Abstract
The mannose-binding lectin gene MANNOSE-BINDING LECTIN 1 (MBL1) is a member of the G-type lectin family and is involved in defense in strawberry (Fragaria × ananassa). Genome-wide identification of the G-type lectin family was carried out in woodland strawberry, F. vesca, and 133 G-lectin genes were found. Their expression profiles were retrieved from available databases and indicated that many are actively expressed during plant development or interaction with pathogens. We selected MBL1 for further investigation and generated stable transgenic FaMBL1-overexpressing plants of F. ×ananassa to examine the role of this gene in defense. Plants were selected and evaluated for their contents of disease-related phytohormones and their reaction to biotic stresses, and this revealed that jasmonic acid decreased in the overexpressing lines compared with the wild-type (WT). Petioles of the overexpressing lines inoculated with Colletotrichum fioriniae had lower disease incidence than the WT, and leaves of these lines challenged by Botrytis cinerea showed significantly smaller lesion diameters than the WT and higher expression of CLASS II CHITINASE 2-1. Our results indicate that FaMBL1 plays important roles in strawberry response to fungal diseases caused by C. fioriniae and B. cinerea.
Keywords: Anthracnose, fungal disease resistance, B-lectin, Botrytis cinerea, Colletotrichum fioriniae, Fragaria × ananassa, G-type lectin, grey mould, phytohormones, strawberry
FaMBL1plays a role in strawberry defense against fungal pathogens, and transgenic octoploid strawberries overexpressing FaMBL1are less susceptible to anthracnose and grey mould.
Introduction
Strawberry (Fragaria × ananassa) is an economically important fruit worldwide and is considered a model plant system for the Rosaceae. It is susceptible to a large number of pathogens including the Colletotrichum acutatum species complex (anthracnose) and Botrytis cinerea (grey mould), which cause enormous economic losses (Guidarelli et al., 2011; Petrasch et al., 2019). Although both fungi can infect the fruits at both unripe and ripe stages, the symptoms appear only on red ripe fruits whilst on white unripe fruits the pathogens become quiescent. Transcriptome analysis of white and red fruits inoculated with C. acutatum have shown that a mannose-binding lectin gene, FaMBL1 (GenBank accession no. KF962716) is the most up-regulated gene in resistant white fruit (Guidarelli et al., 2011). Transient transformation to silence FaMBL1 results in white fruit with an increased susceptibility to C. acutatum (Guidarelli et al., 2014).
The protein encoded by FaMBL1 is composed of an N-terminal signal peptide, a Galanthus nivalis agglutinin-related lectin (GNA) domain, and a Pan-apple (PAN) domain. The GNA domain is the characteristic domain of G-type lectin, which is an important family of plant lectins that have affinity to mannose or mannose-containing N-glycans (Barre et al., 2001). Due to their ability to recognize and bind mannose, G-type lectins have important functions in plant growth and defense. The roles of G-type lectins in resistance to insects, fungi, and bacteria have been described; for example, the expression of G-type lectin genes in potato (Down et al., 1996), maize (Wang et al., 2005), and wheat (Miao et al., 2011) increases resistance to aphids by inhibiting their development and decreasing fecundity. The pepper G-type lectin genes CaMBL1 and CaGLP1 have also been found to be involved in defense against Xanthomonas campestris pv vesicatoria and are required for plant cell death and defense signaling (Hwang and Hwang, 2011; Kim et al., 2015). The G-type lectin gene Pi-d2 from rice cv. Digu provides resistance to Magnaporthe grisea. The transfer of Pi-d2 from the resistant Digu to the susceptible cv. TP309 confers the latter with resistance to Magnaporthe grisea (Chen et al., 2006). Interestingly, the difference between the native Pi-d2 protein forms in Digu versus TP309 is only a single amino acid change (Ile441Met) in the transmembrane (TM) domain. This suggests that the TM domain plays an important role in the ligand recognition and signal transduction (Chen et al., 2006). Another G-type lectin, LORE, from Arabidopsis is composed of GNA, S-locus glycoprotein, PAN, transmembrane, and kinase domains and plays a role in plant innate immunity via sensing the lipopolysaccharide (LPS) of Pseudomonas and Xanthomonas (Ranf et al., 2015). Accordingly, Arabidopsis Col-0 plants pre-treated with Pseudomonas LPS are more resistant to subsequent infection with Pseudomonas. To determine the role of different domains, Ranf et al. (2015) produced the mutants lore-1 and lore-2 by protein truncation after the S-locus glycoprotein domain and substitution of an amino acid (Gly391Glu) in the PAN domain, respectively, and found that LPS-induced resistance was lost in both.
It is well known that the balance of hormonal crosstalk strongly influences the outcome of plant–pathogen interactions (Robert-Seilaniantz et al., 2011), and the contribution of lectins to plant resistance seems to be consistently displayed in a phytohormone-dependent manner (Bonaventure, 2011; Gilardoni et al., 2011; Hwang and Hwang, 2011).
Despite the clear importance of G-type lectins in plant defense responses, there have been few studies examining their expression strawberry (Martínez Zamora et al., 2008; Ma et al., 2021). Recently, with the release of the updated genome annotation and comprehensive gene expression atlas of woodland strawberry, F. vesca (Li et al., 2019), reliable data have become available for genome-wide analysis of G-type lectin genes in strawberry. In this study, we used this genome annotation together with the available transcriptome data to identify the G-lectin gene family members in this species and to analyse their domain arrangements and expression profiles. The results implied the great potential of many G-lectin members in strawberry defense responses and provided the basis for functional characterization of FaMBL1 in cultivated strawberry. To gain insights into the effects of this gene on plant defense, we generated and characterized genetically transformed plants overexpressing FaMBL1.
Materials and methods
Genome-wide analysis of G-type lectin genes in F. vesca
Identification and domain organization of G-type lectins
To identify G-type lectin genes, the GNA domain of FvH4_3g18380 (homolog of FaMBL1) was used as the query for a BLASTp search in the Genome Database for Rosaceae (GDR; https://www.rosaceae.org/) (Jung et al., 2019). The Fragaria vesca Whole Genome v4.0.a2 database was also used (https://www.rosaceae.org/species/fragaria_vesca/genome_v4.0.a2). Results with E-values <1 × 10–6 were considered as G-type lectin protein candidates. With the same settings, a second BLASTp was conducted using a number of variable GNA domains from G-lectin proteins found in the first BLASTp search: FvH4_1g23370, FvH4_2g12390, FvH4_2g14250, FvH4_2g26490, FvH4_2g29050, FvH4_2g33830, FvH4_3g03230, FvH4_3g03301, FvH4_3g03410, FvH4_3g03430, FvH4_3g06140, FvH4_3g15930, FvH4_3g18370, FvH4_3g21270, FvH4_3g43440, FvH4_4g02170, FvH4_5g31680, FvH4_6g00300, FvH4_6g12870, FvH4_6g44106. The domains of each candidate gene were checked manually using the InterProScan website (https://www.ebi.ac.uk/interpro/search/sequence/) (Quevillon et al., 2005). The transmembrane domain was checked using TMHMM Server v. 2.0 (https://dtu.biolib.com/DeepTMHMM) (Krogh et al., 2001).
Chromosome location of G-type lectins
Visualization of the chromosome locations of the G-lectin genes was accomplished using MapGene2Chromosome v.2.0 (http://mg2c.iask.in/mg2c_v2.0/) (Chao et al., 2015). The coordinates of the G-lectin genes on the strawberry genome were obtained from the GDR website (F. vesca v4.0.a2).
Expression profiles of G-type lectin genes
Expression profiles for the F. vesca G-lectin genes were retrieved from the supplementary material provided by Li et al. (2019). The expression levels of the genes in different tissues (flowers, fruits at different developmental stages, seedlings, leaves, meristems, and roots) were used to construct a heatmap using the the R package, ComplexHeatmap (Gu et al., 2016).
To infer the expression of G-lectin genes upon pathogen infection, we used the transcriptome profiles of F. ×ananassa infected by Botrytis cinerea (Xiong et al., 2018; Haile et al., 2019), and of F. vesca infected by Phytophthora cactorum (Toljamo et al., 2016) and by Podosphaera aphanis (Jambagi and Dunwell, 2015) . In addition, we also used transcriptome profiles of F.×ananassa after cold stress (Zhang et al., 2019) and after preharvest application of benzothiadiazole and chitosan (Landi et al., 2017) to obtain G-lectin gene expression profiles in different tissues. For this, the G-lectin genes from the F.×ananassa transcriptome datasets were converted to their F. vesca orthologs.
