Abstract
The human body represents a complex and diverse reservoir of microorganisms. Although the human microbiome remains poorly characterized and understood, it should not be underestimated, since recent studies have highlighted its importance in health. This is especially evident when considering microbiota in the male reproductive system, responsible for men’s fertility and sexual behavior. Therefore, the aim of this systematic review is to provide an overview of the microbial communities of the healthy male genital mucosa and its role in disease. This study was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. The search was limited to the English language and studies published until August 2022 that included culture-independent techniques for microbiome characterization in male genital mucosa. Ten articles were included. The bacterial composition of the male genital mucosa consists of several genera including Prevotella, Finegoldia, Peptoniphilus, Staphylococcus, Corynebacterium, and Anaerococcus, suggesting that the male genital microbiome composition shows similarities with the adjacent anatomical sites and is related with sexual intercourse. Moreover, male circumcision appears to influence the penile microbiome. Despite the lack of knowledge on the male genital mucosa microbiome in disease, it was reported that Staphylococcus warneri and Prevotella bivia were associated with balanoposthitis, whereas Enterobacteriaceae, Prevotella, and Fusobacterium were more abundant in male genital lichen sclerosus. The limited data and paucity of prospective controlled studies highlight the need for additional studies and established criteria for sampling methods and the microbiome assay procedure. Such a consensus would foster the knowledge about the composition of the genital microbiome of healthy males and its role in disease.
Keywords: male genital mucosa microbiome, penis microbiome, reproductive system, urogenital system, genital disease
1. Introduction
The complex and diverse human body microbiome has an enormous potential to influence human pathophysiology. Studies of the human microbiome suggest that several microbial communities play important roles in host homeostasis, regulating health, infection risk, and disease progression [1].
Recent advances in high throughput sequencing technologies, such as next-generation sequencing (NGS) techniques, the whole genome shotgun, and metagenomic sequencing, have been stimulating a new wave of research on the human microbiome [2]. These sequencing analyses allow for a more accurate detection of resident microbial DNA, revealing a considerable diversity of hidden microbes on most exposed surfaces of the human body, including the reproductive system [3].
The microbiome of male genital mucosa is extensively less studied compared to other areas of microbiome research, such as inflammatory diseases [4], skin conditions [5], gut and colorectal cancer [6], prostate cancer [7], and even the female reproductive tract [8]. Nevertheless, emerging evidence indicates that the male genital tract microbiome is a field of increasing scientific interest since it has important implications for the male reproductive health, men’s fertility, and sexual behavior [9,10,11]. Attention is now focused on understanding the alterations in the microbiome in case of dysbiosis [3].
Similar to vaginal mucosa, male genital mucosa is the most significant and adaptable structure in the male reproductive system, capable of mounting a full range of immune responses [12]. Thus, elucidation of the composition of the healthy male genital mucosa microbiome and its variations is of paramount importance. Although some studies have already investigated the role of the male reproductive tract microbiome [3], they are of limited scope and mainly focused on the urine or the coronal sulcus. The lack of comprehensive studies addressing the male genital mucosa microbiome undermines the prognostic determinants that include risk factors such as bacterial and virus infections. Thus, the aim of this systematic review is to identify and summarize studies focused on the microbial community’s composition in the different anatomical parts of the healthy and diseased male genital mucosa.
2. Materials and Methods
This systematic review was executed following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [13]. The methods used, including title and abstract screening, full text review, inclusion, and data extraction criteria, were approved by all the authors.
2.1. Search Strategy
We performed a search in MEDLINE (PubMed), Web of Science, Scopus, and Google Scholar on 2 September 2022. The search strategy included keywords related to the scientific literature concerning the microbiome of the male genital mucosa, such as (alone or in combination): microbiome, male, genital tract, genital mucosa, penis, glans, balanopreputial sulcus, coronal sulcus, prepuce, and foreskin. Supplementary File S1 comprises the complete search strategies for each database.
2.2. Inclusion and Exclusion Criteria
Articles and clinical reports published until August 2022 and written in English were eligible for inclusion. Eligible studies had to be related to microbiome research on the male genital mucosa and include culture-independent techniques for the whole spectrum of detected microbiota.
Reviews, books and book chapters, poster presentations, theses and dissertations, and articles of which only the abstract was available, studies with only culture-dependent techniques, animal studies, and articles written in languages other than English were excluded.
2.3. Study Selection
Titles and/or abstracts of studies retrieved using the above-mentioned search strategy were screened independently by two authors (M.F.M.G. and Â.R.F.) to identify studies eligible for full-text screening. The full texts of these eligible articles were retrieved and independently assessed by the same two authors. Any disagreements concerning the eligibility of studies were resolved through discussion with a third author (C.L.).
2.4. Data Extraction
Data were extracted from the included studies for an assessment of study quality. Predefined extracted information included: field of study; study population; participant demographics and baseline characteristics (including age); study setting (including ethnicity and country); target anatomical organ; study methodology (including sample type method and type of microbiome analysis); main outcomes and limitations.
2.5. Study Quality Assessment
Two authors (M.F.M.G. and Â.R.F.) independently assessed the quality evaluation of individual studies according to the National Institutes of Health (NIH) Study Quality Assessment Tools for observational studies [14] and Cochrane risk-of-bias tool for randomized trials [15]. Disagreements resulting from this process were resolved through discussion with the other authors (A.G.R. and C.L.).
3. Results
3.1. Number of Retrieved Papers
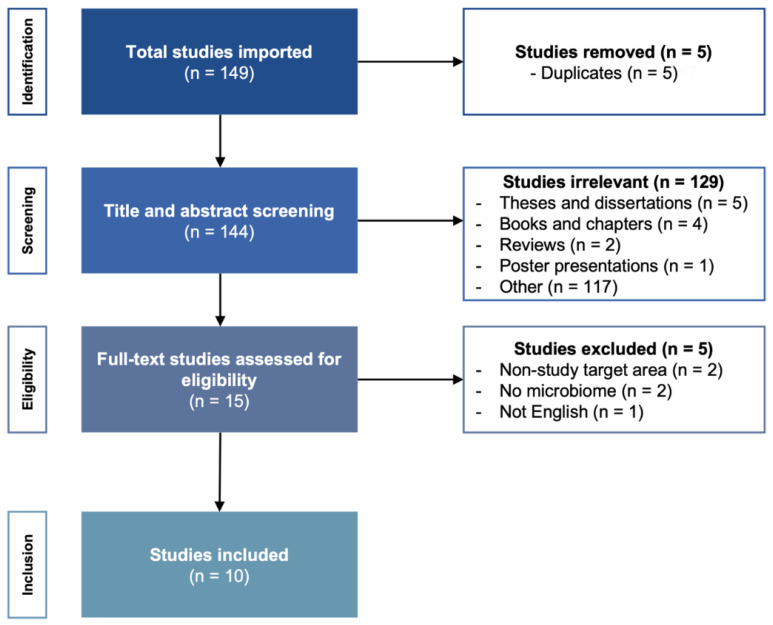
A flowchart of the search strategy and study selection process of the articles is shown in Figure 1. The search yielded 149 articles, which resulted in 144 unique articles after removing duplicates. After screening the titles and abstracts, 129 articles were excluded based on title and study type. After reading the full-text articles, 15 articles were eligible for inclusion, and 5 were excluded (non-study target area, n = 2; no microbiome, n = 2; articles written in languages other than English, n = 1). Table 1 provides an overview of the 10 final selected articles and summarizes the characteristics and reported taxonomic findings.
