Abstract
Myeloid cell leukemia‐1 (MCL1), an antiapoptotic member of the BCL2 family characterized by a short half‐life, functions as a rapid sensor that regulates cell death and other relevant processes that include cell cycle progression and mitochondrial homeostasis. In cancer, MCL1 overexpression contributes to cell survival and resistance to diverse chemotherapeutic agents; for this reason, several MCL1 inhibitors are currently under preclinical and clinical development for cancer treatment. However, the nonapoptotic functions of MCL1 may influence their therapeutic potential. Overall, the complexity of MCL1 regulation and function represent challenges to the clinical application of MCL1 inhibitors. We now summarize the current knowledge regarding MCL1 structure, regulation, and function that could impact the clinical success of MCL1 inhibitors.
Keywords: apoptosis, BCL2 family, cell cycle, MCL1, MCL1 inhibitor
Myeloid cell leukemia‐1 (MCL1) is an antiapoptotic member of the BCL2 family whose dysregulation contributes to cell survival and resistance to diverse chemotherapeutic agents. This review summarizes the current knowledge regarding the complexity of MCL1 regulation, structure, and function that could influence the clinical success of MCL1 inhibitors.
Abbreviations
- A1
BCL2-related protein A1
- BAD
BCL2-associated agonist of cell death
- BAK
BCL2 homologous antagonist/killer
- BAX
BCL2-associated X protein
- BCL2
B-cell lymphoma 2
- BCLB
BCL2-like protein 10
- BCLW
BCL2-like protein 2
- BCL-xL
BCL-extra-large
- BID
BH3-interacting domain death agonist
- BIM
BCL2-like protein 11
- BMF
BCL2-modifying factor
- BOK
BCL-2-related ovarian killer
- MCL1ES
myeloid cell leukemia-1 extra short form
- MCL1L
myeloid cell leukemia-1 long form
- MCL1S
myeloid cell leukemia-1 short form
- NOXA
phorbol-12-myristate-13-acetate-induced protein 1
- PUMA
BCL2-modifying factor
- TIM/TOM
translocase of inner/outer membrane
Introduction
Myeloid cell leukemia‐1 (MCL1), a member of the BCL2 (B‐cell lymphoma 2) family of proteins, was initially identified as an early gene induced in human myeloblastic leukemia (ML‐1) cells upon phorbol ester (TPA) exposure. In this case, MCL1 upregulation triggered the differentiation of ML‐1 cells into monocytes or macrophages; however, MCL1 downregulation induced apoptosis, thereby suggesting a role for MCL1 in cell survival. Protein homology studies provided evidence for a primary function of the BCL2 family of proteins in regulating cell death and, in particular, in mitochondrial outer membrane permeabilization (MOMP) [1, 2, 3].
BCL2 proteins modulate apoptosis, a form of programmed cell death essential for the control of homeostasis, and are characterized by the presence of BCL2 homology (BH) domains and a carboxy‐terminal transmembrane domain (TMD) present in most members. BCL2 proteins are classified into three groups according to their function and the number of BH domains—the antiapoptotic proteins (BCL2, BCL‐xL, BCLW, MCL1, A1, and BCLB), the proapoptotic BH3 ‐only ‘sensor’ proteins (including BAD, BID, BIM, BMF, PUMA, and NOXA), and the proapoptotic ‘executors’ (BOK, BAX, and BAK). A complex network of interactions between proapoptotic and antiapoptotic members of this protein family governs MOMP, which first induces the release of apoptogenic factors such as cytochrome c into the cytosol, where it binds to apoptosis protease‐activating factor 1 (APAF1) and procaspase‐9 in a macromolecular complex known as the apoptosome. Subsequently, activated procaspase‐9 activates the procaspase‐3 effector protease that finally dismantles the cell [4, 5].
MCL1 has three confirmed BH domains (BH1–BH3), a putative BH4 domain, a C‐terminal TMD, and a large N‐terminal region [1–170 amino acids (aa)] (Fig. 1). The large N‐terminal region differs from the other BCL2 family members and contains large numbers of proline (P), glutamic acid (E), serine (S), and threonine (T) residues (or ‘PEST’ sequences). PEST enrichment and arginine pairs represent typical features of labile proteins; consequently, MCL1 displays a short half‐life and is highly regulated, thereby providing a means by which MCL1 can modulate apoptosis in response to rapidly changing cellular contexts. Intense recent efforts have led to the description of new small‐molecule drugs that block MCL1 function in diseases such as cancer, in which MCL1 plays an inhibitory role in apoptosis. Importantly, MCL1 also plays important nonapoptotic roles in mitochondrial homeostasis, embryonic development, autophagy, and cell cycle.
Fig. 1.
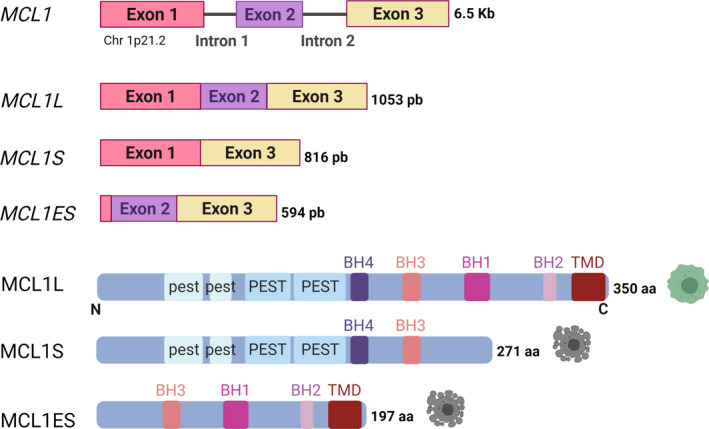
Schematic representation of MCL1 gene, mRNAs, and protein variants. MCL1S and MCL1ES display proapoptotic behavior while MCL1L is antiapoptotic.
Given the availability of tools that modulate MCL1 function, the coming years may provide an avalanche of preclinical studies and clinical trials that will require a deeper molecular understanding of this critical protein.
MCL1 structural features that impact function and treatment strategies
The human MCL1 gene located on chromosome 1p21.2 contains three exons and encodes a 350 aa prosurvival protein called MCL1 or MCL1L [6]. Alternative splicing generates two additional shorter forms, MCL1S and MCL1ES (Fig. 1).
The MCL1S variant (271 aa) lacks exon two and produces a protein without BH1, BH2, and the TMD—as a result, MCL1S behaves like a BH3‐only sensor protein, induces apoptosis, selectively forms heterodimers with MCL1L, and displays a predominantly cytosolic localization [but exists to a lesser extent in the endoplasmic reticulum (ER)] [7, 8]. Thus, a balance between MCL1L/MCL1S expression may regulate the machinery controlling mitochondrial fusion and fission [9].
The shortest MCL1ES variant (1–179 aa) possesses a truncated exon one, which results in the loss of the N‐terminal section of the PEST sequences. MCL1ES binds to MCL1L, localizes to the outer mitochondrial membrane (OMM), and induces BAK‐ and BAX‐independent mitochondrial cell death [10, 11]. Evidence suggests that the formation of MCL1L‐MCL1ES oligomers triggers MOMP and apoptotic signaling.
The first reported structure of MCL1 in solution, corresponding to residues 152–308 of mouse MCL1, was determined in 2005 [12]. Like all MCL1 structures published to date, it lacked the large PEST domain located in the N‐terminal region and the C‐terminal TMD; however, the publication of the human protein structure soon after established a significant similarity to the mouse MCL1 protein [13]. Important differences included a negative charge distribution in the binding groove of the helix α3 region of human MCL1, a region which is positively charged in the mouse protein [14]. Since the publication of these studies, many MCL1 structures complexed with BH3‐only proteins or inhibitors have been described.
Human and mouse MCL1 share a helical core structure consisting of eight helices [in which a set of amphipathic helices surrounds a hydrophobic central helix (α5)] with other BCL2 antiapoptotic proteins. The folded structure of MCL1 generates a hydrophobic region known as the ‘BH3‐binding groove’, which is formed by residues from helices two, three, and four; meanwhile, helices five and eight comprise the groove base (Fig. 2A). The BH3‐binding groove establishes interactions with BH3 domains in other BCL2 protein family members (Fig. 2B). Of note, this interaction has been studied in‐depth due to its relevance as a pharmacological intervention point for the modulation of apoptosis.
Fig. 2.
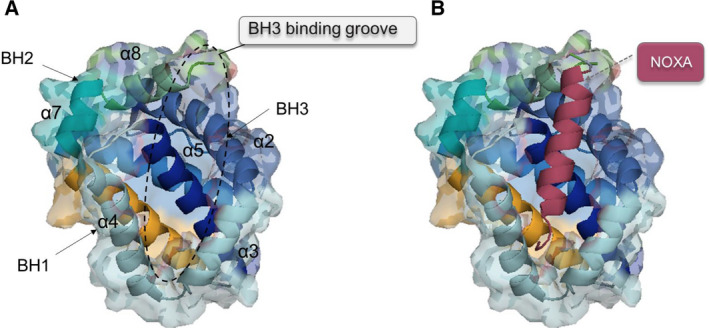
Crystal structure of MCL1. (A) Helical folding of MCL1. Helices (α1–α8) form the helical core where the amphipathic α5 is surrounded by the rest of helices, thereby creating the hydrophobic BH3‐binding groove. (B) Structure of MCL1 in complex with the BH3‐only protein NOXA. Figure was generated using Protein Data Bank (PDB) code 2NLA.
A conserved pattern within BH3 domains, comprising four positions (h1–h4) occupied by hydrophobic residues located on the same face of the helix, controls interactions with the BH3‐binding groove (Fig. 3A). An aspartic acid residue from the BH3 domain forms a salt bridge with arginine 263 of the MCL1‐binding groove [15, 16, 17, 18].
Fig. 3.
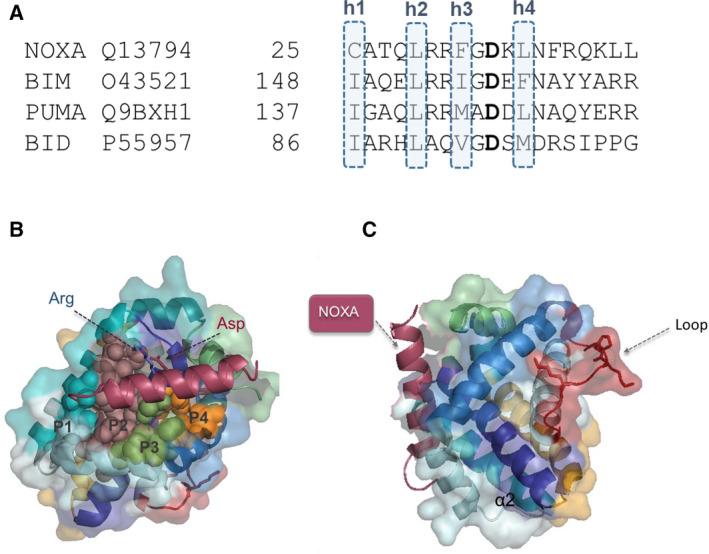
Interaction sites relevant for MCL1 function. (A) Conserved positions of BH3 domains (h1–h4) required to establish interactions with the binding groove. (B) Crystal structure of MCL1 with the P1–P4 pockets of the binding groove highlighted. (C) Crystal structure of MCL1 with the loop relevant for senescence regulation highlighted in red. The figure was generated using PDB code 6YBJ.
The MCL1‐binding groove can also be subdivided into four pockets (P1‐P4) that interact respectively with residues h1‐h4 from BH3‐only proteins (Fig. 3B). Exhaustive structural comparisons of different protein complexes have identified P2 and P3 pockets as the location for the ‘hot spot’ residues for protein–protein interactions in MCL1, which is unlike antiapoptotic proteins such as BCL2 (P2 and P4/P1) or BCL‐xL (P2 and P4) [19, 20]. The structure of MCL1 P2 is shorter and wider compared with the BCL2 P2 [20], while the MCL1 groove displays a more open conformation when compared to other antiapoptotic BCL2 proteins. Moreover, the surface properties of BCL2 family proteins differ significantly despite high levels of structural homology. MCL1 has abundant lysine and histidine residues, which generate an electropositive surface that influences drug interactions with the binding groove. Together, these differences explain the interaction specificities of different BH3‐only proteins and contribute to the design of novel specific MCL1 inhibitors and an appreciation of their mechanism of action.
Song et al. [21] recently identified a conserved domain formed by residues Q221, R222, and N223 (QRN motif) in the BH3 domain of MCL1, that undergoes a conformational switch (to a helix) following NOXA binding that facilitates protein ubiquitination by MCL1 ubiquitin ligase E3 (MULE, also called LASAU1, ARF‐BP1, or HUWE1). Interestingly, binding of the BIM BH3 domain stabilizes the nonhelical structure of this motif to avoid MCL1 ubiquitination and degradation. Thus, inhibitors designed to target the QRN motif could interfere with BH3 binding and facilitate protein degradation, thereby increasing apoptosis.
As noted previously, the MCL1 protein plays additional apoptosis‐independent roles, including the inhibition of senescence. Mutational and structural studies have demonstrated that this senescence‐inhibiting function does not depend on the BH3 binding domain [22]. Instead, a loop formed by G203, P198, K197, and K1974 (Fig. 3C) controls the MCL1‐mediated inhibition of senescence. Interestingly, peptides derived from this so‐called senescence‐regulating domain counteracted MCL1 function in vitro and in vivo in a colon cancer model of doxorubicin‐induced senescence [23].
The complex regulation of MCL1 expression
MCL1 expression is controlled at the transcriptional and post‐transcriptional levels and the translational and post‐translational levels. Overall, MCL1 displays low tissue and cell‐specific expression and broad intracellular localization. While localizing primarily to the mitochondria membranes [24], MCL1 can be found in light membrane fractions, such as the ER [9] and the nucleus [25, 26].
The dysregulation of MCL1 transcription in cancer cells can induce resistance to apoptosis. Cytokines (e.g., IL‐3, 5, and 6), growth factors [e.g., granulocyte‐macrophage colony‐stimulating factor (GM‐CSF), epidermal growth factor (EGF), and vascular endothelial growth factor (VEGF)], interferon proteins, and stress stimuli (e.g., ER stress or hypoxia) can all stimulate the transcriptional upregulation of MCL1 through the direct activity of crucial transcription factors (Fig. 4) [6]. Said factors include signal transducer and activator of transcription (STAT) proteins, cAMP response element‐binding protein (CREB), nuclear factor kappa light chain enhancer of activated B cells (NF‐κβ), hypoxia‐inducible factor 1‐alpha (HIF1α), transcription factor binding purine‐rich sequence (PU.1), Sp/KLF family of transcription factor 1 (SP1), and estrogen receptor alpha (ERα) [27]. MCL1 transcriptional repression following growth factor withdrawal, ultraviolet light exposure, or flavopiridol (a flavonoid alkaloid cyclin‐dependent kinase 9 [CDK9] inhibitor) treatment occurs mainly through the inactivation of the previously mentioned transcription factors or the direct binding of E2F transcription factor 1 (E2F1) to the MCL1 promoter [28, 29].
Fig. 4.
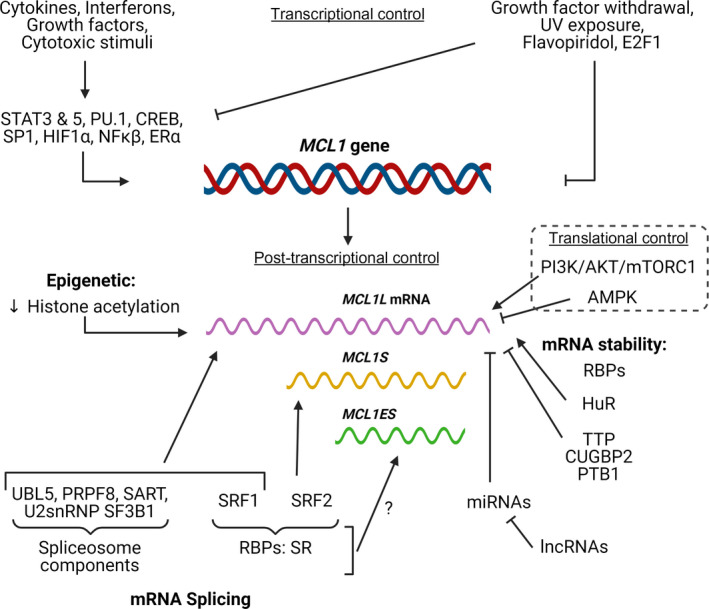
Summary of the main regulators of MCL1 expression at the transcriptional, post‐transcriptional, and translational level.
