Abstract
More than 100 chemical modifications of RNA, termed the epitranscriptome, have been described, most of which occur in prokaryotic and eukaryotic ribosomal, transfer, and noncoding RNA and eukaryotic messenger RNA. DNA and RNA viruses can modify their RNA either directly via genome‐encoded enzymes or by hijacking the host enzymatic machinery. Among the many RNA modifications described to date, four play particularly important roles in promoting viral infection by facilitating viral gene expression and replication and by enabling escape from the host innate immune response. Here, we discuss our current understanding of the mechanisms by which the RNA modifications such as N6‐methyladenosine (m6A), N6,2′‐O‐dimethyladenosine (m6Am), 5‐methylcytidine (m5C), N4‐acetylcytidine (ac4C), and 2′‐O‐methylation (Nm) promote viral replication and/or suppress recognition by innate sensors and downstream activation of the host antiviral response.
This article is categorized under:
RNA in Disease and Development > RNA in Disease
RNA Structure and Dynamics > Influence of RNA Structure in Biological Systems
RNA Evolution and Genomics > RNA and Ribonucleoprotein Evolution
Keywords: immune stimulation, m6A RNA methylation, RNA modifications, viral pathogenesis
Viral RNA modifications and regulation of innate immune sensing.

1. INTRODUCTION
Macromolecules such as proteins, DNA, and RNA bear covalent modifications that regulates their expression and function in a highly dynamic manner. In particular, more than 100 chemical modifications of RNA have been described, most of which affect ribosomal and transfer RNA. In recent years, technological advances in qualitative and quantitative analytical methods have enabled the identification of many less abundant modifications in messenger RNA (mRNA). Although the mechanisms by which some RNA modifications affect biological processes are gradually becoming clearer, little is known about the functions of RNA modifications in the majority of physiological and pathological contexts. Intense research interest in this topic has led to a newly emerging field termed epitranscriptomics (Barbieri & Kouzarides, 2020; Li & Mason, 2014; Roundtree et al., 2017).
The innate immune system has a well characterized role as the first line of defense against invading pathogens, including viruses, bacteria, and parasites. Eukaryotic cells have evolved sensing systems to enable early detection of microbial infection followed by rapid epigenetic and transcriptional inflammatory responses while simultaneously avoiding activation by self‐molecules. The predominant microbe‐specific sensing mechanisms involve recognition of unique molecular components known as pathogen‐associated molecular patterns (PAMPs) by families of cell‐surface and intracellular pattern recognition receptors (PRRs) expressed by cells provoking innate immune response. Upon engagement by PRRs, PAMPs such as microbe‐specific lipoproteins, glycoproteins, and nucleic acids initiate a cascade of downstream signaling events that culminate in proinflammatory gene expression (Jensen & Thomsen, 2012). While all microbes have evolved strategies to evade recognition by the innate immune sensing system, we focus this review on the host–virus interaction.
Covalent modification of RNA is increasingly recognized to be a crucial mechanism by which viruses escape recognition by PRRs and/or interfere with PRR‐activated signaling pathways. Many of the same RNA modifications detected in eukaryotes have been found in genomic and transcribed viral RNA; for example, 2′‐O‐methylation (Nm), N6‐methyladenosine (m6A), 5‐methylcytidine (m5C), and N4‐acetylcytidine (ac4C) have been reported to regulate viral gene expression and replication (Tsai & Cullen, 2020); and m6A and pseudouridine, formed by 5′ ribosylation of uracil, have been demonstrated to significantly impede RNA sensing and pathway activation in innate immune cells (Durbin et al., 2016; Kariko et al., 2005). An increasing body of evidence suggests that modification of both viral RNA and host cellular RNA can greatly influence viral replication and the host innate immune response. Given the global public health threat posed by viral infections for which we have few effective therapies, there is a clear need to increase our understanding of viral RNA modifications and their impact on the virus–host interaction.
In this review, we summarize our current knowledge of the main viral RNA modifications and their effects on PAMP recognition and the innate immune response as well as on viral replication and escape. Consistent with the relative abundance of the literature on Nm and m6A, the review focuses mainly on these two modifications, with shorter sections on other viral RNA modifications. We include examples from a variety of virus families, including those with established global medical importance.
2. INNATE IMMUNE MECHANISMS FOR SENSING VIRAL INFECTION
Cells of the innate immune system, which classically include macrophages, natural killer cells (NKs), dendritic cells (DCs), granulocytes, and other cell types, express families of PRRs that detect PAMPs that commonly occur in microbes. In the case of viral infections, the host PRRs recognize single‐stranded (ss) or double‐stranded (ds) RNA, dsDNA, viral coat proteins, and glycosylated proteins as PAMPs. PRR engagement leads to activation of various signaling pathways that result in transcription of inflammation‐associated cytokines/chemokines, including type I interferons (IFNα/β), which, in turn, trigger activation of the Janus kinase (JAK)‐signal transducer and activator of transcription (STAT) signaling pathway followed by transcription of interferon‐stimulated genes (ISGs). ISGs include an array of antiviral effectors that amplify the innate immune response. Interferon regulatory factor 3 (IRF3), IRF7, NF‐κB, and AP‐1 are the crucial transcription factors associated with expression of IFNs, ISGs, and other proinflammatory genes (Akira et al., 2006; Jensen & Thomsen, 2012; Schneider et al., 2014).
In humans, the major viral‐sensing PRRs are the Toll‐like receptors (TLRs), the retinoic acid‐inducible gene I (RIG‐I)‐like receptors (RLRs), and the nucleotide‐binding oligomerization domain‐containing (NOD)‐like receptors (NLRs). Viral invasion is also sensed by nonreceptor proteins such as cyclic GMP–AMP synthase, a cytosolic DNA sensor; and 2′,5′‐oligoadenylate synthetase (OAS) and the downstream RNase L, which are ISGs that bind to and cleave viral RNA. Notably, these various sensing systems have different subcellular localizations and expression patterns that support efficient detection of multiple viral families.
2.1. Toll‐like receptors
TLRs are a family of noncatalytic cell‐surface and intracellular PRRs. To date, 10 members have been identified in the human genome (TLR1–10) and an additional 3 TLRs are expressed in mice. Five TLRs are known to be involved in viral recognition: TLR7 and TLR8 are endosomal receptors for ssRNA; TLR3 is an endosomal receptor for dsRNA, which can be derived directly from the viral genome of dsRNA viruses or as replication intermediates of ssRNA viruses; and TLR2 and TLR4 are cell‐surface receptors that recognize viral envelope glycoproteins. All of the TLRs are transmembrane glycoproteins and contain N‐terminal PAMP recognition regions and C‐terminal Toll/IL‐1R homology (TIR) domains, which bind to a variety of adaptor molecules to propagate downstream signaling (Akira et al., 2001; Akira & Takeda, 2004; Jensen & Thomsen, 2012).
2.1.1. TLR7 and TLR8
TLR7 is expressed by DCs and B cells, while TLR8 is expressed by DCs and monocytes. Both receptors recognize ssRNA from viruses that invade host cells by endocytosis; these include influenza A virus (IAV), vesicular stomatitis virus, dengue virus (DENV), Sendai virus, and human immunodeficiency virus (HIV) (Diebold et al., 2004; Heil et al., 2004; Jurk et al., 2002; Kane et al., 2011; Wang et al., 2006). Ligand binding induces association of the TLR TIR domain with the adaptor protein myeloid differentiation primary response gene 88 (MyD88), which then recruits two interleukin‐1 receptor‐associated kinases, IRAK‐4 and IRAK‐1. IRAK‐1 is phosphorylated by IRAK‐4 and binds to TRAF6 (tumor necrosis factor [TNF] receptor‐associated factor 6), which contains ubiquitin ligase activity. The IRAK‐1/TRAF6 complex then dissociates from TLR7/8 and TRAF6 polyubiquitinates tumor growth factor beta‐activated kinase 1 (TAK1) and inhibitor of nuclear factor kappa B (IκB) kinase‐γ (IKKγ). Activated IKKγ or TAK1 then phosphorylates and activates IKKβ, which phosphorylates and degrades IκB, allowing the release of the transcriptionally active subunits NF‐κB. NF‐κB can then translocate from the cytosol into the nucleus to initiate gene transcription. Concomitantly, TAK1 also phosphorylates and activates components of the mitogen‐activated protein kinase (MAPK) pathway, which results in translocation of the transcription factor AP‐1 to the nucleus and amplification of inflammatory gene transcription. IRAK1 also phosphorylates IRF7, which, together with IRF3, promotes IFN transcription (Akira & Takeda, 2004; Medzhitov et al., 1998).
2.1.2. TLR3
TLR3 is expressed by both immune and nonimmune cells, including macrophages, B cells, DC cells, NK cells, some epithelial cells, and fibroblasts. TLR3 recognizes dsRNA derived either from the genome of dsRNA viruses, such as the reoviruses, or as a replication intermediate of ssRNA viruses, such as coxsackievirus B3, respiratory syncytial virus (RSV), West Nile virus (WNV), poliovirus, and IAV (Daffis et al., 2008; Gowen et al., 2006; Guillot et al., 2005; Negishi et al., 2008; Oshiumi et al., 2011; Rudd et al., 2005; Wang et al., 2004). Engagement of TLR3 activates downstream signaling pathways that share some similarities with those triggered via TLR7/8 but also include some TLR3‐specific events. First, TLR3 dimerization recruits the adaptor protein Toll/IL‐1R homology domain‐containing adaptor protein inducing IFN‐β (TRIF), rather than MyD88. TRIF then interacts with TRAF6, resulting in the nuclear translocation of NF‐κB and AP‐1, as described above. Second, TRIF can also bind to TRAF3, which recruits IKKε and the kinase TRAF family member‐associated NF‐κB activator (TANK)‐binding kinase‐1 (TBK1). TBK1/IKKε then phosphorylate IRF3 and IRF7, and dimerized IRF3/IRF7 translocate to the nucleus to induce inflammatory cytokine expression (Kawai & Akira, 2006; Meylan et al., 2004).
2.2. Retinoic acid‐inducible gene I (RIG‐I)‐like receptors
RLRs are a three‐member family of cytosolic proteins composed of RIG‐I, melanoma differentiation‐associated antigen 5 (MDA5), and the lesser studied laboratory of genetics and physiology 2 (LGP2). RIG‐I and MDA5 have a central DExD/H‐box (aspartate–glutamate–any amino acid–aspartate/histidine) helicase domain, a C‐terminal domain (CTD), and tandem N‐terminal CARDs (caspase activation and recruitment domains). LGP2 contains the CTD and helicase domain but lacks CARD domains, and it thus acts as a modulator of RIG‐I and MDA5 activity. RLRs are also regulated by the OAS/RNase L sensor system. RIG‐I and MDA5 are expressed in most cell types except for plasmacytoid DCs, and their ability to recognize ss/dsRNA enables them to act as sensors for nearly all of the major virus families (Kang et al., 2002; Kato et al., 2005; Rehwinkel & Gack, 2020).
2.2.1. Retinoic acid‐inducible gene I
In its unbound form, RIG‐I exists in a closed conformation in which the CARD2 domain is bound to part of the central helicase domain. RIG‐I recognizes and is activated by viral RNAs that have three key features: a 5′‐triphosphate or 5′‐phosphate group (5′‐PPP or 5′‐PP), duplex formation with a complementary RNA, and an unmethylated nucleotide at the 5′‐cap (cap1/2). Upon viral RNA binding via the CTD, the RIG‐I helicase domain interacts with the RNA duplex structure, which displaces the CARD2 domain and exposes both CARDs to allow binding of the adaptor protein mitochondrial antiviral‐signaling protein (MAVS), also known as IFN‐β promoter stimulator I (Kato et al., 2005; Loo & Gale Jr., 2011; Rehwinkel & Gack, 2020). The interaction between RIG‐I and MAVS results in the recruitment of other signaling molecules, including TNF receptor‐associated death domain (TRADD), TRAF3, and TANK. In turn, these proteins activate TBK1 and IKKε, which then activate IRF3/IRF7 and induce transcription of IFNα/β and ISGs. The MAVS/TRADD complex can also recruit receptor‐interacting protein 1 (RIP1) and Fas‐associated death domain (FADD) to activate the IKKα/β/γ–IκB–NF‐κB cascade of events that results in increased inflammatory gene transcription (Figure 1) (Jensen & Thomsen, 2012; Loo & Gale Jr., 2011; Rehwinkel & Gack, 2020).
