Abstract
The prevalence of Pneumocystis carinii pneumonia (PCP) in humans caused by more than a single genotype has been reported to range from 10 to 67%, depending on the method used for detection (3, 19). Most coinfections were associated with primary rather than recurrent disease. To better understand the factors influencing the development of coinfections, the time periods between inoculations and the genotype of the infecting organisms were evaluated in the chronically immunosuppressed-inoculated rat model of PCP. P. carinii f. sp. carinii infecting rats differentiated by karyotypic profiles exhibit the same low level of genetic divergence manifested by organisms infecting humans. P. carinii f. sp. carinii karyotype forms 1, 2, and 6 were inoculated into immunosuppressed rats, individually and in dual combinations, spaced 0, 10, and 20 days apart. Infections comprised of both organism forms resulted from admixtures inoculated at the same time. In contrast, coinfections did not develop in most rats, where a 10- or 20-day gap was inserted between inoculations; only the first organism form inoculated was detected by pulsed-field gel electrophoresis in the resultant infection. Organism burdens were reduced with combinations of forms 1 and 2 spaced 20 days apart but not in rats inoculated with forms 1 and 6. A role for the host response in the elimination of the second population and in reduction of the organism burden was suggested by the lack of direct killing of forms 1 and 2 in an in vitro ATP assay, by reduction of the burden by autoclaved organisms, and by the specific reactions of forms 1 and 2 but not forms 1 and 6. These studies showed that the time between inoculations was critical in establishing coinfections and P. carinii f. sp. carinii karyotype profiles were associated with differences in biological responses. This model provides a useful method for the study of P. carinii coinfections and their transmission in humans.
Organisms termed Pneumocystis carinii were placed in the Fungal kingdom more than 10 years ago on the basis of gene sequence comparisons (14, 29), yet much of their basic biological processes remains poorly understood, due in large part to their poor growth outside the mammalian lung environment (11). Recent genetic comparisons (32), as well as animal inoculation studies (16) and antigenic analyses (2, 15), provide strong evidence that “P. carinii” is actually a family of organisms that, although related, exhibit species specificity among its mammalian hosts.
Comparisons at five genetic loci suggest three levels of genetic divergence are present among P. carinii organism populations (30). The highest level of divergence, class III, was observed among Pneumocystis populations isolated from different mammalian hosts and ranged from 5 to 50% divergence in nucleotide sequence at these selected loci, with the internal transcribed sequence (ITS) regions being most divergent. Class II divergence ranged from 4 to 7% for all genes, with a 20 to 30% difference at the ITS regions. Organisms demonstrating class II divergence have been found to infect the same mammalian host but, only two mammalian hosts, the ferret and the rat, have been shown to harbor such divergent populations to date (12). Class I sequences differed by 0 to 0.8% in the four gene sequences and by 2 to 4% in the ITS regions. This low level of divergence has been described for all P. carinii f. sp. hominis populations found in human beings and among several subpopulations of P. carinii f. sp. carinii infecting rats ((12, 30, 32); see also below).
The genetic divergence observed for Pneumocystis organisms within each of the class III and class II levels is equivalent to those differences seen among bona fide species of other microbes (29). Based on these genotypic differences and phenotypic differences (2, 15, 16), the community of Pneumocystis investigators voted to move toward a standardized system of nomenclature recognizing these differences (28). The nomenclature follows the recommendations of the Botanical Code for special physiological forms of fungi and uses a tripartite naming convention (21). Thus, for example, the term P. carinii f. sp. hominis specifies organisms from human beings, the term P. carinii f. sp. mustelae specifies organisms from ferrets, and the term P. carinii f. sp. mus specifies organisms from mice. This convention is followed in the present publication.
P. carinii f. sp. carinii and P. carinii f. sp. ratti are both found in rats. Nevertheless, the two special forms exhibit class II genetic divergence and markedly different serologic responses to anti-P. carinii monoclonal antibodies and polyclonal sera (12, 31), suggesting they may be distinct species (9). Previous karyotyping studies found that these two populations could either coexist within the same rat lung or exist as apparent individual infections, although infection by P. carinii f. sp. ratti alone was far less frequent than infection by P. carinii f. sp. carinii (8, 9).
Subpopulations of P. carinii f. sp. carinii, defined by karyotypic profiles on pulsed-field gels, exhibit a low-level genetic divergence (11, 23) similar to that seen in P. carinii f. sp. hominis isolates (24, 25, 30). In the present study, we used three of these subpopulations to investigate the influence of genetic identity on the establishment of coinfections in immunosuppressed rats. Surveys of several commercial rat colonies identified eight distinct populations of P. carinii f. sp. carinii based on karyotype profiles (8, 9, 11). We have recently identified two additional karyotype forms, forms 9 and 10, from other commercial rat colonies (unpublished data). Forms 1, 2, and 6 were chosen for these experiments because of their distinct karyotypic profiles, facilitating the recognition of coinfections on pulsed-field gels.
MATERIALS AND METHODS
Induction of experimental P. carinii f. sp. carinii infections.
Viral-antibody-negative CD male rats were received in filtered containers at 140 to 160 g from Charles River Breeding Laboratories, Hollister, Calif. Upon receipt, the rats were immediately placed under barrier isolation consisting of sterile polycarbonate shoebox cages with sterile bedding; the cages had been fitted with microisolator tops placed on stainless steel racks within a horizontal flow hood. To reduce the occurrence of infection with other microbial pathogens, the rats received irradiated food (TekLab Irradiated Chow; Harlan Industries, Indianapolis, Ind.) and autoclaved water, into which a sterile solution of cephadrine (Velosef; E. R. Squibb & Sons, Inc., Princeton, N.J.) was injected to achieve a final concentration of 0.200 mg/ml, as described in detail elsewhere (7, 8). After 7 days of acclimation, the rats were started on a regimen of immunosuppression consisting of weekly injections of methylprednisolone acetate (Depo Medrol; The Upjohn Co., Kalamazoo, Mich.) at 4 mg/week. After two injections of methylprednisolone, the rats received intratracheal inoculations of P. carinii organisms or sham inoculations as described below. Immunosuppression was continued throughout the course of the experiment. The time course of the experiment included the initial 2-week immunosuppression, the first inoculation, and an additional 8 weeks of immunosuppression with the second inoculations administered 10 and 20 days after the initial inoculation during this period.
Preparation of inocula.
Cryopreserved and enumerated P. carinii f. sp. carinii preparations were used in these studies. P. carinii f. sp. carinii populations were isolated from individual rats and frozen in RPMI 1640 supplemented with 10% fetal bovine serum (Sigma Chemical Co., St. Louis, Mo.) and 7.5% dimethyl sulfoxide (tissue culture grade; Sigma) as previously described (6). Populations were characterized by electrophoretic karyotyping (8, 9) and sequencing of the mitochondrial large-subunit rRNA (27) and the nuclear small-subunit rRNA (29). Form 1 originated from a Sprague-Dawley rat (2669, O'Fallon Colony; Sasco, Inc., St. Louis, Mo.) which had been cryopreserved for 5 months prior to usage. Form 2 originated from a Sprague-Dawley rat (T5-967; Hilltop Laboratories, Scottsdale, Pa.) and was cryopreserved 17 months prior to usage. Form 6 originated from a Sprague-Dawley rat (T5-999; Charles River Breeding Laboratories [room 064]) and was cryopreserved for 17 months prior to usage. Cryovials containing the populations were removed from liquid nitrogen, rapidly thawed in a 37°C water bath, centrifuged, and reconstituted in 1.0 ml of prewarmed RPMI 1640 tissue culture medium per vial. A 5-μl sample was removed for evaluation of viability by the calcein AM-ethidium homodimer dual fluorescent staining assay (22). Organism preparations were diluted in RPMI 1640 supplemented with 10% fetal bovine serum to a parasite density required for the experimental groups (see below). The fetal bovine serum served to help stabilize the thawed organisms. It did not participate in the reduction of organism burden observed for forms 1 and 2 after a 20-day gap or in the elimination of the second inoculated population since it was common to all inoculations and such reductions did not occur in all cases (e.g., forms 1 and 6) nor in the elimination of second populations (i.e., form 1 was inoculated 10 days after form 2). The target number of organisms was delivered in a volume of 0.2 ml. The numbers of organisms were adjusted to compensate for the loss of viability, as determined by the dual fluorescent assay. The percentage of organisms exhibiting bright green fluorescence without nuclear staining (live) varied between 70 and 90% in the preparations used. After preparation, the inocula were stored on ice prior to inoculations. Inoculations were administered after light anesthesia with halothane using a feeding cannula as described by Boylan and Current (6).