Post‐transcriptional control of MCL1 occurs at two levels—mRNA splicing and mRNA stability. Several spliceosome components and regulatory RNA‐binding proteins (RBPs) modulate MCL1 splicing. As an example, the knockdown of the spliceosome components ubiquitin‐like protein 5 (UBL5), pre‐mRNA‐processing‐splicing factor 8 (PRPF8), squamous cell carcinoma antigen recognized by T cells (SART), and the U2 small nuclear ribonucleoprotein (snRNP) splicing factor 3b subunit 1 (SF3B1) induce the generation of the MCL1S variant, which sensitizes MCL1‐dependent neuroblastomas to treatment with ABT‐737, a small‐molecule drug that inhibits BCL2 and BCL‐xL [30]. Members of the serine/arginine‐rich (SR) protein family of RBPs (SRSF1 and SRSF2) also participate in MCL1 splicing (Fig. 4)—while SRSF1 knockdown increases MCL1S expression, SRSF2 knockdown in renal cancer cells decreases MCL1S expression and inhibits apoptosis [31, 32]. While we understand little regarding the epigenetic regulation of MCL1 expression, Khan et al. [33] have reported that the acetylation of histones H3 and H4 within exon two favors exon skipping and MCL1S expression. Of note, the splicing factors regulating the formation of MCL1ES remain unidentified.
MCL1 mRNA has a short half‐life (two to three hours average), and RNA sequence elements and the binding of RBPs determine stability (Fig. 4). Several cis‐acting elements have been identified in the 3′ untranslated region (UTR) of antiapoptotic BCL2 family members (including CU‐rich, AU‐rich, and GU‐rich elements) that regulate mRNA stability in an RBP‐dependent manner. Human antigen R (HuR) binds to AU‐rich elements in the MCL1 3′ UTR to stabilize mRNA and increase expression in glioma [34]; however, RNA‐destabilizing protein Tristetraprolin (TTP) binds to this same motif to destabilize MCL1 mRNA and decrease expression during bacterial infection [35]. CUG triplet repeat, RNA‐binding protein 2 (CUGBP2) binds to the GU‐rich elements in the MCL1 3′ UTR to stabilize mRNA and inhibit its translation in colon cancer cells [36]. Finally, polypyrimidine tract‐binding protein 1 (PTBP1) binds to CU‐rich elements in the MCL1 3′ UTR to decrease expression in multiple cancer cell lines [37].
miRNAs, small noncoding RNAs (sncRNAs) of around 22 nucleotides, target the 3′ UTR region of mRNAs via base pairing to induce mRNA degradation or translational repression. There currently exist thirteen validated miRNAs that target MCL1 (Table 1). While most miRNAs only target MCL1, miR‐125b, miR‐133a/b, and miR‐153 also target other BCL2 family members [38]. Long noncoding RNAs (lncRNAs), which are generally longer than 200 nucleotides, can regulate gene expression through diverse mechanisms, including the prevention of miRNA binding to their target mRNAs; therefore, lncRNAs can modulate MCL1 mRNA stability. Twelve lncRNAs, including metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1), antisense noncoding RNA in the INK4 locus (ANRIL), H19 imprinted maternally expressed transcript (H19) (Table 1), are currently understood to regulate MCL1 expression [27].
Table 1.
Summary of the main identified ncRNAs regulating MCL1.
| miRNAs | lncRNAs | ||
|---|---|---|---|
| Name | References | Name | References |
| miR‐29 | [173] | MALAT1 | [174, 175] |
| miR‐30 | [176] | ANRIL | [177] |
| miR‐101 | [178] | circHIPK3 | [179] |
| miR‐125b | [180] | H19 | [181] |
| miR‐133a | [182] | HULC | [183] |
| miR‐133b | [184] | LINC00152 | [185, 186] |
| miR‐153 | [187] | MYOSLID | [188] |
| miR‐181 | [189] | PMS2L2 | [190] |
| miR‐193a | [191] | SNHG12 | [192] |
| miR‐302b | [193] | ||
| miR‐320 | [194] | ||
| miR‐512 | [195] | ||
Endoplasmic reticulum stress, ultraviolet light, elevated osmotic pressure, and arsenite treatment also decrease MCL1 levels through the eukaryotic initiation factor 2 (eIF2)‐mediated suppression of mRNA translation [39]. Metabolic sensors such as the mammalian target of rapamycin complex 1 (mTORC1) and AMP‐activated protein kinase (AMPK) also modulate the synthesis of MCL1. MCL1 mRNA is considered a ‘weak mRNA’ as its 5′ UTR possesses a robust secondary structure. Such mRNAs are translated through a regulated cap‐dependent system controlled by the eIF4F protein complex, which, in turn, is regulated by mTORC1 and AMPK. While mTORC1 activity induces MCL1 synthesis [40], AMPK activation and mTORC1 inhibition following a block in glycolysis prompt a decrease in MCL1 synthesis [41].
Post‐translational modifications such as cleavage, phosphorylation, and ubiquitination regulate the availability of MCL1, which is a short‐lived protein (Fig. 5). MCL1 possesses two weak and two strong PEST motifs in the N‐terminal region, which function as signals for degradation and induce rapid turnover. Thus, MCL1 becomes degraded by the proteasomal machinery by both ubiquitin‐independent and ubiquitin‐dependent pathways [42]. The unstructured N‐terminal region of MCL1 promotes proteasome recognition and degradation in a ubiquitin‐independent manner [43]; therefore, proteins interacting with this region may interfere with proteasome binding and contribute to MCL1 stabilization. While the proapoptotic BH3‐only family members [13] and translationally controlled tumor protein (TCTP) [44] have been also identified as MCL1 modulators that function by regulating degradation, however, the mechanisms involved remain incompletely understood.
Fig. 5.
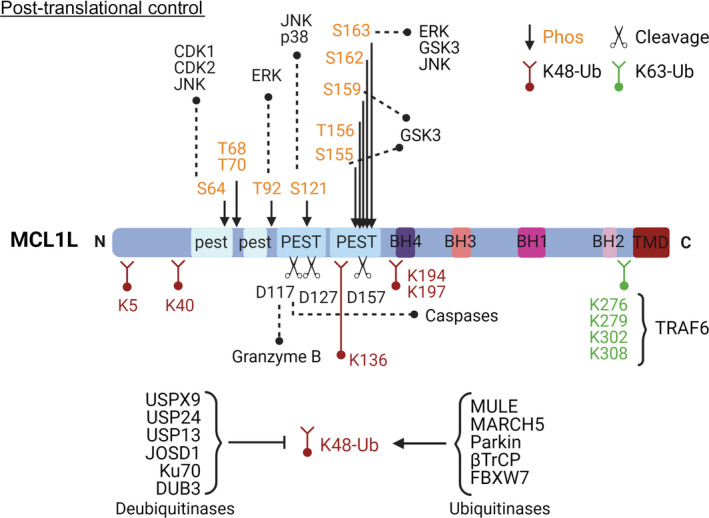
Schematic diagram of MCL1 highlighting the main residues involved in protease cleavage (black), phosphorylation (orange), and ubiquitination regulation (red and green). The main proteins responsible for these post‐translational modifications are also named in the figure.
Ubiquitination plays a vital role in the turnover of MCL1—fourteen Lys (K) residues have been identified as putative ubiquitination sites, and several ubiquitin ligases and deubiquitinases can modulate MCL1 stability. The BH3 domain of MULE, a K48 ubiquitin ligase that polyubiquitinates and targets MCL1 for degradation [45], interacts with the MCL1 BH3 groove and competes with the binding of other BH3‐only proteins. Knockdown of membrane‐associated RING‐CH protein 5 (MARCH5) reduces degradation of MCL1; however, whether this occurs through a direct ubiquitination‐associated mechanism remains unclear. The E3 ubiquitin‐protein ligase parkin (Parkin) also directly ubiquitinates MCL1, as do the E3 ligases beta transducing‐containing protein (β‐TrCP) and F‐Box and WD repeat domain containing 7 (FBXW7) polyubiquitinate MCL1, although they require the previous phosphorylation of MCL1 by glycogen synthase kinase 3 (GSK3) to mediate recognition [46]. The K63 ubiquitin ligase tumor necrosis factor receptor‐associated factor 6 (TRAF6) also regulates MCL1 by preventing its interaction with the proteasome. Of note, all Lys residues subjected to K63 ubiquitination localize to the C‐terminal region of the protein [47]. Several deubiquitinases, including ubiquitin‐specific peptidase 9 X‐linked (USP9X), ubiquitin carboxyl‐terminal hydrolase 13 (USP13), ubiquitin carboxyl‐terminal hydrolase 24 (USP24), ubiquitin carboxyl‐terminal hydrolase 17 (DUB3), Josephin domain containing 1 (JOSD1), and X‐ray repair cross‐complementing protein 6 (Ku70), also play roles in controlling MCL1 ubiquitination [27]; however, we require additional studies to fully understand how the interplay between E3 ligases and deubiquitinases regulates MCL1 levels and function.
The phosphorylation of MCL1, the most common post‐translational modification, usually implies protein degradation, although the role of all phosphorylated residues has yet to be experimentally confirmed [27, 48]. The N‐terminal region of MCL1 contains ten putative phosphorylation sites (Fig. 5) that can be modified in a cell context‐dependent manner by a range of kinases to produce site‐specific outputs. S64 phosphorylation by cyclin‐dependent kinase 1 and 2 (CDK1 and 2) and c‐Jun N‐terminal kinase (JNK) occurs during the G2/M phase of the cell cycle; however, these modifications fail to impact MCL1 half‐life [49]. Extracellular signal‐regulated kinase (ERK)‐mediated phosphorylation of T92 stabilizes the interaction of MCL1 with peptidyl‐prolyl cis/trans isomerase 1 (PIN1), blocks the association of MCL1 with protein phosphatase 2A (PP2A), and precedes phosphorylation of other residues (such as S121, S159, and T163). Overall, T92 phosphorylation is considered a priming signal for the subsequent phosphorylation and degradation of MCL1 [50]. Phosphorylation of T163 by ERK results in the increased stability and antiapoptotic activity of MCL1; however, phosphorylation of T163 combined with the phosphorylation of T92, S121, and S159 targets MCL1 for ubiquitination and proteasomal degradation.
Finally, MCL1 proteolysis and cleavage also occur during apoptosis. Several groups have identified D127, D157, and D117 as cleavage sites in the N terminus of the protein for executioner caspases and Granzyme B (Fig. 5). Although the function of the resultant fragments remains under investigation, MCL1 cleavage impedes interactions with other members of the BCL2 family and compromises antiapoptotic activity [51, 52, 53].
The survival role of MCL1 in apoptosis
As an antiapoptotic BCL2 family member, MCL1 promotes cell survival by interfering in the cascade of the events that cause MOMP and trigger cell death. Under steady‐state conditions, the proapoptotic BAX protein remains in an inactive state in the cytosol, with the C‐terminal domain sequestered within a hydrophobic surface groove [54]. Therefore, antiapoptotic proteins perform two complementary prosurvival functions: binding and neutralizing the BH3‐only ‘sensor’ proteins and inhibiting BAX and BAK effector proteins. When cells sense proapoptotic stimuli, the upregulation of BH3‐only proteins produces changes in interaction equilibria among BCL2 family members. The insertion of BH3‐only proteins into the hydrophobic groove formed by the BH1, BH2, and BH3 domains of antiapoptotic members [12, 55, 56, 57] decreases the proportion of prosurvival proteins available to inhibit BAX/BAK activity. Moreover, BH3‐only proteins interact directly with and activate BAX and BAK [58]. During activation, the C‐terminal hydrophobic domain of BAX becomes released and interacts with the membrane to promote BAX and BAK proteolipid pore formation to induce MOMP, apoptogenic factor release, apoptosome formation, caspase‐3 activation, and apoptosis.
MCL1 prevents cell death by inhibiting the oligomerization of the BAX and BAK effector proteins; thus, MCL1 degradation prompts increased BAK and BAX activity [59]. While studies have established the existence of a direct MCL1–BAK interaction under various conditions, any direct interaction between MCL1 and BAX remains unreported or undetectable [59, 60, 61]. Supporting the relevance of MCL1–BAK interactions, Moulding et al. [62] reported that apoptosis induced by the specific loss of MCL1 expression occurred alongside an increase in BAK expression; however, BAX expression remained unaltered. Additionally, ABT‐737 (a BCL2 and BCL‐xL inhibitor) triggers apoptosis in BAK‐null HCT116 cells; however, BAX‐null HCT116 cells displayed ABT‐737 resistance [63]. BAX and BAK are regulated differently. While BAX is mainly cytosolic in healthy cells and needs to go through conformational changes to localize to the mitochondria, as explained above, BAK resides inactive on the outer membrane of mitochondria thanks to a double negative regulation mechanism. BAK BH3 domain binds the hydrophobic groove of MCL1 and also the groove of BCL‐xL. Release of BAK from both proteins is required for apoptosis [59]. Thus, inhibition of both mechanisms has to be considered for therapeutic modulation of cell death [64].
MCL1 also interacts with BOK [65, 66], another proapoptotic effector of the BCL2 family. Hsu et al. [65] described physical interactions between MCL1 and BOK using double hybrid experiments, while more recent studies confirmed this interaction by coimmunoprecipitation [67]. Significantly, MCL1 interacts with BOK in a unique manner from the other BCL2 family members, using transmembrane fragment interactions [68] to afford control over BOK‐induced apoptosis [67].
MCL1 also associates with the PUMA, BIM, and NOXA BH3‐only ‘sensor’ proteins [13, 69]. Analysis using truncated proteins reported the interaction of MCL1 with BID and BMF, although with significantly lower affinity [70, 71] (Fig. 6). Of note, binding affinity estimations using soluble proteins fail to consider that interactions generally occur within mitochondrial membranes, thereby suggesting the careful consideration of these results. PUMA, BIM, and NOXA interfere with the binding between MCL1 and BAK to promote apoptosis [59, 64, 72, 73]. NOXA binds to MCL1 with a much higher affinity than other antiapoptotic BCL2 family proteins [70]. Additionally, unlike the rest of the BH3‐only proteins, NOXA possesses a C‐terminal region that interacts with the QRN motif in the MCL1 BH3 domain [13, 21]. Therefore, this interaction destabilizes BAK and MCL1 binding and promotes MCL1 degradation. Gomez‐Bougie et al. [74] reported that the USP9X deubiquitinase and MULE ubiquitinase participate in this process, although the precise mechanism involved requires further investigation.
Fig. 6.
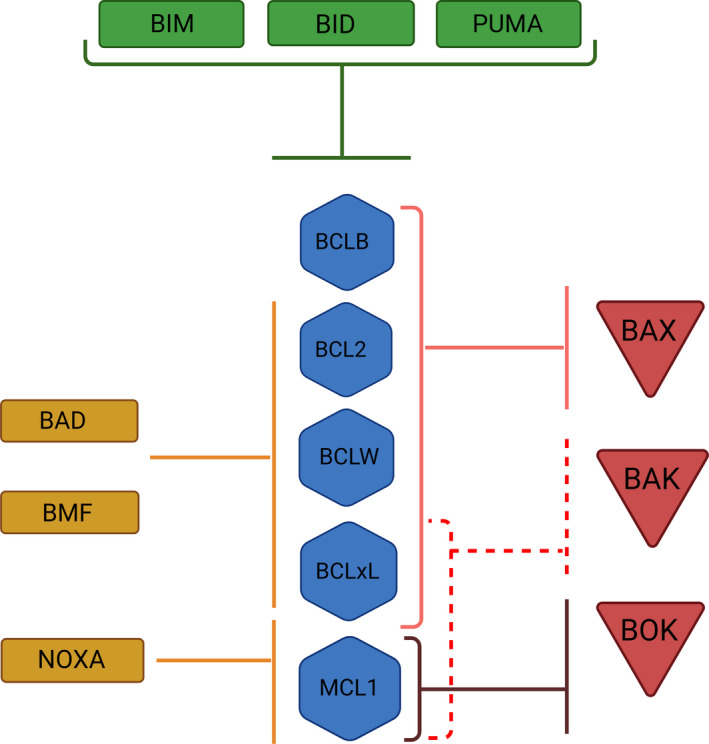
Diagram representing interactions described among the members of the BCL2 family. Red triangles show proapoptotic members; blue hexagons: antiapoptotic group; green rectangles: BH3‐only activators, and orange rectangles: BH3‐only sensors.