FIGURE 1.
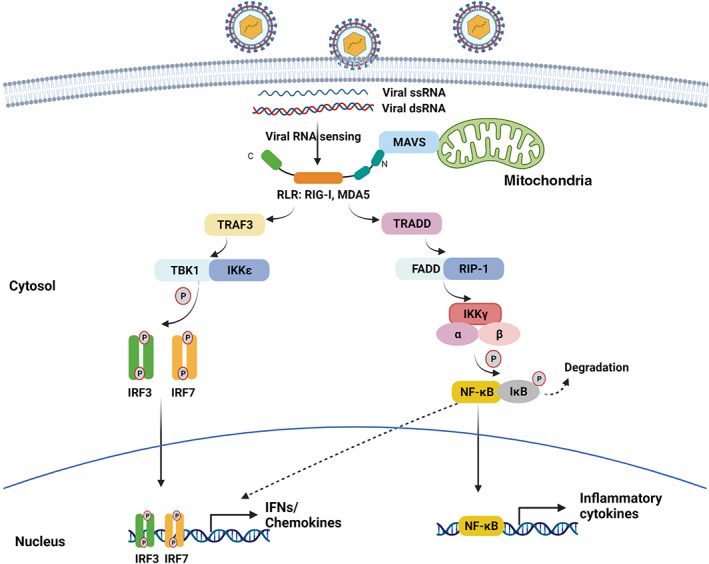
RIG‐I and MDA5 sensing of viral RNA and activation of downstream signaling pathways. Viral ssRNA and dsRNA are sensed by the RLR family members RIG‐I and MDA5. Upon receptor engagement, the N‐terminal CARD domains become exposed and recruit multiple signaling proteins, including the adaptor MAVS. The downstream signaling pathways culminate in activation of the transcription factors IRF3, IRF7, NF‐κB, and AP‐1, which promote transcription of IFNs and other inflammatory mediators
By virtue of its recognition criteria, RIG‐I can bind to multiple ssRNA viruses, including negative‐sense viruses (e.g., Paramyxoviridae, Rhabdoviridae, Orthomyxoviridae, Arenaviridae, Bunyaviridae, and Filoviridae) that form a duplex structure and retain a 5′‐PPP group during replication, as well as positive‐sense viruses (e.g., Flaviviridae, Coronaviridae, Enteroviridae, and Picornaviridae) that are recognized as RNA replication intermediates via their 5′‐PPP or 5′‐PP groups. Interestingly, hepatitis C virus (HCV) RNA is recognized by RIG‐I through both the 5′‐PPP group and a PAMP motif containing uridine‐rich sequence (Fredericksen et al., 2008; Kato et al., 2005; Kell & Gale Jr., 2015; Li et al., 2010; Li et al., 2021; Loo et al., 2008; Yoneyama et al., 2005; Zhou et al., 2010).
2.2.2. Melanoma differentiation‐associated antigen 5
MDA5 shares many structural features with RIG‐I, including the overall protein domain structure composed of tandem CARDs, a helicase domain, and a CTD, as well as recruitment of MAVS/TRAFs/IKKs and downstream IRF/IRF7/NF‐κB‐dependent transcriptional events. Among its unique features, MDA5 has a more open conformation than RIG‐I in the unbound state, and MDA5 recognizes the internal duplex structure of long dsRNAs, which contrasts with binding of RIG‐I to the 5′ terminus of short dsRNAs (Kato et al., 2006; Wu et al., 2013). MDA5 can reportedly recognize both ds/ssRNAs, but it preferentially binds to long dsRNAs lacking 2‐O‐methylated cap1 or two nucleotides. MDA5 is indispensable for the recognition of picornaviruses, such as encephalomyocarditis virus, Theiler's virus, and Mengo virus, and additionally recognizes some viruses also sensed by RIG‐I, including Flaviviridae, Coronaviridae, Picornaviridae, Paramyxoviridae, Reoviridae, and Rhabdoviridae (Faul et al., 2010; Fredericksen et al., 2008; Gitlin et al., 2006, 2010; Kato et al., 2006; Loo et al., 2008; Sen et al., 2011).
2.3. Other viral RNA sensors
OAS/RNase L are non‐PRR sensors that bind and cleave viral RNA to generate small duplex RNAs, which then activate RIG‐I and MDA5. The dsRNA‐dependent RNA‐activated protein kinase PKR is also considered to be a viral RNA sensor by virtue of its binding to dsRNA; moreover, PKR has been shown to play a role in modulating viral replication and the host IFN response (Balachandran et al., 2000; Balachandran & Barber, 2007; Malathi et al., 2010).
NLR family proteins contains N‐terminal caspase recruitment “C” (CARD) or pyrin “P” (PYD) domains, central NOD domain, and C‐terminal leucine‐rich repeat (LRR) motif. NLR proteins are also important viral sensors and induce formation of inflammasome signaling complexes. NLRs members NLRC1 and NLRC2 interact with receptor‐interacting protein kinase 2 (RIPK2) (also known as RICK or RIP2) and induce NF‐κB and mitogen‐activated protein kinase (MAPK) signaling. One of the most studied and well‐known roles for NLRs is activation of inflammasome. Inflammasome is a multiprotein complex which contains NLR family members such as NLR pyrin domain‐containing 3 (NLRP3), adaptor protein apoptosis‐associated speck‐like protein containing caspase recruitment domain (ASC), and the effector protein procaspase‐1. Although multiple viruses were reported to activate NLRs, until recently NLRP1 was reported to be a dsRNA sensor (Bauernfried et al., 2021).The mechanism of NLR activation and inflammasome formation upon viral infection is due to viral RNA sensing by TLRs, RLRs, and the NF‐κB‐mediated upregulation of pro‐IL‐1β and NLRP3; or viral RNA sensing by PKR (Jones et al., 2016; Lupfer & Kanneganti, 2013; Shrivastava et al., 2016). Inflammasomes play a dual role in viral defense by triggering a form of cell death known as pyroptosis and by promoting maturation of proinflammatory cytokines. NLR proteins bind to viral RNA through a C‐terminal leucine‐rich repeat (LRR) motif. NLRs can form several types of inflammasomes depending on the specific NLR sensor, but in general, they all contain the adaptor protein apoptosis‐associated speck‐like protein containing a CARD (ASC) and the effector protease caspase 1. Upon viral binding, NLRP3 interacts with the pyrin N‐terminal homology domain of ASC and procaspase‐1 to form the NLRP3 inflammasome, which promotes maturation of interleukin (IL)‐1β and IL‐18. NOD‐like receptor family, CARD‐containing 2 (NLRC2) also recognizes ssRNA of RSV, IAV, and parainfluenza virus, and activates MAVS‐dependent signaling for inflammatory gene expression (Geddes et al., 2009; Jensen & Thomsen, 2012; Wilmanski et al., 2008).
3. VIRAL RNA MODIFICATIONS THAT CONTRIBUTE TO EVASION OF HOST SENSOR RECOGNITION AND DOWNSTREAM SIGNALING
The strategies employed by viruses to evade the innate immune response can be broadly classified as either enabling escape from PRR recognition or disrupting downstream PRR‐triggered signaling. Upon viral invasion, viruses can hijack cellular components to form vesicle‐like structures, known as replication organelles, that contain the machinery required for replication of the viral genome. Such replication organelles or similar structures shield the viral replication intermediates from recognition by intracellular PRRs. In addition, a cap structure may be added to viral RNA to mimic the host mRNA, and some viruses encode endonucleases that cleave RNA during replication to avoid PRR recognition (Suthar et al., 2013). Viruses can also interfere with PRR‐activated signal transduction downstream of viral recognition. For example, rhinovirus 2A and 3C proteases cleave the adaptor protein MAVS, while the nonstructural protein NS1 of influenza A virus binds to the E3 ligase TRIM25 to block ubiquitination of lysine 63 in RIG‐I, which inhibits RIG‐I function. Many other viruses encode proteins which inhibit IFN production. SARS‐CoV ORF3b, ORF6, and N directly inhibit IFN transcription and production. ZIKV non‐structural proteins inhibit RIG‐I‐induced IFN‐β production by disrupting RIG‐I‐TBK1‐IRF3‐IFN cascade, in which NS1 and evolutionary NS1 mutation A188V, NS2A, NS2B, and NS4B reduce TBK1 phosphorylation; NS4A reduces IRF3 phosphorylation; and NS5 interacts with IRF3 (Kopecky‐Bromberg et al., 2007; Nelemans & Kikkert, 2019; Xia et al., 2018). More recently, it was reported that several severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) nonstructural proteins (NSP1, 3, 12, 13, 4), accessory proteins (ORF3, ORF6), and membrane (M) protein could inhibit IFN‐β promoter activation and production in infected cells (Lei et al., 2020). Not surprisingly, epitranscriptomic regulation of the viral genome has become an important topic of interest. It is now recognized that, in addition to modification of the cap structure, several internal viral RNA modifications also play a role in viral preventing recognition and clearance by the innate immune system (Durbin et al., 2016; Kariko et al., 2005).
3.1. 2′‐ O‐Methylation (Nm)
Nm refers to methylation of the 2′‐hydroxyl group of a ribose in any nucleotide and is catalyzed by a 2′‐O‐methyltransferase (2′‐O‐MTase). In viral RNA, Nm occurs at the 5′‐cap structure and internally, where it plays important roles in RNA stability as well as in sensing by TLRs and RLRs, especially MDA5. As noted above, RLRs discriminate between viral and self RNAs in part by the presence or absence of Nm at the first and second nucleotides adjacent to the cap (cap1/2). Viruses have evolved to evade RNA sensors by various capping mechanisms using their own or host capping machineries, and even by cap snatching from host mRNAs (Decroly et al., 2011). Similarly, some viruses add Nm to their RNA by encoding their own 2′‐O‐MTases while others use host enzymes (Table 1).
TABLE 1.
Effects of viral nm modification on viral replication and immune evasion
| Virus family/virus | Viral enzyme | Host enzyme | Nm location | Effect on viral replication | Effect on host immunity | References |
|---|---|---|---|---|---|---|
| Flaviviridae | ||||||
| WNV | NS5 | Cap1 and internal | Internal Nm: Reduces RNA elongation efficiency and translation, and viral replication | Cap Nm: Involved in innate immune evasion and viral replication (MND) | (Daffis et al., 2010; Szretter et al., 2012; Zhou et al., 2007) | |
| DENV | NS5 | Cap1 and internal | Cap Nm: Promotes replication; internal Nm reduces RNA elongation efficiency, translation, and replication | Cap Nm: Involved in innate immune evasion during early viral infection and replication (MND) | (Chang et al., 2016; Zust et al., 2013) | |
| JEV | NS5 | Cap1 | Promotes replication | Involved in innate immune evasion (MND) | (Li et al., 2013) | |
| ZIKV | NS5 | Cap1 | ND | ND | (Coloma et al., 2016; Zhao et al., 2017) | |
| Coronaviridae | ||||||
| SARS‐CoV | NSP16 | Cap1 | Promotes replication | Involved in innate immune evasion (MND) | (Menachery et al., 2014) | |
| SARS‐CoV‐2 | NSP16 | Cap1 | Promotes replication | Involved in innate immune evasion (need to validate) | (Viswanathan et al., 2021; Wilamowski et al., 2021) | |
| HCoV‐229E | NSP16 | Cap1 | Promotes replication | Involved in innate immune evasion (MND) | (Daffis et al., 2010; Zust et al., 2011) | |
| MHV‐A59 | NSP16 | Cap1 | Promotes replication | Involved in innate immune evasion (MND) | (Daffis et al., 2010; Zust et al., 2011) | |
| Mononegaviridae | ||||||
| Ebola virus | L | Cap1 and internal | ND | ND | (Martin et al., 2018) | |
| Poxviridae | ||||||
| VACV | VP39 | Cap1 | Promotes replication | Involved in innate immune evasion (MND) | (Daffis et al., 2010; Schnierle et al., 1992) | |
| Retroviridae | ||||||
| HIV | FTSJ3 | Internal | Promotes replication | Involved in innate immune evasion (MND) | (Ringeard et al., 2019) | |
Abbreviations: DENV, dengue virus; HCoV‐229E, human coronavirus 229E; HCV, hepatitis C virus; HIV, human immunodeficiency virus; JEV, Japanese encephalitis virus; MHV‐A59, murine hepatitis virus A59; MND, mechanism of immune evasion not determined; ND, not determined; SARS‐CoV (‐2), severe acute respiratory syndrome coronavirus (‐2); VACV, vaccinia virus; WNV, West Nile virus; ZIKV, Zika virus.