Figure 1.
Flowchart of the search strategy for the selection of relevant articles following PRISMA methodology.
Table 1.
Overview of the final selected articles and the characteristics and reported findings. HV = healthy volunteers; CS = coronal sulcus; BP = balanoposthitis; HIV = human immunodeficiency virus; MGLSc = male genital lichen sclerosus; BS = balanopreputial sac; BV = Bacterial vaginosis; ↓ = decrease; ↑ = increase.
| Author | Study Design | Field of Study | Participants | Age (Years) | Ethnicity, Country | Anatomical Site | Sample Type | Microbial Analysis | Key Findings | Limitations | Quality Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Iniesta et al. [16] | Control intervention | Supplementation with a probiotic | 17 couples under artificial reproductive treatment | Couples: 22–40 Male (mean 36) Female (mean 35) |
Caucasian, Spain | Glans | Glans, semen, and blood | 16S rRNA amplification of the V3–V4 region | Treatment with Lactobacillus salivarius PS11610 modified the microbiota composition improving the urogenital tract microbiome, solving the dysbiosis in 88.9% of the couples. Male samples showed higher bacterial diversity at genus level than female samples. Most prevalent genera in glans: Initial: Peptoniphilus, Staphylococcus, Fineglodia, and Corynebacterium; 3 and 6 months: Peptonophilus, Finegoldia, Corynebacterium (↓ Staphylococcus) | Small sample size. No control group treated with placebo. | Fair |
| Li et al. [17] | Case control | Balanoposthitis (BP) | 26 BP uncircumcised 29 HV uncircumcised |
18–65 | Unknown, China | Glans, penis, and prepuce | Swabs | 16S rRNA amplification of the V4 region | Microbiome BP ∼ HV, but ≠ HV with redundant prepuce. Most prevalent species BP: Staphylococcus warneri (with condom use) and Prevotella bivia (without sexual activity). Most prevalent HV: Ezakiella (redundant prepuce), Porphyromonas somerae (normal prepuce). | Small sample size. No ethnicity records. V4 region is considered a relatively low informative region for taxonomic assignment. | Fair |
| Liu et al. [18] | Randomized controlled trial | Circumcision | 77 HV uncircumcised 79 HIV-negative pre- and post-circumcision |
15–49 | Unknown, USA | Coronal sulcus (CS) | Swabs | 16S rRNA amplification of the V3-V6 region | Male circumcision reduced the prevalence and the absolute abundance of CS bacteria and the diversity of microbiota. Day 0: Prevalent but low abundance: Prevotella sp., Clostridiales and Corynebacterium sp. At 5%: Peptoniphilus sp., Anaerococcus sp., Fenigoldia sp., Murdochiella sp., Porphyromonas sp., and Lactobacillus sp. Year 1: Reduction in bacterial load on post-circumcision: Porphymonas sp., Prevotella sp., Negativicoccus sp., Dialister sp., Mobiluncus sp., and 6 genera from Clostridiales family XI. No reduction: Atopolium sp., Sneathia sp., and Megasphaera. More prevalent: Kocuria sp. and Facklamia sp. |
No ethnicity records. | Good |
| Liu et al. [19] | Case control | Circumcision and Human immunodeficiency virus (HIV) |
46 HIV-positive uncircumcised 136 HIV-negative uncircumcised |
15–49 | Unknown, Uganda | Coronal sulcus | Swabs | 16S rRNA amplification of the V3–V4 region | HIV-positive uncircumcised: ↑ penile anaerobes. Most prevalent genera HIV-positive uncircumcised: Prevotella, Dialister, Mobiluncus, Murdochiella, and Peptostreptococcus. | No ethnicity records. | Good |
| Nelson et al. [20] | Observational cohort | Circumcision | 18 HV (6 uncircumcised and 12 circumcised) |
14–17 | Mixed Black, Caucasian, and Latin, USA | Coronal sulcus | Swabs and first catch urine |
16S rRNA amplification of the V1–V3, V3–V5, and V6–V9 regions | Microbiome CS ≠ urine. Most prevalent genera CS: Corynebacteria, Staphylococcus, Anaerococcus, Peptoniphilus, Prevotella, Finegoldia, Porphyromonas, Propionibacterium, and Delftia. Uncircumcised: ↑ Prevotella and Porphyromonas. Most prevalent genera urine: Streptococcus, Lactobacillus, Gardnerella, and Veillonella. | Small sample size. | Fair |
| Plummer et al. [21] | Randomized controlled trial | Bacterial vaginosis under antibiotic treatment | 27 couples | >18 | Unknown, Australia | Penis | Swab and first catch urine | 16S rRNA amplification of the V3–V4 region | Day 0: Male specimens were heterogeneous in composition. Most abundant in penile swab: Corynebacterium, Staphylococcus, Peptoniphilus, and Prevotella. Day 8: Decreased on penile swab taxa—Anaerococcus, Finegoldia, Peptoniphilus, Prevotela spp., and Dialister. (↑ Staphyloccocus) | Small sample size. Self-collected swab. | Fair |