Most studies of the interactions between MCL1 and other BCL2 family members have been conducted with protein fragments and a membranous environment. Efforts made with other members of the family, such as BCL‐xL, to understand membrane interactions [75] may help to better understand the role of MCL1 in the regulation of MOMP and, in particular, the relevance of its interactions with BOK. This knowledge may be especially relevant, as we now appreciate that TMDs of these proteins, far from having a passive role, intervene in those interactions that modulate apoptosis [67, 76, 77].
The nonapoptotic functions of MCL1
Other than apoptosis, MCL1 also plays controlling roles in processes such as embryonic development, mitochondrial homeostasis, mitochondrial bioenergetics, autophagy, cell cycle control, and senescence.
MCL1 in embryonic development
Rinkenberger et al. [78] first revealed the fundamental role of MCL1 in mouse embryonic development by demonstrating preimplantation lethality following MCL1 knockout. MCL1‐null blastocysts failed to display any evidence of apoptosis; however, their delayed maturation and lack of cell culture adhesion indicated a defect in trophectoderm development. Furthermore, the survival of neural precursor cells (NPCs) decreased dramatically in MCL1 conditional knockout embryos, suggesting a role for MCL1 in developmental neurogenesis [79]. Finally, Fogarty et al. established the dependence of NPCs on MCL1 as neurogenesis commences and then on BCL‐xL as neurogenesis proceeds (Table 2). Although this role of MCL1 has been considered independent of its apoptotic function, no strict evidence has been published. Studies showing whether the loss of MCL1 leads to embryonic lethality in the absence of BAX/BAK will be needed for further understanding.
Table 2.
Main identified animal models and their application in the study of MCL1.
| Model type | Application | Reference | ||
|---|---|---|---|---|
| Knockout MCL1 | Absolute KO−/− | Development/Hematopoiesis | Survival of stem cells | [78] |
| Conditional KO−/− tissue‐specific | Hematopoiesis | Maintenance of lymphocytes | [85] | |
| Survival of hematopoietic lineages | [82, 196] | |||
| Megakaryocytic lineage | [83] | |||
| Development | Neurogenesis | [79] | ||
| Pathology | Chronic liver disease and liver tumorigenesis | [121, 122, 123, 197, 198, 199, 200] | ||
| Breast cancer | [201] | |||
| Pancreatic β‐cell survival | [120] | |||
| Neutrophil regulator in cerebral stroke | [118] | |||
| Conditional KO−/− tissue‐specific inducible | Hematopoiesis | Survival of stem cells | [84] | |
| Survival plasma cells | [202] | |||
| Pathology | Acute myeloid leukemia | [203] | ||
| Liver disease | [204] | |||
| Mitochondrial homeostasis and heart disease | [95, 205] | |||
| Conditional KO+/− tissue‐specific inducible | Hematopoiesis | Activated B‐cell survival and B‐cell memory | [206, 207] | |
| Pathology | Lymphomas | [208] | ||
| Neurodegeneration | [209] | |||
| Neutrophil regulator in cerebral stroke | [118] | |||
| Knockin MCL1 | KI human MCL1 | Comparative biology | Orthologous proteins | [210, 211] |
| KI humanized MCL1 mice | Pharmacological role | Drug efficacy | [158] | |
| Breast cancer | [159] | |||
| Pathology | Allergy and inflammation | [119] | ||
| KI degradation resistant | Pharmacological role | Colon cancer | [212] | |
In relation to MCL1 functions during development, studies with knockin and conditional knockout mouse models have also demonstrated the requirement of MCL1 expression for medullary epithelial cell survival, maintenance of thymic architecture, and positive selection in thymocyte differentiation during development [80] (Table 2). The elevated level of MCL1 expression in long‐term hematopoietic stem cells (HSCs) and its subsequent decline in HSC‐derived progenitor populations suggested that the antiapoptotic function of MCL1 may play a significant role during the earliest stages of hematopoiesis [81]. Interestingly, the conditional knockout of MCL1 in a mouse model resulted in rapid, fatal, multilineage hematopoietic failure [82, 83, 84, 85] (Table 2).
MCL1 in mitochondrial homeostasis and bioenergetics
The expression of MCL1 increases during the generation of induced pluripotent stem cells (iPSCs), with MCL1 localizing to the mitochondrial matrix [86]. As MCL1 interacts with mitochondrial fission and fusion regulators, dynamin‐related protein 1 (DRP1), and OPA1 mitochondrial dynamin‐like GTPase (OPA1) (see more below), the regulation of mitochondrial dynamics through MCL1 may help to maintain stem cell pluripotency. Mitochondria lacking MCL1 exhibit alterations in the tubular mitochondrial network, increased reactive oxygen species (ROS) production, defective cristae structure, lower respiratory chain efficiency, and decreased ATP levels and oxygen consumption rate [87]. These observations provide further evidence for the role of MCL1 in the regulation of mitochondrial homeostasis and bioenergetics.
The translocation of MCL1 between the OMM and the mitochondrial matrix [87] occurs through the activity of a translocase (TIM/TOM) complex from the outer/inner membrane that involves the proteolytic cleavage of the first 50–80 aa, creating a truncated N‐terminal form [87, 88, 89]. This ‘matrix form’ of MCL1 promotes oligomerization of the F1 and F0 domains of ATP synthase; however, MCL1 depletion alters ATP synthase oligomerization resulting in a disorganized cristae membrane, reduced ATP levels, lower mitochondrial membrane potential, and elevated ROS production [87]. However, the interaction between MCL1 and ATPase subunits (direct/indirect) remains undefined.
The matrix form of MCL1 also directly interacts with very‐long‐chain acyl‐CoA dehydrogenase (VLCAD), an essential enzyme of the mitochondrial fatty acid β‐oxidation pathway, suggesting a functional role for MCL1 in lipid metabolism. The MCL1–VLCAD complex prevents hyperactivity of the fatty acid β‐oxidation pathway, maintains normal acetyl‐CoA levels, and optimizes substrate consumption. Furthermore, MCL1 interacts with VLCAD in a noncanonical conformation that exposes the MCL1 BH3 helix [90].
As mentioned previously, MCL1 also interacts with mitochondrial proteins related to mitochondrial dynamics and remodeling networks such as DRP1 and OPA1. Thus, MCL1 modulates mitochondrial fission at the OMM and fusion at the IMM [86]. OPA1 locates to the inner mitochondrial membrane (IMM) and regulates mitochondrial fusion and oxidative phosphorylation, while MCL1 stabilizes and maintains OPA1 function. DRP1‐mediated mitochondrial fission in response to stress can be protective or detrimental [91, 92]. MCL1 exercises antiapoptotic activity within the OMM by inhibiting BAK activation to prevent MOMP and by inducing recruitment of DRP1 to the mitochondria. Thus, the MCL1–DRP1 interaction may allow cells to adapt to stress and induce survival independent of its antiapoptotic role [9, 93]. As the MCL1 BH3‐binding groove mediates OPA1 and DRP1 interactions, small‐molecule inhibitors directed against the canonical MCL1 BH3‐binding groove interfere with DRP1–MCL1 and OPA1–MCL1 interactions without influencing fatty acid oxidation [86, 90].
Deficiencies in the respiratory chain and ATP production together with increased ROS production could explain the cardiotoxic phenotype observed following MCL1 deletion in cell and animal models [48, 94]. While cardiotoxicity represents an important consideration when designing MCL1 inhibitors, the selective targeting of MCL1 (and leaving other proteins of the BCL2 family, e.g., BCL‐xL, unaffected) may improve drug tolerability in cardiomyocytes [95] (Table 2).
In agreement with a role in bioenergetics, MCL1 tightly regulates Ca2+ flux. MCL1 interacts with the inositol trisphosphate receptor (IP3R) in the ER through its C‐terminal region with the same affinity as BCL2 and BCL‐xL and increases the release of Ca2+ from the ER to the mitochondria at a rate that improves mitochondrial bioenergetics and inhibits apoptosis [96]. Furthermore, overexpression of the proapoptotic MCL1S isoform prompts mitochondrial hyperpolarization, increased mitochondrial Ca2+ accumulation, and apoptosis due to DRP1‐dependent mitochondrial hyperfusion [9]. Thus, both MCL1 protein content and MCL1L/MCL1S isoform ratio modulate Ca2+ and mitochondrial network homeostasis.
MCL1 also binds to voltage‐gated anion channel protein (VDAC) in the mitochondria to stimulate Ca2+ uptake and, as a consequence, increase ROS generation [97]. Interestingly, Ca2+‐dependent ROS production promotes lung cancer cell migration without affecting cell proliferation [97]. Furthermore, studies reported that ROS production depends on the MCL1–VDAC interaction, as VDAC‐based inhibitor peptides decrease ROS production in cells expressing MCL1; however, MCL1 knockdown failed to induce an effect. These findings are controversial when considering the previously discussed evidence regarding how the MCL1 matrix form impacts ROS production [87]. As there exists evidence that VDAC and IP3R act on the same signaling pathway [98], further investigations will be needed to clarify the connections between IP3R, VDAC, and the OMM‐ and matrix‐forms of MCL1 forms.
In addition to this, Cen et al. [99, 100] described recently the interaction of MCL1 with microtubule‐associated 1A/1B light chain 3A (LC3A) protein to promote mitophagy. The chemical inhibition of MCL1–BAK interaction releases MCL1 allowing MCL1 to interact with LC3A. Interestingly, authors showed that mitophagy could be a potential target for neurodegenerative diseases and in particular, in Alzheimer's disease (AD). In AD, the impaired mitochondria trigger energetic stress, calcium imbalance, and accumulation of amyloid‐β deposits in the brains of AD mouse models. Mitophagy is also compromised in AD and contributes to regulation of named mitochondrial alterations. Further studies are needed to understand the whole picture of MCL1 controlling mitochondria homeostasis.
MCL1 in autophagy
Autophagy, the process of degradation and recycling of intracellular content, intervenes in modulating the equilibria between cell survival or death. The role of MCL1 in autophagy depends on the cellular context. Several studies have described MCL1‐driven inhibition of autophagy through the binding of the BECLIN1 BH3 motif to the BH3 binding groove of MCL1 and the inhibition of autophagosome formation [101]. The ER and mitochondria both provide the lipids required for autophagosome formation [102]; therefore, MCL1 may regulate the mitochondrial pathway of autophagosome formation with evidence for this pathway existing in neurons [103]. The MCL1–BECLIN1 interaction may also explain why the oxidative stress‐triggered increase in BOK expression leads to induction of autophagy in preeclamptic placentas [104]. Increased BOK protein levels can sequester MCL1, thereby releasing BECLIN1 to promote autophagy.
A second explanation for MCL1‐driven autophagy inhibition arises from studies that established how MCL1 downregulation leads to increased BECLIN1 levels and the induction of basal autophagy. MCL1 and BECLIN1 compete to bind to the same region of the USP9X deubiquitinase; therefore, the displacement of BECLIN1 by MCL1 could induce the consequent increase in ubiquitination and proteasomal degradation. The inverse correlation of these proteins supports the development of certain tumors, given that levels of BECLIN1 decrease and MCL1 increase during tumor progression [105]. However, David Vaux work showed that MCL1, among others antiapoptotic proteins, inhibits autophagy pathway by affecting BAX/BAK levels, suggesting an indirect role. In a cellular context where BAX and BAK are not present, inhibition of MCL1 does not increase autophagy rates [106]. In addition, MCL1 exerts a proautophagic function in specific cellular contexts. Thomas et al. [48] reported that MCL1 deletion impairs autophagy in cardiomyocytes, with mitochondrial dysfunction, decreased ATP production, and increased ROS production all observed. Thus, the role of MCL1 in autophagy remains controversial. MCL1 may also control apoptosis and autophagy in a context‐dependent manner, according to cellular conditions and depending on the relative dominance of autophagic or proapoptotic factors.
Nuclear roles of MCL1
MCL1 also localizes to the nucleus and plays prominent regulatory roles during various stages of the cell cycle. MCL1 accumulates during the cell cycle and reaches a peak around the late G2 phase [107], where it stimulates checkpoint kinase 1 (CHK1) phosphorylation, a regulator of DNA damage response [108]. As CHK1 phosphorylation mediates G2/M arrest, MCL1 plays a G2/M checkpoint protein role and provides time for DNA damage repair [26, 109].
A study by Jamil et al. detected the interaction between a short nuclear form of MCL1 with cyclin‐dependent kinase 1 (CDK1), which inhibited kinase activity and cell cycle progression through G2/M phases. Of note, the MCL1–CDK1 association did not depend on interactions of CDK1 with cyclins [110]. Another study established that CDK1 binding to mitotic cyclin B induces the phosphorylation of MCL1 at T92, with phosphorylation and polyubiquitylation promoting MCL1 degradation and inducing apoptosis [107]. Interestingly, if MCL1 levels fall below a critical level as occurs in the mitotic arrest state, cell death occurs via the intrinsic apoptotic pathway [111]. Thus, the inhibition of CDK1 and CHK1 activities makes MCL1 a significant player at the G2/M checkpoint; however, the MCL1S pro‐apoptotic isoform counteracts G2/M checkpoint function, accelerates cell cycle progression through mitosis, and promotes DNA damage accumulation [112]. MCL1 also interacts with the S phase cell cycle regulator proliferating cell nuclear antigen (PCNA) to inhibit cell cycle progression [25].
The interaction between MCL1 and cyclin‐dependent kinase 4 inhibitor C (P18INK4C), an inhibitor of CDK4/6, promotes G1/S progression by avoiding the inhibitory activity of P18INK4C on CDK4/6. Consequently, CDK4/6 interacts with retinoblastoma (RB1) facilitating S phase entry [113]. MCL1 causes depletion of P18INK4C protein levels; therefore, the accumulation of cells in G2/M after MCL1 overexpression may represent a consequence of increased cell cycle entry promoted by MCL1–P18INK4C interactions. Intriguingly, MCL1 interacts with cyclin‐dependent kinase inhibitor 1B (CDKN1B) in the nervous system to control cell cycle progression through the G1 phase to promote the differentiation and survival of NPCs [114].
Widden et al. also described the interaction of MCL1 with p73, which plays a role in modulating the expression of target genes associated with the DNA damage response and cell cycle progression [115]. p73 directly interacts with the MCL1 BH3‐binding pocket in the nucleus to inhibit the DNA‐binding ability of p73. Therefore, MCL1 promotes cell cycle progression by blocking the transcriptional activity of p73 [116].
Overall, MCL1 interacts with various cell cycle proteins to induce divergent outcomes; further research may disentangle the exact mechanisms by which MCL1 modifies cell cycle progression.
Senescence
Finally, MCL1 inhibits chemotherapy‐induced senescence and can enhance tumor growth independent of its apoptotic function. As described before, a domain located outside the classical BH regions mediates this senescence‐inhibiting function, which could also be implicated in, for example, the ability of MCL1 to interfere with ROS production [22, 23, 117]. Overall, the MCL1‐mediated inhibition of apoptosis and senescence through distinct molecular domains should be considered when evaluating current developing MCL1 inhibitors.
MCL1‐related pathologies
Given the wide range of cellular functions associated with MCL1, investigations into a range of pathological processes have also established crucial links to MCL1 dysregulation.