3.1.1. Flaviviridae
Flaviviruses are positive‐strand ssRNA viruses in which 2′‐O‐MTase activity is encoded by the highly conserved nonstructural NS5 protein (Egloff et al., 2002). Nm deposition occurs during cap1 formation and internally, where it reduces viral replication by interfering with RNA elongation efficiency and translation (Dong et al., 2012). The importance of Nm in flavivirus infection has been demonstrated in several studies in which NS5 was mutated. In WNV, for example, mutation of E218A in 2′‐O‐MTase abrogates 2′‐O‐methylation and formation of the RNA cap1 structure, which attenuates viral replication and enhances the innate immune response, and the effects are dependent or independent on Ifit1 in vivo in different studies (Daffis et al., 2010; Szretter et al., 2012; Zhou et al., 2007). Similarly, introduction of E216A into the 2′‐O‐MTase K61‐D146‐K180‐E216 motif, which is crucial for its activity, abrogates cap1 formation and attenuates DENV replication in vitro and in vivo. This mutant is also able to elicit a strong and rapid innate immune response (Chang et al., 2016; Zust et al., 2013). The MTase encoded by Japanese encephalitis virus (JEV) mediates both guanosine N7 methylation and the Nm modification, which were shown to be important for JEV immune evasion and activation of the IFN response in vitro and in vivo (Li et al., 2013). Finally, the MTase encoded by Zika virus (ZIKV) NS5 was reported to be highly similar to the JEV enzyme and to share the same methylation substrates (Coloma et al., 2016; Zhao et al., 2017).
3.1.2. Coronaviridae
Coronaviruses are positive‐strand ssRNA viruses in which 2′‐O‐MTase activity is encoded by NSP16. Not surprisingly, this gene is also highly conserved among the family members. Mutation of SARS‐CoV NSP16 at ORF1a/b attenuates replication of SARS‐CoV virus in vitro and in vivo, in part via promotion of MDA5‐mediated sensing and increased IFIT1 expression (Menachery et al., 2014). In SARS‐CoV‐2, a MTase encoded by NSP16 acts in concert with the cofactor encoded by NSP10 to generate Nm at the first nucleotide adjacent to the cap (Viswanathan et al., 2021; Wilamowski et al., 2021). A D129A mutation in NSP16 of human coronavirus (HCoV) strain 229E was shown to abrogate 2′‐O‐MTase activity, which both attenuated viral replication and amplified the production of IFNs possibly by MDA5 sensing. A similar scenario is induced by D130A mutation of NSP16 in mouse hepatitis virus (MHV) strain A59, where 2′‐O‐MTase activity was abrogated and IFNβ production was enhanced in an MDA5‐dependent manner in vitro and in vivo (Daffis et al., 2010; Zust et al., 2011).
3.1.3. Poxviridae
The Poxviridae family are large dsDNA viruses that replicate in the cytosol. The vaccinia virus protein VP39 is a 2′‐O‐MTase that methylates the 5′ cap of nascent RNA transcripts (Schnierle et al., 1992). A VP39 K175R mutation was shown to attenuate vaccinia replication in macrophages and to render the mutant more sensitive to IFIT2 overexpression (Daffis et al., 2010).
3.1.4. Filoviridae
This is a family of negative‐sense ssRNA viruses that include Ebola virus. The L (large) protein of Ebola virus catalyzes viral transcription and RNA capping. The C‐terminal conserved region VI of L protein is a 2′‐O‐MTase and catalyzes N7‐ and 2′‐O‐methylation of the cap structure as well as 2′‐O‐methylation of internal adenosines (Martin et al., 2018).
3.1.5. Retroviridae
The genomes of the retroviridae family of positive‐strand RNA viruses do not encode 2′‐O‐MTase activity, and they instead exploit host enzymes to perform this RNA modification. The transactivating response [TAR] RNA‐binding protein (TRBP) of HIV recruits the host enzyme FTSJ3 (FtsJ RNA 2′‐O‐methyltransferase 3), which mediates 2′‐O‐methylation of internal residues, mostly adenosines, in the HIV RNA genome. Depletion of FTSJ3 or mutation of the 2′‐O‐MTase KDKE motif leads to diminished 2′‐O‐methylation of HIV RNA, reduced viral replication, increased recognition by MDA5, and enhanced IFNα/β production (Ringeard et al., 2019). The study by Ringeard et al. was the first to report not only that the HIV genome is 2′‐O‐methylated via a host enzyme but also that 2′‐O methylation of internal residues, rather than cap1, is important to escape MDA5‐mediated recognition. Further work will be needed to determine whether this may be a common evasion mechanism for other retroviruses as well as other viral families. Other studies have shown that FTSJ3 knockdown increases RNA polymerase II‐mediated transcription of HIV‐1 and that HIV is also recognized by RIG‐I, suggesting that multiple evasion mechanisms may be employed by HIV (Li et al., 2020; Solis et al., 2011; Table 1).
3.2. N6‐methyladenosine (m6A)
m6A is the most abundant and best studied internal modification of eukaryotic mRNAs, and it has been shown to play crucial roles in multiple physiological and pathological processes. Viral RNA modification by m6A was first detected in the 1970s in several nuclear‐replicating viruses, including IAV, simian virus 40 (SV40), Rous sarcoma virus, avian sarcoma virus, and adenovirus (Dimock & Stoltzfus, 1977; Kane & Beemon, 1985; Krug et al., 1976; Lavi & Shatkin, 1975; Sommer et al., 1976). In recent years, several innovative methods have been introduced that to facilitate m6A detection in RNA; these include m6A RNA immunoprecipitation followed by high‐throughput sequencing (MeRIP‐seq), m6A individual‐nucleotide‐resolution cross‐linking and immunoprecipitation (miCLIP), and photocrosslinking‐assisted m6A sequencing (PA‐m6A‐seq). These techniques allow the location and distribution of viral RNA m6A to be analyzed in detail. m6‐modified adenosines are usually located within a DRACH (D = G/A/U, R = G/A, H = A/U/C) motif, or minimally RAC, where R is a purine for both viral and host RNAs. We and others showed that m6A located in HIV RNA regulates viral nuclear transport, protein synthesis, and replication (Kennedy et al., 2016; Lichinchi, Gao, et al., 2016; Tirumuru et al., 2016); and that m6A is also present in cytoplasm‐replicating viruses such as the Flaviviridae ZIKV, DENV, WNV, yellow fever virus, and HCV (Gokhale et al., 2016; Lichinchi & Rana, 2019; Lichinchi, Zhao, et al., 2016). More recently, m6A was identified in SARS‐CoV‐2 RNA, where it regulates recognition of the virus by RIG‐I (Li et al., 2021). Effects of viral m6A modification on virus replication and immune evasion are summarized in Table 2. Unlike 2′‐O‐MTase, the m6A‐modifying machinery is not encoded within the viral genome. Instead, both the presence and function of viral m6A are solely dependent on host MTases (writers: METTL3/14 complex), demethylases (erasers: ALKBH5, FTO), and adaptors (readers: YTHDF and YTHDC proteins).
TABLE 2.
Effects of viral m6A modification on virus replication and immune evasion
| Virus family | Host writer | Host eraser | Host reader | Effect on viral replication | Effect on host immunity | References |
|---|---|---|---|---|---|---|
| Flaviviridae | ||||||
| HCV | METTL3/14 | FTO | YTHDF1–3 | Host m6A, writers, and readers prevent viral particle production | Viral m6A prevents RIG‐I‐mediated innate sensing and activation | (Gokhale et al., 2016; Kim et al., 2020) |
| DENV, WNV, YFV | ND | ND | ND | ND | ND | (Gokhale et al., 2016) |
| ZIKV | METTL3/14 | ALKBH5/FTO | YTHDF1–3 | Host m6A, writers, and readers prevent viral particle production | ND | (Gokhale et al., 2016; Lichinchi, Zhao, et al., 2016) |
| Coronaviridae | ||||||
| SARS‐CoV‐2 | METTL3/14 | ND | ND | Host m6A and writers promote viral replication | Viral m6A prevents RIG‐I‐mediated innate sensing and activation | (Li et al., 2021; Liu et al., 2021) |
| Picornaviridae | ||||||
| EV71 | METTL3 | FTO | YTHDF1–3 | Host m6A and writer promote viral replication | ND | (Hao et al., 2019) |
| Paramyxoviridae | ||||||
| RSV | METTL3/14 | ALKBH5/FTO | YTHDF1–3 | Host m6A, writers, and readers promote viral replication | ND | (Xue et al., 2019) |
| Orthomyxoviridae | ||||||
| IAV | METTL3/14 | ND | YTHDF1–3 | Host m6A, writers, and readers promote viral gene expression and replication | ND | (Courtney et al., 2017; Krug et al., 1976) |
| Pneumoviridae | ||||||
| HMPV | METTL3/14 | ALKBH5 | YTHDF1–3 | Host m6A, writers, and readers increase viral gene expression and replication | Viral m6A prevents RIG‐I‐mediated innate sensing and activation | (Lu et al., 2020) |
| Retroviridae | ||||||
| HIV | METTL3/14 | ALKBH5/FTO | YTHDF1–3 | Host m6A and writers promote viral gene expression, nuclear export, and replication; readers promote/inhibit replication | Viral m6A prevents RIG‐I‐mediated innate sensing and activation | (Chen et al., 2021; Kennedy et al., 2016; Lichinchi, Gao, et al., 2016; Lu et al., 2018; Tirumuru et al., 2016) |
| MLV | ND | ND | YTHDF2 | Host m6A and reader increase viral replication | ND | (Bondurant et al., 1976; Courtney, Chalem, et al., 2019) |
| RoSV, FeLV | ND | ND | ND | ND | ND | (Furuichi et al., 1975; Kane & Beemon, 1985; Thomason et al., 1976) |
| Polyomaviridae | ||||||
| SV40 | METTL3 | ND | YTHDF2–3 | Host m6A, writer, and readers increase viral gene expression and replication | ND | (Finkel & Groner, 1983; Tsai et al., 2018) |
| Adenoviridae | ||||||
| Ad‐2 | METTL3 | ND | ND | Host m6A and writer promote viral transcript splicing. Depletion of METTL3 reduced viral late genes Hexon, Penton, and Fiber expression and infectious progeny production. | ND | (Hashimoto & Green, 1976; Price et al., 2020) |
| Herpesviridae | ||||||
| HSV‐1 | METTL3/14 | ALKBH5/FTO | YTHDF1–3 | Host m6A, writers, and readers promote viral replication | ND | (Moss et al., 1977; Zhuoying Feng et al., 2021) |
| KSHV | METTL3 | ND | YTHDF2 | Host m6A, writer, and reader promote/inhibit viral replication (cell line dependent) | ND | (Hesser et al., 2018; Tan et al., 2018) |
| EBV | METTL3/14 | ALKBH5 | YTHDF1–3 | Host m6A, writer, and readers promote latent viral gene expression but suppress lytic viral transcript expression and replication | N/D | (Lang et al., 2019; Xia et al., 2021) |
| Hepadnaviridae | ||||||
| HBV | METTL3/14 | FTO | YTHDF2–3 | Host m6A, writer, and reader reduce viral protein expression and pgRNA stability but increase reverse transcription | Viral m6A prevents RIG‐I‐mediated innate sensing and activation | (Imam et al., 2018; Kim et al., 2020) |
Abbreviations: Ad2, adenovirus‐2; DENV, dengue virus; EBV, Epstein–Barr virus; EV71, enterovirus 71; FeLV, feline leukemia virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HMPV, human metapneumovirus; HSV‐1, herpes simplex virus‐1; IAV, influenza A virus; KSHV, Kaposi's sarcoma‐associated herpesvirus; MLV, murine leukemia virus; ND, not determined; RoSV, Rous sarcoma virus' RSV, respiratory syncytial virus; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SV40, simian virus 40; WNV, West Nile virus; YFV, yellow fever virus; ZIKV, Zika virus.
