Abstract
The biological significance of fimbrial phase variation in Salmonella serotypes is currently unknown. Exposure to long polar (LP) fimbriae of Salmonella enterica serotype Typhimurium results in selection against lpf phase ON cells of serotype Enteritidis during a subsequent challenge, suggesting that fimbrial phase variation may be a mechanism to evade cross-immunity between Salmonella serotypes. This notion was tested by assessing the effect of an immune response against serotype Typhimurium LP fimbriae on colonization of mice with a serotype Enteritidis mutant in which the lpf promoter region was replaced with the Escherichia coli lac promoter. During a challenge with a serotype Enteritidis mutant carrying the lac promoter in front of the lpf operon, significantly lower numbers were recovered from organs and feces of mice previously immunized with an lpf phase ON culture of serotype Typhimurium than from mice not previously exposed to LP fimbriae. Immunization with the lpf phase ON culture of serotype Typhimurium elicited antibodies that cross-reacted with a purified gluthathione-S-transferase–LpfA fusion protein of serotype Enteritidis. These data suggested that cross-immunity against LP fimbrial proteins cannot be evaded if phase variation on the transcriptional level is prevented by expressing the lpf operon from the lac promoter. These data hence support the idea that phase variation of LP fimbriae is a mechanism to evade cross-immunity between serotypes Enteritidis and Typhimurium.
Expression of several fimbrial antigens of Salmonella enterica serotype Typhimurium, including type 1 fimbriae, long polar (LP) fimbriae, and plasmid-encoded (PE) fimbriae, is regulated by phase variation (19, 21–23, 28). Conventional wisdom holds that phase variation of surface antigens is a mechanism for immune evasion. However, the significance of fimbrial phase variation in serotype Typhimurium is uncertain because this pathogen is not able to evade immunity against constitutively expressed surface structures, such as the O antigen repeat of its lipopolysaccharide (LPS), and is hence not able to cause recurrent infections.
While serotype Typhimurium is unable to evade an adaptive immune response encountered in a host with recent exposure to this organism, it does evade cross-immunity against Salmonella serotypes expressing different O antigen repeat units (15). For instance, immunization of mice or chickens with S. enterica serotype Enteritidis does not confer protection against challenge with serotype Typhimurium and vice versa, vaccination with serotype Typhimurium does not protect against challenge with serotype Enteritidis (7, 12, 20). The O antigen repeat units of serotypes Enteritidis and Typhimurium share the trisaccharide backbone (mannose-rhamnose-galactose) which is termed the O12 antigen. However, the two serotypes differ in the sugar residues branching from the mannose residues in the trisaccharide backbone of their O antigen repeat units. In serotype Enteritidis, the branching sugar is tyvelose (O9 antigen), while in serotype Typhimurium the O antigen has abequose attached as the side unit to mannose (O4 antigen). Analysis of the serologic response against serotypes Typhimurium and Enteritidis suggests that the O4 antigen and the O9 antigen are immunodominant determinants of their lipopolysaccharides (LPS), respectively. For instance, exposure to serotype Enteritidis elicits higher antibody titers against the O9 antigen than against the O12 antigen (2). Similarly, immunization with serotype Typhimurium results in higher titers against the O4 antigen than the O12 antigen (26).
Antibody titers may not be a reliable indicator for immunity against Salmonella serotypes since cellular immunity is known to contribute to protection. Nonetheless, there is convincing experimental evidence that expression of the O9 or O4 antigens has important consequences for cross-immunity. Immunization of mice with a serotype Enteritidis aroA mutant elicits protection against challenge with a virulent Enteritidis strain (expressing the O9 antigen) but not against a virulent strain of serotype Typhimurium (expressing the O4 antigen). However, cross-protection between both serotypes is observed when mice immunized with the Enteritidis aroA strain are challenged with a virulent serotype Typhimurium mutant that is genetically engineered to express the O9 antigen instead of the O4 antigen (12). These data suggest that O antigen polymorphism is a mechanism for the evasion of cross-immunity between serotypes Enteritidis and Typhimurium.
While serotypes Enteritidis and Typhimurium evade cross-immunity against LPS by O antigen polymorphism, other surface structures are well conserved between these organisms. For instance, the primary structure of LpfA fimbrial proteins of serotypes Typhimurium and Enteritidis differs by only one amino acid. Rabbit serum raised against a purified gluthathione-S-transferase (GST)-LpfA fusion protein is strongly cross-reactive with a purified GST-LpfA fusion protein from serotype Enteritidis (20). Furthermore, immunization of mice with an lpf phase ON culture of serotype Typhimurium results in selection against lpf phase ON variants of serotype Enteritidis during a subsequent challenge. In contrast, no selection against lpf phase ON variants of serotype Enteritidis is observed during a challenge of mice previously immunized with a serotype Typhimurium lpf deletion mutant (20). These data support the idea that cross-immunity elicited by the cross-reactive LpfA fimbrial antigen of serotypes Typhimurium and Enteritidis is evaded by phase variation. However, vaccination with an lpf phase ON culture of serotype Typhimurium does not confer protection against mortality, nor does it reduce organ colonization or fecal counts observed during a challenge with serotype Enteritidis, presumably because lpf phase OFF variants are able to evade cross-immunity (20).
Although the available data are consistent with the idea that lpf phase variation is a mechanism to evade cross-immunity between Salmonella serotypes, this evidence is indirect. Validation of this hypothesis requires direct evidence that a response against LP fimbrial proteins of serotype Typhimurium will result in cross-immunity against a serotype Enteritidis strain which constitutively expresses the lpf operon. To this end, we constructed and characterized a serotype Enteritidis strain in which the lpf promoter region was replaced with the promoter of the Escherichia coli lactose operon.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains TA One Shot and Top 10F′ were purchased from Invitrogen. E. coli strains S17-1 λpir (25) and DH5α (9) have been described previously. Derivatives of serotypes Typhimurium strain ATCC 14028 and serotype Enteritidis strain CDC SSU79998 used in this study are listed in Table 1. All bacteria were routinely cultured in Luria-Bertani (LB) broth or on plates (16). If appropriate, antibiotics were included at the following concentrations: nalidixic acid, 50 μg/ml (LB+Nal); chloramphenicol, 30 μg/ml (LB+Cm); kanamycin, 100 μg/ml (LB+Km); carbenicillin, 100 μg/ml (LB+Cb); and tetracycline, 20 μg/ml (LB+Tc). When required, the Lac indicator 5-bromo-4-chloro-3-indoyl-β-galactopyranoside (X-Gal) was added to LB plates at a final concentration of 40 mg/liter (LB+X-Gal). If appropriate, the alkaline phosphatase indicator 5-bromo-4-chloro-3-indolylphosphate (X-P) was added to LB plates at a final concentration of 40 mg/liter (LB+X-P).
TABLE 1.
