flmB genes are found in a putative operon together with neuA, flmD, and neuB and are involved in LPS O-Ag biosynthesis and probably have a role in flagellum assembly.
Mesophilic Aeromonas has been associated with gastrointestinal and wound infections of healthy humans, and less commonly with septicemias of immunocompromised patients (12). A. caviae, in particular, is reported as the most prevalent paediatric enteropathogenic species of the genus (26, 39).
A number of putative pathogenicity determinants have been reported for aeromonads; these include toxins, adhesins, and invasins (34). There is still little known about their adherence process, although long-wavy pili have been implicated as important colonization factors of A. hydrophila and A. veronii bv. sobria (14). However, many clinical isolates are poorly piliated or nonpiliated (15), and alternative adherence factors such as the lipopolysaccharide O antigen (O-Ag) and the polar flagellum have been suggested (35). Mesophilic aeromonads are usually motile by means of a single polar unsheathed flagellum; this has been proposed to aid adherence to and invasion of fish cell lines by A. hydrophila (24). The lipopolysaccharide (LPS) of the genus has been studied more extensively. Aeromonas LPS has been suggested to follow the characteristics of its counterparts in Escherichia coli and Salmonella enterica (21), with smooth ladder-like patterns predominating among clinical isolates (37). Approximately 100 serogroups have been described for the genus, with AX2, O:3, and O:17 being the commonest among A. caviae isolates (32). For A. hydrophila serogroup O:34, LPS O-Ag has been implicated in in vitro colonization and virulence in fish and mice (1, 22). Despite reports suggesting the involvement of O-Ag and the polar flagellum in Aeromonas colonization, none of the structural or the biosynthetic genes of either have been described to date.
In this study, we describe five genes of A. caviae that belong to a putative operon involved in the biosynthesis of LPS O-Ag and flagellum assembly. We have also investigated the distribution of these genes among the mesophilic Aeromonas species, which allowed us to draw conclusions about the roles of these two surface structures in the adherence of aeromonads to human epithelial cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown on Luria-Bertani (LB) Miller broth and LB Miller agar, while Aeromonas strains were grown either on tryptic soy broth or agar or in brain heart infusion broth (BHIB) (Oxoid). Ampicillin (50 μg/ml), nalidixic acid (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), rifampin (100 μg/ml), and tetracycline (20 μg/ml) were added to the different media when needed.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotypea | Reference(s) or source |
|---|---|---|
| Strains | ||
| A. caviae | ||
| Sch3 | Clinical isolate | 39 |
| Sch3N | Sch3, spontaneous Nalr | This work |
| IAG75 | Sch3NflmD::mini-Tn5Cm, Nalr Cmr | This work |
| IAG570 | Sch3NflmB::mini-Tn5Cm, Nalr Cmr | This work |
| IAG1419 | Sch3NneuA::mini-Tn5Cm, Nalr Cmr | This work |
| IAG1639 | Sch3NflmB::mini-Tn5Cm, Nalr Cmr | This work |
| A. hydrophila | ||
| AH-3 | O:34, wild type | 20 |
| AH-1726 | AH-3flmB::Kmr | This work |
| O:3 | O:3 wild type | 1 |
| AH-1881 | O:3flmB::Kmr | This work |
| O:25 | O:25 wild type | 1 |
| AH-1882 | O:25flmB::Kmr | This work |
| A. veronii bv. sobria | ||
| AH-1 | O:11, wild type | 1 |
| AH-1883 | AH-1flmB::Kmr | This work |
| A. veronii bv. veronii | ||
| AS-28 | O:11 wild type | 1 |
| AH-1884 | AS-28flmB::Kmr | This work |
| E. coli | ||
| BW19851 | RP4-2tet::Mu-1Kan::Tn7 integrant/ΔuidA::pir+recA1 hsdR17 creB510 endA1 zbf-5 thi | 25 |
| S17-1 | hsdR pro recA, RP4-2 in chromosome Km::Tn7 (Tc::Mu) | 30 |
| XL1-Blue | endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 lac | Stratagene |
| DH5α | F−endA hsdR17 (rK− mK+) supE44 thi-1 recA1 gyr-A96 80lacZ | 10 |
| MC106(λpir) | thi thr1 leu6 proA2 his4 argE2 lacY1 galK2 ara14 xyl5 supE44 pir | 28 |
| SM10(λpir) | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr pir | 28 |
| Plasmids | ||
| pUT mini-Tn5Cm | bla ori R6K mob RP4 tnp gene of Tn5-IS50R that lacks NotI site, MCS of M13tg131; 8.7 kb, Ampr Cmr | 6, 11 |
| pUC18 | High-copy-number cloning vector, Ampr | Gibco BRL |
| pUI2224 | pUC18 with 2.2 kb of Sch3N DNA inserted in the HindIII site; 4.9 kb, Ampr | This work |
| pUI2814 | pUC18 with 2.8 kb of Sch3N DNA inserted in the HindIII site; 5.5 kb, Ampr | This work |
| pDSK519 | High-copy-number broad-host-range cloning vector; 8.1 kb, Kmr | 13 |
| pDI2211 | pDSK519 with 2.2 kb of Sch3N DNA inserted in the HindIII site of the polylinker; 10.3 kb, Kmr | This work |
| pDI284 | pDSK519 with 2.8 kb of Sch3N DNA inserted in the HindIII site of the polylinker; 10.9 kb, Kmr | This work |
| pDI54 | pDI2211 with 2.8 kb of Sch3N DNA inserted in the HindIII site downstream of the 2.2-kb insert; 13.1 kb, Kmr | This work |
| pGEM-T | PCR cloning vector, Ampr | Promega |
| pFS100 | pGP704 suicide plasmid, pir dependent | 28 |
| pFS-Flm | pFS100 with an internal fragment (672 bp) of flmB gene | This work |
| pLA-Flm | pLA2917 cosmid with a complete flmB gene from strain AH-3 | This work |
Abbreviations: Amp, ampicillin; Ap, β-lactam antibiotics, including piperacillin; Cm, chloramphenicol; Km, kanamycin; Nal, nalidixic acid; MCS, multiple cloning site.
HEp-2 cell culture and adherence assay.
Tissue culture was maintained as described by Thornley et al. (34). The adherence assay was conducted as a slight modification of that described by Carrello et al. (4). Bacteria were grown statically in BHIB at 37°C, harvested by gentle centrifugation (1,600 × g for 5 min), and resuspended in phosphate-buffered saline (PBS), pH 7.2, at approximately 106 to 107 CFU/ml (A600 ≈ 0.07). The monolayer was infected with 1 ml of the bacterial suspension for 90 min at 37°C in 5% CO2. Following infection, the nonadherent bacteria were removed from the monolayer by three washes with PBS. The remaining adherent bacteria and the monolayers were then fixed in 100% methanol for 5 min. Methanol was removed by washing with PBS, and the HEp-2 cells with the adherent bacteria were stained for 45 min in 10% (vol/vol) Giemsa stain (BDH, Poole, United Kingdom) prepared in Giemsa buffer. The coverslips were air dried, mounted, and viewed by oil immersion under a light microscope at ×1,000 magnification. Twenty HEp-2 cells/coverslip were randomly chosen, and the number of bacteria adhering per HEp-2 cell was recorded. Assays were carried out in duplicates or triplicates.
