Abstract
Cryptococcus neoformans is a pathogenic basidiomycete with a defined sexual cycle involving mating between haploid yeast cells with a transient diploid state. C. neoformans occurs in four predominant serotypes (A, B, C, and D), which represent different varieties or species. Rare clinical and environmental isolates with an unusual AD serotype have been reported and suggested to be diploid. We found by fluorescence-activated cell sorter analysis that serotype AD strains are aneuploid or diploid. PCR analysis with primers specific for serotype A or D alleles of the CNA1, CLA4, and GPA1 genes revealed that both alleles are often present in serotype AD strains. PCR analysis with primers specific for genes in the MATa or MATα mating-type loci revealed that serotype AD strains are heterozygous for the mating-type locus. Interestingly, in several serotype AD strains, the MATα locus was derived from the serotype D parent and the MATa locus was inherited from a serotype A parent that has been thought to be extinct. Basidiospores from a self-fertile serotype AD strain bearing the putative serotype A MATa locus showed a very low viability (∼5%), and no fertile serotype A MATa strain could be recovered. Serotype AD strains were virulent in a murine model. Hybrid AD strains could readily be isolated following a laboratory cross between a serotype A strain and a serotype D strain. In summary, serotype AD strains of C. neoformans are unusual aneuploid or diploid strains that result from matings between serotype A and D strains. Self-fertile isolates fail to undergo normal meiosis because of genetic divergence. Our findings further suggest that serotype A MATa strains may exist in nature.
The basidiomycete Cryptococcus neoformans is an opportunistic human fungal pathogen that causes cryptococcal meningitis predominantly in immunocompromised patients. A well-defined sexual life cycle (20–22, 25) and the establishment of various molecular biological tools make this organism an excellent model system for studies of fungal pathogenicity (8).
The life cycle of C. neoformans is characterized by a dimorphic transition between a haploid yeast form and a dikaryotic filamentous form (1). After a haploid cell senses a cell of opposite mating type (MATa or MATα), the cells produce filament-like structures called conjugation tubes that protrude toward the mating partner. After cell fusion, a dikaryotic mycelium is generated, composed of filaments characterized by unfused parental nuclei and fused clamp connections. Under proper environmental conditions, the tips of these filaments develop into swollen cells termed basidia. Within this new structure, karyogamy occurs followed by meiosis to generate four recombinant haploid nuclei. The haploid nuclei divide mitotically and bud off from the basidium to generate four long chains of basidiospores. These spores germinate and produce vegetative, yeast cells.
The diploid phase of C. neoformans is normally transient, and therefore most environmental and clinical isolates are haploid. Nevertheless, rare diploid isolates have been reported based on assays determining cellular DNA contents (33–35), the analysis of randomly amplified polymorphic DNA markers (4, 37), or isozyme analysis (5). Furthermore, we and others isolated diploid strains following defined genetic crosses (32, 38, 39). Interestingly, most of the environmental and clinical isolates that are thought to be diploid belong to the unusual class of serotype AD strains (5, 34).
C. neoformans has been classified into three varieties based in part on serological differences in capsular antigens. Serotype A and D strains each belong to a separate variety (varieties grubii and neoformans, respectively), whereas serotype B and C strains are classified as a single variety (variety gattii) (16, 23). A fifth, unusual serotype, AD, has also been described but is much less common than the four predominant serotypes. DNA sequence analysis has revealed that serotype B and C strains are phylogenetically much more closely related to each other than to A or D strains, which in turn are estimated to have diverged from each other approximately 18 million years ago (13, 42). Karyotype analysis revealed that the average chromosome number for C. neoformans is between 12 and 13 (28, 29, 41). The smallest chromosomes in variety gattii are 400 to 700 kb in size, whereas the smallest chromosomes of varieties neoformans and grubii are approximately 770 kb. Nucleotide sequence comparison of the URA5 gene revealed ∼8% sequence divergence between variety gattii and variety neoformans or grubii (7, 14) and ∼5% sequence divergence between varieties neoformans and grubii.
Here we have characterized strains of the unusual serotype AD class to establish their origin. By fluorescence-activated cell sorter (FACS) analysis we found that serotype AD strains are diploid or aneuploid (>1n but ≤2n). Second, by PCR analysis we showed that these strains are heterozygous for serotype A- and D-specific alleles and the MATa and MATα mating-type loci. Third, we found that three serotype AD strains were self-fertile and produced filaments, basidia, and basidiospores that germinated poorly (∼5%) to produce progeny that were still diploid or aneuploid. Our findings reveal that serotype AD strains are hybrids produced by crosses between serotype A and D parental strains and suggest that evolutionary divergence and sequence differences prevent proper chromosome segregation during meiosis in AD hybrid strains.
MATERIALS AND METHODS
Strains.
The C. neoformans strains used were clinical isolates ZG287, ZG290, MMRL774, and KW5 from the Duke Medical Center permanent strain collection, strains CBS132 and ATCC 48184 from the American Type Culture Collection, and strains CDC228, CDC304, CDC92-74, and CDC94-383 from the Centers for Disease Control and Prevention. The congenic serotype D laboratory strains JEC20 (MATa) and JEC21 (MATα) and the serotype A strain H99 (MATα) were used as haploid control strains. In addition, several auxotrophic serotype D strains were used in genetic crosses (JEC34 [MATa ura5-1], JEC43 [MATα ura5], JEC170 [MATα lys2 ade2], JEC171 [MATa lys2 ade2], and H99 [MATα 5-FOAr]). Strains CDC228-1 to CDC228-22 were isolated by dissecting basidiospores from the self-fertile strain CDC228.
Media.
Strains were grown and maintained on YPD or YNB medium; mating assays and confrontation assays were conducted on V8 or filamentation agar medium, respectively. For serotype analysis, several strains were grown in liquid Eagle's cell culture medium (Dulbecco's modification; Mediatech Cellgro) supplemented with 25 mM NaHCO3.
Serotyping.
Serotype analysis was performed using the Crypto Check serotyping kit from Iatron Laboratories (Tokyo, Japan). Strains were grown at room temperature on YPD plates for 48 h. Longer incubation times (up to 10 days) normally resulted in stronger agglutination reactions. A small amount of the growth was removed from the medium and resuspended in 50 μl of physiological saline (0.9%). A 5-μl volume of the cell suspension was mixed on a glass slide with 10 μl of each factor serum. After 2 min of gentle agitation, cell agglutination was visible, indicating positive reactions. In some cases, growth of the strains in a capsule-inducing cell culture medium (Eagle's) supplemented with 25 mM NaHCO3 increased the reactivity in serotype analysis. Strains were grown for 3 to 4 days at 30°C on a rotary incubator in 5 ml of liquid medium. The cells were pelleted, washed several times with physiological saline, and resuspended in 50 μl of saline, and antibody reactions were performed as described above. The patterns of agglutination results were interpreted as follows: serotype A strains reacted with antigenic factors 1 and 7, serotype D strains reacted with factors 1 and 8, and serotype AD strains reacted with factors 1, 7, and 8.
