Abstract
Xyloglucan endotransglycosylase/hydrolase (XTH) genes play an important role in plant resistance to abiotic stress. However, systematic studies of the response of Boehmeria nivea (ramie) XTH genes (BnXTHs) to cadmium (Cd) stress are lacking. We sought to identify the BnXTH-family genes in ramie through bioinformatics analyses and to investigate their responses to Cd stress. We identified 19 members of the BnXTH gene family from the ramie genome, referred to as BnXTH1-19, among which BnXTH18 and BnXTH19 were located on no chromosomes and the remaining genes were unevenly distributed across 11 chromosomes. The 19 members were divided into four groups, Groups I/II/IIIA/IIIB, according to their phylogenetic relationships, and these groups were supported by analyses of intron–exon structure and conserved motif composition. A highly conserved catalytic site (HDEIDFEFLG) was observed in all BnXTH proteins. Additionally, three gene pairs (BnXTH6–BnXTH16, BnXTH8–BnXTH9, and BnXTH17–BnXTH18) were obtained with a fragment and tandem-repeat event analysis of the ramie genome. An analysis of cisregulatory elements revealed that BnXTH expression might be regulated by multiple hormones and abiotic and biotic stress responses. In particular, 17 cisregulatory elements related to abiotic and biotic stress responses and 11 cisregulatory elements related to hormone responses were identified. We also found that most BnXTH genes responded to Cd stress, and BnXTH1, BnXTH3, BnXTH6, and BnXTH15 were most likely to contribute to the Cd tolerance of ramie, as evidenced by the substantial increases in expression under Cd treatment. Heterologous expression of BnXTH1, BnXTH6, and BnXTH15 significantly enhanced the Cd tolerance of transgenic yeast cells. These results suggest that the BnXTH gene family is involved in Cd stress responses, laying a theoretical foundation for functional studies of BnXTH genes and the innovative breeding of Cd-tolerant ramie.
Keywords: Boehmeria nivea, XTH gene family, genome-wide identification, Cd stress, expression analysis
1. Introduction
Cadmium (Cd) is a nonessential trace metal with high mobility and toxicity [1]. It accumulates in plants through polluted farmland, soil, and water sources and enters the human body through the food chain, thus affecting human health [2]. According to statistics, more than 5 million soils worldwide, mainly in developing and underdeveloped countries (India, Bangladesh, Pakistan, etc.), are polluted with heavy metals [3]. China has the most serious heavy-metal pollution among these countries, with several “Cd rice” incidents [4]. A survey of cultivated land in China showed that about 2.79 × 109 m2 of agricultural land, accounting for 20% of the total cultivated land area, is polluted with Cd [5,6]. The southern provinces, such as Hunan, Guizhou, Guangdong, Guangxi, and Fujian, have the most Cd pollution mainly because they are mining areas and smelting-process zones of nonferrous metals; their wastewaters are used for irrigation without treatment, polluting the surrounding cultivated soils [7,8,9]. Therefore, Cd pollution poses a major threat to global food security and human health, necessitating urgent mitigation.
Phytoremediation is a better mitigation measure for Cd pollution, involving Cd enrichment and accumulation in plants from contaminated soils [10]. Ramie is a nonedible perennial plant with rapid growth, a high biomass, a high Cd tolerance and enrichment ability, and a high economic value. Therefore, ramie is an ideal plant resource for remediation of Cd-contaminated soils [11]. Recent evidence has shown that cell walls, especially hemicellulose, may affect the Cd tolerance of ramie, since most Cd is enriched in hemicellulose [12]. However, it is not clear how hemicellulose binds to Cd. Therefore, it is necessary to further explore the role and the potential physiological and molecular mechanisms of action of hemicellulose in Cd accumulation and tolerance.
Hemicelluloses can be divided into four classes: xylans, mannans, β-glucans with mixed linkages, and xyloglucans [13]. Xyloglucans are the most abundant hemicellulose in the primary cell walls of dicotyledons and nonGramineae monocotyledons [14]. Their synthesis requires a glycosidic-bond synthase and various glycosyltransferases [15]. Xyloglucan endoglycosidase/hydrolase (XTH), a key xyloglucan-modifying enzyme belonging to the glycoside hydrolase 16 (GH16) family, catalyzes the cleavage and polymerization of xyloglucan molecules, thereby modifying the cellulose–xyloglucan composite structure of the cell wall [16]. XTH exhibits two catalytic functions: xyloglucan endohydrolase (XEH) activity, which catalyzes hydrolysis of xyloglucan, and xyloglucan endotransglucosylase (XET) activity, which cuts and rejoins xyloglucan chains [17].
XTH gene family members have been identified in many species, including Arabidopsis thaliana (33 members) [18], Nicotiana tabacum (56 members) [19], Oryza sativa (29 members) [20], Medicago truncatula (44 members) [21], Brassica rapa (53 members) [22], Poplar spp. (41 members) [23], Glycine max (61 members) [24], and Schima superba (34 members) [25]. Based on their phylogenetic relationships, XTH genes are classified into three major groups: Group I, Group II, and Group III [18]; however, some scholars have further divided Group-III members into groups IIIA and IIIB [20].
Recent research of XTH genes has focused on abiotic stress responses, including osmotic, salt, and low-temperature stress responses. Unique members of the XTH gene family have also been identified in many species in response to abiotic stress. For example, in A. thaliana roots, AtXTH14, AtXTH15, and AtXTH31 are downregulated in response to aluminum (Al) stress [26]. In M. truncatula, 28 MtXTH genes respond to Hg stress, 21 respond to salt stress, and another 21 respond to drought stress [21]. Three homologous genes, CaXTH1, CaXTH2, and CaXTH3, were found to respond to drought, high salt, and low-temperature stress in pepper [27]. The functions of some XTH genes have also been studied. For example, PvXTH9 and PvXTHb were associated with Al accumulation in the cell wall of the common bean [28]. Overexpression of CaXTH3 improves drought and salt tolerance in transgenic tomatoes [29], and overexpression of AtXTH31 improves flooding-stress tolerance in Glycine max [30]. Moreover, AtXTH19 can improve the freezing tolerance of A. thaliana after cold and subzero acclimation [31].
This study identified and analyzed BnXTH genes from the ramie genome to reveal the role of XTH-family genes in response to Cd stress. We also explored the responses of these genes to Cd stress through yeast expression experiments. The findings of this study provide a theoretical basis for functional studies of BnXTH-family genes in ramie.
2. Results
2.1. Identification of Chromosomal Locations and Physicochemical-Property Analysis of the BnXTH Gene Family
A total of 19 BnXTH genes were identified from published ramie genome data. The genes were denoted BnXTH1-19, among which BnXTH6-19 were named based on chromosomal positions (Figure S1 and Table 1). As illustrated in Figure S1, BnXTH18 and BnXTH19 were located on the Scaffold16 fragment and not on the chromosome, which may have been due to the poor assembly of the ramie genome. The other 17 BnXTH genes were unevenly distributed on chromosomes 2, 4, 5, 6, 7, 8, 9, 11, 12, 13, and 14. Chromosomes 1, 3, and 10 had no XTH genes, and chromosome 6 contained the largest number of BnXTH genes (3; 15.79%), followed by chromosomes 4, 5, and 14, which contained two BnXTH genes each. The remaining chromosomes only had one BnXTH member each. This study also found that the number of family genes mapped in the chromosomes had no correlation with chromosome length. To further understand the physical and chemical properties of the BnXTH-protein family, we evaluated each protein’s CDS length, amino acid (aa) number, molecular weight (Mw), isoelectric point (PI), grand average of hydropathicity (GRAVY), and aliphatic index. We also predicted the subcellular locations of these proteins (Table 1). These results showed that out of the 19 BnXTH protein sequences, the shortest was BnXTH10, which was encoded by 264 amino acids, while BnXTH3 was the longest, encoded by 395 amino acids. The Mw of the BnXTHs ranged from 30.812 (BnXTH10) to 40.228 kDa (BnXTH4), while the GRAVY ranged from −0.800 (BnXTH12) to −0.223 (BnXTH5). The aliphatic index of the proteins was between 59.44 (BnXTH12) and 73.39 (BnXTH7), and the PI ranged from 4.64 (BnXTH17) to 9.47 (BnXTH9). Moreover, subcellular localization prediction showed that BnXTH13/17 might have been located in extracellular regions, while BnXTH3/7/9/15/18/19 may have been located in the cell wall or cytoplasm, and the remaining BnXTH proteins may have played roles in the cell wall.
