Abstract
There is an unmet need for novel biomarkers to diagnose and monitor patients with neuroendocrine neoplasms. The EXPLAIN study explores a multi‐plasma protein and supervised machine learning strategy to improve the diagnosis of pancreatic neuroendocrine tumors (PanNET) and differentiate them from small intestinal neuroendocrine tumors (SI‐NET). At time of diagnosis, blood samples were collected and analyzed from 39 patients with PanNET, 135 with SI‐NET (World Health Organization Grade 1–2) and 144 controls. Exclusion criteria were other malignant diseases, chronic inflammatory diseases, reduced kidney or liver function. Prosed Oncology‐II (i.e., OLink) was used to measure 92 cancer related plasma proteins. Chromogranin A was analyzed separately. Median age in all groups was 65–67 years and with a similar sex distribution (females: PanNET, 51%; SI‐NET, 42%; controls, 42%). Tumor grade (G1/G2): PanNET, 39/61%; SI‐NET, 46/54%. Patients with liver metastases: PanNET, 78%; SI‐NET, 63%. The classification model of PanNET versus controls provided a sensitivity (SEN) of 0.84, specificity (SPE) 0.98, positive predictive value (PPV) of 0.92 and negative predictive value (NPV) of 0.95, and area under the receiver operating characteristic curve (AUROC) of 0.99; the model for the discrimination of PanNET versus SI‐NET providing a SEN 0.61, SPE 0.96, PPV 0.83, NPV 0.90 and AUROC 0.98. These results suggest that a multi‐plasma protein strategy can significantly improve diagnostic accuracy of PanNET and SI‐NET.
Keywords: biomarker, diagnosis, machine learning, NET, Plasma proteins

The strength of machine learning analysis of multiple plasma proteins for the detectionof pancreatic neuroendocrine tumors is shown in an AUROC with hightrue positive and low false negative rates.
ROCcurves and the corresponding AUC values for the detection of PanNET.
1. INTRODUCTION
Pancreatic neuroendocrine tumors (PanNET) are rare with an incidence of 0.25–0.8 cases per 100,000 inhabitants, and the incidence is rising. 1 A minority of these tumors produce hormones such as insulin, glucagon and gastrin that give rise to specific clinical symptoms and are classified as functional tumors. Patients often have incurable metastatic disease at diagnosis. 2
Commonly used blood‐based biomarkers for NET are chromogranin A (CgA) and 5‐hydroxyindolicacetic acid (5‐HIAA). According to recent studies, CgA has a limited role as a diagnostic biomarker for well‐differentiated PanNET. 3 , 4 , 5 An analysis from the CLARINET study demonstrated that, at the time of diagnosis, only 49% and 26% of patients with PanNET had a higher than upper limit of normal (ULN) levels of CgA and 5‐HIAA, respectively. 6 In the CLARINET FORTE study, only 22.7% of the PanNET patients had a CgA level > ULN. 7 For non‐functional PanNET pancreatic polypeptide (PP) could be used to assist in the diagnosis of the disease. 8 , 9 , 10 The sensitivity for PP in metastatic disease is in the range 50–80%. 10 , 11 Analyzing levels of multiple mRNA from blood showed to be superior to CgA in the detection of PanNET. 12 , 13 There is clearly an unmet need to identify new biomarkers that could more precisely detect PanNET at an early curable stage, as well as recurrent disease after surgery.
Supervised machine learning (SML) has been used in research for the detection, classification and prognostication of cancer diseases for more than two decades. 14 , 15 , 16 , 17 , 18 We have previously shown that this method improves the diagnostic accuracy of patients with small intestinal neuroendocrine tumors (SI‐NET) at the time of diagnosis, especially in patients with normal CgA levels. 19 These techniques can uncover and recognize patterns and correlations in complex collections of data from biomarkers. The aim of this analysis from the EXPLAIN‐study was to investigate, in a real‐life clinical setting, whether a plasma protein multi‐biomarker strategy, measuring the relative concentrations of 92 cancer‐related plasma proteins at time of the diagnosis, could improve diagnostic accuracy of PanNET and differentiate them from SI‐NET.
2. MATERIALS AND METHODS
This is an exploratory analysis from the ongoing Nordic EXPLAIN study (Exploratory, non‐interventional study for evaluating the diagnostic, prognostic, and response‐predictive value of a multi‐biomarker approach in metastatic GEP NETs [ClinicalTrials.gov: NCT02630654]).
2.1. Patients
In total, 135 patients with SI‐NETs and 39 with PanNET were included in the present study. These patients were referred to the hospitals attending the study because of an already diagnosed NET, or suspicion of having NET based on symptoms and/or radiological findings. All patients went through standard work‐up with computed tomography (CT)/magnetic resonance imaging (MRI)/somatostatin receptor imaging and eventually tissue sampling for final diagnose and grading according to the European neuroendocrine Tumor Society (ENETS) guidelines. Inclusion criteria were provision of written informed consent, metastatic non‐resectable SI‐NET or metastatic non‐resectable PanNET (World Health Organization Grade 1 or 2, up to 20% Ki‐67), aged 18 years or older. Exclusion criteria were previously treated with anti‐proliferative treatments for NET disease (including somatostatin analogues), peptide receptor radionuclide therapy, other malignant diseases, chronic inflammatory diseases, or severe renal or liver dysfunction. Patients who had surgery of the primary tumor, but with residual metastatic disease, were included.
Clinical data were collected at each regular follow up and entered into an electronic case report form (VieDoc, Pharma Consulting Group, Uppsala Sweden). The use of proton pump inhibitors (PPI) was registered. Patients using PPI were not excluded, nor was the PPI treatment stopped before blood sample collection. Radiologic evaluation of metastatic disease at the different centers included contrast enhanced CT, contrast enhanced MRI and somatostatin receptor positron emission tomography/CT imaging.
