Abstract
Signature tagged mutagenesis has recently revealed that the Ssp serine protease (V8 protease) contributes to in vivo growth and survival of Staphylococcus aureus in different infection models, and our previous work indicated that Ssp could play a role in controlling microbial adhesion. In this study, we describe an operon structure within the ssp locus of S. aureus RN6390. The ssp gene encoding V8 protease is designated as sspA, and is followed by sspB, which encodes a 40.6-kDa cysteine protease, and sspC, which encodes a 12.9-kDa protein of unknown function. S. aureus SP6391 is an isogenic derivative of RN6390, in which specific loss of SspA function was achieved through a nonpolar allelic replacement mutation. In addition to losing SspA, the culture supernatant of SP6391 showed a loss of 22- to 23-kDa proteins and the appearance of a 40-kDa protein corresponding to SspB. Although the 40-kDa SspB protein could degrade denatured collagen, our data establish that this is a precursor form which is normally processed by SspA to form a mature cysteine protease. Culture supernatant of SP6391 also showed a new 42-kDa glucosaminidase and enhanced glucosaminidase activity in the 29 to 32 kDa range. Although nonpolar inactivation of sspA exerted a pleiotropic effect, S. aureus SP6391 exhibited enhanced virulence in a tissue abscess infection model relative to RN6390. Therefore, we conclude that SspA is required for maturation of SspB and plays a role in controlling autolytic activity but does not by itself exert a significant contribution to the development of tissue abscess infections.
The serine protease of Staphylococcus aureus strain V8 (Ssp, also known as V8 protease) was one of the first secreted enzymes of S. aureus to be purified and characterized in detail (16). It is a member of the glutamyl endopeptidase family of enzymes (24) and has been widely used in this capacity as a specific tool for determining protein structure. However, its contributions to the growth and survival of S. aureus have not been elucidated. S. aureus is a major cause of infectious morbidity and mortality in both the community and hospital settings (30), and although capable of expressing several different toxins, it is not generally equated with other pathogens that cause illness primarily through the elaboration of specific toxins (17). Rather, the hallmarks of S. aureus disease are its rapid multiplication and induction of inflammation at the site of infection and its ability to disseminate and initiate metastatic infection (50, 51). This is facilitated by an accessory gene regulator locus, agr, which at high cell density is responsible for inducing the expression of secreted toxins and exoenzymes, while simultaneously promoting the reduced expression of cell surface adhesins and colonization factors (18, 38, 41, 48). Therefore, agr-null mutants demonstrate enhanced expression of colonization factors and a pleiotropic defect in expression of secreted virulence factors.
Due to the inability to express secreted virulence factors, agr-null strains of S. aureus exhibit attenuated virulence in several different infection models, and similar observations apply to a second regulatory locus, sar, which is required for optimal transcription of agr (5, 12, 14). However, with few exceptions, the function of individual secreted proteins is less well defined. In this respect, a signature tagged mutagenesis (STM) study has recently indicated that the Ssp serine protease contributes to in vivo growth and survival of S. aureus RN6390 in each of three different infection models (14). Remarkably, although S. aureus secretes numerous toxins and tissue-degrading enzymes, Ssp was the only secreted protein identified by STM as being required for the in vivo growth and survival of S. aureus. Our previous work also implicated a role for Ssp in degrading a cell surface fibronectin (Fn) binding protein (32), suggesting that Ssp could play an important role in controlling the stability and/or processing of cell surface proteins. These findings collectively indicate a significant contribution of the Ssp serine protease towards the growth and survival of S. aureus.
Herein, we present the first application of molecular techniques towards providing a detailed understanding of the functions of a secreted protease of S. aureus. Through sequence analysis of the S. aureus strain COL genome (http://www.tigr.org), we have found that the ssp structural gene (sspA) is followed closely by an open reading frame encoding a cysteine protease, designated sspB. The sspA and sspB proteases are transcribed as an operon, which also includes a third open reading frame sspC, of unknown function. Through construction of a nonpolar allelic replacement mutation, inactivation of sspA was achieved without affecting transcription of sspB or sspC. Loss of the SspA serine protease function resulted in a pleiotropic effect on the profile of secreted proteins, including autolysin activity and proteolytic maturation of the SspB cysteine protease, but did not result in attenuated virulence in a tissue abscess model of infection. Therefore, SspA alone does not exert a significant contribution to tissue abscess infections. Others have shown that SspA is itself expressed as an inactive precursor that is activated by a metalloprotease (15). Therefore, we have defined a step in a cascade pathway of proteolytic activity, where metalloprotease is required for maturation of SspA, which then processes SspB and controls autolytic activity.
Nucleotide sequence accession number.
The nucleotide sequence of the ssp operon has been deposited in GenBank (accession number AF309515).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and recombinant plasmids used in this study are listed in Table 1. S. aureus was cultured at 37°C in brain heart infusion (BHI) broth (Difco, Detroit, Mich.), NZY broth (20), or medium optimized for protease expression (16). Escherichia coli DH5α was grown at 37°C in Luria-Bertani medium (Difco). Culture media were supplemented with agar (15 g·l−1; Difco) where required, and with ampicillin (50 μg·ml−1), erythromycin (10 μg·ml−1), chloramphenicol (5 μg·ml−1), or tetracycline (5 μg·ml−1) when needed for selective purposes. For phenotypic assays, a single-colony inoculum was grown overnight in the appropriate medium on an orbital shaker (250 rpm) and then subcultured into prewarmed medium in Erlenmeyer flasks to achieve an initial optical density at 600 nm (OD600) of 0.1. For assays of Fn binding and coagulase activity, cells were harvested from BHI broth at mid-exponential (2 h) or stationary phase (6 to 7 h). Cell pellets were processed for assay of Fn binding (54), while culture supernatants were frozen at −70°C for coagulase assay. For protease and protein profile determinations, cultures were grown in protease culture medium for 8 or 18 h, and samples of cell culture supernatant were stored at −70°C. For analysis of autolysin profiles, cultures were grown to stationary phase in NZY broth (20), and cell culture supernatant was concentrated 30-fold in a Centricon-3 micro-concentrator (Millipore, Bedford, Mass.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Restriction-deficient strain; host for plasmid vectors | 36 |
| SP4221 | RN4220 containing pVE1C integrated in ssp locus | This study |
| RN6390B | Wild type; high protease, low Fn binding | 38 |
| RN6911 | agrΔ::tetM Tcr; agr-null isogenic derivative of RN6390B | 38 |
| SP6391 | sspA is inactivated by insertion of ermAB; Emr Cms | This study |
| SP6912 | Mutant sspA allele from SP6391 transduced into RN6911; Emr Tcr | This study |
| E. coli DH5α | Host strain for construction of recombinant plasmids | 22 |
| Plasmids | ||
| pBKS+ | E. coli cloning vector; Apr | 52 |
| pUC18 | E. coli cloning vector; Apr | 56 |
| pPQ126 | S. aureus plasmid; source of E194ts temperature-sensitive replication origin | 31 |
| pUC-V8 | 1.3-kb sspA PCR amplicon, cloned at HindIII and XbaI sites of pUC18; Apr | This study |
| pBS-E1 | 1.4-kb erm PCR amplicon, cloned in pBKS at BamHI; Apr, Emr | This study |
| pUC-VE | pUC-V8 digested with BsmI at a unique site in ssp, then converted to blunt ends and ligated to the 1.4-kb erm fragment from pBS-E1; Apr Emr | This study |
| pUC-E194C | PstI-XbaI fragment of pPQ126, containing the E194ts replication origin and chloramphenicol acetyl transferase (cat), cloned in pUC18; Apr Cmr | This study |
| pVE1C | 4.0-kb E194Cat fragment excised from pUC-E194C with PvuII and cloned at SmaI site of pUC-VE; Apr Emr Cmr | This study |
Inactivation of V8 protease (ssp-encoded) structural gene.