3.2.1. Flaviviridae
HCV genome contains m6A modification, while host m6A methyltransferases METTL3/14 and reader proteins YTHDF1‐3 negatively regulate HCV virion production, demethylase FTO has the opposite function. Mutation of m6A sites in the E1 region of the HCV genome, which encodes envelope glycoprotein, increases the interaction between the viral RNA and the HCV structural protein Core protein, which promotes viral particle production (Gokhale et al., 2016). m6A modification of HCV genome also inhibits RIG‐I binding and the antiviral response. Knockdown of host METTL3/14 or YTHDF2/3 increases IFN‐β production and IRF3 phosphorylation (Kim et al., 2020), consistent with a negative immune regulatory role for m6A. m6A modification of ZIKV RNA inhibits viral production; specifically, knockdown of METTL3/14 or YTHDF1‐3 increased ZIKV replication while knockdown of the demethylases ALKBH5 and FTO had the opposite effect (Gokhale et al., 2016; Lichinchi, Zhao, et al., 2016).
3.2.2. Coronaviridae
We recently showed that m6A plays an important role in SARS‐CoV‐2 replication and the host response to the virus. Notably, METLL3 knockdown reduces m6A levels in both viral and host cellular RNA, promotes the production of inflammatory cytokines and chemokines, and METTL3 knockdown reduces viral replication in human Caco‐2 cells.
Consistent with this, either METTL3 depletion or mutagenesis of m6A modified sites in SARS‐CoV‐2 enhanced its recognition by RIG‐I and signaling for NF‐κB activation (Li et al., 2021). The finding that m6A is involved in regulating SARS‐CoV‐2 replication has been independently confirmed by others (Burgess et al., 2021; Liu et al., 2021; Zhang, Hao, et al., 2021).
3.2.3. Picornaviridae
This family consists of positive‐strand RNA viruses. m6A has been detected in the enterovirus type 71 (EV71) genome and shown to play a positive role in viral replication. Specifically, METTL3 knockdown or mutation of m6A sites reduces viral replication, while FTO knockdown has the opposite effect. The role of YTHDF1–3 in EV72 viral replication is less clear, in that the precise effects of protein knockdown on replication have been reported to vary depending on the infected cell type (Hao et al., 2019).
3.2.4. Paramyxoviridae
The Paramyxovirus family are negative‐strand RNA viruses that include RSV. m6A has been detected in both the genome and antigenome of RSV and is known to enhance viral replication in vitro and in vivo. Consistent with this, m6A‐deficient RSV variants display reduced replication in vivo (Xue et al., 2019).
3.2.5. Orthomyxoviridae
Orthomyxoviruses are negative‐sense RNA viruses. IAV was the first virus in which m6A was detected at an internal adenosine residue. IAV contains m6A on both plus and minus strands, and the modification is enriched in regions encoding several structural proteins. m6A has been shown to enhance IAV gene expression and replication in vitro and in vivo. Accordingly, mutation of known m6A sites inhibited hemagglutinin mRNA and protein expression and reduced IAV pathogenicity in mice (Courtney et al., 2017; Krug et al., 1976).
3.2.6. Pneumoviridae
Pneumoviruses have negative‐sense ssRNA genomes. One family member is human metapneumovirus (HMPV), which contains m6A in both the genome and antigenome, with the highest enrichment within the G gene. A study by Lu et al. showed that overexpression of METTL3/14 or YTHDF1‐3 increased viral gene expression and replication. Moreover, mutation of m6A sites or overexpression of ALKBH5 reduced m6A levels, increased expression of IFNβ, and reduce viral replication. Mechanistically, m6A knockdown in HMPV RNA enhanced RIG‐I binding, thereby by increasing activation of downstream proinflammatory signaling pathways in vitro and in vivo (Lu et al., 2020).
3.2.7. Retroviridae
Several groups have explored the functions of m6A in HIV biology. HIV infection induces a significant increase in m6A in both host and viral mRNAs and that m6A plays a positive role in viral propagation. Thus, knockdown of the m6A writers METTL3 and METTL14 or eraser ALKBH5 resulted in reduced or enhanced HIV replication, respectively (Lichinchi, Gao, et al., 2016). Additionally, m6A methylation in the stem–loop region in the Rev response element (RRE) promoted RNA binding of Rev protein and nuclear export of HIV RNA (Lichinchi, Gao, et al., 2016). m6A modification of RRE stem IIB minimally affected thermal stability and structural dynamics of the RNA in vitro, (Chu et al., 2019) suggesting that Rev is able to recognize RNAs with functional group substitutions and subtle structural changes in vivo during the assembly of functional Rev–RRE RNP complexes. However, the effect of m6A modifications on RNA structure and RNP complexes in vivo remains to be determined. In addition, Kennedy et al. reported a positive regulatory role for m6A and readers YTHDF1‐3 in HIV replication in their study of overexpression or knockout YTHDF1–3 or mutation of m6A sites at the 3′‐untranslated region of HIV RNA (Kennedy et al., 2016). However, YTHDF1–3 proteins were reported to inhibit HIV‐1 infection in Jurkat and HeLa cell lines, and primary CD4+ T‐cells. YTHDF proteins bind to m6A and decrease HIV‐1 gag RNA in infected cells and inhibit of HIV‐1 reverse transcription (Lu et al., 2018; Tirumuru et al., 2016). In general, all the studies show disruption of METTL3/14 function decreases expression of HIV‐1 replication, while interference with the erasers ALKBH5 and FTO have the opposite effect (Lichinchi, Gao, et al., 2016; Tirumuru et al., 2016). However, the function of readers YTHDF1–3 in regulating HIV replication is under debate. Depending on the infected cell types, infection conditions and detection stage, YTHDF1–3 were reported to positively or negatively regulate HIV replication (Kennedy et al., 2016; Lu et al., 2018; Tirumuru et al., 2016). In HIV‐1 infected differentiated U937 cells and primary macrophages, the m6A modification in HIV‐1 viral transcripts reduces RLR‐mediated IRF3/7 phosphorylation and signaling, leading to reduced IFNα/β production (Chen et al., 2021).
Studies of another retrovirus, murine leukemia virus, also showed evidence for a positive effect of m6A on the viral replication in that overexpression of YTHDF2 was found to enhance viral replication in 3T3 cells (Bondurant et al., 1976; Courtney, Chalem, et al., 2019). Although m6A has been detected in RNA transcripts of other retroviruses, such as Rous sarcoma virus and feline leukemia virus, the functional consequences of this modification are unclear (Furuichi et al., 1975; Kane & Beemon, 1985; Thomason et al., 1976).
3.2.8. Polyomaviridae
This family of dsDNA viruses includes SV40, a small tumor virus. m6A is present in SV40 mRNA and plays a role in the regulation or viral structural protein expression as well as viral replication. Overexpression of YTHDF2 enhances viral replication and viral structural protein VP1 expression. While knockout of METTL3 or YTHDF2 or inactivation mutation of m6A sites resulted in a reduced expression of the structural proteins VP1 and suppressed viral replication in SV40‐permissive cell line BSC40 cells (Finkel & Groner, 1983; Tsai et al., 2018).
3.2.9. Adenoviridae
This is a large family of dsDNA viruses that are responsible for many human diseases. Adenovirus‐2 transcripts contain m6A, and depletion of METTL3 reduces m6A levels in late transcripts and decreases splicing efficiency. Depletion of METTL3 reduced viral late genes Hexon, Penton, and Fiber expression and infectious progeny production. (Hashimoto & Green, 1976; Price et al., 2020). However, the effects of m6A on the host innate immune response is unknown.
3.2.10. Herpesviridae
Herpes simplex virus type 1 (HSV‐1) is a member of the Herpesviridae family of linear dsDNA viruses. The HSV‐1 genome contains 12 m6A peaks that are important for replication, as demonstrated by a reduction in replication upon METTL3/14 or YTHDF1‐3 depletion and an enhancement of replication upon ALKBH5 or FTO knockdown (Moss et al., 1977; Zhuoying Feng et al., 2021).
Kaposi's sarcoma‐associated herpesvirus (KSHV) viral mRNA transcripts have also harbor m6A, but interestingly, the effects of m6A on viral biology appear to vary depending on the infected cell type. For example, in lines derived from baculovirus‐infected SLK endothelial cells iSLK.219 and iSLK.BAC16 cells, knockdown of METTL3, YTHDF2, or m6A‐abrogating mutation inhibits viral replication, suggesting a beneficial role of m6A for viral replication. However, in the KSHV‐infected B cell line BCBL‐1, the same manipulations amplified viral protein ORF50 expression, implying that m6A inhibits replication. In line with the latter findings, another study showed that YTHDF2 depletion in the KSHV‐infected cell line KiSLK resulted in increased viral lytic replication by impeding KSHV RNA degradation (Hesser et al., 2018; Tan et al., 2018).
RNA modification by m6A has been detected in both latent and lytic transcripts of Epstein–Barr virus (EBV). In one study, METTL14 depletion was shown to reduce m6A levels, leading to a reduction in stability of several latent transcripts, including EBNA1, EBNA3C, and LMP1 (Lang et al., 2019). Another study found that knockdown of METTL3/14 and, especially, YTHDF1 destabilized m6A‐modified BZLF1 and BRLF1 transcripts, leading to a reduction in lytic EBV replication and infection (Lang et al., 2019; Xia et al., 2021).
3.2.11. Hepadnaviridae
Hepatitis B virus (HBV) is the prototypical member of the Hepadnavirus family of dsDNA viruses. This family employs its own reverse‐transcription polymerase to replicate from an RNA intermediate pregenomic RNA (pgRNA) to generate viral DNA. HBV contains m6A in the ε stem‐loop of RNA transcripts and the presence of m6A is associated with a reduction in pgRNA stability and HBs and HBc gene expression. However, m6A was found to positively regulate reverse transcription in HBV (Imam et al., 2018). In addition, m6A modification of HBV pgRNA inhibits RIG‐I binding and dampens the innate immune response; thus, knockdown of METTL3/14 or YTHDF2/3 increases RIG‐I innate sensing and IFN‐β production (Kim et al., 2020).
3.3. Other RNA modifications
3.3.1. 5‐Methylcytidine (m5C)
Methylation of the fifth carbon of cytosine (m5C) is another RNA modification that plays important roles in regulating RNA stability and function. To date, all viruses have been found to lack m5C‐MTase activity, and they instead deposit the modification is mediated by the host enzymes NSUN (NOL1/NOP2/SUN domain) or DNMT2/TRDMT1 (DNA‐MTase 2/tRNA‐MTase 1). Many viruses have been reported to harbor RNA modified by m5C, including Sindbis virus, adenovirus 2, murine leukemia virus, HIV‐1, and—possibly—SARS‐CoV‐2 (Courtney, Chalem, et al., 2019; Courtney, Tsai, et al., 2019; Dubin et al., 1977; Dubin & Stollar, 1975; Sommer et al., 1976; Taiaroa et al., 2020). The MTase NSUN2 is the primary m5C writer for HIV genomic RNA, and NSUN2‐catalyzed m5C modification has been shown to promote replication of both HIV and murine leukemia virus (Courtney, Chalem, et al., 2019; Courtney, Tsai, et al., 2019). Although m5C MTases have been implicated in the regulation of host innate immunity to many viruses, there is currently no direct evident that m5C modification of viral RNA specifically affects the antiviral response.
3.3.2. N4‐Acetylcytidine
The prevalence and function of N4‐acetylated cytidine in viral RNA are currently unclear; however, N4‐acetylcytidine (ac4C) has been shown to enhance both the stability and translation of cellular mRNA (Arango et al., 2018). ac4C has been detected in HIV RNA, where the modification is catalyzed by host N‐acetyltransferase 10. ac4C modification and its writer NAT10 promotes viral RNA stability, gene expression and replication (Tsai et al., 2020).
3.3.3. N6,2′‐O‐dimethyladenosine
The RNA modification N6,2′‐O‐dimethyladenosine (m6Am) localizes at the first transcribed nucleotide adenosine and is mediated by host methyltransferase phosphorylated C‐terminal domain interacting factor 1 (PCIF1), which not only occurs in host RNA but also in viral RNA (Akichika et al., 2019; Boulias et al., 2019; Mauer et al., 2017; Sendinc et al., 2019; Tartell et al., 2021). Recently, negative‐sense RNA viruses which replicate in cytoplasm such as vesicular stomatitis virus (VSV), rabies and measles viruses were found to harbor m6Am modifications at their mRNA cap structure, which is modified by host methyltransferase PCIF1. Intriguingly, PCIF1‐mediated m6Am modification of VSV viral mRNA attenuates the antiviral effects when the infected cells were treated with interferon‐β (IFNB). However, the dampened antiviral effects by viral m6Am modification is not dependent on RIG‐I or IFIT1 sensing, suggesting other virus sensing molecules or mechanism may be involved in the attenuated host innate immune response affected by viral RNA m6Am modification (Tartell et al., 2021).
