Abstract
Isoegomaketone is a water-soluble natural ketone compound that is commonly present in Rabdosia angustifolia and Perilla frutescens. At present, it is known that isoegomaketone has a wide range of pharmacological activity, but there has been no thorough investigation of its potential targets. As a result, we examined the potential targets of isoegomaketone using the network pharmacology approach. In our study, the TCM Database@Taiwan was utilized to search for the chemical formula. The pharmacological characteristics of isoegomaketone were then evaluated in silico using the Swiss Absorption, Distribution, Metabolism, and Excretion (Swiss ADME) and Deep Learning–Acute Oral Toxicity (DL-AOT) methods, and the potential isoegomaketone target genes were identified using a literature study. Additionally, using the clusterProfiler R package 3.8.1, the Gene Ontology (GO) enrichment analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of target genes were performed. In order to obtain the protein interaction network, we simultaneously submitted the targets to the STRING database. After this, we performed molecular docking with respect to targets and isoegomaketone. Finally, we created visual networks of protein–protein interactions (PPI) and examined these networks. Our results showed that isoegomaketone had good drug-likeness, bioavailability, medicinal chemistry friendliness, and acceptable toxicity. Subsequently, through the literature analysis, 48 target genes were selected. The bioinformatics analysis and network analysis found that these target genes were closely related to the biological processes of isoegomaketone, such as atherosclerotic formation, inflammation, tumor formation, cytotoxicity, bacterial infection, virus infection, and parasite infection. These findings show that isoegomaketone may interact with a wide range of proteins and biochemical processes to form a systematic pharmacological network, which has good value for the creation and use of drugs.
Keywords: isoegomaketone, network pharmacology, molecular docking, biological activity, active molecule
1. Introduction
Traditional Chinese medicine contains a large number of active drug molecules [1]. Through the mining of traditional Chinese medicine, the development of drugs can be greatly enriched [2]. Isoegomaketone is a water-soluble natural ketone compound [3]. It is commonly present in traditional Chinese medicines Rabdosia angustifolia and Perilla frutescens [4]. Recent studies have shown that isoegomaketone has various types of biological activity, such as anti-inflammatory, anti-tumor, anti-rheumatoid arthritis, etc. [5,6,7,8,9]. Thus, isoegomaketone is considered to be an important active component of Perilla frutescens and has great drug development potential. However, the potential targets of isoegomaketone and its potential for further development are not yet fully understood. At present, the extraction of potential targets from existing research and the use of computer methods to predict the potential development directions has become the main method [10,11,12]. Compared with traditional methods, this method provides more convenient technology and a clearer direction for drug design and development.
As a result, we applied the network pharmacology approach to comprehensively examine the pharmacological effects of isoegomaketone. Firstly, the Swiss Absorption, Distribution, Metabolism, and Excretion (Swiss ADME) and Deep Learning–Acute Oral Toxicity (DL-AOT) methods were used to evaluate in silico the drug properties of isoegomaketone. Furthermore, we provided candidate target genes through literature analysis. Additionally, Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, and molecular docking were performed on these potential target genes. Finally, we thoroughly demonstrated the probable targets of the compound by creating the isoegomaketone pharmacological interaction network.
2. Method
2.1. Molecular Formula and In Silico Drug Properties of Isoegomaketone
An important database resource that is free to use and readily available is the TCM Database@Taiwan (http://tcm.cmu.edu.tw/ (accessed on 25 March 2022)), which houses sources of chemical and pharmacological information on isoegomaketone [13]. The Swiss ADME (http://www.swissadme.ch/ (accessed on 25 March 2022)), which supports drug development, enables researchers to calculate the physicochemical descriptors and predict the ADME parameters, pharmacokinetic features, drug-like nature, and medicinal chemistry friendliness of one or more small compounds [14]. Thus, in our study, the in silico drug properties of isoegomaketone were analyzed using the Swiss ADME.
The DL-AOT prediction server (http://www.pkumdl.cn:8080/DLAOT/DLAOThome.php (accessed on 25 March 2022)) is a tool used to evaluate the acute oral toxicity (AOT) of small molecules [15]. Thus, in our study, the in silico toxicity (LD50) of isoegomaketone was analyzed using the DL-AOT prediction server.
2.2. Target Gene Screening for Isoegomaketone
All candidate potential targets are derived from literature analysis. Literature analysis was carried out through various academic databases, with the keyword “isoegomaketone” [5,6,16,17,18,19,20,21,22,23,24]. Finally, eleven published, peer-reviewed pharmacological studies on isoegomaketone were selected. All eleven selected pharmacological studies on isoegomaketone demonstrated high research quality and were all carried out via in vivo or in vitro methods.
2.3. Analysis of PPI Network
One of the online databases that compiles all publicly available sources of knowledge of PPI is STRING 11.0 (https://string-db.org/ (accessed on 29 March 2022)), which enables users to supplement the knowledge already known about PPI with computational predictions [25]. We uploaded 48 putative targets of isoegomaketone to the STRING database in order to build a PPI network, with the species defined as “Homo sapiens” and the minimum interaction score set to 0.4. After this, the outcomes were imported into Cytoscape 3.7.2 for visual evaluation. Cytoscape, an open-source software platform for visualizing complex networks, can calculate the parameters of each node in the network diagram, such as the degree, betweenness centrality (BC), and closeness centrality (CC) [26]. We utilized the cytoHubba plug-in topological method to identify the significant protein nodes and subnetworks in the network, after filtering the target nodes by the matching median values of degree values, BC and CC in the PPI network [27].
2.4. Gene Function and Pathway Enrichment Analysis
GO functional annotation analysis is a standard method used to carry out large-scale functional enrichment analyses of genes, and it comprises biological processes (BP), molecular functions (MF), and cellular components (CC) [28]. It is possible to assign functional meanings to genes and genomes at the molecular and higher levels by adopting the widely used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [29]. Using the clusterProfiler R package 3.8.1 with FDR < 0.2 (FDR, false discovery rate) and p value < 0.05, KEGG pathway analysis and GO analysis were carried out, and the most important targets engaged in the pertinent biological processes were examined.
2.5. Compound–Target Molecular Docking
The 2D structure of isoegomaketone, downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/ (accessed on 26 March 2022)) in mol2 format, was imported into the AutoDockTools 1.5.6 software. After checking its spatial structure, adding atomic charges, assigning atomic types, and changing all flexible bonds to rotatable by default, it was saved in pdbqt format as a docking ligand. The 3D structures of target-gene-associated proteins were downloaded from the Uniprot database (https://www.uniprot.org/ (accessed on 26 March 2022)) and the pdb database (https://www.rcsb.org/ (accessed on 26 March 2022)), and they were then saved in pdb format. AutoDockTools 1.5.6 software was used to carry out the removal of origin ligands, removal of water molecules, hydrogenation, charge addition, and various optimizations. The grid box was set as the default value and the final optimized proteins were saved in pdbqt format as docking acceptors. Molecular docking was performed using AutoDock Vina 1.1.2. The protein structure was set during molecular docking to be a rigid macromolecule, and the Genetic Algorithm Parameters algorithm was used. PyMOL was used to illustrate the outcomes for the group in which each protein had the lowest binding energy.