Experimental design.
Two inoculation studies were performed using six rats per group, which provides a power of 0.80, an α value of 0.05 assuming a standard deviation of 0.5, and an expected difference of 1.0 in the organism burden (see the grading system below). Prior to the initiation of the inoculations, all rats had received 2 weeks of immunosuppression. In study 1, for the single inoculation, rats were each inoculated with preparations of 5 × 107 nuclei of P. carinii f. sp. carinii forms 1, 2, or 6. For the simultaneous-coinoculation, two groups received mixtures of 2.5 × 107 nuclei each of forms 1 and 2 or forms 1 and 6. For the 10- day or 20-day delays between inoculations, rats received an initial inocula of 2.5 × 107 nuclei of form 1, 2 or 6 and then a second inocula of either form 1, 2, or 6 after 10 or 20 days. After inoculation, all rats remained on immunosuppression for the duration of the 10-week study. All rats were sacrificed at a single terminal time point 8 weeks after the first inoculation.
A second, more-limited study was conducted to verify the form 1 and 2 results observed in the previous study, to assess the distribution of the forms throughout the lobes of the rat lungs, and to increase the sensitivity of detection of low organism numbers. Study 2 was conducted as described above, except that 2.5 × 107 P. carinii f. sp. carinii were used for all inoculations.
A control group of six rats that received the full regimen of steroids, but no P. carinii f. sp. carinii inoculation, was included in each study. These rats were sacrificed at the completion of each study to ensure that the rats were not latently infected with P. carinii f. sp. carinii (or P. carinii f. sp. ratti) or did not become contaminated during the course of the study. The PCR-based studies (described below) were conducted on the rat lungs from study 2.
Preparation of P. carinii f. sp. carinii organisms from rats.
Rats were sacrificed by carbon dioxide inhalation, and the lungs were removed and processed for pulsed-field gel electrophoresis (PFGE) as described in detail elsewhere (8, 9). Briefly, the lungs from individual rats were homogenized with a laboratory blender (Stomacher 80; Tekmar, Inc., Cincinnati, Ohio). Large particles were removed by sieving the material through sterile gauze, and the homogenates were treated with aqueous ammonium chloride (0.85%, pH 6.8) to lyse the erythrocytes and some host cells. Host cell numbers were further reduced by at least two passes through 10-μm-pore-size filters (Mitex; Millipore Corp., Bedford, Mass.). For the PCR studies described below, a small portion (ca. 0.05 g) of each of the five lobes of the lungs was excised prior to homogenization.
Estimation of organism burden and statistical analyses.
To evaluate the organism burden in the inoculated rats, we used a modification of semiquantitative methods used by other investigators (1, 6). Slides with touch impressions of a medial section of the left lung were stained with a rapid variant of the Giemsa stain (LeukoStat; Fisher Scientific, Cincinnati, Ohio) and evaluated according to the following scale: 0.5+ (at least 1 organism was seen in 30 oil immersion fields and up to an average of 2.2/field, a value corresponding to approximately 1.3 × 104 to 9.7 × 105 organisms/ml); 1+ (2.3 to 22.7 organisms/field or up to 107 organisms/ml); 2+ (23 to 227 organisms/field; up to 108/ml); 3+ (228 to 2,268 organisms/field; up to 109 organisms/ml); and 4+ (>2,269 organisms/field; >1010/ml). Although this system was only semiquantitative, it could differentiate between heavy, moderate, and light infections. A one-way analysis of variance was performed on the mean and standard error of the mean from rats in each inoculation group, and the results were evaluated for significance by the Student Newman-Keuls multiple-comparison test using the GraphPad INSTAT v.3 program. The α-value was set at 0.05; a P value of <0.05 was considered significant.
Preparation of organisms for PFGE.
After release from the lungs, the P. carinii f. sp. carinii organisms were treated with DNase I (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) at 10 μg/ml in a solution of 150 mM NaCl–10 mM MgCl2–10 mM Tris at pH 7.2 for 30 min at 37°C to digest extracellular DNA and then washed once in 250 mM EDTA and twice in 125 mM EDTA (5, 6). Organisms were embedded in low-melting-point agarose (Boehringer Mannheim) at a final concentration of 0.8% in disposable plug molds (Bio-Rad, Hercules, Calif.) or in disposable spectrophotometric cuvettes (Fisher Scientific), depending on the organism densities. Gel-embedded organisms were digested with 0.25 mg of proteinase K (Boehringer Mannheim) per ml in a solution of 1% N-lauroylsarcosine (Sigma)–0.45 M EDTA–0.01 M Tris in a 55°C water bath for 24 to 48 h. Digested samples were stored at 4°C in 0.5 M EDTA.
CHEF conditions.
Gels for contour-clamped homogeneous electrical-field (CHEF) electrophoresis contained 1% FMC SeaKem GTG-agarose (SeaKem, Rockland, Maine) prepared in 0.5× TBE (45 mM Tris HCl, 45 mM boric acid, 1.25 mM EDTA) for a total volume of 200 ml and final dimensions of 14 by 21 cm. Electrophoresis was performed using a Bio-Rad CHEF DR II or CHEF DR III apparatus. Gels were run for 104 to 144 h at 14°C in 0.5× TBE at 3.8 V/cm with a 50-s initial pulse that was gradually increased to 100 s (8, 9). Chromosome-sized DNA bands were visualized by the nucleic acid stain, SYBR-Gold (Molecular Probes, Inc., Eugene, Oreg.).
PCR conditions.
The PCR was performed on lung tissue samples (∼0.05 g) taken from each of the five lung lobes of rats inoculated with forms 1 and 2 individually and in combination at the different time periods (see above) and from uninoculated immunosuppressed rats. The lung lobes were numbered as follows: 1, left lung; 2, cranial lobe of the right lung; 3, medial lobe of the right lung; 4, caudal lobe of the right lung; and 5, accessory lobe. Tissue samples were prepared for the PCR by digestion in lysis buffer (1% sodium dodecyl sulfate, 25 mg of proteinase K per ml) at 55°C overnight and precipitation of the resultant DNA with isopropanol at −20°C. Primers directed to a region upstream of the α-tubulin gene p10 (5′-AAAGATGGTGAATTGTAACTC-3′) and pIV (36) were used in 25-μl reactions amplified under the following conditions: a hot start of 95°C for 5 min, then 40 cycles of incubation at 95°C for 60 s and at 42°C for 60 s, and finally elongation at 72°C for 60 s (23). Amplicons were electrophoresed through 0.7% gels at 60 V for 1 h and visualized by ethidium bromide staining under UV light. Under these conditions, P. carinii f. sp. carinii form 1 DNA produced a product of ca. 520 bp, while form 2 DNA had an approximate size of 600 bp due to the presence of multiple repeats of TAACCCTAA sequences (23).
Evaluations to determine the efficiency of the PCR for form 1 and form 2 templates were conducted. Dilutions of DNA extracted from the same numbers of each organism population were individually amplified with the primers described above and with primers directed to a region in the mitochondrial large ribosomal subunit that is the identical size (340 bp) in each genome (27). Amplicons were visualized by ethidium bromide staining of 1% agarose gels, quantified by densitometric analysis, converted to pixels, and graphed against the number of P. carinii f. sp. carinii in the reaction.
Computer simulation of form 1 and form 2 trajectories.
