Abstract
As representative of the early-divergent groups of angiosperms, Saxifragales is extremely divergent in morphology, comprising 15 families. Within this order, our previous case studies observed significant structural diversities among the plastomes of several lineages, suggesting a possible role in elucidating their deep phylogenetic relationships. Here, we collected 208 available plastomes from 11 constituent families to explore the evolutionary patterns among Saxifragales. With thorough comparisons, the losses of two genes and three introns were found in several groups. Notably, 432 indel events have been observed from the introns of all 17 plastomic intron-containing genes, which could well play an important role in family barcoding. Moreover, numerous heterogeneities and strong intrafamilial phylogenetic implications were revealed in pttRNA (plastomic tRNA) structures, and the unique structural patterns were also determined for five families. Most importantly, based on the well-supported phylogenetic trees, evident phylogenetic signals were detected in combinations with the identified pttRNAs features and intron indels, demonstrating abundant lineage-specific characteristics for Saxifragales. Collectively, the results reported here could not only provide a deeper understanding into the evolutionary patterns of Saxifragales, but also provide a case study for exploring the plastome evolution at a high taxonomic level of angiosperms.
Keywords: Saxifragales, angiosperms, plant evolution, plastome diversity, pttRNAs, microstructural changes, gene loss, intron loss, phylogeny
1. Introduction
Throughout the investigation of plant phylogeny, plastid data play an important role in plant systematics. As one of the most iconic features of plant cells [1], the plastome can be highly informative in reflecting the plant evolution [2]. In higher plants, the plastid is uniparentally inherited and its genome commonly possesses a quadripartite structure, 120–160 kb in size [3]. Furthermore, the substitution rates of plastomic genes lie in the middle between mitochondrial and nuclear substitution [4], although they may vary across different taxa or among genes [5,6,7]. Consequently, these features mentioned above make plastomes a perfect choice for inferring plant phylogeny [8,9]. With the advent of deep sequencing technology, plastome data has gradually served as a valuable routine tool for molecular evolutionary analyses. To date, more than 8600 plastomes are accessible via Genbank.
For angiosperms, there is a general consensus that the organization and gene content of plastomes is greatly conserved [10,11,12,13]. However, along with the rapidly expanding availability of sequence data, many variation events of plastomes have been detected in structures. For instance, total losses of 26 genes and 8 introns of plastomes were observed during the evolution of angiosperms [14]. These losses might be attributed to the transfer of plastomic genes to the nucleus or their function replacement by nuclear genes [15,16,17,18,19]. Additionally, several gene and intron losses, including infA, rpl32, rps16, the 2nd intron of ycf3, etc., were proved to be highly lineage-specific among the investigated species of Malpighiales [20]. In addition, introns of plastomes, with considerable structured sequences, have been proven to undergo complicated evolution processes, resulting in either sequence conservation or mutational hotspots [21]. Most interestingly, some indels of group II introns, such as the petD intron, have been found to be highly useful in the phylogeny of basal angiosperms [22].
Indeed, over the processes of nucleotide mutations, notable structural changes are observed within plastomes. The most conspicuous example is the expansions or contractions of inverted repeats (IRs: IRa and IRb), which could influence not only the overall size of plastome [23,24,25,26], but also the plastomic gene organization [27,28,29]. It is also worth noting that such structural changes have taxonomic significance and can be employed as tools for evolutionary interpretation [30,31,32]. For instance, a unique pattern of rps19 at the junction of LSC and IRb has been documented by several studies in Crassulaceae (Saxifragales) [1,33], which still holds true in a wider sampling of 69 species from this family [34]. Furthermore, a recently discovered structural variation has been attracting attention. As we know, transfer RNA (tRNA), with a characteristic clover leaf-like structure, acts an indispensable part during protein synthesis [35]. However, some variations have been identified by recent studies. More importantly, two current studies from our group strongly suggest that some novel structural variations detected in plastomic tRNAs (pttRNA) might have phylogenetic implications and can be used in potential plant barcoding [34,36].
As a representative of the early-divergent groups of angiosperms [37,38,39,40], the order Saxifragales comprises 15 families and was found to be monophyletic [41,42,43]. Due to the rapid radiation of Saxifragales [44,45], the extreme morphological diversity (trees, herbs, succulents, etc.) posed a major obstacle in earlier phylogenetic studies. As more molecular data were employed, the interfamilial relationships within Saxifragales have been generally well-resolved, with seven primary clades [34,45,46,47]. Saxifragales can serve as a good case for studying the diversification patterns in angiosperms [47]. By far, through careful investigation, several structural variations of plastomes have been observed among species of Saxifragales, such as the losses of infA and rpl32 in Paeoniaceae [1,48], the intron losses of rps16 in the genus Penthorum [1], the intron losses of rpl2 in the genera Saxifraga and Heuchera [1,49], and the family-specific pattern of the IR junction and the phylogenetic informative pttRNAs’ structures in Crassulaceae [34]. In fact, structural variations of plastomes have not been comprehensively and systematically studied within this order so far. Hence, more efforts are needed to address this issue.
To explore the plastomic variations within Saxifragales, we retrieved sequence data from 208 taxa representing 11 families of this order. By comprehensive analyses, we tried to address (1) the gene content variations among the plastomes of Saxifragales, (2) the microstructural changes within introns of plastomic genes, (3) the structural diversifications of pttRNAs, and (4) the patterns of plastomic homoplasy features of this order. Thereby, our findings may facilitate a deep understanding of the evolutionary patterns of Saxifragales, and also provide an excellent case study for plastome evolution at a higher taxonomic level.
2. Results
2.1. Overall Variations of Plastomic Gene Organization among Saxifragales
To obtain a more comprehensive estimation for the plastomic features among Saxifragales, we investigated a total of 208 taxa, which are summarized in Table 1. In general, these plastomes demonstrated a rather conserved pattern, including quadripartite structure, gene order, and GC content, etc. However, several differences were still recognized with respect to genomic size, loss event for both of PCGs (protein-coding gene) and introns, and gene content at IR junctions.
Table 1.
Basic genomic characteristics among the investigated 208 Saxifragales plastomes.
| Taxa | Size (Base Pair, bp) | GC Content (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | LSC | IR | SSC | Total | LSC | IR | SSC | |
| Altingiaceae | 160,641 ± 216 | 88,882–89,162 | 26,274–26,471 | 18,917–19,011 | 37.93 ± 0.03 | 36.04–36.10 | 43.04–43.08 | 32.18–32.42 |
| Cercidiphyllaceae | 159,877 ± 32 | 88,035–88,058 | 26,427–26,434 | 18,973–18,965 | 37.92 ± 0.01 | 36.00 | 43.00 | 32.40 |
| Crassulaceae | 150,690 ± 1013 | 79,465–83,253 | 24,810–25,984 | 16,520–17,111 | 37.75 ± 0.10 | 35.45–36.29 | 42.80–43.31 | 31.09–32.40 |
| Daphniphyllaceae | 160,273 ± 192 | 88,075–88,103 | 26,546–26,605 | 18,970–19,095 | 37.86 ± 0.04 | 36.00–36.10 | 42.90–43.00 | 32.10–32.10 |
| Grossulariaceae | 157,559 ± 313 | 86,812–87,412 | 25,887–26,018 | 18,334–18,562 | 38.13 ± 0.02 | 36.20 | 43.08–43.14 | 33.20–33.40 |
| Haloragaceae | 159,050 ± 781 | 88,165–89,941 | 25,637–25,978 | 18,469–19,000 | 36.73 ± 0.22 | 34.20–35.00 | 42.73–42.88 | 30.20–30.90 |
| Hamamelidaceae | 159,293 ± 477 | 87,102–89,016 | 26,209–26,422 | 18,127–19,173 | 38.00 ± 0.07 | 35.75–36.35 | 43.04–43.22 | 32.27–32.89 |
| Iteaceae | 160,258 | 88,714 | 26,648 | 18,248 | 37.10 | 34.80 | 42.70 | 31.60 |
| Paeoniaceae | 152,834 ± 429 | 84,242–86,057 | 25,246–25,751 | 16,681–17,423 | 38.41 ± 0.05 | 36.61–36.83 | 43.04–43.18 | 32.57–33.02 |
| Penthoraceae | 156,686 | 86,735 | 25,776 | 18,399 | 37.30 | 35.20 | 42.80 | 31.30 |
| Saxifragaceae | 154,057 ± 2863 | 79,310–88,109 | 25,097–26,224 | 15,082–18,447 | 37.77 ± 0.19 | 35.05–36.22 | 42.69–43.28 | 31.16–32.85 |
Primarily, among the 208 plastomes, the ranges of 145,737 (Crassula perforate)–160,861 bp (Altingia excelsa) and 36.48 (Myriophyllum aquaticum)–38.55% (Paeonia brownii) were identified for genome size and GC content, respectively. Notably, with the whole plastome length ranging from 160,401 to 160,861 bp, Altingiaceae (represented by seven taxa) generally exhibits the largest sizes within this order (160,641 ± 216 bp), followed by Daphniphyllaceae and Iteaceae (represented by only one species). As for the GC content, the seven Haloragaceae plastomes featured the lowest value, with an average of 36.73 ± 0.22 % for total genome and 30.20–30.90% for SSC regions. Moreover, several loss events were detected for both plastomic genes and introns. The genes infA and rpl32 were found lost in all the involved 20 Paeoniaceae plastomes. Intron losses were also identified in three genes, i.e., atpF in Pachyphytum compactum, rps16 in Penthorum chinense, and rpl2 in Saxifragaceae (containing 50 taxa).