Recombinant plasmids and nucleotide primers for PCR amplification are listed in Tables 1 and 2, respectively. Construction of the pVE1C vector for inactivation of sspA was performed in E. coli DH5α, as outlined in Table 1. The ssp gene (9) was amplified from S. aureus RN4220 with primers sspA-F1 and sspA-R1 and then cloned in pUC18 using a HindIII site incorporated by the forward primer and an XbaI site at nucleotide 1383 of the ssp sequence, near the 3′ end of the PCR amplicon. The resulting plasmid is designated pUC-V8. The ermAB rRNA methylase of Tn917 (49) was amplified from plasmid pPQ126 (31), with primers Tn917-F and Tn917-R that incorporate terminal BamHI sequences. The 1.4-kb amplicon was cloned in pBKS+, creating pBS-E1. The ermAB fragment was excised from pBSE1, blunt ended with T4 polymerase, and ligated to pUC-V8 at a unique BsmI site at nucleotide 650 of sspA, creating plasmid pUC-VE. A fragment of S. aureus plasmid pPQ126 (31) containing the pE194ts temperature-sensitive plasmid replication origin and chloramphenicol acetyl transferase was excised with PstI and XbaI. After cloning in pUC18, the fragment was excised from the resulting pUC-VE plasmid with PυuII and ligated to SmaI-digested pUC-VE, creating shuttle vector pVE1C.
TABLE 2.
Primers used for amplification of gene fragments and specific probes
| Primer | Sequence | Nucleotide coordinates | Gene (reference) |
|---|---|---|---|
| sspA-F1 | AACAAATACTTTCCATTCGCTC | 107–128 | ssp (9) |
| sspA-F2 | CCAGATGAACCAAATAACCCTGAC | 1272–1295 | ssp (9) |
| sspA-F3 | CATTCGCTCTCAATTCCTTTC | 127–147 | ssp (9) |
| sspA-R1 | AGCTGTTTAGAGTGTGAATCGG | 1573–1552 | ssp (9) |
| sspA-R2 | AATGGTTTAAGGTTACCGCC | 542–523 | ssp (9) |
| sspB-F1 | AAAGCTACGCCTCTACCTGG | 1974–1993 | ssp operon |
| sspB-F2 | ACGGTAAATCACAAGGCAGAG | 2320–2340 | ssp operon |
| sspB-R1 | GTGGGTCATTAGGGTTTTGAG | 2454–2434 | ssp operon |
| sspB-R2 | CATACTGAATCCTGCACACCATGAG | 2189–2165 | ssp operon |
| sspC-F1 | TACGACACAACCAAACTCACAAC | 2695–2718 | ssp operon |
| sspC-R1 | GCGAAGTGCCAATACCTTG | 2965–2947 | ssp operon |
| sspC-R2 | CGTTTGCATGAGGATATGCTG | 2937–2917 | ssp operon |
| Tn917-F | ATTGGCACAAACAGGTAACGG | 256–276 | ermAB (49) |
| Tn917-R | CGGTCGTTTATGGTACCATTC | 1657–1638 | ermAB (49) |
| RNAIII-F | GAAGTAGAACAGCAACGCG | 744–762 | agr locus (37) |
| RNAIII-R | GATCACAGAGATGTGATGG | 1569–1551 | agr locus (37) |
pVE1C was transformed into S. aureus RN4220 by electroporation (36), selecting for erythromycin-resistant (Emr) transformants on BHI agar at 30°C. Mutants containing pVE1C integrated within the sspA allele were then selected by growth at 42°C on BHI agar supplemented with erythromycin (10 μg·ml−1), as described by Greene et al. (19). One mutant designated SP4221 was used as the donor strain for transfer of the mutation to S. aureus strain RN6390 by transduction with phage 85 (36). Emr transductants were replica plated on BHI containing chloramphenicol (5 μg·ml−1), and an Emr Cms transductant was designated SP6391. Southern blotting of HindIII-digested genomic DNA from this strain with probes specific for sspA and ermAB confirmed the expected allelic replacement mutation (data not shown).
Sequence analyses.
Nucleotide sequence data used to identify the ssp operon in S. aureus COL was obtained from The Institute for Genomic Research (http://www.tigr.org). Nucleotide sequence analysis was conducted with the IBI MacVector program (Eastman Kodak, New Haven, Conn.). Homology searches were conducted using the BLAST algorithms (1, 2) provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Protein sequence analysis was performed using programs provided by the ExPasy Proteomics Tools molecular biology server (3), at http://www.expasy.ch/tools.
PCR conditions.
Primers used for PCRs are described in Table 2. DNA segments were amplified in a volume of 25 μl, containing 1.0 ng of template DNA, 1.5 mM MgCl2, 1.5 μM (each) forward and reverse primers, 0.2 mM deoxynucleoside triphosphate mix, and 1.25 U of AmpliTaq DNA polymerase (Roche Canada, Laval, Quebec, Canada), using the buffer as supplied by the manufacturer. Cycling conditions consisted of a 4-min denaturation at 94°C followed by 30 cycles with denaturation at 94°C for 1 min, annealing at 52°C for 2 min, and extension at 72°C for 1 min. Where necessary, PCR products were gel purified using the GeneClean II kit (Bio 101; Vista, Calif.) for use in cloning experiments, or as probes in Southern and Northern blots.
Southern and Northern blotting.
DNA probes for Southern blots were generated by PCR as described above, using primers sspA-F1–sspA-R1 and Tn917-F–Tn917-R (Table 2). Additional probes for Northern blots were generated by PCR using primers sspB-F1–sspB-R1, sspC-F1–sspC-R1, sspA-F3–sspA-R2, and RNAIII-F–RNAIII-R. All probes were labeled with the ECL direct nucleic acid labeling system (Amersham-Pharmacia, Piscataway, N.J.). S. aureus genomic DNA was purified using QIAGEN 100/G genomic tips (Qiagen, Inc., Valencia, Calif.), following the manufacturer's protocol for gram-positive bacteria. RNA was prepared from stationary-phase (8 h) S. aureus cultures grown in protease culture medium, using TRIzol reagent (Gibco/BRL, Gaithersburg, Md.) and the FASTPREP FP120 instrument (Bio 101) as described previously (40). The concentration and purity of each sample were determined by measuring the absorbance at 260 and 280 nm.
For Southern blotting, 1.0 μg of HindIII-digested genomic DNA was electrophoresed through 0.8% (wt·vol−1) agarose containing ethidium bromide (0.5 μg·ml−1), in Tris-acetate-EDTA buffer. DNA was blotted by capillary transfer to Hybond N+ membrane (Amersham) using the alkaline transfer method (29). Northern blot analysis was performed with 10 μg of RNA after electrophoresis in 1.0% (wt·vol−1) agarose containing 0.66 M formaldehyde in morpholine propanesulfonic (MOPS) acid running buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA [pH 7.0]). For estimation of transcript sizes, 10 μg of RNA molecular weight standards (Sigma) ranging in size from 0.2 to 10 kb were electrophoresed next to RNA from S. aureus RN6390. RNA was transferred to a Hybond N+ membrane in 10× SSC buffer (0.15 M Na3-citrate, 1.5 M NaCl [pH 7.0]), and fixed to the membrane by baking at 80°C for 2 h. Processing of the blotted membranes was performed using the reagents and protocols provided with the ECL direct nucleic acid labeling and detection system (Amersham-Pharmacia), using Kodak Biomax ML autoradiography film.