3. Result
3.1. Molecular Formula and In Silico Drug Properties of Isoegomaketone
We obtained the chemical formula for isoegomaketone using the TCM Database@Taiwan database (Figure 1). Through Swiss ADME, we obtained essential ADME-related data on isoegomaketone, and the drug-likeness as well as the medicinal chemistry of isoegomaketone were subsequently evaluated via the classic formula. The results are provided in Table 1. Regarding the drug-likeness, lead-drug likeness, and medicinal chemistry friendliness, the pharmacokinetic characteristics of isoegomaketone were consistent with Lipinski’s rule of five, the Ghose filter, Veber’s Rule, and the RO (3) rule, and the synthetic accessibility was 2.9. Definitions of these rules and parameters can be found in References [30,31,32,33,34].
Figure 1.
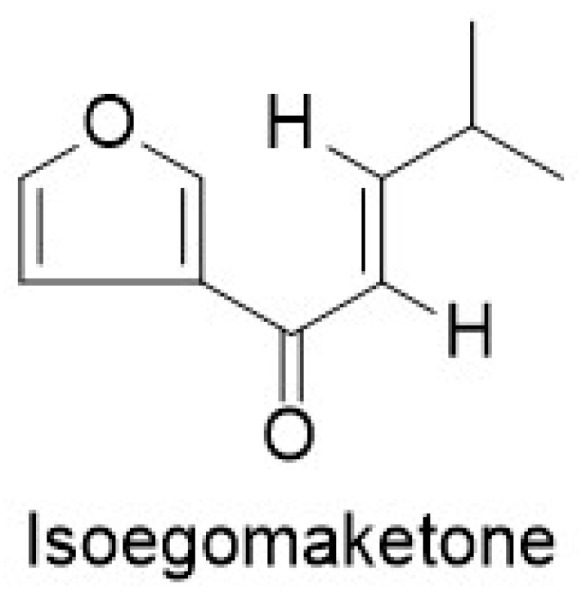
Molecular formula of isoegomaketone.
Table 1.
In silico drug properties of isoegomaketone.
| MW (g/mol) | LogP | nRotB | nHBD | nHBA | MR | TPSA (Å2) |
LogS | GI Absorption | BBB Permeance |
|---|---|---|---|---|---|---|---|---|---|
| 164.20 | 2.40 | 3 | 0 | 2 | 47.7 | 30.21 | −2.39 | High | Yes |
| Lipinski | Ghose | Veber | RO(3) | Synthetic accessibility | |||||
| Yes | Yes | Yes | Yes | 2.89 |
MW = molecular weight, LogP = Log Po/w (iLOGP), nRotB = number of rotatable bonds, nHBD = number of H-bond donors, nHBA = number of H-bond acceptors, MR = molar refractivity, TPSA = topological polar surface area, LogS = Log S (ESOL), GI absorption = gastrointestinal absorption, BBB = blood–brain barrier.
The LD50 value of isoegomaketone was predicted by DL-AOT, as shown in Table 2. The result showed that the LD50 value of isoegomaketone was 31.2 mg/kg, while the evaluation indicated “caution”.
Table 2.
LD50 values of isoegomaketone.
| Compound | LD50 | Toxicity Evaluation |
|---|---|---|
| isoegomaketone | 31.2 | Caution |
Toxicity evaluation classification: (1) danger/poison; (2) warning; (3) caution; (4) none required.
3.2. Candidate Target Genes from Literature Analysis
Through literature analysis, eleven papers were selected. Finally, 48 candidate targets of isoegomaketone were selected for study, as shown in Table 3.
Table 3.
| No. | Gene ID | Gene Name | Gene Description |
|---|---|---|---|
| 1 | ENSG00000275199 | AKT3 | AKT serine/threonine kinase 3 |
| 2 | ENSG00000171791 | BCL2 | BCL2 apoptosis regulator |
| 3 | ENSG00000015475 | BID | BH3 interacting domain death agonist |
| 4 | ENSG00000156709 | AIFM1 | apoptosis inducing factor mitochondria associated 1 |
| 5 | ENSG00000121691 | CAT | catalase |
| 6 | ENSG00000188130 | MAPK12 | mitogen-activated protein kinase 12 |
| 7 | ENSG00000185386 | MAPK11 | mitogen-activated protein kinase 11 |
| 8 | ENSG00000077150 | NFKB2 | nuclear factor kappa B subunit 2 |
| 9 | ENSG00000198793 | MTOR | mechanistic target of rapamycin kinase |
| 10 | ENSG00000125740 | FOSB | FosB proto-oncogene, AP-1 transcription factor subunit |
| 11 | ENSG00000172115 | CYCS | cytochrome c, somatic |
| 12 | ENSG00000007171 | NOS2 | nitric oxide synthase 2 |
| 13 | ENSG00000104856 | RELB | RELB proto-oncogene, NF-kB subunit |
| 14 | ENSG00000132906 | CASP9 | caspase 9 |
| 15 | ENSG00000105221 | AKT2 | AKT serine/threonine kinase 2 |
| 16 | ENSG00000243955 | GSTA1 | glutathione S-transferase alpha 1 |
| 17 | ENSG00000181019 | NQO1 | NAD(P)H quinone dehydrogenase 1 |
| 18 | ENSG00000123080 | CDKN2C | cyclin dependent kinase inhibitor 2C |
| 19 | ENSG00000111537 | IFNG | interferon gamma |