In addition, we also compared the expansion and contraction of IR junctions to the adjacent genes among Saxifragales. Interestingly, as listed in Table S1, both similarities and differences were revealed. By thorough comparison, two features were highly conserved in all 208 plastomes: (1) IRa possessed an expansion to ycf1 (ranging from 400–1870 bp), accordingly resulting in a partial fragment (pseudo-copy) in IRb; and (2) the 3’-terminal 3 bp of trnH-GUG was located in IRa. On the contrary, considerable diversities were harbored by the contraction and expansion to genes of JLB (junction IRb/LSC) and JSB (junction IRb/SSC). For simplicity, six different patterns could be outlined: (1) expansion for JLB to rps19 and contraction for JSB to ndhF (Altingiaceae, Daphniphyllaceae, Grossulariaceae, and Iteaceae), (2) contraction for JSB to rps19 and expansion for JSB to ndhF (Penthoraceae), (3) expansion/contraction to rps19 for JLB and only expansion for JSB to ndhF (Haloragaceae), (4) only expansion (Crassulaceae), or (5) only contraction (Cercidiphyllaceae), or (6) expansion/contraction (Hamamelidaceae, Paeoniaceae and Saxifragaceae) for both junctions of JLB and JSB to these two genes.
Most notably, family-specific features were found in two families: (1) the IRb regions of Crassulaceae all expanded 110 bp to rps19, with only four exceptions (105 bp for Orostachys fimbriata, Phedimus kamtschaticus, P. spurius, and P. takesimense); and (2) the IRa regions of Paeoniaceae all have 1077-bp expansions to ycf1, except for Paeonia emodi (1105 bp) and P. lactiflora (1081 bp).
2.2. Hypervariable Loci Assessment among Saxifragales
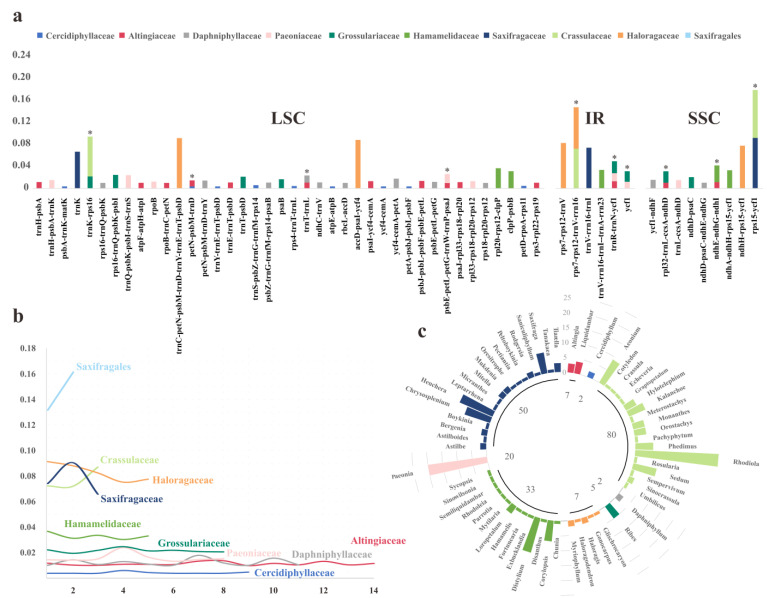
To reach an overall exploration of the sequence variations among Saxifragales plastomes, the DNA polymorphism was evaluated by nucleotide diversity values (π). The comparative analyses were performed in ten groups: nine separate families (excluding Iteaceae and Penthoraceae, that with only one sequence data released, respectively) (Figure 1c), and the whole order of Saxifragales. As detailed in Table S2, the patterns of hypervariable loci generally exhibited a high degree of diversity among the families. As for the number of HPR (highly polymorphic region), Crassulaceae and Saxifragaceae displayed the smallest (three for both), followed by Haloragaceae and Hamamelidaceae with five loci, respectively, while the highest was seen for Altingiaceae (14), followed by Daphniphyllaceae and Cercidiphyllaceae. Excluding the same loci among different groups, we identified a total of 55 HPRs. Of these HPRs, nine (16.4%) and six (10.9%) were located in SSC and IR regions, respectively, with the majority (72.7%) in LSC regions (Figure 1a). Moreover, ten separate HPRs were shared by some families. For instance, one HPR (trnR-ACG-trnN-GUU-ycf1) was possessed by three families (Altingiaceae, Paeoniaceae, and Grossulariaceae). Each of the remaining nine HPRs were observed in two of the families.
Figure 1.
The hotspots patterns among Saxifragales. (a) Distributions of all the identified HPR loci, the HPRs that were shared by multiple families were marked by *; (b) Patterns of π values of different lineages; (c) Taxon samplings of this analysis, with the numbers in the circle indicating the numbers of sampling plastomes in each family, the numbers and heights of columns represented the number of genera involved, and plastomes that contained in each genus.
Most interestingly, the patterns of π values appeared to be the opposite. As outlined in Figure 1b, the four families (Saxifragaceae, Crassulaceae, Haloragaceae, and Hamamelidaceae) with less HPR harbored much higher values—averaging from 0.03294 to 0.08285—than the other five (Grossulariaceae, Paeoniaceae, Daphniphyllaceae, Altingiaceae, and Cercidiphyllaceae) with more loci present as the lower values (0.00417–0.02153). Further, at the order level, only two regions were classified as HPRs, i.e., rpoB (π = 0.13251) and ndhH-rps15-ycf1 (π = 0.16323).
2.3. Microstructural Changes within Plastomic Introns
In total, 432 indel events have been obtained from the aligned intron matrixes of 17 separate plastomic genes (five tRNA genes and 12 PCGs) in the 208 Saxifragales taxa (Table S3). Among them, 221 indels (51.2%) represented deletions, while the remaining 211 (48.8%) were insertions. Moreover, notable heterogeneities of indel sizes have been observed (ranging from 1 to 176 nt), with the largest proportion of 5-nt indels (14.7%), followed by 6-nt (11.6%) and single-base (9.3%). However, overall, these indels were mostly no more than 10 nt in size (79.7%). Additionally, the indels remarkably displayed uneven distributions among the 17 intron-containing genes. In general, the greatest number of indels were harbored by trnK-UUU (62, 14.4%), then by ndhA (40, 9.3%), and rps16 (33, 7.6%). In contrast, three plastomic genes, i.e., rps12, ndhB, and rpl2, showed far fewer indels, with only one (0.2%), four (0.9%), and five (1.2%), respectively.
Strikingly, the indel patterns among Saxifragales exhibited a high potential for assessing phylogenetic relationships at the family level. With the only exception of Hamamelidaceae, all investigated families had multiple family-unique indels. As can be seen from Table 2, Paeoniaceae had the highest number of unique indels (82) compared to other nine families (3–41). Meanwhile, indels from the introns of trnK-UUU, ndhA, and rps16 could successfully distinguish seven families, respectively. Notably, the 3′ introns of rps12 are highly conserved across Saxifragales, with no family-unique indels. Further, in the intron regions of ndhB and trnL-UAA, only one unique indel site was observed in Iteaceae and Crassulaceae, respectively.