S. enterica strains used in this study
| Strain | Description | Reference |
|---|---|---|
| IR715 | Serotype Typhimurium ATCC 14028 Nalr | 27 |
| TN3 | IR715 aroA::Tn10 (Tcr) lpfABCDE::lacZYA (Cmr) | 20 |
| TN4 | IR715 aroA::Tn10 (Tcr) ΔlpfABCDE (Kmr) | 20 |
| AJB589 | IR715 Plpf::lacZYA (Cmr) | This study |
| TN2 | Serotype Enteritidis CDC SSU7998 Nalr | 20 |
| TN5 | TN2 lpfABCDE::lacZYA (Cmr) | 20 |
| TN7 | TN2 Plac::lpfABCDE (Cmr) | This study |
| TN11 | TN2 ΔlpfABCDE (Kmr) | This study |
| TN17 | TN2 phoN::Kmr | This study |
| TN18 | TN2 Plac::lpfABCDE (Cmr) phoN::Kmr | This study |
| TN19 | TN2 phoN::KmrlpfABCDE::phoA (Cbr) | This study |
| TN20 | TN2 Plac::lpfABCDE::phoA (Cmr Cbr) phoN::Kmr | This study |
Construction of mutants.
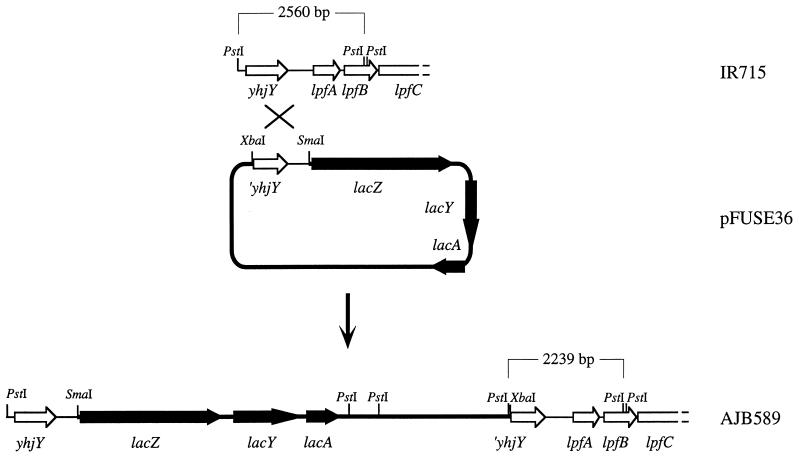
A mutant carrying a transcriptional fusion between the lpf promoter region and the promoterless lacZYA genes was constructed by PCR, amplifying the 1,384-bp fragment located immediately upstream of lpfA using the primers 5′-CCCGGGAATGGAGTGTATAGAGGTGGG-3′ (engineered SmaI site at the 5′ end) and 5′-TCTAGACTGTTGACCTTCAAGACAGATC-3′ (engineered XbaI site at the 5′ end). The PCR product was cloned into plasmid pCRII (Invitrogen), and the insert was excised by SmaI-XbaI digestion and cloned into SmaI-XbaI-digested suicide vector pFUSE (5) to give suicide plasmid pFUSE36 (Fig. 1). E. coli strain S17-1 λpir(pFUSE36) was conjugated with serotype Typhimurium strain IR715 (nalidixic acid-resistant wild type), and exconjugants were selected on LB+Nal+Cm plates. One exconjugant was selected and termed AJB589 (Fig. 1).
FIG. 1.
Construction of a single-copy transcriptional fusion between the lpfA upstream region and lacZYA. A restriction map of the serotype Typhimurium (IR715) lpf promoter region is shown at the top (open arrows indicate genes). Suicide vector pFUSE36 carrying the lpf promoter region cloned in front of the promoterless lacZYA genes (solid arrows) of E. coli is shown in the middle. Integration of pFUSE36 into the serotype Typhimurium (IR715) chromosome by homologous recombination is indicated (cross). The resulting strain (AJB589) carries a transcriptional fusion between the lpfA upstream region and lacZYA, which is shown at the bottom. The sizes of genomic PstI restriction fragments of strains IR715 and AJB589 hybridizing with an lpfA-specific DNA probe are indicated.
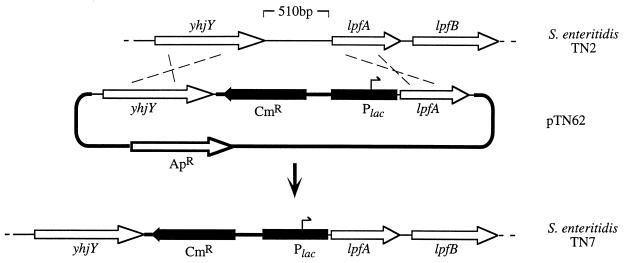
For constructing a serotype Enteritidis mutant carrying the lac promoter in place of the lpf promoter, the lac promoter was PCR amplified from pBluescript SK+ (24) using the primer 5′-CAGTCGACGTTATCCCCTGATTC-3′ (engineered SalI site at the 5′ end) and the T7 primer and cloned into plasmid pCRII (Invitrogen). The insert was excised by SalI-SacI digestion and cloned into SalI-SacI-digested suicide vector pGP704 (14) to give rise to suicide plasmid pTN29. A 1,095-bp DNA fragment extending from 21 bp upstream to 1,074 bp downstream of the lpfA start codon was PCR amplified using the primers 5′-GGAGCTCTTCTGTTATCTACCGTC-3′ (engineered SacI site at the 5′ end) and 5′-CAGAATTCTCCAGCCGTCCATG-3′ (engineered EcoRI site at the 5′ end). The PCR product was cloned into plasmid pCRII (Invitrogen), and the insert was excised by SacI-EcoRI digestion and cloned into SacI-EcoRI-digested pTN29 to give rise to suicide plasmid pTN30. A 1,095-bp DNA fragment extending from bp 1527 to 476 upstream of the lpfA start codon was PCR amplified using the primers 5′-TCGAGATCTTCCTCCTGCATAG-3′ (engineered BglII site at the 5′ end) and 5′-CCACAAGTCGACTTATCACTGCG-3′ (engineered SalI site at the 5′ end). The PCR product was cloned into plasmid pCRII (Invitrogen), and the insert was excised by BglII-SalI digestion and cloned into BglII-SalI-restricted pTN30 to give rise to pTN46. The chloramphenicol resistance gene was PCR amplified from plasmid pACYC184 using the primers 5′-GTCGACCTTAAAAAAATTACGCCCCGC-3′ (engineered SalI site at the 5′ end) and 5′-GTCGACGCCGAATAAATACCTGTGACG-3′ (engineered SalI site at the 5′ end) and cloned into plasmid pCRII (Invitrogen), and the SalI-restricted insert was cloned into SalI-restricted pTN46 to give rise to pTN62 (Fig. 2). E. coli strain S17-1 λpir (pTN62) was conjugated with serotype Enteritidis strain TN2 (nalidixic acid-resistant wild type), and exconjugants were selected on LB+Nal+Cm plates. Exconjugants were patched onto LB+Cb plates, and a mutant which was resistant to chloramphenicol but sensitive to carbenicillin was identified and termed TN7 (Fig. 2).
FIG. 2.