| Price et al. [22] | Randomized controlled trial | Circumcision | 12 HIV-negative pre- and post-circumcision | 15–49 | Unknown, Uganda | Coronal sulcus | Swabs | 16S rRNA amplification of the V4 region | Microbiome post-circumcision: ↓ penile anaerobes. Most prevalent genera pre-circumcision: Anaerococcus, Peptoniphilus, Finegoldia, and Prevotella. Most prevalent genera post-circumcision: Staphylococcus and Corynebacterium. | Small sample size. No ethnicity records. | Poor |
| Storm et al. [23] | Observational cohort | Healthy | 48 HV males 18 HV females |
0–18 | Unknown, USA | Males: urethra, perineum, and foreskin Females: urethra, perineum, and vagina |
Swabs | 16S rRNA amplification of the V4 region | Perineal microbiomes differed significantly by age; urethral and foreskin microbiomes did not. Most common genera foreskin: Prevotella, Staphylococcus, Corynebacterium, Peptoniphilus, Mobiluncus, and Winkia. | Small sample size. No record of the pubertal status of the older cohort. No ethnicity records. V4 region is considered a relatively low informative region for taxonomic assignment. | Fair |
| Watchorn et al. [24] | Case control | Male genital lichen sclerosus (MGLSc) | 20 MGLSc uncircumcised 20 HV uncircumcised |
MGLSs:26–73 HV:19–63 |
Unknown, UK | Balanopreputial sac (BS) (Glans + inner prepuce) |
Swabs and first catch urine |
16S rRNA amplification of the V3–V4 region | Microbiome BS (MGLSc) ∼ urine (MGLSc). Microbiome BS (MGLSc) ≠ balanopreputial sac (HV). Most prevalent genera BS (MGLSc): Enterobacteriaceae, Prevotella, Fusobacterium, and Finegoldia. Most prevalent genera BS (HV): ↑ Finegoldia, Staphyloccocus, Corynebacterium. | Small sample size. No ethnicity records. | Poor |
| Zozaya et al. [25] | Cross-sectional | Bacterial vaginosis (BV) | 65 HV-males (23 circumcised) 65 BV-males (35 circumcised) |
Mean 30.7 | African American, USA | Glans, coronal sulcus, penis | Swab | 16S rRNA amplification of the V4-V6 region | More penile skin diversity of BV-males than normal-males, but urethral diversity did not differ between groups. BV-associated bacteria were more abundant in penile and urethral specimens on BV-males. Most abundant on penile skin of BV-males: Peptoniphilus, Anaerococcus, Pv. 123-f-82, Pv. 123-b-46, Lactobacillus iners, and Pv.123-f-110. Most abundant on penile skin of HV: Peptoniphilus, Anaerococcus, Pv 123-f-82, L. iners, Porphyromonas, and Prevotela disiens. | No records of recruitment duration. | Good |
From the 10 studies that were included in this review, all of them employed 16S rRNA gene amplicon sequencing for analysis of the male genital mucosa microbiome. However, the amplification regions of the 16S rRNA genes were different (V3–V4, n = 4; V4, n = 3; V3–V6, n = 1; V4–V6, n = 1; and combination of V1–V3, V3–V5 and V6–V9, n = 1). None of these studies analyzed the fungal fraction of the microbiota through sequencing of the internal transcribed spacer (ITS) region of the rDNA. In total, the male genital mucosa microbiome analysis has been performed on 697 men (Table 1).
3.2. Pediatric Foreskin Microbiome
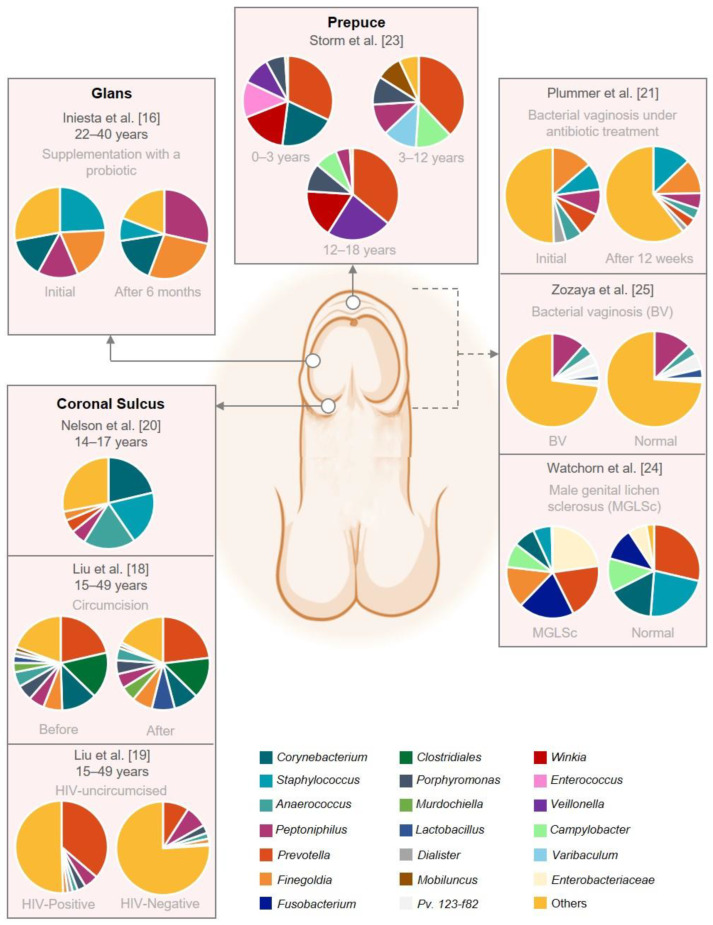
The study by Storm et al. [23] is the only one included in this review that performs the analysis of the foreskin microbiome at the pediatric age. Forty-eight males and twenty-six females less than 18 years old without previous antibiotic exposure were recruited to study the urobiome. In contrast to the female urobiome (urethra, perineum, and vagina), no significant changes were observed in the urethral and foreskin microbiome composition over time in males. Only the perineal microbiome differed significantly between prepubertal/toilet-trained (3–12 years) and post-pubertal (>12 years) males. The foreskin microbiome showed a higher abundance of Prevotella, Staphylococcus, Corynebacterium, Peptoniphilus, Mobiluncus, and Winkia. In the perineum of pre-toilet-trained boys, Veillonella, Bifidobacterium, and Enterococcus genera were abundant. In prepubertal boys, Peptoniphilus sp., Anaerococcus sp., Faecalibacterium sp., and Finegoldia sp. were the most abundant organisms. In post-pubertal boys, Corynebacterium spp. was abundant; and C. tuberculosteraricum and S. aureus were the most common species. This study demonstrated that a core group of urinary bacteria is present in early infancy and changes throughout childhood, with differences between males and females.