Table 2.
Family-specific indels identified from the intron matrix of 17 plastomic genes among Saxifragales.
| Taxa | Total | trnA | trnI | trnK | trnL | trnV | atpF | clpPa | clpPb | ndhA | ndhB | petB | petD | rpl16 | rpl2 | rpoC1 | rps12 | rps16 | ycf3a | ycf3b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alting. | 10/4/14 | - | 1/-/1 | 2/1/3 | - | -/1/1 | 2/-/2 | - | - | - | - | - | 1/-/1 | 1/-/1 | - | 1/-/1 | - | 1/-/1 | -/1/1 | 1/1/2 |
| Cercidi. | 2/3/5 | - | - | - | - | - | - | -/1/1 | - | 1/-/1 | - | - | -/1/1 | - | - | 1/-/1 | - | -/1/1 | - | - |
| Crass. | 26/15/41 | - | 1/-/1 | 2/1/3 | 1/1/2 | -/1/1 | - | 2/2/4 | 5/-/5 | 3/1/4 | - | 1/1/2 | 0/2/2 | 1/1/2 | 1/-/1 | -/1/1 | - | 5/1/6 | 2/2/4 | 2/1/3 |
| Daphni. | 1/2/3 | - | - | -/1/1 | - | - | - | - | -/1/1 | - | - | 1/-/1 | - | - | - | - | - | - | - | - |
| Grossu. | 6/6/12 | - | 1/-/1 | - | - | 1/-/1 | -/1/1 | - | - | -/1/1 | - | - | 3/-/3 | 1/2/3 | - | - | - | -/1/1 | - | -/1/1 |
| Halora. | 5/11/16 | -/1/1 | - | 1/-/1 | - | - | -/2/2 | 1/-/1 | - | 2/3/5 | - | - | - | - | - | -/2/2 | - | -/1/1 | 1/1/2 | -/1/1 |
| Hama. | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Itea. | 2/8/10 | - | - | -/2/2 | - | - | - | - | - | -/1/1 | -/1/1 | - | - | 1/1/2 | - | -/1/1 | - | -/2/2 | - | 1/-/1 |
| Paeonia. | 43/39/82 | 1/-/1 | 3/2/5 | 3/4/7 | - | 2/2/4 | 2/6/8 | 4/3/7 | 2/3/5 | 5/1/6 | - | - | 5/4/9 | 3/4/7 | -/1/1 | 2/3/5 | - | 6/4/10 | 3/1/4 | 2/1/3 |
| Pentho. | 2/5/7 | - | 1/-/1 | -/1/1 | - | 1/-/1 | -/1/1 | -/1/1 | - | -/1/1 | - | -/1/1 | - | - | - | - | - | - | - | - |
| Saxifra. | 4/2/6 | - | - | - | - | - | - | - | 3/-/3 | - | - | -/2/2 | 1/-/1 | - | - | - | - | - | - | - |
2.4. Specific Markers from pttRNAs’ Structural Diversifications
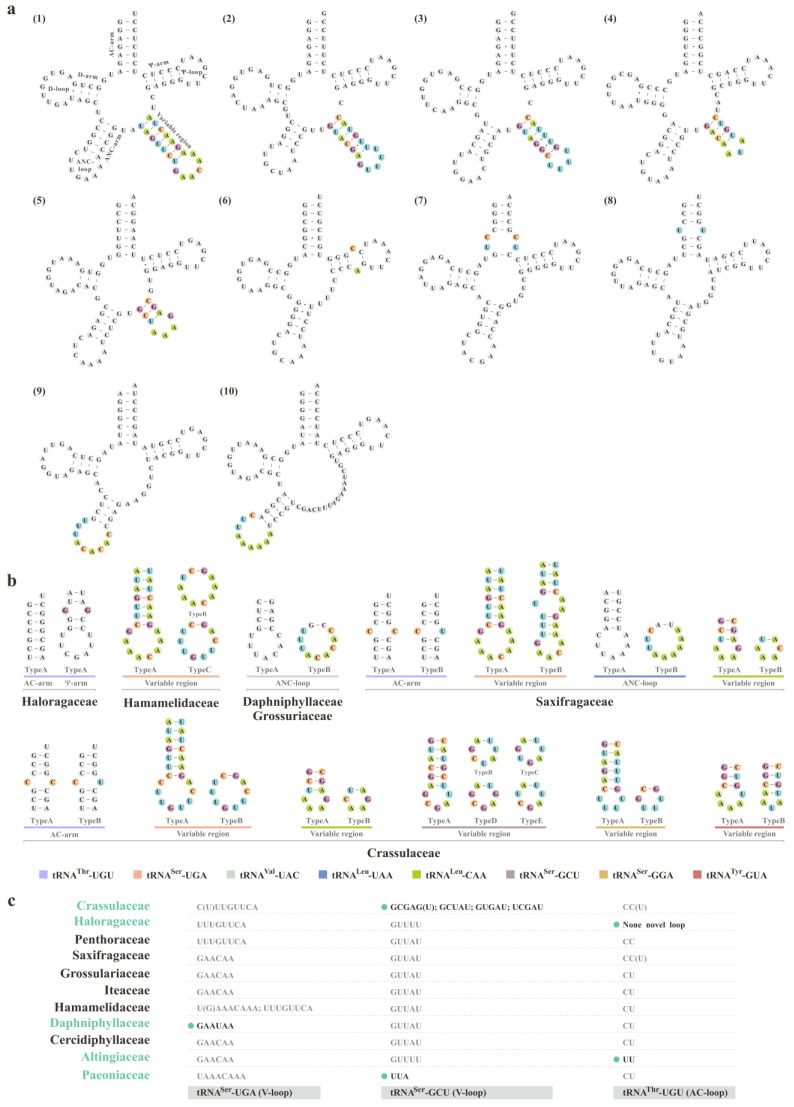
The secondary structures of pttRNAs were predicted within Saxifragales. Among the 7488 pttRNAs investigated in this study, 10 putative non-typical structures were observed for all the constituent families (Figure 2a). In general, four separate categories can be inferred from these unconventional pttRNA structures, including (1) an additional loop at AC-arm (tRNAArg-ACG with a 4-nt loop, tRNAThr-UGU with a 2-nt loop); (2) an expanded 9-nt ANC-loop (tRNAVal-UAC and tRNALeu-UAA); (3) a long variable region at V-arm (tRNALeu-CAA, tRNASer-UGA, tRNASer-GCU, tRNASer-GGA, and tRNATyr-GUA); and (4) an extra loop at Ψ-arm (tRNACys-GCA), respectively.
Figure 2.
The pttRNAs’ structural diversifications among Saxifragales. (a) The predicted secondary structures of pttRNAs from Altingiaceae (as a representative) and the locations of non-typical structures were indicated by color:, (1) tRNASer-UGA, (2) tRNASer-GCU, (3) tRNASer-GGA, (4) tRNATyr-GUA, (5) tRNALeu-CAA, (6) tRNACys-GCA, (7) tRNAArg-ACG, (8) tRNAThr-UGU, (9) tRNAVal-UAC, (10) tRNALeu-UAA; (b) Abundant diversities identified within the 11 investigated families; (c) Unique structural patterns that were detected for five families through three pttRNAs.
To gain deeper insights into the implications harbored by such heterogeneities, overall comparison was further conducted among all the non-typical pttRNA structures mentioned above. Strikingly, we found that these structures could be highly specific to different hierarchies of Saxifragales (i.e., interfamily and intrafamily levels). As for intrafamilial level, nine families (except for Iteaceae and Penthoraceae), which contained more than one sequenced plastome, were considered for further analyses. Among the nine families, three were examined to be strictly conserved for the non-typical structures, while substantial variations were identified in the other six families. Of these, four of the six had significant infra-family differences (tRNAThr-UGU for Haloragaceae, tRNASer-UGA for Hamamelidaceae, and tRNAVal-UAC for Daphniphyllaceae and Grossulariaceae) (Figure 2b). For instance, within the family Grossulariaceae (representing five Ribes taxa), two types of ANC-loop in plastomic tRNAVal-UAC were revealed: (1) type A (typical 7-nt loop) for R. nevadense and R. roezlii and (2) type B (expanded 9-nt loop) for the other three taxa. Most surprisingly, in contrast to other families, we found that Crassulaceae and Saxifragaceae harbored wider intra-family variations, with six and four types of pttRNAs, respectively.
Interestingly, at the interfamilial level, unique structural patterns were also detected for five families through three pttRNAs. Figure 2c summarized the details of the characteristic features: (1) differing from other 10 families, Daphniphyllaceae featured 5′-GAAUAA-3′ in the V-loop of tRNASer-UGA, (2) for tRNASer-GCU, the distinctive 5′-UUA-3′ loop of variable regions was merely identified in all 20 Paeoniaceae taxa, (3) Crassulaceae was the only one that evolved five different types of V-loop in tRNASer-GCU, which were not found in the other families, (4) 5′-UU-3′ at AC-loop of tRNAThr-UGU was unique for Altingiaceae, and (5) 5′-GG-3′ at Ψ-loop (Glischrocaryon aureum) or a non-novel structure (all the remaining six accessible plastomes) of tRNAThr-UGU was observed in Haloragaceae.
2.5. Phyloplastomic Analyses among Saxifragales
To explore the taxonomic relationships within Saxifragales, 79 plastomic PCGs and complete cp genomes (CPGs) from 208 ingroups and three outgroups (Rosids) were employed for analyses, respectively. The phylogenomic inferences, obtained by 77,274-bp (PCG dataset) and 262,199-bp (CPG dataset) concatenated matrixes, respectively, generally yielded highly similar topologies between ML and BI trees (Figure 3 and Figure S1).
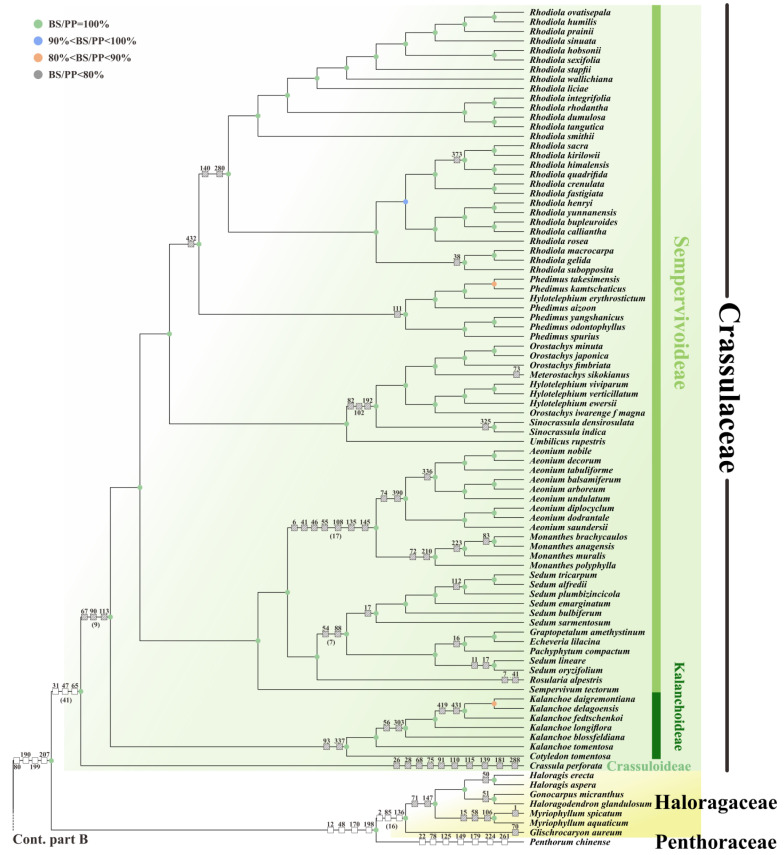
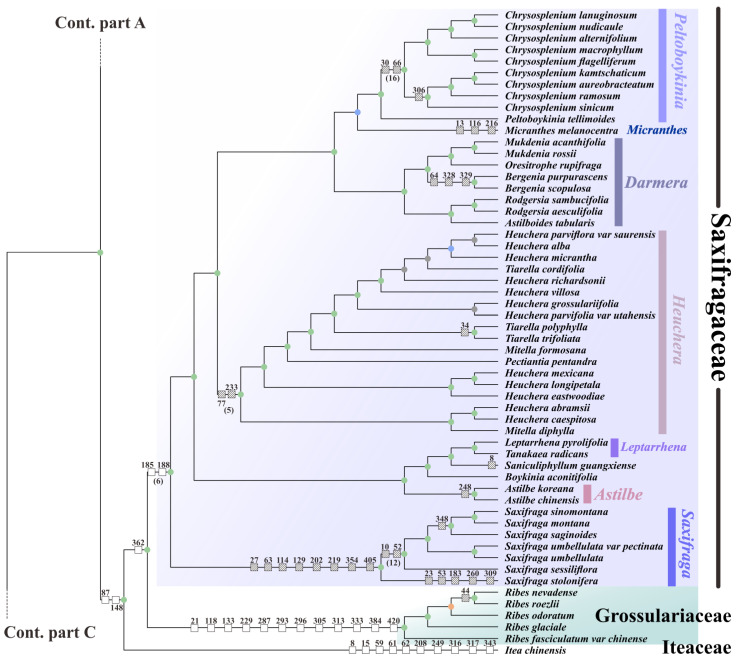
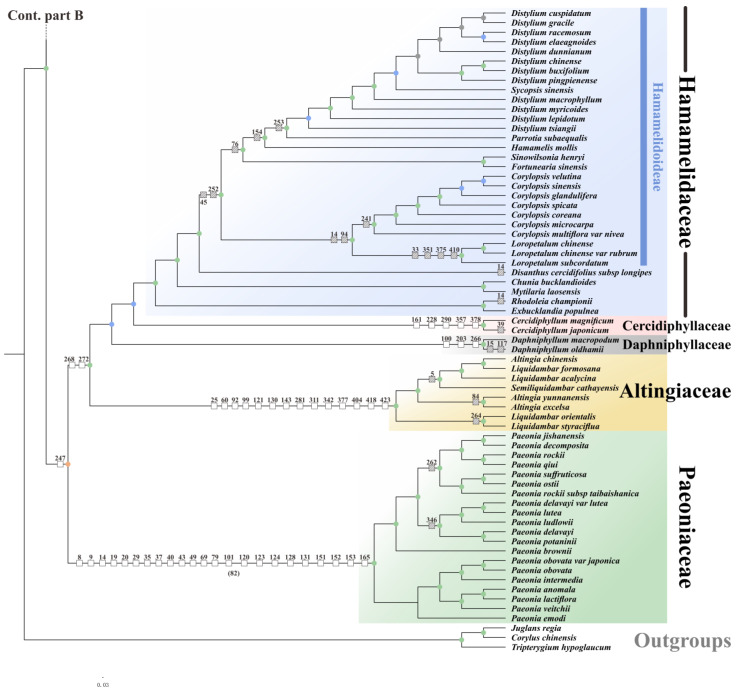
Figure 3.