The relevance of MCL1 in heart disease has emerged as a significant concern regarding the use of MCL1 inhibitors in the clinic. In cardiomyocyte‐based models, MCL1 deletion produces cardiotoxicity, thereby decreasing cell survival and impairing contractile function [94], a phenotype attributed to the roles of MCL1 in the control of mitochondrial homeostasis and autophagy. In agreement with in vitro cell‐based findings, mice lacking cardiomyocyte‐specific MCL1 expression exhibited signs of heart disease, manifested as a loss of cardiac contractility, defects in mitochondrial structure and mitochondrial respiration, and lower ATP production compared with wild‐type [48, 95].
MCL1 also plays a prominent role in the outcome of cerebrovascular disorders. For example, MCL1‐deficient stroke model mice presented with less harmful lesions in the ischemic area with an almost complete absence of neutrophil recruitment compared with wild‐type or heterozygous animals [118]. Therefore, this study provides evidence that small‐molecule inhibitors of MCL1 may represent a possible therapeutic avenue for treating ischemic stroke in human patients. Additionally, Felton et al. [119] linked the overexpression of the human form of MCL1 in mice with the exacerbation of allergic airway inflammation, with increased cellularity of bronchoalveolar lavage fluid, eosinophil number, total protein, and airway mucus production observed.
Analysis in type I diabetes model has uncovered a pancreas‐specific decrease in MCL1 expression [120]. MCL1 knockout mice did not display alterations to the development and function of pancreatic islets; however, the loss of MCL1 expression did affect pancreatic β cells, making them more susceptible to apoptosis mediated by proinflammatory cytokines. β‐cells from the pancreas of MCL1 knockout mice also exhibited hyperglycemia and low pancreatic insulin content after toxic stimuli. In conclusion, maintaining MCL1 homeostasis may represent an exciting strategy for treating type 1 diabetes patients [120].
Chronic liver disease patients suffer from an elevated risk of developing hepatocellular carcinoma, with epithelial cell apoptosis frequently observed in cirrhotic livers [121]. Hypothetically, the genetic depletion of MCL1 may increase apoptosis and inhibit tumorigenesis; however, the livers of mice with a hepatocyte‐specific knockout of MCL1 displayed increased mild tissue fibrosis, oxidative stress, and inflammatory cytokine release, all of which can support cancer development. The role of MCL1 in cell cycle progression, DNA repair, and ROS production may explain the described phenotype [121, 122, 123].
MCL1 dysregulation occurs in many types of cancers and often correlates with poor prognosis and therapeutic resistance. In fact, genomic data analysis from The Cancer Genome Atlas shows high MCL1 protein expression in at least 22 cancer types [124]. Interestingly, MCL1 is located in a focal amplification peak region of the chromosome 1qS with other eight genes that shows amplifications in 10.9% of cancers in multiple tissue types. This is considered a common mechanism for cancers, to increase cell survival that could be targeted. In particular, in lung cancer it has been shown that MCL1 amplification is required for sustained survival and its inhibition delays tumor progression [124, 125].
In colon adenoma and carcinoma patients [126], MCL1 protein levels directly correlate with tumor grade/stage and the presence of metastasis [127]. Interestingly, the MCL1 expression pattern correlates with responses to 5‐fluorouracil (5‐FU)—those patients with perinuclear MCL1 expression responded more frequently to treatment [128]. High MCL1 expression correlates with poor survival for patients with stage III ovarian carcinomas, with diffuse MCL1 expression correlating with advanced clinical stage, high histologic grade, and poor survival [129]. In gastric carcinoma patients, the detection of elevated MCL1 levels by tumor immunohistochemistry correlates with a significantly worse prognosis than patients lacking tumor MCL1 expression [130, 131]. In multiple myeloma, MCL1 mRNA and protein overexpression in patient samples correlated with disease severity and shorter event‐free survival times [132]. The mechanisms that multiple myeloma cells employed to survive involved the stabilization of MCL1 by PP2A‐mediated dephosphorylation [215]. High MCL1 levels appear at high frequency in non–small‐cell lung cancer (NSCLC) cohorts, and retrospective studies have provided evidence of the significantly worse five‐year survival rate in those patients with elevated tumor MCL1 expression [133]. Moreover, NSCLC patients with high MCL1 expression display a high cellular proliferation index and lower overall survival rates than patients with low MCL1 expression [134]. MCL1 expression also correlates with protein kinase B (AKT) activity in NSCLC tumors, with interactions between the MCL1 PEST domain and the pleckstrin homology domain of AKT forming at least part of the underlying mechanism. This interaction leaves the AKT kinase site in an active conformation, thereby promoting cell survival [135]. Interestingly, the MCL1 deletion or inhibition of MCL1 activity (with the MCL1 antagonist S63845) caused a significant reduction in tumorigenesis [124]. MCL1 overexpression also occurs in metastatic malignant melanoma [136]; antisense therapies, gene silencing, or inhibitor administration alone or in combination with other chemotherapeutic agents significantly decreased tumor growth [137, 138, 139, 140]. In breast cancer patients, MCL1 high expression correlates with poor prognosis, high tumor grade, and poor survival [141, 142, 143]. As EGF or ER activation upregulates MCL1 expression [144, 145, 146], targeting MCL1 represents the mechanism of action for many breast cancer‐targeted drugs [147, 148, 149, 150]. In the aggressive triple‐negative breast cancer (TNBC) subtype, inhibiting MCL1 sensitized breast cancer cells to apoptosis by conventional chemotherapy [142, 151].
By contrast, the hepatocyte‐specific loss of MCL1 increased the frequency of liver tumors [152]. This unexpected response depended on the apoptotic function of MCL1, as elimination of BAK decreased tumorigenicity. The increased frequency of tumors appears to correlate with excessive apoptosis in MCL1‐deficient livers, which prompted higher levels of TNF‐α production and increased oxidative stress to sustain a protumorigenic liver environment [121]. Induced hepatocyte compensatory liver regeneration as a consequence of excessive apoptosis contributes to liver carcinogenesis [123]; however, liver cancer patients exhibited MCL1 overexpression [153] and studies have demonstrated that specifically inhibiting MCL1 reduces the survival of hepatocellular cancer stem‐like cells [154]. Therefore, extensive liver cancer studies with novel specific MCL1 inhibitors may support the design of appropriate pharmacological strategies that will eliminate cancer cells but inhibit detrimental hepatic regeneration mechanisms to improve patient outcomes.
In summary, MCL1 participates in cancer progression, malignancy, and therapeutic resistance in several tumor types. Therefore, the clinical development of specific inhibitors will provide an opportunity to understand tumor biology better and, more importantly, provide patients with new therapeutic strategies.
Therapeutical strategies: MCL1 inhibitors in clinical trials
The critical role of MCL1 in different pathological scenarios has driven the search for pharmacological modulators of MCL1 function. Accordingly, the pharmaceutical industry has focused its attention on the prosurvival effect of MCL1 expression in tumor cells and its involvement in chemotherapy resistance, which has led to the discovery and clinical development of selective MCL1 inhibitors that target the BH3 binding groove and modulate interactions with other BCL2 members.
We now summarize the small molecules designed to neutralize the antiapoptotic activity of MCL1. Table 3 summarizes the small‐molecule inhibitors currently under clinical evaluation.
Table 3.
Summary of MCL1 inhibitors under clinical trials.
| Compound, chemical structure, and institution | Potency (TR‐FRET) | Clinical trials a | Study | Administration | Indication | References |
|---|---|---|---|---|---|---|
|
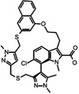
AZD5991 AstraZeneca |
MCL1 K i = 0.2 nm BCL2 K i = 6.8 μm BCL‐xL K i = 18 μm BCLW K i = 25 μm BFL1 K i = 12 μm |
NCT03218683 | Phase I, monotherapy, and combination | IV | Refractory Hematological malignancies | [213] |
|
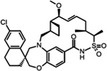
AMG‐176 Amgen |
MCL1 K i = 0.06 nm BCL2 K i = 0.95 μm BCL‐xL K i = 0.7 μm |
NCT02675452 | Phase I, monotherapy | IV | Relapsed or refractory MM/AML | [159] |
| NCT03797261 | Phase I, monotherapy with venetoclax | IV | AML/Lymphoma | |||
|
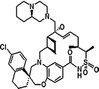
AMG‐397 Amgen |
Not disclosed | NCT03465540 | Phase I, monotherapy | Oral | MM/AML/NHL | [214] |
|
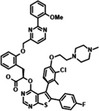
S64315/MIK665 Servier/Novartis |
MCL1 K i = 1.2 nm | NCT02979366 | Phase I, monotherapy | IV | AML/MDS | [156] |
| NCT02992483 | Phase I, monotherapy | IV | MM/Lymphoma | |||
| NCT03672695 | Phase I, combination with venetoclax | IV | AML/MDS | |||
| NCT04629443 | Phase II, combination with azacitidine | IV | AML | |||
| NCT04702425 | Phase I, combination with VOB560 | IV | NHL/AML/MM | |||
|
Structure not disclosed ABBV‐467 AbbVie |
Not disclosed | NCT04178902 | Phase I, monotherapy | IV | MM | Not disclosed |
|
Structure not disclosed PRT1419 Prelude Therapeutics |
Not disclosed | NCT04543305 | Phase I, monotherapy | Oral | Refractory Hematological malignancies | Not disclosed |
| NCT04837677 | Phase I, monotherapy | IV | Relapsed or refractory solid tumors |
From www.ClinicalTrials.gov.
S64315/MIK665
Structure‐guided optimization of S63845, a previously reported small‐molecule MCL1 antagonist [155], led to the development of S64315/MIK665, a potent and selective MCL1 inhibitor. Structural studies of S64315/MIK665 complexed with MCL1 revealed that the molecule fills the P2 pocket and the P4‐P5 region of MCL1 [156]. Compared with its predecessor, S64315/MIK665 demonstrated improved affinity and cell‐based activity. S64315/MIK665 presented low‐affinity binding for BCL2 and BCL‐xL (58 and 237 μm, respectively) [157] and displaced BAX and BAK proteins (but not their antiapoptotic relatives) from MCL1 [156]. As for other MCL1 inhibitors, S64315/MIK665 increased MCL1 protein accumulation in a dose‐dependent manner and induced BAX/BAK‐mediated apoptosis in tumor cells. Hematological cancer‐derived cell lines displayed high susceptibility (IC50 < 100 nm) to S64315/MIK665, while intravenous dosing of S64315/MIK665 for five consecutive days elicited dose‐dependent antitumor activity in a multiple myeloma (MM) xenograft model. S64315/MIK665 also induced similar tumor growth inhibition in animal models treated once a week or treated daily for four weeks, highlighting the safety and tolerability of this drug.
Given that S63845 displayed a higher affinity to human MCL1 than mouse MCL1, Brennan et al. [158] evaluated efficacy and tolerability in a mouse model expressing human MCL1. Engineered mice displayed greater sensitivity to S63845 treatment (as evidenced by its ability to inhibit tumorigenesis) than wild‐type mice but did prompt the transient reduction in B cells in the blood, spleen, and bone marrow. In addition, the study also noted a nonsignificant reduction in neutrophil levels in the bone marrow [158].
Based on its favorable preclinical evaluation, intravenous S64315/MIK665 has entered clinical trials to treat relapsed and/or refractory MM, lymphoma, acute myeloid leukemia (AML), and myelodysplastic syndrome (MDS) (NCT02979366 and NCT02992483). In addition, S64315/MIK665 combined with Venetoclax is also under evaluation for AML treatment (NCT03672695).
AMG‐176 and AMG‐397
Amgen optimized a series of spiromacrocyclic molecules guided by a small‐molecule conformational restriction strategy that led to the development of AMG‐176 [159]. AMG‐176 (and the analog AM‐8621) have a picomolar affinity for human MCL1. AM‐8621 has been used as a molecular tool to characterize the molecular mechanism of AMG‐176 in cellular assays. AM‐8621 disrupted MCL1 interactions with BAK and BIM and induced both MCL1 stabilization and BAX/BAK‐dependent apoptosis. Hematologic cancer cell lines displayed greater sensitivity to AM‐8621 than solid tumor lines, with the most significant sensitivity displayed by MM, AML, B‐cell lymphoma, and subsets of acute lymphocytic leukemia and Burkitt lymphoma cell lines. Breast cancer cells displayed the most significant sensitivity among solid tumor cell lines. In OPM‐2 mouse xenografts (a model of MM), oral administration of AMG‐176 activated BAK, cleaved caspase‐3 and poly (ADP‐ribose) polymerase (PARP), and induced apoptosis and tumor regression.
This study also reported a synergistic effect of AMG‐176 with Venetoclax in an AML orthotopic model, where twice‐weekly AMG‐176 and daily Venetoclax treatments prompted tumor inhibition and regression. Mice carrying human MCL1 were also used to evaluate AMG‐176 efficacy and tolerability. The dosing regimen of AMG‐176 required for tumor growth inhibition resulted in reduced numbers of B cells, monocytes, and neutrophils in blood and bone marrow; however, AMG‐176 failed to induce systemic toxicity based on body weight change. In the orthotopic AML model, combined MCL1 and BCL2 inhibition by AMG‐176 and Venetoclax were well tolerated, and no overt toxicity was determined; however, the study did note a decrease in peripheral blood B cells and monocytes.
Clinical evaluation of AMG‐176 has been initiated in a phase I clinical trial (NCT02675452) in patients with relapsed or refractory MM and AML, and preliminary results have been disclosed [160]. Although the maximum tolerated dose was not reached for relapsed MM patients, predominant side effects included hematological (anemia and neutropenia) and gastrointestinal (nausea and diarrhea) problems. Nevertheless, disease status remained stable in 11 out of 26 patients. Another phase I clinical trial is also currently evaluating AMG‐176 in combination with Venetoclax in patients with AML and non‐Hodgkin lymphoma (NCT03797261).
From the same molecular family as AMG‐176, AMG‐397 (the first oral MCL1 inhibitor used in the clinic) has been evaluated for the treatment of patients with MM, AML, and non‐Hodgkin lymphoma (NCT03465540); however, the FDA halted these trials due to cardiac toxicity (ASHClinicalNews. FDA places trials of MCL1 inhibitor on clinical hold (2019). www.ashclinicalnews.org/news/latest‐and‐greatest/fda‐places‐trials‐MCL1‐inhibitor‐clinical‐hold/). As described previously, MCL1 may be essential for normal mitochondrial activity in cardiomyocytes [95]. The side effects that may relate to AMG‐397 on‐target activity may limit the therapeutic scope of AMG‐397 (and that of other MCL1 inhibitors) [86].
AZD5991
AstraZeneca researchers applied structure‐based drug design to a series of previously reported indole‐2‐carboxylic acids (WO2008130970A1, WO2008131000A2, and Ref. [161]) to develop AZD5991, a selective and potent macrocyclic MCL1 inhibitor. AZD5991 selectively binds to MCL1 over other antiapoptotic proteins due to its specific interactions with and the flexibility of the MCL1 BH3‐binding groove. Furthermore, as reported for other selective MCL1 inhibitors, AZD5991 stabilized MCL1 in a concentration‐dependent manner, disrupted MCL1–BAK interactions, and induced apoptosis in a BAK‐dependent manner.
AZD5991 preferentially induced cell death in hematological cell lines and subsets of breast cancer and NSCLC lines. AZD5991 also promoted apoptosis in MM primary cells and mouse models. Results suggested the overall tolerability of a single intravenous infusion of AZD5991 (i.e., acceptable body weight changes) and demonstrated complete tumor regression at the highest doses. MCL1 inhibition with AZD5991 also exhibited a dose‐dependent antitumor activity in mouse and rat AML models.
The combination of AZD5991 with the proteasome inhibitor bortezomib provided for synergistic antitumor activity in a MM mouse model. In addition, AZD5991 combined with Venetoclax also induced enhanced tumor growth inhibition in subcutaneous MM and AML xenografts.
AZD5991 also demonstrated potent antitumor activity and increased survival in MCL1‐dependent T‐cell lymphoma patient‐derived xenografts in mice when combined with CHOP chemotherapy (cyclophosphamide, doxorubicin hydrochloride, vincristine sulfate, and prednisone) [162].