Whole-cell protein preparation, SDS-PAGE, and immunoblotting.
Whole-cell proteins were obtained from Aeromonas strains grown statically overnight in BHIB at 37°C. Equivalent numbers of cells were harvested by centrifugation, and the cell pellet was resuspended in 50 to 200 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (29) and boiled for 5 min, before adding an equal volume of distilled water and boiling for a further 5 min. The samples were centrifuged at 5,000 × g for 5 min at room temperature, and the supernatants were kept at −20°C until needed.
Protein samples were separated on SDS-polyacrylamide gels (12%) as described by Laemmli (17). For immunoblotting, proteins were transferred onto Hybond-C (Amersham) nitrocellulose membrane. Following transfer, membranes were blocked with 5% skim milk and probed with a polyclonal rabbit anti-polar flagellin antibody (1:500). The unbound antibody was removed by five washes in PBS, and a goat anti-rabbit peroxidase-conjugated secondary antibody (1:1,000) was added. The unbound secondary antibody was washed away with PBS as described for the primary antibody. The bound conjugate was then detected by the addition of 2 ml of 0.5% 4-chloro-1-naphthol (Sigma) prepared in methanol and diluted in 8 ml of PBS containing 50 μl of H2O2 (30% [wt/wt]).
Motility assay.
Freshly grown bacterial colonies were transferred with a sterile toothpick into the center of motility agar (1% tryptone, 0.5% NaCl, 0.25% agar). The plates were incubated face up at 37°C for 16 to 24 h, and motility was assessed by examining the migration of bacteria through the agar from the center towards the periphery of the plate.
LPS extraction and PAGE analysis.
LPS was purified by the method of Westphal and Jann (38). For screening purposes LPS was obtained after proteinase K digestion of whole cells according to the procedure of Darveau and Hancock (5). SDS-PAGE was performed, and LPS bands were detected by the silver staining method of Tsai and Frasch (36).
EM.
Electron microscopy (EM) techniques for visualizing stained whole cells and flagella were previously described (20).
Mini-Tn5Cm mutagenesis.
Conjugal transfer of pUT-mini-Tn5Cm from E. coli BW19851 to A. caviae Sch3N was performed using a filter mating technique. Bacterial conjugation was allowed to proceed for 6 to 8 h at 37°C on sterile nitrocellulose filters (0.45-μm pore size) placed onto an LB agar (LBA) plate. Serial dilutions of the mating mix were plated on LBA supplemented with nalidixic acid and chloramphenicol, the latter added in order to select for mini-Tn5Cm.
General DNA methods.
DNA manipulations were carried out essentially as previously described (29). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers.
Southern blot and dot blot hybridizations.
Southern blotting was performed by capillary transfer (29). For dot blot hybridizations, the DNA was denatured by boiling for 5 min, chilled on ice for another 5 min, and spotted onto Hybond N+ (Amersham) nylon membrane. Probe labeling, hybridization, and detection was carried out using the enhanced chemiluminescence labeling and detection system (Amersham) according to the manufacturer's instructions.
Cloning of DNA flanking mini-Tn5Cm insertions by inverse PCR.
Chromosomal DNA of mini-Tn5Cm mutants IAG75, IAG570, IAG1419, and IAG1639 was digested with PstI, purified, and then ligated overnight at 15°C. Samples of 100 to 200 ng of ligated DNA were then subjected to inverse PCR. The sequences flanking the transposon were amplified by using the primers 5′-AGATCTGATCAAGAGACAG-3′ and 5′-ACTTGTGTATAAGAGTCAG-3′, which are specific to the 19-nucleotide (nt) I end and O end of miniTn5Cm, respectively. This was performed using Pfu DNA polymerase (Stratagene) at 2.5 mM MgCl2 in a Hybaid Omnigene Thermal cycler. Initial DNA denaturation was carried out for 2 min, and amplification reactions were carried out for 25 cycles with denaturation at 95°C for 30 s, primer annealing at 48°C for 1 min, and elongation at 72°C for 8 min. A final elongation step of 10 min at 72°C was also performed. PCR products were ligated into the SmaI site of pUC18 and sequenced.
Nucleotide sequencing and sequence analysis.
Double-stranded DNA sequencing was performed by using the Sanger dideoxy-chain termination method with the ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer). DNA fragments were ligated into pUC18 and sequenced using an ABI PRISM 377 DNA sequencer (Perkin-Elmer Corporation). The 18-mer forward (5′-TGTAAAACGACGGCCAGT-3′) and the 22-mer reverse (5′-TCACACAGGAAACAGCTATGAC-3′) M13 primers were employed in sequencing the ends of the DNA inserts. Following the first sequencing reaction and whenever required, primers were designed until the inserts' sequences were complete. Primers used for DNA sequencing were purchased from Pharmacia LKB Biotechnology. The DNA sequence was translated in all six frames, and all open reading frames (ORFs) greater than 100 bp were inspected. Deduced amino acid sequences were compared with those of DNA translated in all six frames from nonredundant GenBank and EMBL databases by using the BLAST network service at the National Center for Biotechnology Information (NCBI) (2). Multiple sequence alignments were carried out using the Clustal W program (33). Determination of possible terminator sequences was done by using the Terminator program from the Genetics Computer Group package (Madison, Wisconsin) in a VAX 4300. Hydropathy profiles were calculated according to the method of Kyte and Doolittle (16).
Construction of flmB-defined insertion mutants.
To obtain defined insertion mutants in flmB we used a method previously described (28) based on suicide plasmid pSF100. Briefly, an internal amplified fragment of this gene was ligated to vector pGEM-T (Promega) and transformed in E. coli DH5α. The fragment was recovered by restriction digestion; blunt ended with Klenow fragment; and finally ligated to EcoRV-digested, blunt-ended, and dephosphorylated pFS100 and transformed into E. coli MC1061(λpir), selecting for Kmr to generate plasmid pFS-Flm. Plasmid pFS-Flm was isolated and transformed on E. coli SM10(λpir). Plasmid pFS-Flm was transferred by conjugation to different mesophilic Aeromonas sp. rifampin-resistant (Rifr) strains to obtain defined insertion mutants in flmB selecting for Rifr and Kmr.
Nucleotide sequence accession number.
The nucleotide sequence of the genes described here have been assigned the following GenBank accession number: AF126256.
RESULTS
Isolation of nonadherent mini-Tn5Cm mutants of A. caviae strain Sch3N.
Conjugations between A. caviae Sch3N and E. coli BW19851 (pUT-mini-Tn5Cm) were carried out by filter mating. Transconjugants were grown for 48 to 72 h on LBA containing naladixic acid and chloramphenicol and subsequently subcultured and purified. Over 2,000 mutants generated by this method were qualitatively screened for reduced adherence to HEp-2 cells by adherence assay. Fourteen mutant strains consistently exhibiting an average of 15% of the wild-type adherence were isolated. All nonadherent mutants were then analyzed for the presence of the transposon by Southern hybridizations of PstI chromosomal DNA digestions. As no PstI restriction sites were present in the transposon, variable size bands larger than the transposon were observed for each mutant. A single band was detected in every mutant chromosome, indicating single transposon insertions. From the hybridizing PstI bands, the size of Aeromonas DNA flanking the transposons could be estimated.
Preliminary characterization of four nonadherent mutants.