SDS-PAGE, zymography, Western blotting, and N-terminal sequencing.
To determine profiles of secreted proteins, cell culture supernatant was mixed with an equal volume of ice cold 20% (wt·vol−1) trichloroacetic acid (TCA) and incubated on ice for 60 min. After centrifugation, (12,000 × g; 10 min) precipitated proteins were washed in ice-cold 70% (vol·vol−1) ethanol, air dried, and solubilized in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) reducing buffer. The samples were then subjected to SDS-PAGE as described by Laemmli (27). Proteins were visualized by staining with Coomassie blue. For zymography, culture supernatants were subjected to SDS-PAGE using 12% (wt·vol−1) acrylamide Zymogram Ready Gels (Bio-Rad, Hercules, Calif.) containing casein or gelatin (1 mg·ml−1). Following electrophoresis, the gels were shaken gently for 60 min at room temperature, in phosphate-buffered saline (PBS) containing 2.5% (vol·vol−1) Triton X-100 (Sigma). The gels were then incubated overnight at 37°C in buffer containing 50 mM Tris-HCl (pH 7.4), 200 mM NaCl, 5 mM CaCl2, 0.02% (vol·vol−1) Triton X-100, and 1 mM cysteine. The gels were then stained with Coomassie blue dye and destained to reveal zones of protease activity. Where indicated, culture supernatants were preincubated with specific protease inhibitors (DFP, 10 mM; E-64, 10 μM), and the same inhibitors were included in the development buffer. Zymograms for detection of autolysin activity were conducted as described previously (45), using acrylamide gels containing heat-killed and lyophilized Micrococcus luteus (Sigma) or S. aureus RN6390 cells (1 mg·ml−1). Zones of autolytic activity were detected by counterstaining with methylene blue. Sample volumes were adjusted to represent equivalent optical density at 600 nm (OD600) units, based on the cell density of each stationary-phase culture.
V8 protease in culture supernatants was detected by Western immunoblotting. To generate specific antibodies, each of two female New Zealand White rabbits were immunized by subcutaneous injection with 100 μg of purified V8 protease from S. aureus L530 (32), mixed with Freund's complete adjuvant (Sigma). Booster injections were administered at 2-week intervals, consisting of 100 μg of V8 protease mixed with 0.5 ml of Freund's incomplete adjuvant (Sigma). Purified V8 protease was employed for blot purification of monospecific antibodies, using pooled antisera from the second and third booster injections. For Western immunoblots, proteins from stationary-phase culture supernatants were subjected to SDS-PAGE through 12% (wt·vol−1) polyacrylamide gels, and transferred to Immobilon-P membranes (55). Western blotting was performed with blot-purified anti-V8 protease antibody as primary antibody, and alkaline phosphatase-conjugated goat anti-rabbit (heavy and light chain) immunoglobin G (Jackson Immuno Research, West Grove, Pa.) (5,000× dilution) as the secondary antibody. Blots were developed with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium alkaline phosphatase substrates (Bio-Rad).
For N-terminal sequence determination, culture supernatant proteins were precipitated with TCA and subjected to SDS-PAGE as described above, followed by transfer to polyvinylidene difluoride membrane in CAPS buffer, as described previously (57). After light staining, protein bands were excised with a scalpel and submitted to the University of Toronto HSC Biotechnology Center for N-terminal sequencing.
Assay of Fn binding, protease, and coagulase activity.
Assays for binding of 125I-Fn were conducted as described (40), using 5 × 107 S. aureus cells in each binding assay, with 4.5 × 108 non-Fn-binding S. simulans cells as carriers to assist in centrifugation. Labeling of Fn with 125I (NEN-Dupont, Boston, Mass.) was performed using the chloramine-T protocol (25), to a specific activity of approximately 27 MBq·nmol−1. All assays were performed in duplicate within 24 h of the labeling reaction. Binding data are expressed as the percentage of added Fn (50,000 cpm) that was bound by 5 × 107 S. aureus cells. For quantification of protease activity, samples of 18-h culture supernatants were assayed with resorufin-labeled casein (Roche) as previously described (40). The resorufin chromophore solubilized by protease activity was quantified (A574) after precipitation of undigested casein with TCA. Coagulase activity in culture supernatants from exponential (2 h)- or stationary (7 h)-phase cultures of S. aureus grown in BHI was quantified with sterile reconstituted rabbit plasma (BBL, Bedford, Mass.) in 10- by 75-mm glass culture tubes as described previously (40). The coagulase titer was defined as the log2 reciprocal of the highest dilution producing a firm clot. Protease and coagulase assays were both performed in duplicate.
Murine tissue abscess model.
Virulence of S. aureus RN6390 and isogenic derivatives was assessed using a murine tissue abscess model following established protocols (7, 8). Briefly, cells from logarithmic-phase cultures grown in BHI broth were diluted in sterile PBS and mixed with an equivalent volume of sterilized Cytodex beads (Sigma) suspended in PBS at a concentration of 20 μg·ml−1. The suspension (200 μl) was injected subcutaneously into the right flank of each of eight 4-week-old hairless crl:SKH1(hrhr)Br mice (Charles River, Wilmington, Mass.), using a 1-ml tuberculin syringe. The length and width of the lesions were measured daily, and the wound area (A) was determined by the formula A = π(L × W)/2, where L is the longest axis and W is the shortest axis. The mice were sacrificed at 72 h postchallenge.
Statistical analyses.
Data from phenotypic assays and the murine tissue abscess model were subjected to statistical analyses, by both the single-factor analysis of variance and the Bonferroni t test using the SigmaStat software program (Jandel Scientific). P values less than or equal to 0.05 were considered to indicate significant differences.
RESULTS
Analysis of nucleotide sequence downstream of the ssp-encoded serine protease in the genome of S. aureus COL.
A BLAST search (2 May 2000 release) of the S. aureus COL genome identified the ssp serine protease gene on contiguous nucleotide sequence no. 6218. Analysis of the nucleotide sequence downstream of ssp revealed two additional open reading frames (Fig. 1). The serine protease gene designated sspA is followed by sspB, which encodes a hypothetical protein of 393 amino acids. The SignalP program (3) predicts a signal peptide cleavage site after Ala36, creating a mature protein of 40.6 kDa. Amino acids 220 to 393 of SspB possess 47% identity and 64% similarity to staphopain (Fig. 1B), a 23-kDa cysteine protease purified from S. aureus strain V8 (23). The PROSCAN program (3, 4) identified a eukaryotic thiol protease histidine active site consensus pattern (LGHALAVVGNA), spanning amino acids 338 to 348, which is also conserved in staphopain (Fig. 1). Just 37 nucleotides from the stop codon of sspB is a third open reading frame, sspC, which encodes a 109-amino-acid protein with a predicted size of 12.9 kDa. A cytoplasmic localization was predicted for SspC using the Psort program (33), and SspC did not possess significant homology to other known proteins, including complete and incomplete microbial genomes. Following sspC is a hairpin structure, representing a possible transcription termination signal. PCR of genomic DNA with primer pairs sspA-F2–sspB-R2 and sspB-F2–sspC-R2 (Table 2) confirmed that the same order of genes was present on the genome of S. aureus RN6390 (data not shown).
FIG. 1.