Genetic transformation and characterization of FaMBL1 in F. ×ananassa
Transgenic plants and droplet digital PCR
Genetically transformed plants overexpressing FaMBL1 were produced and their copy numbers of the transgene were determined by droplet digital PCR (ddPCR) (Ma et al., 2021). Briefly, the full-length sequence of FaMBL1 was cloned into the pK7WG2 vector (https://gatewayvectors.vib.be/) and the resulting construct overexpressing FaMBL1 was checked by PCR, restriction enzyme digestion, and sequencing of the PCR product (Ma et al., 2021). The resulting vector, 35S::FaMBL1, was introduced into Agrobacterium tumefaciens strain EHA105 using the freeze–thaw shock method (Weigel and Glazebrook, 2006) to generate transgenic plants. Expanded leaves from 3-week-old in vitro elongated strawberry shoots (F. ×ananassa cv. Sveva) were used as starting explants for the genetic transformation trial, following the protocol developed by Cappelletti et al. (2015). The transformed lines were screened by selective media and specific amplicons via PCR (Ma et al., 2021). Three lines (18F6G1, 19F2G1, and 27F8G1) were selected and used for this study.
ddPCR was performed to determine the copy number of the target gene in each transgenic line using the QX100™ Droplet Digital PCR System (Bio-Rad), following the process described in our previous study (Ma et al., 2021). The copy numbers of overexpressing vectors obtained in this study are relative values. The overexpressing lines with different copy numbers were subsequently propagated through stolons in a greenhouse to obtain 30 plants for each transformed line, which were then used in the subsequent experiments. The plants were grown in pots containing soil, with one plant per pot, under natural daylight.
RNA extraction, cDNA synthesis, and quantitative PCR
Young leaves, stolons, and petioles (150 mg per sample) of similar growth stages were sampled from mature transgenic and wild-type (WT) plants and were used for RNA extraction following a rapid CTAB method (Gambino et al., 2008). The obtained RNA was treated using a TURBO DNA-free™ Kit (Invitrogen) for removal of residual DNA. The purified RNA was quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific) and RNA integrity was checked on an agarose gel. First-strand cDNA was synthesized using an ImProm-II™ Reverse Transcription System (Promega).
Quantitative PCR (qPCR) was conducted to evaluate the expression level of FaMBL1 of the overexpressing lines and the WT strawberry plants, according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009). The housekeeping gene ELONGATION FACTOR 1a (FaEF1a, accession no. BK009992.1) was used as the reference (Guidarelli et al., 2011). The primers FaMBL1_CUR FW and FaMBL1_CUR rev were used to amplify FaMBL1 gene. All primers used in this study are listed in Supplementary Table S1 and were designed using the Primer3 software (http://bioinfo.ut.ee/primer3/. qPCR was performed using three biological replicates and two technical replicates, using SYBR™ Green PCR Master Mix (Thermo Fisher) and a Mx3000P qPCR System (Agilent). Gene expression was calculated using a standard curve and normalized by FaEF1a.
Hormone profiling of FaMBL1-overexpressing plants
In order to investigate the contents of defense-related phytohormones in the FaMBL1-overexpressing lines and the WT, phytohormones were extracted as described by Glauser et al. (2014) from leaves propagated from stolons. A young and fully expanded leaf was sampled from each of three plants per line and pooled together as one replicate, and a total of three replicates were used for each line. The samples were frozen immediately in liquid nitrogen and ground into fine powder. Jasmonic acid (JA), jasmonoyl-isoleucine (JA-Ile), salicylic acid (SA), abscisic acid (ABA), and indole-3-acetic acid (IAA) were measured using ultra-high-pressure liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS), according to the protocol described by Glauser et al. (2014), with 50 mg of tissue powder being used for IAA and 100 mg being used for the others. The hormone concentrations were calculated based on calibration curves. Five calibration points were set at 0.1, 0.5, 2, 20, and 100 ng ml−1, and the solutions of calibration points and leaf samples contained each of the labeled internal standards at concentrations of 10 ng ml−1 for d5-JA, d6-ABA, d6-SA, 13C6-JA-Ile, and 1 μg ml−1 for d5-IAA.
Evaluation of susceptibility of FaMBL1-overexpressing plants to fungal diseases
The susceptibility of FaMBL1-overexpressing lines to anthracnose was evaluated using Colletotrichum fioriniae, which belongs to the C. acutatum species complexes (Damm et al., 2012), and it had previously been found to be the most aggressive species to strawberry stocked in our lab. It was cultured on potato dextrose agar (PDA, in Petri dishes). After 10 d, conidia were harvested in distilled water and filtered through three layers of medical gauze. Using a hemocytometer the concentration was adjusted to 2 × 104 conidia ml−1 for use as the inoculum. Plants propagated from stolons were used for C. fioriniae inoculation. Five petioles of similar growth stage were used for each overexpressing line and the WT. Leaves were removed, and both ends of the petioles were then embedded in moist tissue paper to reduce desiccation. A tiny wound was made on each petiole using a sterilized needle and a 10-µl droplet of inoculum was placed on top of the wound. All inoculated petioles were put in a large plastic tray with moist tissue paper on the bottom. The tray was kept on a bench in the laboratory at room temperature. The disease incidence was recorded at 6 days post-inoculation (dpi) and calculated as the number of petioles with anthracnose symptoms divided by the total number of petioles treated with C. fioriniae, and expressed as a percentage. The disease incidence was determined as the mean value of three repeated trials.
The susceptibility of FaMBL1-overexpressing lines to B. cinerea (isolate B05.10) was determined using conidia harvested from 1-week-old B. cinerea cultured on PDA. To stimulate spore germination, a buffer containing 0.5% KH2PO4 and 1% PDA was prepared and the conidia suspension was mixed with buffer at a ratio of 1:1 (v:v) to get 1 × 104 conidia ml−1. The plants used for B. cinerea inoculation were the same as those used for C. fioriniae inoculation, and young, healthy, and fully expanded leaves of similar growth stages were sampled. One leaf (three leaflets) from each of five different plants per line was sampled and surface-sterilized using 1% sodium hypochlorite for 1 min, and then rinsed in sterilized distilled water for 2 min. The leaves were then separated into leaflets and put in Petri dishes (150 × 15 mm) with moist tissue paper on the bottom. Two 7.5-µl droplets of inoculum were placed on the adaxial side of each leaflet, one on each side of the midrib. Distilled water was sprayed into each Petri dishes to keep the humidity high and the dishes were kept on a laboratory bench at room temperature. The lesion diameters on the leaflets were recorded daily from 3–5 dpi. The rate of increase of lesion diameter was used to indicate the rate of disease progression, and was calculated as (diameter5dpi – diameter3dpi)/2. The inoculation trial was repeated three times.
Determination of gene expression levels in leaves inoculated with B. cinerea
To investigate the molecular responses of the FaMBL1-overexpressing lines upon B. cinerea inoculation, we used qPCR to determine the expression levels of pathogenesis-related and hormone-synthesis genes. Six leaves of each transgenic line and the WT were subjected to inoculation with either B. cinerea as described above or a mock solution using the same volume of the buffer alone (0.25% KH2PO4 and 0.5% PDA). Individual leaflets were inoculated, then at 1 dpi they were pooled back together as leaves and frozen immediately in liquid nitrogen, and stored at –80°C until use. The expression of FaMBL1, FaPGIP (EU117215.1), THAUMATIN-LIKE PROTEIN 1b (FaTLP1b, XM_004287756.2), PHENYLALANINE AMMONIA-LYASE 1 (FaPAL1, KX450226.1), CLASS II CHITINASE (FaChi2-1, MK301536.1; FaChi2-2, MF804503.1), ALLENE OXIDE SYNTHASE 1 (FaAOS1, XM_004291875.2), and 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (FaACO, AY706156.1) was measured by qPCR as described above. The primers used for are listed in Supplementary Table S1.
Statistical analysis
Student’s t-test or one-way ANOVA followed by Fisher’s LSD test were conducted using the OriginPro 2018 statistical software (https://www.originlab.com/2018).