Table 1.
Molecular characterization of BnXTH genes.
| Name | Gene Name | Genome Location |
PI | Mw (kDa) | Peptide Residue (aa) | GRAVY | Aliphatic Index | CDS Length (bp) | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| BnXTH1 | Bni05G007034 | Chr5: 9195318–9197779 | 4.83 | 34.757 | 302 | −0.536 | 62.02 | 909 | Cell Wall |
| BnXTH2 | Bni14G018696 | Chr14: 919549–5922548 | 7.60 | 33.076 | 289 | −0.362 | 70.55 | 870 | Cell Wall |
| BnXTH3 | Bni14G018759 | Chr14: 6848323–6851051 | 8.59 | 33.88 | 395 | −0.406 | 69.73 | 1188 | Cell Wall/Cytoplasm |
| BnXTH4 | Bni07G010864 | Chr7: 13012796–13017451 | 8.57 | 40.228 | 358 | −0.259 | 71.42 | 1077 | Cell Wall |
| BnXTH5 | Bni09G013633 | Chr9: 15698329–15700698 | 7.16 | 34.447 | 307 | −0.223 | 71.47 | 924 | Cell Wall |
| BnXTH6 | Bni02G003149 | Chr2: 16541099–16543546 | 6.59 | 34.170 | 303 | −0.559 | 61.82 | 912 | Cell Wall |
| BnXTH7 | Bni04G005825 | Chr4: 11902190–11926110 | 8.77 | 32.334 | 286 | −0.385 | 73.39 | 861 | Cell Wall/Cytoplasm |
| BnXTH8 | Bni04G006001 | Chr4: 13734274–13737675 | 5.61 | 33.550 | 297 | −0.315 | 67.27 | 894 | Cell Wall |
| BnXTH9 | Bni05G007965 | Chr5: 18140865–18142082 | 9.47 | 32.9 | 292 | −0.404 | 65.24 | 879 | Cell Wall/Cytoplasm |
| BnXTH10 | Bni06G008340 | Chr6: 1295993–1298134 | 8.98 | 30.812 | 264 | −0.548 | 67.58 | 795 | Cell Wall |
| BnXTH11 | Bni06G008558 | Chr6: 4531957–4533699 | 6.46 | 35.716 | 304 | −0.509 | 73.39 | 915 | Cell Wall |
| BnXTH12 | Bni06G008923 | Chr6: 8233006–8239486 | 5.74 | 37.169 | 320 | −0.800 | 59.44 | 963 | Cell Wall |
| BnXTH13 | Bni08G012131 | Chr8: 12169484–12170900 | 9.11 | 33.171 | 291 | −0.355 | 73.02 | 876 | Extracell |
| BnXTH14 | Bni09G013009 | Chr9: 8910982–8915126 | 8.20 | 38.519 | 340 | −0.478 | 69.12 | 1023 | Cell Wall |
| BnXTH15 | Bni11G015460 | Chr11: 9722688–9725914 | 9.11 | 31.746 | 279 | −0.323 | 71.58 | 840 | Cell Wall/Cytoplasm |
| BnXTH16 | Bni12G017196 | Chr12: 15189643–15192043 | 9.31 | 34.254 | 299 | −0.396 | 60.40 | 900 | Cell Wall |
| BnXTH17 | Bni13G017592 | Chr13: 6362009–6373079 | 4.64 | 33.589 | 301 | −0.347 | 65.42 | 906 | Extracell |
| BnXTH18 | BniUnG019321 | Sca16: 619625–621073 | 8.69 | 31.917 | 286 | −0.338 | 69.58 | 861 | Cell Wall/Cytoplasm |
| BnXTH19 | BniUnG019322 | Sca16: 639410–654270 | 6.44 | 32.448 | 286 | −0.329 | 70.94 | 861 | Cell Wall/Cytoplasm |
PI, isoelectric point; Mw, molecular weight; aa, amino acid; GRAVY, grand average of hydropathicity.
2.2. Phylogenetic Analysis and Multiple Sequence Alignment of BnXTHs
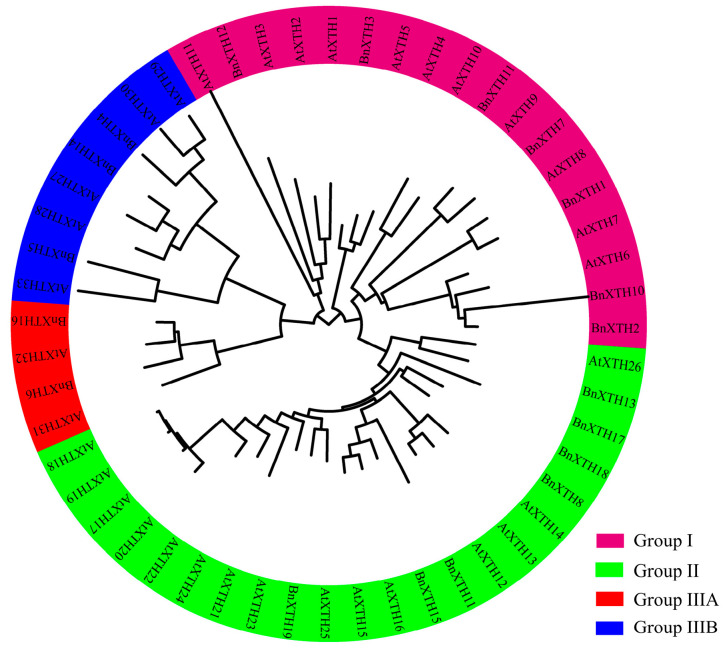
To better understand the evolutionary relationships between BnXTHs and determine their classifications, we used the sequences of the 19 ramie BnXTH genes and the AtXTH family protein sequences of A. thaliana to generate a phylogenetic tree (Figure 1) using the maximum likelihood (ML) method. We divided the BnXTH-family proteins identified in ramie into four subgroups based on a previous classification of the family: namely, Groups I, II, IIIA, and IIIB. The BnXTHs were mainly clustered in Groups I and II, which had 14 members. Among these members, BnXTH1/2/3/7/10/11/12 belonged to Group I, while BnXTH8/9/13/17/15/18/19 belonged to Group II. The remaining BnXTHs (BnXTH4/5/14/6/16) were included in Groups IIIA and IIIB.
Figure 1.
Phylogenetic tree showing the relationships among XTH proteins of Boehmeria nivea and those of Arabidopsis thaliana. The colored arcs show Groups I, II, IIIA, and IIIB. This phylogenetic tree was constructed via the neighbor-joining (NJ) method, using MEGA 6.06 with 1000 bootstrap replicates.