The phylogenetic tree obtained by 79 PCGs among 208 Saxifragales plastomes, with BS and PP values shown by color circles. White boxes indicate the specific indels for deep-level nodes. Lined boxes indicate the specific indels for lower-level nodes. As those clades with too many specific indels were limited in branch lengths, we marked as many indels as possible, and the total number was noted under the corresponding clade.
Significantly, our results recovered two major clades of Saxifragales: (1) Paeoniaceae plus woody group (BS = 97%, PP = 0.99), and (2) “core Saxifragales” (BS = 100%, PP = 0.99). Therein, woody group comprises four families: Altingiaceae + (Daphniphyllaceae + (Cercidiphyllaceae + Hamamelidaceae)) and is sister to Paeoniaceae with relatively high support. In addition, core Saxifragales could be additionally divided into two alliances: Crassulaceae alliance (BS = 100%, PP = 0.99), with Crassulaceae sister to Haloragaceae + Penthoraceae, and Saxifragaceae alliance (BS = 100%, PP = 1.0), including Iteaceae + (Saxifragaceae + Grossulariaceae). Overall, our results were highly congruent with those of former analyses [34,46,47,48]. In general, the 79-PCGs datasets allowed high resolution and support for all the interfamilial relationships, except for those of two clades, i.e., Cercidiphyllaceae + Hamamelidaceae (BS = 65%, PP = 0.91) and Daphniphyllaceae + (Cercidiphyllaceae + Hamamelidaceae) (BS = 55%, PP = 0.92).
Significantly, by further exploration, with the exclusion of Iteaceae and Penthoraceae (represented by one species, respectively), the monophyly of each of the nine families investigated was strongly supported (BS = 100%, PP = 1.0). Moreover, within these families, most of the primary subclades were also well resolved to be monophyletic (BS = 100%, PP = 0.99 or 1.0), such as Sempervivoideae and Kalanchoideae in Crassulaceae; Hamamelidoideae in Hamamelidaceae; as well as Peltoboykinia (including Chrysosplenium and Peltoboykinia), Darmera (including five genera such as Mukdenia and Oresirophe), and Heuchera (including Heuchera and other related four genera) groups in Saxifragaceae, etc. Moreover, several genera turned out to be non-monophyletic, e.g., Altingia and Liquidambar (Altingiaceae), Sedum, Hylotelephium and Orostachys (Crassulaceae), and Heuchera, Tiarella, and Mitella (Saxifragaceae).
3. Discussion
In the present study, a thorough and comprehensive comparison among 208 Saxifragales plastomes was provided with two main aims: (1) reaching an overall understanding of the plastomic evolutionary diversities within this order; and (2) assessing the potential barcoding performance of plastid genomes for not only phylogeny, but also the specific evolved characteristics in plant. Notably, both sequence and structure diversities were clarified across these plastomes, including basic genomic features, gene content, IR boundary patterns, DNA polymorphisms, introns variabilities, structural variations in pttRNAs, and phylogenetic signals and interpretations. Collectively, the results presented here would provide further insights into the evolutionary elucidations for the early-divergent angiosperms, especially for the order Saxifragales.
Despite the quite conserved gene content of angiosperm plastomes, loss events were not rare across their evolutionary history, supposedly after the first endosymbiotic event [14,50]. Jansen et al. [14] sampled 77 PCGs and four rRNAs from each of the 64 plastomes, representing the most major angiosperm groups, and remarkably, this study suggested that all loss events were identified in monocot and eudicot lineages. As one of the largest orders in the core eudicots clade [43], Saxifragales was also found to embody several losses for two genes and three introns of plastomes in our study. Therein, all the 20 investigated Paeoniaceae plastomes lost infA and rpl32, reinforcing the results of our previous works [34,48]. In fact, over the plastomic evolution, independent losses of infA have occurred repeatedly among angiosperms [14,51,52]. Additionally, in these cases, a documented mechanism, lateral gene transfer from plastome to nuclear, has been described for the losses of infA in Rosids [53] and rpl32 in Salicaceae [54,55]. Likewise, the observed three intron losses (atpF, rps16, and rpl2) were also present multiple times [14,49], such as the intron losses of atpF in Malphigiales [56], rps16 in Celastraceae [57], and rpl2 in Lythraceae [58], etc. It is worth noting, by far, that there are three possible pathways for these involved plastomic intron losses: (1) recombination of the RNA-edited intron lacking copy and the initial intact copy (atpF) [56,59]; (2) homologous recombination and reverse transcript mediated mechanism (rps16) [57]; and (3) unequal crossover and gene conversion (rpl2) [60,61,62].
As in earlier studies of molecular phylogeny, only several plastid gene loci were employed, such as matK, rbcL, trnH-psbA, etc. However, some of these core markers have been documented to display low efficiency in resolving many closely related taxa [23,63,64,65,66]. Over the years, as the organelle genomics progressed, it has been well known that plastomes embody numerous potential mutations clustering as “hotspots” across evolution [23,67,68]. Undeniably, delving into the specific HPRs in investigated taxonomic groups is necessary for reaching better barcoding performance [67]. In the case of this work, our results demonstrated the variability patterns of plastomes among the nine families of Saxifragales. Within the plastomes, we observed that LSC regions occupy the major hotspots loci, which was congruent with previous studies, including Liu et al. [69] in Oresitrophe and Mukdenia (Saxifragaceae), Liu et al. [67] in Ormosia (Fabaceae), and Xu et al. [70] in Saccharum (Poaceae). In contrast to LSC and SSC, IR regions generally could accommodate fewer mutations, which might be confined to consistency correction of its two copies [71,72].
In addition, we further assess the applicability of the identified HPRs. Significantly, with the exception of a few uniform sites, apparent substantial inconsistencies were found in comparison with former studies. For example, the results from this work (with 33 taxa) and Wang et al. [73] (with 6 taxa) identified five and seven HPRs in Hamamelidaceae, respectively, only sharing one identical loci (ndhG). Moreover, a similar finding was also found in Grossulariaceae taxa [74]. According to Shahzadi et al. [23], these discrepancies might be caused by different diversity levels of the involved taxa or the impact of analysis approaches. Thus, wider samples and more efforts are needed to clarify the hotspots’ patterns within Saxifragales. Most importantly, plastomic HPRs might serve as potential markers for plant DNA barcoding and phylogenetic inferences.
Intron and IGS regions of plastome sequences, to our best knowledge, could serve as primary sources of phylogeny data [75,76]. Overall, compared to most coding sequences, these two noncoding regions generally possessed higher diversity [22]. It is also worth mentioning that indels, known as the microstructural variabilities, were frequently present at these regions [75]. Yet, among Saxifragales plastomes, the distribution pattern of indels in the noncoding regions were still unclear. Here, with relatively large samples, our thorough analyses allowed determinations of 432 indels within introns. Notably, their size patterns, most no longer than 10 nt, turned out to be consistent with the results of Lohne et al. [22] in the petD intron and petB-petD region among angiosperms. Moreover, as mentioned above, our results found that the 3′-rps12 introns had the rarest indels across Saxifragales. Interestingly, a similar finding was also found by Graham et al. [75] in 3′-rps12 introns of 31 angiosperms. Most significantly, such highly conservative evolution of this intron might be implied by the unique characteristic of rps12. As the sole trans-splicing plastomic gene in plants [77], it has been documented that the unique exon division of rps12 requires conserved intron regions [78]. Above all, by the comparative analyses of plastomes among the constituent families within Saxifragales, abundant indel events were revealed to be highly family-specific, which could well play an important role in plant family level discrimination.
Chloroplast, the metabolic center of higher plants, can involve many fundamental synthesis pathways, such as proteins, lipids, phytohormones, etc. [2]. Encoded by plastomes, pttRNAs act as indispensable parts in translations as the linkage between mRNA and proteins [36,79,80]. As presented by Brennan and Sundaralingam [81], tRNAs could be divided into two categories based on the length of the V-arm: the most common, type I, features a short V-loop (4–5 nt) and type II, with a longer region (10 nt or over), is now thought to be limited to leucine, serine, and tyrosine [36,81,82]. Interestingly, it has been proposed that the bulky V-arm might have an impact on tRNA’s function, by assisting the combination with ribosomes [82]. Several studies have recently explored pttRNAs’ structural variations from type II, and found that they might be highly conserved at relatively low taxonomic ranks, such as two genera of Crassulaceae (Aeonium and Monanthes) [34], Viburnum of Adoxaceae [83] and Bletilla of Orchidaceae [36].