A program written in Fortran 77 was used to simulate different growth models of form 1 and form 2 populations to determine if the lack of the presence of a population could be simply due to differences in growth rates (Jonathan Arnold, Department of Genetics and Computational Biology, University of Georgia). The trajectories of the numbers of form 1 and form 2 were calculated in a simple competition model (Lotka-Volterra) (20) with a common carrying capacity for each form. Doubling times were varied from 6.98 to 698 days, rates of increases ranged from 0.001 to 0.1, lag times were set at 0, 10, and 20 days, and the length of the experiment was set at 10-day increments beginning 10 days and continuing through 60 days.
RESULTS
Electrophoretic karyotypes of P. carinii f. sp. carinii produced by single and coinoculations.
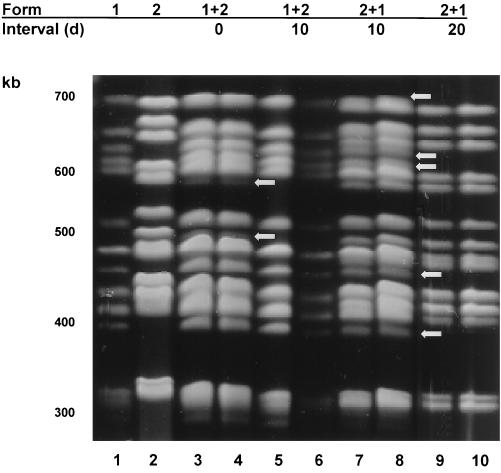
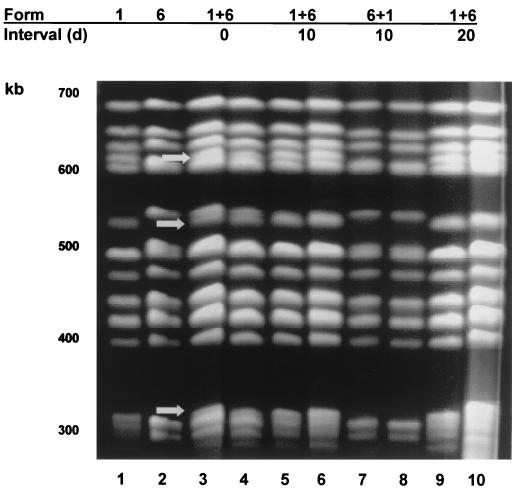
All rats inoculated with form 1, 2, or 6 produced P. carinii f. sp. carinii with the same karyotype profile as that inoculated (Fig. 1 and 2), showing that these inocula were viable and stable upon passage. Inoculation of equal mixtures of P. carinii f. sp. carinii forms 1 and 2 administered at the same time produced infections containing both forms (Fig. 1, lanes 1+2; interval, 0 days). Fewer form 2 organisms were present than form 1 organisms, as evidenced by the bands of lighter intensity of <600 and 500 kb corresponding to the same sized bands in the form 2 control lane (arrows). Forms 1 and 6 administered at the same time and in equivalent numbers produced karyotypes that contained similar numbers of both forms, as evidenced by bands of equal intensity corresponding to each P. carinii f. sp. carinii form in these rats (Fig. 2, lanes 1+6; interval, 0 days). Note the doublet bands at 600 and 550 kb (arrows).
FIG. 1.
Karyotypic profiles of P. carinii f. sp. carinii from rats inoculated with forms 1 and 2 individually and in combinations. The karyotype forms inoculated and the times between inoculations in days (d) are shown at the top of the gel and correspond to the lanes immediately below. Lambda concatamers (48.5-kb increments) were used as size markers (not shown). Lanes: 1, form 1 individually inoculated; 2, form 2 individually inoculated; 3 and 4, forms 1 and 2 inoculated at the same time, arrows indicate bands belonging to form 2; 5 and 6, forms 1 and 2 inoculated 10 days apart; 7 and 8, forms 2 and 1 inoculated 10 days apart, arrows indicate bands of form 1; 9 and 10, forms 2 and 1 inoculated 20 days apart.
FIG. 2.
Karyotypic profiles of P. carinii f. sp. carinii from rats inoculated with forms 1 and 6 individually and in combinations. The karyotype forms inoculated and the times between inoculations in days (d) are shown at the top of the gel and correspond to the lanes immediately below. Lambda concatamers (48.5-kb increments) were used as size markers (not shown). Lanes: 1, form 1 individually inoculated; 2, form 6 individually inoculated; 3 and 4, forms 1 and 6 inoculated at the same time, arrows indicate bands belonging to form 1; 5 and 6, forms 1 and 6 inoculated 10 days apart; 7 and 8, forms 6 and 1 inoculated 10 days apart; 9 and 10, forms 1 and 6 inoculated 20 days apart.
The karyotypes produced from inoculations spaced 10 days apart were that of the first form to be inoculated, with one exception (Fig. 1 and 2, Tables 1 and 2). The single exception occurred in rats that received an initial inoculation of form 2, followed 10 days later by an inoculation of form 1 (Fig. 1, lanes 2+1; 10-day interval). In these rats, P. carinii f. sp. carinii forms 1 and 2 were present in apparently equal amounts despite the gap between inoculations. Note the presence of doublet bands at ca. 700 kb, the seven bands between 680 and 580 kb versus only four bands in this region in the single form 1 or 2 karyotypes, and the bands of 450 and <400 kb, belonging to form 1 (arrows). In rats receiving form one organism followed by a second dose of form 1, 10 or 20 days later, form 1 karyotypes were produced as expected (data not shown). Rats inoculated first with killed form 1 organisms, followed 10 days later by inoculation of form 2, did not produce karyotypes due to the low numbers of organisms. Although sufficient organism numbers were present in rats inoculated first with the live form 1 P. carinii f. sp. carinii, followed by the killed form 2, karyotypes were not discernible due to the abundance of degraded DNA (data not shown). Thus, although the organism burdens in rats coinoculated with live P. carinii f. sp. carinii 10 days apart were equal to those inoculated with a single form of P. carinii f. sp. carinii or with the admixtures (see below), the karyotypes of the resultant organisms causing the infection in most of these rats were comprised of the P. carinii f. sp. carinii organisms first inoculated.
TABLE 1.
Infection scores and karyotype profiles of P. carinii f. sp. carinii forms 1 and 2
| Karyotype form(s) inoculateda | Inoculation interval (days) | Mean infection score (SEM) | Karyotype profileg
|
|
|---|---|---|---|---|
| Major | Minor | |||
| 1 | 0 | 4.00 (0.00) | 1 | – |
| 2 | 0 | 3.75 (0.25) | 2 | – |
| 1+2 | 0 | 4.00 (0.00) | 1 | 2 |
| 1+2 | 10 | 3.20 (0.49) | 1 | – |
| 2+1 | 10 | 3.75 (0.25) | 2 and 1 | – |
| 1+1 | 10 | 3.60 (0.25) | 1 | – |
| 1A+2 | 10 | 0.50b (0.00) | NA | |
| 1+2A | 10 | 2.50c (0.56) | NA | |
| 1+2 | 20 | 1.25d (0.43) | NA | |
| 2+1 | 20 | 2.67 (0.56) | 2 | – |
| 1+1 | 20 | 1.75e (0.48) | 1 | – |
| 1A+2 | 20 | 0.40f (0.10) | NA | |
| 1+2A | 20 | 2.42 (0.61) | 1 | – |
The letter A indicates an autoclaved form given at the same dosage as the live P. carinii.
Significantly different at P <0.001 from forms 1, 2, 1+2, 2+1, and 1+1 and at P < 0.01 from form 1+2A on day 10.
Significantly different at P < 0.05 from form 1.
Significantly different at P < 0.01 from forms 1 and 2.
Significantly different at P < 0.05 from form 1.
Significantly different at P < 0.001 from forms 1 and 2 and at P < 0.05 from form 1+2A and form 2+1.
–, no minor profile was identified; NA, not available due to insufficient organism numbers.
TABLE 2.