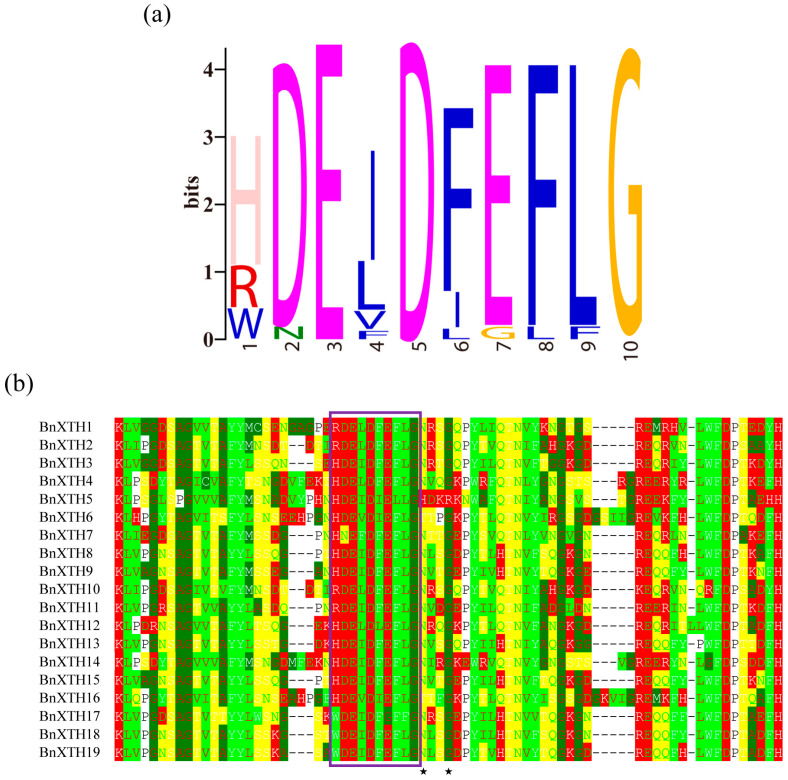
Multiple alignments of the 19 BnXTHs showed that the BnXTH proteins contained a highly conserved catalytic site (HDEIDFEFLG) (Figure 2), with a deviation of one or two amino acids in a few sequences (Figure 2a). Except for the BnXTHs in Group III (BnXTH5/6/16), the active catalytic regions (HDEIDFEFLG) (shown with rectangular purple frames in Figure 2b) of the BnXTHs were adjacent to the N-linked glycosylation site.
Figure 2.
Logo diagram (a) and multiple sequence alignments (b) of the active catalytic regions of BnXTHs. The purple rectangular frames represent active catalytic regions (HDEIDFEFLG), and the asterisks represent N-linked glycosylation sites.
2.3. Structural Analysis of the Conserved Motifs of BnXTHs
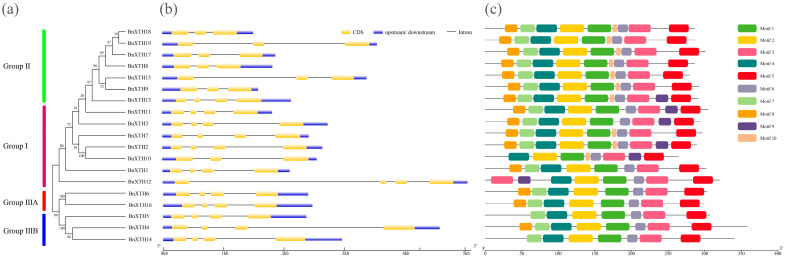
To analyze the gene structures and conserved motifs of BnXTHs, we constructed an evolutionary tree using 19 BnXTH proteins (Figure 3a), which were grouped into four subclasses. Structural analysis of the genomic DNA sequence showed that each BnXTH had two or three introns (Figure 3b) and that the members in Group I, except for BnXTH10, each contained three introns. The Group II members, except for BnXTH13, contained two introns each, while the Group IIIA and Group IIIB members had three introns each. Furthermore, MEME analysis showed that 10 conserved motifs were found in the 19 BnXTH protein sequences (Figure 3c). The amino acid sequence that encoded the protein sequences and the SeqLogo of the 10 conserved motifs are shown in Table S1. Motifs 1, 2, 3, 4, 5, and 6 were abundant in BnXTH proteins, among which motif 2 contained the characteristic active site (HDEIDFEFLG), suggesting that it is the specific motif for the enzymatic reaction of this family of proteins and present in all BnXTHs. We also found that motif 8 seemed unique to Group I, while motif 10 mainly existed in Groups I and II. Thus, these results also support the group-classification results of the phylogenetic tree above.
Figure 3.
Phylogenetic (a), gene-structure (b), and conserved motif (c) analyses of the BnXTH gene family.
2.4. Gene-Duplication Analysis of BnXTHs
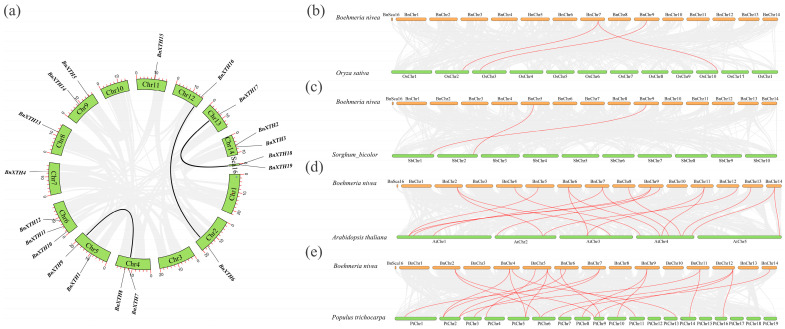
To determine the relationships between BnXTH members, we used the MCScanX method for a collinearity analysis. Six genes (BnXTH6–BnXTH16, BnXTH8–BnXTH9, and BnXTH17–BnXTH18) exhibited complex segmental duplication events (Figure 4a), implying that segmentally duplicated genes may have similar functions regulated via the same biological pathways in ramie. To further evaluate the evolution and development of the BnXTH family, we compared the collinearity of XTH genes between B. nivea and four other plants (O. sativa, S. bicolor, A. thaliana, and P. trichocarpa) (Figure 4b–e). These results showed that three pairs of homologous genes existed between B. nivea and O. sativa, while two pairs existed between B. nivea and S. bicolor. Moreover, 18 pairs of homologous genes were detected between B. nivea and A. thaliana, and 21 pairs were identified between B. nivea and P. trichocarpa (Table S2). These results show that ramie XTHs have less homology with monocotyledons than with dicotyledons, and thus, we speculate that these XTH genes may be involved in differentiation of dicotyledons.
Figure 4.
Chromosomal-level analyses of the BnXTH gene family in the ramie genome. (a) Synteny analysis of BnXTHs. The black lines indicate collinearity blocks and fragment-doubling events. (b–e) Synteny analysis of BnXTHs between Boehmeria nivea and Oryza sativa, Sorghum bicolor, Arabidopsis thaliana, and Populus trichocarpa.
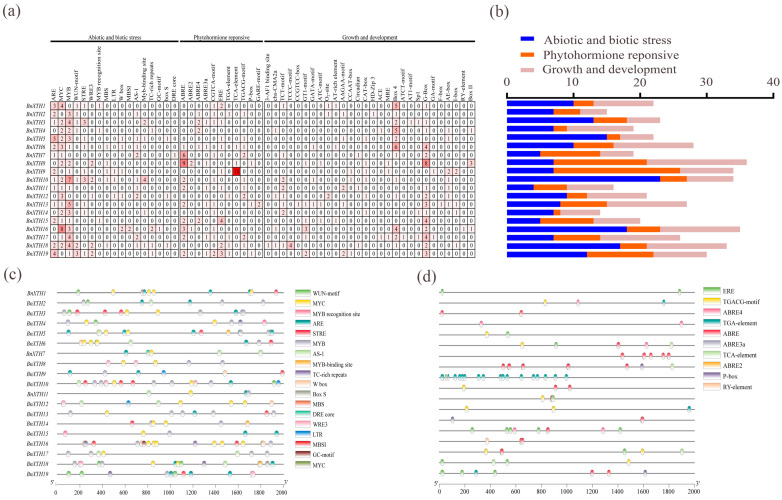
2.5. Ciselement Analysis of the BnXTHs
We used the PlantCARE service to analyze the cisregulatory elements of BnXTHs in the upstream sequences (~2000 bp) of their promoters that were associated with response to abiotic and biotic stress, phytohormone signaling, and plant growth and development. We predicted 56 related cisacting elements (Figure 5 and Table S3), among which 17 were involved in abiotic- and biotic-stress responses. ARE, MYC, STRE, and MBS were the most abundant among the 17 ciselements. Moreover, 11 cisregulatory elements were related to plant hormone responses, including abscisic acid (ABA), gibberellin (GA), salicylic acid (SA), methyl jasmonate (MeJA), and auxin responses. These elements included TGACG motifs, ABREs, TCA elements, TGA elements, and other related ciselements. There were 28 cisregulatory elements related to plant growth and development, among which light-responsive elements, including GT1 motifs, TCCC motifs, GATA motifs, Sp1, and other related elements, were the most abundant.