Mating assays, confrontation assays, and basidiospore dissection.
For mating reactions, strains were pregrown on YPD solid medium for 2 days. The cells were then mixed on V8 agar medium and incubated at 24°C for several days. Filament and basidiospore formation was assessed by light microscopy every other day. In C. neoformans, secretion of pheromone induces specific morphological changes in a mating partner of the opposite mating type. MATα cells exposed to a-pheromone respond by producing thin filament-like structures called conjugation tubes. In contrast, MATa cells exposed to α-pheromone produce fewer conjugation tubes and form unusual enlarged, round, refractile cells. The ability of C. neoformans strains to secrete pheromones or to respond to pheromones was determined by confrontation assays. In confrontation assays, two strains pregrown on YPD medium were streaked onto filament agar medium in two parallel thin lines as close as possible (1 to 2 mm) without touching. Pheromones secreted by one strain diffuse through the agar and induce morphological changes in the confronted strain. Conjugation tube formation and swelling of cells was assessed by light microscopy after 24 and 48 h of incubation at 30°C. To isolate basidiospores, the self-fertile strain CDC228 was grown on V8 medium as described for the mating reactions. Areas showing filament and basidiospore formation were excised from the agar plate, and the basidiospores were transferred to a fresh YPD plate by carefully touching the surface with the agar fragment. Spore dissection was then performed by micromanipulation.
Cell wall staining, nuclear staining, and FACS analysis.
Prior to staining, vegetative cells and filaments were washed with water and fixed in 70% ethanol at 4°C for at least 4 h. Cell wall staining of filaments was performed with Calcofluor White, a dye that is known to stain chitin-rich structures like primary septa. Fixed filaments were washed twice with deionized water and stained with a 1-mg/ml Calcofluor White (Difco) solution for 10 to 15 min. Stained filaments were analyzed for the fusion of clamp cells to the postapical cell, a structure typically associated with septa in filamentous basidiomycetes. To stain nuclei, fixed and washed cells/filaments were incubated with 0.5 mg of ethidium bromide per ml or 10 μg of propidium iodide per ml, two DNA-staining dyes, for 3 to 4 h at 37°C in the dark. Both staining solutions contained 1 mg of RNaseA per ml. Stained cells/filaments were analyzed by fluorescence microscopy to establish whether they were uni- or binucleate. FACS analysis was performed by the method of Tanaka et al. (35). Briefly, strains were grown for 48 h on YPD plates and a small number of the cells were removed, washed, and fixed as described above. The cells were washed twice with NS buffer (10 mM Tris-HCl [pH 7.2], 0.25 M sucrose, 1 mM EDTA, 1 mM MgCl2, 0.1 mM ZnCl2, 0.4 mM phenylmethylsulfonyl fluoride, 7 mM β-mercaptoethanol) and stained with propidium iodide (NS buffer supplemented with 10 μg of propidium iodide per ml and 1 mg of RNase A per ml) for 3 to 4 h at 37°C in the dark. At this point, the cells could be stored at 4°C in the dark for several hours. The cells were washed twice with 15 mM Tris-HCl (pH 8.0) and diluted to about 108 to 109 cells/ml. Before being subjected to FACS analysis, the cells were sonicated with a homogenizer for 10 s. At least 10,000 cells of each strain were analyzed for their DNA content determined by relative fluorescence of stained genomic DNA. The DNA content of diploid cells should display values of 2n and 4n with respect to the 1n and 2n DNA content of haploid strains. In contrast, aneuploid strains have DNA contents between the expected values for haploid and diploid strains.
PCR analysis.
The presence or absence of several known genes or alleles was tested by PCR analysis using gene- and allele-specific primer combinations (sequences are given in Table 1). Before using these primers in our analysis, we tested their specificity on 65 Cryptococcus strains of known serotype and/or mating type (45 serotype A MATα, 15 serotype D MATα, and 5 serotype D MATa strains). Very weak or no cross-reactivity between serotypes or mating types was observed for most primer combinations using 20 to 30 cycles of PCR and the optimal annealing temperature estimated by the overall nucleotide composition of each primer (A or T, 2°C each; G or C, 4°C each). In most cases, weak cross-reactivity could be eliminated by decreasing the PCR cycle numbers and/or increasing the annealing temperatures without the loss of strong, specific signals for the tester strains JEC20, JEC21, H99, and 125.91. Exceptions were the STE20α serotype D-specific primer pair, which showed some cross-reactivity with the serotype A MATα strain H99, and the STE20a serotype A-specific primer pair (see Results).
TABLE 1.
Primers and primer combinations used to identify serotype- and/or mating-type specific genes in serotype AD strains of C. neoformans
| Primer | Primer sequence | Gene or allele |
|---|---|---|
| JOHE1909 | CTCGAGGCTTTCCCCCTTTTT | MFα2 serotype D |
| JOHE1910 | ATTTGAAAAAGAGATCACAGC | |
| JOHE3067 | GATCTGTCTCAGCAGCCAC | STE20a serotype D |
| JOHE3068 | AATATCAGCTGCGCAGGTGA | |
| JOHE3069 | GATTTATCTCAGCAGCCACG | STE20α serotype D |
| JOHE3070 | AAATCGGCTACGGCACGTC | |
| JOHE5169 | TCCACTGGCAACCCTGCGAG | STE20a serotype A |
| JOHE5170 | ATCAGAGACAGAGGAGCAAGAC | |
| JOHE2472 | CCAAAAGCTGATGCTGTGGA | STE20α serotype A |
| JOHE2578 | AGGACATCTATAGCAGAT | STE20α serotype A |
| JOHE1895 | GGGCATATGAGCAGCTTTACACCAGCC | STE11α serotype D |
| JOHE1896 | GGTACCTTAGTTCAGCTTTGTGATTGC | |
| JOHE1671 | CTGAGGAATCTCAAACCAGGGA | STE12α serotype A+D |
| JOHE1672 | CCAGGGCATCTAGAAACAATCG | |
| JOHE1671 | As above | STE12α serotype A |
| JOHE2189 | GCTTCGGATCATACCTAAAA | |
| JOHE3065 | AGTCGGCTATTTCTTATCGTC | CLA4 serotype D |
| JOHE3066 | AATCTGCCCATCCAAACATTG | |
| JOHE3066 | As above | CLA4 serotype A |
| JOHE3236 | GGCTATTTATCAATGGTTAGCGG | |
| JOHE2926 | AAAAAGTTTTCGTTGGTCTTTT | CNA1 serotype D |
| JOHE3238 | CCTCCATCTCGACCTGCCT | |
| JOHE2926 | As above | CNA1 serotype A |
| JOHE3239 | ACTATTATCCTCCATCAACCTTC | |
| JOHE2596 | GCCAGAGAGATTCGATGTTG | GPA1 serotype D |
| JOHE3240 | TCCACCCCATTCATACCCG | |
| JOHE2596 | As above | GPA1 serotype A |
| JOHE3241 | CATCGCTCCACATCTTCGTT |
RESULTS
Confirming serotype AD strain identities by serotyping.