The four mutants IAG75, IAG570, IAG1419, and IAG1639 exhibited an average of 10% of Sch3N adherence (Table 2). The bands obtained on the PstI Southern blots for these four mutants were of similar size (6 to 6.5 kb), and therefore the transposon insertions were thought to be in the same PstI chromosomal DNA fragment, which was estimated around 2.5 to 3.0 kb. Since these mutants also exhibited identical phenotypes (see below) they were chosen for further study.
TABLE 2.
Effect of centrifugation of A. caviae onto HEp-2 cells prior to infection
| Strain | Mean no. of bacteria/HEp-2 cell ± SDa
|
|
|---|---|---|
| Normal | Centrifuged | |
| Sch3N (control) | 23.5 ± 4.8 | 54.55 ± 15.55 |
| IAG75 | 1.7 ± 1.5 (7.2) | 26.8 ± 3.2 (49.1) |
| IAG570 | 1.25 ± 0.9 (5.3) | 10.7 ± 1.9 (19.6) |
| IAG1419 | 3.5 ± 2.7 (14.9) | 18.35 ± 8.35 (33.6) |
| IAG1639 | 3.2 ± 3.15 (13.6) | 6.4 ± 1.25 (11.7) |
Bacteria were grown overnight statically in BHIB at 37°C, and adherence assays were carried out as normally (normal) or after centrifugation of bacteria onto the monolayers (centrifuged). Assays were carried out in duplicates on two separate occasions, and the mean number of adherent bacteria per HEp-2 cell ± standard deviation was recorded for every strain. Values in parentheses are percentages of control results.
Loss of motility and flagellin expression by mutants.
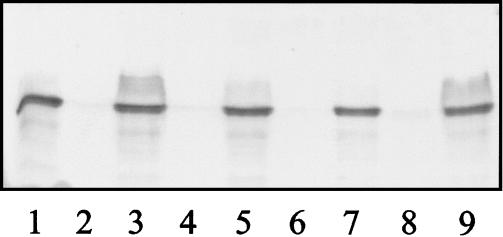
When the mutants were incubated statically in BHIB, they grew at the bottom of the culture as a “loose” pellet and not as a turbid suspension seen for Sch3N. Such a phenotype suggested the loss of motility by the mutants. This was confirmed by the inability of mutant cells to swim in semisolid motility agar and by immunoblotting of the mutant whole-cell protein preparations for the flagellin protein. In contrast to the parental strain, the polar flagellin was not detected in the mutant preparations, an observation suggesting the loss of flagellin protein expression and thus the absence of the polar flagellum filament (Fig. 1).
FIG. 1.
Polar flagellin immunoblots of whole-cell proteins of the mutants IAG75 (lane 2), IAG570 (lane 4), IAG1419 (lane 6), IAG1639 (lane 8), and the respective complemented strains carrying plasmid pDI54 (lanes 3, 5, 7, and 9, respectively). A protein preparation of A. caviae Sch3N was also run as a positive control (lane 1). Proteins were obtained from bacteria grown at 37°C in BHIB and were analyzed by SDS–12% PAGE. They were transferred onto nitrocellulose membranes and immunoblotted with anti-polar flagellin antibodies (1:500).
Effect of centrifugation of mutants onto HEp-2 cells.
To determine whether motility per se was required for adherence, bacteria were centrifuged onto the HEp-2 cells prior to the 90-min infection period. In doing so, the motility defects of the mutants were bypassed. Centrifugation increased adherence of all the strains used (Table 2). Adherence of Sch3N went up from an average of 23.5 to 54.5 bacteria/HEp-2 cell. Centrifugation increased adherence of mutants IAG75 and IAG1419 up to 49.1 and 33.6% of the Sch3N adherence, respectively, whereas adherence of IAG570 and IAG1639 after centrifugation was much lower.
Sequence analysis of loci interrupted by mini-Tn5Cm in mutants IAG75, IAG570, IAG1419, and IAG1639.
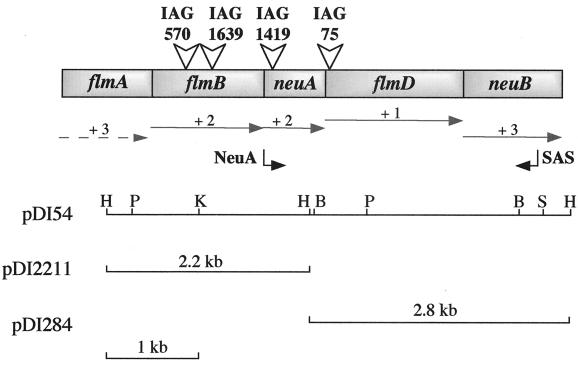
The DNA flanking the transposon in all four mutants was isolated by inverse PCR and then cloned into pUC18 (see Materials and Methods). The PCR products obtained from all four mutants exhibited an identical size, 2.6 kb. Nucleotide sequencing of the four respective PCR products identified three mutated genes clustered on the same PstI fragment. One of the PCR products was then used as a probe to screen a pUC18 HindIII chromosomal DNA library of Sch3N maintained in E. coli XL1-Blue. Subsequently two HindIII fragments of 2,237 and 2,830 bp carrying the wild-type genes were identified and sequenced. Sequence analysis of the two HindIII fragments of total length 5,067 bp identified five ORFs, ORF1 to ORF5, of which ORF1 was incomplete (Fig. 2). To decide on the stop and the start codons of the putative ORFs, the extent of the homology of the putative amino acid sequences to known proteins, the degree of overlap with preceding ORFs, and the presence or otherwise of Shine-Dalgarno sequences were taken into account. The genes appeared to be transcribed in the same direction, and no promoter sequences were identified between the putative ORFs. This suggested that the five genes were clustered into a single operon. Proteins homologous to the putative products of the five ORFs were identified using the BLASTX program (2) of the NCBI database (Table 3).
FIG. 2.
Genetic organization of the A. caviae Sch3N cloned locus. The ∼2.2-kb and the ∼2.8-kb HindIII chromosomal DNA fragments were cloned separately or together into plasmid pDSK519 to give plasmids pDI2211, pDI284, and pDI54 respectively. Predicted ORFs were named after their homologues in other bacterial species. Open arrowheads indicate the site of mini-Tn5Cm insertion in every mutant's chromosome. Horizontal arrows indicating the direction of transcription and the reading frame (+1, +2, +3) are also drawn. The unknown flmA sequence is indicated by a dashed arrow. The approximate hybridization sites of the oligonucleotide primers NeuA and SAS are shown. BglII (B), HindIII (H), KpnI (K), PstI (P), and SacII (S) restriction sites are indicated.
TABLE 3.