A control group (n = 144), age and sex matched with the patient population, were included for biomarker blood sampling. Exclusion criteria for the controls were malignant disease, chronic inflammatory disease, severe renal or hepatic dysfunction. The controls were recruited from the Karolinska University Hospital Clinical Pharmacology Trial Unit.
2.2. Sample collection and biomarker analysis
A study blood sample (4 mL) was collected starting at the first visit before any NET specific treatment was initiated. Thereafter, blood samples were collected at regular follow‐up visits. Samples were collected in tubes containing ethylenediaminetetraacetic acid and placed on ice immediately after sampling. The samples were centrifuged at 2500 × g for 10 min at 4°C. Thereafter plasma was aspirated, aliquoted to new tubes (4 × 0.5 mL) and immediately stored at −80°C. Blood samples for the exploratory plasma protein biomarkers were transported on dry ice to SciLifeLab (Uppsala, Sweden) and analyzed using multiplex proximity extension assay (PEA) by a real‐time polymerase chain reaction (PCR) using the Fluidigm BioMark HD real‐time PCR platform, with the Olink Proseek Oncology II panel (Olink Proteomics; http://www.olink.com) as described previously. 20 , 21 The multiplex PEA was developed from the proximity ligation assay technique reported previously. 20 One pair of antibodies was used to target each specific protein. The antibodies were coupled to complementary oligonucleotides enabling DNA polymerase to amplify the double‐stranded DNA, generating a PCR‐reporter sequence by the proximity‐dependent DNA polymerization event. The panel of proteins was selected by experts at Olink Proteomics after fulfilling several technical as well as biological criteria. The panel was selected on the basis that these proteins previously have been shown to be associated with mechanisms in neoplastic disease and classified according to Uniprot, Human Protein Atlas, Gene Ontology and DisGeNET. 22 , 23
CgA was analyzed centrally at Akademiska Laboratoriet (Uppsala, Sweden) with the NEOLISA CgA assay (Euro Diagnostica). Urine/serum 5‐HIAA was analyzed at each individual hospital using methods according to clinical routine and presented as percentage of upper limit of normal (%ULN). Serum 5‐HIAA was measured at three clinics in Finland, morning urine 5‐HIAA was used in one clinic in Norway and 13 clinics used 24‐h urine samples.
2.3. Statistical analysis
SML techniques: Boosted Tree (BT) (classifier 1), linear discriminant analysis (LDA) (classifier 2) and support vector machine (SVM) (classifier 3) were used to discriminate between PanNET and controls and PanNET and SI‐NET.
Two models were applied using the three classifiers: Model 1 included PanNETs and controls and Model 2 included PanNETs and SI‐NETs. Models 1A and 2A included both the 92 plasma protein biomarkers as well as CgA. In Models 1B and 2B, CgA was excluded from the analysis.
Furthermore, a group of plasma proteins were identified and named “top biomarkers.” Thus, biomarkers with the highest contribution to the fit (i.e., likelihood ratio chi‐square proportion > 0) from each of the three‐fold cross‐validations including all the biomarkers (i.e., 92 plasma proteins including or not CgA) were selected. Then, only the biomarkers identified in two out of the three‐fold cross‐validations were kept. The number of biomarkers included in the “top biomarker” models could vary and are presented in the tables of results. Additional BT models including top biomarkers (including or excluding CgA) were as well performed.
To validate the algorithms, a three‐fold stratified cross‐validation was performed for all the models. Thus, the complete dataset was partitioned into equally (or near equally) sized folds or segments. This gave an approximate split between training and validation sets of 80% and 20%, respectively. The three‐fold cross‐validation strategy was chosen to avoid having small groups of patients in the validation sets. This cross‐validation strategy was applied to all the models and classifiers.
The BT classifier constructs a predictive model by adding a sequence of decision trees where each of the trees fit on the residuals of the previous tree. A maximum of 50 layers and three splits per tree was allowed. SVM is a supervised learning algorithm that is used to classify binary and categorical response data. SVM models classify data by optimizing a hyperplane (linear or not) that separates the classes. This can also be viewed as finding the hyperplane that maximizes the margin between the classes. A linear kernel function with cost parameter = 1 was assumed. Discriminant analysis predicts membership in a group or category based on observed values of several continuous variables. Specifically, discriminant analysis predicts a classification (X) variable (categorical) based on known continuous responses (Y). A linear fitting was assumed. All analyses were performed with JMP PRO, version 15.0 (SAS Institute Inc.).
Performance of the different models and classifiers was evaluated by comparing the following metrics: negative predictive value (NPV), positive predictive value (PPV), sensitivity (SEN), specificity (SPE), misclassification rate (MR) and accuracy (ACC). These metrics were calculated from the Confusion Matrix generated at each fold of the three‐fold cross‐validation. Descriptive statistics (i.e., mean, SD, minimum and maximum values) were calculated for each of the performance metrics. The misclassification rate, comprising the rate for which the response with the highest fitted probability is not the observed category, is also provided. All area under the receiver operating characteristic curve (AUROC) values were calculated with Python, version 3.7.0 (Package sklearn).
2.4. Approval
The study was approved by the regional ethical committee in each participating country (Denmark, Estonia, Finland, Latvia, Lithuania, Norway, and Sweden) and the study complied with the Declaration of Helsinki and Good Clinical Practice guidelines. All patients and the controls provided their written informed consent after receiving a full explanation of the purpose and nature of all of the procedures used.