Here, we provided a comprehensive investigation of pttRNAs focused on the order level. The unique characteristics of type II tRNAs were detected in five pttRNAs from three isotypes in Saxifragales (Leu, Ser, and Tyr). At first, in the current study, we identified strong phylogenetic signals in several families. For instance, by extending our previous work [34] using a larger dataset, we reconfirmed the unique 5′-AUA-3′ V-loop of tRNATyr-GUA in Kalanchoideae (Crassulaceae); all involved Ribes (Grossuriaceae) taxa possessed an expanded ANC-loop in tRNAVal-UAC, except for two closely related species (R. nevadense and R. roezlii, [BS] = 100, [PP] = 1.0) with an ordinary 7-nt loop. Then, within the order Saxifragales, our results clearly demonstrate that Crassulaceae harbors the most extensive diversifications in pttRNAs’ structures. Interestingly, compared to other families, the Crassulaceae clade had considerably longer branch lengths from the early well-supported phylogram of Saxifragales [46,48], indicating that this clade had accumulated more mutations in the process of evolution [84,85,86,87]. Finally, we compared the ANC-loop structures of tRNAVal-UAC and tRNALeu-UAA among Saxifragales, respectively. It was notable that most of the investigated families had an expanded nine-bases loop in the two pttRNAs. However, a typical 7-nt ANC-loop was also identified, which mainly comes from pttRNAs of Saxifragaceae (with the exception of two Saxifraga taxa). It has been proposed that these expanded ANC-loops were attributed to the distal mismatch at the ANC-helix [88,89], perhaps C-U and A-C mismatches for Valine and Leucine, respectively. Most importantly, this unusual structure might influence the step size in translocation and result in a different reading number of bases (three or four) [89,90], which could further generate an impact on the translation. The findings reported here clearly reinforce the high potential role of pttRNA structures in plant taxonomy and DNA barcoding.
Previous efforts have been committed to exploring the backbone phylogeny of Saxifragales. Notably, there are several classic studies in resolving taxonomic relationships within this order, including Fishbein et al. [45], with three plastid and two nuclear genes; Jian et al. [46], based on ten plastomic, four mitochondrial, and two nuclear genes; and Soltis et al. [47], by supermatrix data. In general, these works have collectively resolved parts of the major nodes, such as “core Saxifragales”, Saxifragales, and Crassulaceae alliances. However, by far, the unresolved issues seem to converge upon several families, including Paeoniaceae and the members of woody group, mainly due to the poor support and varied taxonomic positions. For instance, Paeoniaceae was found variedly and weakly a sister to Crassulaceae alliance, Peridiscaceae, or woody group, etc. by different analyses.
In this study, the phyloplastomic analyses yield generally well-supported topologies among 208 Saxifragales taxa. For both the CPG and PCG trees, all the relatively well-supported (BS/PP > 90%) nodes possessed the same topologies, which occupied the majority of all the nodes. Significantly, Paeoniaceae and the woody group formed a relatively well-supported clade in the CPG tree (97% in ML and 0.99 in BI), which received a stronger support not only than the PCG tree, but also than that of our previous work (employing 83 plastomic genes) with 89% in ML and 1.0 in BI [48]. Furthermore, within the woody group, Altingiaceae was sister to (Daphniphyllaceae + (Cercidiphyllaceae + Hamamelidaceae)), differing from (Cercidiphyllaceae + Daphniphyllaceae) + (Hamamelidaceae + Altingiaceae) in Fishbein et al. [91] or (Hamamelidaceae + (Cercidiphyllaceae + Daphniphyllaceae)) + Altingiaceae in Jian et al. [46] and Han et al. [34]. However, all of these relationships received poor support. As Fishbein et al. [45] proposed, the unresolved phylogeny in Saxifragales could be partially explained by the unequal pattern of branch lengths, especially for the placement of Paeoniaceae, which possesses the longest branch within Saxifragales. Moreover, the unclear relationships among the woody group might due to an ancient, rapid radiation [46,48]. Most importantly, our phylogenies might offer an improvement in clarifying the taxonomic position of Paeoniaceae. To better resolve these long-standing enigmas in Saxifragales, further efforts are necessary.
Most interestingly, many evolutionary signals were revealed by the phylogenies in combination with the identified specific characteristics (microstructural changes and pttRNA structures). On one hand, overall, a total of 373 informative indels were marked to the relevant nodes of the PCG tree (Figure 3 and Table S3). First, for the deep-level nodes of Saxifragales, indels were relatively rare, with only nine events: indels No. 80, 190, 199, and 207 for Crassulaceae alliance; No. 87 and 148 for Saxifragaceae alliance; No. 268 and 272 for the woody group; and No. 247 for Paeoniaceae + woody group. Meanwhile, in turn, numerous characteristics were marked at lower-level nodes, which contained not only the family-specific indels (196 indels in total, as summarized in Table 2), but also those specific to the internal nodes within families (148 indels). Examples included the subfamily Hamamelidoideae in Hamamelidaceae (indels No. 45 and 252); the group Heuchera in Saxifragaceae (indels No. 77, 233, 246, 279, and 324); and the genus Rhodiola in Crassulaceae (indels No. 140 and 280), etc. Further, the remaining 29 indels were found to be independent to several leaf nodes. For instance, indel No. 14, an 8-nt SSR insertion, was a homoplastic feature to Disanthus cercidifolius and Rhodoleia championii. In particular, these informative indels seemed to be independently accumulated among different taxa, which supported the expectation that microstructural changes were caused by various mutational processes [22].
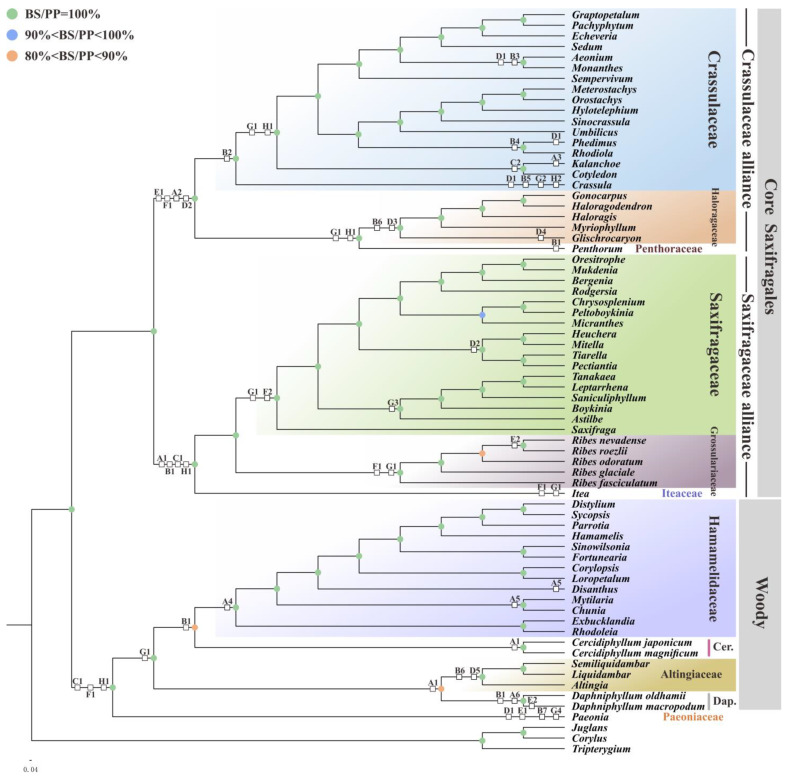
On the other hand, the combination between phylogenies and pttRNAs’ structural diversities allowed rather clear insights into the evolutionary patterns within Saxifragales. Based on the simplified PCG tree, a total of 30 lineage-specific characteristics of Saxifragales were concluded by eight pttRNAs (Figure 4 and Table 3). Among these pttRNAs, multiple heterogeneities were observed in tRNALeu-CAA (four at the V-loops), tRNAThr-UGU (five at the AC-arms), tRNASer-UGA (six at the V-loops), and tRNASer-GCU (seven at the V-loops) across the order. In contrast, tRNATyr-GUA, tRNASer-GGA, tRNALeu-UAA, and tRNAVal-UAC were found to be more conserved, with only two different structures, respectively. Most interestingly, among all the constituent families with multiple samplings, Grossulariaceae, Cercidiphyllaceae, and Daphniphyllaceae maintained the most family-specific characteristics, six for each. There might be two possible processes for such a phenomenon: one is preserving in slow-evolving species and the other is back mutations [92]. To further understand these evolutionary characteristics, more plastomes’ data and ulterior exploration are needed. Overall, the phylogenetic signals inferred here would not only offer potentially specific markers for Saxifragales taxa, but also provide further insights into their plastomic evolution.
Figure 4.
The simplified PCG tree combined with pttRNA structural characteristics. White boxes indicate the specific structures, which are detailed in Table 3. Note that the families Cercidiphyllaceae and Daphniphyllaceae are abbreviated as Cer. and Dap., respectively.
Table 3.