Figure 5.
Prediction of cisregulatory elements of BnXTHs. (a) The number of cisregulatory elements in BnXTH promoters. (b) The number of cisregulatory elements involved in the responses to abiotic and biotic stress, phytohormone signaling, and plant growth and development. (c) Type, quantity, and position of the ciselements of BnXTH promoters in response to biotic and abiotic stress. (d) Type, quantity, and position of the ciselements of the BnXTH promoters in response to phytohormone signaling.
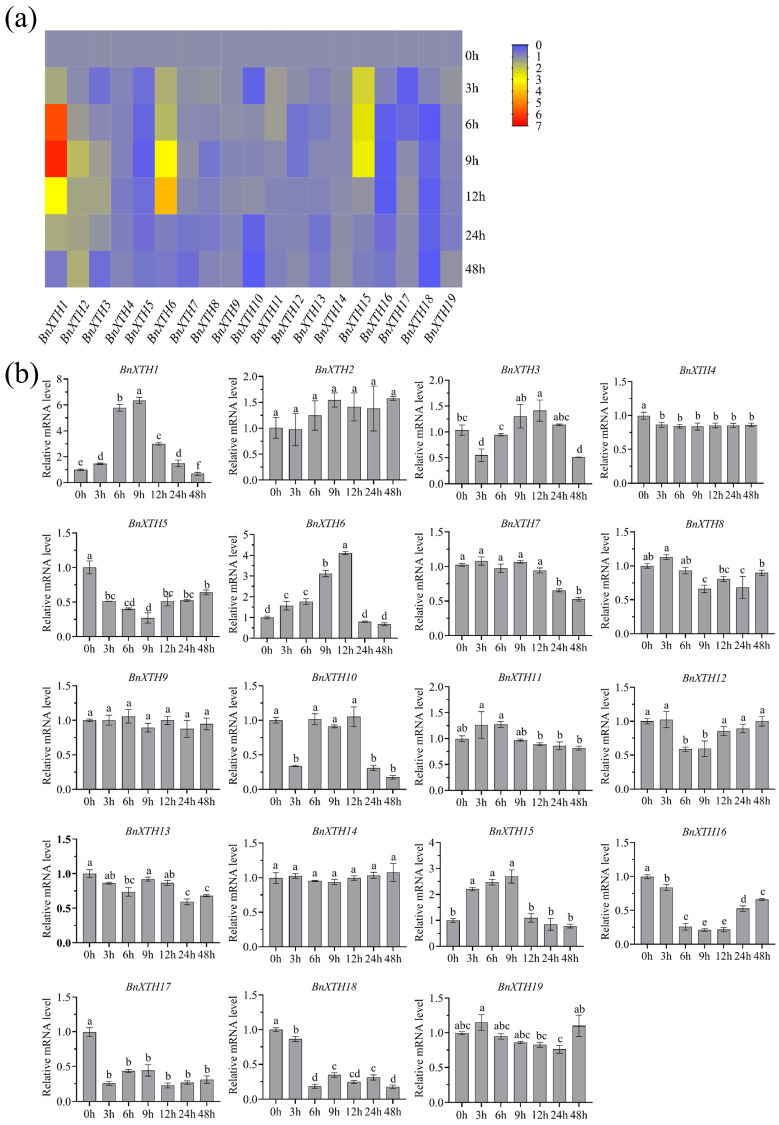
2.6. Expression Analysis of the BnXTH-Family Genes under Cd Treatment
To determine whether BnXTH-family genes are involved in Cd stress responses, we treated ramie seedlings with CdCl2 and sampled the roots at 0, 3, 6, 9, 12, 24, and 48 h after this treatment. The expression levels of the 19 BnXTH genes (Figure 6) were analyzed with RT-qPCR, and these results showed that expression of BnXTH1/6/15, especially BnXTH1, increased significantly under Cd treatment. The expressions of the BnXTH1 genes at 6 h and 9 h after the treatment were 5.78 and 6.34 times higher than that at 0 h, respectively. BnXTH3 was slightly upregulated and had the highest expression at 12 h: 41.22% higher than that at 0 h. Conversely, expression of BnXTH4/9/14/19 genes had no response to the Cd treatment, while the other BnXTH genes exhibited were downregulated under the Cd treatment. In general, the BnXTH-family genes responded differently to Cd stress.
Figure 6.
Expression analysis of the BnXTH genes under Cd stress. (a) Heat map of BnXTH-family genes expression level. (b) Expression of 19 BnXTH genes by qRT-PCR. Ramie seedlings were treated with Cd, and the root samples were collected at 0, 3, 6, 9, 12, 24, and 48 h after treatment. Boehmeria nivea actin (BnActin) was used as the internal control. Data are presented as means ± SD (n = 3). Statistical significance was determined using an LSD test. Different letters indicate significant differences at p ≤ 0.05.
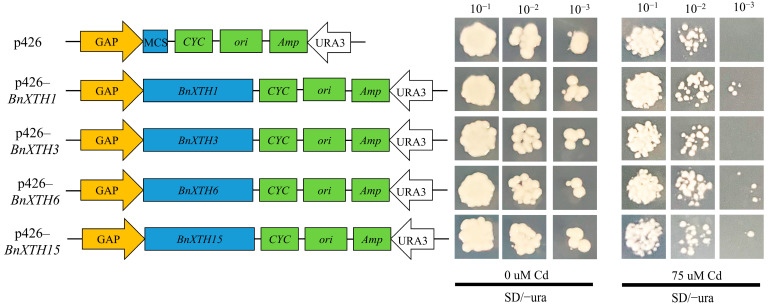
2.7. Functional Analysis of BnXTHs in Yeast
Since the expression levels of BnXTH1/3/6/15 genes were upregulated in response to Cd stress, we selected BnXTH1, BnXTH3, BnXTH6, and BnXTH15 as candidate genes and analyzed their roles in Cd tolerance. The full-length CDSs of BnXTH1, BnXTH3, BnXTH6, and BnXTH15 were cloned and ligated into yeast expression vector p426 GPD and introduced into Cd-sensitive yeast mutant Δyap1. The Cd-tolerance characteristics of transgenic yeast were then analyzed (Figure 7). We found no significant difference in the growth between the BnXTH1/3/6/15 transgenic yeast strains and the empty vector on the SD-URA medium without Cd. However, the growth of the yeast strains that contained the empty vector was significantly lower compared to that in the BnXTH1/6/15-containing yeast strains, particularly the BnXTH1-carrying yeast strains, on the medium that contained 75 μM of Cd. These results suggest that BnXTH1/6/15 genes play important roles in Cd tolerance. However, the growth of the transgenic yeast strain that contained BnXTH3 was not significant under Cd stress, indicating that BnXTH3 had no Cd tolerance. These results show that the BnXTH gene family is related to the Cd tolerance of ramie, and the three Cd-tolerant genes (BnXTH1/6/15) can be used for genetic improvement of Cd tolerance in ramie.
Figure 7.
