Abstract
Ceftazidime/avibactam (CAZ/AVI), a combination of ceftazidime and a novel β‐lactamase inhibitor (avibactam) that has been approved by the U.S. Food and Drug Administration, the European Union, and the National Regulatory Administration in China. CAZ/AVI is used mainly to treat complicated urinary tract infections and complicated intra‐abdominal infections in adults, as well as to treat patients infected with Carbapenem‐resistant Enterobacteriaceae (CRE) susceptible to CAZ/AVI. However, increased clinical application of CAZ/AVI has resulted in the development of resistant strains. Mechanisms of resistance in most of these strains have been attributed to bla KPC mutations, which lead to amino acid substitutions in β‐lactamase and changes in gene expression. Resistance to CAZ/AVI is also associated with reduced expression and loss of outer membrane proteins or overexpression of efflux pumps. In this review, the prevalence of CAZ/AVI‐resistance bacteria, resistance mechanisms, and selection of detection methods of CAZ/AVI are demonstrated, aiming to provide scientific evidence for the clinical prevention and treatment of CAZ/AVI resistant strains, and provide guidance for the development of new drugs.
This article is categorized under:
Infectious Diseases > Molecular and Cellular Physiology
Keywords: ceftazidime/avibactam, efflux pump, outer membrane protein, resistance mechanism, β‐lactamase
The molecular mechanisms to ceftazidime/avibactam resistance.
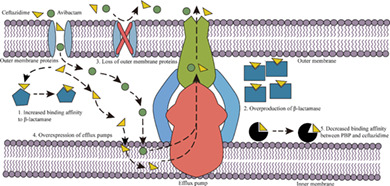
1. INTRODUCTION
Infections of patients with multidrug‐resistant Gram‐negative bacteria (MDRGNB) have become increasingly common. These bacteria have been associated with high morbidity and mortality rates, posing a serious threat to public health worldwide (Orsi et al., 2011). Carbapenems are considered one of the most powerful classes of therapeutic drugs for the treatment of MDRGNB infections; however, their use is limited by the emergence of carbapenemase‐producing bacteria (Daikos & Markogiannakis, 2011). The main carbapenemases produced by bacteria include Klebsiella pneumoniae carbapenemase (KPC), metallo‐β‐lactamase (MBL), and oxacillinase (OXA; Guh et al., 2015; Tamma et al., 2017). Carbapenemase‐resistant bacterial infections, however, may be susceptible to combination of β‐lactam and β‐lactamase inhibitors (BLI) with carbapenemase‐inhibiting activity (Bebrone et al., 2010).
Common classical BLIs in clinical use include clavulanic acid, sulbactam, and tazobactam, all of which have β‐lactam structures. BLIs bind stably and irreversibly to β‐lactamases, initially forming an intermediate (E‐I), which undergoes an additional reaction to form free enzyme (E) and hydrolyzed clavulanic acid (I*), which is known as a “suicide enzyme inhibitor”. These classical BLIs are active against many class A β‐lactamases but do not interact with class B, C, and D β‐lactamases. With the emergence of other carbapenemases, some new non‐β‐lactam inhibitors have been recently approved for clinical use (Drawz et al., 2014). Relebactam is combined with imipenem to treat infections caused by CRE and Pseudomonas aeruginosa, including strains that produce KPC and class C β‐lactamases (Hirsch et al., 2012; Lapuebla et al., 2015; Livermore et al., 2013). Nacubactam, a diazabicyclooctane (DBO) inhibitor combined with meropenem for development, inhibits class A and C serine β‐lactamases (Morinaka et al., 2015). Zidebactam is another DBO inhibitor, combined with cefepime against enterobacteriaceae species and P. aeruginosa that express ESBLs, AmpC, KPC, and OXA‐48 β‐lactamases (D. M. Livermore, Mushtaq, et al., 2017). Vaborbactam, the first boronic acid BLI, is designed specifically to inhibit KPC carbapenemases. And it also has activity against other class A and class C β‐lactamases (Hecker et al., 2015). Meropenem–vaborbactam has shown good in vitro activity against Enterobacter spp., Klebsiella spp., and Citrobacter spp. that produce KPC enzymes (Castanheira et al., 2016; Lomovskaya et al., 2017). Avibactam (AVI) is a novel bridging DBO non‐β‐lactam BLI with weak antibacterial activity (Bonnefoy et al., 2004; Ehmann et al., 2012). However, AVI binds covalently and reversibly to A, C, and some D class serine β‐lactamases to form covalent compounds that inhibit the activity of these enzymes. The active enzyme and AVI can later be regenerated by reversible deacylation and recycling (Bush & Bradford, 2019). However, because the rate of AVI ring opening is much greater than the rate of cyclization, resulting in the maintenance of β‐lactamase inhibition, AVI has a long‐lasting enzyme inhibitory effect (Figure 1; Tooke et al., 2019). Unlike classical BLIs, AVI‐enzyme inhibitor complexes do not generally undergo hydrolysis; AVI undergoes a reaction with KPC‐2 similar to those of clavulanic acid and tazobactam with KPC‐2, with the complexes formed during the reaction between AVI and KPC‐2 able to undergo hydrolysis (Tooke et al., 2019).