The antitumor activity of AZD5991 has also been demonstrated in solid tumor models. AZD5991 restored sensitivity to regorafenib, a multi‐kinase inhibitor for colorectal cancer treatment, during in vitro assays with colorectal carcinoma cell lines [163], while AZD5991 combined with BRAF inhibitors (BRAFi) or MEK inhibitors (MEKi) inhibited the clonogenic survival of melanoma cells. A synergistic effect of between BRAFi/MEKi with AZD5991 was also observed in melanoma animal models, including melanoma xenografts, BRAF‐mutant patient‐derived xenografts, and BRAFi‐ and/or MEKi‐resistant melanomas. In addition, combination therapy induced tumor regression or total growth inhibition and overcame BRAFi‐ and MEKi‐acquired resistance [138].
AZD5991 was selected as a clinical candidate and is currently under evaluation in a phase I clinical trial as a monotherapy for relapsed/refractory chronic lymphocytic leukemia, AML/myelodysplastic syndromes, and MM patients. In addition, a combination therapy with Venetoclax is also being evaluated in patients with relapsed/refractory AML/MDS (NCT03218683).
Two undisclosed MCL1 inhibitors (PRT1419 and ABBV‐467) have also entered clinical trials. Prelude Therapeutics employed a structure‐based design to identify PRT1419, a selective and potent oral MCL1 inhibitor. Once‐weekly dosing of PRT1419 prompted tumor regression in mouse models of MM and AML, and diffuse large B‐cell lymphoma in preclinical studies. Moreover, mouse models display enhanced antitumor activity in combination with Venetoclax in a model of AML. PRT1419 is currently in a phase I clinical trial to treat relapsed/refractory hematologic malignancies (NCT04543305). In addition, AbbVie has also started a phase I clinical trial with ABBV‐467 as monotherapy for MM patients (NCT04178902).
MCL1 therapeutics under development
The Fesik group recently reported on the development of two MCL1 inhibitors: VU661013, which has a picomolar affinity for MCL1 with no detectable binding for BCL‐xL and BCL2, and Compound 42 [164]. VU661013 demonstrated potent cytotoxic activity in AML cell lines, Venetoclax‐resistant AML cells, and AML patient‐derived xenograft models. Furthermore, the combination of Venetoclax with VU661013 further enhanced antitumor activity in murine models of AML [164]. Meanwhile, Compound 42 potently inhibited MCL1 activity and demonstrated in vivo efficacy in xenograft models derived from hematologic and TNBC cell lines [165].
Alternative approaches have also been developed to target the antiapoptotic function of MCL1. For example, complex 14, a copper (II) complex containing β‐carboline ligands, disrupted MCL1 binding to BAX and BAK, induced BAX/BAK‐dependent apoptosis, and inhibited tumor growth in NSCLC xenografts [166].
The downregulation of MCL1 gene expression also represents a promising therapeutic strategy, with flavonoids the most well‐known regulators of MCL1 transcription. Certain flavonoids inhibit CDKs, which ultimately leads to decreased levels of short‐lived proteins such as MCL1. As CDK9 inhibitors, Voruciclib and AZD4573 have proven antitumor activity through indirect MCL1 suppression in hematologic cancer models [167, 168].
Other chemical compounds achieve MCL1 inhibition by blocking mRNA translation. For instance, Norcantharidin upregulated miR‐320d, a negative regulator of MCL1 expression, and induced apoptosis in prostate cancer cells [169]. In addition, simultaneous silencing of MCL1 and the efflux pump P‐glycoprotein by two specific short interfering RNA (siRNAs) in doxorubicin‐resistant breast cancer cells efficiently reduced protein levels and induced significant levels of apoptosis [170].
Proteolysis‐targeting chimeras (PROTACs) represent a powerful technology that can increase the proteasomal degradation of MCL1. PROTACs are engineered bifunctional molecules comprising two components: an MCL1‐binding ligand and an effector ligand that recruits an E3 ubiquitin ligase to trigger proteasomal degradation. A first example employed the A1210477 MCL1 inhibitor as a target ligand and 4‐hydroxythalidomide as an effector ligand to recruit to the E3 ligase cereblon (CRBN) [171], while a second example linked another MCL1 inhibitor to the cereblon–ligand pomalidomide. Both PROTACs induced MCL1 degradation at nanomolar concentrations and induced apoptosis [171].
Finally, cell‐penetrating ‘Alphabodies’, single‐chain polypeptides featuring antiparallel triple‐helix coiled‐coil fold, have been developed to target MCL1. The beta‐helix of the CMPX‐321A and CMPX‐383B Alphabodies bind to the MCL1 BH3 groove to induce selective high‐affinity MCL1 inhibition. As a result, CMPX‐321A displayed cell‐penetrating capacity and induced BAK‐mediated cell death in MCL1‐dependent cancer cell lines, while CMPX‐383B possessed an extended serum half‐life and exhibited a potent antitumor activity that reduced tumor burden in MM xenograft mice [172].
While most of the described strategies targeting MCL1 modulate its antiapoptotic function, there remains the potential to compromise the nonapoptotic functions of MCL1, which remain poorly understood. This risk makes it essential to describe the specific interaction sites involved in MCL1's various roles, design selective MCL1 inhibitors against these interaction sites, and inhibit a specific function of MCL1 without affecting the others, thus preventing potential toxicities related to the unspecific targeting. Deciphering the interacting domains that MCL1 utilizes to execute its prosurvival and nonapoptotic roles represents an important future direction and would allow the development of safe and effective MCL1 inhibitors and the successful clinical translation of anti‐MCL1 therapies.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
All authors wrote, edited, and revised the manuscript.
Acknowledgements
Research in the MO laboratory is funded by the Spanish Ministry of Economy and Competitiveness Grants PID2020‐115048RB‐I00 and SAF2017‐84689‐R and Generalitat Valenciana Grant PROMETEO/2019/065. DL is funded by a predoctoral fellowship (PRDVA21475LEIV) from Spanish Cancer Association (AECC). Figures were created with BioRender.com.
References
- 1. Kozopas KM, Yang T, Buchan HL, Zhou P & Craig RW (1993) MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci USA 90, 3516–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krajewski S, Bodrug S, Krajewska M, Shabaik A, Gascoyne R, Berean K & Reed JC (1995) Immunohistochemical analysis of Mcl‐1 protein in human tissues. Differential regulation of Mcl‐1 and Bcl‐2 protein production suggests a unique role for Mcl‐1 in control of programmed cell death in vivo. Am J Pathol 146, 1309–1319. [PMC free article] [PubMed] [Google Scholar]
- 3. Reynolds JE, Yang T, Qian L, Jenkinson JD, Zhou P, Eastman A & Craig RW (1994) Mcl‐1, a member of the Bcl‐2 family, delays apoptosis induced by c‐Myc overexpression in Chinese hamster ovary cells. Cancer Res 54, 6348–6352. [PubMed] [Google Scholar]
- 4. Liu X, Dai S, Zhu Y, Marrack P & Kappler JW (2003) The structure of a Bcl‐xL/Bim fragment complex: implications for Bim function. Immunity 19, 341–352. [DOI] [PubMed] [Google Scholar]
- 5. Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ et al. (1997) Structure of Bcl‐xL‐Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986. [DOI] [PubMed] [Google Scholar]
- 6. Akgul C, Turner PC, White MR & Edwards SW (2000) Functional analysis of the human MCL‐1 gene. Cell Mol Life Sci 57, 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bae J, Leo CP, Hsu SY & Hsueh AJ (2000) MCL‐1S, a splicing variant of the antiapoptotic BCL‐2 family member MCL‐1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem 275, 25255–25261. [DOI] [PubMed] [Google Scholar]
- 8. Bingle CD, Craig RW, Swales BM, Singleton V, Zhou P & Whyte MK (2000) Exon skipping in Mcl‐1 results in a bcl‐2 homology domain 3 only gene product that promotes cell death. J Biol Chem 275, 22136–22146. [DOI] [PubMed] [Google Scholar]
- 9. Morciano G, Giorgi C, Balestra D, Marchi S, Perrone D, Pinotti M & Pinton P (2016) Mcl‐1 involvement in mitochondrial dynamics is associated with apoptotic cell death. Mol Biol Cell 27, 20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim JH & Bae J (2013) MCL‐1ES induces MCL‐1L‐dependent BAX‐ and BAK‐independent mitochondrial apoptosis. PLoS One 8, e79626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim JH, Sim SH, Ha HJ, Ko JJ, Lee K & Bae J (2009) MCL‐1ES, a novel variant of MCL‐1, associates with MCL‐1L and induces mitochondrial cell death. FEBS Lett 583, 2758–2764. [DOI] [PubMed] [Google Scholar]
- 12. Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC & Hinds MG (2005) Solution structure of prosurvival Mcl‐1 and characterization of its binding by proapoptotic BH3‐only ligands. J Biol Chem 280, 4738–4744. [DOI] [PubMed] [Google Scholar]
- 13. Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DC, Fairlie WD, Hinds MG & Colman PM (2007) Structural insights into the degradation of Mcl‐1 induced by BH3 domains. Proc Natl Acad Sci USA 104, 6217–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu G, Poppe L, Aoki K, Yamane H, Lewis J & Szyperski T (2014) High‐quality NMR structure of human anti‐apoptotic protein domain Mcl‐1(171–327) for cancer drug design. PLoS One 9, e96521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boersma MD, Sadowsky JD, Tomita YA & Gellman SH (2008) Hydrophile scanning as a complement to alanine scanning for exploring and manipulating protein‐protein recognition: application to the Bim BH3 domain. Protein Sci 17, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fogha J, Marekha B, De Giorgi M, Voisin‐Chiret AS, Rault S, Bureau R & Sopkova‐de Oliveira Santos J (2017) Toward understanding Mcl‐1 promiscuous and specific binding mode. J Chem Inf Model 57, 2885–2895. [DOI] [PubMed] [Google Scholar]
- 17. Marimuthu P & Singaravelu K (2018) Prediction of hot spots at myeloid cell leukemia‐1‐inhibitor interface using energy estimation and alanine scanning mutagenesis. Biochemistry 57, 1249–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petros AM, Olejniczak ET & Fesik SW (2004) Structural biology of the Bcl‐2 family of proteins. Biochim Biophys Acta 1644, 83–94. [DOI] [PubMed] [Google Scholar]
- 19. Denis C, Sopkova‐de Oliveira Santos J, Bureau R & Voisin‐Chiret AS (2020) Hot‐spots of Mcl‐1 protein. J Med Chem 63, 928–943. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Z, Yang H, Wu G, Li Z, Song T & Li XQ (2011) Probing the difference between BH3 groove of Mcl‐1 and Bcl‐2 protein: implications for dual inhibitors design. Eur J Med Chem 46, 3909–3916. [DOI] [PubMed] [Google Scholar]
- 21. Song T, Wang Z, Ji F, Feng Y, Fan Y, Chai G, Li X, Li Z & Zhang Z (2016) Deactivation of Mcl‐1 by dual‐function small‐molecule inhibitors targeting the Bcl‐2 homology 3 domain and facilitating Mcl‐1 ubiquitination. Angew Chem Int Ed Engl 55, 14250–14256. [DOI] [PubMed] [Google Scholar]
- 22. Demelash A, Pfannenstiel LW, Tannenbaum CS, Li X, Kalady MF, DeVecchio J & Gastman BR (2015) Structure‐function analysis of the Mcl‐1 protein identifies a novel senescence‐regulating domain. J Biol Chem 290, 21962–21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolesta E, Pfannenstiel LW, Demelash A, Lesniewski ML, Tobin M, Schlanger SE, Nallar SC, Papadimitriou JC, Kalvakolanu DV & Gastman BR (2012) Inhibition of Mcl‐1 promotes senescence in cancer cells: implications for preventing tumor growth and chemotherapy resistance. Mol Cell Biol 32, 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang T, Kozopas KM & Craig RW (1995) The intracellular distribution and pattern of expression of Mcl‐1 overlap with, but are not identical to, those of Bcl‐2. J Cell Biol 128, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fujise K, Zhang D, Liu J & Yeh ET (2000) Regulation of apoptosis and cell cycle progression by MCL1. Differential role of proliferating cell nuclear antigen. J Biol Chem 275, 39458–39465. [DOI] [PubMed] [Google Scholar]
- 26. Jamil S, Stoica C, Hackett TL & Duronio V (2010) MCL‐1 localizes to sites of DNA damage and regulates DNA damage response. Cell Cycle 9, 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Senichkin VV, Streletskaia AY, Gorbunova AS, Zhivotovsky B & Kopeina GS (2020) Saga of Mcl‐1: regulation from transcription to degradation. Cell Death Differ 27, 405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Croxton R, Ma Y, Song L, Haura EB & Cress WD (2002) Direct repression of the Mcl‐1 promoter by E2F1. Oncogene 21, 1359–1369. [DOI] [PubMed] [Google Scholar]
- 29. Rosato RR, Almenara JA, Kolla SS, Maggio SC, Coe S, Gimenez MS, Dent P & Grant S (2007) Mechanism and functional role of XIAP and Mcl‐1 down‐regulation in flavopiridol/vorinostat antileukemic interactions. Mol Cancer Ther 6, 692–702. [DOI] [PubMed] [Google Scholar]
- 30. Laetsch TW, Liu X, Vu A, Sliozberg M, Vido M, Elci OU, Goldsmith KC & Hogarty MD (2014) Multiple components of the spliceosome regulate Mcl1 activity in neuroblastoma. Cell Death Dis 5, e1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gautrey HL & Tyson‐Capper AJ (2012) Regulation of Mcl‐1 by SRSF1 and SRSF5 in cancer cells. PLoS One 7, e51497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kedzierska H, Poplawski P, Hoser G, Rybicka B, Rodzik K, Sokol E, Boguslawska J, Tanski Z, Fogtman A, Koblowska M (2020) Correction: Kedzierska H. et al. Decreased expression of SRSF2 splicing factor inhibits apoptotic pathways in renal cancer. Int. J. Mol. Sci. 2016, 17, 1598. Int J Mol Sci 21, 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khan DH, Gonzalez C, Cooper C, Sun JM, Chen HY, Healy S, Xu W, Smith KT, Workman JL, Leygue E et al. (2014) RNA‐dependent dynamic histone acetylation regulates MCL1 alternative splicing. Nucleic Acids Res 42, 1656–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C & Nabors LB (2011) The RNA‐binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res 9, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebner F, Sedlyarov V, Tasciyan S, Ivin M, Kratochvill F, Gratz N, Kenner L, Villunger A, Sixt M & Kovarik P (2017) The RNA‐binding protein tristetraprolin schedules apoptosis of pathogen‐engaged neutrophils during bacterial infection. J Clin Invest 127, 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Subramaniam D, Natarajan G, Ramalingam S, Ramachandran I, May R, Queimado L, Houchen CW & Anant S (2008) Translation inhibition during cell cycle arrest and apoptosis: Mcl‐1 is a novel target for RNA binding protein CUGBP2. Am J Physiol Gastrointest Liver Physiol 294, G1025–G1032. [DOI] [PubMed] [Google Scholar]