3. RESULTS
Baseline characteristics of the three study groups are given in Table 1. Baseline chromogranin A (nmol L–1) values as mean (SD) and median (range) were: for controls, 4.4 (4.8) and 3.3 (2–43); for SI‐NET, 46.7 (85.5) and 12.0 (2–620); and, for PanNET, 55.8 (145.4) and 8.8 (2–785).
TABLE 1.
Clinical characteristics for PanNET, SI‐NET and controls at the time of diagnosis
| Characteristic | PanNETs (n = 39) | SI‐NETs (n = 135) | Controls (n = 144) |
|---|---|---|---|
| Age (years), median (range) | 65 (39–82) | 66 (38–89) | 67 (36–84) |
| Sex, n (%) female | 20 (51%) | 57 (42%) | 61 (42%) |
| Distant metastasis, n (%) | |||
| No | 5 (13%) | 31 (23%) | NA |
| Yes | 32 (78%) | 91 (67%) | |
| Liver metastases | 32 (78%) | 85 (63%) | |
| Missing data | 2 (9%) | 13 (10%) | |
| Lymph node metastasis, n (%) | |||
| No | 16 (41%) | 5 (3.7%) | NA |
| Yes | 21 (54%) | 120 (88.9%) | |
| Missing data | 2 (5%) | 10 (7.4%) | |
| Ki‐67 (%), median (range) | 7.0 (1.0–10) | 3.0 (0.3–19.2) | NA |
| NET Grade, n (%) | |||
| G1 | 14 (36%) | 61 (45.2%) | NA |
| G2 | 22 (56%) | 69 (51.1%) | |
| Missing data | 3 (8%) | 5 (3.7%) | |
| Urine/serum 5‐HIAA (% ULN), median (range) | 62 (44–100) a | 168 (16–5953) b | NA |
Abbreviations: 5‐HIAA, 5‐hydroxyindolicacetic acid; NA, not applicable; NET, neuroendocrine tumor; Pan, pancreatic; SI, small intestinal.
n = 6.
n = 83.
Altogether, 33 (24%) patients with SI‐NET and 5 (13%) patients with PanNET had primary tumour surgery prior to study inclusion. At the time of diagnosis (baseline visit), 78% of the SI‐NET and 57% of the PanNET cohort had CgA levels > ULN. Eighteen (46%) patients with PanNET had less than three liver metastases and nine (23%) patients had no liver metastases. In this non‐functional PanNET patient cohort, only a minority (15%) of patients had any type of daily symptoms that could be related to the NET. In the SI‐NET cohort, 48% of the patients had at least one daily symptom that could be tumour related, such as flushing, diarrhoea or abdominal pain
3.1. Biomarker analysis
Results from Model 1 (PanNET vs. controls) performance metrics are presented in Table 2. BT and SVM yielded very similar results in terms of accuracy (ACC) performance for both models including the 92 plasma proteins and CgA [ACC Model 1A (mean [SD]), BT = 0.91 (0.04); SVM = 0.94 (0.01)] or after excluding CgA (ACC Model 1B, BT = 0.91 [0.03]; SVM = 0.94 [0.01]). LDA showed lower predictive performance (ACC = 0.82 [0.06]) for both Model 1A and 1B. Mean misclassification rate of the classifiers ranged from 6% to 18%.
TABLE 2.
Performance metrics from Model 1A and 1B (PanNET vs. controls)
| Model 1A | Model 1B | |||||||
|---|---|---|---|---|---|---|---|---|
| 92 plasma biomarkers and CgA | Top biomarkers a | 92 plasma biomarkers | Top biomarkers b | |||||
| Metrics Mean (SD) | BT | SVM | LDA | BT | BT | SVM | LDA | BT |
| PPV | 0.86 (0.15) | 0.90 (0.09) | 0.59 (0.20) | 0.91 (0.16) | 0.86 (0.15) | 0.90 (0.09) | 0.58 (0.18) | 0.92 (0.07) |
| NPV | 0.93 (0.03) | 0.95 (0.03) | 0.92 (0.04) | 0.93 (0.06) | 0.92 (0.02) | 0.95 (0.03) | 0.92 (0.04) | 0.95 (0.05) |
| SEN | 0.71 (0.08) | 0.80 (0.09) | 0.72 (0.08) | 0.74 (0.16) | 0.68 (0.04) | 0.82 (0.09) | 0.72 (0.08) | 0.84 (0.15) |
| SPE | 0.97 (0.03) | 0.98 (0.02) | 0.86 (0.10) | 0.98 (0.04) | 0.97 (0.03) | 0.98 (0.02) | 0.85 (0.09) | 0.98 (0.02) |
| ACC | 0.91 (0.04) | 0.94 (0.01) | 0.82 (0.06) | 0.92 (0.05) | 0.91 (0.03) | 0.94 (0.01) | 0.82 (0.06) | 0.94 (0.06) |
| AUC | 0.99 (0.01) | 0.99 (0.01) | 0.96 (0.01) | 0.99 (0.01) | 0.99 (0.01) | 0.99 (0.01) | 0.96 (0.01) | 0.99 (0.01) |
| MR | 0.08 (0.03) | 0.06 (0.01) | 0.18 (0.06) | 0.08 (0.05) | 0.09 (0.03) | 0.06 (0.01) | 0.17 (0.07) | 0.06 (0.06) |
Abbreviations: ACC, accuracy; AUC, area under the curve; BT, boosted tree; CgA, chromogranin A; LDA, linear discriminant analysis; MR, misclassification rate; NPV, negative predictive value; PPV, positive predictive value; SEN, sensitivity; SPE, specificity; SVM, supported vector machine.