| 20 | ENSG00000105851 | PIK3CG | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| 21 | ENSG00000142208 | AKT1 | AKT serine/threonine kinase 1 |
| 22 | ENSG00000112062 | MAPK14 | mitogen-activated protein kinase 14 |
| 23 | ENSG00000156711 | MAPK13 | mitogen-activated protein kinase 13 |
| 24 | ENSG00000213366 | GSTM2 | glutathione S-transferase mu 2 |
| 25 | ENSG00000130522 | JUND | JunD proto-oncogene, AP-1 transcription factor subunit |
| 26 | ENSG00000120889 | TNFRSF10B | TNF receptor superfamily member 10b |
| 27 | ENSG00000164305 | CASP3 | caspase 3 |
| 28 | ENSG00000051382 | PIK3CB | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
| 29 | ENSG00000162924 | REL | REL proto-oncogene, NF-kB subunit |
| 30 | ENSG00000115415 | STAT1 | signal transducer and activator of transcription 1 |
| 31 | ENSG00000109320 | NFKB1 | nuclear factor kappa B subunit 1 |
| 32 | ENSG00000141510 | TP53 | tumor protein p53 |
| 33 | ENSG00000107643 | MAPK8 | mitogen-activated protein kinase 8 |
| 34 | ENSG00000197448 | GSTK1 | glutathione S-transferase kappa 1 |
| 35 | ENSG00000116044 | NFE2L2 | nuclear factor, erythroid 2 like 2 |
| 36 | ENSG00000124762 | CDKN1A | cyclin dependent kinase inhibitor 1A |
| 37 | ENSG00000127666 | TICAM1 | Toll-like receptor adaptor molecule 1 |
| 38 | ENSG00000087088 | BAX | BCL2-associated X, apoptosis regulator |
| 39 | ENSG00000173039 | RELA | RELA proto-oncogene, NF-kB subunit |
| 40 | ENSG00000175197 | DDIT3 | DNA damage inducible transcript 3 |
| 41 | ENSG00000121879 | PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| 42 | ENSG00000177606 | JUN | Jun proto-oncogene, AP-1 transcription factor subunit |
| 43 | ENSG00000136244 | IL6 | interleukin 6 |
| 44 | ENSG00000143799 | PARP1 | poly(ADP-ribose) polymerase 1 |
| 45 | ENSG00000115966 | ATF2 | activating transcription factor 2 |
| 46 | ENSG00000108691 | CCL2 | C-C motif chemokine ligand 2 |
| 47 | ENSG00000171608 | PIK3CD | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| 48 | ENSG00000064012 | CASP8 | caspase 8 |
3.3. Construction of the PPI Network and Screening of Key Targets
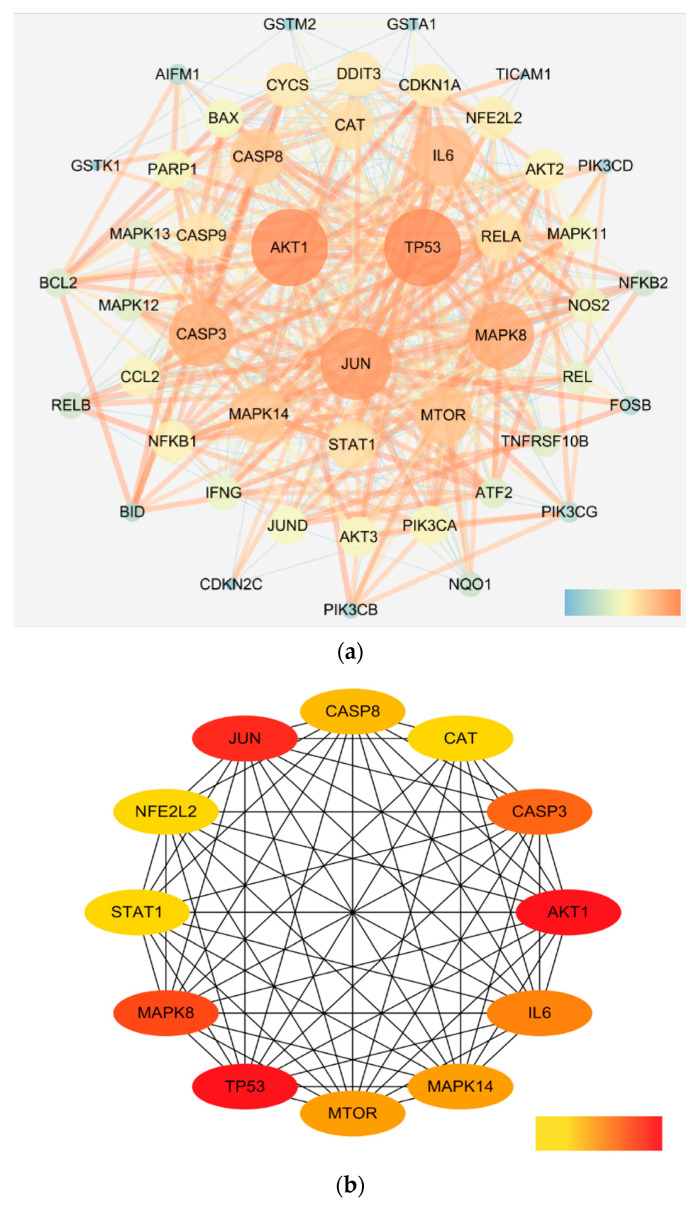
In order to obtain the PPI network association, we imported the 48 target genes into the STRING database and chose Homo sapiens as the organism. The free nodes were then eliminated, and the data were loaded into Cytoscape 3.7.2 for visualization (Figure 2a). The circle’s size and color varied with the degree value, while the width and color of the edges varied with the total score. The image shows 48 targets and 485 interactions, indicating that the more projected disease-related targets there are, the more likely it is that there are successful interactions between those targets. Calculated by the Network Analyzer plug-in in Cytoscape, the average closeness centrality (CC) was 0.63, the average betweenness centrality (BC) was 0.01, and the average degree value (DV) of the node was 19.1. Twelve of them had DV, BC, and CC values that were higher than the norm, while AKT1, TP53, JUN, MAPK8, CASP3, IL6, MTOR, and MAPK14 all displayed high connection degrees that were greater than 30. These could be the key target proteins that are the focus of isoegomaketone. The subnetwork of these key targets is shown in Figure 2b. Table S1 displays the specific node properties in detail.
Figure 2.
(a). Protein interaction network of 48 candidate targets. (b). Protein interaction network diagram of 12 key targets.