Infection scores and karyotype profiles of P. carinii f. sp. carinii forms 1 and 6
| Karyotype form(s) inoculated | Inoculation interval (days) | Mean infection score (SEM) | Karyotype profile
|
|
|---|---|---|---|---|
| Major | Minor | |||
| 1 | 0 | 4.00 (0.00) | 1 | –a |
| 6 | 0 | 4.00 (0.00) | 6 | – |
| 1+6 | 0 | 4.00 (0.00) | 1 and 6 | – |
| 1+6 | 10 | 3.83 (0.17) | 1 | – |
| 6+1 | 10 | 3.60 (0.25) | 6 | – |
| 1+6 | 20 | 3.33 (0.42) | 1 | – |
–, no minor profile was identified.
Karyotypes from rats that received inoculations 20 days apart followed the same pattern as most of the infections resulting from the 10-day gap (Fig. 1 and 2). The initial P. carinii f. sp. carinii form inoculated was the only form detected by PFGE in all cases, even with the combination of form 2 followed by form 1 that produced the mixed karyotype when administered 10 days apart. Two inoculations with live form 1 organisms produced karyotypes of form 1 (data not shown), but those rats receiving killed form 1 organisms prior to form 2 did not produce a karyotype due to the low organism number. Form 1 karyotypes were produced from rats that received live form 1 as the initial inocula, followed 20 days later by autoclaved form 2 organisms (data not shown). Most of the rats inoculated with combinations of live forms 1 and 2 had reduced organism burdens but nonetheless were able to produce discernible karyotypes.
Organism burdens resulting from single and coinoculations.
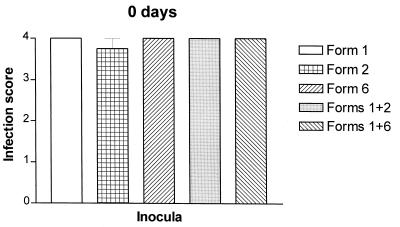
Rats inoculated with equivalent numbers of P. carinii f. sp. carinii form 1, 2, or 6 developed fulminant infection with no significant differences in organism burden (Fig. 3), indicating that the inocula used in these studies were similar in viability and did not have dramatically disparate growth rates. Coinoculations of forms 1 and 2 or forms 1 and 6 at the same number of total organisms as the individual inocula (5 × 107) produced infections with the same organism burdens as those produced by inoculation with a single form, showing that these admixtures had no deleterious effect on organism burdens (Fig. 3, Tables 1 and 2).
FIG. 3.
Organism burdens from rats inoculated with individual populations and admixed combinations of P. carinii f. sp. carinii karyotype forms. Semiquantitative estimates of organism burdens from rats inoculated with individual karyotype forms 1, 2, or 6 and combinations of forms 1 and 2 and forms 1 and 6 administered at the same time and in equal numbers. Organism burdens were determined as averages and standard errors of the mean of infections scored according to a semiquantitative system described in Materials and Methods.
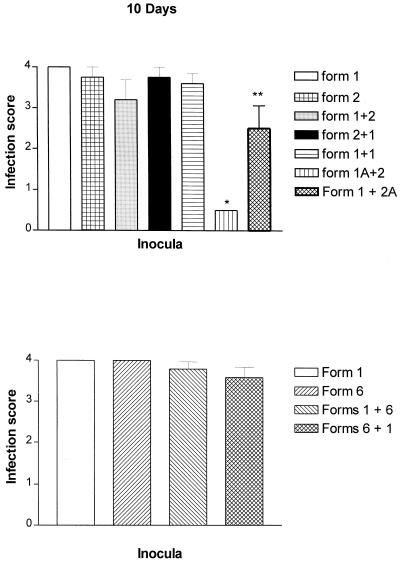
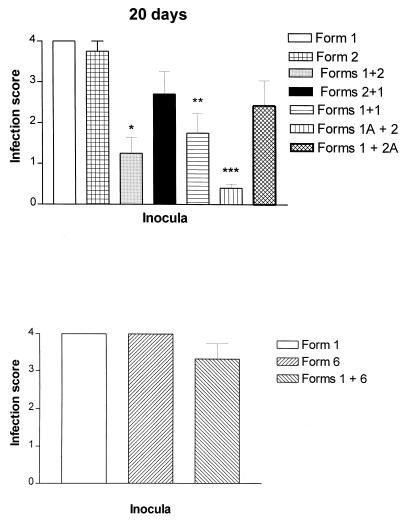
No significant differences in the infection scores were observed among the combinations when a gap of 10 days was introduced between inoculations (Fig. 4, Table 1). However, spacing of the inoculations 20 days apart produced lower organism burdens in rats receiving a combination of form 1 followed by form 2 (Fig. 5, form 1+2; Table 1) but not in rats receiving form 1 followed by form 6 (Fig. 5, form 1+6; Table 2). Although a trend toward reduction was observed in rats inoculated first with form 2 and then form 1, it did not reach significance. Thus, differences in organism burdens were associated with the different P. carinii f. sp. carinii karyotype combinations inoculated.
FIG. 4.
Organism burdens from rats inoculated with combinations of karyotype forms 10 days apart. Semiquantitative estimates of organism burdens from rats inoculated with combinations of karyotype forms in which the second population was given 10 days after the first population. Asterisks indicate significant differences as detailed in Table 2. Organism burdens were determined as averages and standard errors of the mean of infections scored according to a semiquantitative system described in Materials and Methods.
FIG. 5.
Organism burdens from rats inoculated with combinations of karyotype forms 20 days apart. Semiquantitative estimates of organism burdens from rats inoculated with combinations of karyotype forms in which the second population was given 20 days after the first population. Asterisks indicate significant differences as detailed in Table 2. Organism burdens were determined as averages and standard errors of the mean of infections scored according to a semiquantitative system described in Materials and Methods.
A role for the host immune response in the reductions was suggested by the results of control groups. Three control groups were included for inoculations spaced 10 and 20 days apart: (i) two inoculations of live P. carinii f. sp. carinii form 1 organisms from the same cryopreserved batch at each interval (1+1); (ii) inoculation of autoclaved form 1 organisms prior to that of live form 2 P. carinii f. sp. carinii (1A+2); and (iii) inoculation of autoclaved form 2 organisms after live form 1 P. carinii f. sp. carinii (1+2A). These three groups were included to evaluate the specificity of any observed host response and the requirement of live or killed organisms in this response. Inoculation of live form 1 into rats inoculated with form 1 organisms 10 days beforehand produced no difference in the organism burden (Fig. 4, form 1+1), yet the spacing of this same inoculation 20 days apart resulted in decreased organism burdens (Fig. 5, form 1+1). Significant decreases in the level of infection also were observed in rats receiving autoclaved form 1 organisms 10 and 20 days prior to the live form 2 inocula (Fig. 4 and 5, form 1A+2) and in rats receiving the killed form 2 organisms 10 days after the live form 1 inocula (Fig. 4, form 1+2A). The administration of killed form 2 organisms 20 days after live form 1 inoculation also resulted in a trend toward decreased burdens. Recall that reductions in organism burden only occurred when live inoculations were spaced 20 days apart and only with specific pairs of populations, either forms 1 and 2 or forms 1 and 1. Thus, killed organisms were able to elicit protection more rapidly than were live organisms.
Detection of forms 1 and 2 by PCR.