CgA.log, chromogranin A log; MAD.homolog.5, mothers against decapentaplegic homolog 5; MetAP.2, methionine aminopeptidase 2; CDKN1A, cyclin‐dependent kinase inhibitor 1; CPE, carboxypeptidase E; HK8, kallikrein‐8; VIM, vimentin; LYN, tyrosine‐protein kinase Lyn.
MAD.homolog.5, mothers against decapentaplegic homolog 5; CDKN1A, cyclin‐dependent kinase inhibitor 1; CPE, carboxypeptidase E; HK8, kallikrein‐8; ITGB5, integrin beta‐5; ESM.1, GDSL esterase/lipase ESM1; CEACAM5, carcinoembryonic antigen‐related cell adhesion molecule 5; LYN, tyrosine‐protein kinase Lyn; VIM, vimentin; TRAIL, TNF‐related apoptosis‐inducing ligand.
The best accuracy performance to classify between PanNET versus SI‐NET (Model 2) (Table 3) was achieved with SVM; thus for both models including the 92 plasma proteins and CgA (Model 2A, ACC = 0.91 [0.07]) or excluding CgA (Model 2B, ACC = 0.93 [0.05]). Similar accuracy was yielded for both BT and LDA models including the 92 plasma proteins and CgA (ACC Model 2A, BT = 0.84 [0.03]; LDA = 0.83 [0.05]) or excluding CgA (ACC Model 2B, BT = 0.83 [0.02]; LDA = 0.83 [0.04]). Mean misclassification rate of the classifiers ranged from 7% to 17%.
TABLE 3.
Performance metrics from Model 2A and 2B (PanNET vs. SI‐NET)
| Model 2A | Model 2B | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 92 plasma biomarkers and CgA | Top biomarkers a | 92 plasma biomarkers | Top biomarkers b | ||||||
| Metrics Mean (SD) | BT | SVM | LDA | BT | BT | SVM | LDA | BT | |
| PPV | 0.76 (0.13) | 0.88 (0.14) | 0.63 (0.13) | 0.83 (0.11) | 0.72 (0.12) | 0.97 (0.05) | 0.67 (0.17) | 0.86 (0.17) | |
| NPV | 0.86 (0.04) | 0.92 (0.08) | 0.89 (0.08) | 0.90 (0.02) | 0.85 (0.04) | 0.93 (0.07) | 0.89 (0.07) | 0.89 (0.01) | |
| SEN | 0.43 (0.11) | 0.71 (0.27) | 0.63 (0.26) | 0.61 (0.03) | 0.43 (0.11) | 0.73 (0.24) | 0.63 (0.25) | 0.55 (0.05) | |
| SPE | 0.96 (0.04) | 0.97 (0.03) | 0.89 (0.07) | 0.96 (0.02) | 0.95 (0.03) | 0.99 (0.01) | 0.69 (0.43) | 0.97 (0.03) | |
| ACC | 0.84 (0.03) | 0.91 (0.07) | 0.83 (0.05) | 0.88 (0.01) | 0.83 (0.02) | 0.93 (0.05) | 0.83 (0.04) | 0.88 (0.02) | |
| AUC | 0.96 (0.03) | 0.99 (0.01) | 0.95 (0.02) | 0.98 (0.02) | 0.97 (0.02) | 0.98 (0.01) | 0.94 (0.04) | 0.97 (0.01) | |
| MR | 0.16 (0.03) | 0.09 (0.07) | 0.17 (0.05) | 0.12 (0.01) | 0.17 (0.01) | 0.07 (0.05) | 0.17 (0.04) | 0.12 (0.02) | |
Abbreviations: ACC, accuracy; AUC, area under the curve; BT, boosted tree; CgA, chromogranin A; CgA.log, chromogranin A log; GPC1, glypican‐1; GZMB, granzyme B; GZMH, granzyme H; LDA, linear discriminant analysis; MR, misclassification rate; NPV, negative predictive value; PPV, positive predictive value; S100A4, protein S100‐A4; SEN, sensitivity; SPARC, SPARC; SPE, specificity; SVM, supported vector machine.
CPE, carboxypeptidase E; MAD.homolog.5, mothers against decapentaplegic homolog 5; GPNMB, transmembrane glycoprotein NMB; LYN, tyrosine‐protein kinase Lyn;
GPNMB, transmembrane glycoprotein NMB; MAD.homolog.5, mothers against decapentaplegic homolog 5; CPE, carboxypeptidase E; LYN, tyrosine‐protein kinase Lyn; IL6, interleukin‐6; SPARC, SPARC; GPC1, glypican‐1; S100A4, protein S100‐A4; GZMH.
The best BT model for classifying PanNET versus controls was obtained when CgA was excluded from the model and only the top biomarkers (Table 2) were used: SEN = 0.84 (0.15), SPE = 0.98 (0.15), PPV = 0.92 (0.07) and NPV = 0.95 (0.05) (Table 2). Figure 1 shows performance metrics box‐plots for BT Model 1B (best model) including top biomarkers (plasma proteins without CgA).
FIGURE 1.