3.4. GO Enrichment Analysis
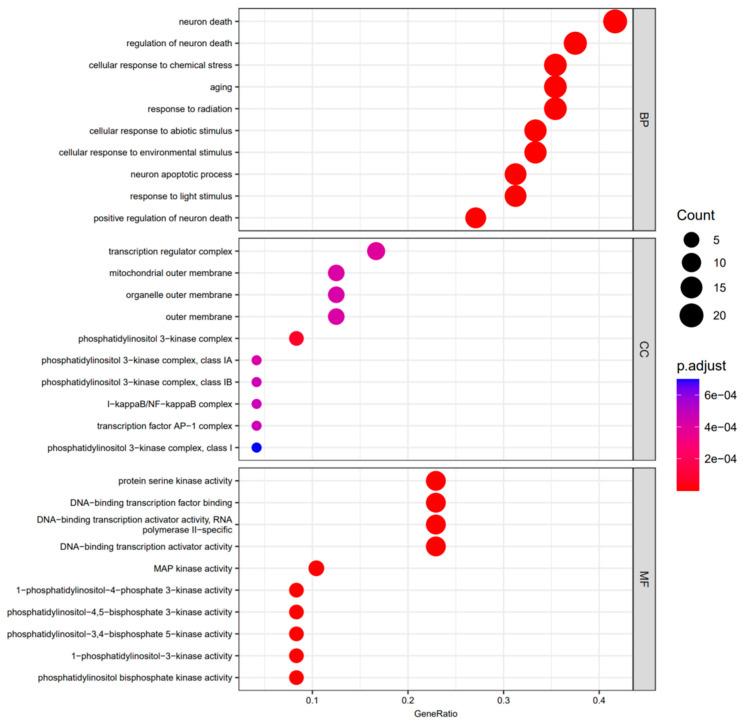
We performed GO enrichment analysis on the 48 chosen target genes in order to further examine them. The majority of the target genes were identified as being engaged in biological processes (BP), cellular components (CC), and molecular functions (MF) according to GO enrichment analysis. BP enrichment mainly involved the following target genes: neuron death (20/48), regulation of neuron death (18/48), cellular response to chemical stress (17/48), aging (17/48), response to radiation (17/48), cellular response to abiotic stimulus (16/48), cellular response to environmental stimulus (16/48), neuron apoptotic process (15/48), response to light stimulus (15/48), and positive regulation of neuron death (13/48). The following target genes were mostly associated with CC enrichment: transcription regulator complex (8/48), mitochondrial outer membrane (6/48), organelle outer membrane (6/48), outer membrane (6/48), phosphatidylinositol 3-kinase complex (4/48) and protein–DNA complex (4/48). Target genes for MF enrichment included the following in particular: protein serine kinase activity (11/48), DNA-binding transcription factor binding (11/48), DNA-binding transcription activator activity (11/48), and MAP kinase activity (5/48). All the results are shown in Figure 3.
Figure 3.
GO enrichment summary of the target genes. The results of biological process (BP), cellular component (CC), and molecular function (MF) are separately shown. The ratio of target genes is represented by the height of the bar.
3.5. KEGG Enrichment Analysis
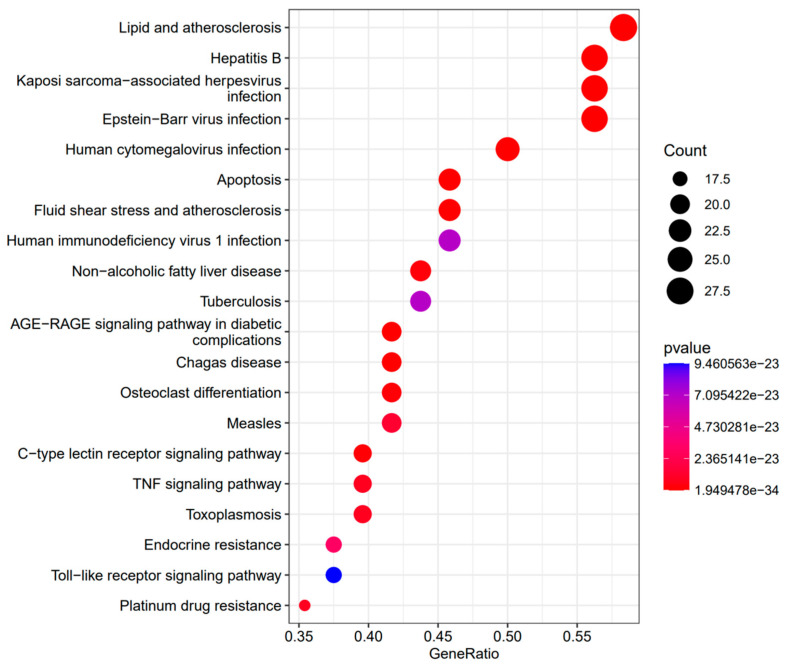
We performed KEGG enrichment analysis of these target genes concurrently. KEGG pathway enrichment analysis revealed that 48 possible target genes were enriched and that there were significant relationships between the target genes and 194 signal pathways (FDR ≤ 0.05). The top 20 pathways with the highest enrichment ratios are demonstrated in Figure 4. The pathway with the largest count and lowest p value was the lipid and atherosclerosis pathway.
Figure 4.
KEGG enrichment analysis of target genes.
3.6. Results of Molecular Docking
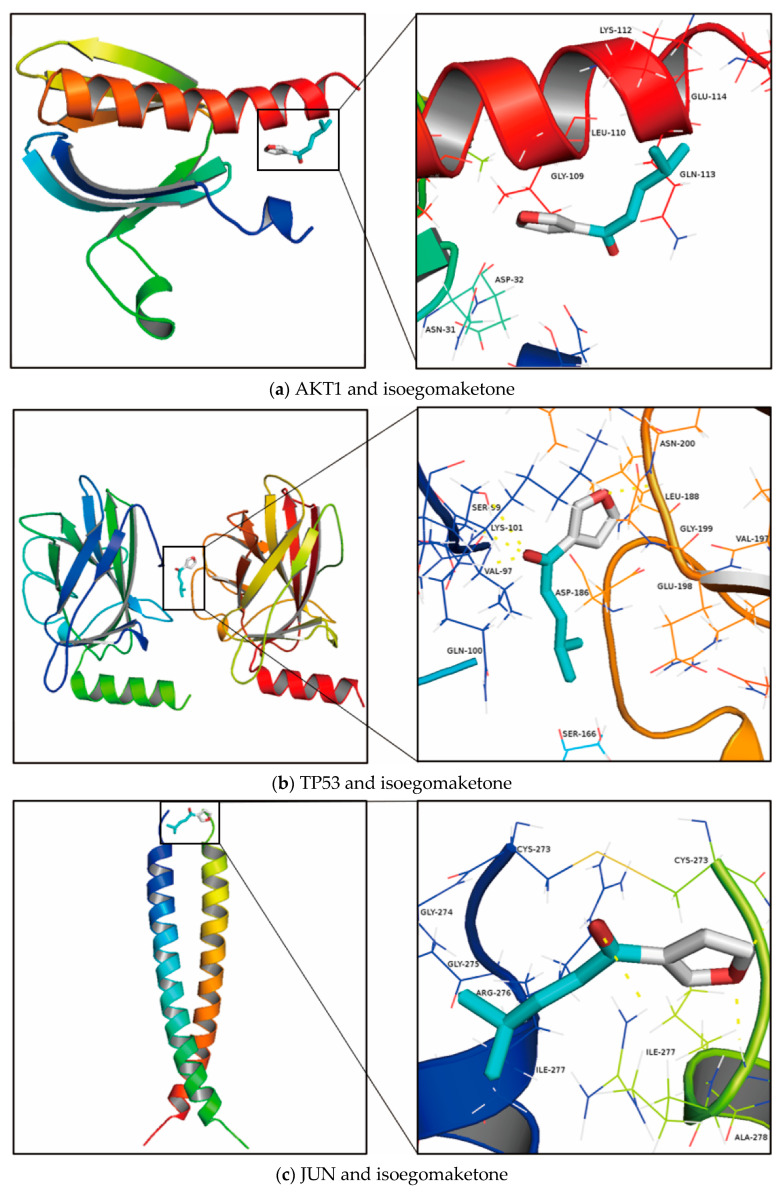
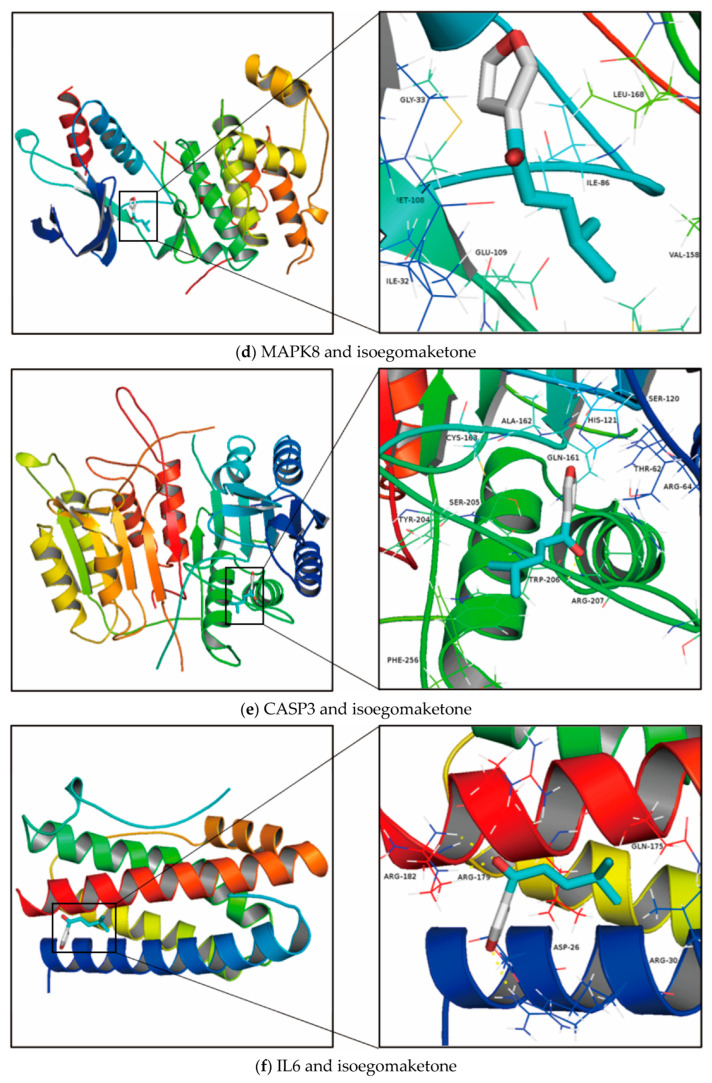
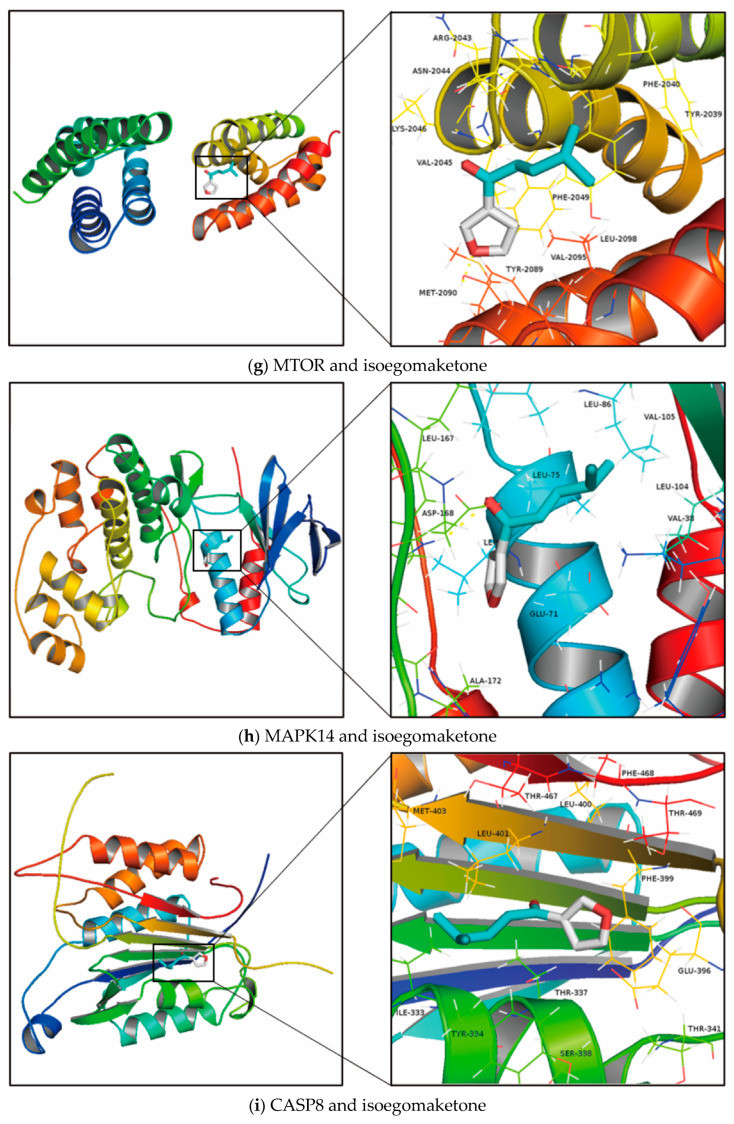
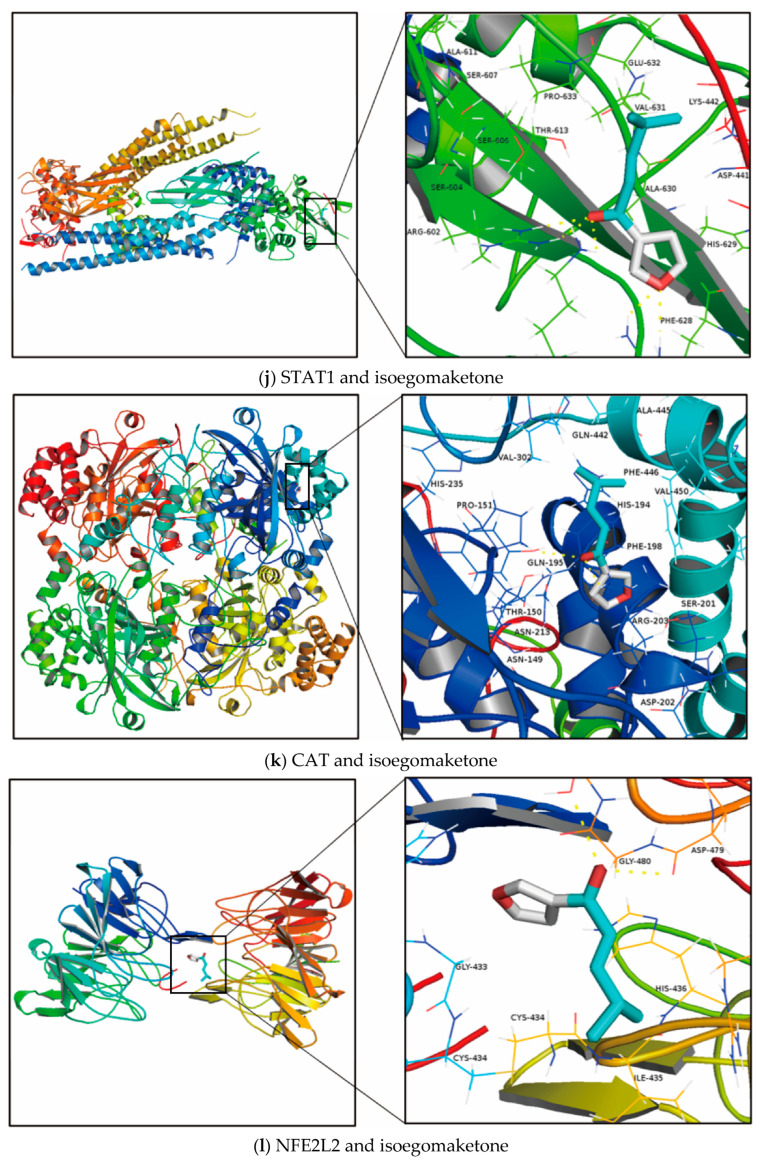