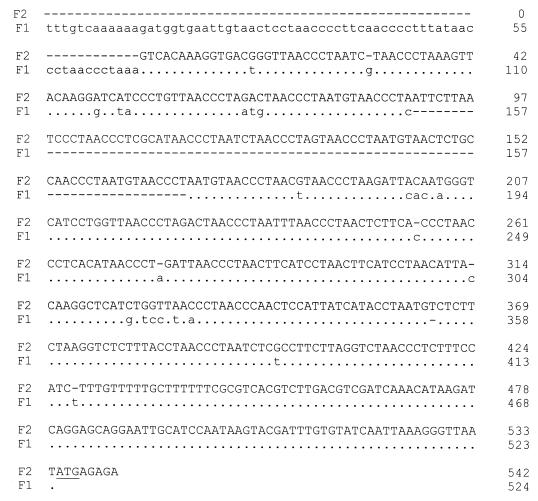
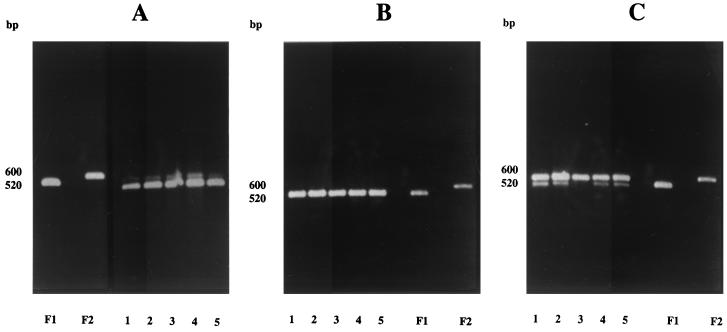
Previous studies in our laboratories have shown the lower limit of detection for PFGE to be 5 × 106 P. carinii f. sp. carinii nuclei per lane by visualization of the bands stained with SYBR-Gold. To detect the presence of lower numbers of P. carinii f. sp. carinii in the lungs of inoculated rats and to assess the distribution of organisms within the lungs, a PCR-based assay was used in a second inoculation experiment. Forms 1 and 2 were chosen for this analysis because of the availability of a rapid PCR method that could differentiate the two forms and because of the unique interactions demonstrated by karyotyping. Products derived from amplification of an area upstream of the α-tubulin gene have previously been shown to differ in size for P. carinii f. sp. carinii form 1 and 2 populations due to several copies of a telomere-like sequence repeat in form 2 that is not present in form 1 (23). The sequences of this region in the genomes of the two forms are shown in Fig. 6. All five lobes of the infected rat lungs from at least two rats per time point were sampled to provide information on the distribution of the populations. In rats that were coinoculated with an admixture of forms 1 and 2, amplicons corresponding to products from both forms were present in each of the five lobes (Fig. 7A, lanes 1 to 5 [each lane corresponds to a rat lung lobe as defined in Materials and Methods and in the figure legend]). Form 2 amplicons were less intense than those from form 1, suggesting that there may have been fewer of the form 2 organisms, similar to results from the first experiment revealed by CHEF gel analyses (Fig. 1, lanes 1+2; 0-day interval). Amplicons from form 2 populations were not detected in rat lungs inoculated with form 1 followed 10 days later by form 2 inoculations (Fig. 7B). The lack of detection of form 2 populations by the PCR suggests that the organisms were in much lower abundance than were the form 1 organisms, if at all present. In contrast, amplicons corresponding to forms 1 and 2 were apparent in rats that were inoculated with form 2 followed 10 days later by form 1 (Fig. 7C), which again corresponded to the results of the karyotypic profiles.
FIG. 6.
Nucleotide alignment of the upstream region of the α-tubulin gene, the “α-repeat.” F1, form 1; F2, form 2 (P. carinii f. sp. carinii). Dots indicate the identity, and dashes indicate the gaps that were introduced for maximal alignment. The underline indicates the start codon of the α-tubulin gene (23).
FIG. 7.
Distribution of P. carinii f. sp. carinii forms 1 and 2 in the lobes of rats detected by amplification of the α-repeat region. (A) F1, amplicon from rat lung inoculated with form 1; F2, amplicon from rat lung inoculated with form 2; lanes 1 to 5, amplicons from individual lobes of a single rat inoculated with forms 1 and 2 at the same time. In all panels, the lane numbers correspond to the different lung lobes as described in Materials and Methods. (B) Lanes 1 to 5, amplicons from individual lobes of a single rat inoculated with form 1, followed 10 days later with form 2, and from rats inoculated with individual populations of form 1 (F1) or form 2 (F2). (C) Lanes 1 to 5, amplicons from individual lobes of a single rat inoculated with form 2, followed 10 days later by form 1, and from rats inoculated with individual populations of form 1 (F1) or form 2 (F2).
Although PCR is expected to have a much greater sensitivity than many detection methods, including CHEF analysis, some factors, such as differences in amplification efficiency, can reduce the sensitivity of the reaction. Since both the PCR data and the CHEF karyotypes predicted that form 2 organisms would be absent in some rats, we conducted experiments to evaluate the amplification efficiency of form 1 and form 2 templates with the primers directed to the α-repeat region and the mitochondrial large ribosomal subunit. At nuclei numbers of 10,000, the form 2 efficiency of amplification was only 70% of that for form 1. More-dramatic decreases were observed with numbers lower than 10,000. In contrast, primers directed to the mitochondrial large ribosomal subunit did not produce a difference in efficiency. The lower efficiency of the α-repeat primers was likely due to the addition of the ∼80-bp telomere-like repeat in the form 2 genome, making it 20% longer than the form 1 product. Approximately 2% of the infected rat lung was sampled for PCR processing (0.05 g of a 2.50-g lung). At least 10,000 form 2 organisms would have to be present to be detected if an equal or greater number of form 1 organisms were also present. If equally dispersed throughout the lung at these levels, then a total estimated organism burden of form 2 in these lungs would be 500,000; a number insufficient to be detected by CHEF gel analysis. Thus, we can assume that the form 2 organisms, if present, were present in numbers no greater than 500,000 per lung, a number far lower than the 25,000,000 organisms inoculated.
ATP levels of form 1 and 2 organisms mixed in vitro.
In some fungal systems, the addition of a second fungal population to a host or artificial medium colonized by another population with a similar genetic background results in a destructive process causing organism loss in one or both populations (4, 17). Since most P. carinii f. sp. carinii forms inoculated 10 or 20 days after the first inoculation were not detectable by PFGE or PCR (form 2), we postulated that there may be a direct inhibitory effect of one organism population on the other. Experiments were performed to assess the effect on the viability of 2 forms of P. carinii f. sp. carinii mixed in vitro. The viability was determined by measurement of cellular ATP using a bioluminescent assay (7, 10). There is no long-term culture in which detectable and consistent ATP levels of P. carinii f. sp. carinii can be measured over time. The ATP bioluminescent assay is a short-term cell-free assay routinely used in our laboratory for the screening of candidate anti-P. carinii agents and has been shown to be of predictive value for efficacy in animal models (10). Spacing of the introduction of one form onto another was reduced to a 3-day gap since the ATP levels gradually decrease after 7 days. However, the same organism numbers as those inoculated in vivo were used in the in vitro study. Forms 1 and 2 were selected for the in vitro study since both PCR and karyotype data on both forms were available from the in vivo studies. Form 1 and form 2 inoculated individually at these organism densities increased their ATP levels from 35 to 50% throughout 5 days with a slight decrease beginning at day 6 (Table 3, groups 1 to 4). Statistical analyses revealed no difference in the slopes of these lines. Mixing equal numbers of form 1 and 2 prior to culture (group 5) did not result in any decrease in ATP, a result which would have been expected if there were a deleterious effect of one form on another. Rather, this group achieved ATP levels almost identical to the mathematical sums of groups 1 and 2 and to the ATP measurements of groups 3 and 4 inoculated with 5 × 107 organisms each, thus indicating a simple additive effect of the two forms in group 5. The addition of form 2 to cultures of form 1 after 3 days resulted in an increase of ATP concordant with an increase in organisms (group 6). Likewise, when form 1 organisms were added to cultures of form 2, a similar increase in ATP was observed (group 7). Thus, a direct effect on the viability of one form on another was not observed in this in vitro situation.
TABLE 3.