We obtained a collection of 10 strains that were previously reported to be serotype AD. Based on serum reactivity, 7 of the 10 strains that we investigated were in fact serotype AD (Table 2). Strain CDC92-74 typed as serotype D, and strain ZG287 was untypeable even after growth in Eagle's cell culture medium, which induces capsule production and usually facilitates serotyping. The stock strain CBS132, which is deposited as a holotype D strain at the American Type Culture Collection and the Centraalbureau voor Schimmelcultures but was typed as serotype AD by others (18, 19), typed as serotype A in our analysis. Changes in serotype from AD to A or D alone may result from instability of diploid strains during prolonged propagation, as has been suggested by Brandt et al. (5).
TABLE 2.
Fertility, serotype, and ploidy of serotype AD strains of C. neoformans
| Strain | MFα2 | STE11α | STE12α | STE20 D | CLA4 | GPA1 | CNA1 | FACS | serotype | Fertility |
|---|---|---|---|---|---|---|---|---|---|---|
| ZG287 | — | — | A | a | A | A+D | A | 1–2n | uta | Sterile |
| ZG290 | D | D | D | α | A+D | A | A | 2n | AD | Sterile |
| MMRL774 | — | — | A | a | A+D | A | A+D | 2n | AD | Sterile |
| KW5 | — | — | A | a | A | A+D | A+D | 2n | AD | Self (FA)a |
| ATCC 48184 | D | D | D | α | A+D | A+D | D | 2n | AD | Sterile |
| CBS132 | — | — | A | a | A+D | A | D | 2n | A | Sterile |
| CDC94-383 | — | — | — | a | A+D | A+D | A+D | 1–2n | AD | Sterile |
| CDC92-74 | D | D | D | α | A+D | A | A+D | 1–2n | D | Sterile |
| CDC228 | D | D | D | α | A+D | A+D | A+D | 2n | AD | Self |
| CDC304 | D | D | D | α | A+D | A+D | A+D | 1–2n | AD | Self |
self, self-fertile; FA, filament agar medium; ut, untypeable. —, not detectable.
Serotype AD strains are diploid or aneuploid by FACS analysis.
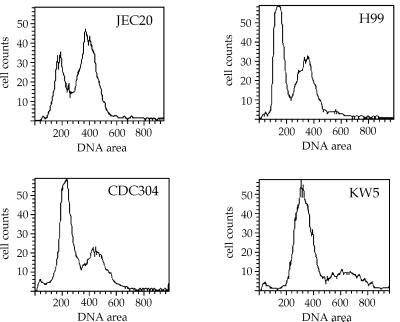
Fluorescence microscopy of propidium iodide-stained vegetative cells revealed that all 10 serotype AD isolates were uninucleate (data not shown). FACS analysis revealed that strains ZG290, MMRL774, KW5, CDC228, CBS132, and ATCC 48184 displayed DNA contents expected for diploid strains (Fig. 1; Table 2). For strains CBS132 and ATCC 48184, similar findings have been reported previously by Tanaka et al. (34). In contrast, strains ZG287, CDC92-74, CDC94-383, and CDC304 were aneuploid and had DNA contents intermediate between the known haploid tester strains H99, JEC20, and JEC21 and the expected and experimentally determined value for known diploid strains (32).
FIG. 1.
Serotype AD strains are aneuploid or diploid by flow cytometry. The haploid strains JEC20 (serotype D) and H99 (serotype A) and 10 serotype AD strains were subjected to DNA content analysis by FACS by the method of Tanaka et al. (35) (see Materials and Methods). In comparison to the haploid tester strains JEC20 and H99, six AD strains exhibited the DNA contents expected for diploid strains (strain KW5 is shown as an example) whereas four strains had DNA contents between 1n and 2n (shown for strain CDC304). For a summary, see Table 2. The DNA content of cells was measured by determining the relative fluorescence intensities.
PCR analysis reveals that serotype AD strains are diploid or aneuploid.
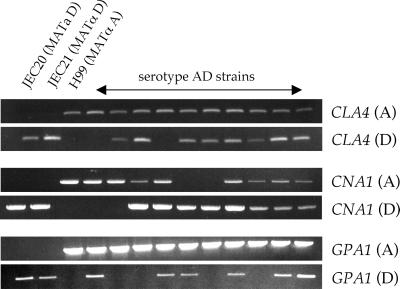
We further examined the ploidy of serotype AD strains using PCR to test for the presence or absence of serotype A- or D-specific alleles of the CLA4, GPA1 (2, 36), and CNA1 (12, 27) genes. As mentioned above, genes of serotype A and D vary in their nucleotide sequence by up to 5%. Based on this sequence divergence, we designed primers to specifically amplify the serotype A or D alleles of the CLA4, GPA1, and CNA1 genes. In general, one of the primers was nonspecific and hybridized to both the serotype A and D alleles while the other primer was designed to divergent regions of the two alleles to preferentially amplify only the serotype A or D allele. The CLA4 unique primers were designed to an exon-intron region showing a divergence of several base pairs in the two CLA4 alleles, and the differences were positioned toward the 3′ end of the primers. The CNA1 and GPA1 serotype A- and D-specific primers took advantage of small (4- to 9-bp) deletions in the promoter or nontranslated leader regions of these genes. As shown in Fig. 2 and summarized in Table 2, all of the analyzed strains were heterozygous for at least one of the CLA4, GPA1, or CNA1 loci. In particular, strains CDC94-383, CDC228, and CDC304 contained the serotype A and D alleles of all three genes. Several strains, including ZG287 and CBS132, exhibited loss of heterozygosity for at least one of the marker genes. Taken together, these findings based on PCR and FACS analysis indicate that many of the serotype AD isolates are diploid for many but not all loci in the genome.