Properties of five putative ORFs of A. caviae Sch3N
| ORF (nt)a | Size (aa)b | Molecular mass (kDa)a | Homologous proteinc | Homologue function | % Identityc | % Similarityc | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| 1 (621)d | 206 | FlaA1, H. pylori | Flagellar filament assembly | 45 | 61 | AE000595 | |
| FlmA, C. crescentus | Flagellar filament assembly | 42 | 57 | U27301 | |||
| SpsDe, Methanococcus jannaschii | Spore coat polysaccharide biosynthesis | 39 | 60 | U67549 | |||
| 2 (1,164) | 387 | 43 | FlmB, C. crescentus | Flagellar filament assembly | 40 | 57 | U27301 |
| SpsCe Bacillus subtilis | Spore coat polysaccharide biosynthesis | 38 | 59 | P39623 | |||
| SpsCe M. jannaschii | Spore coat polysaccharide biosynthesis | 36 | 60 | U67549 | |||
| 3 (687) | 228 | 25.8 | NeuA, H. pylori | CMP-NeuNAc synthetase | 37 | 53 | AE000550 |
| NeuA, E. coli | CMP-NeuNAc synthetase; capsule biosynthesis | 31 | 50 | P13266 | |||
| NeuA, N meningitidis | CMP-NeuAc synthetase; capsule biosynthesis | 27 | 45 | Q57385 | |||
| PtmB, C. coli | Flagellin glycosylation | 27 | 45 | U25992 | |||
| 4 (1,518) | 505 | 56.8 | FlmD, C. crescentus | Flagellar filament assembly | 27 | 40 | U27302 |
| SpsGe, M. jannaschii | Spore coat polysaccharide biosynthesis | 24 | 41 | U67549 | |||
| SpsHe, B. subtilis | Spore coat polysaccharide biosynthesis | 23 | 40 | Z99123 | |||
| 5 (1,059) | 352 | 38.6 | NeuB, C. jejuni | Sialic acid synthetase | 40 | 55 | AJ000855 |
| SpsE, H. pylori | Spore coat polysaccharide biosynthesis | 39 | 59 | AE000538 | |||
| SpsEe, M. jannaschii | Spore coat polysaccharide biosynthesis | 36 | 53 | U67549 | |||
| SpsEeB. subtilis | Spore coat polysaccharide biosynthesis | 31 | 50 | P39625 | |||
| NeuB, Streptococcus agalactiae | Sialic acid synthetase; capsule biosynthesis | 32 | 50 | ABO17355 | |||
| NeuB, E. coli | Sialic acid synthetase; capsule biosynthesis | 32 | 52 | UO5248 |
ORFs and molecular masses were determined by ORF finder of NCBI.
aa, amino acids.
Homologous proteins and identity and similarity scores over the homologous regions were determined by the BLASTX (2) program of NCBI.
Incomplete ORF.
Hypothetical protein.
ORF1 (FlmA homologue).
Only the 3′-end nucleotide sequence (nt 1 to 623) was obtained for ORF1, which was located upstream of the remaining four ORFs. Its deduced amino acid sequence was found to be most similar to FlaA1 and FlmA of Helicobacter pylori and Caulobacter crescentus respectively. FlmA was recently proposed to be essential for flagellar filament assembly through flagellin or flagellar protein modification (18). These proteins have recently been included in the Pseudomonas aeruginosa WbpM subfamily 2, a large group of proteins with diverse functions involved in exopolysaccharide biosynthesis (3). Interestingly, homologues of FlmA have been included in the general protein glycosylation system of Campylobacter jejuni (31).
ORF2 (FlmB homologue, nt 632 to 1795).
ORF2 started 8 nt downstream of ORF1, with the alternative start codon GTG. It encoded a protein of 387 amino acids with a predicted molecular mass of 43 kDa. Transposon insertions in both mutants IAG570 and IAG1639 mapped within this ORF, between nt 1002 and 1003 and 1222 and 1223, respectively. The deduced amino acid sequence of ORF2 was most similar (40% identity) to the flagellar protein FlmB of C. crescentus. FlmB was also encoded by the operon that encoded FlmA, and again was proposed to be required for flagellar filament assembly (18). Moreover, FlmB homologues in C. jejuni (PglE) also belong to the general protein glycosylation system (31).
ORF3 (NeuA homologue, nt 1796 to 2482).
ORF3 was the smallest of the complete ORFs identified. It encoded a putative protein of 228 amino acids with a predicted molecular mass of 25.8 kDa. Mutant IAG1419 carried the transposon insertion near the 5′ end of this ORF, between nt 1887 and 1888. ORF3 started immediately downstream of ORF2 and thus was transcribed in the same reading frame. Its predicted amino acid sequence matched a series of CMP-NeuNAc synthetases (NeuA) of H. pylori, E. coli, and Neisseria meningitidis required for the condensation of N-acetylneuraminic acid (NeuNAc) and CTP into CMP-NeuNAc. These enzymes were previously shown to be required for the biosynthesis of polysialic acid capsules of E. coli K1 (40) and N. meningitidis group B (7). The fourth-highest homology was to the recently identified PtmB of Campylobacter coli VC167, which was proposed to be required for the sialylation of the polar flagellins (9).
ORF4 (FlmD homologue, nt 2488 to 4005).
ORF4 started 6 nt downstream of ORF3 and encoded a putative protein of 505 amino acids with a predicted molecular mass of 56.8 kDa. The transposon insertion of IAG75 mapped between nucleotides 2549 and 2550 and would have therefore inserted just inside the 5′ end of the putative gene. The deduced amino acid sequence of ORF4 was found mainly similar to the flagellar protein FlmD of C. crescentus and matched the deduced amino acid sequence of ORF4 throughout its length. Similar to FlmA and FlmB, FlmD was thought to be required for flagellar filament assembly, but the gene encoding this protein mapped with FlmC in a separate operon (18).
ORF5 (NeuB homologue, nt 3993 to 5051).
ORF5 overlapped with ORF4 and started 13 nt within it. It encoded a putative protein of 352 amino acids with a predicted molecular mass of 38.6 kDa. Its deduced amino acid sequence was found to be homologous to a series of spore coat polysaccharide biosynthetic proteins and sialic acid synthetases (NeuB) from different bacterial species. Three homologues of each protein were identified in the top six matches, and all matched the entire length of the A. caviae putative protein. Recently, one of the three neuB genes possessed by C. jejuni was shown to be required for motility and flagellum production (19).
LPS extraction and PAGE analysis.
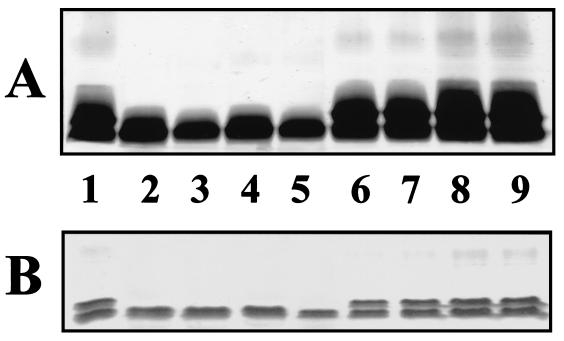
Following the database homology results, the possibility that the mutants carried defects in polysaccharide biosynthesis was investigated. As the A. caviae strain studied did not produce spores or a capsule, the LPS was examined. Strain Sch3N and the mutants IAG75, IAG570, IAG1419, and IAG1639 were grown overnight in BHIB at 37°C, and LPS was extracted and analyzed by PAGE (Fig. 3). As can be observed strain Sch3N was able to exhibit a smooth LPS (O-Ag+) while the mutants were unable to do it (O-Ag−) (Fig. 3A). Furthermore, when we studied the LPS core, we observed (Fig. 3B) that the LPS of the mutants lacks one of the bands always present in the LPS of the wild-type strain. The highest-migrating band, the one lost in the LPS of the mutants, could be part of the outer-core LPS.
FIG. 3.