3.3. Circumcision and Coronal Sulcus Microbiome
Three studies investigated the effect of the male circumcision on the coronal sulcus microbiome of healthy men [18,20,22]. Nelson et al. [20] compared the microbiome of the coronal sulcus and urine from six uncircumcised and twelve circumcised adolescent men between 14 and 17 years old. Differences in the coronal sulcus and urine microbiome composition were found, the coronal sulcus microbiome being more stable and strongly influenced by circumcision. The coronal sulcus contained high proportions of the genera Corynebacteria, Staphylococcus, Anaerococcus, Peptoniphilus, Prevotella, Finegoldia, and Porphyromonas, while in urine, Streptococcus, Lactobacillus, Gardnerella, and Veillonella were the most predominant genera. Some taxa including Prevotella and Porphyromonas were more abundant in uncircumcised men but were not the main components of the coronal sulcus microbiome. The authors also detected bacterial vaginosis associated taxa, including Atopobium sp., Megasphaera sp., Mobiluncus sp., Prevotella sp., and Gemella sp., in the coronal sulcus from sexually experienced and inexperienced men.
Price et al. [22] characterized the coronal sulcus microbiome before and after circumcision from twelve HIV-negative participants aged 15–49 years during a randomized trial of male circumcision for HIV prevention. Price and colleagues reported that the coronal sulcus microbiome before circumcision was more heterogeneous than after circumcision and similar to several core community types observed in the vagina. The most prevalent genera in the coronal sulcus before circumcision were Anaerococcus, Peptoniphilus, Finegoldia, and Prevotella. After circumcision, Staphylococcus and Corynebacterium were the most prevalent genera. In addition, the authors observed a decrease in anaerobic bacteria due to the elimination of anoxic microenvironments under the foreskin. They concluded that circumcision alters the coronal sulcus microbiome, and anaerobic and vaginosis associated bacteria are the most abundant before circumcision due to the existence of the moist anoxic microenvironment of the subpreputial space. Additionally, Liu et al. [18] also evaluated the coronal sulcus microbiome of seventy-nine HIV-negative men (15–49 years) randomly assigned to receive male circumcision and seventy-seven men that remained uncircumcised. Significant changes were observed in the coronal sulcus bacterial load, i.e., male circumcision was associated with a decrease in the coronal sulcus bacterial load relative to uncircumcised men. The prevalence and absolute abundance of 15 coronal sulcus bacteria, among which 12 were anaerobes, decreased significantly in the circumcised men, whereas aerobes increased after circumcision. This reduction includes species of Porphymonas, Prevotella, Negativicoccus, Dialister, Mobiluncus, and six genera from Clostridiales family XI. No significant decrease was observed in other anaerobe species such as Atopobium sp., Sneathia sp., and Megasphaera sp. after circumcision. Seven coronal sulcus bacteria were found to become more prevalent after circumcision; e.g., the aerobic Kocuria spp. and the facultative anaerobic Facklamia spp. were found to be more prevalent exclusively among circumcised males. The authors concluded that the bacterial changes identified in circumcised and uncircumcised HIV-negative men may play an important role in the HIV risk reduction conferred by male circumcision.
To test the hypothesis that penile anaerobe abundance directly promotes HIV risk by inducing a proinflammatory response in the foreskin, Liu et al. [19] used a case-control study. The authors compared the microbiome and cytokine levels in the penile coronal sulcus in 46 uncircumcised men who seroconverted and 136 uncircumcised men who remained HIV seronegative (controls) during a randomized-controlled trial of medical male circumcision in Rakai, Uganda. Total penile bacterial loads were similar in males HIV-positive and HIV-negative. However, males who acquired HIV had significantly higher abundances of penile anaerobes than men who remained HIV negative during the trial, including Prevotella, Dialister, Mobiluncus, Murdochiella, and Peptostreptococcus genera. Without any adjustment for risk factors, the authors found a consistent relationship between anaerobe abundance and HIV seroconversion among 5 of the 10 anaerobic genera. After adjustment, the association between the abundance of anaerobic bacteria and the odds of HIV seroconversion strengthened. Species associated with the greatest increased risk of HIV seroconversion were Prevotella, followed by Dialister, and six other genera of anaerobic bacteria (Peptoniphilus, Finegoldia, Porphyromonas, Mobiluncus, Peptostreptococcus, and Murdochiella). The authors concluded that uncircumcised men who became HIV infected during a 2-year clinical trial had higher levels of penile anaerobes in comparison with uncircumcised men who remained HIV negative.
3.4. Male Genital Mucosa Microbiome and Bacterial Vaginosis
Two studies investigated the role of the microbiome of male genital mucosa associated with bacterial vaginosis. Zozaya et al. [25] examined the microbiome composition of genital bacteria in monogamous couples, including penile skin (glans, the coronal sulcus, and the shaft of the penis) and urethral specimens from predominantly African American men whose sexual partners were diagnosed with bacterial vaginosis, as defined by the Nugent and Amsel score [26]. Penile skin diversity of sixty-five males (23 circumcised and 42 uncircumcised) in the bacterial vaginosis-couples group and sixty-five control males (partners of women without bacterial vaginosis) (35 circumcised and 30 uncircumcised) were analyzed. The authors observed that the penile skin diversity of male partners of women with bacterial vaginosis was significantly higher than that of control males, but urethral diversity did not differ between groups. Moreover, the results showed that in bacterial vaginosis-couples, the penile skin communities were significantly more similar to the vaginal communities of their sexual partner. The authors concluded that sexual transmission of bacterial vaginosis associated bacteria is a common occurrence during sex. No clear separation was found between the penile skin microbiome of circumcised and uncircumcised men, for both males’ partners groups. In general, the penile skin microbiome in this study showed a higher abundance of Peptoniphilus sp., Anaerococcus sp., Pv. 123-f-82, and Lactobacillus iners in both males’ groups.
Plummer et al. [21] observed that concurrent partner treatment for bacterial vaginosis significantly altered the composition of the genital microbiome of both partners in 27 couples. After 12 weeks of antibiotic administration, a reduction in bacterial vaginosis associated bacteria was observed. The abundance of 11 taxa, including the genera Anaerococcus, Finegoldia, Peptoniphilus, Prevotela, and Dialister, reduced significantly, while Staphyloccocus sp. significantly increased during treatment. The authors concluded that prescription of antibiotic treatment for male partners may be a strategy to achieve a sustained bacterial vaginosis cure to improve reproductive and sexual health for women.
3.5. The Microbiome in Genital Mucosa Inflammation
Two recent studies investigated the role of the microbiome in balanoposthitis and lichen sclerosus inflammation that affect both the glans penis and prepuce, comparing patients to controls. The study conducted by Li et al. [17] found no significant differences in the genital mucosa microbiome between 26 men with balanoposthitis and 29 healthy men. However, differences were observed between men with balanoposthitis and healthy men with redundant prepuce. The dominant species in men with balanoposthitis were Staphylococcus warneri and Prevotella bivia, which were positively correlated with disease severity. In healthy men with redundant prepuce, the most prevalent species were Ezakiella sp. and Porphyromonas somerae.