Results
Genome-wide analysis of G-type lectin genes in F. vesca
Identification and classification
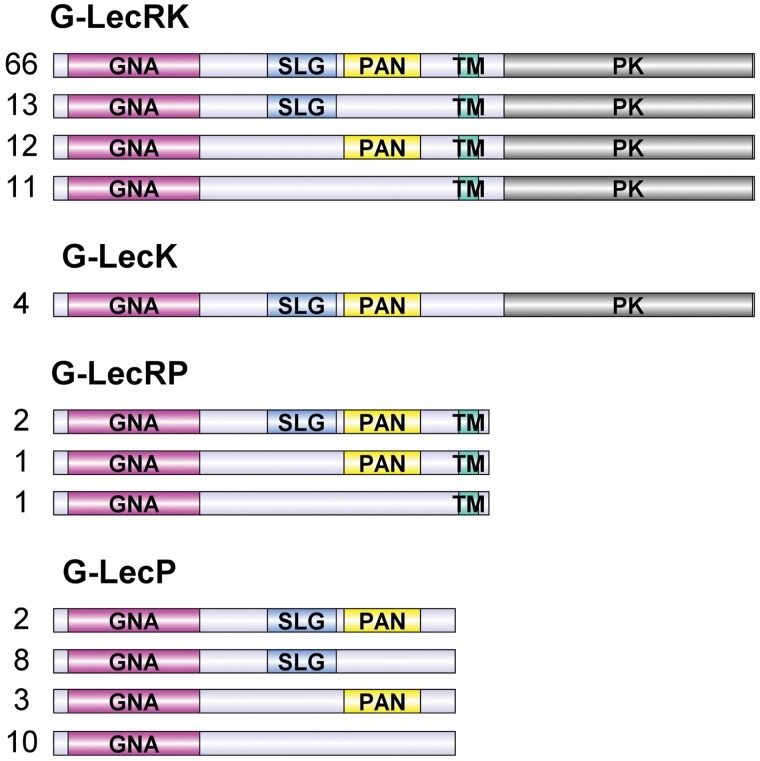
A total of 133 proteins with Galanthus nivalis agglutinin (GNA-)related lectin domains were found in F. vesca. Among these, 102 containing protein kinase (PK) and transmembrane (TM) domains were classified as G-type lectin receptor kinases (G-LecRKs); 23 lacking both domains were classified as G-type lectin proteins (G-LecPs); four lacking PK but retaining the TM domain were grouped as G-type lectin receptor proteins (G-LecRPs); and four missing the TM domain but containing the PK domain were classified as G-type lectin kinases (G-LecKs). Besides the TM and PK domains, most of the G-type lectins in F. vesca also contained domains such as S-locus glycoprotein and/or Pan-apple (PAN) that are involved in self-incompatibility and protein–protein interactions, respectively. Thus, F. vesca G-lectins showed multiple domain arrangements (Fig. 1).
Fig. 1.
Domain arrangements and classification of G-type lectin proteins in Fragaria vesca according to a BLASTp search of the Genome Database for Rosaceae. G-LecRK, G-type lectin receptor kinases; G-LecK, G-type lectin kinases; G-LecRP, G-type lectin receptor proteins; and G-LecP, G-type lectin proteins. GNA, Galanthus nivalis agglutinin-related lectin domain; SLG, S-locus glycoprotein domain; PAN, pan-apple domain; PK, protein kinase domain; TM, transmembrane domain. The numbers of corresponding F. vesca G-type lectin genes are indicated.
For convenience and clarity with regards to the F. vesca G-lectin genes, we propose a nomenclature based on the classification of G-lectin genes and their chromosome locations (Supplementary Table S2). According to this scheme, ‘Fve’ at the beginning of the name indicates that the gene is from F. vesca (Jung et al., 2015), and then ‘GLRK’, ‘GLRP’, ‘GLP’, and ‘GLK’ represent G-LecRK, G-LecRP, G-LecP, and G-LecK, respectively. The number of the chromosome where the gene is located is then given, followed by a sequential number according to the chromosome location of each type of G-lectin gene on this chromosome. Thus, FvH4_3g18380 (the homolog of FaMBL1) becomes FveGLP3.7, i.e. it encodes a G-LecP, it is located on chromosome 3, and it is the seventh G-LecP on this chromosome. This nomenclature for F. vesca G-type lectins is used hereafter in this paper.
Chromosome locations of G-type lectin genes
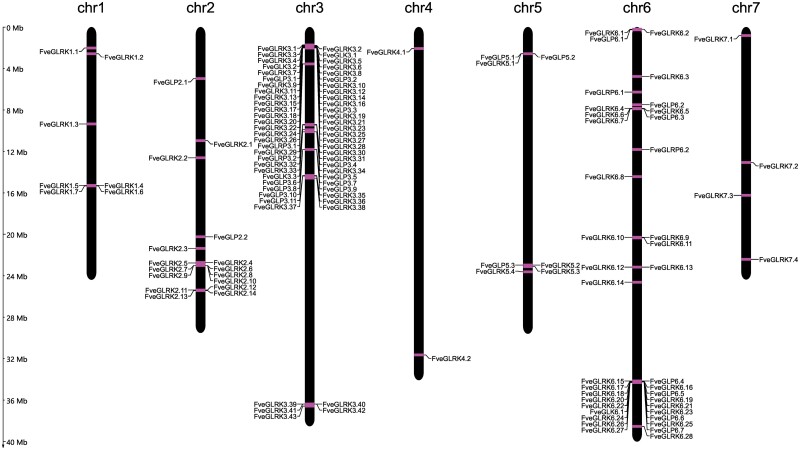
In order to visualize the chromosome locations, the G-lectin genes were mapped to the F. vesca genome (Fig. 2). G-LecRKs were found to be distributed on all the chromosomes, whereas the majority of G-LecRKs were mapped on chromosomes 3 and 6, G-LecPs were mapped on chromosomes 2, 3, 5, and 6, and G-LecRPs were found only on chromosomes 3 and 6.
Fig. 2.
Chromosome localizations of G-type lectin genes in Fragaria vesca. The coordinates of the genes on the strawberry genome were obtained from the Genome Database for Rosaceae (F. vesca v4.0.a2).
Expression profiles of G-lectin genes based on RNA-seq datasets
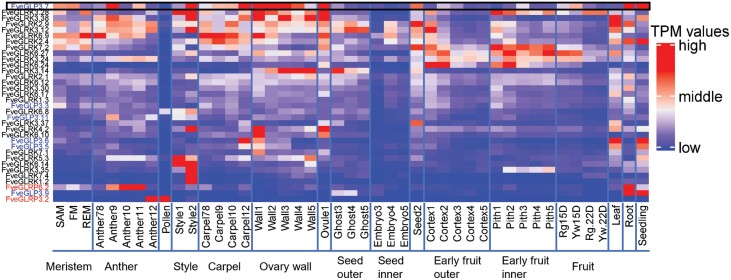
The expression profiles of G-lectin genes were analysed in different tissues and at different developmental stages based on an available strawberry RNA-seq dataset (Li et al., 2019). This showed that G-type lectin genes display a wide range of transcription levels, with some genes highly expressed in various tissues including the style, ovary wall, leaves, and roots, while others are completely silenced (Fig. 3; Supplementary Fig. S1). A few genes appeared to be specifically expressed only in one or two tissues, such as FveGLRP3.2 for example, which was highly expressed in pollen and anthers, but not in the other tissues. In contrast, FveGLP3.7 (the homolog of FaMBL1) showed active expression in many tissues during development, such as in seedlings, roots, and the ovary wall (Fig. 3).
Fig. 3.
Heatmap of the most expressed G-type lectin genes in Fragaria vesca. Expression profiles were retrieved from the database reported by Li et al. (2019). G-type lectin receptor kinases (G-LecRKs) are named in black, G-type lectin proteins (G-LecPs) are in blue, and G-type lectin receptor proteins (G-LecRPs) are in red. The expression levels are shown as transcripts per million reads (TPM) and the value for each gene was scaled before constructing the heatmap. The expression profile of FveGLP3.7 (the homolog of FaMBL1) is highlighted by the black border. SAM, shoot apical meristem; FM, flower meristem; REM, receptacle meristem. For other tissues, the numbers following the name indicate the developmental stage, where ‘78’ indicates stages 7–8 for anthers and carpels. Ghost, endosperm and seed coat; Cortex, outer part of the early fruit; Rg.15D, green fruit stage of cv. Ruegen; Yw.15D, green fruit stage of cv. Yellow Wonder 5AF7; Rg.22D, white fruit stage of Ruegen; Yw.22D, white fruit stage of Yellow Wonder 5AF7.The complete heatmap of all the FveG-lectin genes is shown in Supplementary Fig. S1.