Analysis of LPS isolated from A. caviae Sch3N (lane 1) and the four mutants IAG75, IAG570, IAG1419, and IAG1639 before (lanes 2, 3, 4, and 5, respectively) and after (lanes 6, 7, 8, and 9, respectively) complementation with plasmid pDI54. (A) LPS extracted and analysed by SDS-PAGE (12%) according to Darveau and Hancock (5) and silver stained (36). (B) The same LPS analyzed by Tricine-SDS-PAGE for LPS oligosaccharides (23).
Complementation analysis of mutants.
The 2.2- and the 2.8-kb HindIII DNA fragments that carried wild-type copies of the mutated genes were expressed in the mutants in trans, in an attempt to complement the defects caused by the transposon insertions. The two fragments were ligated separately or together into the HindIII site of the broad-host-range plasmid pDSK519 (13) in an orientation that allowed expression from the lac promoter. The resulting pDSK519 derivatives carrying the 2.2 kb, the 2.8 kb, or the total of ∼5 kb were designated pDI2211, pDI284, and pDI54, respectively. E. coli S17-1 transformants each carrying one of the three plasmids were subsequently obtained. The plasmids were then introduced separately into mutants IAG75, IAG570, IAG1419, and IAG1639 by bacterial conjugation (see Materials and Methods), and the wild-type genes were expressed from the lac promoter in trans. Transconjugant strains grown on both kanamycin and chloramphenicol were isolated from every conjugation experiment. Upon HindIII and PstI digestion, all three plasmids isolated from the Aeromonas transconjugants gave the same restriction patterns as the original plasmids maintained in E. coli S17-1. Adherence, motility, and LPS phenotypes of the resulting 12 transconjugant strains were examined (Table 4).
TABLE 4.
Phenotypic characteristics of original and complemented A. caviae Sch3N mutants
| Strain or plasmid and mutant | Adherencea | Motilityb | Polar flagellin immunoblot | LPSc |
|---|---|---|---|---|
| Sch3N | 17.4 ± 11.3 | + | + | S |
| IAG75 (flmD) | 2.1 ± 1 | − | − | R |
| IAG570 (flmB) | 1.3 ± 0.5 | − | − | R |
| IAG1419 (neuA) | 1.1 ± 0.5 | − | − | R |
| IAG1639 (flmB) | 3.3 ± 1.3 | − | − | R |
| pDI221 (flmB+) | ||||
| IAG75 (flmD) | 1.6 ± 0.6 | − | − | R |
| IAG570 (flmB) | 1.5 ± 0.6 | − | − | R |
| IAG1419 (neuA) | 0.5 ± 0.2 | − | − | R |
| IAG1639 (flmB) | 3 ± 1.3 | − | − | R |
| pDI284 (flmD+ neuB+) | ||||
| IAG75 (flmD) | 15 ± 10.4 | + | + | S |
| IAG570 (flmB) | 0.7 ± 0.3 | − | − | R |
| IAG1419 (neuA) | 0.9 ± 0.5 | − | − | R |
| IAG1639 (flmB) | 0.6 ± 0.4 | − | − | R |
| pDI54 (flmB+ neuA+ flmD+ neuB+) | ||||
| IAG75 (flmD) | 11.9 ± 5 | + | + | S |
| IAG570 (flmB) | 16.7 ± 7.8 | + | + | S |
| IAG1419 (neuA) | 17 ± 4.4 | + | + | S |
| IAG1639 (flmB) | 14.1 ± 3.6 | + | + | S |
Mean number of adherent bacteria per HEp-2 cell ± standard deviation.
Bacteria were inoculated in motility agar (1% tryptone, 0.05% NaCl, 0.25% agar) for 16 to 24 h. +, fully motile, bacteria cover most of the plate; −, nonmotile, no migration from the site of inoculation.
S, smooth; R, rough.
Adherence.
The ability of the four mutant strains carrying either of the three pDSK519-derived vectors, in addition to Sch3N and the four original nonadherent transposon mutants, to adhere to HEp-2 cells was tested by adherence assay. All mutants harboring pDI2211 (flmB+) remained nonadherent, with their mean adhesion values ranging between 0.5 to 3 adherent bacteria/HEp-2 cell. Similar low mean adhesion values of 0.6 to 0.9 adherent bacteria/HEp-2 cell were recorded for IAG570 (flmB), IAG1419 (neuA), and IAG1639 (flmB) carrying pDI284 (flmD+ neuB+). In contrast, IAG75 (flmD) harboring pDI284 (flmD+ neuB+) and all four mutants carrying pDI54 (flmB+ neuA+ flmD+ neuB+) exhibited dramatically increased adherence, with mean adhesion values of 15 and 12 to 17 adherent bacteria/HEp-2 cell, respectively.
Motility and flagellin expression.
Motility of all four mutants expressing either of the three plasmids was tested in motility agar. Again only strain IAG75 expressing pDI284 and all four mutants expressing pDI54 regained their ability to swim in the semisolid motility agar, whereas the rest of the mutants remained nonmotile. To confirm the motility assays results, the whole-cell proteins of the 12 transconjugant mutant strains, in addition to the respective proteins of the four original nonadherent mutant strains and Sch3N, were immunoblotted for the polar flagellin protein. Polar flagellin proteins of an identical size to those of the parental strain were detected in the preparations of the adherent transconjugant strains, whereas no flagellin proteins were detected in the preparations of the nonadherent transconjugants and the original transposon mutant strains (Fig. 1).
LPS analysis.
The profiles of all the mutants harboring pDI2211 and those of mutants IAG570, IAG1419, and IAG1639 harboring pDI284 remained unchanged (data not shown). The remaining mutants which were complemented for adherence, flagellin expression, and motility exhibited fully complemented LPS profiles (Fig. 3). Specifically, these mutants regained the O-Ag as well as the highest-migrating band of the outer-core LPS.
Distribution of flmA, flmB, neuA, flmD, and neuB in mesophilic aeromonads.