Watchorn et al. [24] observed differences in the bacterial composition in the balanopreputial sac in 20 uncircumcised men aged 26–73 years with male genital lichen sclerosus (MGLSc) compared to 20 healthy uncircumcised men aged 19–63 years. Enterobacteriaceae, Prevotella, and Fusobacterium were identified as the most abundant taxa in the MGLSc group, but not in healthy men. In the control group, the most dominant genera were Finegoldia, Staphyloccocus, and Corynebacterium. The relative abundance of Finegoldia was lower in the MGLSc group than in the control group, and Fusobacterium was higher in the MGLSc group than in the control group.
3.6. Influence of Probiotic Supplementation on Glans Microbiome
A study carried out by Iniesta et al. [16] evaluated the effect of Ligilactobacillus salivarius PS116610 on the microbial composition of the urogenital tract in seventeen Caucasian (Spanish) infertile couples aged 20–40 years and under assisted reproduction treatment diagnosed with bacterial dysbiosis. Glans swabs from enrolled males were analyzed at the beginning of the study and after 3 and 6 months of treatment. Unlike the vagina microbiome, no significant changes were observed in the microbiome composition before and after the probiotic treatment in males, although a slight decrease in urogenital pathogens was registered. The glans microbiome from enrolled males is mainly composed by Peptoniphilus sp., Finegoldia sp., Corynebacterium sp., and Staphylococcus sp. The authors concluded that probiotic supplementation with L. salivarius PS116610 in couples with idiopathic infertility under assisted reproduction treatment improved the urogenital tract microbiome, solving the dysbiosis in 88.9% of the couples.
4. Discussion
To our knowledge, this is the first study that assesses the microbiome of healthy and diseased male genital mucosa. Due to the advances in high-throughput sequencing technologies, bioinformatic tools, and mass spectrometry techniques, omics approaches have enhanced our ability to characterize the diversity and function of microbiome. Some of the meta-omics technologies (metagenomics, metatranscriptomics, metaproteomics, and metametabolomics) have been used for clinical diagnosis in various diseases [27,28,29,30,31]. The 16S rRNA gene has been most frequently targeted due to its presence in all prokaryotes. However, to characterize the full genetic content of a community, metagenomic studies go beyond the 16S rRNA gene [27]. Here, all studies performed 16S rRNA sequencing, but focusing on different male age groups, diseases, and conditions. Studies ranged from childhood to early elderly, male genital inflammation (balanoposthitis and male genital lichen sclerosus), female genital inflammation (bacterial vaginosis), male circumcision, and HIV.
One of the main findings of this study is that there is very limited knowledge on the male genital mucosa (prepuce, glans, and coronal sulcus) microbiome. Several bacterial taxa have been indicated as resident microbiota on male genital mucosa such as Prevotela, Finegoldia, Peptoniphilus, Staphylococcus, Corynebacterium, and Anaerococcus. The higher prevalence of such taxa suggests its possible provenance from other sites, such as superficial and sebaceous skin, the inguinal region, the gut, or even vaginal associated taxa [8,32]. The genus Prevotella is the only strict anaerobe, frequently found in the gut [33], and it is negatively associated with sperm mobility [34]. Species of Finegoldia, Peptoniphilus, and Anaerococcus have been reported as colonizers of the skin and mucosal surfaces, such as the mouth, upper respiratory tract, gastrointestinal tract, and female genitourinary tract [35]. Notably, in the healthy male genital mucosa, these species were also found. Furthermore, coryneform bacteria were also identified in many studies on the male urogenital tract [36]. Frequently, these bacteria tend to be overlooked as commensals, but some authors associated these microorganisms with infection [17,24,37].
The representation of the prepuce, glans, and coronal sulcus microbiome provided in Figure 2 suggests that different bacteria can colonize male genital anatomic sites and determine site-specific microbiota. The microbiome of the glans and coronal sulcus appears to be similar in all studies included in this review. However, the only study available on the microbiome of the prepuce focused on subjects under 18 years old [23]. The authors stated that the most abundant microorganisms were the bacterial genera Prevotella, Staphylococcus, Corynebacterium, Peptoniphilus, Mobiluncus, and Winkia. We should highlight that the reporting of one study hampers a comprehensive comparison of the microbiome of the prepuce in older men. Moreover, this study analyzed a small sample size and did not report the pubertal status of an older cohort, which should be encouraged in future studies.
Figure 2.
Graphical representation of the male genital mucosa microbiome composition. This figure was based on eight studies that reported the abundance on genus level. The remaining studies did not have raw abundance data available. The figure was created with BioRender.com (accessed on 16 September 2022).
One of the key points of this review was to identify the microbial community’s composition in the different anatomical parts of the healthy and diseased male genital mucosa. Therefore, the current study confirmed the previous findings in which the male genital mucosa microbiome can be influenced by many factors, such as physical barriers, inflammation, infection, interaction within adjacent niches, antimicrobial peptides, and lipids [16,38,39]. Thus, changes in host-genital microbiome interactions may be linked to disease development [23].
Male circumcision is one of the most common surgical procedures in the world, and, in some situations, this type of surgery is necessary for medical reasons [40]. The four studies included in this review that investigated the effect of the male circumcision on the coronal sulcus microbiome share similar conclusions. Male circumcision appears to impact the coronal sulcus microbiome due to the elimination of the anoxic subpreputial microenvironment. Prepuce removal unbalances the anaerobic sites; consequently, the abundance of anaerobic bacteria decreases, exerting a selective pressure for aerobic bacteria, along with bacterial competition [18,22]. Moreover, higher levels of penile anaerobes detected in uncircumcised men were associated with higher production of immune factors that recruit HIV target cells [19] and the abundance of vaginal-associated taxa [20]. Moreover, the studies by Zozaya et al. [25] and Plummer et al. [21] showed that sexual transmission of vaginal-associated bacteria, such as bacterial vaginosis, is a common occurrence; organisms might be exchanged during sexual intercourse. In fact, some studies already showed that sexual history could be a determinant of the penis microbiome composition. Dong et al. [41] and Nelson et al. [42] showed that the penis and the urethra can be colonized by a variety of bacterial vaginosis associated taxa.