FIG. 2.
Serotype AD strains of C. neoformans are heterozygous for the CLA4, CNA1, and GPA1 genes. Haploid serotype D strains JEC20 and JEC21, the haploid serotype A strain H99, and serotype AD strains (from left to right, ZG287, ZG290, MMRL774, KW5, ATCC48184, CBS132, CDC94-383, CDC92-74, CDC228, and CDC304) were analyzed by PCR for the presence or absence of serotype-specific alleles of the CLA4, CNA1, and GPA1 genes. For a summary, see Table 2.
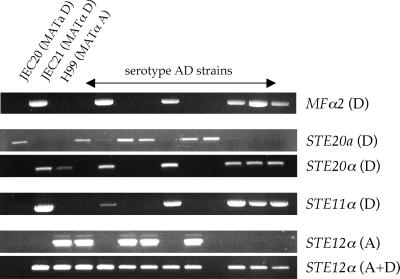
In addition to analyzing the unusual serotype AD strains for heterozygosity at the CLA4, CNA1, and GPA1 genes, we analyzed their genome for heterozygosity at the mating-type locus through the mating-type-specific genes MFα2, STE11α, STE12α (9, 40, 43), and STE20a/α. The mating-type locus was of particular interest because only MATα isolates of serotype A strains have been reported. Therefore, models in which serotype AD strains arose by crosses between serotype A and D strains would predict that the MATα locus was inherited from a serotype A strain. Four of the serotype AD strains tested clearly showed heterozygosity for the mating-type locus (Fig. 3). In these four strains (ZG287, MMRL774, KW5, and CBS132), the serotype D MATa locus and the serotype A MATα locus were present. One other strain, CDC94-383, carries the serotype D MATa locus and either lacks the corresponding chromosome containing the MATα locus or has a deletion in the mating-type locus (Fig. 3, STE12α; Table 2). Surprisingly, the remaining five strains (ZG290, ATCC 48184, CDC228, CDC304, and CDC92-74) inherited the MATα locus from the serotype D mating partner (Fig. 3). This finding implies that the MATa locus of these strains was inherited from a serotype A MATa parental strain, which has been thought to be extinct.
FIG. 3.
Serotype AD strains of C. neoformans are heterozygous at the mating-type locus. Haploid serotype D strains JEC20 and JEC21, the haploid serotype A strain H99, and serotype AD strains (from left to right, ZG287, ZG290, MMRL774, KW5, ATCC48184, CBS132, CDC94-383, CDC92-74, CDC228, and CDC304) were analyzed by PCR for the presence or absence of mating-type- and/or serotype-specific alleles of the MFα2, STE20, STE11α, and STE12α genes. For a summary, see Table 2.
The serotype A MATa locus is present in some serotype AD strains.
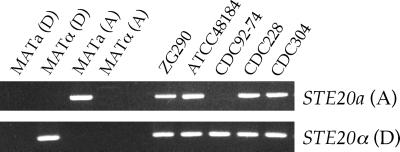
During the process of analyzing the MATa mating-type locus of serotype D in C. neoformans, we recently isolated a novel STE20a allele from a clinical serotype A isolate (K. B. Lengeler, P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman, submitted for publication). With primers specific for this novel serotype A STE20a allele, we found that four strains (ZG290, ATCC 48184, CDC228, and CDC304) contain at least part of the serotype A MATa locus, as was predicted based on the initial PCR analysis (Fig. 4). Because these primers showed weak cross-reactivity with some MATα strains of various serotypes, we cloned and sequenced the 867-bp PCR products. This analysis revealed that the sequences were ∼99% identical to the original STE20a serotype A allele. These findings further confirm that C. neoformans var. grubii MATa strains exist in nature and have intercrossed with C. neoformans var. neoformans MATα strains.
FIG. 4.
Several unusual serotype AD strains of C. neoformans have inherited the serotype A MATa mating-type locus. PCR analysis was conducted with genomic DNA isolated from strains JEC20 (MATa, serotype D), JEC21 (MATα, serotype D), 125.91 (MATa, serotype A), H99 (MATα, serotype A), and five unusual serotype AD strains using primers specific to the serotype D STE20α and serotype A STE20a genes. PCRs were run on separate agarose gels, and DNA was stained with ethidium bromide.
Mating and filamentation properties of serotype AD strains.
The diploid strains were further assessed by testing their ability to mate and filament. Of the 10 strains, 7 were completely sterile and showed no mating reaction with MATa or MATα strains (H99, JEC20, and JEC21), even after several weeks of incubation on V8 or filamentation agar mating medium (Table 2). In addition, no mating was observed with auxotrophic mating partners, which typically exhibit enhanced mating. The seven sterile serotype AD strains failed to respond to either MFa or MFα mating pheromone in confrontation assays and also failed to produce pheromones and induce conjugation tubes or haploid fruiting in MATa or MATα confronting cells. This finding suggests that pheromone expression may be repressed or defective in serotype AD strains.
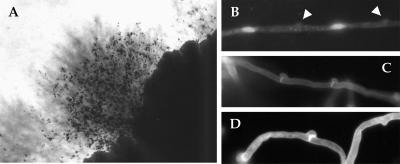
Three serotype AD strains (KW5, CDC228, and CDC304) were self-fertile and produced filaments and basidia when incubated alone on either V8 or filamentation agar medium (Fig. 5A). The filaments produced consisted of uninucleate cells (Fig. 5B), as previously observed with diploid strains produced by defined genetic crosses (32). That serotype AD strains produce uninucleate filament cells demonstrates that these strains are diploid and not heterokaryons. In contrast to congenic diploid strains but similar to mating reactions, filaments produced by the serotype AD strain CDC228 contained both unfused and a few fused clamp connections (Fig. 5C and D). The three self-fertile serotype AD strains produced no more filaments when confronted or coincubated with MATa or MATα cells and did not stimulate conjugation tube formation in either MATa or MATα cells. Thus, the self-fertile isolates are sterile and may not produce pheromones. Finally, the self-fertile strains produced filaments on YNB plates at 24°C but not at 37°C, indicating that these strains are thermally dimorphic, as we recently showed to be the case with congenic diploid strains of C. neoformans (32).
FIG. 5.
Self-fertile serotype AD strains are uninucleate and produce filaments with unfused and fused clamp connections. (A) Self-fertile strains CDC228 and KW5 were self-fertile on V8 medium and produced filaments and basidia after several days of incubation. (B) Staining with ethidium bromide revealed that filament cells are uninucleate. Clamp connections are indicated (arrowheads), determining cell boundaries. (C and D) In addition, Calcofluor White staining showed that clamp connections were mainly unfused (C), although a few fused clamp connections were also present (D).