The evolutionary signals identified from the structures of pttRNAs among Saxifragales.
| Types of pttRNAs | Specific Structures | ||
|---|---|---|---|
| A | tRNASer-UGA (V-loop) | 1 | 5’-GAACAA-3’ |
| 2 | 5’-UUUGUUCA-3’ | ||
| 3 | 5’-CUUGUUCA-3’ | ||
| 4 | 5’-GAAACAAA-3’ | ||
| 5 | 5’-UAAACAAA-3’ | ||
| 6 | 5’-GAAUAA-3’ | ||
| B | tRNASer-GCU (V-loop) | 1 | 5’-GUUAU-3’ |
| 2 | 5’-GCGAU-3’ | ||
| 3 | 5’-UCGAU-3’ | ||
| 4 | 5’-GCUAU-3’ | ||
| 5 | 5’-GUGAU-3’ | ||
| 6 | 5’-GUUUU-3’ | ||
| 7 | 5’-UUA-3’ | ||
| C | tRNATyr-GUA (V-loop) | 1 | 5’-AUA-3’ |
| 2 | 5’-AAAAU-3’ | ||
| D | tRNAThr-UGU (AC-loop) | 1 | 5’-CU-3’ |
| 2 | 5’-CC-3’ | ||
| 3 | No additional loop | ||
| 4 | 5’-GG-3’ at T-arm | ||
| 5 | 5’-UU-3’ | ||
| E | tRNAVal-UAC (ANC-loop) | 1 | Expanded 9-nt loop |
| 2 | Typical 7-nt loop | ||
| F | tRNALeu-UAA (ANC-loop) | 1 | Expanded 9-nt loop |
| 2 | Typical 7-nt loop | ||
| G | tRNALeu -CAA (V-loop) | 1 | 5’-AAAG -3 |
| 2 | 5’-CAAG-3′ | ||
| 3 | 5’-AAAC-3′ | ||
| 4 | 5’-AAAU-3’ | ||
| H | tRNASer -GGA (V-loop) | 1 | 5’-UUUU-3’ |
| 2 | 5’-GUUU-3’ | ||
4. Materials and Methods
4.1. Data Retrieval of Plastomes within Saxifragales
To reach the comprehensive comparison among Saxifragales plastomes, our dataset comprised all the available sequences from 208 species in 11 families, covering all seven major lineages of this order. Notably, 14 sequences were generated by our previous work (Table S4). After retrieval from NCBI, the annotations of all the sequence data were carefully checked. Therein, GeSeq [93] and CPGAVAS2 [94] were employed to examine potential annotation errors. Further, we manually modified the resulting annotated genes by BLAST [95]. Finally, the overall map of plastomes was presented using Chloroplot [96].
4.2. Comparative Analyses of the Sequence Variations among the Plastomes
Subsequently, comprehensive analyses of plastomic variation were performed among the processed datasets. First, all the intron-containing plastomic genes were extracted and Bioedit was employed to select and trim the coding regions for further analyses. Then, MAFFT version 7.505 [97] was used to align the intron sequences among Saxifragales. Based on this, we made more elaborate modifications by Bioedit [98] according to the principles described by Borsch et al. [99]. For instance, the single-positional nucleotide adjacent to an entire indel was insufficient to be identified as an independent event. To avoid such misjudgments, the aligned gaps were all manually modified at the same column [22,100].
Additionally, the highly polymorphic regions (HPR) among Saxifragales plastomes were also explored at the intra- and interfamily levels. The aligned plastome sequences were, respectively, imported into the sliding window analysis of DnaSP 6 [101] using the parament settings described by Han et al. [34]. The nucleotide divergence (Pi) values were then estimated. The hotspot loci were further determined according to the criteria of Bi et al. [102].
4.3. Comparative Analyses of the Structural Diversifications among the Plastomes
To explore evolutionary implications of plastomic structures, the patterns of IR boundaries were analyzed by IRscope [103]. After verifying the gene annotations, we counted and compared the sizes of extension and contraction of each IR region. Moreover, all pttRNAs of samplings were extracted. Then, secondary structure predictions were performed with tRNAscan-SE v.2.0.3 [104].
4.4. Phyloplastomic Reconstruction among Saxifragales
For deeper insights into the taxonomic relationships within Saxifragales, phyloplastomic analyses were implemented for all taxa investigated here. Three species of Rosids, a large clade closely related to Saxifragales [41,105], were selected as outgroups. Two separate datasets were employed for analyses: (1) 79 plastomic PCGs and (2) complete chloroplast genomes (CPGs). The preparations of phylogenetic datasets were conducted by DAMBE for the retrieval of all protein-coding genes (PCGs) [106], MAFFT for alignment [97], and SequenceMatrix for concatenation [107].
After that, phylogenetic trees were built by two methods: maximum-likelihood (ML) and Bayesian inference (BI). Firstly, the ML trees were inferred with RAxML 8.2.12 [108] by conducting 50 runs and 1000 bootstrap replicates under the GTRCAT model. Moreover, we checked the bootstrap convergence by the “-I autoMRE” command in RAxML. Secondly, a Bayesian phylogenetic analysis was carried out by MrBayes 3.2.7a [109]. Under the optimal models calculated with ModelTest-NG [110], the phylogenetic inferences were generated using two independent runs, each with four Markov chains for 20 million generations (sampling every 1000 generations). Then, the convergence was confirmed by Tracer 1.7.1 [111].
5. Conclusions
In the current study, by relatively wide sampling, the comprehensive diversities among the 208 plastomes from 11 families of Saxifragales were thoroughly explored. Several loss events were observed from two genes and three introns, in particular, the intron loss of atpF in Pachyphytum compactum was first reported here. Then, we further investigated the gene content at IR boundaries, DNA polymorphism, indels in the introns of all 17 intron-containing genes, and the pttRNA secondary-structure diversities. Significantly, abundant phylogenetic implications were revealed from them, suggesting that they have strong potential roles in serving as specific markers for Saxifragales. Moreover, our phylogenetic interpretations, based on two datasets (CPGs and PCGs), generally well recovered the internal branching patterns in this order with high resolution. More importantly, the combined phylogenies with indels and pttRNA structural features could provide further insights into the evolutionary patterns among Saxifragales plastomes. Therein, Grossulariaceae, Cercidiphyllaceae, and Daphniphyllaceae were found to retain the most plesiomorphic features. Collectively, our results presented here will facilitate the understanding of the plastome evolution in Saxifragales, and accordingly, provide a case study for comparative plastomics among the early-divergent angiosperms.
Acknowledgments
We kindly acknowledge the anonymous reviewers for the fruitful and critical comments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11243544/s1, Figure S1: Phylogenetic tree obtained by CPGs among 208 Saxifragales plastomes. The BS and PP values of each node were labeled (* denoted 100% bootstrap or 1.00 PP, with the omission of those <50% bootstrap or < 0.5 PP); Table S1: The gene distribution at the IR junctions of Saxifragales plastomes involved in this study; Table S2: Hypervariable loci identified from the plastomes of Saxifragales involved in this study; Table S3: List of microstructural changes that were marked from the indel matrix; Table S4: List of the plastomes of Saxifragales involved in this study.
Author Contributions
Conceptualization, X.K.; methodology, S.H.; software, H.D., S.Z., and J.Y.; validation, D.B. and L.W.; formal analysis, S.Z.; investigation, R.Y., J.G., and Y.Y.; resources, L.W.; data curation, S.H.; writing—original draft preparation, S.H.; writing—review and editing, X.K.; supervision, X.K.; project administration, X.K.; funding acquisition, D.B., L.W., and X.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Opening Foundation of National Engineering Laboratory of Soil Pollution Control and Remediation Technologies, and Key Laboratory of Heavy Metal Pollution Prevention & Control, Ministry of Agriculture and Rural Affairs, grant number NEL&MARA-003, and the Basic Research Program (Natural Science Foundation) of Jiangsu Province, grant number BK20211078.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dong W., Xu C., Cheng T., Zhou S. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS ONE. 2013;8:e77965. doi: 10.1371/journal.pone.0077965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniell H., Lin C.-S., Yu M., Chang W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer J.D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe K.H., Li W.-H., Sharp P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C.-S., Lai Y.-T., Lin C.-P., Wang Y.-N., Chaw S.-M. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: Selection toward a lower-cost strategy. Mol. Phylogenet. Evol. 2009;52:115–124. doi: 10.1016/j.ympev.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y., Zou P., Jiang N., Fang Y., Liu G. Comparative analysis of the complete chloroplast genomes of nine Paphiopedilum species. Front. Genet. 2021;12:772415. doi: 10.3389/fgene.2021.772415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y.-Y., Yang J.-X., Bai M.-Z., Zhang G.-Q., Liu Z.-J. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021;21:248. doi: 10.1186/s12870-021-03053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henriquez C.L., Abdullah, Ahmed I., Carlsen M.M., Zuluaga A., Croat T.B., McKain M.R. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae) Genomics. 2020;112:2349–2360. doi: 10.1016/j.ygeno.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y., Liu F., Yang D.-G., Li W., Zhou X.-J., Pei X.-Y., Liu Y.-G., He K.-L., Zhang W.-S., Ren Z.-Y. Comparative chloroplast genomics of Gossypium species: Insights into repeat sequence variations and phylogeny. Front. Plant Sci. 2018;9:376. doi: 10.3389/fpls.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wicke S., Schneeweiss G.M., Depamphilis C.W., Müller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer J.D. Evolution of chloroplast and mitochemdrial DNA in plants and algae. In: MacIntyre R., editor. Molecular Evolutionary Genetics. Plenum Press; New York, NY, USA: 1985. pp. 131–240. [Google Scholar]
- 12.Wu S., Chen J., Li Y., Liu A., Li A., Yin M., Shrestha N., Liu J., Ren G. Extensive genomic rearrangements mediated by repetitive sequences in plastomes of Medicago and its relatives. BMC Plant Biol. 2021;21:421. doi: 10.1186/s12870-021-03202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravi V., Khurana J., Tyagi A., Khurana P. An update on chloroplast genomes. Plant Syst. Evol. 2008;271:101–122. doi: 10.1007/s00606-007-0608-0. [DOI] [Google Scholar]