Replacement of the yhjY-lpfA intergenic region with the E. coli lac promoter using allelic exchange. Hatched lines indicate homologous recombination events between the genome of serotype Enteritidis strain TN2 (top) and suicide plasmid pTN62 (middle) which give rise to the genetic organization in serotype Enteritidis strain TN7 (bottom). Arrows indicate the positions and orientations of genes. A black bar indicates the position of the lac promoter region (Plac). Cm, chloramphenicol resistance gene; Ap, ampicillin resistance gene.
A serotype Enteritidis mutant carrying a deletion of the lpf operon was generated as previously described for constructing a serotype Typhimurium lpf deletion mutant using suicide plasmid pMS1208 (20). Plasmid pMS1208 confers carbenicillin resistance. Furthermore, pMS1208 contains a kanamycin resistance cassette (KIXX, Pharmacia) that is flanked on one side by a 1.5-kb DNA region upstream of lpfA and on the other side by a 1.2-kb DNA region downstream of lpfE (20). Upon conjugative transfer, homologous recombination of the lpfA upstream and the lpfE downstream DNA regions in pMS1208 with the homologous DNA regions in the serotype Enteritidis (TN2) chromosome will result in deletion of the lpf operon and insertion of the kanamycin resistance gene. E. coli strain S17-1 λpir(pMS1208) was conjugated with serotype Enteritidis strain TN2, and exconjugants were selected on LB+Nal+Km plates. Exconjugants were patched onto LB+Cb plates, and a mutant which was resistant to kanamycin but sensitive to carbenicillin was identified and termed TN11.
An alkaline phosphatase-negative strain of serotype Enteritidis was constructed by insertional inactivation of the phoN gene. The phoN gene of serotype Enteritidis was PCR amplified using the primer pair 5′-GACTCTAGAATAACCGTCCGGGAAATG-3′ (engineered XbaI site at the 5′ end) and 5′-TAACCCGGGATTTGGTGGAGAGTG-3′ (engineered SmaI site at the 5′ end). The PCR product was digested with SmaI and XbaI and cloned into EcoRV-XbaI-restricted suicide vector pGP704 (14), giving rise to plasmid pTN102. A kanamycin resistance cassette (KSAC; Pharmacia) digested with SacI was cloned into plasmid pTN102 after linearizing it at the SacI restriction site that is present in the phoN open reading frame. The resulting plasmid, pTN104, was introduced into serotype Enteritidis strains TN2 (wild type) and TN7 (Plac::lpfABCDE) by conjugation with E. coli strain S17-1 λpir, and exconjugants were selected on LB+Nal+Km plates. Exconjugants were patched onto LB+Cb plates, and mutants which were resistant to kanamycin (KSAC) but sensitive to carbenicillin (pGP704) were identified and termed TN17 (phoN) and TN18 (phoN Plac::lpfABCDE).
To verify that expression of the lpf operon in strain TN18 (phoN Plac::lpfABCDE) was not regulated by phase variation, a transcriptional fusion between lpfE and the phoA gene of E. coli was constructed. The lpfE gene was PCR amplified using the primer pair 5′-GCTCTAGACCGATGGCGTAAA-3′ (engineered XbaI site at the 5′ end) and 5′-AGGATCCGCCCTCAGTGATTATTCGTATG-3′ (engineered BamHI site at the 5′ end), digested with XbaI and BamHI, and cloned into XbaI-BamHI-digested plasmid pUJ10 (8) to give rise to plasmid pTN106. An XhoI-XbaI fragment of pTN106 containing a promoterless phoA gene fused immediately downstream to the stop codon of lpfE was cloned into SalI-XbaI-digested suicide vector pGP704 to give rise to plasmid pTN107. Plasmid pTN104 was introduced into serotype Enteritidis strains TN17 (phoN) and TN18 (phoN Plac::lpfABCDE) by conjugation with E. coli strain S17-1 λpir. Exconjugants were selected on LB+Nal+Cb plates and termed TN19 (phoN lpfABCDE::phoA) and TN20 (phoN Plac::lpfABCDE::phoA).
Mutations in AJB589, TN7, and TN11 were confirmed by Southern hybridization. Isolation of genomic DNA and Southern transfer of DNA onto a nylon membrane were performed as recently described (1). Labeling of nucleotide probes and detection of hybrids were performed using the labeling and detection kit (nonradioactive) from NEN. Deletion of the lpf operon was confirmed using EcoRI-digested genomic DNA of TN11 and its parent (TN2) and a DNA probe specific for lpfCD (the insert of plasmid pMS1039) (4) and an lpfA-specific DNA probe. Replacement of the lpf promoter region with a DNA region containing a chloramphenicol resistance gene and the lac promoter was confirmed using EcoRI-digested genomic DNA of TN7 and its parent (TN2) and a DNA probe generated by labeling the chloramphenicol resistance gene excised by SalI restriction of pTN62. Insertion of pFUSE36 into the chromosome was confirmed by performing Southern hybridization with PstI-digested genomic DNA of strain AJB589 and its parent (IR715). The lpfA-specific DNA probe used in this Southern hybridization has been described previously (3).
The yhjY-lpfA intergenic region was PCR amplified from strain TN7 using the primers 5′-TCGAGATCTTCCTCCTGCATAG-3′ and 5′-CAGAATTCTCCAGCCGTCCATG-3′. To determine the orientation of the chloramphenicol resistance gene and to confirm the presence of the lac promoter, the nucleotide sequence of the PCR product was analyzed. Nucleotide sequence analysis was performed by the Gene Technology Laboratory at Texas A&M University. Nucleotide sequences were analyzed using the MacVector 6.0.1 software package (Oxford Molecular Group).
Animal experiments.
Female (8- to 10-week-old) CBA/J mice were used throughout this study. Fresh fecal pellets were taken from mice prior to infection, suspended in phosphate-buffered saline (PBS), and spread on LB plates containing the appropriate antibiotics to ensure that the indigenous microflora was antibiotic sensitive. Prior to immunization or infection of mice, all bacteria were cultured as static overnight cultures in LB broth. In all experiments the bacterial titer of the inoculum was determined by spreading serial 10-fold dilutions on LB plates containing the appropriate antibiotics. After inoculation of mice, fresh fecal pellets were collected at least three times per week and suspended in PBS, and serial 10-fold dilutions were spread onto LB agar plates containing the appropriate antibiotics. For TN3 (lpfABCDE::lacZYA aroA::Tn10), agar plates were supplemented with X-Gal to quantify bacteria containing the lpf operon in the phase ON or phase OFF expression state.
For competitive infection experiments, groups of six mice were inoculated intragastrically at a total dose of 109 CFU/animal. The inoculum consisted of a 1:1 mixture of two bacterial strains that were distinguishable by their antibiotic resistance profiles. Mice were euthanized at 5 days postinfection, Peyer's patches, mesenteric lymph nodes, and spleens were collected and homogenized in 5 ml of PBS using a Stomacher (Tekmar, Cincinatti, Ohio), and serial 10-fold dilutions were spread onto LB agar plates containing the appropriate antibiotics. Data were normalized by dividing the output ratio (CFU of the mutant/CFU of the wild type) by the input ratio (CFU of the mutant/CFU of the wild type). All data were converted logarithmically prior to the calculation of averages and statistical analysis. A Student's t test was used to determine whether the mutant/wild-type ratio recovered from infected organs or fecal pellets was significantly different from the mutant/wild-type ratio present in the inoculum.