3.3.4. RNA editing
In addition to the viral RNA modifications discussed above, RNA editing is an important mechanism by which genomic and transcriptomic viral RNA is modified. RNA editing refers to the insertion, deletion, or substitution of the original A, U, C, and G with modified nucleotides, and there is evidence that the process plays key roles in both viral replication and host sensing. The main editing mechanisms are uridine‐to‐5‐ribosyl uracil (pseudouridine) editing, which is catalyzed by pseudouridine synthases; adenosine‐to‐inosine (A‐to‐I) editing, which is catalyzed by the deaminase ADAR (adenosine deaminase acting on RNA); and cytidine‐to‐uridine (C‐to‐U) editing, catalyzed by the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide) family of cytidine deaminases (Bishop et al., 2004; Gelinas et al., 2011; Netzband & Pager, 2020).
Pseudouridine, the most abundant RNA modification, occurs in many viruses, including the flaviviruses ZIKV, DENV, and HCV; Picornaviridae family poliovirus (PV), and retroviruses such as HIV‐1. Pseudouridine is thought to enhance the stability and translational capacity of mRNA and to diminish RIG‐I‐mediated recognition of RNA in vitro and in vivo. However, more work is needed to firmly establish the beneficial versus detrimental effects of viral pseudouridine modification (Durbin et al., 2016; Kariko et al., 2008; McIntyre et al., 2018; Netzband & Pager, 2020).
ADAR1‐mediated A‐to‐I editing has been shown to promote the replication of various RNA viruses, including HIV and hepatitis delta (HDV), vesicular stomatitis (VSV), and measles viruses, via a mechanism involving inhibition of the IFN‐induced kinase PKR (Gelinas et al., 2011). In addition, A‐to‐I conversion was also found in SARS‐CoV‐2 viral RNA isolated from bronchoalveolar lavage fluids from COVID‐19 patients (Di Giorgio et al., 2020). Furthermore, in mouse embryos, an increase in inosine content in cellular RNA has been shown to inhibit MDA5‐mediated recognition of viral RNA (Gelinas et al., 2011; Liddicoat et al., 2015; Mannion et al., 2014).
C‐to‐U editing by the ISG APOBEC3 is known to play an antiviral role in retroviruses such as HIV by generating mutations or inhibiting reverse transcription (Bishop et al., 2004; Stavrou et al., 2014; Stavrou & Ross, 2015). APOBEC3A and APOBEC3G inhibited retrovirus M‐MLV viral infection in vivo. APOBEC3G was involved deamination of viral genome, while APOBEC3A affected host cells. HIV Vif protein blocked the anti‐viral activity of APOBEC3G protein (Bishop et al., 2004; Stavrou et al., 2014; Stavrou & Ross, 2015). The editing also involves and regulates innate and adaptive immune response. APOBEC3‐mediated inhibition of reverse transcriptions reduces STING‐mediated innate immune sensing and IFN‐induced antiviral response. Furthermore, Higher uracil content in HIV transcripts edited by APOBEC3G‐mediated cytidine deamination activated base‐excision–repair pathway, and then induce NKG2D ligand expression and enhance NK cell mediated cell lysis (Bishop et al., 2004; Stavrou & Ross, 2015). In addition, C‐to‐U changes were also found in SARS‐CoV‐2 viral RNA isolated from bronchoalveolar lavage fluids from COVID‐19 patients (Di Giorgio et al., 2020).
4. RNA MODIFICATIONS OF HOST GENES
Although we focus on the regulation of innate immune sensing and activation by viral genome RNA modifications here, another important regulatory pathway of host innate immune response is through RNA modification of host cell mRNA. There are multiple studies reported that m6A modification can directly regulate host gene expression such as IFNB, PTEN to modulate innate immune response (Kim et al., 2021;Rubio et al., 2018; Winkler et al., 2019). For example, IFNB gene was reported to have m6A modification and its expression was directly regulated by m6A methyltransferase METTL3, METTL14 and reader YTHDF2. During HCMV viral infection, depletion of METTL3/14, IFNB mRNA expression was stabilized and ISGs were stimulated (Rubio et al., 2018; Winkler et al., 2019). As both viruses and host mRNA utilize host m6A methyltransferases to deposit m6A on viral genome or host mRNA, therefore when evaluating the effects of certain modification on host innate immune response, both viral RNA and host RNA should be considered during experiments relying on depleting host enzyme machinery. However, there are multiple ways to distinguish whether the effects on innate immune response are dependent on viral genome or host gene RNA modification. For example, if innate immune responses are mediated through viral RNA modification affecting PRR sensing, the upstream event are directly affected including sensor binding to the viral RNA leading to conformational changes, which are followed by downstream signaling pathway activation, IFN and inflammation cytokine expression and ISGs stimulation. Such broad immune responses are not affected when host gene mRNA modification regulates specific steps downstream of PRR sensing.
Viruses may also have evolved mechanisms to suppress host methyltransferases in order to regulate expression of pro‐ or antiviral genes. During HIV and ZIKA infections, drastic changes in m6A modified host mRNAs and gene expressions were detected (Lichinchi, Gao, et al., 2016; Lichinchi, Zhao, et al., 2016). Recent studies show that HIV infection causes a dramatic decrease in m6Am of cellular mRNAs (Zhang, Kang, et al., 2021). In T cells, 2237 m6Am modified genes were identified and 854 were affected by HIV infection. In addition, PCIF1 methyltransferase function restricts HIV replication and HIV viral protein R (Vpr) interacts with PCIF1 and induces PCIF1 ubiquitination and degradation (Zhang, Kang, et al., 2021). In summary, regulation of RNA modifications by viruses reprograms host cell gene expression, which may provide new mechanisms to modulate host–virus interactions and antiviral immunity.
5. CONCLUSIONS AND PERSPECTIVE
Viral epitranscriptomics is an emerging field that has great relevance for understanding fundamental aspects of viral biology and the host antiviral response. In this review, we have summarized the most important modifications in viral RNA, touched on their functions with respect to viral replication and host sensing and signaling, and provided examples of the virus families impacted by each modification (Figure 2). We have focused on the regulatory role of viral RNA modifications on PRRs sensing especially RLRs since many studies have conducted experiments in the cell lines relying on the RLRs to facilitate innate sensing. However, other PRRs, as listed above, also play important roles in innate sensing and RNA modifications of exogenous RNA can also affect the innate sensing of those PRRs. For example, TLR3, TLR7 and TLR8 which are expressed in dendritic cells (DCs) recognize synthetic RNAs and stimulate downstream innate immune signaling pathway, while the sensing of exogenous RNA and downstream innate immune pathway activation was significantly impaired when the RNAs contained N6‐methyaladanosine (m6A), 5‐methylcytidine (m5C), 2‐thiouridine, or pseudouridine (U) modifications (Kariko et al., 2005). In future, examination of the role of viral RNA modification in PRRs sensing and innate immune pathway activation in primary cells and in vivo models will be of great importance to delineate the exact role of RLR, TLR or NLR in this process.
FIGURE 2.
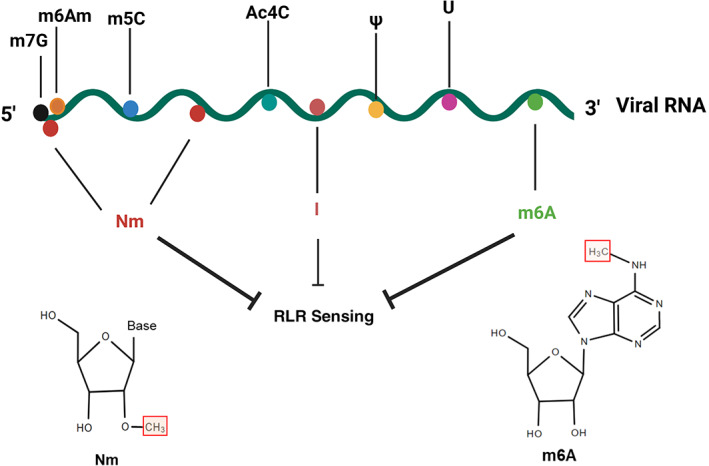
Viral RNA modifications involved in innate immune sensing. Among the various viral RNA modifications, nm, A‐to‐I editing, and m6A have been reported to inhibit RLR‐mediated sensing of viral RNA. ac4C, N4‐acetylcytidine; I, inosine; m5C, 5‐methylcytidine; m6A, N6‐methyladenosine (m6A); m6Am, N6,2′‐O‐dimethyladenosine; m7G, 7‐methylguanosine; Nm, 2′‐ O‐methylation (Nm); U, uracil; Ψ, pseudouridine
While technology advances will undoubtedly provide further insights into the functions of the modifications described here, it is safe to assume that additional RNA modifications that affect viral replication and host immunity remain to be discovered. This is an exciting and fast‐moving field, and there is much to learn about the viral and host proteins that write, read, and erase specific modifications, including whether they could be amenable to manipulated pharmacologically. However, caution will be needed in this respect because many viruses are reliant on host RNA‐modifying enzymes, and knockdown of some viral enzymes have different effects on replication depending on the cell types.
Another fertile topic for future studies is the interplay between different viral RNA modifications and their effects on the viral life cycle and host antiviral response. It seems likely that the nature and level of various viral RNA modifications will fluctuate at different stages of the viral life cycle. Therefore, it will be important to obtain more precise qualitative and quantitative characterizations of each viral RNA modification through the infection cycle, and to uncover the precise effects on induction or evasion of the host innate immune system. Finally, identifying the mutual influences of epitranscriptomic marks between virus and host RNA will be crucial to fully understand their implications for human health and disease.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Na Li: Data curation (lead); investigation (equal); methodology (equal); writing – original draft (lead). Tariq Rana: Conceptualization (lead); funding acquisition (lead); investigation (equal); project administration (lead); resources (lead); supervision (lead); writing – review and editing (lead).
RELATED WIREs ARTICLES
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health (NIH) grants (AI125103, DA046171, and DA049524). We apologize to authors whose seminal work was not cited due to space limitations.
Li, N. , & Rana, T. M. (2022). Regulation of antiviral innate immunity by chemical modification of viral RNA . Wiley Interdisciplinary Reviews: RNA, 13(6), e1720. 10.1002/wrna.1720
Edited by: Jeff Wilusz, Editor‐in‐Chief
Funding information National Institutes of Health, Grant/Award Numbers: AI125103, DA046171, DA049524
DATA AVAILABILITY STATEMENT
“Data sharing is not applicable to this article as no new data were created or analyzed in this study.”