- 14.Jansen R.K., Cai Z., Raubeson L.A., Daniell H., Depamphilis C.W., Leebens-Mack J., Müller K.F., Guisinger-Bellian M., Haberle R.C., Hansen A.K. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen R.K., Saski C., Lee S.-B., Hansen A.K., Daniell H. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 2011;28:835–847. doi: 10.1093/molbev/msq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S., Ruhlman T.A., Weng M.-L., Hajrah N.H., Sabir J.S., Jansen R.K. Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of Geranium. Genome Biol. Evol. 2017;9:1766–1780. doi: 10.1093/gbe/evx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S., An B., Park S. Recurrent gene duplication in the angiosperm tribe Delphinieae (Ranunculaceae) inferred from intracellular gene transfer events and heteroplasmic mutations in the plastid matK gene. Sci. Rep. 2020;10:2720. doi: 10.1038/s41598-020-59547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S., Park S. Large-scale phylogenomics reveals ancient introgression in Asian Hepatica and new insights into the origin of the insular endemic Hepatica maxima. Sci. Rep. 2020;10:16288. doi: 10.1038/s41598-020-73397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrestha B., Gilbert L.E., Ruhlman T.A., Jansen R.K. Rampant nuclear transfer and substitutions of plastid genes in Passiflora. Genome Biol. Evol. 2020;12:1313–1329. doi: 10.1093/gbe/evaa123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claude S.-J., Park S., Park S. Gene loss, genome rearrangement, and accelerated substitution rates in plastid genome of Hypericum ascyron (Hypericaceae) BMC Plant Biol. 2022;22:135. doi: 10.1186/s12870-022-03515-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelchner S.A. The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann. Mo. Bot. Gard. 2000;87:482–498. doi: 10.2307/2666142. [DOI] [Google Scholar]
- 22.Lohne C., Borsch T. Molecular evolution and phylogenetic utility of the petD group II intron: A case study in basal angiosperms. Mol. Biol. Evol. 2005;22:317–332. doi: 10.1093/molbev/msi019. [DOI] [PubMed] [Google Scholar]
- 23.Shahzadi I., Abdullah, Mehmood F., Ali Z., Ahmed I., Mirza B. Chloroplast genome sequences of Artemisia maritima and Artemisia absinthium: Comparative analyses, mutational hotspots in genus Artemisia and phylogeny in family Asteraceae. Genomics. 2020;112:1454–1463. doi: 10.1016/j.ygeno.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Xu Q., Xiong G., Li P., He F., Huang Y., Wang K., Li Z., Hua J. Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: Origin and evolution of allotetraploids. PLoS ONE. 2012;8:e37128. doi: 10.1371/journal.pone.0037128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menezes A., Resende-Moreira L.C., Buzatti R.S., Nazareno A.G., Carlsen M., Lobo F.P., Kalapothakis E., Lovato M.B. Chloroplast genomes of Byrsonima species (Malpighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018;8:2210. doi: 10.1038/s41598-018-20189-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehmood F., Shahzadi I., Waseem S., Mirza B., Ahmed I., Waheed M.T. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): Comparative analyses and identification of mutational hotspots. Genomics. 2020;112:581–591. doi: 10.1016/j.ygeno.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Park S., An B., Park S. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. Sci. Rep. 2018;8:13568. doi: 10.1038/s41598-018-31938-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weng M.L., Ruhlman T.A., Jansen R.K. Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 2017;214:842–851. doi: 10.1111/nph.14375. [DOI] [PubMed] [Google Scholar]
- 29.Guisinger M.M., Kuehl J.V., Boore J.L., Jansen R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y.-D., Jansen R. Characterization and phylogenetic distribution of a chloroplast DNA rearrangement in the Berberidaceae. Plant Syst. Evol. 1994;193:107–114. doi: 10.1007/BF00983544. [DOI] [Google Scholar]
- 31.Plunkett G.M., Downie S.R. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Syst. Bot. 2000;25:648–667. doi: 10.2307/2666726. [DOI] [Google Scholar]
- 32.Wang R.-J., Cheng C.-L., Chang C.-C., Wu C.-L., Su T.-M., Chaw S.-M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang H., Zhang L., Xie H., Liu J., Xi Z., Xu X. The conservation of chloroplast genome structure and improved resolution of infrafamilial relationships of crassulaceae. Front. Plant Sci. 2021;12:631884. doi: 10.3389/fpls.2021.631884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han S., Bi D., Yi R., Ding H., Wu L., Kan X. Plastome evolution of Aeonium and Monanthes (Crassulaceae): Insights into the variation of plastomic tRNAs, and the patterns of codon usage and aversion. Planta. 2022;256:35. doi: 10.1007/s00425-022-03950-y. [DOI] [PubMed] [Google Scholar]
- 35.Holley R.W., Apgar J., Everett G.A., Madison J.T., Marquisee M., Merrill S.H., Penswick J.R., Zamir A. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 36.Han S., Wang R., Hong X., Wu C., Zhang S., Kan X. Plastomes of Bletilla (Orchidaceae) and Phylogenetic Implications. Int. J. Mol. Sci. 2022;23:10151. doi: 10.3390/ijms231710151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magallon S., Crane P.R., Herendeen P.S. Phylogenetic pattern, diversity, and diversification of eudicots. Ann. Mo. Bot. Gard. 1999;86:297–372. doi: 10.2307/2666180. [DOI] [Google Scholar]
- 38.Moody M.L., Les D.H. Phylogenetic systematics and character evolution in the angiosperm family Haloragaceae. Am. J. Bot. 2007;94:2005–2025. doi: 10.3732/ajb.94.12.2005. [DOI] [PubMed] [Google Scholar]
- 39.Moore M.J., Soltis P.S., Bell C.D., Burleigh J.G., Soltis D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soltis D., Smith S., Cellinese N., Refulio-Rodriquez N., Olmstead R., Crawley S., Black C., Diouf D., Hilu K., Latvis M. Inferring angiosperm phylogeny: A 17-gene analysis. Am. J. Bot. 2011;98:704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- 41.Chase M.W., Christenhusz M.J., Fay M.F., Byng J., Judd W., Soltis D., Mabberley D., Sennikov A., Soltis P. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181:1–20. [Google Scholar]
- 42.Savolainen V., Chase M.W., Hoot S.B., Morton C.M., Soltis D.E., Bayer C., Fay M.F., De Bruijn A.Y., Sullivan S., Qiu Y.-L. Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences. Syst. Biol. 2000;49:306–362. doi: 10.1093/sysbio/49.2.306. [DOI] [PubMed] [Google Scholar]
- 43.Soltis P.S., Soltis D.E. The origin and diversification of angiosperms. Am. J. Bot. 2004;91:1614–1626. doi: 10.3732/ajb.91.10.1614. [DOI] [PubMed] [Google Scholar]
- 44.Soltis D.E., Soltis P.S. Phylogenetic relationships in Saxifragaceae sensu lato: A comparison of topologies based on 18S rDNA and rbcL sequences. Am. J. Bot. 1997;84:504–522. doi: 10.2307/2446027. [DOI] [PubMed] [Google Scholar]
- 45.Fishbein M., Hibsch-Jetter C., Soltis D.E., Hufford L. Phylogeny of Saxifragales (angiosperms, eudicots): Analysis of a rapid, ancient radiation. Syst. Biol. 2001;50:817–847. doi: 10.1080/106351501753462821. [DOI] [PubMed] [Google Scholar]
- 46.Jian S., Soltis P.S., Gitzendanner M.A., Moore M.J., Li R., Hendry T.A., Qiu Y.L., Dhingra A., Bell C.D., Soltis D.E. Resolving an ancient, rapid radiation in Saxifragales. Syst. Biol. 2008;57:38–57. doi: 10.1080/10635150801888871. [DOI] [PubMed] [Google Scholar]
- 47.Soltis D.E., Mort M.E., Latvis M., Mavrodiev E.V., O’Meara B.C., Soltis P.S., Burleigh J.G., Rubio de Casas R. Phylogenetic relationships and character evolution analysis of Saxifragales using a supermatrix approach. Am. J. Bot. 2013;100:916–929. doi: 10.3732/ajb.1300044. [DOI] [PubMed] [Google Scholar]
- 48.Ding H., Zhu R., Dong J., Bi D., Jiang L., Zeng J., Huang Q., Liu H., Xu W., Wu L., et al. Next-generation genome sequencing of Sedum plumbizincicola sheds light on the structural evolution of plastid rRNA operon and phylogenetic implications within saxifragales. Plants. 2019;8:386. doi: 10.3390/plants8100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Downie S.R., Olmstead R.G., Zurawski G., Soltis D.E., Soltis P.S., Watson J.C., Palmer J.D. Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: Molecular and phylogenetic implications. Evolution. 1991;45:1245–1259. doi: 10.2307/2409731. [DOI] [PubMed] [Google Scholar]
- 50.Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeon J.H., Kim S.C. Comparative analysis of the complete chloroplast genome sequences of three closely related east-asian wild roses (Rosa sect. Synstylae; Rosaceae) Genes. 2019;10:23. doi: 10.3390/genes10010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filip E., Skuza L. Horizontal gene transfer involving chloroplasts. Int. J. Mol. Sci. 2021;22:4484. doi: 10.3390/ijms22094484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Millen R.S., Olmstead R.G., Adams K.L., Palmer J.D., Lao N.T., Heggie L., Kavanagh T.A., Hibberd J.M., Gray J.C., Morden C.W., et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cusack B.P., Wolfe K.H. When gene marriages don’t work out: Divorce by subfunctionalization. Trends Genet. 2007;23:270–272. doi: 10.1016/j.tig.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Ueda M., Fujimoto M., Arimura S.-i., Murata J., Tsutsumi N., Kadowaki K.-I. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene. 2007;402:51–56. doi: 10.1016/j.gene.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 56.Daniell H., Wurdack K.J., Kanagaraj A., Lee S.B., Saski C., Jansen R.K. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron. Theor. Appl. Genet. 2008;116:723–737. doi: 10.1007/s00122-007-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu C., Tembrock L.R., Zheng S., Wu Z. The complete chloroplast genome of Catha edulis: A comparative analysis of genome features with related species. Int. J. Mol. Sci. 2018;19:525. doi: 10.3390/ijms19020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu C., Ma L., Wu Z., Chen K., Wang Y. Comparative analyses of chloroplast genomes from 22 Lythraceae species: Inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 2019;19:281. doi: 10.1186/s12870-019-1870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jansen R.K., Ruhlman T.A. Plastid genomes of seed plants. In: Bock R., Knoop V., editors. Genomics of Chloroplasts and Mitochondria. Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes) Volume 35. Springer; Dordrecht, The Netherlands: 2012. pp. 103–126. [Google Scholar]