Comparing the present results of the male genital mucosa microbiome with the recent systematic review by Pagan et al. [8] of the vulvar microbiome, the taxa present on the vulva are equivalent to some of the most prevalent taxa of male genital mucosa, such as the genera Corynebacterium, Staphylococcus, and Prevotella, but with the exception of Lactobacillus. In fact, Lactobacillus are well-known lactic-acid-producing bacteria that colonize the female genital tract [43]. The authors suggest that these bacteria might be exhibited in vaginal, cutaneous, and intestinal niches. As mentioned above, the male genital mucosa microbiome can be influenced by adjacent or nearby niches, which may include the perineum, inguinal region, and intestine. Both male and female genital mucosa are characterized as a moist environment [8,18] creating the favorable conditions for certain microorganisms. Therefore, the understanding of shared genital microbiota between sexual partners might be a milestone in genitourinary diseases.
Nevertheless, several limitations of the current literature can be identified. Firstly, the sample population of practically all studies is rather low (ranging between 12 to 50 participants), with the exception of three (out of ten) studies that encompass more than 130 participants per study [18,19,25]. In addition, participants’ ethnicity data are most often absent. Three studies identified the ethnicity record of participants: Caucasian [16], African American [25], and one study included a diverse population, i.e., Mixed Black, Caucasian, and Latin [20]. Furthermore, most of these studies focused on a specific sample of individuals enrolled in medical clinics with different specialties, such as in sexually transmitted disease clinics, HIV prevention, or in dermatology clinics. Moreover, the current literature includes different male age groups. Only two studies addressed the analysis of the microbiome at the pediatric age [20,23] but with different purposes. Nelson et al. [20] evaluated the bacterial communities of the coronal sulcus and distal urethra of circumcised and uncircumcised adolescent males, and Storm et al. [23] studied the urethral and foreskin microbiome composition over time in males aged 0 to 18 years. Therefore, results from these studies should be carefully interpreted, and type I errors should be noted for future research.
Moreover, different sample collection methods and the possible risk of contamination during sampling prevented direct comparison between studies. It is worth noting that, when the catheterized urine technique is applied, there is a reduction in contamination by the urethra, avoiding the overrepresentation of microorganisms [23]. Genital hygiene and frequency of sexual intercourse should also be considered. Self-hygiene prior to sample collection can add bias to microbiome studies as well as self-collected samples, such as in the study by Plummer et al. [21] whose males self-collected a cutaneous penile swab.
Notably, none of the ten studies under the scope of this systematic review investigated the mycobiome, virome, and parasitome in male genital mucosa. All studies analyzed the bacteriome through 16S rRNA gene amplicon sequencing. The usefulness of the 16s rRNA gene sequencing technique is well known [44]. However, its range to the genus level is a limitation [21], and the choice of the hypervariable region target of the 16s rRNA gene can lead to a discrepancy in microbial quantification and influences the outcomes. As mentioned by Li et al. [17], the V4 region is considered a relatively low informative region for taxonomic assignment. Thus, choosing the ideal 16S rRNA hypervariable region will depend on the bacterial composition according to the studied environment [45]. For example, Kameoka et al. [46] showed that the V1–V2 region is more precise than V3–V4 for the gut microbiome of Japanese individuals, while Hoffman et al. [47] found that the V1–V3 and V2–V3 regions are preferred on the female urobiome. Additionally, Graspeuntner et al. [48] recognized that the V1–V3 region allows for a more complete assessment of the cutaneous and vaginal analysis. Furthermore, based on systematic comparisons of all nine 16S rRNA hypervariable regions, Heidrich et al. [45] concluded that V1–V2 is more suitable for male urinary microbiota profiling. To our knowledge and given the few available studies about the male genital mucosa microbiome, there is no information yet concerning the best 16S rRNA hypervariable region for microbial characterization of the prepuce, glans, and coronal sulcus. Interestingly, there was an enormous discrepancy in the 16S rRNA hypervariable region used in the studies included in this review. For example, to evaluate the glans and prepuce microbiome, Watchorn et al. [24] and Plummer et al. [21] used primers for the V3–V4 region, while Zozaya et al. [25] used the V4–V6 region. The same incongruity was observed for coronal sulcus microbiome studies, in which Liu et al. [19] used the V3–V4 region, and Liu et al. [18] used the V3–V6 region.
5. Conclusions
This systematic review represents the first report that encompasses the composition of microbial communities in the different anatomical parts of the healthy and diseased male genital mucosa. Following the analysis of the selected studies, we concluded that the knowledge about the microbiome of the male genital mucosa is still very limited. Besides that, some limitations can also be found across these studies, namely the small size of samples, the lack of ethnicity records, different methodological approaches (e.g., self-collected swab), and different microbiome analysis methods (e.g., 16S rRNA hypervariable regions). Therefore, these limitations can bias the results and comparisons between studies.
The microbiome composition of the male genital mucosa shows a high diversity of commensals organisms from the adjacent anatomical sites (perineum, skin, gut) or even urine. In addition, due to different types of sexual activities, the penis and urethra can be colonized by a variety of microorganisms, including bacterial vaginosis associated taxa as a result from partnered sexual activity. We cannot rule out that some of these microorganisms may play a specific role in the local milieu, providing the male genital mucosa a distinct signature that should be better elucidated in further studies. Acknowledging the microbiome of male genital mucosa is essential to understand and determine the interaction and role of the different microorganisms in male reproductive health, men’s fertility, and sexual behavior. Thus, changes in the microbiome composition may represent risk factors for microbial infection.
Future studies unraveling the mycobiome, virome, and parasitome on male genital mucosa are highly needed and should be addressed due to the prevalence of sexual transmitted fungal, viral and parasite infections, such as candidosis, trichomoniasis, HIV, and herpes. Moreover, studies comparing the sequencing performance of different hypervariable regions of 16S rRNA are necessary to identify which primer sets and combinations are more suitable for each anatomical part of the male genital mucosa. Despite the research motivation, a holistic and integrated approach combining other culture-independent techniques, such as pyrosequencing, whole metagenome sequencing and quantitative real-time PCR, and other meta-omics technologies are essential to ensure the accuracy of the results. This elucidates potential biomarkers intended for the diagnosis, prevention, and management of infections by screening for microbiological risk factors.
Acknowledgments
The authors acknowledge the individual post-doctoral research grant offered to support the project to M.F.M.G. and the research grant to Â.R.F.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10122312/s1, File S1: Search strategy.