Nucleotide and deduced amino acid sequence of the sspB and sspC genes from S. aureus strain COL (A) and alignment of the C-terminal region of SspB with the staphopain cysteine protease (23) purified from S. aureus strain V8 (B). (A) The sspA open reading frame encoding V8 protease terminates at nucleotide 1364, and the nucleotide coordinates of the ssp operon are additive to the original ssp sequence of S. aureus strain V8 (9). The 3′ end of the published ssp sequence terminates at nucleotide 1634 of the ssp operon, indicated by a bullet. Stop codons are indicated by asterisks. The predicted signal peptide cleavage site in the SspB amino acid sequence is indicated by a downward-pointing arrow, and the experimentally determined N-terminal sequence obtained from the 41-kDa SspB protein secreted by S. aureus SP6391 is represented by a solid underbar. The dashed underbar represents the N-terminal sequence of a 22-kDa protein secreted in increased abundance in a sarA mutant of S. aureus 8325-4 (10). An upward-pointing arrow indicates a potential processing site for cleavage of SspB at Glu219 by SspA to form a mature cysteine protease. The double underbar following the sspC open reading frame indicates a hairpin structure, representing a putative transcriptional termination signal. (B) The arrow points to an active site cysteine residue in staphopain, which has been determined by crystallography to bind the cysteine protease inhibitor, E-64 (23). The boxed and shaded residues in both panels represent a pattern of amino acids that is identified as a eukaryotic thiol protease histidine active site. Identical amino acids are indicated by colons, and lowercase letters represent nonidentical amino acids.
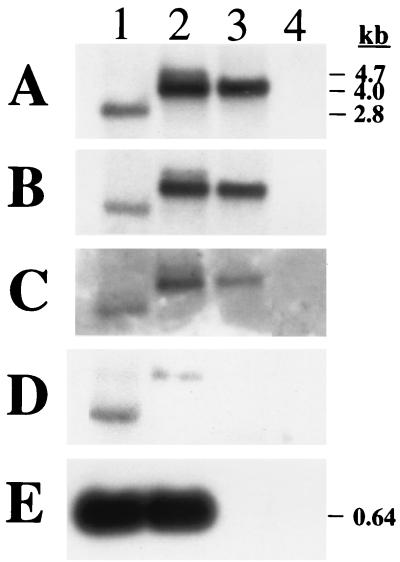
Northern blot analysis of the ssp transcript in S. aureus RN6390 and isogenic mutants.
The organization of the sspA, sspB, and sspC genes shown in Fig. 1 is suggestive of an operon structure. Consequently, it was necessary to inactivate sspA in S. aureus RN6390 using a nonpolar allelic replacement strategy. Northern blot analysis provided proof that the ssp genes are transcribed as an operon and that inactivation of sspA was achieved without affecting transcription of downstream genes (Fig. 2). RNA from S. aureus RN6390 displayed a 2.8-kb transcript that hybridized with probes specific for sspA, sspB, or sspC (Fig. 2A to C, lane 1). This is in agreement with the translation start codon of sspA and the stop codon of sspC being separated by 2.64 kb. Although sspA was disrupted by insertion of ermAB in SP6391, probes specific for sspA, sspB, or sspC each detected two RNA transcripts of 4.0 and 4.7 kb (Fig. 2A to C, lane 2). The same pattern was also observed with a probe specific for ermAB (data not shown). These findings suggest that transcription initiated from both the native sspA promoter and the ermAB cassette continues through the ssp operon.
FIG. 2.
Northern blots of RNA (10 μg) extracted from S. aureus RN6390 (lane 1), SP6391 (sspA::ermAB [lane 2]), SP6912 (agr sspA::ermAB [lane 3]), and RN6911 (agr [lane 4]). The probes used correspond to sspA (A), sspB (B), sspC (C), the 5′ end of sspA prior to the ermAB insertion point (D), or RNAIII (E).
This was confirmed by transducing the inactivated sspA allele (sspA::ermAB) of SP6391 into the agr-null strain S. aureus RN6911, creating strain SP6912 (agrΔ::tetM sspA::ermAB). In this situation, strain SP6912 displayed a single 4.0-kb ssp transcript (Fig. 2A to C, lane 3), whereas no ssp transcript was detected in the agr-null parent strain RN6911 (Fig. 2A to C, lane 4). Both strains maintained the same defect in transcription of RNAIII (Fig. 2E, lanes 3 and 4), due to the deletion of the agr locus in S. aureus RN6911. Therefore, the 4.0-kb ssp transcript in SP6912 and SP6391 originates from the ermAB cassette, which permits transcription of sspB and sspC independently of agr function. To confirm that the larger 4.7-kb ssp transcript in SP6391 originates from the native sspA promoter, a Northern blot was performed with a probe obtained by PCR with primer pair sspA-F3 and sspA-R2 (Table 2), spanning the 5′ end of sspA prior to the ermAB insertion point. As expected, this probe hybridized only to the 4.7-kb transcript of SP6391 and did not detect any transcript in SP6912. These data demonstrate that sspABC is transcribed as an operon and that nonpolar inactivation of sspA has been achieved in strain SP6391.
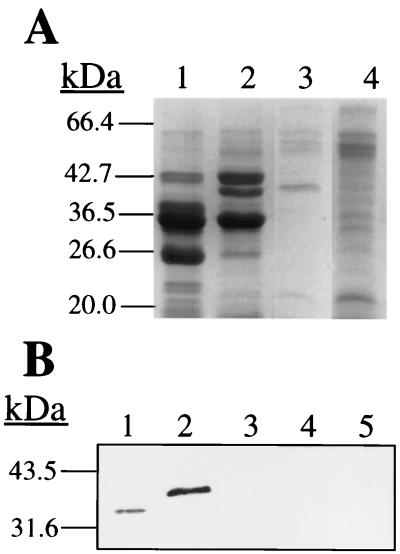
Inactivation of sspA exerts a pleiotropic effect on the profile of secreted proteins and proteases.
SDS-PAGE analysis was performed to assess the affect of the sspA mutation on the profile of secreted proteins (Fig. 3A). Culture supernatant of S. aureus SP6391 (sspA::ermAB) possessed a new 40-kDa protein that was not observed in RN6390. Relative to RN6390, culture supernatant of SP6391 also exhibited increased abundance of a protein doublet at approximately 43 kDa, decreased abundance of proteins at 37 and 27 kDa, and loss of at least two minor protein bands at 22 to 23 kDa (lane 2). The new 40-kDa protein in SP6391 corresponds to the expected size of SspB after processing at a predicted signal peptidase cleavage site, and N-terminal sequencing of this protein yielded the sequence DSHSKQLEINV, which matches perfectly with the predicted N terminus of secreted SspB (Fig. 1A). A protein of the same size was also present in the culture supernatant of SP6912 (agrΔ::tetM sspA::ermAB) (lane 3) and had the same N-terminal sequence DSHSKQLEIN, confirming its identity as SspB. This protein was not expressed in the agr-null parent strain RN6911. Therefore, inactivation of sspA results in accumulation of the 40-kDa SspB protein in the culture supernatant of SP6391, and transduction of the mutant allele into the agr-null strain S. aureus RN6911 permits this protein to be expressed and secreted independently of agr function.
FIG. 3.
SDS-PAGE (A) and Western immunoblot (B) of secreted proteins from S. aureus RN6390 and isogenic derivatives. The molecular masses of protein standards are indicated to the left of each gel. (A) Lane 1, S. aureus RN6390; lane 2, SP6391; lane 3, SP6912; lane 4, RN6911. The amount of protein loaded represents the equivalent of 2.3 OD600 units (lanes 1 and 2) or 6.8 OD600 units (lanes 3 and 4) of original culture. Secreted proteins are visualized by staining with Coomassie blue dye. (B) Purified V8 protease (60 ng) from S. aureus L530 (lane 1) or supernatant corresponding to 0.03 OD600 units of stationary phase culture from S. aureus RN6390 (lane 2), SP6391 (lane 3), SP6912 (lane 4), or RN6911 (lane 5) was subjected to SDS-PAGE (10% [wt·vol−1] polyacrylamide), transferred to Immobilon-P membrane, and probed with blot-purified polyclonal anti-V8 protease antibody.