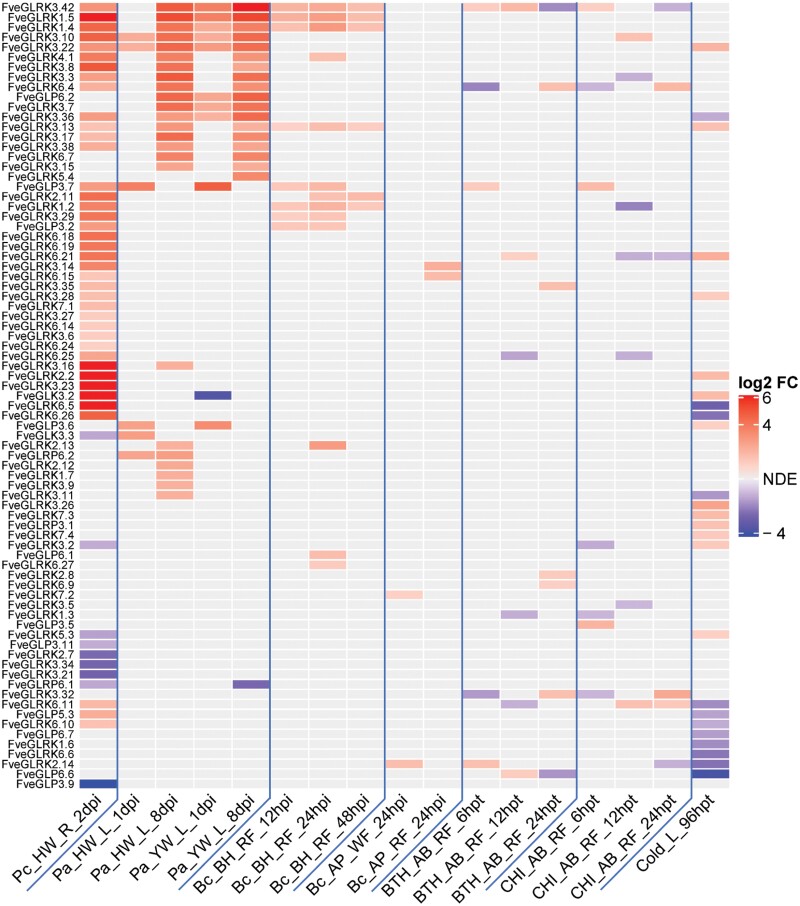
Differences in expression of G-lectin genes were found in the interactions of strawberry with B. cinerea, Podosphaera aphanis, and Phytophthora cactorum (Fig. 4). Around 50 G-lectin genes appeared differentially expressed when the roots were infected with P. cactorum, with most of them being up-regulated. Several were also found to be up-regulated in response to Po. aphanis infection at 8 dpi. Compared to these two pathogens, few G-lectin genes were transcriptionally altered upon B. cinerea infection. In addition, some G-lectin genes appeared to be regulated by the plant resistance elicitors benzothiadiazole and chitosan. Interestingly, the homolog of FaMBL1, FveGLP3.7, was up-regulated upon infection by B. cinerea, Ph. cactorum, and Po. aphanis as well as by benzothiadiazole and chitosan. In terms of abiotic stress, cold stress caused both up- and down-regulation of several G-lectin genes (Fig. 4).
Fig. 4.
Differently expressed G-lectin genes in Fragaria vesca and F. ×ananassa after exposure to either pathogen infection, resistance inducers, or cold treatment. For sources of data see the Methods section. Data are expressed as log2 FC (fold-change); NDE, no differential expression. Pc, Phytophthora cactorum; Pa, Podosphaera aphanis; Bc, Botrytis cinerea; BTH, benzothiadiazole; CHI, chitosan; Cold, cold stress; HW, strawberry cv. Hawaii 4 (F. vesca); YW, cv. Yellow Wonder 5AF7 (F. vesca); BH, cv. Benihoppe (F. ×ananassa); AP, cv. Alpine (F. vesca); AB, cv. Alba (F. ×ananassa); R, root; L, leaf; RF, red fruit; WF, white fruit; h/dpi, hours/days post-inoculation; hpt, hours post-treatment.
Functional characterization of MBL1 in F. ×ananassa
Transgenic lines and their copy numbers
The data reported above together with previously published results (Guidarelli et al., 2014) suggest the importance of the G-type lectin gene family in strawberry defense against pathogens, especially FveGLP3.7 (homolog of FaMBL1). Hence, we used Agrobacterium-mediated transformation of octoploid strawberry (F.×ananassa, cv. Sveva) (Ma et al., 2021) in order to obtain plants stably overexpressing FaMBL1. Three lines (18F6G1, 19F2G1, and 27F8G1) were selected and used for this study. Using ddPCR (Ma et al., 2021), and we determined that the relative copy numbers for 18F6G1, 19F2G1, and 27F8G1 were 6, 2.5, and 1, respectively.
FaMBL1 expression levels in the transgenic lines
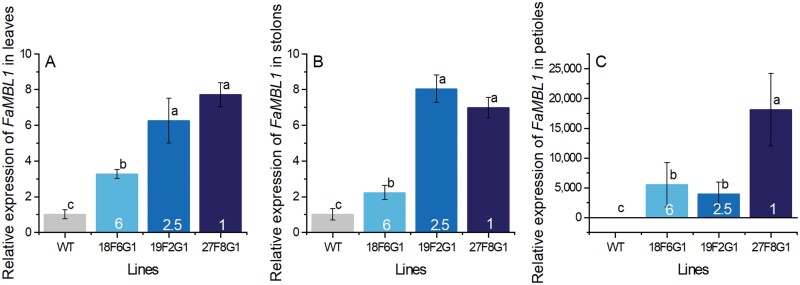
All three of the selected transgenic lines showed significantly higher FaMBL1 expression compared to the WT in leaves, stolons, and petioles (Fig. 5). The lowest expression levels were generally found in 18F6G1 and the highest in 27F8G1. FaMBL1 was barely expressed in the petioles of WT plants (Supplementary Fig. S2) whereas expression was observed in the petioles of the three transgenic lines, thereby indicating the success of the transformation (Fig. 5C).
Fig. 5.
Relative expression of FaMBL1 in different tissues of wild-type strawberry (Fragaria × ananassa) and MBL1-overexpressing lines. Samples were taken from mature plants of the wild-type (WT) and the three overexpressing lines (18F6G1, 19F2G1, and 27F8G1) and expression was determined by RT qPCR with EF1a as the reference gene. Expression is relative to that of the WT, the value of which was set as 1. (A) Leaves, (B) stolons, and (C) petioles. The copy number of FaMBL1 in each transgenic line is indicated. Data are means (±SD) of three biological replicates. Different letters indicate significant differences among means as determined using one-way ANOVA and Fisher’s LSD test (P<0.05).
There was a lack of correlation between the transgene copy number and the FaMBL1 expression level in the three overexpressing lines. For instance, 27F8G1 (copy number 1) had higher expression levels in all the tissues tested than 18F6G1 (copy number 6; Fig. 5). This suggested that RNAi-mediated suppression of transcript expression might have occurred in those lines containing higher FaMBL1 copy numbers (Butaye et al., 2005).
Phytohormone contents in FaMBL1-overexpressing lines
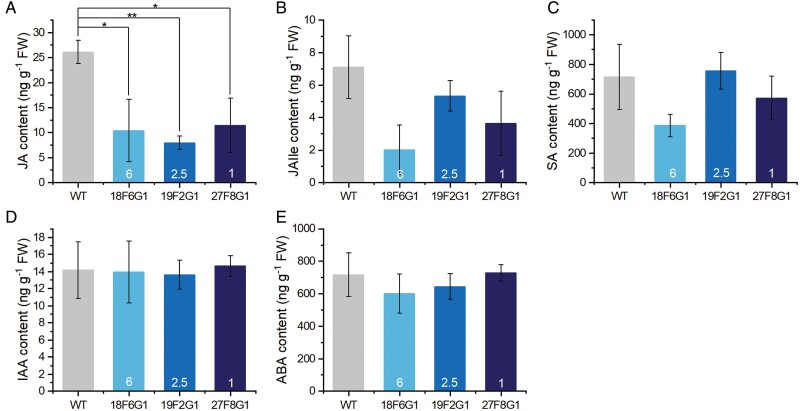
The contents of JA were lower in the overexpressing lines compared to the WT (Fig. 6A), but the effect was reduced and less clear for conjugated JA (Fig. 6B). The contents of IAA, ABA, and SA in the transgenic lines were about the same as those in the WT (Fig. 6C–E) Thus, FaMBL1 could be directly or indirectly involved in the regulation of JA concentration in strawberry.