Three groups of eight mice were used for an immunization experiment. One group served as the naive control, while the remaining two groups were immunized with serotype Typhimurium strains TN3 (lpfABCDE::lacZYA aroA::Tn10) or TN4 (ΔlpfABCDE aroA::Tn10). Immunization was performed intragastrically at a dose of approximately 109 CFU/animal contained in a 0.2-ml volume. Immunized mice were then boosted with the same vaccine strain at 14 days postimmunization. At 14 days postbooster immunization, mice were anesthetized using Rodent cocktail (42.8 mg of ketamine plus 8.6 mg of xylazine plus 1.4 mg of acepromazine; administer 0.5 to 0.7 ml/kg of body weight intraperitoneally), 0.05 ml of blood was collected by periorbital bleed, and samples from each immunization group were pooled. Prior to challenge, fresh fecal pellets were collected from immunized mice to identify animals that had developed long-term carriage, and these animals were removed from the experiment. All three groups were challenged with serotype Enteritidis strain TN7 at day 80 postimmunization. The infection was monitored for 28 days postchallenge by collecting fresh fecal pellets. Statistical analysis of the data was performed using the Wilcoxon rank sum test. The immunization experiment was repeated with three groups of five mice. Immunization and challenge were performed as described above, but mice were euthanized at 8 days postchallenge and Peyer's patches, mesenteric lymph nodes, and spleens were collected. Organs were homogenized in 5 ml of PBS using a Stomacher (Tekmar), and serial 10-fold dilutions were spread onto LB agar plates containing the appropriate antibiotics. A Student's t test was used to determine whether the numbers of bacteria recovered from infected organs were significantly different between experimental groups.
ELISA.
Plasmid pTN66 contains a fragment of the serotype Enteritidis lpfA gene encoding the C-terminal 162 amino acids of LpfA (corresponding to the mature protein after cleavage of the signal peptide) cloned into plasmid pGEX-4T-2 to generate a translational fusion between GST and LpfA (20). Serotype Enteritidis GST-LpfA fusion protein was prepared from E. coli strain Top10F′ (pTN66) as described previously (20). The concentration of purified GST-LpfA protein was determined using a Bradford assay kit (Bio-Rad). Enzyme-linked immunosorbent assay (ELISA) analysis was performed by using a protocol described previously (29). In brief, 96-well polySorp ELISA plates (Nunc) were coated with antigen by adding GST-LpfA of serotype Enteritidis (20 μg/well) to each well. To remove antibodies that were not directed against LpfA, the serum collected from mice was absorbed with strain Top10F′(pGEX-T4-2) grown in LB supplemented with 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) using a protocol described previously (10). Antigen-coated plates were blocked with 0.2 ml of 3% Blotto (3% powdered skim milk, 0.04% antifoam A, 0.05% Tween 20, 0.1% sodium azide in PBS) for 4 h at 37°C. Blocked plates were then washed once with H2O, and mouse serum (0.05 ml/well) was added to antigen-coated plates in duplicate two fold serial dilutions with 3% Blotto as the diluent. The plates were incubated overnight and washed 10 times with H2O. Binding of mouse serum was detected by using goat anti-mouse immunoglobulin G (IgG)-alkaline phosphatase (AP) conjugate (Sigma) diluted 1:1,000 in 3% Blotto. Plates were incubated with secondary antibodies overnight at 37°C and then washed 10 times with H2O. Detection was performed with 0.05 ml of p-nitrophenylphosphate (1 mg/ml; Sigma) diluted in glycine buffer (0.1 M glycine, 1 mM MgCl2, 1 mM ZnCl2, pH 10.4). Reactions were stopped by addition of 0.05 ml of 0.1 M EDTA.
To remove antibodies that were not directed against LpfA, serum collected from a rabbit immunized with GST-LpfA fusion protein (20) was absorbed with a serotype Enteritidis strain in which the entire lpf operon was removed by deletion (TN11) using a protocol described previously (10). ELISA with whole serotype Enteritidis cells was performed by adjusting bacterial cultures to a final protein concentration of 2 mg/ml (Bradford assay) and adding 50 μl of the bacterial suspension to each well. After addition of 20 μl of a 1% sodium azide solution (in PBS) to each well, 96-well polySorp ELISA plates (Nunc) were allowed to dry overnight at 37°C. Antigen-coated plates were blocked with 0.2 ml of 3% Blotto for 4 h at 37°C. Blocked plates were then washed once with H2O, and rabbit serum (0.05 ml/well) was added to antigen-coated plates in duplicate twofold serial dilutions with 3% Blotto as the diluent. The plates were incubated overnight and washed ten times with H2O. Binding of rabbit serum was detected using goat anti-rabbit IgG-AP conjugate (Sigma) diluted 1:1,000 in 3% Blotto. Plates were incubated with secondary antibodies overnight at 37°C and then washed 10 times with H2O. Detection was performed as described above.
The ELISA procedure described by Smith and coworkers (26) was modified and used for the detection of serum antibodies directed against LPS from serotype Typhimurium and serotype Enteritidis. Purified LPS of serotype Typhimurium and serotype Enteritidis prepared by phenol extraction (purchased from Sigma) was resuspended in coating buffer (0.1 M sodium carbonate, 1.0 M NaCl, pH 9.6), and 96-well polySorp ELISA plates (Nunc) were coated with antigen by adding 5 μg to each well. Plates were then covered and incubated overnight at 37°C. The plates were emptied and blocked for 30 min in blocking buffer (PBS plus 0.05% Tween 20 plus 1% bovine serum albumin), and mouse serum (0.05 ml/well) was added to antigen-coated plates in duplicate twofold serial dilutions with blocking buffer as the diluent and incubated for 1 at 37°C. Plates were then washed three times in washing buffer (PBS plus 0.05% Tween 20), and binding of mouse serum was detected by using goat anti-mouse IgG-AP conjugate or goat anti-mouse IgA-AP conjugate (Sigma) diluted 1:1,000 in blocking buffer. Plates were incubated with secondary antibodies for 1 h at 37°C and then washed three times with washing buffer. Detection was performed as described above.
In all ELISA experiments, titers were expressed as the inverse of the highest dilution that gave an A405 value in an ELISA plate reader (MK700 microplate reader; Dynatech) that was above background levels detected in wells that were not incubated with primary serum.
RESULTS
Construction of a serotype Enteritidis strain expressing the lpf operon from the lac promoter.
The goal of this study was to construct and characterize a serotype Enteritidis strain in which expression of LP fimbriae is not regulated by phase variation. Since the mechanism of LP fimbrial phase variation is currently unknown, it was not straightforward to construct a strain in which the lpf operon was locked ON. To overcome this problem, the promoter region of the lpf operon was replaced with the promoter of the lactose (lac) operon of E. coli to eliminate phase variation of LP fimbriae.