REFERENCES
- Akichika, S. , Hirano, S. , Shichino, Y. , Suzuki, T. , Nishimasu, H. , Ishitani, R. , Sugita, A. , Hirose, Y. , Iwasaki, S. , Nureki, O. , & Suzuki, T. (2019). Cap‐specific terminal N (6)‐methylation of RNA by an RNA polymerase II‐associated methyltransferase. Science, 363(6423), eaav0080. [DOI] [PubMed] [Google Scholar]
- Akira, S. , & Takeda, K. (2004). Toll‐like receptor signalling. Nature Reviews Immunology, 4, 499–511. [DOI] [PubMed] [Google Scholar]
- Akira, S. , Takeda, K. , & Kaisho, T. (2001). Toll‐like receptors: Critical proteins linking innate and acquired immunity. Nature Immunology, 2, 675–680. [DOI] [PubMed] [Google Scholar]
- Akira, S. , Uematsu, S. , & Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell, 124, 783–801. [DOI] [PubMed] [Google Scholar]
- Arango, D. , Sturgill, D. , Alhusaini, N. , Dillman, A. A. , Sweet, T. J. , Hanson, G. , Hosogane, M. , Sinclair, W. R. , Nanan, K. K. , Mandler, M. D. , Fox, S. D. , Zengeya, T. T. , Andresson, T. , Meier, J. L. , Coller, J. , & Oberdoerffer, S. (2018). Acetylation of Cytidine in mRNA promotes translation efficiency. Cell, 175, 1872–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran, S. , & Barber, G. N. (2007). PKR in innate immunity, cancer, and viral oncolysis. Methods in Molecular Biology, 383, 277–301. [DOI] [PubMed] [Google Scholar]
- Balachandran, S. , Roberts, P. C. , Brown, L. E. , Truong, H. , Pattnaik, A. K. , Archer, D. R. , & Barber, G. N. (2000). Essential role for the dsRNA‐dependent protein kinase PKR in innate immunity to viral infection. Immunity, 13, 129–141. [DOI] [PubMed] [Google Scholar]
- Barbieri, I. , & Kouzarides, T. (2020). Role of RNA modifications in cancer. Nature Reviews Cancer, 20, 303–322. [DOI] [PubMed] [Google Scholar]
- Bauernfried, S. , Scherr, M. J. , Pichlmair, A. , Duderstadt, K. E. , & Hornung, V. (2021). Human NLRP1 is a sensor for double‐stranded RNA. Science, 371(6528), eabd0811. [DOI] [PubMed] [Google Scholar]
- Bishop, K. N. , Holmes, R. K. , Sheehy, A. M. , & Malim, M. H. (2004). APOBEC‐mediated editing of viral RNA. Science, 305, 645. [DOI] [PubMed] [Google Scholar]
- Bondurant, M. , Hashimoto, S. , & Green, M. (1976). Methylation pattern of genomic RNA from Moloney murine leukemia virus. Journal of Virology, 19, 998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias, K. , Toczydlowska‐Socha, D. , Hawley, B. R. , Liberman, N. , Takashima, K. , Zaccara, S. , Guez, T. , Vasseur, J. J. , Debart, F. , Aravind, L. , Jaffrey, S. R. , & Greer, E. L. (2019). Identification of the m(6)Am methyltransferase PCIF1 reveals the location and functions of m(6)Am in the transcriptome. Molecular Cell, 75, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, H. M. , Depledge, D. P. , Thompson, L. , Srinivas, K. P. , Grande, R. C. , Vink, E. I. , Abebe, J. S. , Blackaby, W. P. , Hendrick, A. , Albertella, M. R. , Kouzarides, T. , Stapleford, K. A. , Wilson, A. C. , & Mohr, I. (2021). Targeting the m(6)A RNA modification pathway blocks SARS‐CoV‐2 and HCoV‐OC43 replication. Genes & Development, 35, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D. C. , Hoang, L. T. , Mohamed Naim, A. N. , Dong, H. , Schreiber, M. J. , Hibberd, M. L. , Tan, M. J. A. , & Shi, P. Y. (2016). Evasion of early innate immune response by 2′‐O‐methylation of dengue genomic RNA. Virology, 499, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Kumar, S. , Espada, C. E. , Tirumuru, N. , Cahill, M. P. , Hu, L. , He, C. , & Wu, L. (2021). N6‐methyladenosine modification of HIV‐1 RNA suppresses type‐I interferon induction in differentiated monocytic cells and primary macrophages. PLoS Pathogens, 17, e1009421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, C. C. , Liu, B. , Plangger, R. , Kreutz, C. , & Al‐Hashimi, H. M. (2019). m6A minimally impacts the structure, dynamics, and Rev ARM binding properties of HIV‐1 RRE stem IIB. PLoS One, 14, e0224850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coloma, J. , Jain, R. , Rajashankar, K. R. , Garcia‐Sastre, A. , & Aggarwal, A. K. (2016). Structures of NS5 Methyltransferase from Zika virus. Cell Reports, 16, 3097–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney, D. G. , Chalem, A. , Bogerd, H. P. , Law, B. A. , Kennedy, E. M. , Holley, C. L. , & Cullen, B. R. (2019). Extensive epitranscriptomic methylation of a and C residues on murine leukemia virus transcripts enhances viral gene expression. MBio, 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney, D. G. , Kennedy, E. M. , Dumm, R. E. , Bogerd, H. P. , Tsai, K. , Heaton, N. S. , & Cullen, B. R. (2017). Epitranscriptomic enhancement of influenza a virus gene expression and replication. Cell Host & Microbe, 22, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney, D. G. , Tsai, K. , Bogerd, H. P. , Kennedy, E. M. , Law, B. A. , Emery, A. , Swanstrom, R. , Holley, C. L. , & Cullen, B. R. (2019). Epitranscriptomic addition of m(5)C to HIV‐1 transcripts regulates viral gene expression. Cell Host & Microbe, 26, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis, S. , Samuel, M. A. , Suthar, M. S. , Gale, M., Jr. , & Diamond, M. S. (2008). Toll‐like receptor 3 has a protective role against West Nile virus infection. Journal of Virology, 82, 10349–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis, S. , Szretter, K. J. , Schriewer, J. , Li, J. , Youn, S. , Errett, J. , Lin, T. Y. , Schneller, S. , Zust, R. , Dong, H. , Thiel, V. , Sen, G. C. , Fensterl, V. , Klimstra, W. B. , Pierson, T. C. , Buller, R. M. , Gale Jr, M. , Shi, P. Y. , & Diamond, M. S. (2010). 2′‐O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature, 468, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly, E. , Ferron, F. , Lescar, J. , & Canard, B. (2011). Conventional and unconventional mechanisms for capping viral mRNA. Nature Reviews. Microbiology, 10, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio, S. , Martignano, F. , Torcia, M. G. , Mattiuz, G. , & Conticello, S. G. (2020). Evidence for host‐dependent RNA editing in the transcriptome of SARS‐CoV‐2. Science Advances, 6, eabb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold, S. S. , Kaisho, T. , Hemmi, H. , Akira, S. , & Reis e Sousa, C. (2004). Innate antiviral responses by means of TLR7‐mediated recognition of single‐stranded RNA. Science, 303, 1529–1531. [DOI] [PubMed] [Google Scholar]
- Dimock, K. , & Stoltzfus, C. M. (1977). Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry, 16, 471–478. [DOI] [PubMed] [Google Scholar]
- Dong, H. , Chang, D. C. , Hua, M. H. , Lim, S. P. , Chionh, Y. H. , Hia, F. , Lee, Y. H. , Kukkaro, P. , Lok, S. M. , Dedon, P. C. , & Shi, P. Y. (2012). 2′‐O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathogens, 8, e1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin, D. T. , & Stollar, V. (1975). Methylation of Sindbis virus “26S” messenger RNA. Biochemical and Biophysical Research Communications, 66, 1373–1379. [DOI] [PubMed] [Google Scholar]
- Dubin, D. T. , Stollar, V. , Hsuchen, C. C. , Timko, K. , & Guild, G. M. (1977). Sindbis virus messenger RNA: The 5′‐termini and methylated residues of 26 and 42 S RNA. Virology, 77, 457–470. [DOI] [PubMed] [Google Scholar]
- Durbin, A. F. , Wang, C. , Marcotrigiano, J. , & Gehrke, L. (2016). RNAs containing modified nucleotides fail to trigger RIG‐I conformational changes for innate immune signaling. MBio, 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff, M. P. , Benarroch, D. , Selisko, B. , Romette, J. L. , & Canard, B. (2002). An RNA cap (nucleoside‐2′‐O‐)‐methyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. The EMBO Journal, 21, 2757–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul, E. J. , Wanjalla, C. N. , Suthar, M. S. , Gale, M. , Wirblich, C. , & Schnell, M. J. (2010). Rabies virus infection induces type I interferon production in an IPS‐1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathogens, 6, e1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Zhou, F. , Tan, M. , Wang, T. , Chen, Y. , Xu, W. , Li, B. , Wang, X. , Deng, X. , & He, M.‐L. (2021). Targeting m6A modification inhibits herpes virus 1 infection. Genes & Diseases. 10.1016/j.gendis.2021.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, D. , & Groner, Y. (1983). Methylations of adenosine residues (m6A) in pre‐mRNA are important for formation of late simian virus 40 mRNAs. Virology, 131, 409–425. [DOI] [PubMed] [Google Scholar]
- Fredericksen, B. L. , Keller, B. C. , Fornek, J. , Katze, M. G. , & Gale, M., Jr. (2008). Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG‐I and MDA5 signaling through IPS‐1. Journal of Virology, 82, 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi, Y. , Shatkin, A. J. , Stavnezer, E. , & Bishop, J. M. (1975). Blocked, methylated 5′‐terminal sequence in avian sarcoma virus RNA. Nature, 257, 618–620. [DOI] [PubMed] [Google Scholar]
- Geddes, K. , Magalhaes, J. G. , & Girardin, S. E. (2009). Unleashing the therapeutic potential of NOD‐like receptors. Nature Reviews Drug Discovery, 8, 465–479. [DOI] [PubMed] [Google Scholar]
- Gelinas, J. F. , Clerzius, G. , Shaw, E. , & Gatignol, A. (2011). Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA‐activated protein kinase. Journal of Virology, 85, 8460–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin, L. , Barchet, W. , Gilfillan, S. , Cella, M. , Beutler, B. , Flavell, R. A. , Diamond, M. S. , & Colonna, M. (2006). Essential role of mda‐5 in type I IFN responses to polyriboinosinic:Polyribocytidylic acid and encephalomyocarditis picornavirus. Proceedings of the National Academy of Sciences of the United States of America, 103, 8459–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin, L. , Benoit, L. , Song, C. , Cella, M. , Gilfillan, S. , Holtzman, M. J. , & Colonna, M. (2010). Melanoma differentiation‐associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathogens, 6, e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale, N. S. , McIntyre, A. B. R. , McFadden, M. J. , Roder, A. E. , Kennedy, E. M. , Gandara, J. A. , Hopcraft, S. E. , Quicke, K. M. , Vazquez, C. , Willer, J. , Ilkayeva, O. R. , Law, B. A. , Holley, C. L. , Garcia‐Blanco, M. A. , Evans, M. J. , Suthar, M. S. , Bradrick, S. S. , Mason, C. E. , & Horner, S. M. (2016). N6‐methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host & Microbe, 20, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen, B. B. , Hoopes, J. D. , Wong, M. H. , Jung, K. H. , Isakson, K. C. , Alexopoulou, L. , Flavell, R. A. , & Sidwell, R. W. (2006). TLR3 deletion limits mortality and disease severity due to Phlebovirus infection. Journal of Immunology, 177, 6301–6307. [DOI] [PubMed] [Google Scholar]
- Guillot, L. , Le Goffic, R. , Bloch, S. , Escriou, N. , Akira, S. , Chignard, M. , & Si‐Tahar, M. (2005). Involvement of toll‐like receptor 3 in the immune response of lung epithelial cells to double‐stranded RNA and influenza a virus. The Journal of Biological Chemistry, 280, 5571–5580. [DOI] [PubMed] [Google Scholar]
- Hao, H. , Hao, S. , Chen, H. , Chen, Z. , Zhang, Y. , Wang, J. , Wang, H. , Zhang, B. , Qiu, J. , Deng, F. , & Guan, W. (2019). N6‐methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Research, 47, 362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, S. I. , & Green, M. (1976). Multiple methylated cap sequences in adenovirus type 2 early mRNA. Journal of Virology, 20, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil, F. , Hemmi, H. , Hochrein, H. , Ampenberger, F. , Kirschning, C. , Akira, S. , Lipford, G. , Wagner, H. , & Bauer, S. (2004). Species‐specific recognition of single‐stranded RNA via toll‐like receptor 7 and 8. Science, 303, 1526–1529. [DOI] [PubMed] [Google Scholar]