Fig. 6.
Concentrations of defense-related phytohormones in leaves of wild-type strawberry (Fragaria × ananassa) and MBL1-overexpressing lines. Samples were taken from mature plants of the wild-type (WT) and the three overexpressing lines (18F6G1, 19F2G1, and 27F8G1) and concentrations were determined using UHPLC-MS/MS analysis. (A) Jasmonic acid (JA), (B) jasmonoyl-isoleucine (JA-Ile), (C) salicylic acid (SA), (D) indole-3-acetic acid (IAA), and (E) abscisic acid (ABA). The copy number of FaMBL1 in each transgenic line is indicated. Data are means (±SD) of three biological replicates. Significant differences between different lines were determined using one-way ANOVA and Fisher’s LSD test: *P<0.05; **P<0.01.
Response of FaMBL1-overexpressing lines to fungal inoculation
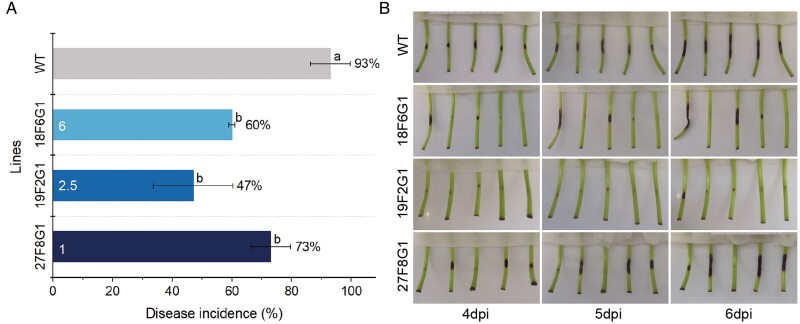
We found that C. fioriniae became quiescent after infection on strawberry leaves and they appeared symptomless, hence making them unsuitable for evaluation of susceptibility. In contrast, petioles showed uniform and reproducible anthracnose symptoms after inoculation, and hence we used them to examine the responses of the FaMBL1-overexpressing lines. The disease incidence was significantly lower in overexpressing lines as compared to the WT at 6 dpi (Fig. 7A), suggesting that the overexpression of FaMBL1 decreases the susceptibility of strawberry to C. fioriniae. Where infection occurred, petioles showed anthracnose symptoms at 4 dpi and this was followed by rapid progress of disease at 5 dpi and 6 dpi, when the symptoms were more apparent (Fig. 7B).
Fig. 7.
Progression of anthracnose disease on petioles of wild-type strawberry (Fragaria × ananassa) and MBL1-overexpressing lines. Petioles of the wild-type (WT) and the three overexpressing lines (18F6G1, 19F2G1, and 27F8G1) were inoculated with a suspension of conidia of Colletotrichum fioriniae. (A) Disease incidence on the petioles at 6 days post-inoculation (dpi). The copy number of FaMBL1 in each transgenic line is indicated. Petioles with lesions longer than 3 mm were regarded as successfully infected, and incidence was calculated as the number of petioles with symptoms divided by the total number of petioles treated with C. fioriniae. Data are means (±SE) of three experiments, each of which consisted five petioles of each line. Different letters indicate significant differences among means as determined using one-way ANOVA and Fisher’s LSD test (P<0.05). (B) Images of inoculated petioles of the WT and FaMBL1-overexpressing lines at 4–6 dpi in one representative experiment.
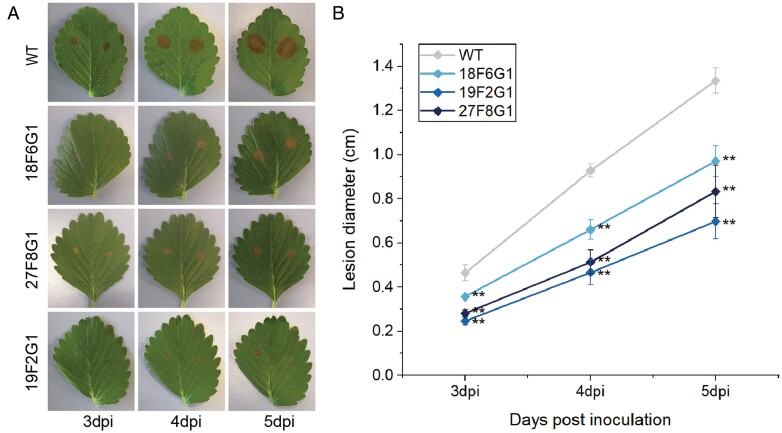
Whilst B. cinerea can infect both vegetative and reproductive tissues of strawberry, the greatest economic loss due to this pathogen is the result of fruit infection. However, infected vegetative tissues are an important source of inoculum, and improving its resistance is indispensable for the management of B. cinerea. Moreover, infection trials using leaves show uniform disease symptoms and reproducible data, which is ideal for evaluation of resistance. Hence, we tested the responses of the FaMBL1-overexpressing lines to B. cinerea on detached leaflets. We found that necrotic lesions started to be apparent at 3 dpi in both the transgenic lines and the WT (Fig. 8A); however, the disease progression was greater in the WT. The rate increase of lesion size was significantly higher in the WT than in the FaMBL1-overexpressing lines (Fig. 8B), indicating that they were less susceptible to B. cinerea.
Fig. 8.
Progression of grey mould disease on detached leaflets of wild-type strawberry (Fragaria × ananassa) and MBL1-overexpressing lines. Leaflets of the wild-type (WT) and the three overexpressing lines (18F6G1, 19F2G1, and 27F8G1) were inoculated with a suspension of conidia of Botrytis cinera. (A) Appearance of lesions at 3–5 days post-inoculation (dpi). (B) Increase in lesion size with time. Data are means (±SD) of three experiments, each of which consisted of five leaves separated into leaflets. Significant differences were determined using one-way ANOVA and Fisher’s LSD test (**P<0.01).
Expression of defense-related genes after B. cinerea inoculation
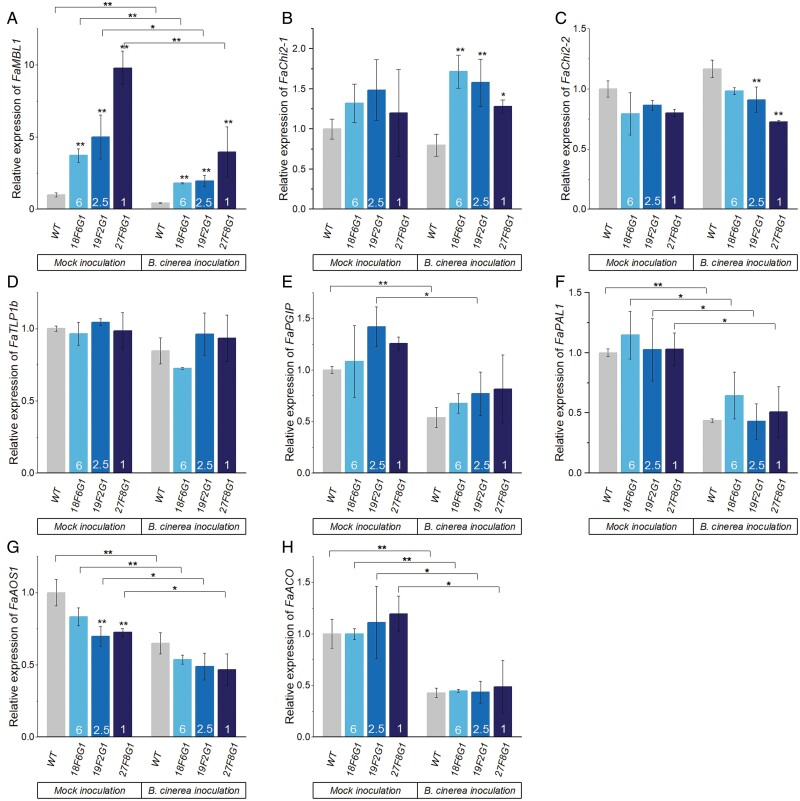
To gain insights into the role of FaMBL1 in strawberry defense against B. cinerea, we examined its relative expression together with those of other defense-related genes in the leaves of the overexpressing lines and the WT at 24 h after inoculation. We selected genes that have previously been reported to be induced upon B. cinerea infection, namely those encoding CLASS II CHITINASE (FaChi2-1 and FaChi2-2), POLYGALACTURONASE-INHIBITING PROTEIN (FaPGIP), PHENYLALANINE AMMONIA-LYASE 1 (FaPAL1), THAUMATIN-LIKE PROTEIN 1b (FaTLP1b), 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (FaACO), AND ALLENE OXIDE SYNTHASE 1 (FaAOS1) (Mehli et al., 2005; Nagpala et al., 2016; Jia et al., 2021; Lee et al., 2021). Whilst FaMBL1 expression was reduced in both the overexpressing lines and the WT upon B. cinerea infection, it remained significantly higher in the overexpressing lines (Fig. 9A). The expression of FaChi2-1 did not differ among the overexpressing lines and the WT in the absence of inoculation (Fig. 9B); however, after B. cinerea infection, its expression was significantly induced only in the overexpressing lines (Fig. 9B). The overexpressing lines 19F2G1 and 27F8G1 showed significant down-regulation of FaChi2-2 compared with the WT in infected plants (Fig. 9C) and significant down-regulation of FaAOS1 compared with the WT in the absence of infection (Fig. 9G). No other genes showed any significant differences in expression between the FaMBL1-overexpressing lines and the WT in the absence of infection. In contrast, in the presence of B. cinerea the expression of the defense-related genes FaPGIP, FaPAL1, FaAOS1 (involved in JA synthesis), and FaACO (involved in ET synthesis) was down-regulated in both the overexpressing and WT plants (Fig. 9E–H).