ATP levels of forms 1 and 2 coinoculated in vitro
| P. carinii form(s) (no. of nuclei) | Group | ATP levels (mean relative light units ± SD) at:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | ||
| Form 1 (2.5 × 107) | 1 | 25,434 ± 1,764 | 31,182 ± 1,734 | 31,840 ± 2,860 | 35,639 ± 6,446 | 34,443 ± 3,476 | 22,770 ± 3,480 | 15,170 ± 1,093 |
| Form 2 (2.5 × 107) | 2 | 17,082 ± 1,742 | 17,801 ± 814 | 22,592 ± 2,720 | 22,831 ± 1,367 | 22,760 ± 1,391 | 18,804 ± 673 | 17,538 ± 997 |
| Form 1 (5.0 × 107) | 3 | 43,603 ± 3,062 | 43,977 ± 3,868 | 57,858 ± 3,500 | 59,660 ± 3,036 | 65,390 ± 6,747 | 56,490 ± 3,326 | 39,923 ± 3,772 |
| Form 2 (5.0 × 107 | 4 | 31,833 ± 1,869 | 32,771 ± 2,168 | 40,460 ± 1,453 | 42,303 ± 1,826 | 48,039 ± 5,966 | 41,430 ± 3,051 | 36,185 ± 4,258 |
| Forms 1 and 2 (2.5 × 107 each) | 5 | 37,960 ± 1,699 | 44,857 ± 3,253 | 54,236 ± 4,700 | 56,379 ± 4,821 | 57,605 ± 3,799 | 51,368 ± 3,251 | 48,723 ± 4,657 |
| Form 1 (2.5 × 107) and form 2 (2.5 × 107) at day 3 | 6 | 27,698 ± 2,214 | 28,053 ± 1,475 | 33,910 ± 6,518a | 47,268 ± 4,328 | 47,725 ± 2,215 | 47,017 ± 5,466 | 46,500 ± 2,876 |
| Form 2 (2.5 × 107) and form 1 (2.5 × 107) at day 3 | 7 | 16,729 ± 840 | 19,081 ± 1,411 | 20,655 ± 1,366a | 39,887 ± 1,986 | 45,223 ± 2,686 | 43,512 ± 2,258 | 41,364 ± 5,772 |
Second inoculation of 2.5 × 107 P. carinii; ATP content reflects organism pools prior to inoculation.
DISCUSSION
Two general conclusions can be drawn from these studies. First, the time between inoculations of the two P. carinii f. sp. carinii forms was critical in determining the organism population(s) that ultimately caused the pneumonia. Coinfections were only established when equal numbers of the two organism forms were administered at the same time. When gaps of 10 or 20 days were introduced between these inoculations, the first organism form inoculated became the causative agent of infection in all but one case. Second, reductions in organism burdens resulted from a variety of factors, including the viability of organism inoculations, the karyotypes of the inoculated organisms, and the amount of time between inoculations.
The preponderance of the first population inoculated was not due to differential growth rates or to a direct effect of one population on the other.
A 10-day gap between all but one set of paired inoculations was sufficient to produce an end infection consisting only of the first population inoculated, as detected by karyotypic analysis. These results showed that the second inocula did not contribute to the organism burden and in fact were in numbers below 5 million, the minimum number of organisms needed for visualization with nucleic acid stains on the pulsed-field gel. These results cannot be explained by a simple difference in viability among the inocula, since burdens were similar in rats receiving live inoculations of forms 1, 2, and 6 as individual inocula and as paired admixtures. Likewise, it is unlikely that the 10- or 20-day delay in growth contributed to the absence of the second population. Each of the inocula contained 2.5 × 107 organisms, sufficient in itself to produce a visible karyotype. If one assumes for argument's sake that only 10% of the organisms were able to grow, due to host response or the loss of viability, and a lengthy doubling time of 1 week, the elimination of 10 days from the total 8-week run of the experiment would produce approximately 3 × 108 P. carinii f. sp. carinii, an amount more than sufficient to produce a visible karyotype. Elimination of 20 days of growth time would permit replication to a level of 8 × 107, which would also be detectable on the pulsed-field gels. The same argument can be made for disparate growth rates of the different populations. Even if the growth rates were disparate, the slower-replicating population should still be able to be visualized on the pulsed-field gels since the band patterns are sufficiently discriminating. In addition, the growth rates could not be dramatically different, since the P. carinii f. sp. carinii populations administered as individual inoculations produced equivalent infection burdens after a growth period of 8 weeks.
A mathematical model was constructed to further validate this conclusion. Using form 1 and form 2 as the test populations, a model based on the Lotka-Volterra paradigm (20) was used to calculate the resultant population numbers of forms 1 and 2 from logistic growth curves as trajectories, assuming uniform competition between the forms and similar carrying capacities of each population. Similar carrying capacities can be assumed from the organism burdens for each form shown in Table 1 and Fig. 3, and was set at 1010 organisms, a figure based on previous enumeration studies in our laboratory. For all of the calculated scenarios, forms 1 and 2 remained above the limits of detection by CHEF analysis, suggesting that extrinsic factors were influencing the observed population decreases.
A more compelling argument for an active suppression of growth rather than disparate growth rates can be found in the organism burdens in rats receiving form 1 first and then form 2 after 20 days. If there was simply a difference in growth rate, one would have anticipated an organism burden similar to that of the control rats that received form 1 alone. However, a dramatic decrease in organism burden was observed in these rats, suggesting a more complicated mechanism. This dramatic reduction in organism burden was not observed in coinoculations of forms 1 and 6, further suggesting that this mechanism may be specific for the form pairs inoculated.
If, therefore, the organism burden and identity of the infecting population were not due to a difference in growth rate and appeared to be specific to the organism forms inoculated, an alternative explanation might be the active suppression of one population by another. Such interactions are commonly reported in other fungi, including Neurospora crassa and other ascomycetes (4, 17) and in some myxomycetes (5). In those cases, the fungal population which has established growth actively inhibits coinfection by another strain of the same fungus. We posited that a similar mechanism may have been involved in the exclusion of incoming populations of P. carinii attempting to colonize the same rat lung. We tested this hypothesis by exposing the form 1 and form 2 populations to one other in an in vitro setting, but we found no detrimental effects on viability, as determined by ATP levels. Rather, an additive effect in which both populations contributed to the ATP levels was observed. These results suggested that a direct detrimental effect of one population on the other was not a likely explanation for the predominance of the first population over the second.
Potential role for the host.
Lack of evidence supporting a direct effect of one population on another, protective effects of autoclaved organism inoculations, and an apparently specific response with successive inoculations of live forms 1 and 2 or forms 1 and 1 with 20-day gaps strongly suggests a role for the host immune response in these findings. A simple explanation for the majority of these observations could be framed in the context of a changing immune response in the chronically immunosuppressed rat model of infection. The first inoculation of a P. carinii f. sp. carinii form occurred after 2 weeks of corticosteroid treatment in a P. carinii f. sp. carinii-naive host. The host is sufficiently immunosuppressed to permit replication of the organisms and subsequent colonization of the lung alveoli by attachment to the type I pneumocytes. In nonimmunocompromised antigen-naive rats, an immune response usually takes 10 days to 3 weeks after antigen exposure to develop (P. Mirley [Charles River Laboratories, Wilmington, Mass.], personal communication). By extrapolation to our model, one would predict that the second inoculation 10 days after the first would provide organisms easily accessible to the cellular mediators of a developing, yet truncated immune response (due to the effects of the corticosteroids), thus permitting eradication. The first population may withstand this response by its privileged position in the alveolar hypophase and tight adherence to the type I pneumocytes or because of the immature immune response. After 20 days, an immune response to P. carinii f. sp. carinii would have had time to develop, even in the immunosuppressed host (36), and inoculation of the second organism population could then result in a more dramatic immune response not only resulting in elimination of the incoming organisms but also of some of the resident organisms if the two populations were sufficiently similar in antigenic composition.