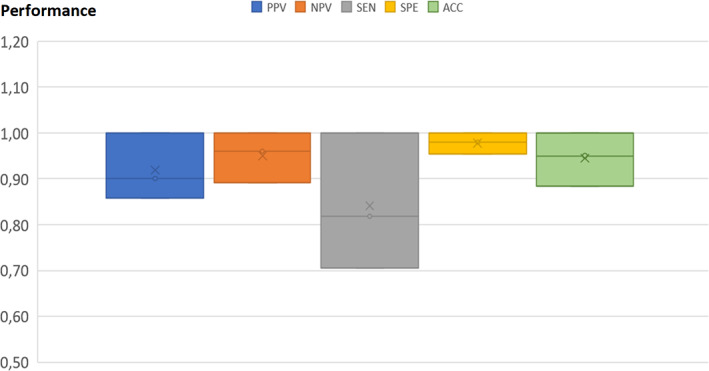
Box‐plots for the best boosted tree model discriminating pancreatic neuroendocrine tumors from controls. ACC, accuracy; NPV, negative predictive value; PPV, positive predictive value; SEN, sensitivity; SPE, specificity
Receiver operating characteristic curve and corresponding area under curve values (AUROC) generated for the detection of PanNET versus controls using the BT Model (1B) was 0.99 (0.01) (Figure 2).
FIGURE 2.
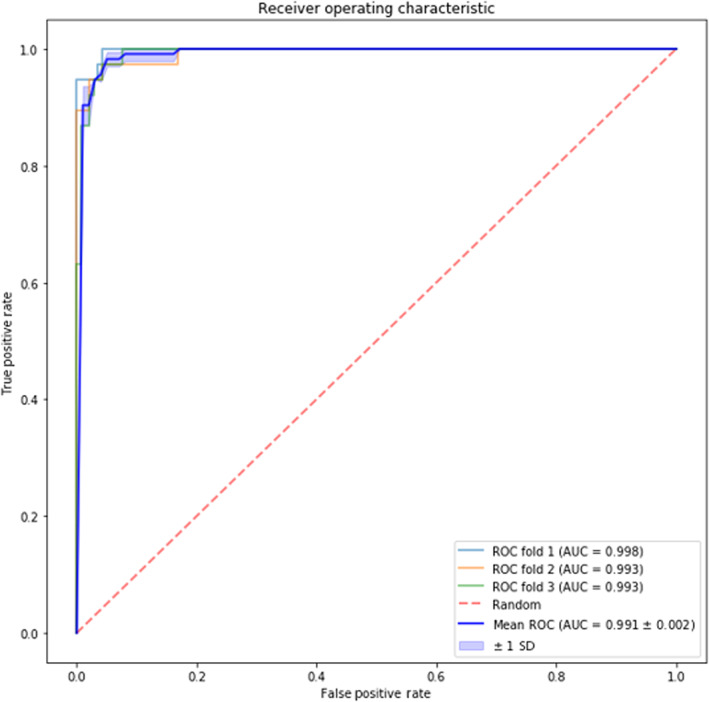
Receiver operating characteristic (ROC) curves and the corresponding area under the curve (AUC) values generated from the best boosted tree model including the control group and patients with pancreatic neuroendocrine tumors with the top ranked identified plasma protein biomarkers
When discriminating between PanNET and SI‐NET populations (Model 2), more variable results and somewhat lower performance metrics were observed compared to detecting PanNET versus controls (Table 3 and Figure 3). This was found both when all 92 plasma proteins were included in the models with CgA (Model 2A) or when CgA was excluded (Model 2B). A better predictive performance was obtained when only the top biomarkers (Table 3) were included in the BT Model, SEN = 0.61 (0.03), SPE = 0.96 (0.02), PPV = 0.83 (0.11) and NPV = 0.90 (0.02) (Table 3). Figure 2 shows performance metrics box‐plots for BT Model 2A (best model) including top biomarkers (plasma proteins with CgA). CgA was more important for detecting SI‐NET than PanNET (both for the present study and the previous by Kjellman et al 19 ). Receiver operating characteristic curve and the corresponding area under curve values (i.e., AUROC) generated from the BT model analysis of detecting PanNET versus SI‐NET was 0.98 (0.02) and 0.97 (0.01) for top biomarker models.
FIGURE 3.
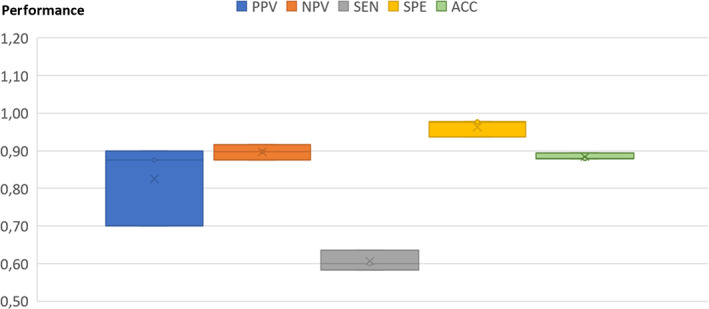
Box‐plots for the best boosted tree model discriminating pancreatic neuroendocrine tumors from small intestinal neuroendocrine tumors. ACC, accuracy; NPV, negative predictive value; PPV, positive predictive value; SEN, sensitivity; SPE, specificity.
Several plasma proteins were identified to be important for the detection of PanNET and discrimination versus SI‐NET: such as carboxypeptidase E (CPE), mothers against decapentaplegic homolog 5 (MAD homolog 5), transmembrane glycoprotein NMB (GPNMB) and tyrosine‐protein kinase Lyn (LYN). The plasma proteins with highest contributions to the BT models (Model 1A and Model 2A) are presented in Tables 2 and 3.
CgA used alone for detecting PanNET had a sensitivity of 41%, a specificity of 94%, a PPV of 64% and a NPV of 84%.