Serotype AD strains are virulent in a murine model.
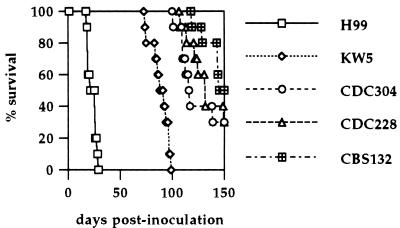
Most unusual serotype AD strains cited in the literature are clinical isolates. Nevertheless, we wanted to examine whether serotype AD isolates are virulent in a standard murine model. A total of 5 × 104 yeast cells of each of the self-fertile strains (KW5, CDC228, and CDC304), the holotype strain CBS132, and the well-characterized serotype A strain H99 were used to infect 10 female A/Jcr mice by nasal inhalation. Survival was monitored for 150 days postinoculation. We found that serotype AD strains were moderately virulent in comparison to the serotype A strain H99 (Fig. 6). Infection with the serotype A strain H99 resulted in 100% lethal infections by day 30 postinoculation. In contrast, all four serotype AD strains were less virulent. Strain KW5 resulted in 100% lethal infection by day 100, whereas strains CDC304, CDC228, and CBS132 caused 50% lethal infection by days 116, 132, and 145, respectively.
FIG. 6.
Serotype AD strains are virulent in a murine model. Ten female A/Jcr mice each were infected with 5 × 104 cells of one of the serotype AD strains CBS132, KW5, CDC228, and CDC304 or the serotype A strain H99 by nasal inhalation and monitored for survival postinoculation. The strains were pregrown in liquid YPD medium overnight at 30°C and washed with phosphate-buffered saline several times prior to infection.
Self-fertile strain CDC228 produces viable diploid spores.
Most genetic analysis with C. neoformans is performed with serotype D strains because of the existence of a congenic pair of MATa and MATα strains (17, 24). On the other hand, serotype A strains are often more virulent in patients and in animal model systems. One drawback to studies with serotype A strains is the lack of a congenic MATa strain. The identification of a MATa serotype A strain would therefore be of great importance since it would allow the generation of congenic MATa and MATα serotype A strains.
Our PCR analysis indicated that four of the serotype AD strains studied here inherited the MATa locus from a serotype A strain. One such strain, CDC228, is self-fertile, 2n by FACS analysis, and heterozygous for every gene analyzed by PCR. We took advantage of the self-fertile phenotype of the CDC228 strain in an attempt to isolate a haploid or aneuploid serotype A MATa strain. Of 428 dissected basidiospores, only 22 germinated to produce viable colonies for an overall survival rate of approximately 5.5%, which is considerably lower than the germination rate of basidiospores isolated from defined genetic crosses (>80%).
By PCR analysis, all 22 progeny strains were still heterozygous for the CLA4 and GPA1 serotype A and D alleles (Table 3). Most of the segregants still contained the serotype D MATα locus like the parental strain CDC228, although segregant CDC228-2 appeared to have a deletion in the STE20 gene. One segregant, CDC228-4, lacked the MATα locus from serotype D and may contain the desired serotype A MATa locus. Ten segregants were self-fertile on V8 plates, which fell into two different classes. Eight segregants were self-fertile. The remaining two segregants were self-fertile on V8 medium alone, but filament formation was inhibited by coincubation with MATa or MATα cells (JEC20 or JEC21, respectively). Two strains mated with both tester strains (bimaters) but did not produce filaments on their own. In addition, three strains were sterile, including the presumptive serotype A MATa strain CDC228-4. The remaining seven segregants mated as MATα cells (Table 3). Serotype analysis revealed that the presumptive serotype A MATa strain CDC228-4 was untypeable whereas five of the seven MATα strains reacted strongly with serum factors 1 and 8 and are therefore serotype D. Two MATα strains, CDC228-9 and CDC228-16 showed very weak reactivity with serum factor 7 and strong reactivity with factors 1 and 8, indicating that they are serotype AD. These findings suggest that serotype may be linked to the MAT locus.
TABLE 3.
Mating and ploidy analysis of 22 isolates derived from basidiospores of the self-fertile serotype AD strain CDC228
| Spore | CLA4 | GPA1 | STE20α | STE12α | MFα2 | Matinga
|
||
|---|---|---|---|---|---|---|---|---|
| JEC20 | JEC21 | Self | ||||||
| CDC228-1 | A+D | A+D | + | + | + | + | + | w |
| CDC228-2 | A+D | A+D | − | + | + | + | + | w |
| CDC228-3 | A+D | A+D | + | + | + | + | + | − |
| CDC228-4 | A+D | A+D | − | − | − | − | − | − |
| CDC228-5 | A+D | A+D | + | + | + | − | − | w |
| CDC228-6 | A+D | A+D | + | + | + | w | w | + |
| CDC228-7 | A+D | A+D | + | + | + | + | + | w |
| CDC228-8 | A+D | A+D | + | + | + | + | − | − |
| CDC228-9 | A+D | A+D | + | + | + | + | − | − |
| CDC228-10 | A+D | A+D | + | + | + | + | − | − |
| CDC228-11 | A+D | A+D | + | + | + | + | − | − |
| CDC228-12 | A+D | A+D | + | + | + | + | + | w |
| CDC228-13 | A+D | A+D | + | + | + | w | w | − |
| CDC228-14 | A+D | A+D | + | + | + | − | − | w |
| CDC228-15 | A+D | A+D | + | + | + | + | − | − |
| CDC228-16 | A+D | A+D | + | + | + | + | − | − |
| CDC228-17 | A+D | A+D | + | + | + | + | − | − |
| CDC228-18 | A+D | A+D | + | + | + | + | + | + |
| CDC228-19 | A+D | A+D | + | + | + | − | − | − |
| CDC228-20 | A+D | A+D | + | + | + | − | − | − |
| CDC228-21 | A+D | A+D | + | + | + | + | + | + |
| CDC228-22 | A+D | A+D | + | + | + | + | + | + |
| CDC228 | A+D | A+D | + | + | + | + | + | + |
w, weak filamentation; −, no filamentation; +, filamentation.
In summary, the serotype AD strain CDC228 sporulated to produce few viable spores, many of which were still diploid or aneuploid, self-fertile, and serotype AD, suggesting that meiosis does not occur properly in this strain.
AD hybrids result from defined laboratory crosses.
