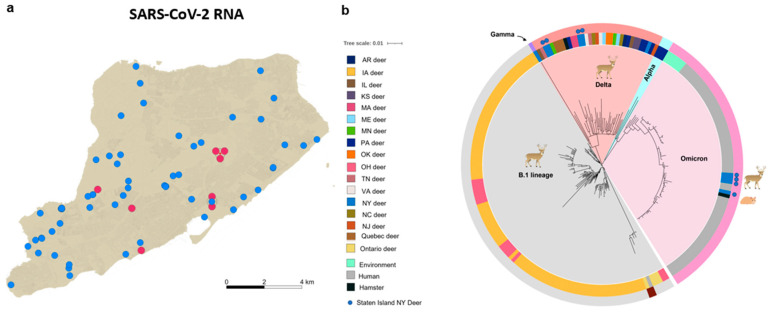
Figure 1.
Distribution and whole-genome single nucleotide polymorphism (SNP)-based phylogenies of SARS-CoV-2 recovered from white-tailed deer on Staten Island, New York: (a) the spatial distribution of the collection sites of nasal and tonsillar swabs from white-tailed deer that were tested for the presence of SARS-CoV-2 viral RNA; red circles show sites where swabs were positive for SARS-CoV-2 viral RNA, and blue-filled circles show swabs that were negative; (b) whole-genome sequences of eight newly characterized white-tailed-deer-origin SARS-CoV-2 genomes were analyzed in the context of 135 publicly available white-tailed-deer-origin SARS-CoV-2 isolates, and 63 arbitrarily selected SARS-CoV-2 Omicron genomes circulating amongst humans in New York City during this same time period, as well as representative isolates from the environment or from Syrian hamsters (SI, Table S2); the genome sequences were screened for quality, for SNP positions called against the SARS-CoV-2 reference genome (NC_045512), and for SNP alignments used to generate a maximum-likelihood phylogenetic tree, using RAxML.