- 37. Cui J & Placzek WJ (2016) PTBP1 modulation of MCL1 expression regulates cellular apoptosis induced by antitubulin chemotherapeutics. Cell Death Differ 23, 1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cui J & Placzek WJ (2018) Post‐transcriptional regulation of anti‐apoptotic BCL2 family members. Int J Mol Sci 19, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fritsch RM, Schneider G, Saur D, Scheibel M & Schmid RM (2007) Translational repression of MCL‐1 couples stress‐induced eIF2 alpha phosphorylation to mitochondrial apoptosis initiation. J Biol Chem 282, 22551–22562. [DOI] [PubMed] [Google Scholar]
- 40. Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT et al. (2008) mTORC1 promotes survival through translational control of Mcl‐1. Proc Natl Acad Sci USA 105, 10853–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pradelli LA, Beneteau M, Chauvin C, Jacquin MA, Marchetti S, Munoz‐Pinedo C, Auberger P, Pende M & Ricci JE (2010) Glycolysis inhibition sensitizes tumor cells to death receptors‐induced apoptosis by AMP kinase activation leading to Mcl‐1 block in translation. Oncogene 29, 1641–1652. [DOI] [PubMed] [Google Scholar]
- 42. Stewart DP, Koss B, Bathina M, Perciavalle RM, Bisanz K & Opferman JT (2010) Ubiquitin‐independent degradation of antiapoptotic MCL‐1. Mol Cell Biol 30, 3099–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Asher G, Reuven N & Shaul Y (2006) 20S proteasomes and protein degradation “by default”. BioEssays 28, 844–849. [DOI] [PubMed] [Google Scholar]
- 44. Liu H, Peng HW, Cheng YS, Yuan HS & Yang‐Yen HF (2005) Stabilization and enhancement of the antiapoptotic activity of mcl‐1 by TCTP. Mol Cell Biol 25, 3117–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong Q, Gao W, Du F & Wang X (2005) Mule/ARF‐BP1, a BH3‐only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl‐1 and regulates apoptosis. Cell 121, 1085–1095. [DOI] [PubMed] [Google Scholar]
- 46. Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, Lee DF, Liu JC, Zhong Q, Wang X et al. (2007) Degradation of Mcl‐1 by beta‐TrCP mediates glycogen synthase kinase 3‐induced tumor suppression and chemosensitization. Mol Cell Biol 27, 4006–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choi YB & Harhaj EW (2014) HTLV‐1 tax stabilizes MCL‐1 via TRAF6‐dependent K63‐linked polyubiquitination to promote cell survival and transformation. PLoS Pathog 10, e1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thomas LW, Lam C & Edwards SW (2010) Mcl‐1; the molecular regulation of protein function. FEBS Lett 584, 2981–2989. [DOI] [PubMed] [Google Scholar]
- 49. Kobayashi S, Lee SH, Meng XW, Mott JL, Bronk SF, Werneburg NW, Craig RW, Kaufmann SH & Gores GJ (2007) Serine 64 phosphorylation enhances the antiapoptotic function of Mcl‐1. J Biol Chem 282, 18407–18417. [DOI] [PubMed] [Google Scholar]
- 50. Hailey DW, Rambold AS, Satpute‐Krishnan P, Mitra K, Sougrat R, Kim PK & Lippincott‐Schwartz J (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clohessy JG, Zhuang J & Brady HJ (2004) Characterisation of Mcl‐1 cleavage during apoptosis of haematopoietic cells. Br J Haematol 125, 655–665. [DOI] [PubMed] [Google Scholar]
- 52. Han J, Goldstein LA, Gastman BR, Rabinovitz A & Rabinowich H (2005) Disruption of Mcl‐1.Bim complex in granzyme B‐mediated mitochondrial apoptosis. J Biol Chem 280, 16383–16392. [DOI] [PubMed] [Google Scholar]
- 53. Wang T, Yang Z, Zhang Y, Zhang X, Wang L, Zhang S & Jia L (2018) Caspase cleavage of Mcl‐1 impairs its anti‐apoptotic activity and proteasomal degradation in non‐small lung cancer cells. Apoptosis 23, 54–64. [DOI] [PubMed] [Google Scholar]
- 54. Ashkenazi A & Dixit VM (1998) Death receptors: signaling and modulation. Science 281, 1305–1308. [DOI] [PubMed] [Google Scholar]
- 55. Birkinshaw RW & Czabotar PE (2017) The BCL‐2 family of proteins and mitochondrial outer membrane permeabilisation. Semin Cell Dev Biol 72, 152–162. [DOI] [PubMed] [Google Scholar]
- 56. Hockenbery D, Nunez G, Milliman C, Schreiber RD & Korsmeyer SJ (1990) Bcl‐2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature 348, 334–336. [DOI] [PubMed] [Google Scholar]
- 57. Suzuki M, Youle RJ & Tjandra N (2000) Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 103, 645–654. [DOI] [PubMed] [Google Scholar]
- 58. Denisov AY, Madiraju MS, Chen G, Khadir A, Beauparlant P, Attardo G, Shore GC & Gehring K (2003) Solution structure of human BCL‐w: modulation of ligand binding by the C‐terminal helix. J Biol Chem 278, 21124–21128. [DOI] [PubMed] [Google Scholar]
- 59. Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM & Huang DC (2005) Proapoptotic Bak is sequestered by Mcl‐1 and Bcl‐xL, but not Bcl‐2, until displaced by BH3‐only proteins. Genes Dev 19, 1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi YX, Sneed T et al. (2006) Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT‐737 in acute myeloid leukemia. Cancer Cell 10, 375–388. [DOI] [PubMed] [Google Scholar]
- 61. Zhai D, Jin C, Huang Z, Satterthwait AC & Reed JC (2008) Differential regulation of Bax and Bak by anti‐apoptotic Bcl‐2 family proteins Bcl‐B and Mcl‐1. J Biol Chem 283, 9580–9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moulding DA, Giles RV, Spiller DG, White MR, Tidd DM & Edwards SW (2000) Apoptosis is rapidly triggered by antisense depletion of MCL‐1 in differentiating U937 cells. Blood 96, 1756–1763. [PubMed] [Google Scholar]
- 63. Wang C & Youle RJ (2012) Predominant requirement of Bax for apoptosis in HCT116 cells is determined by Mcl‐1's inhibitory effect on Bak. Oncogene 31, 3177–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gelinas C & White E (2005) BH3‐only proteins in control: specificity regulates MCL‐1 and BAK‐mediated apoptosis. Genes Dev 19, 1263–1268. [DOI] [PubMed] [Google Scholar]
- 65. Hsu SY, Kaipia A, McGee E, Lomeli M & Hsueh AJ (1997) Bok is a pro‐apoptotic Bcl‐2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti‐apoptotic Bcl‐2 family members. Proc Natl Acad Sci USA 94, 12401–12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Inohara N, Ekhterae D, Garcia I, Carrio R, Merino J, Merry A, Chen S & Nunez G (1998) Mtd, a novel Bcl‐2 family member activates apoptosis in the absence of heterodimerization with Bcl‐2 and Bcl‐XL. J Biol Chem 273, 8705–8710. [DOI] [PubMed] [Google Scholar]
- 67. Lucendo E, Sancho M, Lolicato F, Javanainen M, Kulig W, Leiva D, Duart G, Andreu‐Fernandez V, Mingarro I & Orzaez M (2020) Mcl‐1 and Bok transmembrane domains: unexpected players in the modulation of apoptosis. Proc Natl Acad Sci USA 117, 27980–27988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stehle D, Grimm M, Einsele‐Scholz S, Ladwig F, Johanning J, Fischer G, Gillissen B, Schulze‐Osthoff K & Essmann F (2018) Contribution of BH3‐domain and transmembrane‐domain to the activity and interaction of the pore‐forming Bcl‐2 proteins Bok, Bak, and Bax. Sci Rep 8, 12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Day CL, Smits C, Fan FC, Lee EF, Fairlie WD & Hinds MG (2008) Structure of the BH3 domains from the p53‐inducible BH3‐only proteins Noxa and Puma in complex with Mcl‐1. J Mol Biol 380, 958–971. [DOI] [PubMed] [Google Scholar]
- 70. Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA & Letai A (2006) Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL‐2 family members. Cancer Cell 9, 351–365. [DOI] [PubMed] [Google Scholar]
- 71. Ku B, Liang C, Jung JU & Oh BH (2011) Evidence that inhibition of BAX activation by BCL‐2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res 21, 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Huang K, O'Neill KL, Li J, Zhou W, Han N, Pang X, Wu W, Struble L, Borgstahl G, Liu Z et al. (2019) BH3‐only proteins target BCL‐xL/MCL‐1, not BAX/BAK, to initiate apoptosis. Cell Res 29, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Senft D, Weber A, Saathoff F, Berking C, Heppt MV, Kammerbauer C, Rothenfusser S, Kellner S, Kurgyis Z, Besch R et al. (2015) In non‐transformed cells Bak activates upon loss of anti‐apoptotic Bcl‐XL and Mcl‐1 but in the absence of active BH3‐only proteins. Cell Death Dis 6, e1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gomez‐Bougie P, Menoret E, Juin P, Dousset C, Pellat‐Deceunynck C & Amiot M (2011) Noxa controls Mule‐dependent Mcl‐1 ubiquitination through the regulation of the Mcl‐1/USP9X interaction. Biochem Biophys Res Commun 413, 460–464. [DOI] [PubMed] [Google Scholar]
- 75. Bleicken S, Hantusch A, Das KK, Frickey T & Garcia‐Saez AJ (2017) Quantitative interactome of a membrane Bcl‐2 network identifies a hierarchy of complexes for apoptosis regulation. Nat Commun 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Andreu‐Fernandez V, Sancho M, Genoves A, Lucendo E, Todt F, Lauterwasser J, Funk K, Jahreis G, Perez‐Paya E, Mingarro I et al. (2017) Bax transmembrane domain interacts with prosurvival Bcl‐2 proteins in biological membranes. Proc Natl Acad Sci USA 114, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Todt F, Cakir Z, Reichenbach F, Youle RJ & Edlich F (2013) The C‐terminal helix of Bcl‐x(L) mediates Bax retrotranslocation from the mitochondria. Cell Death Differ 20, 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rinkenberger JL, Horning S, Klocke B, Roth K & Korsmeyer SJ (2000) Mcl‐1 deficiency results in peri‐implantation embryonic lethality. Genes Dev 14, 23–27. [PMC free article] [PubMed] [Google Scholar]
- 79. Fogarty LC, Flemmer RT, Geizer BA, Licursi M, Karunanithy A, Opferman JT, Hirasawa K & Vanderluit JL (2019) Mcl‐1 and Bcl‐xL are essential for survival of the developing nervous system. Cell Death Differ 26, 1501–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jain R, Sheridan JM, Policheni A, Heinlein M, Gandolfo LC, Dewson G, Smyth GK, Sansom SN, Fu NY, Visvader JE et al. (2017) A critical epithelial survival axis regulated by MCL‐1 maintains thymic function in mice. Blood 130, 2504–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Turnis ME, Kaminska E, Smith KH, Kartchner BJ, Vogel P, Laxton JD, Ashmun RA, Ney PA & Opferman JT (2021) Requirement for antiapoptotic MCL‐1 during early erythropoiesis. Blood 137, 1945–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dzhagalov I, St John A & He YW (2007) The antiapoptotic protein Mcl‐1 is essential for the survival of neutrophils but not macrophages. Blood 109, 1620–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kodama T, Hikita H, Kawaguchi T, Shigekawa M, Shimizu S, Hayashi Y, Li W, Miyagi T, Hosui A, Tatsumi T et al. (2012) Mcl‐1 and Bcl‐xL regulate Bak/Bax‐dependent apoptosis of the megakaryocytic lineage at multistages. Cell Death Differ 19, 1856–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Opferman JT, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K & Korsmeyer SJ (2005) Obligate role of anti‐apoptotic MCL‐1 in the survival of hematopoietic stem cells. Science 307, 1101–1104. [DOI] [PubMed] [Google Scholar]
- 85. Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC & Korsmeyer SJ (2003) Development and maintenance of B and T lymphocytes requires antiapoptotic MCL‐1. Nature 426, 671–676. [DOI] [PubMed] [Google Scholar]
- 86. Rasmussen ML, Kline LA, Park KP, Ortolano NA, Romero‐Morales AI, Anthony CC, Beckermann KE & Gama V (2018) A non‐apoptotic function of MCL‐1 in promoting pluripotency and modulating mitochondrial dynamics in stem cells. Stem Cell Reports 10, 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Perciavalle RM, Stewart DP, Koss B, Lynch J, Milasta S, Bathina M, Temirov J, Cleland MM, Pelletier S, Schuetz JD et al. (2012) Anti‐apoptotic MCL‐1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol 14, 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Germain M & Duronio V (2007) The N terminus of the anti‐apoptotic BCL‐2 homologue MCL‐1 regulates its localization and function. J Biol Chem 282, 32233–32242. [DOI] [PubMed] [Google Scholar]
- 89. Huang CR & Yang‐Yen HF (2010) The fast‐mobility isoform of mouse Mcl‐1 is a mitochondrial matrix‐localized protein with attenuated anti‐apoptotic activity. FEBS Lett 584, 3323–3330. [DOI] [PubMed] [Google Scholar]
- 90. Escudero S, Zaganjor E, Lee S, Mill CP, Morgan AM, Crawford EB, Chen J, Wales TE, Mourtada R, Luccarelli J et al. (2018) Dynamic regulation of long‐chain fatty acid oxidation by a noncanonical interaction between the MCL‐1 BH3 helix and VLCAD. Mol Cell 69, 729–743.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Coronado M, Fajardo G, Nguyen K, Zhao M, Kooiker K, Jung G, Hu DQ, Reddy S, Sandoval E, Stotland A et al. (2018) Physiological mitochondrial fragmentation is a normal cardiac adaptation to increased energy demand. Circ Res 122, 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Youle RJ & van der Bliek AM (2012) Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Moyzis AG, Lally NS, Liang W, Leon LJ, Najor RH, Orogo AM & Gustafsson AB (2020) Mcl‐1‐mediated mitochondrial fission protects against stress but impairs cardiac adaptation to exercise. J Mol Cell Cardiol 146, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Guo L, Eldridge S, Furniss M, Mussio J & Davis M (2018) Role of Mcl‐1 in regulation of cell death in human induced pluripotent stem cell‐derived cardiomyocytes in vitro. Toxicol Appl Pharmacol 360, 88–98. [DOI] [PubMed] [Google Scholar]
- 95. Wang X, Bathina M, Lynch J, Koss B, Calabrese C, Frase S, Schuetz JD, Rehg JE & Opferman JT (2013) Deletion of MCL‐1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev 27, 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Eckenrode EF, Yang J, Velmurugan GV, Foskett JK & White C (2010) Apoptosis protection by Mcl‐1 and Bcl‐2 modulation of inositol 1,4,5‐trisphosphate receptor‐dependent Ca2+ signaling. J Biol Chem 285, 13678–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Huang H, Shah K, Bradbury NA, Li C & White C (2014) Mcl‐1 promotes lung cancer cell migration by directly interacting with VDAC to increase mitochondrial Ca2+ uptake and reactive oxygen species generation. Cell Death Dis 5, e1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. De Stefani D, Bononi A, Romagnoli A, Messina A, De Pinto V, Pinton P & Rizzuto R (2012) VDAC1 selectively transfers apoptotic Ca2+ signals to mitochondria. Cell Death Differ 19, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cen X, Chen Y, Xu X, Wu R, He F, Zhao Q, Sun Q, Yi C, Wu J, Najafov A et al. (2020) Pharmacological targeting of MCL‐1 promotes mitophagy and improves disease pathologies in an Alzheimer's disease mouse model. Nat Commun 11, 5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cen X, Xu X & Xia H (2021) Targeting MCL1 to induce mitophagy is a potential therapeutic strategy for Alzheimer disease. Autophagy 17, 818–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi‐Harel S, Hirsch JA, Stein R & Pinkas‐Kramarski R (2007) Differential interactions between Beclin 1 and Bcl‐2 family members. Autophagy 3, 561–568. [DOI] [PubMed] [Google Scholar]