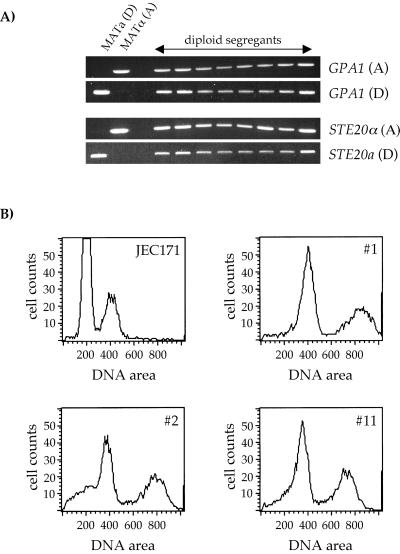
As mentioned above, serotype AD strains are thought to have arisen by crosses between serotype A and D strains in nature. We addressed this hypothesis by testing if AD hybrid strains can be generated by defined crosses in the laboratory. The serotype D strain JEC171 (MATa ade2 lys2) was crossed with a 5-fluoroorotic acid (5-FOA)-resistant mutant of the serotype A strain H99 (MATα), and recombinant, prototrophic basidiospores were selected on YNB medium. A total of 48 prototrophic segregants were than analyzed for filamentation on YNB medium at 24°C, and 46 isolates exhibited temperature-dependent dimorphism characteristic of diploid strains in C. neoformans (32). Yeast cells of 18 isolates, including the two nonfilamenting strains, were stained with propidium iodide to determine ploidy by FACS analysis. All 18 strains were uninucleate by fluorescence microscopy and aneuploid or near diploid by FACS analysis (Fig. 7B). The first DNA peaks of the AD hybrid strains were between the 1n and 2n DNA contents for the two haploid tester strains H99 (5-FOAr) and JEC171 (DNA area values, 200 and 400, respectively). Eight isolates were subjected to further analysis. By PCR, these eight strains were heterozygous and contained both the serotype A and D alleles of the common GPA1 gene and the mating-type-specific genes STE20α (serotype A) and STE20a (serotype D) (Fig. 7A). Surprisingly, by antibody reactions, none of these strains were serotype AD: six strains were serotype D, one strain was untypeable, and one strain even showed both B and D reactivity. Overall, our data show that hybrid AD strains can easily be generated in defined laboratory crosses. These AD hybrid strains are self-fertile, thermally dimorphic, and diploid by FACS and PCR analysis. In addition, the results obtained from serotyping the hybrid AD strains suggest that serotype D is dominant in C. neoformans.
FIG. 7.
AD diploid strains can be isolated following a defined genetic cross. Serotype A strain H99 (5-FOAr) was crossed to the serotype D strain JEC171 (MATa lys2 ade2) on V8 agar medium. A total of 48 recombinant prototrophic progeny were isolated by plating basidiospores on YNB agar medium. (A) The genomic DNA of eight progeny was isolated (isolates 1, 2, 3, 4, 11, 23, 28, and 40) and subjected to PCR analysis using primers to the serotype A- or D-specific alleles of the gene GPA1 or the mating-type- and serotype-specific alleles STE20a (serotype D) and STE20α (serotype A). Control DNA was derived from strains JEC20 (MATa, serotype D) and H99 (MATα, serotype A). (B) Yeast cells of 18 prototrophic isolates were stained with propidium iodide as described (see Materials and Methods) and analyzed by FACS. The results shown are for the haploid parental strain JEC171 and three progeny (isolates 1, 2, and 11). All 18 progeny showed two peaks in DNA content that correspond to 2n and 4n amounts of DNA compared to the 1n and 2n contents of the haploid parental strain JEC171.
DISCUSSION
Although C. neoformans is predominantly a haploid organism, diploid strains have been generated through defined genetic crosses (32, 38, 39). In addition, environmental and clinical isolates have been identified that show some characteristics of diploids, and most of these strains are serotype AD (4, 5, 33–35, 37). In this report we show that serotype AD strains are probably aneuploids or near diploids. First, by FACS analysis, 4 of 10 strains had total DNA contents in between those of haploid and diploid strains; the remaining six strains had near-diploid DNA contents. Second, by PCR analysis, serotype AD strains are heterozygous for many but not all genetic markers analyzed. Five strains that appeared diploid by FACS analysis carried only a serotype A or serotype D allele of at least one marker gene, which may reflect either chromosome loss or gene conversion. We conclude that some serotype AD isolates are aneuploid, and, based on our data, aneuploidy or diploidy is a consistent explanation for the other isolates.
We and others have recently shown that stable diploid strains can be isolated from defined genetic crosses. That environmental and clinical serotype AD isolates are aneuploids rather than true diploids is probably due to genome instabilities in C. neoformans. Franzot et al. (15) and Viviani et al. (37) have recently shown that C. neoformans can undergo phenotypic as well as genotypic changes during prolonged propagation and improper storage. One phenotype that shows high variability in serotype AD strains is serotype. This may be the reason why the stock strain CBS132 serotyped differently in our studies from in other studies. Genotypic changes in diploid cells may include structural chromosomal aberrations and even loss of whole chromosomes.
In addition, meiotic events may contribute to genome instability in C. neoformans. A high variability in karyotype was found among the progeny of a defined cross with respect to each other as well as in comparison to the parental strains (4). This may explain our finding that the progeny of the self-fertile diploid strain CDC228 were highly variable in genotype and phenotype. Interestingly, all of the progeny isolated were still heterozygous by PCR analysis. It therefore might be that, in contrast to the diploid strains generated by defined genetic crosses between congenic serotype D strains, serotype AD diploids are unable to properly execute meiosis. In addition to self-fertile or sterile isolates, several segregants mated as MATα cells and were serotype D. This implies that some serotype D MATα isolates found in nature might in fact be aneuploids instead of true haploid strains. Interestingly, Tolkacheva et al. found during the isolation of the G-protein α subunit GPA1 from C. neoformans that strain ATCC 42163 contained two different alleles of this gene (36). Strains ATCC 42163 (mating type α) and ATCC 42164 (mating type a) were both isolated from a self-fertile isolate (ATCC 34868) reported to be serotype A, but strains ATCC 42164 and ATCC 34868 both typed as serotype D in our studies. The presence of both mating types in the progeny of strain ATCC 34868 indicates that this strain probably is an aneuploid or diploid strain. This fact and the finding that two copies of GPA1 were present in strain ATCC 42163 suggest that the progeny of strain ATCC 34868 are still aneuploid or diploid. This clearly demonstrates that careful strain characterization of environmental and clinical isolates used in laboratory experiments is an important issue in C. neoformans research.