Using a strain carrying an lpfABCDE::lacZYA reporter gene fusion, we have previously demonstrated that expression of the lpf operon is regulated by phase variation (21). To determine whether fimbrial phase variation is mediated by a promoter located upstream of lpfA, we constructed a strain (AJB589) in which the promoterless lacZYA reporter genes were integrated into the bacterial chromosome and directly fused to the yhjY-lpfA intergenic region (proximal to lpfA) (Fig. 1). Strain AJB589 yielded both Lac+ and Lac− colony phenotypes on LB+X-Gal agar plates. Alternation between Lac+ (phase ON) and Lac− (phase OFF) phenotypes occurred by a heritable phase variation mechanism, since inoculation of broth cultures with bacteria picked from a Lac+ colony gave rise to a considerably higher proportion of Lac+ colonies than inoculation with bacteria picked from a Lac− colony. These data suggested that expression of the lpf operon was driven by a phase-variable promoter located upstream of the lpfA start codon.
The yhjY-lpfA intergenic region (bp 21 to 476 upstream of lpfA) of serotype Enteritidis strain TN2 was replaced by allelic exchange with a DNA region containing a chloramphenicol resistance cassette and the lac promoter using suicide plasmid pTN62 (Fig. 2). Allelic exchange in the resulting strain (TN7) was confirmed by Southern blot analysis using a DNA probe specific for the chloramphenicol resistance gene (data not shown). Furthermore, the yhjY-lpfA intergenic region of strain TN7 was PCR amplified and cloned, and its sequence was determined to verify the presence of the lac promoter upstream of lpfA. No mutations were detected in the sequence of the chloramphenicol resistance gene or the lac promoter.
Characterization of a serotype Enteritidis strain expressing the lpf operon from the lac promoter.
The phoN gene in serotype Enteritidis strains TN2 (wild type) and TN7 (Plac::lpfABCDE) was inactivated by inserting a kanamycin resistance cassette, and the resulting strains were termed TN17 (phoN) and TN18 (phoN Plac::lpfABCDE), repectively. To study expression of the lpf operon on the transcriptional level, a promoterless phoA reporter gene was inserted immediately downstream of the stop codon of lpfE in strains TN17 (phoN) and TN18 (phoN Plac::lpfABCDE) to give rise to strains TN19 (phoN lpfABCDE::phoA) and TN20 (phoN Plac::lpfABCDE::phoA), respectively. Strain TN19 (phoN lpfABCDE::phoA) yielded both PhoA+ and PhoA− colony phenotypes on LB+X−P agar plates. In contrast, strain TN20 (phoN Plac::lpfABCDE::phoA) only gave rise to PhoA+ colonies on LB+X−P agar plates, confirming that phase variation at the transcriptional level had been eliminated by replacing the lpf promoter region with the lac promoter.
Serotype Enteritidis strain TN7 (Plac::lpfABCDE) was analyzed by ELISA to verify that LpfA fimbrial protein was expressed in this strain. The reactivity of TN7 with rabbit anti-LpfA serum was compared to that of an isogenic serotype Enteritidis strain carrying a deletion of the lpf operon. The entire lpf operon was deleted from serotype Enteritidis strain TN2 by allelic exchange as previously described for serotype Typhimurium (20). The resulting serotype Enteritidis strain was termed TN11, and the absence of sequences hybridizing with lpfA was confirmed by Southern hybridization (data not shown). Rabbit serum raised against purified GST-LpfA fusion protein of serotype Typhimurium has been described previously (20) and was absorbed with serotype Enteritidis strain TN11 to remove antibodies that were not directed against LP fimbrial proteins. Expression of LpfA in serotype Enteritidis strains TN7 (Plac::lpfABCDE) and TN11 (ΔlpfABCDE) was assessed by ELISA using absorbed rabbit anti-GST-LpfA serum. Rabbit anti-GST-LpfA serum contained 16-fold-higher IgG titers against strain TN7 (Plac::lpfABCDE) than against strain TN11 (ΔlpfABCDE), confirming expression of LpfA in strain TN7 (Table 2).
TABLE 2.
Anti-LpfA antibodies present in rabbit or murine serum
| Serum | Antigen | IgG titer |
|---|---|---|
| Rabbit anti-GST-LpfA | Enteritidis TN7 (Plac::lpfABCDE) | 2,560 |
| Rabbit anti-GST-LpfA | Enteritidis TN11 (ΔlpfABCDE) | 160 |
| Mouse anti-Typhimurium TN3 (lpfABCDE::lacZYA) | Purified Enteritidis GST-LpfA | 480 |
| Mouse anti-Typhimurium TN4 (ΔlpfABCDE) | Purified Enteritidis GST-LpfA | 10 |
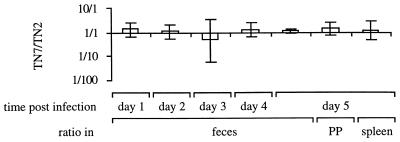
Since the lac promoter may be expressed in different host environments and at different levels than the native lpf promoter, we next addressed the question of whether the presence of the lac promoter in front of the lpf operon resulted in attenuation of serotype Enteritidis for mouse virulence. For this purpose, we compared the ability of TN7 (Plac::lpfABCDE) to colonize murine tissues during a competitive infection with the isogenic wild type (TN2). A group of five mice were infected intragastrically with a 1:1 mixture of TN7 and TN2. Bacteria were recovered from fecal pellets for 5 days postinfection, and on the fifth day bacteria were recovered from organs. The TN7/TN2 ratios recovered from feces or infected organs were not significantly different from the ratio present in the inoculum (Fig. 3). These data suggested that expression of the lpf operon from the lac promoter did not result in marked attenuation of serotype Enteritidis for mouse virulence.
FIG. 3.
Ratio of mutant to wild type recovered from mice infected with an equal mixture of serotype Enteritidis strain TN7 (Plac::lpfABCDE) and the isogenic wild type (TN2). Data are given as means ± standard deviation. PP, Peyer's patches.
Cross-immunity between serotype Typhimurium and serotype Enteritidis strain TN7.
The immune response generated during vaccination with a live attenuated lpf phase ON Typhimurium strain is sufficient to cause selection against lpf phase ON variants of serotype Enteritidis during a subsequent challenge (20). However, similar total numbers of serotype Enteritidis are recovered from naive mice and mice previously exposed to a live attenuated lpf phase ON variant of serotype Typhimurium. These data can be explained by assuming that serotype Enteritidis lpf phase OFF variants can evade immunity against LP fimbrial proteins, giving rise to similar bacterial loads in organs and feces of naive animals and animals previously exposed to LP fimbriae. Thus, phase variation of the lpf operon may be a mechanism to evade cross-immunity between Salmonella serotypes. This hypothesis implies that in case fimbrial phase variation is prevented, the total numbers of serotype Enteritidis recovered from animals with immunity to serotype Typhimurium LP fimbrial proteins during a challenge would be significantly reduced compared to the numbers recovered from naive control mice. A vaccination and challenge study was performed to test this hypothesis. Since naive susceptible mice challenged with virulent serotype Enteritidis die before adaptive immunity is fully induced (usually between days 6 and 10 postinfection), colonization levels cannot be compared with those observed in immune animals at later time points. To avoid this pitfall, we performed immunization studies with a resistant mouse lineage, CBA/J. CBA/J mice do not succumb to oral challenge with virulent serotype Enteritidis. Thus, the bacterial load in naive and vaccinated groups can be compared until bacteria are cleared from the animals, which may take more than 4 weeks.