In general, the likelihood of interaction decreases with the energy of conformational stability of the ligand and receptor. There is yet no single standard for the target screening of active compounds, and, at present, binding energy ≤ −5.0 kcal/mol is typically used as the basis for screening. The findings of molecular docking revealed that 6 of the 12 chosen target proteins exhibited an affinity for isoegomaketone that was less than −5.0 kcal/mol. All docking results are shown in Table 4 and Figure 5.
Table 4.
Compound–target molecular docking binding energy.
| No. | Target | PDB ID | Binding Energy (kcal/mol) |
|---|---|---|---|
| 1 | AKT1 | 2uzs | −3.8 |
| 2 | TP53 | 6shz | −5.2 |
| 3 | JUN | 1jun | −3.7 |
| 4 | MAPK8 | 1ukh | −5.2 |
| 5 | CASP3 | 1cp3 | −5.3 |
| 6 | IL6 | 1alu | −4.7 |
| 7 | MTOR | 1aue | −4.6 |
| 8 | MAPK14 | 1a9u | −5.5 |
| 9 | CASP8 | 1qtn | −5.4 |
| 10 | STAT1 | 1yvl | −4.3 |
| 11 | CAT | 1dgf | −6.0 |
| 12 | NFE2L2 | 3zgc | −3.8 |
Figure 5.
Images of the nine best docking results: (a) AKT1, (b) TP53, (c) JUN, (d) MAPK8, (e) CASP3, (f) IL6, (g) MTOR, (h) MAPK14, (i) CASP8, (j) STAT1, (k) CAT, (l) NFE2L2.
4. Discussion
Currently, network pharmacology is being used with increasing frequency in the development and utilization of novel drugs. It converts the “single target, single drug” concept of drug research into a “network target, multicomponent treatment” model [35]. As a result, in this study, we built a multidimensional network through target prediction and protein interaction networks based on the network pharmacology and molecular docking research methodologies to clarify the probable targets of isoegomaketone.
Drug-likeness is recognized as a guideline for chemical library screening optimization in the area of drug development [36]. Lipinski’s rule of five, the Ghose filter, and Veber’s Rule are some of the most widely used rules of thumb. The main purpose of these tests is to determine whether a substance may be used as a drug or whether a substance, having pharmacological or biological activity, can be converted into an oral drug for human use. All of these formulas evaluate specific values of a given compound, including logP, TPSA, MW, MR, nHBA, nRotB, and nHBD. According to Table 1, isoegomaketone, with MW ≤ 500, logP < 5, nRotB ≤ 10, nHBD ≤ 5, nHBA ≤ 10, MR ≤ 140, and TPSA < 140 Å, perfectly passes the test of Lipinski’s rule of five, the