- Hesser, C. R. , Karijolich, J. , Dominissini, D. , He, C. , & Glaunsinger, B. A. (2018). N6‐methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma‐associated herpesvirus infection. PLoS Pathogens, 14, e1006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam, H. , Khan, M. , Gokhale, N. S. , McIntyre, A. B. R. , Kim, G. W. , Jang, J. Y. , Kim, S. J. , Mason, C. E. , Horner, S. M. , & Siddiqui, A. (2018). N6‐methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proceedings of the National Academy of Sciences of the United States of America, 115, 8829–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S. , & Thomsen, A. R. (2012). Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. Journal of Virology, 86, 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J. D. , Vance, R. E. , & Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science, 354, aaf6395. [DOI] [PubMed] [Google Scholar]
- Jurk, M. , Heil, F. , Vollmer, J. , Schetter, C. , Krieg, A. M. , Wagner, H. , Lipford, G. , & Bauer, S. (2002). Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R‐848. Nature Immunology, 3, 499. [DOI] [PubMed] [Google Scholar]
- Kane, M. , Case, L. K. , Wang, C. , Yurkovetskiy, L. , Dikiy, S. , & Golovkina, T. V. (2011). Innate immune sensing of retroviral infection via toll‐like receptor 7 occurs upon viral entry. Immunity, 35, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, S. E. , & Beemon, K. (1985). Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Molecular and Cellular Biology, 5, 2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, D. C. , Gopalkrishnan, R. V. , Wu, Q. , Jankowsky, E. , Pyle, A. M. , & Fisher, P. B. (2002). mda‐5: An interferon‐inducible putative RNA helicase with double‐stranded RNA‐dependent ATPase activity and melanoma growth‐suppressive properties. Proceedings of the National Academy of Sciences of the United States of America, 99, 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko, K. , Buckstein, M. , Ni, H. , & Weissman, D. (2005). Suppression of RNA recognition by toll‐like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity, 23, 165–175. [DOI] [PubMed] [Google Scholar]
- Kariko, K. , Muramatsu, H. , Welsh, F. A. , Ludwig, J. , Kato, H. , Akira, S. , & Weissman, D. (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular Therapy, 16, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, H. , Sato, S. , Yoneyama, M. , Yamamoto, M. , Uematsu, S. , Matsui, K. , Tsujimura, T. , Takeda, K. , Fujita, T. , Takeuchi, O. , & Akira, S. (2005). Cell type‐specific involvement of RIG‐I in antiviral response. Immunity, 23, 19–28. [DOI] [PubMed] [Google Scholar]
- Kato, H. , Takeuchi, O. , Sato, S. , Yoneyama, M. , Yamamoto, M. , Matsui, K. , Uematsu, S. , Jung, A. , Kawai, T. , Ishii, K. J. , Yamaguchi, O. , Otsu, K. , Tsujimura, T. , Koh, C. S. , Reis e Sousa, C. , Matsuura, Y. , Fujita, T. , & Akira, S. (2006). Differential roles of MDA5 and RIG‐I helicases in the recognition of RNA viruses. Nature, 441, 101–105. [DOI] [PubMed] [Google Scholar]
- Kawai, T. , & Akira, S. (2006). Innate immune recognition of viral infection. Nature Immunology, 7, 131–137. [DOI] [PubMed] [Google Scholar]
- Kell, A. M. , & Gale, M., Jr. (2015). RIG‐I in RNA virus recognition. Virology, 479–480, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, E. M. , Bogerd, H. P. , Kornepati, A. V. , Kang, D. , Ghoshal, D. , Marshall, J. B. , Poling, B. C. , Tsai, K. , Gokhale, N. S. , Horner, S. M. , & Cullen, B. R. (2016). Posttranscriptional m(6)A editing of HIV‐1 mRNAs enhances viral gene expression. Cell Host & Microbe, 19, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G. W. , Imam, H. , Khan, M. , Mir, S. A. , Kim, S. J. , Yoon, S. K. , Hur, W. , & Siddiqui, A. (2021). HBV‐induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology, 73, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G. W. , Imam, H. , Khan, M. , & Siddiqui, A. (2020). N (6)‐methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG‐I signaling. The Journal of Biological Chemistry, 295, 13123–13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky‐Bromberg, S. A. , Martinez‐Sobrido, L. , Frieman, M. , Baric, R. A. , & Palese, P. (2007). Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. Journal of Virology, 81, 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug, R. M. , Morgan, M. A. , & Shatkin, A. J. (1976). Influenza viral mRNA contains internal N6‐methyladenosine and 5′‐terminal 7‐methylguanosine in cap structures. Journal of Virology, 20, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, F. , Singh, R. K. , Pei, Y. , Zhang, S. , Sun, K. , & Robertson, E. S. (2019). EBV epitranscriptome reprogramming by METTL14 is critical for viral‐associated tumorigenesis. PLoS Pathogens, 15, e1007796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi, S. , & Shatkin, A. J. (1975). Methylated simian virus 40‐specific RNA from nuclei and cytoplasm of infected BSC‐1 cells. Proceedings of the National Academy of Sciences of the United States of America, 72, 2012–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, X. , Dong, X. , Ma, R. , Wang, W. , Xiao, X. , Tian, Z. , Wang, C. , Wang, Y. , Li, L. , Ren, L. , Guo, F. , Zhao, Z. , Zhou, Z. , Xiang, Z. , & Wang, J. (2020). Activation and evasion of type I interferon responses by SARS‐CoV‐2. Nature Communications, 11, 3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Liu, Y. , & Zhang, X. (2010). Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG‐I and MDA5. Journal of Virology, 84, 6472–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Hui, H. , Bray, B. , Gonzalez, G. M. , Zeller, M. , Anderson, K. G. , Knight, R. , Smith, D. , Wang, Y. , Carlin, A. F. , & Rana, T. M. (2021). METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS‐CoV‐2 infection. Cell Reports, 35(109), 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , & Mason, C. E. (2014). The pivotal regulatory landscape of RNA modifications. Annual Review of Genomics and Human Genetics, 15, 127–150. [DOI] [PubMed] [Google Scholar]
- Li, S. H. , Dong, H. , Li, X. F. , Xie, X. , Zhao, H. , Deng, Y. Q. , Wang, X. Y. , Ye, Q. , Zhu, S. Y. , Wang, H. J. , Zhang, B. , Leng, Q. B. , Zuest, R. , Qin, E. D. , Qin, C. F. , & Shi, P. Y. (2013). Rational design of a flavivirus vaccine by abolishing viral RNA 2′‐O methylation. Journal of Virology, 87, 5812–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Hajian, C. , & Greene, W. C. (2020). Identification of unrecognized host factors promoting HIV‐1 latency. PLoS Pathogens, 16, e1009055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi, G. , Gao, S. , Saletore, Y. , Gonzalez, G. M. , Bansal, V. , Wang, Y. , Mason, C. E. , & Rana, T. M. (2016). Dynamics of the human and viral m(6)A RNA methylomes during HIV‐1 infection of T cells. Nature Microbiology, 1(16), 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi, G. , & Rana, T. M. (2019). Profiling of N(6)‐methyladenosine in zika Virus RNA and host cellular mRNA. Methods in Molecular Biology, 1870, 209–218. [DOI] [PubMed] [Google Scholar]
- Lichinchi, G. , Zhao, B. S. , Wu, Y. , Lu, Z. , Qin, Y. , He, C. , & Rana, T. M. (2016). Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host & Microbe, 20, 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat, B. J. , Piskol, R. , Chalk, A. M. , Ramaswami, G. , Higuchi, M. , Hartner, J. C. , Li, J. B. , Seeburg, P. H. , & Walkley, C. R. (2015). RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science, 349, 1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Xu, Y. P. , Li, K. , Ye, Q. , Zhou, H. Y. , Sun, H. , Li, X. , Yu, L. , Deng, Y. Q. , Li, R. T. , Cheng, M. L. , He, B. , Zhou, J. , Li, X. F. , Wu, A. , Yi, C. , & Qin, C. F. (2021). The m(6)A methylome of SARS‐CoV‐2 in host cells. Cell Research, 31, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, Y. M. , Fornek, J. , Crochet, N. , Bajwa, G. , Perwitasari, O. , Martinez‐Sobrido, L. , Akira, S. , Gill, M. A. , Garcia‐Sastre, A. , Katze, M. G. , & Gale, M. (2008). Distinct RIG‐I and MDA5 signaling by RNA viruses in innate immunity. Journal of Virology, 82, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, Y. M. , & Gale, M., Jr. (2011). Immune signaling by RIG‐I‐like receptors. Immunity, 34, 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M. , Zhang, Z. , Xue, M. , Zhao, B. S. , Harder, O. , Li, A. , Liang, X. , Gao, T. Z. , Xu, Y. , Zhou, J. , Feng, Z. , Niewiesk, S. , Peeples, M. E. , He, C. , & Li, J. (2020). N(6)‐methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG‐I. Nature Microbiology, 5, 584–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W. , Tirumuru, N. , St Gelais, C. , Koneru, P. C. , Liu, C. , Kvaratskhelia, M. , He, C. , & Wu, L. (2018). N(6)‐methyladenosine‐binding proteins suppress HIV‐1 infectivity and viral production. The Journal of Biological Chemistry, 293, 12992–13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer, C. , & Kanneganti, T. D. (2013). The expanding role of NLRs in antiviral immunity. Immunological Reviews, 255, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi, K. , Saito, T. , Crochet, N. , Barton, D. J. , Gale, M., Jr. , & Silverman, R. H. (2010). RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA, 16, 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion, N. M. , Greenwood, S. M. , Young, R. , Cox, S. , Brindle, J. , Read, D. , Nellaker, C. , Vesely, C. , Ponting, C. P. , McLaughlin, P. J. , Jantsch, M. F. , Dorin, J. , Adams, I. R. , Scadden, A. D. , Ohman, M. , Keegan, L. P. , & O'Connell, M. A. (2014). The RNA‐editing enzyme ADAR1 controls innate immune responses to RNA. Cell Reports, 9, 1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, B. , Coutard, B. , Guez, T. , Paesen, G. C. , Canard, B. , Debart, F. , Vasseur, J. J. , Grimes, J. M. , & Decroly, E. (2018). The methyltransferase domain of The Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Research, 46, 7902–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer, J. , Luo, X. , Blanjoie, A. , Jiao, X. , Grozhik, A. V. , Patil, D. P. , Linder, B. , Pickering, B. F. , Vasseur, J. J. , Chen, Q. , Gross, S. S. , Elemento, O. , Debart, F. , Kiledjian, M. , & Jaffrey, S. R. (2017). Reversible methylation of m(6)Am in the 5′ cap controls mRNA stability. Nature, 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre, W. , Netzband, R. , Bonenfant, G. , Biegel, J. M. , Miller, C. , Fuchs, G. , Henderson, E. , Arra, M. , Canki, M. , Fabris, D. , & Pager, C. T. (2018). Positive‐sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Research, 46, 5776–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov, R. , Preston‐Hurlburt, P. , Kopp, E. , Stadlen, A. , Chen, C. , Ghosh, S. , & Janeway, C. A., Jr. (1998). MyD88 is an adaptor protein in the hToll/IL‐1 receptor family signaling pathways. Molecular Cell, 2, 253–258. [DOI] [PubMed] [Google Scholar]
- Menachery, V. D. , Yount, B. L., Jr. , Josset, L. , Gralinski, L. E. , Scobey, T. , Agnihothram, S. , Katze, M. G. , & Baric, R. S. (2014). Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′‐o‐methyltransferase activity. Journal of Virology, 88, 4251–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan, E. , Burns, K. , Hofmann, K. , Blancheteau, V. , Martinon, F. , Kelliher, M. , & Tschopp, J. (2004). RIP1 is an essential mediator of toll‐like receptor 3‐induced NF‐kappa B activation. Nature Immunology, 5, 503–507. [DOI] [PubMed] [Google Scholar]
- Moss, B. , Gershowitz, A. , Stringer, J. R. , Holland, L. E. , & Wagner, E. K. (1977). 5′‐Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. Journal of Virology, 23, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi, H. , Osawa, T. , Ogami, K. , Ouyang, X. , Sakaguchi, S. , Koshiba, R. , Yanai, H. , Seko, Y. , Shitara, H. , Bishop, K. , Yonekawa, H. , Tamura, T. , Kaisho, T. , Taya, C. , Taniguchi, T. , & Honda, K. (2008). A critical link between toll‐like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proceedings of the National Academy of Sciences of the United States of America, 105, 20446–20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelemans, T. , & Kikkert, M. (2019). Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses, 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzband, R. , & Pager, C. T. (2020). Epitranscriptomic marks: Emerging modulators of RNA virus gene expression. Wiley Interdisciplinary Reviews: RNA, 11, e1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi, H. , Okamoto, M. , Fujii, K. , Kawanishi, T. , Matsumoto, M. , Koike, S. , & Seya, T. (2011). The TLR3/TICAM‐1 pathway is mandatory for innate immune responses to poliovirus infection. Journal of Immunology, 187, 5320–5327. [DOI] [PubMed] [Google Scholar]