FIGURE 1.
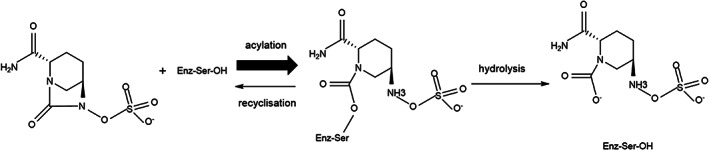
Schematic representation of the acylation, recyclization, and hydrolysis reactions between a serine β‐lactamase and avibactam
Ceftazidime/Avibactam (CAZ/AVI) is the first drug approved by the U.S. Food and Drug Administration for the treatment of CRE. Although CAZ is easily hydrolyzed by β‐lactamase when used alone (Figure 2), this combination, which is administered intravenously in a 4:1 ratio by weight (2.0 g CAZ/0.5 g AVI), has good antibacterial activity against carbapenemase‐producing Gram‐negative bacteria (Shirley, 2018). In October 2015, a KPC‐3‐producing strain of K. pneumoniae isolated from a blood culture of a 62‐year‐old woman not treated with CAZ/AVI was found to be resistant to CAZ/AVI by antimicrobial susceptibility testing, making this the first reported case of CAZ/AVI resistance (Humphries et al., 2015). Since then, many reports have described bacterial strains resistant to CAZ/AVI (Giddins et al., 2018; Winkler, Papp‐Wallace, & Bonomo, 2015). This review summarized the prevalence, resistant mechanism, and detection methods of CAZ/AVI‐resistant Enterobacteriaceae, P. aeruginosa, and A. baumannii, especially the influence of β‐lactamase variation on MIC values of CAZ/AVI, aiming at establishing the scientific and reasonable diagnosis and treatment plan, according to corresponding molecular mechanisms. It provides a theoretical basis for the rational use of CAZ/AVI in the treatment of bacterial infectious diseases in clinical practice.
FIGURE 2.
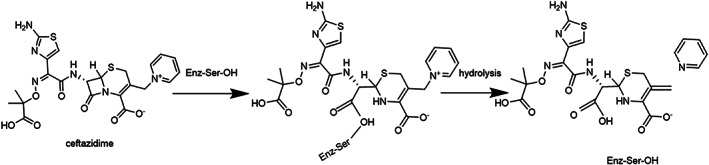
Example of an irreversible reaction between a β‐lactam and a nucleophilic serine enzyme. The figure shows the reaction of a penicillin with a penicillin binding protein (transpeptidase) to yield a stable acyl‐enzyme complex, which subsequently undergoes slow hydrolysis.
2. PREVALENCE OF CAZ/AVI‐RESISTANT BACTERIA
2.1. Global resistance surveillance
According to the 2012–2016 ATLAS (Antimicrobial Testing Leadership and Surveillance), Global Resistance Surveillance reported that the overall resistance rate of Gram‐negative bacteria to CAZ/AVI was low (0.2%–8.1%). The resistance rate of Enterobacteriaceae to CAZ/AVI was less than 2.2%, with 0.2% for Escherichia coli, 1.2% for K. pneumoniae, and 2.2% for Enterobacter cloacae. While among CRE, the resistance rate of Carbapenem‐resistant K. pneumoniae (CRKP), Carbapenem‐resistant E. coli (CREC) and Carbapenem‐resistant E. cloacae were 14.4%, 27.7%, and 57.7%, respectively. The overall resistance rate of P. aeruginosa to CAZ/AVI was significantly higher than that of Enterobacteriaceae at 8.1%; Carbapenem‐resistant P. aeruginosa (CRPA) showed 25.5% resistance to CAZ/AVI. In addition, the resistance rate of CAZ/AVI varied greatly among Enterobacteriaceae and P. aeruginosa producing different carbapenemases: among Enterobacteriaceae, KPC‐producing strains were resistant to CAZ/AVI at 1.5%, and NDM‐producing strains at 99.6%, while among P. aeruginosa, KPC‐producing strains were resistant to CAZ/AVI at 35.7% and NDM‐producing strains at 97% (Table 1; Kiratisin et al., 2021; H. Zhang, Xu, et al., 2020).
TABLE 1.