Functional analysis of BnXTH genes through heterologous expression in yeast. (a) Schematic diagram of the constructed yeast vector. p426 represents the empty vector, while p426–BnXTH1, p426–BnXTH3, p426–BnXTH6, and p426–BnXTH15 represent the recombinant p426 vectors that contained the BnXTH1, BnXTH3, BnXTH6, and BnXTH15 genes, respectively. (b) Cd-tolerance assay of BnXTH genes in transgenic yeast (Δyap1). The yeast cells were treated with 0 μM and 75 μM of Cd in an SD-URA medium for 3 days at 30 °C.
3. Discussion
3.1. Evolutionary Characteristics of the BnXTH Gene Family in Ramie
A total of 19 BnXTH genes were identified in ramie, compared with the 33 family members in A. thaliana and 29 in O. sativa, suggesting that there are many fewer members of the BnXTH gene family in ramie. This may have been due to the incomplete assembly of the genome, resulting in a failure to identify other members of the BnXTH gene family or a loss of several BnXTH genes in the genome [32]. This result is consistent with those of a previous study that showed that Brassica oleracea contains fewer XTH family members than those in Brassica rapa [22]. These phylogenetic results showed that BnXTHs were relatively conserved at the DNA and protein levels and could be divided into four subclasses (Figure 1), similar to those of other plants [33,34]. There were 14 BnXTH members in Groups I and II, accounting for 73.68% of all members. When only BnXTH-family proteins were used to construct an evolutionary tree, members of Groups I and II clustered together, making it difficult to distinguish them (Figure 3a). This was similar to the findings of previous studies, which reported no significant difference between members of Groups I and II, suggesting that the two can be combined into a larger group: that is, Group I/II [35,36]. Furthermore, a multiple sequence alignment showed that the members of Groups I, II, and IIIB (except for BnXTH5) were located near the XET catalytic site (HDEIDFEFLG) and had typical N-glycosylation residues, while Group IIIA members lacked a consensual N-glycosylation site. The same phenomenon was reported in A. thaliana [37,38]. Additionally, variation of the conserved catalytic motif (HDEIDFEFLG) was observed in BnXTH proteins, consistently with previous studies involving plants, such as A. thaliana [18] and P. trichocarpa [14]. Variation in this motif may affect its enzyme catalytic activity, which necessitates further studies of the enzyme activity and other functions of these motif-variant genes.
3.2. Evolutionary Analysis of the BnXTH Gene Family
It is speculated that A. thaliana experienced genome-wide duplication thrice in the past 250 million years, while O. sativa experienced genome-wide duplication about 70 million years ago [39]. As early as 1970, scholars proposed that gene replication is the basic mechanism through which genes obtain new functions, enabling one gene to maintain its original function and another to change its function [40]. Gene duplications are also considered one of the main forces of evolution and expansion of gene families [41]. Previous studies of XTH-family genes have shown that gene-fragment-repetition events have occurred in tobacco [19], soybeans [30], and other plants. Gene-duplication phenomena also occurred in the BnXTH gene family of ramie, with fragment-repetition events of the three pairs of isogenous genes (BnXTH6–BnXTH16, BnXTH8–BnXTH9, and BnXTH17–BnXTH18) (Figure 4a). This indicates that gene replication plays a certain role in evolution of BnXTH genes but is not the main driver of the expansion of the BnXTH gene family. The sequence similarity between the BnXTH8–BnXTH9 and BnXTH17–BnXTH18 gene pairs was significantly high (Figure 3a), and these two gene pairs exhibited similar expression patterns under Cd stress (Figure 6). Although the BnXTH6–BnXTH16 gene pair also had high sequence similarity, its expression patterns were completely different under Cd stress. Similar observations were reported in B. rapa, where the BraA.XTH32.a–BraA.XTH32.c pair and the BraA.XTH14.a–BraA.XTH14.b pair showed similar expression patterns, while BraA.XTH23.a and BraA.XTH23.b showed different expression patterns [22]. This may have been because duplicate genes underwent nonfunctionalization, neofunctionalization, or subfunctionalization during evolution, resulting in similar or different gene-expression patterns [42]. There were few collinear gene pairs between ramie and monocotyledonous plants (O. sativa and S. bicolor) compared to the gene pairs detected between ramie and dicotyledonous plants (A. thaliana and P. trichocarpa). This indicates that numerous XTH gene variations and replications may occur in dicotyledonous plants during evolution. This evolution phenomenon was also found in the CAX-family genes of P. trichocarpa [43].
3.3. BnXTH Gene Response to Cd Stress
Abiotic stress can lead to transcript-level changes in XTH genes. For example, expression of AtXTH14 and AtXTH15 decreased significantly under Al stress, resulting in reduced XET activity and thus enhancing the Al tolerance of A. thaliana [26]. Additionally, expression of PeXTH was significantly upregulated in the roots and leaves of P. euphratica under Cd stress [44]. The current study found that the BnXTH gene family responded to Cd stress, under which BnXTH1, BnXTH3, BnXTH6, and BnXTH15 were upregulated, while BnXTH5, BnXTH16, BnXTH17, and BnXTH18 were significantly downregulated. Similar contrasting expression patterns of this gene family in response to abiotic stress have been reported in other plants. For example, expression of CsXTH1, CsXTH4, CsXTH6, and CsXTH7 was upregulated, while that of CsXTH3 was downregulated, in Camellia sinensis under fluorine stress [33]. Furthermore, we also found that BnXTH-family proteins have different subcellular localizations; for example, most BnXTH-family proteins were located in the cell wall, while BnXTH13 and BnXTH17 were located in the extracellular region. This may have been due to the expression-pattern diversity of the XTH gene family [18]. The differences in the subcellular localizations and expressions of proteins in the same family lead to differences in gene function [45], indicating that the different members of the BnXTH gene family exhibit different functions.
Expression of a gene often depends on the regulation of its upstream promoter [46]; thus, it is particularly important to analyze the upstream promoter sequences of a gene. The sequence analysis of the upstream promoter sequence of the BnXTH-family gene showed that the promoter of the BnXTH-family gene contained several cisacting elements, such as MYB, ABRE, AS-1, STRE, and MBS, that were involved in biotic and abiotic stress responses. MYB, MBS, and other ciselements are the binding sites of MYB transcription factors, which regulate defense responses by binding to the MBS elements on target genes [47]. Some hormone response elements, such as EREs and ABREs, are also involved in biotic and abiotic stress responses, whereby ABREs play important roles in response to abiotic stress [48,49]. These elements ensure that BnXTH genes are rapidly induced under stressful conditions.
Excessive absorption of heavy metals by plants causes serious toxicity to those plants [50]. Cell walls, especially hemicellulose, are reportedly the key Cd storage areas in plants [12]. In A. thaliana, phosphorus-deficiency tolerance significantly reduced hemicellulose content in the cell wall and alleviated Cd toxicity [51]. Heterologous expression of PeXTH in tobacco increased the root length and fresh weight of transgenic plants by enhancing their tolerance of Cd [44]. Similarly, in this study, the tolerance analysis of the transgenic yeast showed that heterologous expression of BnXTH1, BnXTH6, and BnXTH15 under Cd stress could enhance the Cd tolerance of yeast cells (Figure 7). These results suggest that the BnXTH gene family is involved in Cd stress responses.
4. Materials and Methods
4.1. Identification and Analysis of BnXTH-Family Genes in the Ramie Genome
There are two conserved domains in XTH proteins: the Glyco_hydro_16 domain (PF00722) and the XET_C domain (PF06955) [22]. We generated a hidden Markov model (HMM) file of these two conserved domains using the Pfam database (https://Pfam.xfam.org/ (accessed on 13 October 2021)) [52]. The ramie genome was analyzed using HMMER v3.3.2 (Howard Hughes Medical Institute, Washington, DC, USA) [53], which identified candidate genes. Redundant sequences were manually removed, and the ramie BnXTH genes were finally obtained.