Author Contributions
Conceptualization, C.L. and A.G.R.; methodology, M.F.M.G. and Â.R.F.; formal analysis, M.F.M.G. and Â.R.F.; investigation, M.F.M.G. and Â.R.F.; data curation, M.F.M.G., Â.R.F., and C.L.; writing—original draft preparation, M.F.M.G. and Â.R.F.; writing—review and editing, M.F.M.G., A.G.R., and C.L.; supervision, C.L.; project administration, C.L. and A.G.R. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was part of the project NORTE-01-0145-FEDER-000057 (SexHealth & Prostate Cancer, Psychobiological Determinants of Sexual Health in Men with Prostate Cancer), financial support from the Horizon Europe programme and Norte 2020.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Waldman A.J., Balskus E.P. The human microbiota, infectious disease, and global health: Challenges and opportunities. ACS Infect. Dis. 2018;4:14–26. doi: 10.1021/acsinfecdis.7b00232. [DOI] [PubMed] [Google Scholar]
- 2.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koedooder R., Mackens S., Budding A., Fares D., Blockeel C., Laven J., Schoenmakers S. Identification and evaluation of the microbiome in the female and male reproductive tracts. Hum. Reprod. Update. 2019;25:298–325. doi: 10.1093/humupd/dmy048. [DOI] [PubMed] [Google Scholar]
- 4.Slingerland A.E., Schwabkey Z., Wiesnoski D.H., Jenq R.R. Clinical evidence for the microbiome in inflammatory diseases. Front. Immunol. 2017;8:400. doi: 10.3389/fimmu.2017.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreno B., Araviiskaia E., Berardesca E., Gontijo G., Sanchez Viera M., Xiang L.F., Martin R., Bieber T. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016;30:2038–2047. doi: 10.1111/jdv.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saus E., Iraola-Guzman S., Willis J.R., Brunet-Vega A., Gabaldon T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol. Asp. Med. 2019;69:93–106. doi: 10.1016/j.mam.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katongole P., Sande O.J., Joloba M., Reynolds S.J., Niyonzima N. The human microbiome and its link in prostate cancer risk and pathogenesis. Infect. Agent Cancer. 2020;15:53. doi: 10.1186/s13027-020-00319-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pagan L., Ederveen R.A.M., Huisman B.W., Schoones J.W., Zwittink R.D., Schuren F.H.J., Rissmann R., Piek J.M.J., Van Poelgeest M.I.E. The human vulvar microbiome: A systematic review. Microorganisms. 2021;9:2568. doi: 10.3390/microorganisms9122568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altmae S., Franasiak J.M., Mandar R. The seminal microbiome in health and disease. Nat. Rev. Urol. 2019;16:703–721. doi: 10.1038/s41585-019-0250-y. [DOI] [PubMed] [Google Scholar]
- 10.Baud D., Pattaroni C., Vulliemoz N., Castella V., Marsland B.J., Stojanov M. Sperm microbiota and its impact on semen parameters. Front. Microbiol. 2019;10:234. doi: 10.3389/fmicb.2019.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuddenham S., Ravel J., Marrazzo J.M. Protection and risk: Male and female genital microbiota and sexually transmitted infections. J. Infect. Dis. 2021;223:S222–S235. doi: 10.1093/infdis/jiaa762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen P.V., Kafka J.K., Ferreira V.H., Roth K., Kaushic C. Innate and adaptive immune responses in male and female reproductive tracts in homeostasis and following HIV infection. Cell. Mol. Immunol. 2014;11:410–427. doi: 10.1038/cmi.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Heart Lung and Blood Institute . Study Quality Assessment Tools. NHLBI; Bethesda, MD, USA: 2019. [Google Scholar]
- 15.Sterne J.A.C., Savovic J., Page M.J., Elbers R.G., Blencowe N.S., Boutron I., Cates C.J., Cheng H.Y., Corbett M.S., Eldridge S.M., et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366:l4898. doi: 10.1136/bmj.l4898. [DOI] [PubMed] [Google Scholar]
- 16.Iniesta S., Esteban S., Armijo O., Lobo S., Manzano S., Espinosa I., Cardenas N., Bartha J.L., Jimenez E. Ligilactobacillus salivarius PS11610 exerts an effect on the microbial and immunological profile of couples suffering unknown infertility. Am. J. Reprod. Immunol. 2022;88:e13552. doi: 10.1111/aji.13552. [DOI] [PubMed] [Google Scholar]
- 17.Li M., Mao J.X., Jiang H.H., Huang C.M., Gao X.H., Zhang L. Microbiome profile in patients with adult balanoposthitis: Relationship with redundant prepuce, genital mucosa physical barrier status and inflammation. Acta Derm. Venereol. 2021;101:adv00466. doi: 10.2340/00015555-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C.M., Hungate B.A., Tobian A.A., Serwadda D., Ravel J., Lester R., Kigozi G., Aziz M., Galiwango R.M., Nalugoda F., et al. Male circumcision significantly reduces prevalence and load of genital anaerobic bacteria. mBio. 2013;4:e00076. doi: 10.1128/mBio.00076-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C.M., Prodger J.L., Tobian A.A.R., Abraham A.G., Kigozi G., Hungate B.A., Aziz M., Nalugoda F., Sariya S., Serwadda D., et al. Penile anaerobic dysbiosis as a risk factor for HIV infection. mBio. 2017;8:e00996-17. doi: 10.1128/mBio.00996-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson D.E., Dong Q., Van der Pol B., Toh E., Fan B., Katz B.P., Mi D., Rong R., Weinstock G.M., Sodergren E., et al. Bacterial communities of the coronal sulcus and distal urethra of adolescent males. PLoS ONE. 2012;7:e36298. doi: 10.1371/journal.pone.0036298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plummer E.L., Vodstrcil L.A., Doyle M., Danielewski J.A., Murray G.L., Fehler G., Fairley C.K., Bulach D.M., Garland S.M., Chow E.P.F., et al. A prospective, open-label pilot study of concurrent male partner treatment for bacterial vaginosis. mBio. 2021;12:e0232321. doi: 10.1128/mBio.02323-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price L.B., Liu C.M., Johnson K.E., Aziz M., Lau M.K., Bowers J., Ravel J., Keim P.S., Serwadda D., Wawer M.J., et al. The effects of circumcision on the penis microbiome. PLoS ONE. 2010;5:e8422. doi: 10.1371/journal.pone.0008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storm D.W., Copp H.L., Halverson T.M., Du J., Juhr D., Wolfe A.J. A Child’s urine is not sterile: A pilot study evaluating the Pediatric Urinary Microbiome. J. Pediatr. Urol. 2022;18:383–392. doi: 10.1016/j.jpurol.2022.02.025. [DOI] [PubMed] [Google Scholar]