We reasoned that cross-immunity between serotypes Typhimurium and Enteritidis elicited by exposure to LP fimbrial proteins could be assessed by comparing the responses elicited by vaccination with an lpf phase ON culture of serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) with that induced by immunization with an isogenic strain (TN4) expressing all antigens except LP fimbriae (aroA Δlpf). Groups of eight mice were immunized intragastrically with either an lpf phase ON culture of serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) or a culture of serotype Typhimurium strain TN4 (aroA Δlpf). Both vaccination groups were boosted 14 days postimmunization with the same strain. Prior to challenge, two and three animals were removed from the groups of mice immunized with strains TN4 and TN3 respectively, because the animals had developed long-term carriage of the vaccine strain. To allow nonspecific immune mechanisms, such as macrophage activation, to return to their normal levels, challenge with serotype Enteritidis strain TN7 (Plac::lpfABCDE) was performed at 80 days post-booster immunization. In addition, a group of eight naive mice of the same age were challenged with the same dose of TN7.
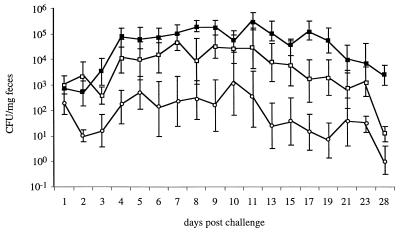
Fecal shedding of serotype Enteritidis strain TN7 (Plac::lpfABCDE) was monitored by determining bacterial numbers recovered from fecal pellets of mice collected on days 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 13, 15, 17, 19, 21, 23, and 28 postchallenge. Numbers recovered from fecal pellets varied over a wide range, which may reflect differences between animals as well as the fact that Salmonella serotypes tend to be shed intermittently with the feces. The average numbers of serotype Enteritidis TN7 recovered from feces of mice immunized with serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) were consistently lower than those recovered from the naive control group (Fig. 4). The difference was largest on day 19 postchallenge, when the average number of serotype Enteritidis TN7 recovered from naive mice was approximately 5,500-fold higher than that recovered from mice immunized with strain TN3 (aroA lpfABCDE::lacZYA). Statistical analysis revealed that for days 2, 4, 6, 7, 8, 9, 11, 13, 15, 17, 19, 21, 23, and 28 postchallenge, the numbers of serotype Enteritidis TN7 recovered from mice immunized with serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) were significantly lower than those recovered from the naive group (P < 0.05).
FIG. 4.
Recovery of serotype Enteritidis strain TN7 (Plac::lpfABCDE) from feces during a challenge of naive CBA/J mice and mice immunized with serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) or TN4 (aroA Δlpf). The standard errors and the average numbers of strain TN7 recovered from feces of naive animals (solid squares) and mice previously vaccinated with strain TN3 (open circles) or TN4 (open squares) are shown.
Mice immunized with serotype Typhimurium strain TN4 (aroA Δlpf) shed serotype Enteritidis strain TN7 (Plac::lpfABCDE) on average at lower numbers than naive mice but at higher numbers than mice immunized with serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) (Fig. 4). Statistical analysis revealed that the numbers of serotype Enteritidis TN7 recovered from mice immunized with serotype Typhimurium strain TN4 (aroA Δlpf) were significantly lower than those recovered from naive mice at days 17 and 28 postchallenge (P < 0.05). The numbers of serotype Enteritidis TN7 recovered from mice immunized with serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) were significantly lower than those recovered from mice immunized with serotype Typhimurium strain TN4 (aroA Δlpf) on days 2, 7, and 17 postchallenge (P < 0.05).
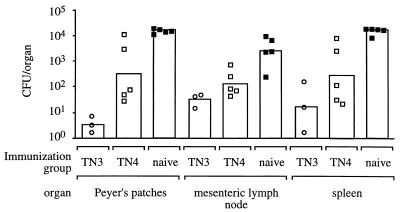
To assess whether reduced fecal shedding was indicative of colonization of murine organs, the experiment was repeated with groups of five mice. Prior to challenge, two animals were removed from the group of mice immunized with strain TN3 because the animals had developed long-term carriage of the vaccine strain. At 8 days post challenge with serotype Enteritidis strain TN7 (Plac::lpfABCDE), mice were euthanized, and bacteria in organs were enumerated (Fig. 5). Overall, the results were similar to those obtained during recovery of serotype Enteritidis strain TN7 from fecal pellets. Mice immunized with serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) contained significantly fewer CFU of TN7 in Peyer's patches (P < 0.001) and mesenteric lymph nodes (P < 0.005) than naive mice. Furthermore, bacterial numbers recovered from Peyer's patches of mice immunized with serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) were significantly lower than those recovered from mice immunized with serotype Typhimurium strain TN4 (aroA Δlpf) (P < 0.25). Although numbers of serotype Enteritidis strain TN7 recovered from organs of mice immunized with serotype Typhimurium strain TN4 (aroA Δlpf) were on average lower than those recovered from naive mice, this difference was not statistically significant (P > 0.1). In summary, these data showed that a serotype Enteritidis strain expressing LP fimbriae from the lac promoter (TN7) colonized mice previously exposed to serotype Typhimurium expressing LP fimbriae in significantly lower numbers than mice not previously exposed to serotype Typhimurium LP fimbriae.
FIG. 5.
Recovery of serotype Enteritidis strain TN7 (Plac::lpfABCDE) at day 8 post challenge from organs of mice that were either naive (solid squares) or previously immunized with serotype Typhimurium strain TN3 (open circles) or TN4 (open squares). Each circle or square represents data for an individual animal. Average numbers of bacteria recovered from an organ are indicated by a bar.
Serological response of mice immunized with serotype Typhimurium.
The results of the challenge experiments suggested that mice previously exposed to serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) developed an immune response against LP fimbrial proteins which resulted in reduced fecal and organ colonization of serotype Enteritidis strain TN7 (Plac::lpfABCDE) (Fig. 4 and 5). To confirm this assumption experimentally, antibody titers against purified GST-LpfA fusion protein of serotype Enteritidis generated during vaccination with serotype Typhimurium strains TN3 (aroA lpfABCDE::lacZYA) or TN4 (aroA Δlpf) were determined by ELISA (Table 2). The serum of mice immunized with an lpf phase ON culture of serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) contained 48-fold higher titers against serotype Enteritidis GST-LpfA fusion protein than the serum of mice immunized with serotype Typhimurium strain TN4 (aroA Δlpf). The serum of naive mice did not react with the fusion protein (data not shown). These data supported the idea that LpfA was expressed during vaccination with serotype Typhimurium strain TN3 and elicited an immune response that resulted in cross-reactivity with serotype Enteritidis LpfA.