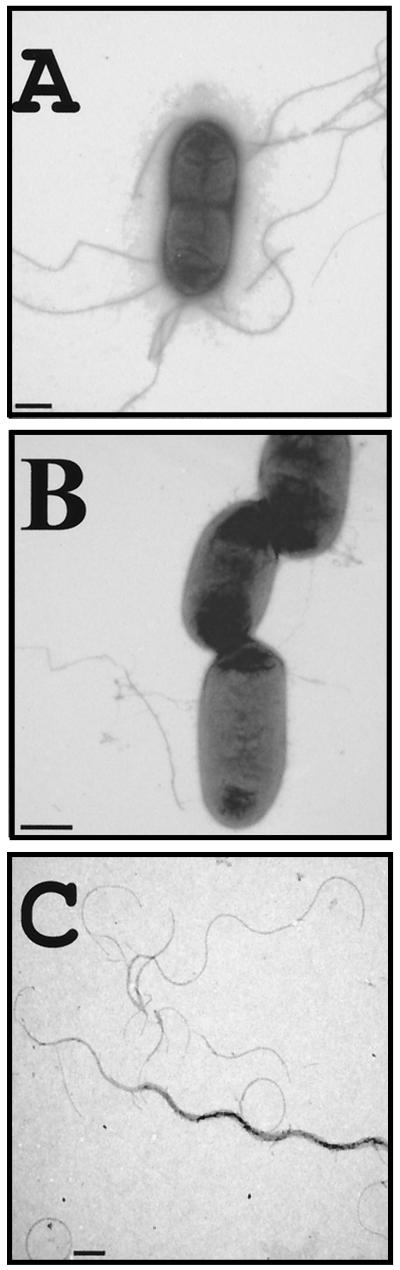
Using five separate PCR-generated probes to each of the A. caviae Sch3N genes, we investigated the distribution of these genes among 20 mesophilic Aeromonas spp. (including A. hydrophila, A. veronii bv. veronii, and A. veronii bv. sobria), by dot blot hybridization. Both flmA and flmB probes hybridized to the chromosomal DNA of all the strains tested, whereas no hybridization was observed for the probes to neuA, neuB, or flmD. Additionally, the oligonucleotide primers NeuA, 5′-GACTCATATGAATATTGCCATCATCCC-3′, and SAS, 5′-CTTTACATAACGCAGCAA-3′, were used to amplify the 2,998-bp neuA-flmD-neuB region from A. caviae Sch3N by PCR. This PCR product was used as a probe to screen 19 strains of A. caviae by Southern hybridization. However, only the chromosomal DNA of A. caviae Sch3N hybridized with the probe. Oligonucleotides flmB2, 5′-TCTGATTTTCTAACTCAGGG-3′ (initial base 67), and flmB4, 5′-GTCATTCGGTAGTTAAAGCC-3′ (final base 739), were then used to amplify an internal fragment (672 bp) of both the A. caviae Sch3N and A. hydrophila AH-3 flmB genes; these were subsequently confirmed by sequencing. The amplified fragment from strain AH-3 was ligated into the suicide plasmid pFS100 to generate plasmid pFS-FlmB, in order to obtain defined insertion mutants as previously described (28). Using this plasmid we created defined flmB insertion mutants in A. hydrophila AH-3 as well as in four other mesophilic Aeromonas strains (A. hydrophila serogroup O:3 and O:25, A. veronii bv. sobria AH-1 [serogroup O:11], and A. veronii bv. veronii AS-28 [serogroup O:11]). All of these mutants were nonmotile when assayed on semisolid motility agar and lacked polar flagella, while their lateral flagella were not attached to the cell surface (Fig. 4 shows a comparison between AH-3 and a flmB-defined insertion mutant from the same strain). However, no changes could be observed in the LPS profiles of any of these strains.
FIG. 4.
EM of A. hydrophila motile strain AH-3 (wild type) (A), and nonmotile mutant AH-1726 (flmB) derived from AH-3(B) grown on motility agar for 12 h at 30°C. Samples were picked and gently resuspended before being adsorbed on the grid. As can be observed, loose flagella are frequently observed (C) on preparations of mutant strain AH-1726, while nothing similar could be observed on wild-type strain AH-3. Bar, 0.5 μm.
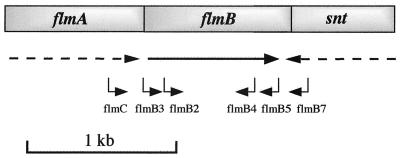
Taking advantage of a previously constructed genomic library of strain AH-3 (1) we complemented the FlmB mutant of this strain (AH-1726, Kmr) selecting for Kmr Tcr clones that were motile on semisolid motility agar. Plasmid pLA-Flm, which complemented the mutation, was sequenced, and two incomplete ORFs (ORF1 and ORF3) and one complete ORF (ORF2) were identified. The deduced amino acid sequence of ORF1 was clearly homologus to that of FlmA, while ORF2 was a homologue of FlmB. ORF3 is transcribed in the opposite direction, and its deduced amino acid sequence was found to be similar to a putative sugar nucleotidyl transferase (Snt) from C. jejuni (Fig. 5). Oligonucleotide primers were designed that hybridized to the edges of ORF1 and ORF2, flmC 5′-GAGAAGTTGCATGAAGTGATG-3′ and flmB5 5′-GTCATGCGGTAGTTGAAACC-3′. These amplified a single fragment of 976 bp from A. hydrophila AH-3 in a PCR. Single bands were also obtained in PCRs using chromosomal DNA from the Aeromonas strains: A. caviae Sch3N, A. hydrophila serogroups O:3 and O:25, A. veronii bv. sobria AH-1 (serogroup O:11), and A. veronii bv. veronii AS-28 (serogroup O:11). When oligonucleotide primers were designed that hybridized to the edges of ORF2 and ORF3, flmB3 5′-TCCGATTTTCTGACTCAGGG-3′ and flmB7 5′-GGGGAAATTGATTCACC-3′ (Fig. 5), again a single band was amplified by PCR in all strains tested with the notable exception of A. caviae Sch3N. Both the PCR and Southern hybridization results suggest that for the mesophilic aeromonads, flmA and flmB are usually found together upstream of snt and not in a putative operon along with neuA-flmD-neuB as is the case for A. caviae Sch3N.
FIG. 5.
Genetic organization of the A. hydrophila flm locus. Predicted ORFs were named after their homologues in other bacterial species. Horizontal arrows indicate the direction of transcription, and partial ORFs are indicated by dashed arrows. The approximate hybridization sites of the oligonucleotide primers flmC, flmB3, flmB2, flmB4, flmB5, and flmB7 are shown.
All of the FlmB-defined insertion mutants exhibited reduced adherence to HEp-2 cells that could either be fully restored by complementation with pLA-Flm or by centrifugation onto the monolayer (Table 5). After complementation with pLA-Flm, all the mutants were motile and expressed polar or lateral flagella depending on the medium fluidity, similar to their respective wild types.
TABLE 5.
Adhesion and effect of centrifugation of different Aeromonas strains onto HEp-2 cells prior to infection
| Strain | Mean no. of bacteria/ HEp-2 cell ± SDa
|
|
|---|---|---|
| Normal | Centrifuged | |
| A. hydrophila AH-3 (wild type) | 18.3 ± 2.2 | 31.6 ± 3.0 |
| FlmB insertion mutant from AH-3 (AH-1726) | 9.4 ± 1.2 (51) | 29.6 ± 3.2 (93) |
| AH-1726 complemented with pLA-Flm | 18.0 ± 2.5 | 32.5 ± 2.3 |
| A. hydrophila O:3 (wild type) | 21.6 ± 2.4 | 42.7 ± 4.3 |
| FlmB insertion mutant from O:3 (AH-1881) | 10.1 ± 1.9 (46) | 40.8 ± 5.1 (95) |
| AH-1881 complemented with pLA-Flm | 21.9 ± 3.2 | 42.1 ± 5.2 |
| A. hydrophila O:25 (wild type) | 16.0 ± 2.4 | 29.3 ± 2.8 |
| FlmB insertion mutant from O:25 (AH-1882) | 7.7 ± 1.3 (48) | 28.6 ± 2.9 (96) |
| AH-1882 complemented with pLA-Flm | 15.7 ± 2.6 | 29.9 ± 3.1 |
| A. veronii bv. sobria AH-1 (wild type) | 32.3 ± 3.9 | 71.4 ± 10.7 |
| FlmB insertion mutant from AH-1 (AH-1883) | 18.1 ± 2.2 (56) | 67.3 ± 9.9 (94) |
| AH-1883 complemented with pLA-Flm | 31.6 ± 4.4 | 70.3 ± 9.2 |
| A. veronii bv. veronii AS-28 (wild type) | 31.7 ± 4.6 | 66.8 ± 10.2 |
| FlmB insertion mutant from AS-28 (AH-1884) | 15.5 ± 3.4 (48) | 63.3 ± 9.1 (94) |
| AH-1884 complemented with pLA-Flm | 32.4 ± 3.8 | 65.9 ± 10.0 |
See footnote a to Table 2.