- 102. Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G & Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3‐phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS, Opferman JT & Slack RS (2011) MCL‐1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J 30, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kalkat M, Garcia J, Ebrahimi J, Melland‐Smith M, Todros T, Post M & Caniggia I (2013) Placental autophagy regulation by the BOK‐MCL1 rheostat. Autophagy 9, 2140–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Elgendy M, Ciro M, Abdel‐Aziz AK, Belmonte G, Dal Zuffo R, Mercurio C, Miracco C, Lanfrancone L, Foiani M & Minucci S (2014) Beclin 1 restrains tumorigenesis through Mcl‐1 destabilization in an autophagy‐independent reciprocal manner. Nat Commun 5, 5637. [DOI] [PubMed] [Google Scholar]
- 106. Lindqvist LM, Heinlein M, Huang DC & Vaux DL (2014) Prosurvival Bcl‐2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc Natl Acad Sci USA 111, 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Harley ME, Allan LA, Sanderson HS & Clarke PR (2010) Phosphorylation of Mcl‐1 by CDK1‐cyclin B1 initiates its Cdc20‐dependent destruction during mitotic arrest. EMBO J 29, 2407–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jamil S, Mojtabavi S, Hojabrpour P, Cheah S & Duronio V (2008) An essential role for MCL‐1 in ATR‐mediated CHK1 phosphorylation. Mol Biol Cell 19, 3212–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pawlikowska P, Leray I, de Laval B, Guihard S, Kumar R, Rosselli F & Porteu F (2010) ATM‐dependent expression of IEX‐1 controls nuclear accumulation of Mcl‐1 and the DNA damage response. Cell Death Differ 17, 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jamil S, Sobouti R, Hojabrpour P, Raj M, Kast J & Duronio V (2005) A proteolytic fragment of Mcl‐1 exhibits nuclear localization and regulates cell growth by interaction with Cdk1. Biochem J 387, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Clarke PR, Allan LA & Skowyra A (2018) Timed degradation of Mcl‐1 controls mitotic cell death. Mol Cell Oncol 5, e1516450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Streletskaia AY, Senichkin VV, Prikazchikova TA, Zatsepin TS, Zhivotovsky B & Kopeina GS (2020) Upregulation of Mcl‐1S causes cell‐cycle perturbations and DNA damage accumulation. Front Cell Dev Biol 8, 543066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Whitaker RH & Placzek WJ (2020) MCL1 binding to the reverse BH3 motif of P18INK4C couples cell survival to cell proliferation. Cell Death Dis 11, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hasan SM, Sheen AD, Power AM, Langevin LM, Xiong J, Furlong M, Day K, Schuurmans C, Opferman JT & Vanderluit JL (2013) Mcl1 regulates the terminal mitosis of neural precursor cells in the mammalian brain through p27Kip1. Development 140, 3118–3127. [DOI] [PubMed] [Google Scholar]
- 115. Levrero M, De Laurenzi V, Costanzo A, Gong J, Melino G & Wang JY (1999) Structure, function and regulation of p63 and p73. Cell Death Differ 6, 1146–1153. [DOI] [PubMed] [Google Scholar]
- 116. Widden H, Kaczmarczyk A, Subedi A, Whitaker RH & Placzek WJ (2020) MCL1 binds and negatively regulates the transcriptional function of tumor suppressor p73. Cell Death Dis 11, 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Demelash A, Pfannenstiel LW, Liu L & Gastman BR (2017) Mcl‐1 regulates reactive oxygen species via NOX4 during chemotherapy‐induced senescence. Oncotarget 8, 28154–28168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Frieler RA, Chung Y, Ahlers CG, Gheordunescu G, Song J, Vigil TM, Shah YM & Mortensen RM (2017) Genetic neutrophil deficiency ameliorates cerebral ischemia‐reperfusion injury. Exp Neurol 298, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Felton JM, Dorward DA, Cartwright JA, Potey PM, Robb CT, Gui J, Craig RW, Schwarze J, Haslett C, Duffin R et al. (2020) Mcl‐1 protects eosinophils from apoptosis and exacerbates allergic airway inflammation. Thorax 75, 600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Meyerovich K, Violato NM, Fukaya M, Dirix V, Pachera N, Marselli L, Marchetti P, Strasser A, Eizirik DL & Cardozo AK (2017) MCL‐1 is a key antiapoptotic protein in human and rodent pancreatic beta‐cells. Diabetes 66, 2446–2458. [DOI] [PubMed] [Google Scholar]
- 121. Hikita H, Kodama T, Shimizu S, Li W, Shigekawa M, Tanaka S, Hosui A, Miyagi T, Tatsumi T, Kanto T et al. (2012) Bak deficiency inhibits liver carcinogenesis: a causal link between apoptosis and carcinogenesis. J Hepatol 57, 92–100. [DOI] [PubMed] [Google Scholar]
- 122. Hikita H, Takehara T, Shimizu S, Kodama T, Li W, Miyagi T, Hosui A, Ishida H, Ohkawa K, Kanto T et al. (2009) Mcl‐1 and Bcl‐xL cooperatively maintain integrity of hepatocytes in developing and adult murine liver. Hepatology 50, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Nozaki Y, Hikita H, Tanaka S, Fukumoto K, Urabe M, Sato K, Myojin Y, Doi A, Murai K, Sakane S et al. (2021) Persistent hepatocyte apoptosis promotes tumorigenesis from diethylnitrosamine‐transformed hepatocytes through increased oxidative stress, independent of compensatory liver regeneration. Sci Rep 11, 3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Munkhbaatar E, Dietzen M, Agrawal D, Anton M, Jesinghaus M, Boxberg M, Pfarr N, Bidola P, Uhrig S, Hockendorf U et al. (2020) MCL‐1 gains occur with high frequency in lung adenocarcinoma and can be targeted therapeutically. Nat Commun 11, 4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M et al. (2010) The landscape of somatic copy‐number alteration across human cancers. Nature 463, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pennarun B, Kleibeuker JH, Boersma‐van Ek W, Kruyt FA, Hollema H, de Vries EG & de Jong S (2013) Targeting FLIP and Mcl‐1 using a combination of aspirin and sorafenib sensitizes colon cancer cells to TRAIL. J Pathol 229, 410–421. [DOI] [PubMed] [Google Scholar]
- 127. Henderson‐Jackson EB, Helm J, Ghayouri M, Hakam A, Nasir A, Leon M, Bui M, Yeatman T & Coppola D (2010) Correlation between Mcl‐1 and pAKT protein expression in colorectal cancer. Int J Clin Exp Pathol 3, 768–774. [PMC free article] [PubMed] [Google Scholar]
- 128. Backus HH, van Riel JM, van Groeningen CJ, Vos W, Dukers DF, Bloemena E, Wouters D, Pinedo HM & Peters GJ (2001) Rb, mcl‐1 and p53 expression correlate with clinical outcome in patients with liver metastases from colorectal cancer. Ann Oncol 12, 779–785. [DOI] [PubMed] [Google Scholar]
- 129. Shigemasa K, Katoh O, Shiroyama Y, Mihara S, Mukai K, Nagai N & Ohama K (2002) Increased MCL‐1 expression is associated with poor prognosis in ovarian carcinomas. Jpn J Cancer Res 93, 542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Likui W, Qun L, Wanqing Z, Haifeng S, Fangqiu L & Xiaojun L (2009) Prognostic role of myeloid cell leukemia‐1 protein (Mcl‐1) expression in human gastric cancer. J Surg Oncol 100, 396–400. [DOI] [PubMed] [Google Scholar]
- 131. Maeta Y, Tsujitani S, Matsumoto S, Yamaguchi K, Tatebe S, Kondo A, Ikeguchi M & Kaibara N (2004) Expression of Mcl‐1 and p53 proteins predicts the survival of patients with T3 gastric carcinoma. Gastric Cancer 7, 78–84. [DOI] [PubMed] [Google Scholar]
- 132. Wuilleme‐Toumi S, Robillard N, Gomez P, Moreau P, Le Gouill S, Avet‐Loiseau H, Harousseau JL, Amiot M & Bataille R (2005) Mcl‐1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia 19, 1248–1252. [DOI] [PubMed] [Google Scholar]
- 133. Nakano T, Go T, Nakashima N, Liu D & Yokomise H (2020) Overexpression of antiapoptotic MCL‐1 predicts worse overall survival of patients with non‐small cell lung cancer. Anticancer Res 40, 1007–1014. [DOI] [PubMed] [Google Scholar]
- 134. Wen Q, Zhan Y, Zheng H, Zang H, Luo J, Zhang Y, Wang W, Feng J, Lu J, Chen L et al. (2019) Elevated expression of mcl‐1 inhibits apoptosis and predicts poor prognosis in patients with surgically resected non‐small cell lung cancer. Diagn Pathol 14, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chen G, Park D, Magis AT, Behera M, Ramalingam SS, Owonikoko TK, Sica GL, Ye K, Zhang C, Chen Z et al. (2019) Mcl‐1 interacts with Akt to promote lung cancer progression. Cancer Res 79, 6126–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wong RP, Khosravi S, Martinka M & Li G (2008) Myeloid leukemia‐1 expression in benign and malignant melanocytic lesions. Oncol Rep 19, 933–937. [PubMed] [Google Scholar]
- 137. Chetoui N, Sylla K, Gagnon‐Houde JV, Alcaide‐Loridan C, Charron D, Al‐Daccak R & Aoudjit F (2008) Down‐regulation of mcl‐1 by small interfering RNA sensitizes resistant melanoma cells to fas‐mediated apoptosis. Mol Cancer Res 6, 42–52. [DOI] [PubMed] [Google Scholar]
- 138. Sale MJ, Minihane E, Monks NR, Gilley R, Richards FM, Schifferli KP, Andersen CL, Davies EJ, Vicente MA, Ozono E et al. (2019) Targeting melanoma's MCL1 bias unleashes the apoptotic potential of BRAF and ERK1/2 pathway inhibitors. Nat Commun 10, 5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Thallinger C, Wolschek MF, Wacheck V, Maierhofer H, Gunsberg P, Polterauer P, Pehamberger H, Monia BP, Selzer E, Wolff K et al. (2003) Mcl‐1 antisense therapy chemosensitizes human melanoma in a SCID mouse xenotransplantation model. J Invest Dermatol 120, 1081–1086. [DOI] [PubMed] [Google Scholar]
- 140. Wolter KG, Verhaegen M, Fernandez Y, Nikolovska‐Coleska Z, Riblett M, de la Vega CM, Wang S & Soengas MS (2007) Therapeutic window for melanoma treatment provided by selective effects of the proteasome on Bcl‐2 proteins. Cell Death Differ 14, 1605–1616. [DOI] [PubMed] [Google Scholar]
- 141. Ding Q, He X, Xia W, Hsu JM, Chen CT, Li LY, Lee DF, Yang JY, Xie X, Liu JC et al. (2007) Myeloid cell leukemia‐1 inversely correlates with glycogen synthase kinase‐3beta activity and associates with poor prognosis in human breast cancer. Cancer Res 67, 4564–4571. [DOI] [PubMed] [Google Scholar]
- 142. Goodwin CM, Rossanese OW, Olejniczak ET & Fesik SW (2015) Myeloid cell leukemia‐1 is an important apoptotic survival factor in triple‐negative breast cancer. Cell Death Differ 22, 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Petry IB, Fieber E, Schmidt M, Gehrmann M, Gebhard S, Hermes M, Schormann W, Selinski S, Freis E, Schwender H et al. (2010) ERBB2 induces an antiapoptotic expression pattern of Bcl‐2 family members in node‐negative breast cancer. Clin Cancer Res 16, 451–460. [DOI] [PubMed] [Google Scholar]
- 144. Booy EP, Henson ES & Gibson SB (2011) Epidermal growth factor regulates Mcl‐1 expression through the MAPK‐Elk‐1 signalling pathway contributing to cell survival in breast cancer. Oncogene 30, 2367–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Carpenter RL & Lo HW (2013) Regulation of apoptosis by HER2 in breast cancer. J Carcinog Mutagen 2013, 3. 10.4172/2157-2518.S7-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Schacter JL, Henson ES & Gibson SB (2014) Estrogen regulation of anti‐apoptotic Bcl‐2 family member Mcl‐1 expression in breast cancer cells. PLoS One 9, e100364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Floros KV, Jacob S, Kurupi R, Fairchild CK, Hu B, Puchalapalli M, Koblinski JE, Dozmorov MG, Boikos SA, Scaltriti M et al. (2021) Targeting transcription of MCL‐1 sensitizes HER2‐amplified breast cancers to HER2 inhibitors. Cell Death Dis 12, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Henson ES, Hu X & Gibson SB (2006) Herceptin sensitizes ErbB2‐overexpressing cells to apoptosis by reducing antiapoptotic Mcl‐1 expression. Clin Cancer Res 12, 845–853. [DOI] [PubMed] [Google Scholar]
- 149. Thrane S, Pedersen AM, Thomsen MB, Kirkegaard T, Rasmussen BB, Duun‐Henriksen AK, Laenkholm AV, Bak M, Lykkesfeldt AE & Yde CW (2015) A kinase inhibitor screen identifies Mcl‐1 and Aurora kinase A as novel treatment targets in antiestrogen‐resistant breast cancer cells. Oncogene 34, 4199–4210. [DOI] [PubMed] [Google Scholar]
- 150. Ye T, Wei X, Yin T, Xia Y, Li D, Shao B, Song X, He S, Luo M, Gao X et al. (2014) Inhibition of FGFR signaling by PD173074 improves antitumor immunity and impairs breast cancer metastasis. Breast Cancer Res Treat 143, 435–446. [DOI] [PubMed] [Google Scholar]
- 151. Merino D, Whittle JR, Vaillant F, Serrano A, Gong JN, Giner G, Maragno AL, Chanrion M, Schneider E, Pal B et al. (2017) Synergistic action of the MCL‐1 inhibitor S63845 with current therapies in preclinical models of triple‐negative and HER2‐amplified breast cancer. Sci Transl Med 9, eaam7049. [DOI] [PubMed] [Google Scholar]
- 152. Weber A, Boger R, Vick B, Urbanik T, Haybaeck J, Zoller S, Teufel A, Krammer PH, Opferman JT, Galle PR et al. (2010) Hepatocyte‐specific deletion of the antiapoptotic protein myeloid cell leukemia‐1 triggers proliferation and hepatocarcinogenesis in mice. Hepatology 51, 1226–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sieghart W, Losert D, Strommer S, Cejka D, Schmid K, Rasoul‐Rockenschaub S, Bodingbauer M, Crevenna R, Monia BP, Peck‐Radosavljevic M et al. (2006) Mcl‐1 overexpression in hepatocellular carcinoma: a potential target for antisense therapy. J Hepatol 44, 151–157. [DOI] [PubMed] [Google Scholar]
- 154. Zhang H, Li G, Chen G, Zhang Y, Pan J, Tang H, Li J, Guo W & Zhang S (2019) Targeting Mcl‐1 inhibits survival and self‐renewal of hepatocellular cancer stem‐like cells. Clin Res Hepatol Gastroenterol 43, 292–300. [DOI] [PubMed] [Google Scholar]
- 155. Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin‐Braizat G, Chanrion M, Kelly GL, Gong JN, Moujalled DM et al. (2016) The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482. [DOI] [PubMed] [Google Scholar]
- 156. Szlavik Z, Csekei M, Paczal A, Szabo ZB, Sipos S, Radics G, Proszenyak A, Balint B, Murray J, Davidson J et al. (2020) Discovery of S64315, a potent and selective Mcl‐1 inhibitor. J Med Chem 63, 13762–13795. [DOI] [PubMed] [Google Scholar]
- 157. Murray JB, Davidson J, Chen I, Davis B, Dokurno P, Graham CJ, Harris R, Jordan A, Matassova N, Pedder C et al. (2019) Establishing drug discovery and identification of hit series for the anti‐apoptotic proteins, Bcl‐2 and Mcl‐1. ACS Omega 4, 8892–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Brennan MS, Chang C, Tai L, Lessene G, Strasser A, Dewson G, Kelly GL & Herold MJ (2018) Humanized Mcl‐1 mice enable accurate preclinical evaluation of MCL‐1 inhibitors destined for clinical use. Blood 132, 1573–1583. [DOI] [PubMed] [Google Scholar]
- 159. Caenepeel S, Brown SP, Belmontes B, Moody G, Keegan KS, Chui D, Whittington DA, Huang X, Poppe L, Cheng AC et al. (2018) AMG 176, a selective MCL1 inhibitor, is effective in hematologic cancer models alone and in combination with established therapies. Cancer Discov 8, 1582–1597. [DOI] [PubMed] [Google Scholar]