The loss of SspA expression was confirmed by a Western immunoblot, using antibodies specific for SspA (Fig. 3B). Purified SspA from S. aureus L530 (lane 1) migrated slightly faster than an immunoreactive protein of S. aureus RN6390 (lane 2), which was not present in culture supernatant of SP6391 (lane 2). Therefore, the SspA serine protease of S. aureus RN6390 is slightly larger than that of S. aureus strain L530 and is not expressed in S. aureus SP6391.
The doublet of proteins at 43 kDa in S. aureus RN6390 (Fig. 3A, lane 1) yielded N-terminal sequences of LKANQVQPLNKYP and AT(Y?)KAKDDQTRA(V?)V. The former matches perfectly with amino acids 294 to 306 of the S. aureus glycerol ester hydrolase precursor (28). Signal from the latter sequence was weaker but resembled amino acids 281 to 289 (ta-KAKDDQT) of a triacylglycerol hydrolase from S. aureus (35). Proteins in the same 43-kDa size range were more abundant in the culture supernatant of S. aureus SP6391 (Fig. 3A, lane 2). Lipases of S. aureus are secreted as larger precursor forms of approximately 82 kDa that undergo proteolytic maturation to form mature proteins of 40 to 45 kDa (46). As loss of SspA function did not result in the appearance of unprocessed lipase, it appears that SspA is not required for the proteolytic maturation of secreted lipases. However, the increased abundance in culture supernatant from SP6391 of proteins corresponding in size to the mature lipase enzymes suggests that SspA may influence the stability of secreted lipase.
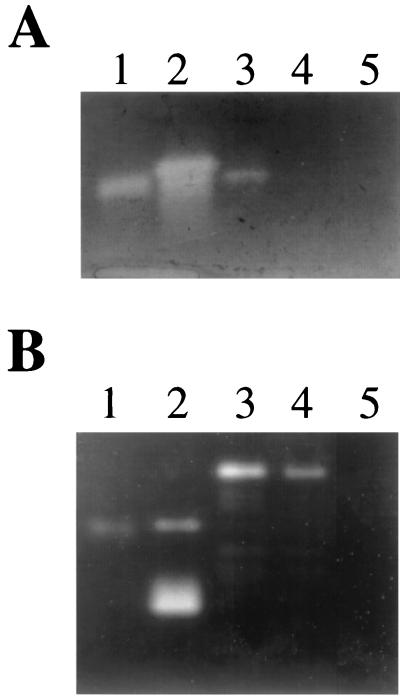
Protease zymogram analyses of S. aureus RN6390 and isogenic derivatives.
Zymogram analyses were conducted to examine the effect of sspA inactivation on the profile of secreted proteases. In zymogram gels containing casein, S. aureus RN6390 exhibited a doublet of closely migrating proteases (Fig. 4A, lane 2). The more active and slower-migrating protease was absent from culture supernatant of S. aureus SP6391 (lane 3), and as in the Western immunoblot, the serine protease of S. aureus RN6390 was slightly larger than purified SspA from S. aureus L530 (lane 1). Although the 40-kDa SspB protein was present only in the culture supernatant of SP6391 and SP6912 (Fig. 3A), no new zones of protease activity were detected on casein zymograms (Fig. 4A, lanes 3 and 4). However, in zymograms containing gelatin (Fig. 4B), SP6391 and SP6912 each exhibited a higher mass protease activity (lanes 3 and 4) that was not present in either RN6390 or RN6911 (lanes 2 and 5). Furthermore, a low-molecular-mass gelatinase of RN6390 was absent from culture supernatant of SP6391. When the samples were pretreated with E-64, a specific inhibitor of cysteine protease activity, the lower-mass gelatinase of RN6390 and the higher-mass activity in SP6391 and RN6912 were no longer detected (data not shown). Therefore, inactivation of sspA has resulted in the loss of a low-molecular-mass cysteine protease and the appearance of a higher-molecular-mass protease, representing the 40-kDa SspB protein.
FIG. 4.
Zymogram detection of secreted protease activity after electrophoresis in 12% acrylamide gels containing casein (A) or gelatin (B) as substrate. (A) For the casein zymogram, sample loading consisted of purified SspA (200 ng) from S. aureus L530 (lane 1) or supernatant from stationary-phase cultures of S. aureus RN6390 (lane 2), SP6391 (lane 3), SP6912 (lane 4), or RN6911 (lane 5). The amount of supernatant applied was equivalent to 0.012 OD600 units of stationary-phase culture for lane 2 (RN6390) or 0.03 OD600 units in lanes 3, 4, and 5. (B) The loading order is the same as that in panel A, except that lane 1 contained 600 ng of purified SspA and lanes 2 to 5 each contained 0.03 OD600 units of stationary phase culture supernatant. Protease activity appears as a clear zone against a Coomassie blue-stained background.
These observations suggest that the SspA serine protease is required for proteolytic maturation of the 40-kDa form of SspB. Accordingly, when culture supernatant from SP6912 (agr sspA::ermAB) was treated with purified SspA prior to zymogram analysis, the precursor form of SspB was processed to form lower-molecular-mass gelatinase activities (Fig. 5). Furthermore, as evident from Fig. 3A, inactivation of sspA resulted in loss of two proteins of 22 to 23 kDa, and appearance of the 40-kDa SspB protein in culture supernatant of SP6391. These observations were also reflected in the profile of secreted gelatinase activities (Fig. 4B). Cumulatively, these data indicate that SspB is expressed as a 40-kDa precursor protein, which is processed by SspA to form a mature cysteine protease of 22 to 23 kDa.
FIG. 5.
Zymogram analysis demonstrating conversion of SspB into lower-molecular-mass forms by treatment with purified SspA. The assay was performed as described above for Fig. 4B. Lane 1, 150 ng of V8 protease in PBS containing 1 mM cysteine; lane 2, 150 ng V8 protease pretreated with 10 mM DFP serine protease inhibitor; lane 3, 0.025 OD600 units of stationary-phase culture supernatant from SP6912; lane 4, supernatant from 0.08 OD600 units of SP6912 culture, incubated for 60 min in PBS containing 300 ng of purified V8 protease and 1 mM cysteine, followed by addition of 10 mM DFP; lane 5, identical to lane 4, except that DFP treatment was omitted. The higher-molecular-mass activity in lanes 1 and 2 containing purified SspA represents a shift in mobility that is observed when the sample is incubated in a reducing agent prior to electrophoresis. Coomassie blue staining of SDS-PAGE gels containing reduced or nonreduced SspA has confirmed that SspA exhibits faster migration when nonreduced (data not shown).