4. DISCUSSION
In the present study, we used a proximity extension assay to investigate 92 plasma proteins known to be associated with malignancy in general. After analyzing plasma, collected prior to any NET specific treatment being initiated, SML methods were able, with high sensitivity and specificity, to discriminate patients with PanNET from controls based on the presence of specific proteins. We were also able to differentiate patients with PanNET from patients with SI‐NET. Our data support data from a previous study, conducted using the same methodology, where the top 12 of the 92 proteins, also used in the present study, differentiated SI‐NET from controls. 19 This indicates that the findings from this multi‐biomarker strategy could be applied also on patients with neuroendocrine tumors from other primaries.
All three different SML algorithms generated comparable results. Both BT and SVM provided robust performance in terms of AUROC and accuracy. This strengthens the validity of our multi‐biomarker strategy approach. Although SVM had, in some cases, better performance than BT, BT was chosen to identify biomarkers with the highest contribution to the classification model. One of the drawbacks of SVM is that it does not provide an explanation or a compressible justification for the knowledge it learns, and this limits the practical application. 24
Our model performs clearly better than previously published analyses of single biomarkers such as plasma 5‐HIAA, PP and CgA in patients with NET. 25 Our results confirm other studies showing relatively low sensitivity and specificity of CgA for the detection of PanNET. 3 , 4 CgA as a single biomarker has a medium high sensitivity and specificity for the detection of SI‐NET in the range 0.7–0.8. 25 , 26 Only 57% of the PanNET patients in the present study had CgA levels > ULN, and CgA did not classify as one of the top 10 biomarkers for the detection of PanNET. In addition, it was only the seventh most important biomarker for discriminating between PanNET and SI‐NET in the best models. We found that other proteins, such as CPE and MAD homolog 5, strongly contributed both to the detection of PanNET and in discriminating PanNET from SI‐NET. Carboxypeptidase E is a metallo‐carboxypeptidase involved in the biosynthesis of peptide hormones. MAD homolog 5 is a transcription regulatory protein involved in the signaling pathway by which transforming growth factor‐beta inhibits proliferation. An additional protein of interest is LYN, which contributed to improve diagnostic accuracy for PanNET. Additionally, LYN was also highly important in detecting SI‐NET. 19 Tyrosine‐protein kinase Lyn belongs to a group of proteins often targeted in cancer treatment. Dasatinib, nintedanib and bosutinib are tyrosine kinase inhibitors with an inhibitory effect on LYN and are approved for the treatment of different cancers (DrugBank.com). These findings imply that other not yet fully characterized proteins have the potential for becoming important biomarkers for neuroendocrine tumors.
In the present study, despite the rather small sample size of PanNETs, mainly as a result of its rarity, we were able to generate predictive models with good classification performances and to identify novels proteins associated with both NET subgroups. A more balanced situation regarding groups size would have been desirable and probably could have provided even more robust predictions.
The NETest is a validated and commercially available test that has shown high diagnostic accuracy for neuroendocrine tumors based on measurement of neuroendocrine tumor gene expression in blood. 12 Whether the plasma protein measurement approach presented in the present study will be developed into a diagnostic test, and how it potentially will compare with the NETest, is not possible to assess at present.
The “real‐life clinical design”, the large number of patients with NET, centralized analysis of CgA and biomarkers, as well as controls with similar age and sex profile, are the main strengths of the present study. A model using multiple plasma proteins and machine learning has not been tested to discriminate between patients with NET and other benign conditions that could display similar symptoms. Likewise, the plasma proteins used in our study could also be elevated in other malignant diseases than neuroendocrine tumors. We have, however, shown in the present study that the EXPLAIN strategy can differentiate between two closely related NET types. Further studies are needed to explore whether this strategy can discriminate PanNET from other malignancies and benign diseases as diseases of the pancreas.
The EXPLAIN study, of which these results are a part, comprises an exploratory study. The 92 plasma proteins analyzed were selected because of their known association with malignant tumors. Their possible association with neuroendocrine tumors was not known at the beginning of the study. As shown in our results, the different proteins had variable strength in detecting PanNET and discriminating PanNET from SI‐NET. Discovering new plasma proteins more strongly associated with neuroendocrine tumors, as well as subtypes of neuroendocrine tumors, could further strengthen our model and increase the diagnostic accuracy and specificity towards other diseases.
In the PanNET cohort, most patients had a significant tumor burden easily detected on imaging. We do not know how this multi‐protein approach would perform in patients with only a small primary or only modest affection of regional lymph nodes. Likewise, we do not know how this test would perform when applied after presumed curative surgery to detect recurrence. Currently, this model is not developed for clinical routine and further work is needed to enable the entry of this new biomarker test.
Nevertheless, this multi‐biomarker strategy is valuable in research and produces a vast number of biomarker data providing opportunities for identifying novel biomarker plasma proteins. The plasma protein assay technique used has high sensitivity and specificity based on two specific antibodies and the PCR, and only a drop of dried blood on a paper is needed to perform the analysis. 27 This enables cheap and easy collection and transport of samples. The applicability and utility in clinical routine should be validated in larger populations in the future. Ongoing analysis will evaluate the possible value of these biomarkers with respect to predicting treatment response and risk of progression.
In conclusion, the present study demonstrates that a multi‐biomarker plasma protein strategy and SML significantly improves detection of PanNET and discriminates PanNET from controls, as well as SI‐NET, at the time of diagnosis in a real‐world setting. This approach can provide a new tool in the work up of a patient with known, or suspected, neuroendocrine tumors and has the potential to be used in the follow‐up of patients after radical resection of neuroendocrine tumors. The assay would have to be validated in additional prospective studies.