The corresponding BnXTH gene locations were obtained from the annotation file of the ramie genome and visualized on chromosomes via MG2C v2.1 online software (http://mg2c.iask.in/mg2c_v2.1/ (accessed on 20 October 2021)) [54]. ExPASy software (https://web.expasy.org/protparam/ (accessed on 20 October 2021)) [55] was used to predict the physical and chemical properties of the selected BnXTH-family members. These properties included each gene’s PI, Mw, GRAVY, and aliphatic index. Furthermore, Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/ (accessed on 20 September 2022)) [56] was used to predict the subcellular localization of the BnXTH-family proteins.
4.2. Sequence Alignment and Phylogenetic Analyses
The XTH protein sequence of A. thaliana was searched for in the NCBI protein database (http://www.ncbi.nlm.nih.gov/protein/ (accessed on 21 September 2022)), and coding sequences (CDSs) of BnXTH-family genes were used to generate BnXTH protein sequences. A neighbor-joining (NJ) phylogenetic tree based on full-length sequences of AtXTHs and BnXTHs was constructed via MEGA 6.0 [57] using 1000 bootstrap replicates. A multiple sequence alignment of all BnXTH proteins was then conducted with Clustal (version:X 2.0, University College Dublin, Dublin, Ireland) [58].
4.3. Gene Structure and Motif Composition Analysis
The genomic sequences and CDSs of the BnXTH-family genes were extracted from the ramie genome, and structures of the BnXTH-family genes were constructed using GSDS 2.0 online software (http://gsds.gao-lab.org/ (accessed on 22 September 2022)) [59]. MEME online software (https://meme-suite.org/meme/tools/meme (accessed on 22 September 2022)) [60] was then used to analyze the conserved motifs of the BnXTH proteins, after which the conserved sites were set at 6–50 and the conserved-motif number parameter was set to 10. These structures were then visualized using TBtools (version: v1.098774, South China Agricultural University, Guangzhou, China) [61].
4.4. Analysis of Gene Duplication Events and Collinearity of the BnXTHs
We used a multiple-collinearity scanning toolkit (MCScanX) and TBtools software plug-ins to analyze gene duplication events of BnXTH in the ramie genome and the collinearity of the XTH genes between Boehmeria nivea and O. sativa, Sorghum bicolor, A. thaliana, and Populus trichocarpa. A collinearity graph was then generated using TBtools.
4.5. Analysis of the BnXTH Gene Promoter
The 2000 bp sequence upstream of the BnXTH gene was predicted using the PlantCARE database [62], and its cisregulatory elements were analyzed. These cisregulatory elements were then counted and classified according to the functional effects on the promoter. Thereafter, TBtools was used to visualize results and generate heat maps.
4.6. RT-qPCR Analysis of BnXTH Expression under Cd Stress
The “Xiangzhu No. 3” ramie material used in this study was provided by our research group at Hunan Agricultural University. Terminal buds were cultured in a half-strength Hoagland nutrient solution for 3 weeks, after which the roots of ramie seedlings with the same growth rate were collected. After 1 week of culturing in the half-strength Hoagland nutrient solution, the ramie seedlings were treated with 50 μM of CdCl2, and the root samples were collected at 0, 3, 6, 9, 12, 24, and 48 h after the treatment. The samples were frozen in liquid nitrogen and preserved at −80 °C. The growth conditions of the ramie were as follows: 14 h day/10 h night photoperiod, day and night temperature of 26/24 °C, relative humidity of 60%, and light intensity of 20,000 lux.
Total RNA was extracted according to the instructions of the plant RNA extraction kit (Vazyme, Nanjing, China). The extracted RNA was reverse-transcribed into cDNA using a reverse transcription kit (Vazyme, Nanjing, China) with primers that were designed using Premier 5.0 and synthesized via Sangon Biotech (Shanghai, China). The primer sequences are presented in Table S4. The synthesized cDNA was used as a template for qPCR analysis on the Bio-Rad CFX96 instrument (Bio-Rad, Hercules, CA, USA) using the AceQ® Universal SYBR qPCR kit (Vazyme, Nanjing, China) and the BnActin gene as the control. Each sample was quantified in triplicate, and the relative quantitation of each gene was conducted using the 2−ΔΔCt method [21].
4.7. Functional Analysis of Cd-Induced Expression of BnXTHs in Yeast
The BnXTH1, BnXTH3, BnXTH6, and BnXTH15 genes, which were shown to respond to Cd stress, were introduced into the yeast cells for functional analysis. Briefly, the ramie cDNA was used as a template for PCR, using the primers shown in Table S4. The PCR conditions and procedures were as described by Jiang et al. [14]. Thereafter, the PCR products were cloned into the pEASY-blunt vector (TransGen Biotech Company, Beijing, China) and sequenced at Sangon Biotech (Shanghai, China), followed by subsequent cloning into the p426 GPD vector at the SmaI/SalI sites. The p426–BnXTH1/3/6/15 recombinant vector and the empty p426 vector were introduced into a mutant Saccharomyces cerevisiae yeast strain, Δyap1, which lacked transcriptional regulatory protein YAP-1 for Cd tolerance. For the Cd-tolerance analysis, the transgenic yeast cells were inoculated into a liquid synthetic dropout medium without uracil (SD-URA) and incubated at 30 °C on a shaker at 200 rpm until OD600 = 1.0 was reached. The precipitate was collected via centrifugation at 10,000 rpm for 1min, followed by suspension in ddH2O. The suspension was diluted to 10−1, 10−2, and 10−3 times its original state, and 2 µL of the 10−2 dilution was cultured as droplets on solid SD-URA media that contained 0 and 75 μM of CdCl2. The plates were incubated at 30 °C for 3 days, and the cultures were observed and photographed.
4.8. Statistical Analysis
All data are presented as means ± SD. The data were analyzed with one-way analysis of variance (ANOVA), followed by an LSD post hoc test using SAS 9.4 (SAS Institute, Cary, NC, USA) at a p ≤ 0.05 significance level.
5. Conclusions
This study identified 19 BnXTHs and evaluated their evolution, phylogeny, chromosomal locations, gene duplications, and cisregulatory elements. RT-qPCR results showed that most BnXTH genes responded to Cd stress. Many cisregulatory elements in BnXTH gene promoters were related to abiotic and biotic stress responses. In summary, under Cd stress, transcription factors that are located upstream of BnXTHs are triggered to bind to the cisregulatory elements that are upstream of BnXTHs and regulate expression of BnXTH genes to enhance the Cd tolerance of ramie. Additionally, functional analysis of heterologous expression in yeast showed that BnXTH1, BnXTH6, and BnXTH15 may be involved in Cd tolerance. However, further validation, using transgenic methods, in ramie or other plants is needed. These results improved our understanding of the BnXTH gene family and laid a foundation for exploration of the function of BnXTH genes in Cd tolerance and enrichment in ramie.
Acknowledgments
We thank two reviewers for their helpful suggestions for this manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232416104/s1.