Although cross protection between serotypes Typhimurium and Enteritidis is not observed during oral infection of BALB/c mice (20), a modest reduction in numbers of serotype Enteritidis strain TN7 was noted when data for naive mice and mice previously immunized with serotype Typhimurium strain TN4 (aroA Δlpf) were compared (Fig. 4 and 5). The O antigens of serotypes Enteritidis and Typhimurium differ with respect to their immunodominant epitopes (O9 and O4, respectively) but possess an identical trisaccharide backbone (the O12 antigen). The degree of cross-reactivity between LPS of serotypes Typhimurium and Enteritidis with serum from immunized mice was assessed by ELISA (Table 3). Titers against homologous (serotype Typhimurium) LPS were consistently higher than titers against heterologous (serotype Enteritidis) LPS. These data thus confirmed that LPS of serotypes Typhimurium and Enteritidis differ in their immunodominant epitopes. However, substantial cross-reactivity between LPS of serotypes Typhimurium and Enteritidis was detected with serum raised against serotype Typhimurium, presumably because of the shared antigens (O12, core oligosaccharide, or lipid A).
TABLE 3.
Serological response of mice immunized with serotype Typhimurium against purified LPS in ELISA
| LPS | Titer in serum of mice
|
|||||
|---|---|---|---|---|---|---|
| Serotype Typhimurium vaccinated
|
Naive
|
|||||
| TN3 (aroA lpfABCDE::lacZYA)
|
TN4 (aroA Δlpf)
|
IgG | IgA | |||
| IgG | IgA | IgG | IgA | |||
| Typhimurium | 1,600 | 120 | 3,200 | 160 | <10 | <10 |
| Enteritidis | 600 | <10 | 2,400 | 60 | <10 | <10 |
DISCUSSION
Based on their immunodominant O antigens, Salmonella serotypes are traditionally classified into serogroups (13). S. enterica serotype Typhimurium is a member of serogroup B, which is characterized by LPS containing the O4 antigen. S. enterica serotype Enteritidis, on the other hand, expresses the O9 antigen, the group-specific antigen of serogroup D1. Immunization with a Salmonella serotype elicits high antibody titers against its group-specific O antigen (e.g., O4 or O9), while reactivity to LPS of other serogroups is usually low (26). For instance, the serum of chickens vaccinated with S. enterica serotype Gallinarum (serogroup D1) contains antibody titers against LPS of serogroup D1 that are 2,048-fold, 512-fold, and 16-fold higher than those against LPS of serogroups C1 (serotype Virchow), C2 (serotype Hadar), and B (serotype Typhimurium), respectively (2). The relatively high level of cross-reactivity between LPS of serogroups D1 and B detected with chicken sera is due to the shared O12 antigen (6). However, antibody titers against the O4 or O9 antigen are commonly higher than those directed against the shared O12 antigen (12, 18, 26). Similarly, we found that immunization of mice with serotype Typhimurium elicited between 1.3- and 12-fold-higher titers against its own LPS (O-antigen formula O4, 12) than against LPS of serotype Enteritidis (O-antigen formula O9, 12) (Table 3).
Although the differences in antibody titers against LPS of serogroups B and D1 are modest, the group-specific O4 and O9 antigens are important targets for protective immunity (12). In contrast, expression of the O12 antigen by serotypes Enteritidis and Typhimurium does not result in cross-immunity in genetically susceptible (BALB/c) mice) (11, 12, 20). Using genetically resistant mice (CBA/J), we found that on 2 of 18 occasions on which bacterial numbers in fecal pellets were enumerated (days 17 and 28 postchallenge), serotype Enteritidis strain TN7 was recovered in significantly lower numbers from animals vaccinated with serotype Typhimurium (TN4) than from naive animals (Fig. 3). This low degree of cross-immunity between serotypes Typhimurium and Enteritidis observed in CBA/J mice at late time points may be due to expression of cross-reactive antigens, such as the O12 antigen. However, since BALB/c mice succumb to infection with serotype Enteritidis within 6 to 10 days, cross-immunity in CBA/J mice indicated by reduced numbers recovered from feces at days 17 and 28 postinfection may not be indicative of protection that can be observed in BALB/c mice. Our results therefore do not contradict previous reports which show that BALB/c mice vaccinated with serotype Typhimurium (serogroup B) are not protected against challenge with serotype Enteritidis (serogroup D1) (11, 12, 20).
To assess the role of LP fimbrial phase variation in evading cross-immunity between serotypes Typhimurium and Enteritidis, we constructed a serotype Enteritidis strain (TN7) in which the yhjY-lpfA intergenic region was replaced by the lac promoter (Fig. 2). Expression of LpfA by TN7 (Plac::lpfABCDE) was confirmed by ELISA (Table 2). Constitutive expression of type 1 fimbriae reduces the ability of E. coli to colonize mice. For instance, during competitive infection experiments, an E. coli mutant in which expression of type 1 fimbriae is locked ON colonizes the mouse large intestine at lower levels than the isogenic wild type in which expression of the fim operon is regulated by phase variation (17). In contrast, serotype Enteritidis strain TN7 (Plac::lpfABCDE) was recovered at similar numbers as the isogenic wild type (TN2) from murine feces and organs during competitive infection experiments (Fig. 3). These data suggested that expression of the lpf operon from the lac promoter did not markedly reduce the ability of serotype Enteritidis to colonize the intestine or internal organs of mice.
We have previously shown that immunization of mice with an lpf phase ON culture of serotype Typhimurium results in selection against lpf phase ON variants of serotype Enteritidis during a subsequent challenge. However, the total numbers of serotype Enteritidis recovered during the challenge from fecal pellets or organs of mice immunized with an lpf phase ON culture of serotype Typhimurium are not significantly different from those recovered from a naive control group, presumably because phase OFF variants are able to evade cross-immunity (20). These data provide indirect evidence that fimbrial phase variation is a mechanism to evade cross-immunity between serotypes Typhimurium and Enteritidis. Furthermore, this hypothesis predicts that cross-immunity between serotypes Typhimurium and Enteritidis would be observed if a promoter that does not undergo phase variation drove expression of LP fimbriae. To test this prediction, mice were immunized with an lpf phase ON culture of serotype Typhimurium (TN3) and then challenged with serotype Enteritidis strain TN7 (Plac::lpfABCDE). On 14 of the 18 occasions on which bacterial numbers in fecal pellets were enumerated postinfection, the total numbers of serotype Enteritidis strain TN7 recovered from mice immunized with serotype Typhimurium strain TN3 were significantly lower than those recovered from a naive control group. Statistically significant differences were detected as early as 2 days postchallenge, and the difference was largest on day 19 postchallenge, when the average number of serotype Enteritidis TN7 recovered from naive mice was approximately 5,500-fold higher that that recovered from mice immunized with serotype Typhimurium strain TN3 (aroA lpfABCDE::lacZYA) (Fig. 4). Furthermore, significantly smaller numbers of serotype Enteritidis strain TN7 (Plac::lpfABCDE) were recovered at 8 days postinfection from Peyer's patches (P < 0.001) and mesenteric lymph nodes (P < 0.005) of mice immunized with an lpf phase ON culture of serotype Typhimurium (TN3) than from the organs of naive mice (Fig. 5). In contrast, the numbers of serotype Enteritidis strain TN7 recovered from organs of mice immunized with a Typhimurium lpf deletion mutant (TN4) were not significantly different from those recovered from naive mice (P > 0.1). Serum of mice immunized with an lpf phase ON culture of serotype Typhimurium (TN3) reacted with GST-LpfA fusion protein of serotype Enteritidis, suggesting in vivo expression of LpfA by the vaccine strain (TN3) (Table 2). These data further suggested that exposure to LP fimbrial proteins of serotype Typhimurium elicited the production of antibodies with cross-reactivity against LP fimbrial proteins of serotype Enteritidis.