This scenario is supported by an understanding of the immunological changes in the corticosteroid-treated rat model of P. carinii pneumonia and by recent reports implicating CD8+ cells as mediators of lung injury. Walzer et al. (33) tracked the generalized depletion of lymphocytes and inversion of the CD4+/CD8+ lymphocyte ratios in the chronic corticosteroid-treated rat model of P. carinii pneumonia. After 3 weeks of immunosuppression, the lymphocyte numbers are <1,000/mm3 and the CD4+/CD8+ ratio has fallen from 2.4 in the nonimmunosuppressed state to 1.0. At 4 weeks of immunosuppression, this ratio drops to 0.6. In the present study, the second inoculation of the 10-day gap coincides with 3 weeks of immunosuppression (2 weeks prior and 1 week after the first inoculation), while the 20-day gap period coincides with 4 weeks of immunosuppression. When the second P. carinii f. sp. carinii population was introduced 10 days after the first, there was likely a sufficient immune response that could eliminate these organisms prior to their initiating colonization of the alveolar lining cells. However, after a gap of 20 days, the CD8+ cell population was the predominant cell type. Rats receiving inoculations of form 1 and 2 combinations may have had organism burdens reduced by a specific inflammatory response exacerbated by the CD8+ cells. A recent report by Wright et al. (35) implicated the CD8+ cell as a major factor in immune-mediated lung injury in CD4+-depleted mice infected with P. carinii f. sp. mus. These effects were more pronounced in later infection than during an earlier phase of infection, a result similar to our own findings. Although the experimental design used by Wright et al. was not identical to ours, the role of the host immune response in mediating the infective process in immune-deficient rodents is supported.
Harmsen et al. reported a somewhat similar observation of a reduction in organism burden mediated by two inoculations of P. carinii f. sp. mus in a mouse model of pneumocystosis (18). These investigators showed that intratracheal inoculation of mice with live P. carinii f. sp. mus could reduce the infection that followed when the same mice were depleted of CD4+ cells and reinoculated with P. carinii f. sp. mus.
Results from inoculation of paired form 1 and form 2 live and autoclaved organisms support the role of the immune response in the reduction of organism burdens and in the elimination of secondary populations. Not only were these autoclaved organisms easily accessible to the immune system, they likely evoked a more vigorous response due to exposure of additional antigens as a result of the autoclaving process.
An unique aspect of the present study was the use of characterized organisms which were found to play a significant role in the outcome of the infections. The reduction in burden severity was most clear upon coinoculations of live forms 1 and 2 with a gap of 20 days between inoculations. Organism burdens were not significantly reduced by paired inoculations of forms 1 and 6, but a clear dominance in karyotype was observed by the population that was first inoculated. These data would be consistent with the hypothesis described above such that the second inoculation was easily accessible to the immune system but, in this case, did not initiate a specific immune response as seen for forms 1 and 2. The role for a specific immune response in burden reduction is supported additionally by results in rats given live form 1 20 days after the initial form 1 inoculation.
Failure to detect form 2 organisms by PCR suggests elimination rather than growth inhibition.
The absence of a P. carinii f. sp. carinii form on a PFGE gel does not necessarily reflect the absence of organisms, since the level of sensitivity on these gels is estimated to be 5 million organisms (M. Cushion, unpublished data). PCR was used to evaluate the presence of organisms in rat lungs which were not visible on PFGE gels. Form 2 amplicons could be detected in preparations in which the form 2 karyotype was visible, e.g., inoculations of equal numbers of forms 1 and 2 administered at the same time or a singular inoculation of form 2. However, no amplicons resulted from preparations in which the form 2 population could not be visualized by PFGE. These results show that the form 2 organisms not only failed to grow but were also present in far fewer numbers than when inoculated, suggesting the elimination of this population.
Coinfections of forms 2 and 1 were not due to physical separation within the lung.
The ability of the form 1 population to exist as a coinfection when inoculated 10 days after form 2 was puzzling in light of the results from all other coinoculations. The same result was reproduced in a second study using different preparations of form 1 and 2 organisms and thus was not a spurious finding. We postulated that perhaps the two forms colonized different lobes of the rat lungs and might coexist as physically isolated colonies. However, studies to assess the extent of colonization of the lobes of the lungs by forms 1 and 2 showed that both populations were able to migrate throughout the entire lung by the termination of the study when they were inoculated as admixtures or when form 1 followed form 2. Thus, it appeared these two populations under this specific inoculation regimen were able to establish infection in the rat lungs and could coexist within the same lobes of the lung. Further studies will be required to understand the conditions permitting form 1 and form 2 coexistence. However, such findings contribute to our growing appreciation of the diversity of members of this family of fungal pathogens, their interactions within the host, and their ability to cause infection. This result also illustrates the differences in biological properties that can now be associated with karyotype profiles. Only forms 2 and 1 were able to produce this coinfection. Forms 1 and 6 did not.
Factors permitting coinfection in the rat have relevance to human infection.
The P. carinii f. sp. hominis populations infecting human beings exhibit levels of polymorphisms in the large-subunit mitochondrial rRNA (24) and in the intergenic regions of the nuclear rRNA locus (25, 26) similar to those differences observed between the P. carinii f. sp. carinii karyotype forms used in the present study. Whether the organism strains coinfecting human beings also differ in karyotypic profiles is not known, since most samples (usually bronchoalveolar lavage fluids) from humans do not supply the necessary numbers of organisms needed for visualization of the chromosomes by PFGE.
Our data suggest that establishment of successful coinfections is dependent upon the genotype of organism populations and the timing of their introduction into the lung, with some mediation of outcome by the host. Reports of the prevalence of infections in humans containing more than one strain range from 10 to 69% (3, 19), depending on the method used for detection. Furthermore, Beard et al. reported the presence of multiple genotypes associated with primary infection but not recurrent P. carinii f. sp. hominis pneumonia (3). The studies presented here experimentally reproduced coinfections similar to those occurring in humans and extended our understanding of the factors leading to infection with more than a single P. carinii population. Previous colonization with one P. carinii f. sp. carinii prevented the growth of a second population in all but one case. Based on our studies, it would be reasonable to hypothesize that the lack of multiple genotypes in recurrent human infection may be mediated by the host response to organisms of the primary infection. Coinfections could be established when P. carinii f. sp. carinii populations were inoculated at the same time, suggesting that primary human infections may arise from the acquisition of multiple genotypes within a relatively short time frame or even as a transmission of multiple genotypes from a single source.
In the present study, we have shown that the establishment of coinfections in immunosuppressed rats was dependent on the identity of the populations used for the coinoculations and on the time between those inoculations. The inhibition of growth of some karyotype forms by others suggests that an active process by the host, but not by the organisms themselves, may be involved. Such findings have implications for attempts to inoculate organisms into rats previously colonized with another population and should be considered in the experimental design of a study, especially those measuring the immune response. These studies also revealed clear biological differences among P. carinii f. sp. carinii defined by karyotype profiles, thus associating phenotype with genotype. Inoculations of defined P. carinii f. sp. carinii karyotype forms should facilitate experimental approaches for defining the transmission and acquisition of P. carinii f. sp. hominis in humans.
ACKNOWLEDGMENTS
This work was supported by grants RO1 AI29839 (M.T.C.) and RO1 AI36701 (J.R.S.) from the National Institutes of Health.
We thank Jonathan Arnold, University of Georgia, for his invaluable assistance in calculating the logistical growth curves of forms 1 and 2.