AUTHOR CONTRIBUTIONS
Magnus Kjellman: Conceptualization; investigation; methodology; writing – review and editing. Ulrich Peter Knigge: Investigation; methodology; writing – review and editing. Henning Gronbaek: Investigation; methodology; writing – review and editing. Camilla Schalin‐Jantti: Investigation; writing – review and editing. Staffan Welin: Investigation; writing – review and editing. Halfdan Sorbye: Investigation; writing – review and editing. Maria del Pilar Schneider: Methodology; project administration; software; validation; visualization; writing – original draft; writing – review and editing.
CONFLICTS OF INTEREST
ETE: Research grants from Novartis, consulting fees from Ipsen and speaker honoraria from Novartis, Ipsen and Pfizer. MK: Grants from Ipsen, consulting fees from Ipsen and Novartis Healthcare. UK: Grants from Ipsen and Novartis Healthcare, consulting fees from Ipsen. HG: Research grants from the NOVO Nordisk Foundation, Intercept, Abbvie, and consulting fees from Ipsen and Novartis. CSJ: Grants from Ipsen and Pfizer, consulting fees from Ipsen. HS: Research grant from Amgen. Consulting fees from Novartis, Pfizer, Keocyt, AstraZeneca, Hutchinson, Bayer and ITM. Honoraria from: Novartis, Roche, Ipsen, Merck, Bayer, SAM Nordic and Pierre Fabre. HW: No conflicts of interest. AKE: No conflicts of interest. MTJ: No conflicts of interest. SW: Consulting fees from Ipsen and Novartis Healthcare. JAS: Consulting fee from Novartis. SM: No conflicts of interest. TE: Speaker honoraria from Ipsen, AstraZeneca, Novo Nordisk Pharma, consulting fee from Amgen, educational grants by Novartis, Ipsen and MSD Finland to the employer institution for international symposia. FL: Speaker honorarium from Novartis. KL: No conflicts of interest. FS: No conflicts of interest. GW: No conflicts of interest. GP: speaker honoraria from Ipsen. RJ: speaker honoraria from Ipsen, Novartis, Merck Serono, Bristol Myers Squibb, MSD, Servier. TS: speaker honoraria from Ipsen. IK: No conflicts of interest. RS: No conflicts of interest. EB: speaker honoraria from Ipsen. MPS and RB: Ipsen employees.
ACKNOWLEDGMENTS
This study was sponsored by Ipsen. We thank all of the patients who made this study possible through their participation in the study. We thank Torbjörn Ström at IPSEN Nordic and Karin Becker (both at IPSEN at the time the study was conducted) for study monitoring. We thank Daniel Seisdedos at Pharma Consulting Group Uppsala Sweden for data management. We thank Dr Nabil Al‐Tawil at the Karolinska University Hospital Clinical Pharmacology Trial Unit, Stockholm Sweden for recruiting sex and age matched controls. We also thank all of the additional physicians and nurses at the 20 hospitals who have contributed to make this study possible. The Nordic NET Biomarker Group: E. Thiis‐Evensen (Oslo University Hospital, Oslo, Norway). M. Kjellman (Karolinska Hospital, Stockholm, Sweden). U. Knigge (Rigshospitalet, Copenhagen, Denmark). S. Welin (Akademiska Hospital, Uppsala, Sweden). H. Gronbaek (Aarhus University Hospital, Aarhus, Denmark). H. Sorbye (Haukeland University Hospital, Bergen, Norway). M. T. Joergensen (Odense University Hospital, Odense, Denmark). A. K. Elf (Sahlgrenska Hospital, Gothenburg, Sweden). R. Belusa (Previously at Ipsen Nordic, Stockholm, Sweden). C. Schalin‐Jäntti (Helsinki University Hospital, Helsinki, Finland). M. T. Jørgensen (Odense University Hospital, Odense, Denmark). H. Waldum (St Olav University Hospital, Trondheim, Norway). J. A. Søreide (Stavanger University Hospital. Stavanger, Norway). S. Metso (Tampere University Hospital, Tampere, Finland). T. Ebeling (Oulu University Hospital, Oulu, Finland). F. Lindberg (Norrland University Hospital, Umeå, Sweden). K. Landerholm (Ryhov County Hospital, Jönköping, Sweden). G. Wallin (Örebro University Hospital, Örebro, Sweden). F. Salem (Skåne University Hospital, Lund, Sweden). G. Purkalne (Pauls Stradiņš Clinical University Hospital, Riga, Latvia). I. Kudaba (Riga East University Hospital, Riga, Latvia). R. Janciauskiene (Lithuanian University of Health Sciences, Kaunas, Lithuania). T. Suuroja (North‐Estonian Regional Hospital, Tallinn Estonia). Edita Baltruškevičienė (National Cancer Institute, Vilnius, Lithuania).
Thiis‐Evensen E, Kjellman M, Knigge U, et al. Plasma protein biomarkers for the detection of pancreatic neuroendocrine tumors and differentiation from small intestinal neuroendocrine tumors. J Neuroendocrinol. 2022;34(7):e13176. doi: 10.1111/jne.13176
Funding information Ipsen Pharma
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the Sponsor, [Ipsen], upon reasonable request.