Author Contributions
Y.-S.M.: methodology, investigation, formal analysis, writing—original draft preparation; H.-D.J., L.Z., X.-Y.L., X.-C.L., Y.-Y.T., Y.Z., P.-L.H. and H.-C.X.: formal analysis and visualization; Y.-C.J.: conceptualization, supervision, and project administration. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant No. 31872877 and grant No. 32071940), the National Key R&D Program of China (grant No. 2019YFD1002205-3), and the Research and Development Projects in the Key Area of Hunan Province (grant No. 2019NK206102 and grant No. 2020NK2028).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Pinto A.P., Mota A.M., de Varennes A., Pinto F.C. Influence of organic matter on the uptake of cadmium, zinc, copper and iron by sorghum plants. Sci. Total Environ. 2004;326:239–247. doi: 10.1016/j.scitotenv.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Liu M., Liu X., Kang J., Korpelainen H., Li C. Are males and females of Populus cathayana differentially sensitive to Cd stress? J. Hazard. Mater. 2020;393:122411. doi: 10.1016/j.jhazmat.2020.122411. [DOI] [PubMed] [Google Scholar]
- 3.Palansooriya K.N., Shaheen S.M., Chen S.S., Tsang D.C.W., Hashimoto Y., Hou D., Bolan N.S., Rinklebe J., Ok Y.S. Soil amendments for immobilization of potentially toxic elements in contaminated soils: A critical review. Environ. Int. 2020;134:105046. doi: 10.1016/j.envint.2019.105046. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J., Yang W., Zhang S., Yang T., Liu Q., Dong J., Fu H., Mao X., Liu B. Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice. 2018;11:1–15. doi: 10.1186/s12284-018-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y., Cheng H., Tao S. The Challenges and Solutions for Cadmium-contaminated Rice in China: A Critical Review. Environ. Int. 2016;92–93:515–532. doi: 10.1016/j.envint.2016.04.042. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Y.X., Du W.X., Fang X.Z., Zhang L.L., Jin C.W. Knockdown of BTS may provide a new strategy to improve cadmium-phytoremediation efficiency by improving iron status in plants. J. Hazard. Mater. 2020;384:121473. doi: 10.1016/j.jhazmat.2019.121473. [DOI] [PubMed] [Google Scholar]
- 7.Qin G., Niu Z., Yu J., Li Z., Ma J., Xiang P. Soil heavy metal pollution and food safety in China: Effects, sources and removing technology. Chemosphere. 2021;267:129205. doi: 10.1016/j.chemosphere.2020.129205. [DOI] [PubMed] [Google Scholar]
- 8.Ali A., Guo D., Li Y., Shaheen S.M., Wahid F., Antoniadis V., Abdelrahman H., Al-Solaimani S.G., Li R., Tsang D.C., et al. Streptomyces pactum addition to contaminated mining soils improved soil quality and enhanced metals phytoextraction by wheat in a green remediation trial. Chemosphere. 2021;273:129692. doi: 10.1016/j.chemosphere.2021.129692. [DOI] [PubMed] [Google Scholar]
- 9.Kong F., Chen Y., Huang L., Yang Z., Zhu K. Human health risk visualization of potentially toxic elements in farmland soil: A combined method of source and probability. Ecotoxicol. Environ. Saf. 2021;211:111922. doi: 10.1016/j.ecoenv.2021.111922. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham S.D., Berti W.R., Huang J.W. Phytoremediation of contaminated soils. Trends Biotechnol. 1995;13:393–397. doi: 10.1016/S0167-7799(00)88987-8. [DOI] [Google Scholar]
- 11.Wang X., Liu Y., Zeng G., Chai L., Song X., Min Z., Xiao X. Subcellular distribution and chemical forms of cadmium in Bechmeria nivea (L.) Gaud. Environ. Exp. Bot. 2008;62:389–395. doi: 10.1016/j.envexpbot.2007.10.014. [DOI] [Google Scholar]
- 12.Ma Y., Jie H., Tang Y., Xing H., Jie Y. The Role of Hemicellulose in Cadmium Tolerance in Ramie (Boehmeria nivea (L.) Gaud.) Plants. 2022;11:1941. doi: 10.3390/plants11151941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosgrove D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Y., Li Y., Lu C., Tang Y., Jiang X., Gai Y. Isolation and characterization of Populus xyloglucan endotransglycosylase/hydrolase (XTH) involved in osmotic stress responses. Int. J. Biol. Macromol. 2020;155:1277–1287. doi: 10.1016/j.ijbiomac.2019.11.099. [DOI] [PubMed] [Google Scholar]
- 15.Chabi M., Goulas E., Leclercq C.C., de Waele I., Rihouey C., Cenci U., Day A., Blervacq A.-S., Neutelings G., Duponchel L., et al. A Cell Wall Proteome and Targeted Cell Wall Analyses Provide Novel Information on Hemicellulose Metabolism in Flax. Mol. Cell. Proteom. 2017;16:1634–1651. doi: 10.1074/mcp.M116.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Sandt V.S.T., Suslov D., Verbelen J.-P., Vissenberg K. Xyloglucan Endotransglucosylase Activity Loosens a Plant Cell Wall. Ann. Bot. 2007;100:1467–1473. doi: 10.1093/aob/mcm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X.F., Shi Y.Z., Lei G.J., Fry S.C., Zhang B.C., Zhou Y.H., Braam J., Jiang T., Xu X.Y., Mao C.Z., et al. XTH31, Encoding an in Vitro XEH/XET-Active Enzyme, Regulates Aluminum Sensitivity by Modulating in Vivo XET Action, Cell Wall Xyloglucan Content, and Aluminum Binding Capacity in Arabidopsis. Plant Cell. 2012;24:4731–4747. doi: 10.1105/tpc.112.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokoyama R., Nishitani K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001;42:1025–1033. doi: 10.1093/pcp/pce154. [DOI] [PubMed] [Google Scholar]
- 19.Wang M., Xu Z., Ding A., Kong Y. Genome-Wide Identification and Expression Profiling Analysis of the Xyloglucan Endotransglucosylase/Hydrolase Gene Family in Tobacco (Nicotiana tabacum L.) Genes. 2018;9:273. doi: 10.3390/genes9060273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama R., Rose J.K., Nishitani K. A Surprising Diversity and Abundance of Xyloglucan Endotransglucosylase/Hydrolases in Rice. Classification and Expression Analysis. Plant Physiol. 2004;134:1088–1099. doi: 10.1104/pp.103.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xuan Y., Zhou Z.S., Li H.B., Yang Z.M. Identification of a group of XTHs genes responding to heavy metal mercury, salinity and drought stresses in Medicago truncatula. Ecotoxicol. Environ. Saf. 2016;132:153–163. doi: 10.1016/j.ecoenv.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Wu D., Liu A., Qu X., Liang J., Song M. Genome-wide identification, and phylogenetic and expression profiling analyses of XTH gene families in Brassica rapa L. and Brassica oleracea L. BMC Genom. 2020;21:782. doi: 10.1186/s12864-020-07153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisler-Lee J., Geisler M., Coutinho P.M., Segerman B., Nishikubo N., Takahashi J., Aspeborg H., Djerbi S., Master E., Andersson-Gunnerås S., et al. Poplar Carbohydrate-Active Enzymes. Gene Identification and Expression Analyses. Plant Physiol. 2006;140:946–962. doi: 10.1104/pp.105.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nawaz M.A., Rehman H.M., Imtiaz M., Baloch F.S., Lee J.D., Yang S.H., Lee S.I., Chung G. Systems Identification and Characterization of Cell Wall Reassembly and Degradation Related Genes in Glycine max (L.) Merill, a Bioenergy Legume. Sci. Rep. 2017;7:10862. doi: 10.1038/s41598-017-11495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z., Zhang R., Zhou Z. The XTH Gene Family in Schima superba: Genome-Wide Identification, Expression Profiles, and Functional Interaction Network Analysis. Front. Plant Sci. 2022;13:911761. doi: 10.3389/fpls.2022.911761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J.L., Zhu X.F., Peng Y.X., Zheng C., Li G.X., Liu Y., Shi Y.Z., Zheng S.J. Cell Wall Hemicellulose Contributes Significantly to Aluminum Adsorption and Root Growth in Arabidopsis. Plant Physiol. 2011;155:1885–1892. doi: 10.1104/pp.111.172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho S.K., Kim J.E., Park J.A., Eom T.J., Kim W.T. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 2006;580:3136–3144. doi: 10.1016/j.febslet.2006.04.062. [DOI] [PubMed] [Google Scholar]