Fig. 9.
Relative expression of defense-response genes in leaves of wild-type strawberry (Fragaria × ananassa) and MBL1-overexpressing lines challenged with Botrytis cinerea at 1 d post-inoculation. Samples were taken from mature plants of the wild-type (WT) and the three overexpressing lines (18F6G1, 19F2G1, and 27F8G1) and expression was determined by RT qPCR with EF1a as the reference gene. Expression is relative to that of the WT, the value of which was set as 1. Relative expression of (A) MANNOSE-BINDING LECTIN 1 (FaMBL1), (B) CLASS II CHITINASE 1 (FaChi2-1), (C) CLASS II CHITINASE 2 (FaChi2-2), (D) THAUMATIN-LIKE PROTEIN 1b (FaTLP1b), (E) POLYGALACTURONASE-INHIBITING PROTEIN (FaPGIP), (F) PHENYLALANINE AMMONIA-LYASE 1 (FaPAL1), (G) ALLENE OXIDE SYNTHASE 1 (FaAOS1), and (H) 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (FaACO). The copy number of FaMBL1 in each transgenic line is indicated. Data are means (±SD) of three biological replicates. Significant differences between means were determined using one-way ANOVA and Fisher’s LSD test: *P<0.05; **P<0.01.
Discussion
Anthracnose (Colletotrichum acutatum species complex) and grey mould (Botrytis cinerea) are two of the most destructive strawberry fungal diseases. Increasing plant resistance is one of the most sustainable and effective strategies for management; however, the high levels of heterozygosity and polyploidy in strawberry increase the complexity of applying traditional breeding methods (Nehra et al., 1990; Limera et al., 2017; Mezzetti et al., 2018), and hence the importance of genetic transformation techniques for investigating resistance genes.
G-type lectins are reported to play roles in plant defense against biotic and abiotic stresses (Ghequire et al., 2012; Siripipatthana et al., 2015). In woodland strawberry, F. vesca, we found 133 genes belonging to the G-lectin family (Fig. 1). The FveG-lectin proteins encoded by these genes showed multiple domain arrangements, which creates a fairly high degree of protein diversity and provides flexible adaptation to changing environments (Kersting et al., 2012). According to the transcriptome data, many G-lectin genes are actively expressed in different tissues at different developmental stages of strawberries (Fig. 3; Supplementary Fig. S1). Moreover, many G-lectins in strawberry actively respond to pathogens and elicitors, and some appear to respond to biotic stresses. Notably, red fruits infected with B. cinerea have been found to show different expression profiles of G-lectin genes at 24 h post-infection (hpi; Fig. 4). In particular, FveGLP3.7, the FaMBL1 homolog, is up-regulated in response to B. cinerea in the octoploid red fruit of F. ×ananassa cv. Benihoppe (BH in Fig. 4) but not in diploid fruit of cv. Alpine (AP). On the other hand, we found that expression of FaMBL1 was down-regulated in leaves of octoploid cv. Sveva infected with B. cinerea at 24 hpi (Fig. 9A), suggesting that the regulation of this gene in response to infection is not only cultivar-specific but also dependent on the tissue type. The resistance to B. cinerea that we observed in the leaves of transgenic plants overexpressing FaMBL1 (Fig. 8) could be due to signaling pathways downstream of FaMBL1 that are still to be elucidated. Given that the homolog of FaMBL1, FveGLP3.7, is up-regulated upon infection by the biotroph Podopshaera aphanis and the hemibiotroph Phytophtora cactorum (Fig.4), which are fungal pathogens with different infection strategies to B. cinerea, it is most likely that FaMBL1 acts as a key regulator in strawberry response to pathogens, with different expression pathways depending on the specific biotic stress (Guidarelli et al., 2014).
Given the potential of FaMBL1 in defense, we have previously produced genetically transformed F. ×ananassa plants overexpressing FaMBL1 to confirm its role as defense protein (Ma et al., 2021). We obtained three overexpressing lines with different copy numbers and increased FaMBL1 expression in leaves, stolons, and petioles compared to the WT (Fig. 5). The differences in insertion copy number were not reflected in the expression levels in the different overexpressing lines. Thus, line 18F6G1 that possesses six copies had lower expression of FaMBL1 than line 27F8G1 that has only one copy. This is possibly due to the epigenetic silencing mechanism activated by plants in response to multiple transgene copies (Vaucheret et al., 1998). Notably, FaMBL1 was not expressed in petioles of the WT (Fig. 5C; Supplementary Fig. S2), whereas ectopic expression was observed in the petioles of transgenic plants driven the by 35S promoter. The decreased susceptibility to C. fioriniae observed in the petioles of transgenic plants therefore further support the role of FaMBL1 in resistance to anthracnose disease.
Plant hormones play important roles in modulating plant resistance and susceptibility to pathogens (Robert-Seilaniantz et al., 2011). G-type lectins in other species have been reported to be involved in phytohormone-related defense pathways. For example, the pepper G-lectin gene CaMBL1 has been found to be indispensable for the defense response against Xanthomonas campestris pv vesicatoria, with transient expression inducing the accumulation of SA and the activation of defense-related genes (Hwang and Hwang, 2011). In this study, the JA content was reduced in the FaMBL1-overexpressing lines compared to the WT (Fig. 6A), suggesting that FaMBL1 might participate in the modulation of JA concentration. JA is a hormone that is well known for its role as an elicitor of plant responses to pathogens, wounding, and herbivores (Antico et al., 2012). A number of studies have consistently reported that JA-dependent signaling can trigger the expression of defense-related effector genes and that JA-deficient mutants display increased susceptibility to infection by necrotrophic fungi (see review by Antico et al., 2012). However, there are also several studies that have shown that some mutations causing deficiency in JA biosynthesis display increased resistance to some necrotrophic fungi (Antico et al., 2012). Taken together, these observations reveal the complexity of the JA-dependent regulation of plant responses.
The decreased JA production that we observed in the FaMBL1-overexpressing lines might ultimately be related to the resistance to fungal pathogens that we observed; however, the data that we obtained are not sufficient to confirm this hypothesis. Further experiments are needed to establish a relationship between FaMBL1 expression and JA-mediated defense responses, using exogenous applications of inducers or inhibitors and quantification of the hormones in infected tissues.