REFERENCES
- 1. Leoncini E, Boffetta P, Shafir M, et al. Increased incidence trend of low‐grade and high‐grade neuroendocrine neoplasms. Endocrine. 2017;58:368‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyar Cetinkaya R, Aagnes B, Thiis‐Evensen E, et al. Trends in incidence of neuroendocrine neoplasms in Norway: a report of 16,075 cases from 1993 through 2010. Neuroendocrinology. 2017;104:1‐10. [DOI] [PubMed] [Google Scholar]
- 3. Pulvirenti A, Rao D, Mcintyre CA, et al. Limited role of chromogranin a as clinical biomarker for pancreatic neuroendocrine tumors. HPB. 2019;21:612‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marotta V, Zatelli MC, Sciammarella C, et al. Chromogranin a as circulating marker for diagnosis and management of neuroendocrine neoplasms: more flaws than fame. Endocr‐Relat Cancer. 2018;25:R11‐R29. [DOI] [PubMed] [Google Scholar]
- 5. Baekdal J, Krogh J, Klose M, et al. Limited diagnostic utility of chromogranin a measurements in workup of neuroendocrine tumors. Diagnostics (Basel); 2020, 10(11), 881. doi: 10.3390/diagnostics10110881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pavel ME, Phan AT, Wolin EM, et al. Effect of Lanreotide depot/autogel on urinary 5‐Hydroxyindoleacetic acid and plasma chromogranin a biomarkers in nonfunctional metastatic Enteropancreatic neuroendocrine tumors. The Oncologist. 2019;24:463‐474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pavel ME, Ćwikła JB, Lombard‐Bohas J, et al. Efficacy and safety of high‐dose lanreotide autogel in patients with progressive pancreatic or midgut neuroendocrine tumours: CLARINET FORTE phase 2 study results. Eur J Cancer. 2021;157:403‐414. [DOI] [PubMed] [Google Scholar]
- 8. Adrian TE, Uttenthal LO, Williams SJ, et al. Secretion of pancreatic polypeptide in patients with pancreatic endocrine tumors. N Engl J Medicine. 1986;315:287‐291. [DOI] [PubMed] [Google Scholar]
- 9. Panzuto F, Severi C, Cannizzaro R, et al. Utility of combined use of plasma levels of chromogranin A and pancreatic polypeptide in the diagnosis of gastrointestinal and pancreatic endocrine tumors. J Endocrinol Invest. 2004;27:6‐11. [DOI] [PubMed] [Google Scholar]
- 10. Walter T, Chardon L, Chopin‐laly X, et al. Is the combination of chromogranin a and pancreatic polypeptide serum determinations of interest in the diagnosis and follow‐up of gastro‐entero‐pancreatic neuroendocrine tumours? Eur J Cancer. 2012;48:1766‐1773. [DOI] [PubMed] [Google Scholar]
- 11. Metz DC, Jensen RT. Gastrointestinal neuroendocrine tumors: pancreatic endocrine tumors. Gastroenterology. 2008;135:1469‐1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Modlin IM, Kidd M, Bodei L, et al. The clinical utility of a novel blood‐based multi‐transcriptome assay for the diagnosis of neuroendocrine tumors of the gastrointestinal tract. Am J Gastroenterol. 2015;110:1223‐1232. [DOI] [PubMed] [Google Scholar]
- 13. Modlin IM, Kidd M, Malczewska A, et al. The NETest: the clinical utility of multigene blood analysis in the diagnosis and Management of Neuroendocrine Tumors. Endocrinol Metab Clin North Am. 2018;47:485‐504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furey TS, Cristianini N, Duffy N, et al. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16:906‐914. [DOI] [PubMed] [Google Scholar]
- 15. Adam B‐L, Qu Y, Davis JW, et al. Serum protein fingerprinting coupled with a pattern‐matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609‐3614. [PubMed] [Google Scholar]
- 16. Glotsos D, Tohka J, Ravazoula P, et al. Automated diagnosis of brain tumours astrocytomas using probabilistic neural network clustering and support vector machines. Int J Neural Syst. 2005;15:1‐11. [DOI] [PubMed] [Google Scholar]
- 17. Statnikov A, Wang L, Aliferis CF. A comprehensive comparison of random forests and support vector machines for microarray‐based cancer classification. BMC Bioinform. 2008;9:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kourou K, Exarchos TP, Exarchos KP, et al. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kjellman M, Knigge U, Welin S, et al. A plasma protein biomarker strategy for detection of small intestinal neuroendocrine tumors. Neuroendocrinology. 2021;111:840‐849. doi:. doi: 10.1159/000510483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lundberg M, Thorsen SB, Assarsson E, et al. Multiplexed homogeneous proximity ligation assays for high‐throughput protein biomarker research in serological material. Mol Cell Proteomics. 2011;10:M110.004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Assarsson E, Lundberg M, Holmquist G, et al. Homogenous 96‐plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mahboob S, Ahn SB, Cheruku HR, et al. A novel multiplexed immunoassay identifies CEA, IL‐8 and prolactin as prospective markers for Dukes' stages A‐D colorectal cancers. Clin Proteom. 2015;12:10. doi: 10.1186/s12014-015-9081-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneiderova P, Pika T, Gajdos P, et al. Serum protein fingerprinting by PEA immunoassay coupled with a pattern‐recognition algorithms distinguishes MGUS and multiple myeloma. Oncotarget. 2017;8:69408‐69421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barakat N, Bradley AP. Rule extraction from support vector machines: a review. Neurocomputing. 2010;74:178‐190. [Google Scholar]
- 25. Oberg K, Krenning E, Sundin A, et al. A Delphic consensus assessment: imaging and biomarkers in gastroenteropancreatic neuroendocrine tumor disease management. Endocr Connect. 2016;5:174‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dam G, Grønbæk H, Sorbye H, et al. Prospective study of chromogranin a as a predictor of progression in patients with pancreatic, small‐intestinal, and unknown primary neuroendocrine tumors. Neuroendocrinology. 2020;110:217‐224. [DOI] [PubMed] [Google Scholar]
- 27. Björkesten J, Enroth S, Shen Q, et al. Stability of proteins in dried blood spot biobanks. Mol Cell Proteom. 2017;16:1286‐1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the Sponsor, [Ipsen], upon reasonable request.