DISCUSSION
In this study we used the HEp-2 cell model to initially identify the genes involved in the adherence mechanism of A. caviae Sch3, a strain able to exhibit similar patterns of diffuse adherence to the human epithelial cell lines HEp-2 and Caco-2 (34). Preliminary data generated in our laboratory during this and previous studies (34) suggested the involvement of the polar flagellum in the adherence of A. caviae Sch3. Moreover, phenotypic characterization of the nonadherent transposon mutants of A. caviae Sch3N IAG75, IAG570, IAG1419, and IAG1639 strengthened this view. The wild-type copies of the genes mutated in these transposon mutants were cloned and sequenced and were shown to be clustered in a putative operon involved in LPS O-Ag biosynthesis and possibly flagellum assembly. The products of the five putative A. caviae genes (ORF1 to ORF5) were similar to a series of bacterial polysaccharide biosynthesis proteins, although the products of ORF1, ORF2, and ORF4 were most similar to the flagellar proteins FlmA, FlmB, and FlmD of C. crescentus, respectively. Flm proteins were reported to be involved in flagellar filament assembly of Caulobacter, possibly through glycosylation of the flagellin or another flagellar protein(s) (18). However, the C. crescentus flm mutants do not exhibit any LPS defects (18), in contrast to those of A. caviae. As the A. caviae Sch3N transposon mutations affect the LPS O-Ag, an alternative explanation for the loss of flagella could be the rough phenotype which has been reported for LPS core mutants (rfa) of E. coli (27). Such mutations occur in the LPS core and are thought to destabilize in a pleiotropic way the outer membrane and affect outer membrane protein insertion and O-Ag attachment. Mutations in the flm locus of A. caviae could possibly be affecting the insertion of another adhesin into the outer membrane. However, this putative adhesin did not appear to be an outer membrane protein as the outer membrane protein profiles of the four mutations did not differ from the wild types when analyzed on polyacrylamide gels (data not shown).
In order to try to explain this phenomenon we investigated the presence of these genes (flmA, flmB, neuA, flmD, and neuB) in different mesophilic Aeromonas spp., including the strain A. hydrophila AH-3 (serogroup O:34). We used this strain in particular because the genes responsible for the O:34 LPS antigen biosynthesis have been cloned and sequenced and the LPS composition is known (unpublished results). The flmA- and flmB-like genes were found in all mesophilic Aeromonas strains (A. hydrophila, A. caviae, A. veronii bv. veronii, and A. veronii bv. sobria) when tested by dot blot hybridization, while only A. caviae Sch3N contained the neuA-, flmD-, and neuB-like genes. This suggests that A. caviae Sch3N is different from most of the other mesophilic aeromonads (including A. caviae) and that a genetic rearrangement has probably occurred in this strain to incorporate flmA and flmB in a putative operon along with neuA, flmD, and neuB.
Using PCR we were able to amplify an internal fragment of the flmB-like gene from A. hydrophila AH-3; this allowed us to generate a defined insertion mutant in this strain. The flmB mutant of strain AH-3 lacked flagella (polar and lateral flagella) and therefore was nonmotile. It was clear by EM that these mutants were able to produce flagella but unable to assemble them on the cell. However, no changes were seen in the LPS of this mutant, a situation similar to that of Caulobacter and the other mesophilic aeromonad flmB mutants created in this study but different from that observed for A. caviae Sch3N.
After complementation of the flmB mutant of strain AH-3, it was clear that flmA and flmB are transcribed in the same direction, and no neuA-like gene could be found downstream of flmB. Instead, a gene similar to a putative sugar nucleotidyl transferase from C. jejuni, one that is transcribed in the opposite direction, was present. This could be a possible reason that no changes in the LPS profile were observed in the flmB mutant of AH-3. A similar situation was observed for the flmB mutants of the four other mesophilic Aeromonas strains.
In the case of A. caviae Sch3N the phenotype of the original and the complemented mutants did not allow us to assess the separate roles of the polar flagellum and the LPS O-Ag in adherence. Either of the two or both could be responsible for the in vitro adherence of A. caviae Sch3N. The ability of the complemented motile mutants to adhere to HEp-2 cells and the increased adherence seen after the centrifugation of bacteria onto the monolayers suggested the involvement of both the polar flagellum and motility in adherence. Furthermore, the approximate 50% inhibition of adherence produced by the polar flagellin antibodies (data not shown) also supported the importance of motility and flagella in the process. We previously indicated that the polar flagellum and motility are required for adherence to, and invasion of, fish cell lines by A. hydrophila serogroup O:34, as nonflagellate Tn5 mutants lost adherence and invasiveness; however, the mutated genes were not identified (24). The LPS O-Ag has also been described as an adhesin in Aeromonas. Ourselves and other workers have associated LPS O-Ag expression with adherence of A. hydrophila and A. veronii bv. sobria clinical isolates to HEp-2 cells (1, 8, 23). Furthermore, LPS O-Ag transposon mutants of A. hydrophila O:34 were shown to be unable to colonize the germfree chicken gut model (22). Nevertheless, the flmB mutations in A. hydrophila AH-3 and the other mesophilic aeromonads (excluding A. caviae Sch3N), allowed us to conclude that the flagellum is essential for HEp-2 cell colonization, because these strains were nonadherent and nonmotile and lacked flagella but retained complete LPS profiles. However, there was only a 60% decrease in adherence for the A. hydrophila AH-3 flmB mutant, compared to the 80 to 90% decrease in adherence observed for A. caviae Sch3N flm locus transposon mutants. Additionally, the A. hydrophila AH-3 flmB mutation could be partially rescued, and adherence to wild-type levels could be recovered by centrifugation of the bacteria onto the monolayer. This was not the case for the A. caviae mutants, for which centrifugation alone only restored the adherence to 12 to 50% of the wild-type levels. This is most probably due to the A. caviae mutants' lacking two adhesins: flagellar and LPS O-Ag. After centrifugation the mutations in flmB (IAG570 and IAG1639) caused the greatest loss of HEp-2 cell colonization. This could be possibly due to FlmB being required for the expression of another adhesin or acting as an adhesin itself, although the outer membrane protein profiles of the flmB mutants did not differ from those of the other mutants or that of the wild types (data not shown). From all the results we can conclude (i) that flagella and possibly the LPS O antigen are surface structures of mesophilic Aeromonas involved in adherence to human epithelial cells; (ii) flmA and flmB are genes widely distributed in mesophilic Aeromonas and are involved in flagellum assembly, and thus adherence; and (iii) in A. caviae Sch3N the flmA and flmB genes are located in a putative operon together with neuA, flmD, and neuB. These genes perform a role in LPS O-Ag biosynthesis and are likely to be involved flagellum assembly.
ACKNOWLEDGMENTS
We thank Ann Cooke and Margaret Lee for their help with tissue culture and Ali Rabaan for the gift of the flagellin antibodies. We thank Maite Polo for her technical assistance.
Part of this work was supported by grants from DGICYT and Plan Nacional de I+D (Ministerio de Educación y Cultura, Spain).