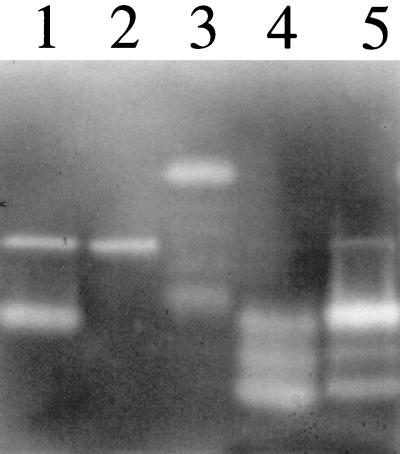
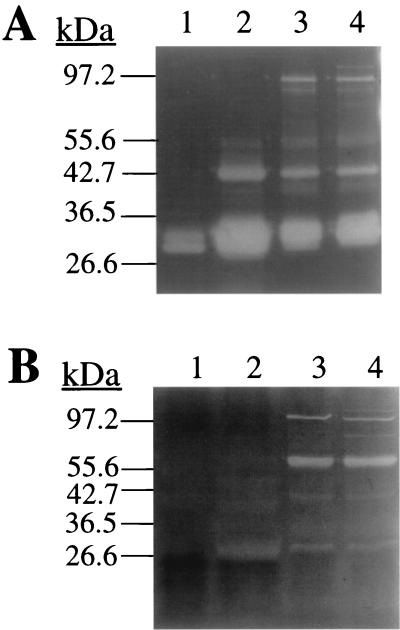
Inactivation of sspA results in an altered profile of secreted autolysins.
Through sequence analysis of the partially complete S. aureus COL genome (http://www.tigr.org), we have discovered that the atl gene encoding a major autolysin activity of S. aureus is located 5 kb upstream of the ssp operon. Atl is expressed as a 138-kDa precursor protein that is processed by proteolytic activity to release glucosaminidase (GL) and amidase (AM) domains of 54 and 63 kDa, respectively (39). To determine if autolytic activity is influenced by the sspA mutation, zymogram analyses of culture supernatants were conducted using gels containing M. luteus or S. aureus cells, to detect GL (Fig. 6A) and AM (Fig. 6B) activity, respectively. Compared to RN6390 (Fig. 6A, lane 1), S. aureus SP6391 (lane 2) and RN6911 (lane 4) both exhibited enhanced GL activity in the 30-kDa size range, together with the appearance of an active 43-kDa GL and a less active 56-kDa GL, which were not present in RN6390 (Fig. 6A). Therefore, loss of SspA function has altered the profile of secreted GL activities, resembling that of the agr-null strain, RN6911. In addition, RN6911 exhibited at least three GL activities in the high-molecular-mass range (∼97 kDa) that were not evident in either SP6391 or RN6390.
FIG. 6.
Detection of secreted GL (A) and AM (B) activity by electrophoresis of culture supernatants in 10% acrylamide gels containing heat-killed M. luteus (A) or S. aureus RN6390 (B) cells (1 mg·ml−1). Lane 1, S. aureus RN6390; lane 2, SP6391; lane 3, SP6912; lane 4, RN6911. Sample loading was equivalent to either 0.58 OD600 units (A) or 3.0 OD600 units (B) of stationary-phase culture supernatant. Autolytic activity appears as a clear zone against a methylene blue-stained background. The molecular masses of protein standards are indicated to the left of each gel.
Although S. aureus SP6391 exhibited a number of weak AM activities ranging in size from 29- to 56-kDa that were not evident in RN6390 (Fig. 6B), loss of SspA function did not have the same impact on AM profiles as it did on GL activity. When the autolysin profiles of RN6911 (agr) and SP6912 (agr sspA::ermAB) were compared, RN6911 exhibited a 105-kDa autolysin that was active on both GL and AM substrates and also a faint 73-kDa AM activity that were not detected in SP6912. As these two strains differ only in the sspA::ermAB mutation that results in secretion of the SspB cysteine protease, this result suggests that SspB may process the high-molecular-mass precursor forms of specific autolysins.
Quantitative assessment of virulence and virulence factor expression.
Total protease activity in culture supernatant of S. aureus SP6391 was diminished by more than 60% relative to RN6390, as determined with resorufin-labeled casein (Table 3). Culture supernatant of S. aureus RN6911 was devoid of protease activity, consistent with the deletion of the agr locus in this strain. Although the protease activity of SP6912 was numerically 10-fold greater than that of RN6911 (0.039 versus 0.004), it was still very low. Therefore, as also evident from zymogram analysis, this protease is active on gelatin but does not exhibit appreciable activity towards casein. S. aureus strains RN6390 and SP6391 both showed growth phase-dependent reduction in coagulase titer and Fn binding and did not differ significantly from one another in either of these activities. Therefore, inactivation of sspA function did not affect the stability of these proteins. Results from Fn binding and coagulase assays of RN6911 were as expected for the agr-null mutant phenotype of this strain.
TABLE 3.
Phenotypic properties of S. aureus RN6390 and isogenic derivativesa
| Strain | Protease activity (n = 4) at 18 h | Coagulase titer (n = 3) at:
|
% Fn binding (n = 2) at:
|
||
|---|---|---|---|---|---|
| 2 h | 7 h | 2 h | 6 h | ||
| RN6390 | 0.830 ± 0.04 | 4.3 ± 0.7 | 1.0 ± 1.0 | 6.4 ± 0.4 | 0.8 ± 0.1 |
| RN6911 | 0.004 ± 0.002* | 4.7 ± 0.3 | 4.7 ± 0.3* | 17.1 ± 0.1* | 5.3 ± 0.3* |
| SP6391 | 0.310 ± 0.06* | 3.7 ± 1.2 | 0.7 ± 0.7 | 4.1 ± 1.6 | 1.2 ± 0.3 |
| SP6912 | 0.039 ± 0.002* | NDb | ND | ND | ND |
Protease activity (A574), coagulase titer (log2), and percent binding of 125I-Fn were determined for cultures harvested in exponential growth phase (2 h) or stationary phase (6, 7, or 18 h.). Values are reported as the mean of n independent assays ± standard error of the mean. Values that are significantly different from S. aureus RN6390, as established by the Bonferroni t test (P < 0.05) are indicated by asterisks.
ND, not determined.
When the virulence of RN6390 and that of SP6391 were assessed using a murine abscess infection model, mice challenged with SP6391 exhibited a larger area of tissue abscess at each of 24, 48, and 72 h postchallenge compared to S. aureus RN6390 (Table 4). This difference was statistically significant at 72 h but not at 24 or 48 h. Although the mice challenged with SP6391 received a larger challenge inoculum, these data were also reproducible at smaller challenge doses. Therefore, loss of serine protease function has not impaired the ability of SP6391 to form a tissue abscess.
TABLE 4.
Surface area of tissue abscess at indicated time points following challenge with S. aureus RN6390 and isogenic derivatives
| Strain | Inoculum (CFU) | Mean surface area ± SEM (mm2) of tissue abscess at:
|
||
|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||
| RN6390 | 3.6 × 106 | 454.9 ± 62.2 | 399.6 ± 54.1 | 245.9 ± 35.9 |
| RN6911 | 2.6 × 106 | No visible lesion | No visible lesion | No visible lesion |
| SP6391 | 6.5 × 106 | 564.2 ± 53.0 | 536.9 ± 55.2 | 376.2 ± 35.7a |
Significant difference compared to RN6390 (P < 0.05).
DISCUSSION
We have identified an operon structure encoding the secreted serine and cysteine proteases SspA and SspB of S. aureus. The organization of the operon and our experimental findings indicate a sequential pathway in which the SspA serine protease is required for proteolytic maturation of SspB. This conclusion is supported by a study on the phenotype of a sarA mutation in S. aureus. Although the SarA protein is required for optimal transcription of the agr locus (11, 13), it also represses expression of secreted proteases. Inactivation of sarA in S. aureus 8325-4 resulted in the appearance in the culture supernatant with elevated levels of V8 protease (SspA), a putative metalloprotease, and a 22-kDa protein designated P4 (10). The reported N-terminal sequence of P4 matches perfectly with amino acids 231 to 250 of SspB (Fig. 1A), suggesting that the 22-kDa P4 protein originated from processing of the 40-kDa precursor form of SspB after Glu219. This is consistent with the SspA serine protease's exhibiting a high specificity for cleavage on the carboxyl-side of glutamic acid residues (24). These observations, combined with the results from our present study, define a requirement for the SspA serine protease in proteolytic maturation of the 40-kDa SspB cysteine protease. To our knowledge, this is the first report of an operon structure containing two tandem protease genes, in which the activity of the first encoded protease is required to process the second.