- 28.Zhang M., Ma Y., Horst W.J., Yang Z.-B. Spatial–temporal analysis of polyethylene glycol-reduced aluminium accumulation and xyloglucan endotransglucosylase action in root tips of common bean (Phaseolus vulgaris) Ann. Bot. 2016;118:1–9. doi: 10.1093/aob/mcw062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi J.Y., Seo Y.S., Kim S.J., Kim W.T., Shin J.S. Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. Dotaerang) Plant Cell Rep. 2011;30:867–877. doi: 10.1007/s00299-010-0989-3. [DOI] [PubMed] [Google Scholar]
- 30.Song L., Valliyodan B., Prince S., Wan J., Nguyen H.T. Characterization of the XTH Gene Family: New Insight to the Roles in Soybean Flooding Tolerance. Int. J. Mol. Sci. 2018;19:2705. doi: 10.3390/ijms19092705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi D., Johnson K.L., Hao P., Tuong T., Erban A., Sampathkumar A., Bacic A., Iii D.P.L., Kopka J., Kuroha T., et al. Cell wall modification by the xyloglucan endotransglucosylase/hydrolase XTH19 influences freezing tolerance after cold and sub-zero acclimation. Plant Cell Environ. 2021;44:915–930. doi: 10.1111/pce.13953. [DOI] [PubMed] [Google Scholar]
- 32.Fan P., Nie L., Jiang P., Feng J., Lv S., Chen X., Bao H., Guo J., Tai F., Wang J., et al. Transcriptome Analysis of Salicornia europaea under Saline Conditions Revealed the Adaptive Primary Metabolic Pathways as Early Events to Facilitate Salt Adaptation. PLoS ONE. 2013;8:e80595. doi: 10.1371/journal.pone.0080595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Z., Cui C., Xing A., Xu X., Sun Y., Tian Z., Li X., Zhu J., Wang G., Wang Y. Identification and response analysis of xyloglucan endotransglycosylase/hydrolases (XTH) family to fluoride and aluminum treatment in Camellia sinensis. BMC Genom. 2021;22:761. doi: 10.1186/s12864-021-08056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiika R.J., Wei J., Cui G., Ma Y., Yang H., Duan H. Transcriptome-wide characterization and functional analysis of Xyloglucan endo-transglycosylase/hydrolase (XTH) gene family of Salicornia europaea L. under salinity and drought stress. BMC Plant Biol. 2021;21:1–15. doi: 10.1186/s12870-021-03269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumann M.J., Eklof J.M., Michel G., Kallas A.M., Teeri T.T., Czjzek M., Brumer H., III. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell. 2007;19:1947–1963. doi: 10.1105/tpc.107.051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michailidis G., Argiriou A., Darzentas N., Tsaftaris A. Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes from allotetraploid (Gossypium hirsutum) cotton and its diploid progenitors expressed during fiber elongation. J. Plant Physiol. 2009;166:403–416. doi: 10.1016/j.jplph.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Eklöf J.M., Brumer H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010;153:456–466. doi: 10.1104/pp.110.156844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaewthai N., Gendre D., Eklöf J.M., Ibatullin F.M., Ezcurra I., Bhalerao R.P., Brumer H. Group III-A XTH genes of Arabidopsis encode predominant xyloglucan endohydrolases that are dispensable for normal growth. Plant Physiol. 2013;161:440–454. doi: 10.1104/pp.112.207308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirakawa Y. Evolution of meristem zonation by CLE gene duplication in land plants. Nat. Plants. 2022;8:735–740. doi: 10.1038/s41477-022-01199-7. [DOI] [PubMed] [Google Scholar]
- 40.Panchy N., Lehti-Shiu M., Shiu S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou W., Chen J., Meng L., Chen D., He H., Ye G. The Rice Cation/H+ Exchanger Family Involved in Cd Tolerance and Transport. Int. J. Mol. Sci. 2021;22:8186. doi: 10.3390/ijms22158186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch M., Conery J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 43.He F., Shi Y.-J., Li J.-L., Lin T.-T., Zhao K.-J., Chen L.-H., Mi J.-X., Zhang F., Zhong Y., Lu M.-M., et al. Genome-wide analysis and expression profiling of Cation/H+ exchanger (CAX) family genes reveal likely functions in cadmium stress responses in poplar. Int. J. Biol. Macromol. 2022;204:76–88. doi: 10.1016/j.ijbiomac.2022.01.202. [DOI] [PubMed] [Google Scholar]
- 44.Han Y., Sa G., Sun J., Shen Z., Zhao R., Ding M., Deng S., Lu Y., Zhang Y., Shen X., et al. Overexpression of Populus euphratica xyloglucan endotransglucosylase/hydrolase gene confers enhanced cadmium tolerance by the restriction of root cadmium uptake in transgenic tobacco. Environ. Exp. Bot. 2014;100:74–83. doi: 10.1016/j.envexpbot.2013.12.021. [DOI] [Google Scholar]
- 45.Wang H.-L., Zhang Y., Wang T., Yang Q., Yang Y., Li Z., Li B., Wen X., Li W., Yin W., et al. An Alternative Splicing Variant of PtRD26 Delays Leaf Senescence by Regulating Multiple NAC Transcription Factors in Populus. Plant Cell. 2021;33:1594–1614. doi: 10.1093/plcell/koab046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Y., Avramova Z., Fromm M. Two Distinct Roles of ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1) at Promoters and within Transcribed Regions of ATX1-Regulated Genes. Plant Cell. 2011;23:350–363. doi: 10.1105/tpc.110.080150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shan T., Rong W., Xu H., Du L., Liu X., Zhang Z. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci. Rep. 2016;6:28777. doi: 10.1038/srep28777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He F., Niu M.-X., Feng C.-H., Li H.-G., Su Y., Su W.-L., Pang H., Yang Y., Yu X., Wang H.-L., et al. PeSTZ1 confers salt stress tolerance by scavenging the accumulation of ROS through regulating the expression of PeZAT12 and PeAPX2 in Populus. Tree Physiol. 2020;40:1292–1311. doi: 10.1093/treephys/tpaa050. [DOI] [PubMed] [Google Scholar]
- 49.Yu Y., Guo D.-L., Li G., Yang Y., Zhang G., Li S., Liang Z. The grapevine R2R3-type MYB transcription factor VdMYB1 positively regulates defense responses by activating the stilbene synthase gene 2 (VdSTS2) BMC Plant Biol. 2019;19:478. doi: 10.1186/s12870-019-1993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z., Yang F., Liu J.-L., Wu H.-T., Yang H., Shi Y., Liu J., Zhang Y.-F., Luo Y.-R., Chen K.-M. Heavy metal transporters: Functional mechanisms, regulation, and application in phytoremediation. Sci. Total. Environ. 2021;809:151099. doi: 10.1016/j.scitotenv.2021.151099. [DOI] [PubMed] [Google Scholar]
- 51.Zhu X.F., Lei G.J., Jiang T., Liu Y., Li G.X., Zheng S.J. Cell wall polysaccharides are involved in P-deficiency-induced Cd exclusion in Arabidopsis thaliana. Planta. 2012;236:989–997. doi: 10.1007/s00425-012-1652-8. [DOI] [PubMed] [Google Scholar]
- 52.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C., Paladin L., Raj S., Richardson L.J., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potter S.C., Luciani A., Eddy S.R., Park Y., López R., Finn R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chao J., Kong Y., Wang Q., Sun Y., Gong D., Lv J., Liu G. Mapgene2chrom, a tool to draw gene physical map based on perl and svg languages. Hereditas. 2015;37:91–97. doi: 10.16288/j.yczz.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 55.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou K.C., Shen H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE. 2010;5:e11335. doi: 10.1371/journal.pone.0011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 59.Hu B., Jin J., Guo A.-Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:w202–w208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen C.J., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y.H., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 62.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.