REFERENCES
- 1.Aguilar A, Merino S, Rubirés X, Tomás J M. The influence of osmolarity on lipopolysaccharide and virulence of Aeromonas hydrophila serotype O:34 strains grown at 37°C. Infect Immun. 1997;65:1245–1250. doi: 10.1128/iai.65.4.1245-1250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Stephen F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrows L L, Urbanic R V, Lam J S. Functional conservation of the polysaccharide biosynthetic protein WbpM and its homologues in Pseudomonas aeruginosa and other medically significant bacteria. Infect Immun. 2000;68:931–936. doi: 10.1128/iai.68.2.931-936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrello A, Silburn K A, Budden J R, Chang B J. Adhesion of clinical and environmental Aeromonas isolates to HEp-2 cells. J Med Microbiol. 1988;26:19–27. doi: 10.1099/00222615-26-1-19. [DOI] [PubMed] [Google Scholar]
- 5.Darveau R P, Hancock R E W. Procedure for the isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol. 1983;155:831–838. doi: 10.1128/jb.155.2.831-838.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards U, Muller A, Hammerschmidt S, Gerardy-Schahn R, Frosch M. Molecular analysis of the biosynthetic pathway of the α-2,8 polysialic acid capsule by Neisseria meningitidis serogroup B. Mol Microbiol. 1994;14:141–149. doi: 10.1111/j.1365-2958.1994.tb01274.x. [DOI] [PubMed] [Google Scholar]
- 8.Francki K T, Chang B J. Variable expression of O-antigen and the role of lipopolysaccharide as an adhesin in Aeromonas sobria. FEMS Microbiol Lett. 1994;122:97–102. doi: 10.1111/j.1574-6968.1994.tb07150.x. [DOI] [PubMed] [Google Scholar]
- 9.Guerry P, Doig P, Alm R A, Burr D H, Kinsella N, Trust T J. Identification and characterisation of genes required for post-translational modification of Campylobacter coli VC167 flagellin. Mol Microbiol. 1996;19:369–378. doi: 10.1046/j.1365-2958.1996.369895.x. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 11.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janda J M, Abbott S L. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin Infect Dis. 1998;27:332–344. doi: 10.1086/514652. [DOI] [PubMed] [Google Scholar]
- 13.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 14.Kirov S M. Adhesion and piliation of Aeromonas spp. Med Microbiol Lett. 1993;2:274–280. [Google Scholar]
- 15.Kirov S M, Jacobs I, Hayward L J, Hapin R H. Electron microscopic examination of factors influencing the expression of filamentous surface structures on clinical and environmental isolates of Aeromonas veronii biotype sobria. Microbiol Immunol. 1995;39:329–338. doi: 10.1111/j.1348-0421.1995.tb02209.x. [DOI] [PubMed] [Google Scholar]
- 16.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Leclerc G, Wang S, Ely B. A new class of Caulobacter crescentus flagellar genes. J Bacteriol. 1998;180:5010–5019. doi: 10.1128/jb.180.19.5010-5019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linton D, Karlyshev A V, Hitchen P G, Morris H R, Dell A, Gregson N A, Wren B W. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol Microbiol. 2000;35:1120–1134. doi: 10.1046/j.1365-2958.2000.01780.x. [DOI] [PubMed] [Google Scholar]
- 20.Merino S, Camprubí S, Tomás J M. Isolation and characterization of bacteriophage PM3 from Aeromonas hydrophila, the bacterial receptor for which is the monopolar flagellum. FEMS Microbiol Lett. 1990;69:277–282. doi: 10.1016/0378-1097(90)90080-a. [DOI] [PubMed] [Google Scholar]
- 21.Merino S, Rubirés X, Knokel S, Tomás J M. Emerging pathogens: Aeromonas spp. Int J Food Microbiol. 1995;28:157–168. doi: 10.1016/0168-1605(95)00054-2. [DOI] [PubMed] [Google Scholar]
- 22.Merino S, Rubirés X, Aguilar A, Guillot J F, Tomás J M. The role of the O-Ag lipopolysaccharide on the colonisation in vivo of the germ free chicken gut by Aeromonas hydrophila serogroup O:34. Microb Pathog. 1996;20:325–333. doi: 10.1006/mpat.1996.0031. [DOI] [PubMed] [Google Scholar]
- 23.Merino S, Rubirés X, Aguilar A, Tomás J M. The O:34-antigen lipopolysaccharide as an adhesin in Aeromonas hydrophila. FEMS Microbiol Lett. 1996;139:97–101. doi: 10.1111/j.1574-6968.1996.tb08186.x. [DOI] [PubMed] [Google Scholar]
- 24.Merino S, Rubirés X, Aguilar A, Tomás J M. The role of flagella and motility in the adherence and invasion to fish cell lines by Aeromonas hydrophila serogroup O:34 strains. FEMS Microbiol Lett. 1997;151:213–217. doi: 10.1111/j.1574-6968.1997.tb12572.x. [DOI] [PubMed] [Google Scholar]
- 25.Metcalf W W, Jiang W, Wanner B L. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6Kγ origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 26.Namdari H, Bottone E J. Microbiologic and clinical evidence supporting the role of Aeromonas caviae as pediatric enteric pathogen. J Clin Microbiol. 1990;28:837–840. doi: 10.1128/jcm.28.5.837-840.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker C T, Kloser A W, Schnaitman C A, Stein M A, Gottesman S, Gibson B W. Role of rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992;174:2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubirés X, Saigí F, Piqué N, Climent N, Merino S, Albertí S, Tomás J M, Regué M. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J Bacteriol. 1997;179:7581–7586. doi: 10.1128/jb.179.23.7581-7586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Simon R, Priefer V, Puhler A. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 31.Szymanski C M, Yao R, Ewing C P, Trust T J, Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;35:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 32.Thomas L V, Gross R J, Cheasty T, Rowe B. Extended serogrouping scheme for motile, mesophilic Aeromonas species. J Clin Microbiol. 1990;28:980–984. doi: 10.1128/jcm.28.5.980-984.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thornley J P, Shaw J G, Gryllos I A, Eley A. Adherence of Aeromonas caviae to human cell lines HEp-2 and Caco-2. J Med Microbiol. 1996;45:445–451. doi: 10.1099/00222615-45-6-445. [DOI] [PubMed] [Google Scholar]
- 35.Thornley J P, Shaw J G, Gryllos I A, Eley A. Virulence properties of clinically significant Aeromonas species. Rev Med Microbiol. 1997;8:61–72. [Google Scholar]
- 36.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharide in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 37.Tso M D, Dooley J S G. Temperature-dependent protein and lipopolysaccharide expression in clinical Aeromonas isolates. J Med Microbiol. 1995;42:32–38. doi: 10.1099/00222615-42-1-32. [DOI] [PubMed] [Google Scholar]
- 38.Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 39.Wilcox M H, Cook A M, Eley A, Spencer R C. Aeromonas spp. as a potential cause of diarrhoea in children. J Clin Pathol. 1992;45:959–963. doi: 10.1136/jcp.45.11.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zapata G, Vann W F, Aaronson W, Lewis M S, Moos M. Sequence of the cloned Escherichia coli K1 CMP-N-acetylneuraminic acid synthetase. J Biol Chem. 1989;264:14769–14774. [PubMed] [Google Scholar]