Others have shown that the V8 protease (SspA) is secreted as an inactive precursor form, which is processed by the metalloprotease aureolysin to form the mature serine protease (15). Aureolysin is also expressed as a precursor form (47), but its mechanism of maturation is not known. Therefore, there is a cascade pathway in the maturation of the secreted proteases of S. aureus, where the aureolysin metalloprotease activates the SspA serine protease, which in turn is required for proteolytic maturation of the SspB cysteine protease. It is not known if similar pathways exist in other gram-positive pathogens, but there is an interesting parallel with the cotranscribed gelE gelatinase and sprE serine protease of Enterococcus faecalis (44). A BLAST search reveals that gelE and sprE exhibit the greatest homology towards the aureolysin and SspA proteases respectively of S. aureus. Therefore, the gelE-sprE transcriptional unit of E. faecalis may represent another example of an operon structure in which the first encoded protease serves to process the second.
A gene encoding an N-acetylmuramidase activity is upstream of the gelE-sprE operon of E. faecalis (44), and searching of the S. aureus COL genome reveals that the major atl autolysin of S. aureus is also upstream of the ssp operon. Oshida et al. have shown that processing of the high-molecular-mass form of Atl can be blocked with protease inhibitors (39). However, loss of SspA function did not result in the appearance of an autolysin corresponding in size to the 138-kDa precursor form of Atl. Therefore, it appears that SspA is not required for maturation of the Atl autolysin. Our finding that loss of SspA resulted in enhanced levels of a 29- to 32-kDa GL and appearance of a 43-kDa GL suggests that SspA functions to control autolytic activity. This is consistent with a report in which protease-hyperproducing strains of Bacillus subtilis exhibited decreased rates of peptidoglycan turnover, while cultures supplemented with a serine protease inhibitor demonstrated an increased rate of cell wall turnover (26). However, it is not clear from our data if SspA serves to inactivate autolytic function or if the 43-kDa GL appearing in the culture supernatant of SP6391 is due to another mechanism. In this respect, inactivation of the lrgAB operon in S. aureus RN6390 resulted in the appearance of 43 to 56-kDa autolytic activities (20). It was proposed that the LrgAB proteins act as antiholins, thereby prohibiting the export of autolysins. Therefore, proteases could influence autolytic activity at multiple levels, including inactivation of autolytic activity, or by controlling the stability of holin and antiholin proteins that moderate autolysin secretion.
Enhanced autolytic activity may have contributed to the slight increase in virulence of S. aureus SP6391 in the tissue abscess infection model, perhaps by promoting the release of proinflammatory cell wall material. This result differs from a finding in which STM was applied towards S. aureus RN6390. In this study, a Tn917 insertion in the ssp-encoded serine protease resulted in moderate attenuation of virulence in tissue abscess and burn wound infection models and strong attenuation in a systemic infection model (14). Possibly, the ssp mutant obtained by STM exerted a polar effect on transcription of genes downstream of sspA. Therefore, the attenuated virulence attributed to SspA may have been due to the loss of function conferred by sspB and sspC. Similar considerations affected the determination of phenotype attributed the gelE gelatinase of E. faecalis. Initially, inactivation of gelE was reported to result in a significantly delayed time to death in a peritonitis model of infection (53). However, a subsequent study revealed that inactivation of gelE exerted a polar effect on the cotranscribed sprE serine protease (44). Therefore, the virulence phenotype could not be attributed to gelE alone.
Except for its homology to the staphopain cysteine protease purified from S. aureus strain V8 as noted in Fig. 1, SspB exhibits no significant homology towards any other protein. The 40-kDa precursor form of SspB is active on gelatin, suggesting that proteolytic processing is not an absolute requirement for activity. This differs from the 40-kDa SpeB cysteine protease of Streptococcus pyogenes, which is secreted as an inactive zymogen that undergoes autocatalytic conversion to form a mature cysteine protease (21). In contrast, the periodontain and gingipain cysteine proteases of Porphyromonas gingivalis are expressed as active precursors that undergo proteolytic maturation (42, 43). As with SspB, periodontain was active on gelatin zymograms but exhibited no activity towards casein. Periodontain also exhibited no activity towards a number of other structured proteins (34), with the exception of its ability to cleave and inactivate plasma α1-proteinase inhibitor (α1-PI). The α1-PI normally functions to limit tissue destruction during inflammatory responses by moderating the activity of the large quantities of human neutrophil elastase and cathepsin G released by degranulating neutrophiles. Consequently, inactivation of α1-PI by periodontain was proposed to promote the destructive inflammation associated with acute P. gingivalis gingivitis. Others have reported that V8 protease (SspA) of S. aureus can cleave α1-PI and that the cleaved product was a potent chemoattractant of human neutrophiles (6). However, our data did not support a role for SspA in promoting the inflammation associated with tissue abscess formation.
The SspA-defective mutant also did not exhibit enhanced Fn binding, as predicted from our previous work in which purified V8 protease promoted the rapid loss of cell-surface fibronectin binding protein when added to early-exponential-phase cultures of S. aureus clinical isolates (32). Possibly, growth phase-dependent loss of cell surface Fn-binding protein may represent the combined effect of two or more proteases. In this respect, we have shown that loss of SspA function promoted a pleiotropic phenotype, including failure to process the secreted precursor form of SspB, and appearance of new autolytic activities in the culture supernatant. Therefore, the secreted proteases of S. aureus may cumulatively exert a profound effect on the growth and physiology of this diverse microbial pathogen. Studies are in progress to precisely define the functions of the SspB cysteine protease and SspC, which is predicted to reside in the cytoplasm and exhibits no significant homology to any other known protein.
ACKNOWLEDGMENTS
This work was funded by Medical Research Council operating grant MOP-12669. K.R. is the recipient of an Ontario Graduate Scholarship award.