- 24.Watchorn R.E., Van den Munckhof E.H.A., Quint K.D., Eliahoo J., de Koning M.N.C., Quint W.G.V., Bunker C.B. Balanopreputial sac and urine microbiota in patients with male genital lichen sclerosus. Int. J. Dermatol. 2021;60:201–207. doi: 10.1111/ijd.15252. [DOI] [PubMed] [Google Scholar]
- 25.Zozaya M., Ferris M.J., Siren J.D., Lillis R., Myers L., Nsuami M.J., Eren A.M., Brown J., Taylor C.M., Martin D.H. Bacterial communities in penile skin, male urethra, and vaginas of heterosexual couples with and without bacterial vaginosis. Microbiome. 2016;4:16. doi: 10.1186/s40168-016-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sha B.E., Chen H.Y., Wang Q.J., Zariffard M.R., Cohen M.H., Spear G.T. Utility of Amsel criteria, Nugent score, and quantitative PCR for Gardnerella vaginalis, Mycoplasma hominis, and Lactobacillus spp. for diagnosis of bacterial vaginosis in human immunodeficiency virus-infected women. J. Clin. Microbiol. 2005;43:4607–4612. doi: 10.1128/JCM.43.9.4607-4612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karaduta O., Dvanajscak Z., Zybailov B. Metaproteomics—An advantageous option in studies of host-microbiota interaction. Microorganisms. 2021;9:980. doi: 10.3390/microorganisms9050980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buffet-Bataillon S., Rizk G., Cattoir V., Sassi M., Thibault V., Del Giudice J., Gangneux J.P. Efficient and quality-optimized metagenomic pipeline designed for taxonomic classification in routine microbiological clinical tests. Microorganisms. 2022;10:711. doi: 10.3390/microorganisms10040711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Cobas A.E., Gosalbes M.J., Friedrichs A., Knecht H., Artacho A., Eismann K., Otto W., Rojo D., Bargiela R., Von Bergen M., et al. Gut microbiota disturbance during antibiotic therapy: A multi-omic approach. Gut. 2013;62:1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu I.W., Gao S.S., Chou H.C., Yang H.Y., Chang L.C., Kuo Y.L., Vinh Dinh M.C., Chung W.H., Yang C.W., Lai H.C., et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics. 2020;10:5398. doi: 10.7150/thno.41725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Y., Ramnarine V.R., Bell R., Volik S., Davicioni E., Hayes V.M., Ren S., Collins C.C. Metagenomic and metatranscriptomic analysis of human prostate microbiota from patients with prostate cancer. BMC Genom. 2019;20:146. doi: 10.1186/s12864-019-5457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Z., Tseng C.H., Pei Z., Blaser M.J. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ley R.E. Prevotella in the gut: Choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016;13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 34.Campisciano G., Iebba V., Zito G., Luppi S., Martinelli M., Fischer L., De Seta F., Basile G., Ricci G., Comar M. Lactobacillus iners and gasseri, Prevotella bivia and HPV belong to the microbiological signature negatively affecting human reproduction. Microorganisms. 2020;9:39. doi: 10.3390/microorganisms9010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy E.C., Frick I.M. Gram-positive anaerobic cocci-commensals and opportunistic pathogens. FEMS Microbiol. Rev. 2013;37:520–553. doi: 10.1111/1574-6976.12005. [DOI] [PubMed] [Google Scholar]
- 36.Mandar R. Microbiota of male genital tract: Impact on the health of man and his partner. Pharmacol. Res. 2013;69:32–41. doi: 10.1016/j.phrs.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Dutta S., Sengupta P., Izuka E., Menuba I., Jegasothy R., Nwagha U. Staphylococcal infections and infertility: Mechanisms and management. Mol. Cell. Biochem. 2020;474:57–72. doi: 10.1007/s11010-020-03833-4. [DOI] [PubMed] [Google Scholar]
- 38.Fischer C.L. Antimicrobial activity of host-derived lipids. Antibiotics. 2020;9:75. doi: 10.3390/antibiotics9020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baurecht H., Ruhlemann M.C., Rodriguez E., Thielking F., Harder I., Erkens A.S., Stolzl D., Ellinghaus E., Hotze M., Lieb W., et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J. Allergy Clin. Immunol. 2018;141:1668–1676. doi: 10.1016/j.jaci.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 40.Iacob S.I., Feinn R.S., Sardi L. Systematic review of complications arising from male circumcision. BJUI Compass. 2022;3:99–123. doi: 10.1002/bco2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Q., Nelson D.E., Toh E., Diao L., Gao X., Fortenberry J.D., Van der Pol B. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS ONE. 2011;6:e19709. doi: 10.1371/journal.pone.0019709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson D.E., Van Der Pol B., Dong Q., Revanna K.V., Fan B., Easwaran S., Sodergren E., Weinstock G.M., Diao L., Fortenberry J.D. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS ONE. 2010;5:e14116. doi: 10.1371/journal.pone.0014116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X., Da M., Zhang W., Qi Q., Zhang C., Han S. Role of Lactobacillus in cervical cancer. Cancer Manag. Res. 2018;10:1219–1229. doi: 10.2147/CMAR.S165228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janda J.M., Abbott S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heidrich V., Inoue L.T., Asprino P.F., Bettoni F., Mariotti A.C.H., Bastos D.A., Jardim D.L.F., Arap M.A., Camargo A.A. Choice of 16S ribosomal RNA primers impacts male urinary microbiota profiling. Front. Cell. Infect. Microbiol. 2022;12:862338. doi: 10.3389/fcimb.2022.862338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kameoka S., Motooka D., Watanabe S., Kubo R., Jung N., Midorikawa Y., Shinozaki N.O., Sawai Y., Takeda A.K., Nakamura S. Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1–V2 and V3–V4 primer sets. BMC Genom. 2021;22:527. doi: 10.1186/s12864-021-07746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman C., Siddiqui N.Y., Fields I., Gregory W.T., Simon H.M., Mooney M.A., Wolfe A.J., Karstens L. Species-level resolution of female bladder microbiota from 16S rRNA amplicon sequencing. mSystems. 2021;6:e00518–e00521. doi: 10.1128/mSystems.00518-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graspeuntner S., Loeper N., Kunzel S., Baines J.F., Rupp J. Selection of validated hypervariable regions is crucial in 16S-based microbiota studies of the female genital tract. Sci. Rep. 2018;8:9678. doi: 10.1038/s41598-018-27757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.