Ghose filter, and Veber’s rule. Typically, a molecule that satisfies two of the three rules can be considered to fit the bioavailability of drugs. Thus, this study strongly indicates that isoegomaketone has excellent bioavailability in drugs, making it plausible that it is an active oral substance in humans. In addition, according to Table 1, isoegomaketone was predicted to have high gastrointestinal absorption, and it was also predicted to be able to permeate the blood–brain barrier. The LogS value for isoegomaketone is −2.39, which indicates its solubility in water. Moreover, isoegomaketone complies with the RO (3) rule and has good synthetic accessibility, which suggests its potential as a lead drug. The LD50 value of isoegomaketone was predicted to be 31.2 mg/kg, as shown in Table 2. According to DL-AOT, compounds are divided into four categories, namely “danger/poison”, “warning”, “caution”, and “none required”, and isoegomaketone is placed in the “caution” group, meaning that it is predicted to have no apparent toxicity to normal cells. All of these results indicate that isoegomaketone is likely to be a good candidate for drug development.
As indicated in Table 3, 48 possible isoegomaketone targets were found in the literature analysis. The STRING database was then used to examine the 48 possible targets, and there were 12 nodes whose degree value, betweenness centrality, and closeness centrality were all greater than the average, most of which (such as AKT1, TP53, JUN, MAPK8, CASP3, IL6, MTOR, MAPK14) are involved in the process of apoptosis and the stress response. These selected isoegomaketone targets suggest that isoegomaketone may have potential anti-inflammatory, anti-tumor, and cytotoxic activity. We also performed molecular docking to further investigate any potential interactions between isoegomaketone and these targets. With a decrease in the conformational energy of the ligand binding to the receptor, there is a higher chance of interaction. If −5.0 kcal/mol is utilized as the screening criterion, isoegomaketone may interact directly with these targets, since 6 out of the 12 targets had binding energies that were less than −5.0 kcal/mol [37,38,39,40].
Most of them are associated with anti-inflammatory, anti-tumor, and cytotoxic responses. They might, thus, be the main targets of isoegomaketone’s pharmacological activity.