In summary, we showed that exposure to LP fimbrial proteins during infection with serotype Typhimurium resulted in selection against a serotype Enteritidis strain expressing the lpf operon from the lac promoter, giving rise to significantly reduced bacterial numbers in feces and organs. These data directly support the hypothesis that LP fimbrial phase variation is a mechanism to evade cross-immunity between S. enterica serotypes Typhimurium and Enteritidis.
ACKNOWLEDGMENTS
We thank Renée Tsolis for helpful suggestions during this work and Robert Kingsley and Andrea Holland for comments on the manuscript.
Work in A.B.'s laboratory is supported by the Texas Advanced Research (Technology) Program under grant number 000089-0051-1999, Public Health Service grant AI40124, and Public Health Service grant AI44170.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 2.Barrow P A, Berchieri A, Jr, al-Haddad O. Serological response of chickens to infection with Salmonella gallinarum-S. pullorum detected by enzyme-linked immunosorbent assay. Avian Dis. 1992;36:227–236. [PubMed] [Google Scholar]
- 3.Bäumler A J, Gilde A J, Tsolis R M, van der Velden A W M, Ahmer B M M, Heffron F. Contribution of horizontal gene transfer and deletion events to the development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J Bacteriol. 1997;179:317–322. doi: 10.1128/jb.179.2.317-322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäumler A J, Heffron F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol. 1995;177:2087–2097. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler A J, Tsolis R M, van der Velden A W M, Stojiljkovic I, Anic S, Heffron F. Identification of a new iron regulated locus of Salmonella typhi. Gene. 1996;193:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- 6.Chart H, Rowe B, Baskerville A, Humphrey T J. Serological response of chickens to Salmonella enteritidis infection. Epidemiol Infect. 1990;104:63–71. doi: 10.1017/s0950268800054534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper G L, Venables L M, Woodward M J, Hormaeche C E. Vaccination of chickens with strain CVL30, a genetically defined Salmonella enteritidis aroA live oral vaccine candidate. Infect Immun. 1994;62:4747–4754. doi: 10.1128/iai.62.11.4747-4754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.deLorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant S G N, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber A, Zingales B. Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. BioTechniques. 1995;19:28–30. [PubMed] [Google Scholar]
- 11.Hormaeche C E, Joysey H S, Desilva L, Izhar M, Stocker B A. Immunity conferred by Aro− Salmonella live vaccines. Microb Pathog. 1991;10:149–158. doi: 10.1016/0882-4010(91)90075-l. [DOI] [PubMed] [Google Scholar]
- 12.Hormaeche C E, Mastroeni P, Harrison J A, Demarco de Hormaeche R, Svenson S, Stocker B A. Protection against oral challenge three months after i.v. immunization of BALB/c mice with live Aro Salmonella typhimurium and Salmonella enteritidis vaccines is serotype (species)-dependent and only partially determined by the main LPS O antigen. Vaccine. 1996;14:251–259. doi: 10.1016/0264-410x(95)00249-z. [DOI] [PubMed] [Google Scholar]
- 13.Kelterborn E. Salmonella-species: first isolations, names and occurrence. S. Leipzig, Germany: Hirzel Verlag; 1967. [Google Scholar]
- 14.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 15.Kingsley R A, Bäumler A J. Host adaptation and the emergence of infectious disease: the Salmonella paradigm. Mol Microbiol. 2000;36:1006–1014. doi: 10.1046/j.1365-2958.2000.01907.x. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis T, Sambrook J, Fritsch E F. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.McCormick B A, Klemm P, Krogfelt K A, Burghoff R L, Pallesen L, Laux D C, Cohen P S. Escherichia coli F-18 phase locked ‘on’ for expression of type 1 fimbriae is a poor colonizer of the streptomycin-treated mouse large intestine. Microb Pathog. 1993;14:33–43. doi: 10.1006/mpat.1993.1004. [DOI] [PubMed] [Google Scholar]
- 18.Nicholas R A, Cullen G A. Development and application of an ELISA for detecting antibodies to Salmonella enteritidis in chicken flocks. Vet Rec. 1991;128:74–76. doi: 10.1136/vr.128.4.74. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson B, Low D. DNA methylation-dependent regulation of Pef expression in Salmonella typhimurium. Mol Microbiol. 2000;35:728–742. doi: 10.1046/j.1365-2958.2000.01743.x. [DOI] [PubMed] [Google Scholar]
- 20.Norris T L, Baumler A J. Phase variation of the lpf operon is a mechanism to evade cross-immunity between Salmonella serotypes. Proc Natl Acad Sci USA. 1999;96:13393–13398. doi: 10.1073/pnas.96.23.13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norris T L, Kingsley R A, Bäumler A J. Expression and transcriptional control of the Salmonella typhimurium lpf fimbrial operon by phase variation. Mol Microbiol. 1998;29:311–320. doi: 10.1046/j.1365-2958.1998.00934.x. [DOI] [PubMed] [Google Scholar]
- 22.Old D C, Corneil I, Gibson L F, Thomson A D, Duguid J P. Fimbriation, pellicle formation and the amount of growth of salmonellas in broth. J Gen Microbiol. 1968;51:1–16. doi: 10.1099/00221287-51-1-1. [DOI] [PubMed] [Google Scholar]
- 23.Old D C, Duguid J P. Selective outgrowth of fimbriate bacteria in static liquid medium. J Bacteriol. 1970;103:447–456. doi: 10.1128/jb.103.2.447-456.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Short J M, Fernandez J M, Sorge J A, Huse W D. λZAP: a bacteriophage expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 26.Smith B P, Dilling G W, House J K, Konrad H, Moore N. Enzyme-linked immunosorbent assay for Salmonella serology using lipopolysaccharide antigen. J Vet Diagn Investig. 1995;7:481–487. doi: 10.1177/104063879500700410. [DOI] [PubMed] [Google Scholar]
- 27.Stojiljkovic I, Bäumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA, cchB, eutE, eutJ, eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swenson D L, Clegg S. Identification of ancillary fim genes affecting fimA expression in Salmonella typhimurium. J Bacteriol. 1992;174:7697–7704. doi: 10.1128/jb.174.23.7697-7704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valentine P J, Devore B P, Heffron F. Identification of three highly attenuated Salmonella typhimurium mutants that are more immunogenic and protective in mice than a prototypical aroA mutant. Infect Immun. 1998;66:3378–3383. doi: 10.1128/iai.66.7.3378-3383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]