- 60.Dujon B. Group I introns as mobile genetic elements: Facts and mechanistic speculations—A review. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- 61.Fink G.R. Pseudogenes in yeast? Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-X. [DOI] [PubMed] [Google Scholar]
- 62.Gu C., Tembrock L.R., Johnson N.G., Simmons M.P., Wu Z. The complete plastid genome of Lagerstroemia fauriei and loss of rpl2 intron from Lagerstroemia (Lythraceae) PLoS ONE. 2016;11:e0150752. doi: 10.1371/journal.pone.0150752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henriquez C.L., Abdullah, Ahmed I., Carlsen M.M., Zuluaga A., Croat T.B., McKain M.R. Molecular evolution of chloroplast genomes in Monsteroideae (Araceae) Planta. 2020;251:72. doi: 10.1007/s00425-020-03365-7. [DOI] [PubMed] [Google Scholar]
- 64.Schroeder H., Hoeltken A., Fladung M. Differentiation of Populus species using chloroplast single nucleotide polymorphism (SNP) markers–essential for comprehensible and reliable poplar breeding. Plant Biol. 2012;14:374–381. doi: 10.1111/j.1438-8677.2011.00502.x. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y., Zhang L., Liu Z., Luo K., Chen S., Chen K. Species identification of Rhododendron (Ericaceae) using the chloroplast deoxyribonucleic acid psbA-trnH genetic marker. Pharmacogn. Mag. 2012;8:29–36. doi: 10.4103/0973-1296.93311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang N., Erickson D.L., Ramachandran P., Ottesen A.R., Timme R.E., Funk V.A., Luo Y., Handy S.M. An analysis of Echinacea chloroplast genomes: Implications for future botanical identification. Sci. Rep. 2017;7:216. doi: 10.1038/s41598-017-00321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu H., Su Z., Yu S., Liu J., Yin X., Zhang G., Liu W., Li B. Genome comparison reveals mutation hotspots in the chloroplast genome and phylogenetic relationships of Ormosia species. Biomed. Res. Int. 2019;2019:7265030. doi: 10.1155/2019/7265030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong W., Liu J., Yu J., Wang L., Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu L., Wang Y., He P., Li P., Lee J., Soltis D.E., Fu C. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genom. 2018;19:235. doi: 10.1186/s12864-018-4633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu F., He L., Gao S., Su Y., Li F., Xu L. Comparative analysis of two sugarcane ancestors Saccharum officinarum and S. spontaneum based on complete chloroplast genome sequences and photosynthetic ability in cold stress. Int. J. Mol. Sci. 2019;20:3828. doi: 10.3390/ijms20153828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng J.Y., Zhang X.S., Zhang D.G., Wang Y., Deng T., Huang X.H., Kuang T.H., Zhou Q. Newly reported chloroplast genome of Sinosenecio albonervius Y. Liu & Q. E. Yang and comparative analyses with other Sinosenecio species. BMC Genom. 2022;23:639. doi: 10.1186/s12864-022-08872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Z., Bielawski J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang N., Chen S., Xie L., Wang L., Feng Y., Lv T., Fang Y., Ding H. The complete chloroplast genomes of three Hamamelidaceae species: Comparative and phylogenetic analyses. Ecol. Evol. 2022;12:e8637. doi: 10.1002/ece3.8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L., Liang J., Sa W., Wang L. Sequencing and comparative analysis of the chloroplast genome of Ribes odoratum provide insights for marker development and phylogenetics in Ribes. Physiol. Mol. Biol. Plants. 2021;27:81–92. doi: 10.1007/s12298-021-00932-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Graham S.W., Reeves P.A., Burns A.C., Olmstead R.G. Microstructural changes in noncoding chloroplast DNA: Interpretation, evolution, and utility of indels and inversions in basal angiosperm phylogenetic inference. Int. J. Plant Sci. 2000;161:S83–S96. doi: 10.1086/317583. [DOI] [Google Scholar]
- 76.Shaw J., Lickey E.B., Schilling E.E., Small R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- 77.Vogel J., Borner T. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 2002;21:3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim K.-J., Lee H.-L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 79.Wang N., Dong W.L., Zhang X.J., Zhou T., Huang X.J., Li B.G., Liu J.N., Ma X.F., Li Z.H. Evolutionary characteristics and phylogeny of cotton chloroplast tRNAs. Planta. 2021;254:116. doi: 10.1007/s00425-021-03775-1. [DOI] [PubMed] [Google Scholar]
- 80.Zhang T.T., Yang Y., Song X.Y., Gao X.Y., Zhang X.L., Zhao J.J., Zhou K.H., Zhao C.B., Li W., Yang D.G., et al. Novel structural variation and evolutionary characteristics of chloroplast tRNA in Gossypium plants. Genes. 2021;12:822. doi: 10.3390/genes12060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brennan T., Sundaralingam M. Structure, of transfer RNA molecules containing the long variable loop. Nucleic Acids Res. 1976;3:3235–3252. doi: 10.1093/nar/3.11.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dock-Bregeon A.C., Westhof E., Giege R., Moras D. Solution structure of a tRNA with a large variable region: Yeast tRNASer. J. Mol. Biol. 1989;206:707–722. doi: 10.1016/0022-2836(89)90578-0. [DOI] [PubMed] [Google Scholar]
- 83.Zhong Q.Y., Fu X.G., Zhang T.T., Zhou T., Yue M., Liu J.N., Li Z.H. Phylogeny and evolution of chloroplast tRNAs in Adoxaceae. Ecol. Evol. 2021;11:1294–1309. doi: 10.1002/ece3.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Omilian A.R., Taylor D.J. Rate acceleration and long-branch attraction in a conserved gene of cryptic daphniid (Crustacea) species. Mol. Biol. Evol. 2001;18:2201–2212. doi: 10.1093/oxfordjournals.molbev.a003767. [DOI] [PubMed] [Google Scholar]
- 85.Page R.D. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinform. 2003;1:6.2.1–6.2.15. doi: 10.1002/0471250953.bi0602s01. [DOI] [PubMed] [Google Scholar]
- 86.Mardulyn P. Trees and/or networks to display intraspecific DNA sequence variation? Mol. Ecol. 2012;21:3385–3390. doi: 10.1111/j.1365-294X.2012.05622.x. [DOI] [PubMed] [Google Scholar]
- 87.Elliott M.J., Knerr N.J., Schmidt-Lebuhn A.N. Choice between phylogram and chronogram can have a dramatic impact on the location of phylogenetic diversity hotspots. J. Biogeogr. 2018;45:2190–2201. doi: 10.1111/jbi.13399. [DOI] [Google Scholar]
- 88.Sumner-Smith M., Hottinger H., Willis I., Koch T.L., Arentzen R., Söll D. The sup8 tRNALeu gene of Schizosaccharomyces pombe has an unusual intervening sequence and reduced pairing in the anticodon stem. Mol. Gen. Genet. 1984;197:447–452. doi: 10.1007/BF00329941. [DOI] [PubMed] [Google Scholar]
- 89.Curran J.F., Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987;238:1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- 90.Atkins J., Weiss R., Thompson S., Gesteland R.F. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: Programmed reading frame shifts and hops. Annu. Rev. Genet. 1991;25:201–228. doi: 10.1146/annurev.ge.25.120191.001221. [DOI] [PubMed] [Google Scholar]
- 91.Fishbein M., Soltis D.E. Further resolution of the rapid radiation of Saxifragales (angiosperms, eudicots) supported by mixed-model Bayesian analysis. Syst. Bot. 2004;29:883–891. doi: 10.1600/0363644042450982. [DOI] [Google Scholar]
- 92.Wagele J.W., Mayer C. Visualizing differences in phylogenetic information content of alignments and distinction of three classes of long-branch effects. BMC Evol. Biol. 2007;7:147. doi: 10.1186/1471-2148-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi L., Chen H., Jiang M., Wang L., Wu X., Huang L., Liu C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019;47:W65–W73. doi: 10.1093/nar/gkz345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zheng S., Poczai P., Hyvönen J., Tang J., Amiryousefi A. Chloroplot: An online program for the versatile plotting of organelle genomes. Front. Genet. 2020;11:576124. doi: 10.3389/fgene.2020.576124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hall T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 99.Borsch T., Hilu K., Quandt D., Wilde V., Neinhuis C., Barthlott W. Noncoding plastid trnT-trnF sequences reveal a well resolved phylogeny of basal angiosperms. J. Evol. Biol. 2003;16:558–576. doi: 10.1046/j.1420-9101.2003.00577.x. [DOI] [PubMed] [Google Scholar]
- 100.Simmons M.P., Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 2000;49:369–381. doi: 10.1093/sysbio/49.2.369. [DOI] [PubMed] [Google Scholar]
- 101.Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 102.Bi Y., Zhang M.F., Xue J., Dong R., Du Y.P., Zhang X.H. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci. Rep. 2018;8:1184. doi: 10.1038/s41598-018-19591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Amiryousefi A., Hyvönen J., Poczai P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 104.Chan P.P., Lin B.Y., Mak A.J., Lowe T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zeng L., Zhang N., Zhang Q., Endress P.K., Huang J., Ma H. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol. 2017;214:1338–1354. doi: 10.1111/nph.14503. [DOI] [PubMed] [Google Scholar]
- 106.Xia X., Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 107.Vaidya G., Lohman D.J., Meier R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 108.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Darriba D., Posada D., Kozlov A.M., Stamatakis A., Morel B., Flouri T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020;37:291–294. doi: 10.1093/molbev/msz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.