Infection with B. cinerea has been reported to induce the expression of CLASS II CHITINASE (FaChi2-1 and FaChi2-2), POLYGALACTURONASE-INHIBITING PROTEIN (FaPGIP), PHENYLALANINE AMMONIA-LYASE 1 (FaPAL1), THAUMATIN-LIKE PROTEIN 1b (FaTLP1b), 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (FaACO), and ALLENE OXIDE SYNTHASE 1 (FaAOS1) and to contribute to strawberry resistance against this pathogen (Mehli et al., 2005; Nagpala et al., 2016; Jia et al., 2021; Lee et al., 2021). After strawberry infection with C. acutatum, uncoupling between SA and JA accumulation and the induction of important SA- and JA-related genes has also been reported, including PATHOGENESIS-RELATED PROTEIN 1 (FaPR1-1), LIPOXYGENASE-2 (FaLOX2), JASMONATE RESISTANT-1 (FaJAR1), and PLANT DEFENSIN-1 (FaPDF1) (Amil-Ruiz et al., 2016). We assessed the expression of some of these genes in the leaves of the FaMBL-overexpressing lines at 24 h after B. cinerea inoculation and found that they showed higher expression of FaChi2-1 than the WT (Fig. 9B). FaChi is one of the most abundant classes of strawberry pathogenesis-related genes and has hydrolytic activity against fungal cell walls (Amil-Ruiz et al., 2011). FaChi2-1 has been found to be involved in defense responses against both anthracnose (Tortora et al., 2012) and grey mould (Mehli et al., 2005). Overall, the higher expression of FaMBL1 in the overexpressing lines can be associated with the higher expression of FaChi2-1 at the early stage of infection by B. cinerea. The rice G-type lectin gene, OslecRK, which is consistently associated with resistance to M. grisea, Xanthomonas oryzae, and brown planthoppers, influences the expression of defense-related genes such as PR1a, lipoxygenase, and chalcone synthase (Cheng et al., 2013). Interestingly, we found that the expression of FaAOS1, a key enzyme in JA biosynthesis, was significantly lower in the 19F2G1 and 27F8G1 transgenic lines compared to the WT in the absence of infection (Fig. 9G), and this could be related to the lower JA contents in the overexpressing lines compared to the WT (Fig. 6A). In contrast, in 18F6G1, where the level of expression of FaMBL1 was lower with respect to the other lines (Fig. 5), the expression of FaAOS1 was not significantly different to that the WT (Fig. 9G), supporting the hypothesis of an involvement of FaMBL1 in downstream regulation. Similarly, the expression of FaChi2-2 was not significantly different in 18F6G1 (Fig. 9C). More studies are needed to clarify the involvement of FaMBL1 in the regulation of downstream expression of defense genes.
In conclusion, G-type lectins form a large gene family in F. vesca and have exploitable potential for strawberry defense against biotic stresses. We found that transgenic octoploid strawberries overexpressing FaMBL1 were less susceptible to the fungal diseases anthracnose and grey mould. The association that we observed between FaMBL1 and JA-dependent signaling pathways should be further investigated in order to obtain more evidence of the defense role of FaMBL1. This gene could be used in future trials for improving resistance in economically important strawberry cultivars through full transgenic, cisgenic, or genome-editing approaches.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Expression profiles of G-type lectin genes in different tissues of Fragaria vesca and at different developmental stages.
Fig. S2. Relative expression of FaMBL1 in different tissues of wild-type Fragaria × ananassa.
Table S1. List of primers used in this study.
Table S2. Proposed nomenclature for G-type lectin genes in Fragaria vesca.
Glossary
Abbreviations
- ABA
abscisic acid
- GNA
Galanthus nivalis agglutinin-related lectin
- G-LecK
G-type lectin kinase
- G-LecP
G-type lectin protein
- G-LecRK
G-type lectin receptor kinase
- G-LecRP
G-type lectin receptor protein
- IAA
indole-3-acetic acid
- JA
jasmonic acid
- JA-Ile
jasmonoyl-isoleucine
- PAN
Pan-apple
- PDA
potato dextrose agar
- PK
protein kinase
- SA
salicylic acid
- TM
transmembrane.
Contributor Information
Lijing Ma, Department of Agricultural and Food Science, DISTAL, Alma Mater Studiorum - University of Bologna, Bologna, Italy.
Zeraye Mehari Haile, Department of Agricultural and Food Science, DISTAL, Alma Mater Studiorum - University of Bologna, Bologna, Italy; Plant Protection Research Division of Melkasa Agricultural Research Center, Ethiopian Institute of Agricultural Research (EIAR), Addis Ababa, Ethiopia.
Silvia Sabbadini, Department of Agricultural, Food and Environmental Sciences, Università Politecnica delle Marche, Ancona, Italy.
Bruno Mezzetti, Department of Agricultural, Food and Environmental Sciences, Università Politecnica delle Marche, Ancona, Italy.
Francesca Negrini, Department of Agricultural and Food Science, DISTAL, Alma Mater Studiorum - University of Bologna, Bologna, Italy.
Elena Baraldi, Department of Agricultural and Food Science, DISTAL, Alma Mater Studiorum - University of Bologna, Bologna, Italy.
Fabrizio Costa, University of Trento, Italy.
Author contributions
EB, FN, and LM conceived and designed the research; LM and FN conducted the bioinformatic analysis; SS conducted the genetic transformation experiments; ZHM supervised the resistance evaluation and data analysis; LM performed the other experiments and wrote the original manuscript; all authors revised and approved the manuscript; EB and BM obtained the funding for this research.
Conflict of interest
The authors declare that they have no conflicts of interest in relation to this work.
Funding
This research was funded by the MIUR-PRIN2017 National Program via grant N.20173LBZM2-Micromolecule, and the PRIMA-Partnership for Research and Innovation in the Mediterranean Area 2019-2022 MEDBERRY project. LM was supported by the China Scholarship Council during her PhD.
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.
References
- Amil-Ruiz F, Blanco-Portales R, Muñoz-Blanco J, Caballero JL.. 2011. The strawberry plant defense mechanism: a molecular review. Plant & Cell Physiology 52, 1873–1903. [DOI] [PubMed] [Google Scholar]
- Amil-Ruiz F, Garrido-Gala J, Gadea J, et al. 2016. Partial activation of SA- and JA-defensive pathways in strawberry upon Colletotrichum acutatum interaction. Frontiers in Plant Science 7, 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antico CJ, Colon C, Banks T, Ramonell KM.. 2012. Insights into the role of jasmonic acid-mediated defenses against necrotrophic and biotrophic fungal pathogens. Frontiers in Biology 7, 48–56. [Google Scholar]
- Barre A, Bourne Y, Van Damme EJM, Peumans WJ, Rougé P.. 2001. Mannose-binding plant lectins: different structural scaffolds for a common sugar-recognition process. Biochimie 83, 645–651. [DOI] [PubMed] [Google Scholar]
- Bonaventure G. 2011. The Nicotiana attenuata LECTIN RECEPTOR KINASE 1 is involved in the perception of insect feeding. Plant Signaling and Behavior 6, 2060–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Butaye KMJ, Cammue BPA, Delauré SL, De Bolle MFC.. 2005. Approaches to minimize variation of transgene expression in plants. Molecular Breeding 16, 79–91. [Google Scholar]
- Cappelletti R, Sabbadini S, Mezzetti B.. 2015. Strawberry (Fragaria×ananassa). In: Wang K, ed. Agrobacterium Protocols. New York: Springer, 217–227. [DOI] [PubMed] [Google Scholar]
- Chao J, Kong Y, Wang Q, Sun Y, Gong D, Lv J, Liu G.. 2015. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi chuan 37, 91–97. [DOI] [PubMed] [Google Scholar]
- Chen X, Shang J, Chen D, et al. 2006. A B-lectin receptor kinase gene conferring rice blast resistance. Plant Journal 46, 794–804. [DOI] [PubMed] [Google Scholar]
- Cheng X, Wu Y, Guo J, Du B, Chen R, Zhu L, He G.. 2013. A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant Journal 76, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, Crous PW.. 2012. The Colletotrichum acutatum species complex. Studies in Mycology 73, 37–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down RE, Gatehouse AMR, Hamilton WDO, Gatehouse JA.. 1996. Snowdrop lectin inhibits development and decreases fecundity of the glasshouse potato aphid (Aulacorthum solani) when administered in vitro and via transgenic plants both in laboratory and glasshouse trials. Journal of Insect Physiology 42, 1035–1045. [Google Scholar]
- Glauser G, Vallat A, Balmer D.. 2014. Hormone profiling. In: Sanchez-Serrano J, Salinas J, eds. Arabidopsis protocols. Methods in Molecular Biology, vol 1062. Totowa, NJ: Humana Press, 597–608. [DOI] [PubMed] [Google Scholar]