- 160. Spencer A, Seth Rosenberg A, Jakubowiak A, Raje N, Chatterjee M, Trudel S, Bahlis NJ, Siegel DS, Wilop S, Harrison SJ et al. (2019) A phase 1, first‐in‐human study of AMG 176, a selective MCL‐1 inhibitor, in patients with relapsed or refractory multiple myeloma. Clin Lymphoma Myeloma Leuk 19, 53–54.30301673 [Google Scholar]
- 161. Friberg A, Vigil D, Zhao B, Daniels RN, Burke JP, Garcia‐Barrantes PM, Camper D, Chauder BA, Lee T, Olejniczak ET et al. (2013) Discovery of potent myeloid cell leukemia 1 (Mcl‐1) inhibitors using fragment‐based methods and structure‐based design. J Med Chem 56, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Koch R, Christie AL, Crombie JL, Palmer AC, Plana D, Shigemori K, Morrow SN, Van Scoyk A, Wu W, Brem EA et al. (2019) Biomarker‐driven strategy for MCL1 inhibition in T‐cell lymphomas. Blood 133, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Song X, Shen L, Tong J, Kuang C, Zeng S, Schoen RE, Yu J, Pei H & Zhang L (2020) Mcl‐1 inhibition overcomes intrinsic and acquired regorafenib resistance in colorectal cancer. Theranostics 10, 8098–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Ramsey HE, Fischer MA, Lee T, Gorska AE, Arrate MP, Fuller L, Boyd KL, Strickland SA, Sensintaffar J, Hogdal LJ et al. (2018) A novel MCL1 inhibitor combined with venetoclax rescues venetoclax‐resistant acute myelogenous leukemia. Cancer Discov 8, 1566–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Lee T, Christov PP, Shaw S, Tarr JC, Zhao B, Veerasamy N, Jeon KO, Mills JJ, Bian Z, Sensintaffar JL et al. (2019) Discovery of potent myeloid cell leukemia‐1 (Mcl‐1) inhibitors that demonstrate in vivo activity in mouse xenograft models of human cancer. J Med Chem 62, 3971–3988. [DOI] [PubMed] [Google Scholar]
- 166. Lu X, Liu YC, Orvig C, Liang H & Chen ZF (2020) Discovery of a copper‐based Mcl‐1 inhibitor as an effective antitumor agent. J Med Chem 63, 9154–9167. [DOI] [PubMed] [Google Scholar]
- 167. Cidado J, Boiko S, Proia T, Ferguson D, Criscione SW, San Martin M, Pop‐Damkov P, Su N, Roamio Franklin VN, Chilamakuri SR et al. (2020) AZD4573 is a highly selective CDK9 inhibitor that suppresses MCL‐1 and induces apoptosis in hematologic cancer cells. Clin Cancer Res 26, 922–934. [DOI] [PubMed] [Google Scholar]
- 168. Dey J, Deckwerth TL, Kerwin WS, Casalini JR, Merrell AJ, Grenley MO, Burns C, Ditzler SH, Dixon CP, Beirne E et al. (2017) Voruciclib, a clinical stage oral CDK9 inhibitor, represses MCL‐1 and sensitizes high‐risk Diffuse Large B‐cell Lymphoma to BCL2 inhibition. Sci Rep 7, 18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lin CL, Chen CM, Lin CL, Cheng CW, Lee CH & Hsieh YH (2017) Norcantharidin induces mitochondrial‐dependent apoptosis through Mcl‐1 inhibition in human prostate cancer cells. Biochim Biophys Acta Mol Cell Res 1864, 1867–1876. [DOI] [PubMed] [Google Scholar]
- 170. Aliabadi HM, Mahdipoor P & Uludag H (2013) Polymeric delivery of siRNA for dual silencing of Mcl‐1 and P‐glycoprotein and apoptosis induction in drug‐resistant breast cancer cells. Cancer Gene Ther 20, 169–177. [DOI] [PubMed] [Google Scholar]
- 171. Wang Z, He N, Guo Z, Niu C, Song T, Guo Y, Cao K, Wang A, Zhu J, Zhang X et al. (2019) Proteolysis targeting chimeras for the selective degradation of Mcl‐1/Bcl‐2 derived from nonselective target binding ligands. J Med Chem 62, 8152–8163. [DOI] [PubMed] [Google Scholar]
- 172. Pannecoucke E, Van Trimpont M, Desmet J, Pieters T, Reunes L, Demoen L, Vuylsteke M, Loverix S, Vandenbroucke K, Alard P et al. (2021) Cell‐penetrating Alphabody protein scaffolds for intracellular drug targeting. Sci Adv 7, eabe1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Mott JL, Kobayashi S, Bronk SF & Gores GJ (2007) mir‐29 regulates Mcl‐1 protein expression and apoptosis. Oncogene 26, 6133–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Wang H, Wang L, Zhang G, Lu C, Chu H, Yang R & Zhao G (2018) MALAT1/miR‐101‐3p/MCL1 axis mediates cisplatin resistance in lung cancer. Oncotarget 9, 7501–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Wang SH, Zhang WJ, Wu XC, Weng MZ, Zhang MD, Cai Q, Zhou D, Wang JD & Quan ZW (2016) The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL‐1 expression by sponging miR‐363‐3p in gallbladder cancer. J Cell Mol Med 20, 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Li XH, Ha CT & Xiao M (2016) MicroRNA‐30 inhibits antiapoptotic factor Mcl‐1 in mouse and human hematopoietic cells after radiation exposure. Apoptosis 21, 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Liu B, Cao W & Xue J (2019) LncRNA ANRIL protects against oxygen and glucose deprivation (OGD)‐induced injury in PC‐12 cells: potential role in ischaemic stroke. Artif Cells Nanomed Biotechnol 47, 1384–1395. [DOI] [PubMed] [Google Scholar]
- 178. Liu X, Tang H, Chen J, Song C, Yang L, Liu P, Wang N, Xie X, Lin X & Xie X (2015) MicroRNA‐101 inhibits cell progression and increases paclitaxel sensitivity by suppressing MCL‐1 expression in human triple‐negative breast cancer. Oncotarget 6, 20070–20083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Chen D, Lu X, Yang F & Xing N (2019) Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR‐193a‐3p and regulating MCL1 expression. Cancer Manag Res 11, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Gong J, Zhang JP, Li B, Zeng C, You K, Chen MX, Yuan Y & Zhuang SM (2013) MicroRNA‐125b promotes apoptosis by regulating the expression of Mcl‐1, Bcl‐w and IL‐6R. Oncogene 32, 3071–3079. [DOI] [PubMed] [Google Scholar]
- 181. Pan Y, Zhang Y, Liu W, Huang Y, Shen X, Jing R, Pu J, Wang X, Ju S, Cong H et al. (2019) LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL‐1 via miR‐29b‐3p. Cell Death Dis 10, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y, Wang Z, Wang Z, Cheng P, Tong D et al. (2013) MicroRNA‐133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl‐xL and Mcl‐1. Bone 56, 220–226. [DOI] [PubMed] [Google Scholar]
- 183. Yin D, Li Y, Fu C & Feng Y (2018) Pro‐angiogenic role of LncRNA HULC in microvascular endothelial cells via sequestrating miR‐124. Cell Physiol Biochem 50, 2188–2202. [DOI] [PubMed] [Google Scholar]
- 184. Crawford M, Batte K, Yu L, Wu X, Nuovo GJ, Marsh CB, Otterson GA & Nana‐Sinkam SP (2009) MicroRNA 133B targets pro‐survival molecules MCL‐1 and BCL2L2 in lung cancer. Biochem Biophys Res Commun 388, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Chen P, Fang X, Xia B, Zhao Y, Li Q & Wu X (2018) Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR‐125b with MCL‐1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med 7, 4530–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Huang Y, Luo H, Li F, Yang Y, Ou G, Ye X & Li N (2018) LINC00152 down‐regulated miR‐193a‐3p to enhance MCL1 expression and promote gastric cancer cells proliferation. Biosci Rep 38, BSR20171607. 10.1042/BSR20171607 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 187. Xu J, Liao X & Wong C (2010) Downregulations of B‐cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG‐05MG. Int J Cancer 126, 1029–1035. [DOI] [PubMed] [Google Scholar]
- 188. Han Y, Wu N, Jiang M, Chu Y, Wang Z, Liu H, Cao J, Liu H, Xu B & Xie X (2019) Long non‐coding RNA MYOSLID functions as a competing endogenous RNA to regulate MCL‐1 expression by sponging miR‐29c‐3p in gastric cancer. Cell Prolif 52, e12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Zimmerman EI, Dollins CM, Crawford M, Grant S, Nana‐Sinkam SP, Richards KL, Hammond SM & Graves LM (2010) Lyn kinase‐dependent regulation of miR181 and myeloid cell leukemia‐1 expression: implications for drug resistance in myelogenous leukemia. Mol Pharmacol 78, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Li X, Yu M, Chen L, Sun T, Wang H, Zhao L & Zhao Q (2019) LncRNA PMS2L2 protects ATDC5 chondrocytes against lipopolysaccharide‐induced inflammatory injury by sponging miR‐203. Life Sci 217, 283–292. [DOI] [PubMed] [Google Scholar]
- 191. Kwon JE, Kim BY, Kwak SY, Bae IH & Han YH (2013) Ionizing radiation‐inducible microRNA miR‐193a‐3p induces apoptosis by directly targeting Mcl‐1. Apoptosis 18, 896–909. [DOI] [PubMed] [Google Scholar]
- 192. Zhou B, Li L, Li Y, Sun H & Zeng C (2018) Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR‐320a/MCL1 axis. Biomed Pharmacother 106, 850–857. [DOI] [PubMed] [Google Scholar]
- 193. Khodayari N, Mohammed KA, Lee H, Kaye F & Nasreen N (2016) MicroRNA‐302b targets Mcl‐1 and inhibits cell proliferation and induces apoptosis in malignant pleural mesothelioma cells. Am J Cancer Res 6, 1996–2009. [PMC free article] [PubMed] [Google Scholar]
- 194. Zhang T, Zou P, Wang T, Xiang J, Cheng J, Chen D & Zhou J (2016) Down‐regulation of miR‐320 associated with cancer progression and cell apoptosis via targeting Mcl‐1 in cervical cancer. Tumour Biol 37, 8931–8940. [DOI] [PubMed] [Google Scholar]
- 195. Saito Y, Suzuki H, Tsugawa H, Nakagawa I, Matsuzaki J, Kanai Y & Hibi T (2009) Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA‐512‐5p with downregulation of Mcl‐1 in human gastric cancer cells. Oncogene 28, 2738–2744. [DOI] [PubMed] [Google Scholar]
- 196. Steimer DA, Boyd K, Takeuchi O, Fisher JK, Zambetti GP & Opferman JT (2009) Selective roles for antiapoptotic MCL‐1 during granulocyte development and macrophage effector function. Blood 113, 2805–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Hikita H, Kodama T, Tanaka S, Saito Y, Nozaki Y, Nakabori T, Shimizu S, Hayashi Y, Li W, Shigekawa M et al. (2015) Activation of the mitochondrial apoptotic pathway produces reactive oxygen species and oxidative damage in hepatocytes that contribute to liver tumorigenesis. Cancer Prev Res (Phila) 8, 693–701. [DOI] [PubMed] [Google Scholar]
- 198. Hikita H, Takehara T, Shimizu S, Kodama T, Shigekawa M, Iwase K, Hosui A, Miyagi T, Tatsumi T, Ishida H et al. (2010) The Bcl‐xL inhibitor, ABT‐737, efficiently induces apoptosis and suppresses growth of hepatoma cells in combination with sorafenib. Hepatology 52, 1310–1321. [DOI] [PubMed] [Google Scholar]
- 199. Kodama T, Hikita H, Kawaguchi T, Saito Y, Tanaka S, Shigekawa M, Shimizu S, Li W, Miyagi T, Kanto T et al. (2013) The Bcl‐2 homology domain 3 (BH3)‐only proteins Bim and bid are functionally active and restrained by anti‐apoptotic Bcl‐2 family proteins in healthy liver. J Biol Chem 288, 30009–30018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Vick B, Weber A, Urbanik T, Maass T, Teufel A, Krammer PH, Opferman JT, Schuchmann M, Galle PR & Schulze‐Bergkamen H (2009) Knockout of myeloid cell leukemia‐1 induces liver damage and increases apoptosis susceptibility of murine hepatocytes. Hepatology 49, 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Campbell KJ, Dhayade S, Ferrari N, Sims AH, Johnson E, Mason SM, Dickson A, Ryan KM, Kalna G, Edwards J et al. (2018) MCL‐1 is a prognostic indicator and drug target in breast cancer. Cell Death Dis 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Peperzak V, Vikstrom I, Walker J, Glaser SP, LePage M, Coquery CM, Erickson LD, Fairfax K, Mackay F, Strasser A et al. (2013) Mcl‐1 is essential for the survival of plasma cells. Nat Immunol 14, 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Glaser SP, Lee EF, Trounson E, Bouillet P, Wei A, Fairlie WD, Izon DJ, Zuber J, Rappaport AR, Herold MJ et al. (2012) Anti‐apoptotic Mcl‐1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev 26, 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Saito Y, Hikita H, Nozaki Y, Kai Y, Makino Y, Nakabori T, Tanaka S, Yamada R, Shigekawa M, Kodama T et al. (2019) DNase II activated by the mitochondrial apoptotic pathway regulates RIP1‐dependent non‐apoptotic hepatocyte death via the TLR9/IFN‐beta signaling pathway. Cell Death Differ 26, 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Thomas RL, Roberts DJ, Kubli DA, Lee Y, Quinsay MN, Owens JB, Fischer KM, Sussman MA, Miyamoto S & Gustafsson AB (2013) Loss of MCL‐1 leads to impaired autophagy and rapid development of heart failure. Genes Dev 27, 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Grabow S, Delbridge AR, Aubrey BJ, Vandenberg CJ & Strasser A (2016) Loss of a single Mcl‐1 allele inhibits MYC‐driven lymphomagenesis by sensitizing pro‐B cells to apoptosis. Cell Rep 14, 2337–2347. [DOI] [PubMed] [Google Scholar]
- 207. Vikstrom I, Carotta S, Luthje K, Peperzak V, Jost PJ, Glaser S, Busslinger M, Bouillet P, Strasser A, Nutt SL et al. (2010) Mcl‐1 is essential for germinal center formation and B cell memory. Science 330, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Kelly GL, Grabow S, Glaser SP, Fitzsimmons L, Aubrey BJ, Okamoto T, Valente LJ, Robati M, Tai L, Fairlie WD et al. (2014) Targeting of MCL‐1 kills MYC‐driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev 28, 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Ekholm‐Reed S, Baker R, Campos AR, Stouffer D, Henze M, Wolf DA, Loring JF, Thomas EA & Reed SI (2019) Reducing Mcl‐1 gene dosage induces dopaminergic neuronal loss and motor impairments in Park2 knockout mice. Commun Biol 2, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Zhou P, Levy NB, Xie H, Qian L, Lee CY, Gascoyne RD & Craig RW (2001) MCL1 transgenic mice exhibit a high incidence of B‐cell lymphoma manifested as a spectrum of histologic subtypes. Blood 97, 3902–3909. [DOI] [PubMed] [Google Scholar]
- 211. Zhou P, Qian L, Bieszczad CK, Noelle R, Binder M, Levy NB & Craig RW (1998) Mcl‐1 in transgenic mice promotes survival in a spectrum of hematopoietic cell types and immortalization in the myeloid lineage. Blood 92, 3226–3239. [PubMed] [Google Scholar]
- 212. Tong J, Wang P, Tan S, Chen D, Nikolovska‐Coleska Z, Zou F, Yu J & Zhang L (2017) Mcl‐1 degradation is required for targeted therapeutics to eradicate colon cancer cells. Cancer Res 77, 2512–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Tron AE, Belmonte MA, Adam A, Aquila BM, Boise LH, Chiarparin E, Cidado J, Embrey KJ, Gangl E, Gibbons FD et al. (2018) Discovery of Mcl‐1‐specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat Commun 9, 5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Caenepeel S, Karen R, Belmontes B, Verlinsky A, Tan H, Yang Y, Chen X, Li K, Allen J, Wahlstrom J, Canon J, Coxon A & Hughes PE. (2020). Abstract 6218: discovery and preclinical evaluation of AMG 397, a potent, selective and orally bioavailable MCL1 inhibitor. Cancer Res 80, 6218. [Google Scholar]
- 215. Slomp A, Moesbergen LM, Eldering E, Kersten MJ, Minnema MC & Peperzak V. (2021). Phosphatase PP2A enhances MCL‐1 protein half‐life in multiple myeloma cells. Cell Death Dis 12, 229. 10.1038/s41419-020-03351-7 [DOI] [PMC free article] [PubMed] [Google Scholar]