- Price, A. M. , Hayer, K. E. , McIntyre, A. B. R. , Gokhale, N. S. , Abebe, J. S. , Della Fera, A. N. , Mason, C. E. , Horner, S. M. , Wilson, A. C. , Depledge, D. P. , & Weitzman, M. D. (2020). Direct RNA sequencing reveals m(6)A modifications on adenovirus RNA are necessary for efficient splicing. Nature Communications, 11, 6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel, J. , & Gack, M. U. (2020). RIG‐I‐like receptors: Their regulation and roles in RNA sensing. Nature Reviews Immunology, 20, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringeard, M. , Marchand, V. , Decroly, E. , Motorin, Y. , & Bennasser, Y. (2019). FTSJ3 is an RNA 2′‐O‐methyltransferase recruited by HIV to avoid innate immune sensing. Nature, 565, 500–504. [DOI] [PubMed] [Google Scholar]
- Roundtree, I. A. , Evans, M. E. , Pan, T. , & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, R. M. , Depledge, D. P. , Bianco, C. , Thompson, L. , & Mohr, I. (2018). RNA m(6)A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes & Development, 32, 1472–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, B. D. , Burstein, E. , Duckett, C. S. , Li, X. , & Lukacs, N. W. (2005). Differential role for TLR3 in respiratory syncytial virus‐induced chemokine expression. Journal of Virology, 79, 3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, W. M. , Chevillotte, M. D. , & Rice, C. M. (2014). Interferon‐stimulated genes: A complex web of host defenses. Annual Review of Immunology, 32, 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnierle, B. S. , Gershon, P. D. , & Moss, B. (1992). Cap‐specific mRNA (nucleoside‐O2′‐)‐methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proceedings of the National Academy of Sciences of the United States of America, 89, 2897–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, A. , Pruijssers, A. J. , Dermody, T. S. , Garcia‐Sastre, A. , & Greenberg, H. B. (2011). The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS‐1, RIG‐I, MDA‐5, and IRF3. Journal of Virology, 85, 3717–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendinc, E. , Valle‐Garcia, D. , Dhall, A. , Chen, H. , Henriques, T. , Navarrete‐Perea, J. , Sheng, W. , Gygi, S. P. , Adelman, K. , & Shi, Y. (2019). PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Molecular Cell, 75, 620–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava, G. , Leon‐Juarez, M. , Garcia‐Cordero, J. , Meza‐Sanchez, D. E. , & Cedillo‐Barron, L. (2016). Inflammasomes and its importance in viral infections. Immunologic Research, 64, 1101–1117. [DOI] [PubMed] [Google Scholar]
- Solis, M. , Nakhaei, P. , Jalalirad, M. , Lacoste, J. , Douville, R. , Arguello, M. , Zhao, T. , Laughrea, M. , Wainberg, M. A. , & Hiscott, J. (2011). RIG‐I‐mediated antiviral signaling is inhibited in HIV‐1 infection by a protease‐mediated sequestration of RIG‐I. Journal of Virology, 85, 1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, S. , Salditt‐Georgieff, M. , Bachenheimer, S. , Darnell, J. E. , Furuichi, Y. , Morgan, M. , & Shatkin, A. J. (1976). The methylation of adenovirus‐specific nuclear and cytoplasmic RNA. Nucleic Acids Research, 3, 749–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou, S. , Crawford, D. , Blouch, K. , Browne, E. P. , Kohli, R. M. , & Ross, S. R. (2014). Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo. PLoS Pathogens, 10, e1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou, S. , & Ross, S. R. (2015). APOBEC3 proteins in viral immunity. Journal of Immunology, 195, 4565–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar, M. S. , Aguirre, S. , & Fernandez‐Sesma, A. (2013). Innate immune sensing of flaviviruses. PLoS Pathogens, 9, e1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter, K. J. , Daniels, B. P. , Cho, H. , Gainey, M. D. , Yokoyama, W. M. , Gale, M., Jr. , Virgin, H. W. , Klein, R. S. , Sen, G. C. , & Diamond, M. S. (2012). 2′‐O methylation of the viral mRNA cap by West Nile virus evades ifit1‐dependent and ‐independent mechanisms of host restriction in vivo. PLoS Pathogens, 8, e1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiaroa, G. R. , Featherstone, L. , Pitt, M. , Caly, L. , Druce, J. , Purcell, D. , Harty, L. , Tran, T. , Roberts, J. , Scott, N. , Catton, M. , Williamson, D. , Coin, L. , & Duchene, S. (2020). Direct RNA sequencing and early evolution of SARS‐CoV‐2. bioRxiv Preprints, PPR116527. 10.1101/2020.03.05.976167 [DOI] [Google Scholar]
- Tan, B. , Liu, H. , Zhang, S. , da Silva, S. R. , Zhang, L. , Meng, J. , Cui, X. , Yuan, H. , Sorel, O. , Zhang, S. W. , Huang, Y. , & Gao, S. J. (2018). Viral and cellular N(6)‐methyladenosine and N(6),2′‐O‐dimethyladenosine epitranscriptomes in the KSHV life cycle. Nature Microbiology, 3, 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartell, M. A. , Boulias, K. , Hoffmann, G. B. , Bloyet, L. M. , Greer, E. L. , & Whelan, S. P. J. (2021). Methylation of viral mRNA cap structures by PCIF1 attenuates the antiviral activity of interferon‐beta. Proceedings of the National Academy of Sciences of the United States of America, 118, e2025769118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason, A. R. , Brian, D. A. , Velicer, L. F. , & Rottman, F. M. (1976). Methylation of high‐molecular‐weight subunit RNA of feline leukemia virus. Journal of Virology, 20, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru, N. , Zhao, B. S. , Lu, W. , Lu, Z. , He, C. , & Wu, L. (2016). N(6)‐methyladenosine of HIV‐1 RNA regulates viral infection and HIV‐1 Gag protein expression. eLife, 5, e15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, K. , Courtney, D. G. , & Cullen, B. R. (2018). Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathogens, 14, e1006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, K. , & Cullen, B. R. (2020). Epigenetic and epitranscriptomic regulation of viral replication. Nature Reviews Microbiology, 18, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, K. , Jaguva Vasudevan, A. A. , Martinez Campos, C. , Emery, A. , Swanstrom, R. , & Cullen, B. R. (2020). Acetylation of cytidine residues boosts HIV‐1 gene expression by increasing viral RNA stability. Cell Host & Microbe, 28, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan, T. , Misra, A. , Chan, S. H. , Qi, S. , Dai, N. , Arya, S. , Martinez‐Sobrido, L. , & Gupta, Y. K. (2021). A metal ion orients SARS‐CoV‐2 mRNA to ensure accurate 2′‐O methylation of its first nucleotide. Nature Communications, 12, 3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. P. , Liu, P. , Latz, E. , Golenbock, D. T. , Finberg, R. W. , & Libraty, D. H. (2006). Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. Journal of Immunology, 177, 7114–7121. [DOI] [PubMed] [Google Scholar]
- Wang, T. , Town, T. , Alexopoulou, L. , Anderson, J. F. , Fikrig, E. , & Flavell, R. A. (2004). Toll‐like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nature Medicine, 10, 1366–1373. [DOI] [PubMed] [Google Scholar]
- Wilamowski, M. , Sherrell, D. A. , Minasov, G. , Kim, Y. , Shuvalova, L. , Lavens, A. , Chard, R. , Maltseva, N. , Jedrzejczak, R. , Rosas‐Lemus, M. , Saint, N. , Foster, I. T. , Michalska, K. , Satchell, K. J. F. , & Joachimiak, A. (2021). 2′‐O methylation of RNA cap in SARS‐CoV‐2 captured by serial crystallography. Proceedings of the National Academy of Sciences of the United States of America, 118, e2100170118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanski, J. M. , Petnicki‐Ocwieja, T. , & Kobayashi, K. S. (2008). NLR proteins: Integral members of innate immunity and mediators of inflammatory diseases. Journal of Leukocyte Biology, 83, 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, R. , Gillis, E. , Lasman, L. , Safra, M. , Geula, S. , Soyris, C. , Nachshon, A. , Tai‐Schmiedel, J. , Friedman, N. , Le‐Trilling, V. T. K. , Trilling, M. , Mandelboim, M. , Hanna, J. H. , Schwartz, S. , & Stern‐Ginossar, N. (2019). m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nature Immunology, 20, 173–182. [DOI] [PubMed] [Google Scholar]
- Wu, B. , Peisley, A. , Richards, C. , Yao, H. , Zeng, X. , Lin, C. , Chu, F. , Walz, T. , & Hur, S. (2013). Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell, 152, 276–289. [DOI] [PubMed] [Google Scholar]
- Xia, H. , Luo, H. , Shan, C. , Muruato, A. E. , Nunes, B. T. D. , Medeiros, D. B. A. , Zou, J. , Xie, X. , Giraldo, M. I. , Vasconcelos, P. F. C. , Weaver, S. C. , Wang, T. , Rajsbaum, R. , & Shi, P. Y. (2018). An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nature Communications, 9, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, T. L. , Li, X. , Wang, X. , Zhu, Y. J. , Zhang, H. , Cheng, W. , Chen, M. L. , Ye, Y. , Li, Y. , Zhang, A. , Dai, D. L. , Zhu, Q. Y. , Yuan, L. , Zheng, J. , Huang, H. , Chen, S. Q. , Xiao, Z. W. , Wang, H. B. , Roy, G. , … Zeng, M. S. (2021). N(6)‐methyladenosine‐binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Reports, 22, e50128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, M. , Zhao, B. S. , Zhang, Z. , Lu, M. , Harder, O. , Chen, P. , Lu, Z. , Li, A. , Ma, Y. , Xu, Y. , Liang, X. , Zhou, J. , Niewiesk, S. , Peeples, M. E. , He, C. , & Li, J. (2019). Viral N(6)‐methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nature Communications, 10, 4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama, M. , Kikuchi, M. , Matsumoto, K. , Imaizumi, T. , Miyagishi, M. , Taira, K. , Foy, E. , Loo, Y. M. , Gale, M., Jr. , Akira, S. , Yonehara, S. , Kato, A. , & Fujita, T. (2005). Shared and unique functions of the DExD/H‐box helicases RIG‐I, MDA5, and LGP2 in antiviral innate immunity. Journal of Immunology, 175, 2851–2858. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Kang, Y. , Wang, S. , Gonzalez, G. M. , Li, W. , Hui, H. , Wang, Y. , & Rana, T. M. (2021). HIV reprograms host m(6)am RNA methylome by viral Vpr protein‐mediated degradation of PCIF1. Nature Communications, 12, 5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Hao, H. , Ma, L. , Zhang, Y. , Hu, X. , Chen, Z. , Liu, D. , Yuan, J. , Hu, Z. , & Guan, W. (2021). Methyltransferase‐like 3 modulates severe acute respiratory syndrome coronavirus‐2 RNA N6‐methyladenosine modification and replication. mBio, 12, e0106721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Yi, G. , Du, F. , Chuang, Y. C. , Vaughan, R. C. , Sankaran, B. , Kao, C. C. , & Li, P. (2017). Structure and function of the Zika virus full‐length NS5 protein. Nature Communications, 8(14), 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S. , Cerny, A. M. , Zacharia, A. , Fitzgerald, K. A. , Kurt‐Jones, E. A. , & Finberg, R. W. (2010). Induction and inhibition of type I interferon responses by distinct components of lymphocytic choriomeningitis virus. Journal of Virology, 84, 9452–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Ray, D. , Zhao, Y. , Dong, H. , Ren, S. , Li, Z. , Guo, Y. , Bernard, K. A. , Shi, P. Y. , & Li, H. (2007). Structure and function of flavivirus NS5 methyltransferase. Journal of Virology, 81, 3891–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust, R. , Cervantes‐Barragan, L. , Habjan, M. , Maier, R. , Neuman, B. W. , Ziebuhr, J. , Szretter, K. J. , Baker, S. C. , Barchet, W. , Diamond, M. S. , Siddell, S. G. , Ludewig, B. , & Thiel, V. (2011). Ribose 2′‐O‐methylation provides a molecular signature for the distinction of self and non‐self mRNA dependent on the RNA sensor Mda5. Nature Immunology, 12, 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust, R. , Dong, H. , Li, X. F. , Chang, D. C. , Zhang, B. , Balakrishnan, T. , Toh, Y. X. , Jiang, T. , Li, S. H. , Deng, Y. Q. , Ellis, B. R. , Ellis, E. M. , Poidinger, M. , Zolezzi, F. , Qin, C. F. , Shi, P. Y. , & Fink, K. (2013). Rational design of a live attenuated dengue vaccine: 2′‐O‐Methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathogens, 9, e1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
“Data sharing is not applicable to this article as no new data were created or analyzed in this study.”