To elucidate the potential biological activity of isoegomaketone based on gene functions and signaling pathways, we further conducted GO analysis and KEGG pathway analysis of these key targets with the clusterProfiler R package 3.8.1. According to the analysis of BP items, isoegomaketone is closely associated with the following: neuron death, regulation of neuron death, cellular response to chemical stress, aging, response to radiation, cellular response to abiotic stimulus, etc. The results of KEGG pathway enrichment analysis show that the lipid and atherosclerosis pathway is highly enriched. It should be mentioned that no previous studies have clarified the relationship between isoegomaketone and atherosclerosis. The result indicates that isoegomaketone may play a comprehensive role in preventing and treating atherosclerosis, as pathways such as the lipid and atherosclerosis pathway and fluid shear stress and atherosclerosis pathway are involved. In addition, the anti-inflammatory activity of isoegomaketone may also be involved. Further in vitro and in vivo experimental validation could be carried out. Moreover, we found that 9 of the top 20 enriched KEGG pathways were related to bacterial, viral, and parasite infections, such as hepatitis B, Kaposi sarcoma-associated herpesvirus, Epstein–Barr virus, human cytomegalovirus, human immunodeficiency virus 1, tuberculosis, Chagas disease (American trypanosomiasis), measles, and toxoplasmosis. According to these findings, isoegomaketone could possess antiviral, antibacterial, and parasitic action. Some pathways are related to inflammatory diseases—for example, the AGE-RAGE signaling pathway in non-alcoholic fatty liver disease and diabetic complications—which is in accordance with a previous in vivo study [9]. In addition, the KEGG enrichment results also include the TNF signaling pathway, Toll-like receptor signaling pathway, C-type lectin receptor signaling pathway, and other immuno-inflammatory-response-related pathways. As a result, our research revealed that isoegomaketone may be involved by preventing an overactive immune response, an influx of inflammatory substances, and the growth of tumor cells. These findings are similar to some of the in vitro studies of isoegomaketone that have reported its anti-inflammatory, anti-tumor, and cytotoxic activity [41]. Therefore, on one hand, it suggests the accuracy and rationality of our GO and KEGG research. On the other hand, the result further reveals the anti-inflammatory, anti-tumor, and cytotoxic activity target of isoegomaketone, suggesting the potential of isoegomaketone as an anti-inflammatory, anti-tumor, and cytotoxic drug. Taking these results together, we created a drug–target pathway network diagram that more obviously showed that isoegomaketone could exert a variety of pharmacological effects.
However, there are limitations to our research. The most significant is that, as mentioned above, our target screening was obtained through a large number of previously published studies. These provide high-quality, real-world evidence, based, in part, on in vivo and in vitro studies [5,6,16,17,18,19,20,21,22,23,24]. However, although our study provides some prospective evidence through the in silico method, some results have not yet been suitably empirically validated. Therefore, real-world validation using in vivo or in vitro methods based on the current prospective results is highly desirable.
5. Conclusions
Our research revealed that isoegomaketone has a wide range of pharmacological effects. At the same time, we investigated isoegomaketone’s potential targets, which may be used to further create secure and efficient anti-atherosclerosis, anti-inflammatory, anti-tumor, cytotoxic, antiviral, antibacterial, and anti-parasite medications. Our work offers a new perspective on the study, creation, and clinical use of isoegomaketone.
Acknowledgments
The authors would like to acknowledge Can Cao from Beijing University of Chinese Medicine and Tongjie Guo from Qiqihar Medical University for their comprehensive assistance with the molecular docking method.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/life12122115/s1, Table S1. Topological parameters of key targets in protein interaction network.
Author Contributions
Conceptualization, J.Z. and R.W.; data curation, J.Z. and R.W.; formal analysis, J.Z. and R.W.; funding acquisition, R.W.; investigation, J.Z. and R.W.; writing—original draft, J.Z.; writing—review and editing, R.W., Y.Q. and C.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that there are no conflict of interest regarding the publication of this paper.
Funding Statement
This study was funded by Shanghai Jiao Tong University School of Medicine.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Xu H., Zhang Y., Wang P., Zhang J., Chen H., Zhang L., Du X., Zhao C., Wu D., Liu F., et al. A comprehensive review of integrative pharmacology-based investigation: A paradigm shift in traditional Chinese medicine. Acta Pharm. Sin. B. 2021;11:1379–1399. doi: 10.1016/j.apsb.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X. New concepts and approaches for drug discovery based on traditional Chinese medicine. Drug Discov. Today Technol. 2006;3:247–253. doi: 10.1016/j.ddtec.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Başer K., Demirci B., Dönmez A. Composition of the essential oil of Perilla frutescens (L.) Britton from Turkey. Flavour Fragr. J. 2003;18:122–123. doi: 10.1002/ffj.1174. [DOI] [Google Scholar]
- 4.Ha T.J., Lee M.-H., Lee J.H. Comparison of antioxidant activities and volatile components using GC/MS from leaves of Korean purple perilla (Perilla frutescens) grown in a greenhouse. Food Sci. Biotechnol. 2015;24:1979–1986. doi: 10.1007/s10068-015-0261-2. [DOI] [Google Scholar]
- 5.Lee J.-H., Cho H.-D., Jeong I.-Y., Lee M.-K., Seo K.-I. Sensitization of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-resistant primary prostate cancer cells by isoegomaketone from Perilla frutescens. J. Nat. Prod. 2014;77:2438–2443. doi: 10.1021/np500452e. [DOI] [PubMed] [Google Scholar]
- 6.Park Y.D., Jin C.H., Choi D.S., Byun M.-W., Jeong I.Y. Biological evaluation of isoegomaketone isolated from Perilla frutescens and its synthetic derivatives as anti-inflammatory agents. Arch. Pharmacal Res. 2011;34:1277–1282. doi: 10.1007/s12272-011-0806-8. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y.-R., Nam B., Han A.-R., Kim J.-B., Jin C.H. Isoegomaketone from Perilla frutescens (L.) Britt stimulates MAPK/ERK pathway in human keratinocyte to promote skin wound healing. Evid.-Based Complement. Altern. Med. 2021;2021:6642606. doi: 10.1155/2021/6642606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin C.H., So Y., Nam B., Han S.N., Kim J.-B. Isoegomaketone alleviates the development of collagen antibody-induced arthritis in male Balb/c mice. Molecules. 2017;22:1209. doi: 10.3390/molecules22071209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.So Y., Jo Y.H., Nam B.M., Lee S.Y., Kim J.-B., Kang S.-Y., Jeong H.G., Jin C.H. Anti-obesity effect of isoegomaketone isolated from Perilla frutescens (L.) Britt. cv. Leaves. Korean J. Pharmacogn. 2015;46:283–288. [Google Scholar]