One hypothesis consistent with all the data is that serotype AD strains have arisen from crosses between haploid serotype A and D strains. The results from our PCR analysis provide further support for this hypothesis and show that AD strains contain both serotype A- and D-specific alleles for most markers tested. In addition, we found that hybrid AD strains could readily be generated following defined genetic crosses of serotype A and D strains. Hybrid AD strains were uninucleate, aneuploid, or diploid by FACS analysis and were heterozygous at the GPA1 gene locus as well as at the mating-type-specific STE20 gene locus. C. neoformans var. neoformans and grubii are thought to have diverged 10 to 20 million years ago and are known to have quite large differences in genome structure and sequence. Crosses between members of these varieties generate few viable spores, similar to the ratio of viable to inviable basidiospores we isolated from the self-fertile strain CDC228. Reduced spore viability is probably due to production of aneuploid spores as a result of improper alignment of chromosomes during meiosis, leading to defects in chromosome segregation or chromosome separation.
We have recently shown that congenic diploid strains are thermally dimorphic and grow as yeasts at 37°C yet spontaneously filament and sporulate at 24°C (32). Of 48 hybrid AD strains isolated following a defined cross between serotype A and D strains, 46 exhibited this thermally dimorphic phenotype; however, only 3 of 10 serotype AD strains that were clinical or environmental isolates were self-filamenting, and all 3 were thermally dimorphic. Because diploid and heterokaryon strains grow very slowly in the filamentous state, mutations or chromosome loss events that prevent filamentous growth may confer a selective growth advantage in serotype AD strains, explaining why most serotype AD strains are aneuploid and no longer self-fertile. This may also contribute to genome instability and plasticity in this lineage of C. neoformans.
Our findings suggest that serotype A MATa strains still exist. Several serotype AD diploid strains contained the serotype D MATα locus, implying that the MATa locus was inherited from the serotype A parent. In fact, we could directly demonstrate that four serotype AD strains contain the novel STE20a serotype A allele recently identified in our laboratory (Lengeler et al., submitted). This result further supports the idea that serotype A MATa strains still exist in nature. The finding that a putative serotype A MATa progeny from the self-fertile AD diploid strain CDC228 (CDC228-4) was sterile may explain why serotype A MATa strains have not been identified thus far. Studies are under way in our laboratory to identify fertile haploid serotype A MATa strains for use in the construction of congenic MATa and MATα serotype A strains.
Most serotype AD strains are clinical isolates. We tested several serotype AD strains in a murine virulence model and found that serotype AD strains are moderately virulent in comparison to the well-characterized serotype A strain H99. Further studies with defined parental A and D haploid strains and with hybrid AD strains are required to fully establish the effect of serotype AD and ploidy on virulence.
Finally, most epidemiological and molecular diagnostic studies with C. neoformans require the analysis of complex data generated either by multilocus enzyme electrophoresis (5, 6) or randomly amplified polymorphic DNA techniques (3, 6, 11, 26, 30, 31). In addition, these techniques are in general not able to distinguish between mating types, which have to be analyzed by time-consuming crosses in which many strains are sterile. Molecular analysis of mating-type loci in C. neoformans provides a new approach. Chaturvedi et al. recently reported that they were able to analyze the mating type, ploidy, and variety of C. neoformans strains based on the sequence divergence of C. neoformans within the mating pheromones (10). Our studies further demonstrate that the serotype, mating type, and ploidy of C. neoformans strains can be readily established by rapid PCR analysis. Further work on the mating-type loci in different varieties of C. neoformans will provide additional sequence information, which should enable the design of primers that are specific for all mating types and varieties present in C. neoformans.
ACKNOWLEDGMENTS
We thank Jianping Xu for strains, Rey A. Sia for assistance in the early stages of this project, and John Perfect for advice and encouragement. We also thank Carol Newlon and John McCusker for comments on the manuscript.
This work was supported in part by NIAID RO1 grants AI39115 and AI42159 to Joseph Heitman and PO1 grant AI44975 to the Duke University Mycology Research Unit. Gary M. Cox is a Burroughs Wellcome new investigator in Molecular Pathogenic Mycology. Joseph Heitman is a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alspaugh J A, Davidson R C, Heitman J. Morphogenesis of Cryptococcus neoformans. In: Ernst J F, Schmidt A, editors. Dimorphism in human pathogenic and apathogenic yeasts, vol. 5. Contributions in Microbiology. Switzerland: Basel; 2000. pp. 217–238. [DOI] [PubMed] [Google Scholar]
- 2.Alspaugh J A, Perfect J R, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki F H, Imai T, Tanaka R, Mikami Y, Taguchi H, Nishimura N F, Nishimura K, Miyaji M, Schreiber A Z, Branchini M L. New PCR primer pairs specific for Cryptococcus neoformans serotype A or B prepared on the basis of random amplified polymorphic DNA fingerprint pattern analyses. J Clin Microbiol. 1999;37:315–320. doi: 10.1128/jcm.37.2.315-320.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boekhout T, Belkum A V. Variability of karyotypes and RAPD types in genetically related strains of Cryptococcus neoformans. Curr Genet. 1997;32:203–208. doi: 10.1007/s002940050267. [DOI] [PubMed] [Google Scholar]
- 5.Brandt M E, Bragg S L, Pinner R W. Multilocus enzyme typing of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2819–2823. doi: 10.1128/jcm.31.10.2819-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt M E, Hutwagner L C, Kuykendall R J, Pinner R W. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. The Cryptococcal Disease Active Surveillance Group. J Clin Microbiol. 1995;33:1890–1895. doi: 10.1128/jcm.33.7.1890-1895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadevall A, Freundlich L F, Marsh L, Scharff M D. Extensive allelic variation in Cryptococcus neoformans. J Clin Microbiol. 1992;30:1080–1084. doi: 10.1128/jcm.30.5.1080-1084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 9.Chang Y C, Wickes B L, Miller G F, Penoyer L A, Kwon-Chung K J. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J Exp Med. 2000;191:871–882. doi: 10.1084/jem.191.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaturvedi S, Rodeghier B, Fan J, McClelland C M, Wickes B L, Chaturvedi V. Direct PCR of Cryptococcus neoformans MATα and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J Clin Microbiol. 2000;38:2007–2009. doi: 10.1128/jcm.38.5.2007-2009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cogliati M, Allaria M, Tortorano A M, Viviani M A. Genotyping Cryptococcus neoformans var. neoformans with specific primers designed from PCR-fingerprinting bands sequenced using a modified PCR-based strategy. Med Mycol. 2000;38:97–103. doi: 10.1080/mmy.38.2.97.103. [DOI] [PubMed] [Google Scholar]