REFERENCES
- 1.Bartlett M S, Fishman J A, Durkin M M, Queener S F, Smith J W. Pneumocystis carinii: improved models to study efficacy of drugs for treatment or prophylaxis of Pneumocystis pneumonia in the rat (Rattus spp.) Exp Parasitol. 1990;70:100–106. doi: 10.1016/0014-4894(90)90089-u. [DOI] [PubMed] [Google Scholar]
- 2.Bauer N L, Paulsrud J R, Bartlett M S, Smith J W, Wilde C E. Pneumocystis carinii organisms obtained from rats, ferret, and mice are antigenically different. Infect Immun. 1993;61:1315–1319. doi: 10.1128/iai.61.4.1315-1319.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard C B, Carter J L, Keely S P, Huang L, Pieniazek N J, Moura I N S, Roberts J M, Hightower A W, Bens M S, Freeman A R, Lee S, Stringer J R, Duchin J S, del Rios C, Rimland D, Baughman R P, Levy D A, Dietz V J, Simon P, Navin T R. Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg Infect Dis. 2000;6:265–272. doi: 10.3201/eid0603.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bégueret J, Turcq B, Clavé C. Vegetative incompatibility in filamentous fungi: het genes begin to talk. Trends Genet. 1994;10:441–445. doi: 10.1016/0168-9525(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 5.Betterley D A, Collins O R. Vegetative incompatibility and myxomycete biology. Mycologia. 1984;76:785–792. [Google Scholar]
- 6.Boylan C J, Current W L. Improved rat model of Pneumocystis carinii pneumonia: induced laboratory infections in Pneumocystis-free animals. Infect Immun. 1992;60:1589–1597. doi: 10.1128/iai.60.4.1589-1597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, Cushion M T. Use of an ATP bioluminescent assay to evaluate viability of Pneumocystis carinii from rats. J Clin Microbiol. 1994;32:2791–2800. doi: 10.1128/jcm.32.11.2791-2800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cushion M T, Kaselis M, Stringer S L, Stringer J R. Genetic stability and diversity of Pneumocystis carinii infecting rat colonies. Infect Immun. 1993;61:4801–4813. doi: 10.1128/iai.61.11.4801-4813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cushion M T, Zhang J, Kaselis M, Giuntoli D, Stringer S L, Stringer J R. Evidence for two genetic variants of Pneumocystis carinii coinfecting laboratory rats. J Clin Microbiol. 1993;31:1217–1223. doi: 10.1128/jcm.31.5.1217-1223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cushion M T, Chen F, Kloepfer N. A cytotoxicity assay for evaluation of candidate anti-Pneumocystis agents. Antimicrob Agents Chemother. 1997;41:379–384. doi: 10.1128/aac.41.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cushion M T. Pneumocystis carinii. In: Balows A, Sussman M, editors. Topley and Wilson's microbiology and microbial infections. 9th ed. Vol. 4. London, England: Arnold Publishers; 1998. pp. 645–683. [Google Scholar]
- 12.Cushion M T. Genetic heterogeneity of rat-derived Pneumocystis. FEMS Immunol Med Microbiol. 1998;22:51–58. doi: 10.1111/j.1574-695X.1998.tb01186.x. [DOI] [PubMed] [Google Scholar]
- 13.Cushion M T, Orr S, Arnold J. Interactions between 2 Pneumocystis populations within the same host. J Eukaryot Microbiol. 1997;44:9S. doi: 10.1111/j.1550-7408.1997.tb05739.x. [DOI] [PubMed] [Google Scholar]
- 14.Edman J C, Kovacs J A, Masur H, Edman U. Ribosomal RNA sequences show Pneumocystis carinii to be a member of the fungi. Nature. 1988;334:519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- 15.Gigliotti F. Host species-specific antigenic variation of a mannosylated surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;165:329–336. doi: 10.1093/infdis/165.2.329. [DOI] [PubMed] [Google Scholar]
- 16.Gigliotti F, Harmsen A G, Haidaris C G, Haidaris P J. Pneumocystis carinii is not universally transmitted between mammalian species. Infect Immun. 1993;61:2886–2890. doi: 10.1128/iai.61.7.2886-2890.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glass N L, Kuldau G A. Mating type and vegetative incompatibility in filamentous ascomycetes. Annu Rev Phytopathol. 1992;30:201–224. doi: 10.1146/annurev.py.30.090192.001221. [DOI] [PubMed] [Google Scholar]
- 18.Harmsen A G, Chen W, Gigliotti F. Active immunity to Pneumocystis carinii reinfection in T-cell depleted mice. Infect Immun. 1995;63:2391–2395. doi: 10.1128/iai.63.7.2391-2395.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser P M, Blanc D S, Bille J, Francioli P. Typing methods to approach Pneumocystis carinii genetic heterogeneity. FEMS Immunol Med Microbiol. 1998;22:27–35. doi: 10.1111/j.1574-695X.1998.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 20.Hutchinson G E. An introduction to population ecology. New Haven, Conn: Yale University Press; 1979. [Google Scholar]
- 21.Jeffrey C. Biological nomenclature. 3rd ed. New York, N.Y: Edward Arnold; 1989. [Google Scholar]
- 22.Kaneshiro E S, Wyder M A, Wu Y-P, Cushion M T. Reliability of calcein acetoxymethylester and ethidium homodimer or propidium iodide for assessment of microbial viability. J Microbiol Methods. 1993;17:1–16. [Google Scholar]
- 23.Keely S P, Cushion M T, Stringer J R. Stability of four genetic loci in Pneumocystis carinii f. sp. carinii. J Eukaryot Microbiol. 1996;43:49S. doi: 10.1111/j.1550-7408.1996.tb04984.x. [DOI] [PubMed] [Google Scholar]
- 24.Keely S P, Stringer J R. Sequences of Pneumocystis carinii f. sp. hominis strains associated with recurrent pneumonia vary at multiple loci. J Clin Microbiol. 1997;35:2745–2747. doi: 10.1128/jcm.35.11.2745-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C H, Helweg-Larsen J, Tang X, Jin S, Li B, Bartlett M S, Lu J J, Lundgren B, Lundgren J D, Olsson M, Lucas S B, Roux P, Cargnel A, Atzori C, Matos O, Smith J W. Update on Pneumocystis carinii f. sp. hominis typing based on nucleotide sequence variations in internal transcribed spacer regions of rRNA genes. J Clin Microbiol. 1998;36:734–741. doi: 10.1128/jcm.36.3.734-741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J J, Bartlett M S, Smith J W, Lee C H. Typing of Pneumocystis carinii strains with type-specific oligonucleotide probes derived from nucleotide sequences of internal transcribed spacers of rRNA genes. J Clin Microbiol. 1995;33:2973–2977. doi: 10.1128/jcm.33.11.2973-2977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pixley F J, Wakefield A E, Banerji S, Hopkin J M. Mitochondrial gene sequences show fungal homology for Pneumocystis carinii. Mol Microbiol. 1991;5:1347–1351. doi: 10.1111/j.1365-2958.1991.tb00781.x. [DOI] [PubMed] [Google Scholar]
- 28.Pneumocystis Working Group. Nomenclature of Pneumocystis. J Eukaryot Microbiol. 1994;41:121–122. [Google Scholar]
- 29.Stringer S L, Stringer J R, Blase M A, Walzer P D, Cushion M T. Pneumocystis carinii: sequence from ribosomal RNA implies a close relationship with fungi. Exp Parasitol. 1989;68:450–461. doi: 10.1016/0014-4894(89)90130-6. [DOI] [PubMed] [Google Scholar]
- 30.Stringer J R. Pneumocystis carinii: what is it exactly? Clin Microbiol Rev. 1996;9:489–498. doi: 10.1128/cmr.9.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasquez J, Linke M J, Smulian A G, Cushion M T. Antigenic differences associated with genetically distinct Pneumocystis carinii. Infect Immun. 1996;64:290–297. doi: 10.1128/iai.64.1.290-297.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakefield A E. Genetic heterogeneity in Pneumocystis carinii: an introduction. FEMS Immunol Med Microbiol. 1998;22:5–14. doi: 10.1111/j.1574-695X.1998.tb01182.x. [DOI] [PubMed] [Google Scholar]
- 33.Walzer P D, LaBine M, Redington T J, Cushion M T. Lymphocyte changes during chronic administration of and withdrawal from corticosteroids: relation to Pneumocystis carinii pneumonia. J Immunol. 1984;133:2502–2508. [PubMed] [Google Scholar]
- 34.Weisbroth S H, Geistfeld J, Weisbroth S P, Williams B, Feldman S H, Linke M J, Orr S, Cushion M T. Latent Pneumocystis carinii infection in commercial rat colonies: comparison of inductive immunosuppressants plus histopathology, PCR, and serology as detection methods. J Clin Microbiol. 1999;37:1441–1446. doi: 10.1128/jcm.37.5.1441-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright T W, Gigliotti F, Finkelstein J N, McBride J T, An C L, Harmsen A G. Immune-mediated inflammation directly impairs pulmonary function, contributing to the pathogenesis of Pneumocystis carinii pneumonia. J Clin Investig. 1999;104:1307–1323. doi: 10.1172/JCI6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Cushion M T, Stringer J R. Molecular characterization of a novel repetitive element from Pneumocystis carinii from rats. J Clin Microbiol. 1993;31:244–248. doi: 10.1128/jcm.31.2.244-248.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]