- 10.Guan M., Guo L., Ma H., Wu H., Fan X. Network pharmacology and molecular docking suggest the mechanism for biological activity of rosmarinic acid. Evid.-Based Complement. Altern. Med. 2021;2021:5190808. doi: 10.1155/2021/5190808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Hung A., Yang A.W.H. Herb-target virtual screening and network pharmacology for prediction of molecular mechanism of Danggui Beimu Kushen Wan for prostate cancer. Sci. Rep. 2021;11:1–15. doi: 10.1038/s41598-021-86141-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 13.Chen C.Y.-C. TCM Database@ Taiwan: The world’s largest traditional Chinese medicine database for drug screening in silico. PLoS ONE. 2011;6:e15939. doi: 10.1371/journal.pone.0015939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y., Pei J., Lai L. Deep learning based regression and multiclass models for acute oral toxicity prediction with automatic chemical feature extraction. J. Chem. Inf. Model. 2017;57:2672–2685. doi: 10.1021/acs.jcim.7b00244. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y. Ph.D. Thesis. Southern Medical University; Guangzhou, China: 2013. Exploration of the Effect and Mechanism of Radiosensitization of Isoegomaketone on Hepatocellular Carcinoma Cells. [Google Scholar]
- 17.Wu G., Huan X., Wu X., Tian J., Liu J., Liu F., Yao X. The preliminary study for radiotherapy sensitization effect of isoegomaketone on human colorectal cancer xenograft tumor in nude mice. J. Multidiscip. Cancer Manag. (Electron. Version) 2020;6:109–112. [Google Scholar]
- 18.Yang F.C., Wang Y.J. The effect of radiosensitization of isoegomaketone on lung cancer cells and the involvement of endoplasmic reticulum stress. J. Clin. Exp. Med. 2016;15:1151–1154. [Google Scholar]
- 19.Cho B.O., Jin C.H., Park Y.D., Ryu H.W., Byun M.W., Seo K.I., Jeong I.Y. Isoegomaketone induces apoptosis through caspase-dependent and caspase-independent pathways in human DLD1 cells. Biosci. Biotechnol. Biochem. 2011;75:1306–1311. doi: 10.1271/bbb.110088. [DOI] [PubMed] [Google Scholar]
- 20.Kwon S.-J., Lee J.-H., Moon K.-D., Jeong I.-Y., Ahn D.-U., Lee M.-K., Seo K.-I. Induction of apoptosis by isoegomaketone from Perilla frutescens L. in B16 melanoma cells is mediated through ROS generation and mitochondrial-dependent,-independent pathway. Food Chem. Toxicol. 2014;65:97–104. doi: 10.1016/j.fct.2013.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Kwon S.-J., Lee J.-H., Moon K.-D., Jeong I.-Y., Yee S.-T., Lee M.-K., Seo K.-I. Isoegomaketone induces apoptosis in SK-MEL-2 human melanoma cells through mitochondrial apoptotic pathway via activating the PI3K/Akt pathway. Int. J. Oncol. 2014;45:1969–1976. doi: 10.3892/ijo.2014.2598. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Huang X., Han J., Zheng W., Ma W. Extract of Perilla frutescens inhibits tumor proliferation of HCC via PI3K/AKT signal pathway. Afr. J. Tradit. Complement. Altern. Med. 2013;10:251–257. doi: 10.4314/ajtcam.v10i2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin C.H., Lee H.J., Park Y.D., Choi D.S., Kim D.S., Kang S.Y., Seo K.I., Jeong I.Y. Isoegomaketone inhibits lipopolysaccharide-induced nitric oxide production in RAW 264.7 macrophages through the heme oxygenase-1 induction and inhibition of the interferon-beta-STAT-1 pathway. J. Agric. Food Chem. 2010;58:860–867. doi: 10.1021/jf9033333. [DOI] [PubMed] [Google Scholar]
- 24.Jin C.H., So Y.K., Han S.N., Kim J.B. Isoegomaketone Upregulates Heme Oxygenase-1 in RAW264.7 Cells via ROS/p38 MAPK/Nrf2 Pathway. Biomol. Ther. 2016;24:510–516. doi: 10.4062/biomolther.2015.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Wei S., Niu S., Ma X., Li H., Jing M., Zhao Y. Network pharmacology prediction and molecular docking-based strategy to explore the potential mechanism of Huanglian Jiedu Decoction against sepsis. Comput. Biol. Med. 2022;144:105389. doi: 10.1016/j.compbiomed.2022.105389. [DOI] [PubMed] [Google Scholar]
- 27.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8((Suppl. 4)):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 31.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 32.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 33.Congreve M., Carr R., Murray C., Jhoti H. A ‘rule of three’ for fragment-based lead discovery? Drug Discov. Today. 2003;8:876–877. doi: 10.1016/S1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 34.Fukunishi Y., Kurosawa T., Mikami Y., Nakamura H. Prediction of synthetic accessibility based on commercially available compound databases. J. Chem. Inf. Model. 2014;54:3259–3267. doi: 10.1021/ci500568d. [DOI] [PubMed] [Google Scholar]
- 35.Zhang R., Zhu X., Bai H., Ning K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019;10:123. doi: 10.3389/fphar.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di L., Kerns E.H., Carter G.T. Drug-like property concepts in pharmaceutical design. Curr. Pharm. Des. 2009;15:2184–2194. doi: 10.2174/138161209788682479. [DOI] [PubMed] [Google Scholar]
- 37.Zhuang Y., Zhang X., Luo S., Wei F., Song Y., Lin G., Yao M., Gong A. Exploring the Molecular Mechanism of Zhi Bai Di Huang Wan in the Treatment of Systemic Lupus Erythematosus Based on Network Pharmacology and Molecular Docking Techniques. Processes. 2022;10:1914. doi: 10.3390/pr10101914. [DOI] [Google Scholar]
- 38.Huang X., Rehman H.M., Szöllősi A.G., Zhou S. Network Pharmacology-Based Approach Combined with Bioinformatic Analytics to Elucidate the Potential of Curcumol against Hepatocellular Carcinoma. Genes. 2022;13:653. doi: 10.3390/genes13040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai L., Zhang G., Wan X. Network Pharmacology and Molecular Docking Analysis Reveal Insights into the Molecular Mechanism of Shengma-Gegen Decoction on Monkeypox. Pathogens. 2022;11:1342. doi: 10.3390/pathogens11111342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou W., Wang J., Wu Z., Huang C., Lu A., Wang Y. Systems pharmacology exploration of botanic drug pairs reveals the mechanism for treating different diseases. Sci. Rep. 2016;6:36985. doi: 10.1038/srep36985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R., Zhang Q., Feng C., Zhang J., Qin Y., Meng L. Advances in the Pharmacological Activities and Effects of Perilla Ketone and Isoegomaketone. Evid.-Based Complement. Alternat. Med. 2022;2022:8809792. doi: 10.1155/2022/8809792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are included within the article.