- 12.Cruz M C, Sia R A L, Olson M, Cox G M, Heitman J. Comparison of the roles of calcineurin in physiology and virulence in serotype D and serotype A strains of Cryptococcus neoformans. Infect Immun. 2000;68:982–985. doi: 10.1128/iai.68.2.982-985.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan M, Currie B P, Gutell R R, Ragan M A, Casadevall A. The 16S-like, 5.8S and 23S-like rRNAs of the two varieties of Cryptococcus neoformans: sequence, secondary structure, phylogenetic analysis and restriction fragment polymorphisms. J Med Vet Mycol. 1994;32:163–180. doi: 10.1080/02681219480000231. [DOI] [PubMed] [Google Scholar]
- 14.Franzot S P, Fries B C, Cleare W, Casadevall A. Genetic relationship between Cryptococcus neoformans var. neoformans strains of serotypes A and D. J Clin Microbiol. 1998;36:2200–2204. doi: 10.1128/jcm.36.8.2200-2204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franzot S P, Mukherjee J, Cherniak R, Chen L C, Hamdan J S, Casadevall A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franzot S P, Salkin I F, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heitman J, Allen B, Alspaugh J A, Kwon-Chung K J. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet Biol. 1999;28:1–5. doi: 10.1006/fgbi.1999.1155. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda R, Nishikawa A, Shinoda T, Fukazawa Y. Chemical characterization of capsular polysaccharide from Cryptococcus neoformans serotype A-D. Microbiol Immunol. 1985;29:981–991. doi: 10.1111/j.1348-0421.1985.tb02962.x. [DOI] [PubMed] [Google Scholar]
- 19.Kabasawa K, Itagaki H, Ikeda R, Shinoda T, Kagaya K, Fukazawa Y. Evaluation of a new method for identification of Cryptococcus neoformans which uses serologic tests aided by selected biological tests. J Clin Microbiol. 1991;29:2873–2876. doi: 10.1128/jcm.29.12.2873-2876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon-Chung K J. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:821–833. [PubMed] [Google Scholar]
- 21.Kwon-Chung K J. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- 22.Kwon-Chung K J. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:942–946. [PubMed] [Google Scholar]
- 23.Kwon-Chung K J, Bennett J E. Medical mycology. Malvern, Pa: Lea & Febiger; 1992. pp. 397–446. [Google Scholar]
- 24.Kwon-Chung K J, Edman J C, Wickes B L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon-Chung K J, Popkin T J. Ultrastructure of septal complex in Filobasidiella neoformans (Cryptococcus neoformans) J Bacteriol. 1976;126:524–528. doi: 10.1128/jb.126.1.524-528.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer W, Marszewska K, Amirmostofian M, Igreja R P, Hardtke C, Methling K, Viviani M A, Chindamporn A, Sukroongreung S, John M A, Ellis D H, Sorrell T C. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–1799. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1790::AID-ELPS1790>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Odom A, Muir S, Lim E, Toffaletti D L, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perfect J R, Ketabchi N, Cox G M, Ingram C W, Beiser C L. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol. 1993;31:3305–3309. doi: 10.1128/jcm.31.12.3305-3309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perfect J R, Magee B B, Magee P T. Separation of chromosomes of Cryptococcus neoformans by pulsed field gel electrophoresis. Infect Immun. 1989;57:2624–2627. doi: 10.1128/iai.57.9.2624-2627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiss E, Tanaka K, Bruker G, Chazalet V, Coleman D, Debeaupuis J P, Hanazawa R, Latge J P, Lortholary J, Makimura K, Morrison C J, Murayama S Y, Naoe S, Paris S, Sarfati J, Shibuya K, Sullivan D, Uchida K, Yamaguchi H. Molecular diagnosis and epidemiology of fungal infections. Med Mycol. 1998;36:249–257. [PubMed] [Google Scholar]
- 31.Ruma P, Chen S C, Sorrell T C, Brownlee A G. Characterization of Cryptococcus neoformans by random DNA amplification. Lett Appl Microbiol. 1996;23:312–316. doi: 10.1111/j.1472-765x.1996.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 32.Sia R A, Lengeler K B, Heitman J. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol. 2000;29:153–163. doi: 10.1006/fgbi.2000.1192. [DOI] [PubMed] [Google Scholar]
- 33.Takeo K, Tanaka R, Taguchi H, Nishimura K. Analysis of ploidy and sexual characteristics of natural isolates of Cryptococcus neoformans. Can J Microbiol. 1993;39:958–963. doi: 10.1139/m93-144. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka R, Nishimura K, Miyaji M. Ploidy of serotype AD strains of Cryptococcus neoformans. J Med Mycol. 1999;40:31–34. doi: 10.3314/jjmm.40.31. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka R, Taguchi H, Takeo K, Miyaji M, Nishimura K. Determination of ploidy in Cryptococcus neoformans by flow cytometry. J Med Vet Mycol. 1996;34:299–301. [PubMed] [Google Scholar]
- 36.Tolkacheva T, McNamara P, Piekarz E, Courchesne W. Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein α-subunit homolog. Infect Immun. 1994;62:2849–2856. doi: 10.1128/iai.62.7.2849-2856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viviani M A, Wen H, Roverselli A, Caldarelli-Stefano R, Cogliate M, Ferrante P, Tortorano A M. Identification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD. J Med Vet Mycol. 1997;35:355–360. [PubMed] [Google Scholar]
- 38.Whelan W L, Kwon-Chung K J. Genetic complementation in Cryptococcus neoformans. J Bacteriol. 1986;166:924–929. doi: 10.1128/jb.166.3.924-929.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White C W, Jacobson E S. Occurrence of diploid strains of Cryptococcus neoformans. J Bacteriol. 1985;161:1231–1232. doi: 10.1128/jb.161.3.1231-1232.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickes B L, Edman U, Edman J C. The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- 41.Wickes B L, Moore T D E, Kwon-Chung K J. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology. 1994;140:543–550. doi: 10.1099/00221287-140-3-543. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Vilgalys R J, Mitchell T G. Multiple gene genealogies reveal dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol Ecol. 2000;9:1471–1482. doi: 10.1046/j.1365-294x.2000.01021.x. [DOI] [PubMed] [Google Scholar]
- 43.Yue C, Cavallo L M, Alspaugh J A, Wang P, Cox G M, Perfect J R, Heitman J. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]