- Gambino G, Perrone I, Gribaudo I.. 2008. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochemical Analysis 525, 520–525. [DOI] [PubMed] [Google Scholar]
- Ghequire MGK, Loris R, De Mot R.. 2012. MMBL proteins: from lectin to bacteriocin. Biochemical Society Transactions 40, 1553–1559. [DOI] [PubMed] [Google Scholar]
- Gilardoni PA, Hettenhausen C, Baldwin IT, Bonaventure G.. 2011. Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell 23, 3512–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Guidarelli M, Carbone F, Mourgues F, Perrotta G, Rosati C, Bertolini P.. 2011. Colletotrichum acutatum interactions with unripe and ripe strawberry fruits and differential responses at histological and transcriptional levels. Plant Pathology 60, 685–697. [Google Scholar]
- Guidarelli M, Zoli L, Orlandini A, Bertolini P, Baraldi E.. 2014. The mannose-binding lectin gene FaMBL1 is involved in the resistance of unripe strawberry fruits to Colletotrichum acutatum. Molecular Plant Pathology 15, 832–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile ZM, Nagpala-De Guzman EG, Moretto M, Sonego P, Engelen K, Zoli L, Moser C, Baraldi E.. 2019. Transcriptome profiles of strawberry (Fragaria vesca) fruit interacting with Botrytis cinerea at different ripening stages. Frontiers in Plant Science 10, 1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IS, Hwang BK.. 2011. The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiology 155, 447–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambagi S, Dunwell JM.. 2015. Global transcriptome analysis and identification of differentially expressed genes after infection of Fragaria vesca with powdery mildew (Podosphaera aphanis). Transcriptomics 3, 1000106. [Google Scholar]
- Jia S, Wang Y, Zhang G, Yan Z, Cai Q.. 2021. Strawberry FaWRKY25 transcription factor negatively regulated the resistance of strawberry fruits to Botrytis cinerea. Genes 12, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Bassett C, Bielenberg DG, Cheng CH, Dardick C, Main D, Meisel L, Slovin J, Troggio M, Schaffer RJ.. 2015. A standard nomenclature for gene designation in the Rosaceae. Tree Genetics and Genomes 11, 108. [Google Scholar]
- Jung S, Lee T, Cheng CH, et al. 2019. 15 years of GDR: new data and functionality in the Genome Database for Rosaceae. Nucleic Acids Research 47, D1137–D1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersting AR, Bornberg-Bauer E, Moore AD, Grath S.. 2012. Dynamics and adaptive benefits of protein domain emergence and arrangements during plant genome evolution. Genome Biology and Evolution 4, 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NH, Lee DH, Choi DS, Hwang BK.. 2015. The pepper GNA-related lectin and PAN domain protein gene, CaGLP1, is required for plant cell death and defense signaling during bacterial infection. Plant Science 241, 307–315. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Landi L, De Miccolis Angelini RM, Pollastro S, Feliziani E, Faretra F, Romanazzi G.. 2017. Global transcriptome analysis and identification of differentially expressed genes in strawberry after preharvest application of benzothiadiazole and chitosan. Frontiers in Plant Science 8, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Lee JG, Min K, Choi JH, Lim S, Lee EJ.. 2021. Transcriptome analysis of the fruit of two strawberry cultivars ‘sunnyberry’ and ‘kingsberry’ that show different susceptibility to Botrytis cinerea after harvest. International Journal of Molecular Sciences 22, 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Pi M, Gao Q, Liu Z, Kang C.. 2019. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Horticulture Research 6, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limera C, Sabbadini S, Sweet JB, Mezzetti B.. 2017. New biotechnological tools for the genetic improvement of major woody fruit species. Frontiers in Plant Science 8, 1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Gebremichael D, Mehari Z, Negrini F, Sabbadini S, Mezzetti B, Baraldi E.. 2021. A transgenic approach to investigate the role of a mannose binding lectin gene on strawberry (Fragaria ×ananassa) resistance to fungal pathogens. Acta Horticulturae, 1–6. [Google Scholar]
- Martínez Zamora MG, Castagnaro AP, Díaz Ricci JC.. 2008. Genetic diversity of Pto-like serine/threonine kinase disease resistance genes in cultivated and wild strawberries. Journal of Molecular Evolution 67, 211–221. [DOI] [PubMed] [Google Scholar]
- Mehli L, Kjellsen TD, Dewey FM, Hietala AM.. 2005. A case study from the interaction of strawberry and Botrytis cinerea highlights the benefits of comonitoring both partners at genomic and mRNA level. New Phytologist 168, 465–474. [DOI] [PubMed] [Google Scholar]
- Mezzetti B, Giampieri F, Zhang Y, Zhong C.. 2018. Status of strawberry breeding programs and cultivation systems in Europe and the rest of the world. Journal of Berry Research 8, 205–221. [Google Scholar]
- Miao J, Wu Y, Xu W, Hu L, Yu Z, Xu Q.. 2011. The impact of transgenic wheat expressing GNA (snowdrop lectin) on the aphids Sitobion avenae, Schizaphis graminum, and Rhopalosiphum padi. Environmental Entomology 40, 743–748. [DOI] [PubMed] [Google Scholar]
- Nagpala EG, Guidarelli M, Gasperotti M, Masuero D, Bertolini P, Vrhovsek U, Baraldi E.. 2016. Polyphenols variation in fruits of the susceptible strawberry cultivar Alba during ripening and upon fungal pathogen interaction and possible involvement in unripe fruit tolerance. Journal of Agricultural and Food Chemistry 64, 1869–1878. [DOI] [PubMed] [Google Scholar]
- Nehra NS, Chibbar RN, Kartha KK, Datla RSS, Crosby WL, Stushnoff C.. 1990. Genetic transformation of strawberry by Agrobacterium tumefaciens using a leaf disk regeneration system. Plant Cell Reports 9, 293–298. [DOI] [PubMed] [Google Scholar]
- Petrasch S, Knapp SJ, Van Kan JAL, Blanco-Ulate B.. 2019. Grey mould of strawberry, a devastating disease caused by the ubiquitous necrotrophic fungal pathogen Botrytis cinerea. Molecular Plant Pathology 20, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R.. 2005. InterProScan: protein domains identifier. Nucleic Acids Research 33, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Gisch N, Schäffer M, et al. 2015. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nature Immunology 16, 426–433. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG.. 2011. Hormone crosstalk in plant disease and defense: more than just JASMONATE-SALICYLATE antagonism. Annual Review of Phytopathology 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Siripipatthana P, Phaonakrop N, Roytrakul S, Senawong G, Mudalige-Jayawickrama RG, Sattayasai N.. 2015. The first trimeric Galanthus nivalis agglutinin-related lectin of Orchidaceae was found in Dendrobium pendulum: purification, characterization, and effects of stress factors. Plant Cell Reports 34, 1253–1262. [DOI] [PubMed] [Google Scholar]
- Toljamo A, Blande D, Kärenlampi S, Kokko H.. 2016. Reprogramming of strawberry (Fragaria vesca) root transcriptome in response to Phytophthora cactorum. PLoS ONE 11, e0161078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortora ML, Díaz-Ricci JC, Pedraza RO.. 2012. Protection of strawberry plants (Fragaria ananassa Duch.) against anthracnose disease induced by Azospirillum brasilense. Plant and Soil 356, 279–290. [Google Scholar]
- Vaucheret H, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Mourrain P, Palauqui JC, Vernhettes S.. 1998. Transgene-induced gene silencing in plants. Plant Journal 16, 651–659. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang K, Sun X, Tang K, Zhang J.. 2005. Enhancement of resistance to aphids by introducing the snowdrop lectin gene gna into maize plants. Journal of Biosciences 30, 627–638. [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J.. 2006. Transformation of agrobacterium using the freeze-thaw method. Cold Spring Harbor Protocols 2006, doi: 10.1101/pdb.prot4666. [DOI] [PubMed] [Google Scholar]
- Xiong JS, Zhu HY, Bai YB, Liu H, Cheng ZM.. 2018. RNA sequencing-based transcriptome analysis of mature strawberry fruit infected by necrotrophic fungal pathogen Botrytis cinerea. Physiological and Molecular Plant Pathology 104, 77–85. [Google Scholar]
- Zhang Y, Zhang Y, Lin Y, Luo Y, Wang X, Chen Q, Sun B, Wang Y, Li M, Tang H.. 2019. A transcriptomic analysis reveals diverse regulatory networks that respond to cold stress in strawberry (Fragaria×ananassa). International Journal of Genomics 2019, 7106092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.