We thank Christine Watt for technical assistance. Sequencing of the S. aureus COL genome was accomplished with support from the National Institutes of Allergy and Infectious Diseases and the Merck Genome Research Institute.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel R D, Bairoch A, Hochstrasser D F. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 4.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balaban N, Goldkorn T, Nhan R T, Dang L B, Scott S, Ridgley R M, Rasooly A, Wright S C, Larrick J W, Rasooly R, Carlson J R. Autoinducer of virulence as a target for vaccine and therapy against Staphylococcus aureus. Science. 1998;280:438–440. doi: 10.1126/science.280.5362.438. [DOI] [PubMed] [Google Scholar]
- 6.Baran K, Gorka M, Potempa J, Porwit-Bobr Z. Chemoattractant activity of Staphylococcus aureus serine proteinase modified human plasma alpha-1-proteinase inhibitor. Antonie Leeuwenhoek. 1989;56:361–365. doi: 10.1007/BF00443750. [DOI] [PubMed] [Google Scholar]
- 7.Betschel S D, Borgia S M, Barg N L, Low D E, de Azavedo J C. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect Immun. 1998;66:1671–1679. doi: 10.1128/iai.66.4.1671-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunce C, Wheeler L, Reed G, Musser J, Barg N. Murine model of cutaneous infection with gram-positive cocci. Infect Immun. 1992;60:2636–2640. doi: 10.1128/iai.60.7.2636-2640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmona C, Gray G L. Nucleotide sequence of the serine protease gene of Staphylococcus aureus, strain V8. Nucleic Acids Res. 1987;15:6757. doi: 10.1093/nar/15.16.6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan P F, Foster S J. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung A L, Bayer M G, Heinrichs J H. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J Bacteriol. 1997;179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung A L, Eberhardt K J, Chung E, Yeaman M R, Sullam P M, Ramos M, Bayer A S. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Investig. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung A L, Projan S J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J Bacteriol. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulter S N, Schwan W R, Ng E Y, Langhorne M H, Ritchie H D, Westbrock-Wadman S, Hufnagle W O, Folger K R, Bayer A S, Stover C K. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 15.Drapeau G R. Role of metalloprotease in activation of the precursor of staphylococcal protease. J Bacteriol. 1978;136:607–613. doi: 10.1128/jb.136.2.607-613.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drapeau G R, Boily Y, Houmard J. Purification and properties of an extracellular protease of Staphylococcus aureus. J Biol Chem. 1972;247:6720–6726. [PubMed] [Google Scholar]
- 17.Fattom A I, Naso R. Staphylococcal vaccines: a realistic dream. Ann Med. 1996;28:43–46. doi: 10.3109/07853899608999073. [DOI] [PubMed] [Google Scholar]
- 18.Gillaspy A F, Lee C Y, Sau S, Cheung A L, Smeltzer M S. Factors affecting the collagen binding capacity of Staphylococcus aureus. Infect Immun. 1998;66:3170–3178. doi: 10.1128/iai.66.7.3170-3178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene C, McDevitt D, Francois P, Vaudaux P E, Lew D P, Foster T J. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol Microbiol. 1995;17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 20.Groicher K H, Firek B A, Fujimoto D F, Bayles K W. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–1801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gubba S, Low D E, Musser J M. Expression and characterization of group A Streptococcus extracellular cysteine protease recombinant mutant proteins and documentation of seroconversion during human invasive disease episodes. Infect Immun. 1998;66:765–770. doi: 10.1128/iai.66.2.765-770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann B, Schomburg D, Hecht H J. Crystal structure of a thiol proteinase from Staphylococcus aureus V-8 in the E-64 inhibitor complex. Acta Crystallogr. 1993;49(Suppl.):102. [Google Scholar]
- 24.Houmard J, Drapeau G R. Staphylococcal protease: a proteolytic enzyme specific for glutamoyl bonds. Proc Natl Acad Sci USA. 1972;69:3506–3509. doi: 10.1073/pnas.69.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter W M. Radioimmunoassay. In: Weir D M, editor. Handbook of experimental immunology. Oxford, United Kingdom: Blackwell Scientific Publications; 1978. pp. 14.1–14.40. [Google Scholar]
- 26.Jolliffe L K, Doyle R J, Streips U N. Extracellular proteases modify cell wall turnover in Bacillus subtilis. J Bacteriol. 1980;141:1199–1208. doi: 10.1128/jb.141.3.1199-1208.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee C Y, Iandolo J J. Lysogenic conversion of staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J Bacteriol. 1986;166:385–391. doi: 10.1128/jb.166.2.385-391.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J K, Parker B, Kowalic T. Rapid alkaline blot-transfer of viral dsRNAs. Anal Biochem. 1987;163:210–218. doi: 10.1016/0003-2697(87)90115-1. [DOI] [PubMed] [Google Scholar]
- 30.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 31.Luchansky J B, Benson A K, Atherly A G. Construction, transfer and properties of a novel temperature-sensitive integrable plasmid for genomic analysis of Staphylococcus aureus. Mol Microbiol. 1989;3:65–78. doi: 10.1111/j.1365-2958.1989.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 32.McGavin M J, Zahradka C, Rice K, Scott J E. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun. 1997;65:2621–2628. doi: 10.1128/iai.65.7.2621-2628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 34.Nelson D, Potempa J, Kordula T, Travis J. Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J Biol Chem. 1999;274:12245–12251. doi: 10.1074/jbc.274.18.12245. [DOI] [PubMed] [Google Scholar]
- 35.Nikoleit K, Rosenstein R, Verheij H M, Gotz F. Comparative biochemical and molecular analysis of the Staphylococcus hyicus, Staphylococcus aureus and a hybrid lipase. Indication for a C-terminal phospholipase domain. Eur J Biochem. 1995;228:732–738. [PubMed] [Google Scholar]
- 36.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 37.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 38.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oshida T, Sugai M, Komatsuzawa H, Hong Y M, Suginaka H, Tomasz A. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc Natl Acad Sci USA. 1995;92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papakyriacou H, Vaz D, Simor A, Louie M, McGavin M J. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J Infect Dis. 2000;181:990–1000. doi: 10.1086/315342. [DOI] [PubMed] [Google Scholar]
- 41.Patel A H, Kornblum J, Kreiswirth B, Novick R, Foster T J. Regulation of the protein A-encoding gene in Staphylococcus aureus. Gene. 1992;114:25–34. doi: 10.1016/0378-1119(92)90703-r. [DOI] [PubMed] [Google Scholar]
- 42.Pavloff N, Potempa J, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 43.Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thogersen I B, Enghild J J, Travis J. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998;273:21648–21657. doi: 10.1074/jbc.273.34.21648. [DOI] [PubMed] [Google Scholar]
- 44.Qin X, Singh K V, Weinstock G M, Murray B E. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun. 2000;68:2579–2586. doi: 10.1128/iai.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qoronfleh M W, Wilkinson B J. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of beta-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob Agents Chemother. 1986;29:250–257. doi: 10.1128/aac.29.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rollof J, Normark S. In vivo processing of Staphylococcus aureus lipase. J Bacteriol. 1992;174:1844–1847. doi: 10.1128/jb.174.6.1844-1847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabat A, Kosowska K, Poulsen K, Kasprowicz A, Sekowska A, van Den Burg B, Travis J, Potempa J. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect Immun. 2000;68:973–976. doi: 10.1128/iai.68.2.973-976.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saravia-Otten P, Muller H P, Arvidson S. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J Bacteriol. 1997;179:5259–5263. doi: 10.1128/jb.179.17.5259-5263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw J H, Clewell D B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985;164:782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheagren J N. Inflammation induced by Staphylococcus aureus. In: Gallen J I, Goldstein I M, Snyderman R, editors. Inflammation: basic principles and clinical correlates. New York, N.Y: Raven Press; 1988. pp. 829–840. [Google Scholar]
- 51.Sheagren J N. Staphylococcus aureus. The persistent pathogen. N Engl J Med. 1984;310:1368–1373. doi: 10.1056/NEJM198405243102107. [DOI] [PubMed] [Google Scholar]
- 52.Short J M, Fernandez J M, Sorge J A, Huse W D. Lambda ZAP: a bacteriophage expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7587. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh K V, Qin X, Weinstock G M, Murray B E. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178:1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 54.Sun Q, Smith G M, Zahradka C, McGavin M J. Identification of D motif epitopes in Staphylococcus aureus fibronectin-binding protein for the production of antibody inhibitors of fibronectin binding. Infect Immun. 1997;65:537–543. doi: 10.1128/iai.65.2.537-543.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 57.Yuen S W, Chui A H, Wilson K J, Yuan P M. Microanalysis of SDS-PAGE electroblotted proteins. BioTechniques. 1989;7:74–82. [PubMed] [Google Scholar]