Resistance rate of pathogens against CAZ/AVI
| Project | Region/country | Pathogen | R% | Charecteristics of pathogen | MIC50 (mg/L) | MIC90 (mg/L) | MIC range (mg/L) | Reference |
|---|---|---|---|---|---|---|---|---|
| INFORM (2012–2015) | Asia‐Pacific countries | Enterobacteriaceae | 1.0 | 0.12 | 0.5 | ≤0.015 to >128 | (Karlowsky et al., 2018) | |
| Enterobacteriaceae | 98.6 | MBL positive | >128 | >128 | 2 to >128 | |||
| Enterobacteriaceae | 0.2 | MBL negative | 0.12 | 0.5 | ≤0.015 to >128 | |||
| E. coli | 0.1 | 0.12 | 0.25 | ≤0.015 to >128 | ||||
| E. coli | 0.1 | MBL negative | 0.12 | 0.25 | ≤0.015 to >128 | |||
| K. pneumoniae | 1.7 | 0.12 | 0.5 | ≤0.015 to >128 | ||||
| K. pneumoniae | 0.3 | MBL negative | 0.12 | 0.5 | ≤0.015 to >128 | |||
| K. oxytoca | 1.2 | 0.12 | 0.25 | ≤0.015 to >128 | ||||
| K. oxytoca | 0.0 | MBL negative | 0.12 | 0.25 | ≤0.015 to 16 | |||
| Enterobacter spp | 1.9 | 0.25 | 1 | ≤0.015 to >128 | ||||
| Enterobacter spp | 0.3 | MBL negative | 0.25 | 1 | ≤0.015 to 64 | |||
| P. aeruginosa | 7.4 | 2 | 8 | 0.03 to >128 | ||||
| P. aeruginosa | 3.9 | MBL negative | 2 | 8 | 0.03 to >128 | |||
| INFORM (2012–2015) | Latin American | Enterobacteriaceae | 0.3 | 0.12 | 0.5 | ≤0.015 to >128 | (Karlowsky et al., 2019) | |
| Enterobacteriaceae | 0.1 | MBL negative | 0.12 | 0.5 | ≤0.015 to >128 | |||
| Enterobacteriaceae | 94.1 | MBL positive | 64 | >128 | 0.12 to >128 | |||
| E. coli | 0.1 | 0.12 | 0.25 | ≤0.015 to 32 | ||||
| E. coli | 0.0 | MBL negative | 0.12 | 0.25 | ≤0.015 to 8 | |||
| K. pneumoniae | 0.5 | 0.25 | 1 | ≤0.015 to >128 | ||||
| K. pneumoniae | 0.0 | MBL negative | 0.12 | 1 | ≤0.015 to >128 | |||
| P. aeruginosa | 12.6 | 2 | 16 | 0.12 to >128 | ||||
| P. aeruginosa | 7.2 | MBL negative | 2 | 8 | 0.12 to >128 | |||
| P. aeruginosa | 94.5 | MBL positive | 32 | >128 | 2 to >128 | |||
| INFORM (2012–2015) | European countries | Enterobacterales | 0.6 | 0.12 | 0.5 | (K. M. Kazmierczak, de Jonge, et al., 2018a) | ||
| Enterobacterales | 90.8 | MBL‐positive | >128 | >128 | 1 to >128 | |||
| P. aeruginosa | 7.6 | 2 | 8 | 0.03 to >128 | (K. M. Kazmierczak, de Jonge, et al., 2018b) | |||
| P. aeruginosa | 97.2 | MBL‐positive | 32 | >128 | 2 to >128 | |||
| ATLAS (2012–2016) | All regions | E. coli | 0.2 | 0.12 | 0.25 | 0.015–256 | (H. Zhang, Xu, et al., 2020) | |
| E. coli | 27.7 | CR | 0.5 | 256 | 0.03–256 | |||
| K. pneumoniae | 1.2 | 0.12 | 1 | 0.015–256 | ||||
| K. pneumoniae | 14.4 | CR | 1 | 256 | 0.015–256 | |||
| E. cloacae | 2.2 | 0.25 | 1 | 0.015–256 | ||||
| E. cloacae | 57.7 | CR | 128 | 256 | 0.06–256 | |||
| P. aeruginosa | 8.1 | 2 | 8 | 0.015–256 | ||||
| P. aeruginosa | 25.5 | CR | 4 | 64 | 0.015–256 | |||
| A. baumannii | 32 | 128 | 0.03–256 | |||||
| A. baumannii | CR | 64 | 256 | 0.06–256 | ||||
| INFORM (2015–2017) | Asia‐Pacific region | Enterobacterales | 1.9 | 0.12 | 0.5 | ≤ 0.015 to ≥ 256 | (Ko & Stone, 2020) | |
| Enterobacterales | 0.0 | MBL negative | 1 | 2 | 0.12–4 | |||
| Enterobacterales | 100.0 | MBL positive | ≥ 256 | ≥ 256 | 32 to ≥ 256 | |||
| E. coli | 0.6 | 0.12 | 0.25 | ≤ 0.015 to ≥ 256 | ||||
| E. coli | 100.0 | CR and MBL positive | ≥ 256 | ≥ 256 | 64 to ≥ 256 | |||
| K. pneumoniae | 2.5 | 0.12 | 0.5 | ≤ 0.015 to ≥ 256 | ||||
| K. pneumoniae | 100.0 | CR and MBL positive | ≥ 256 | ≥ 256 | 32 to ≥ 256 | |||
| P. aeruginosa | 7.3 | 2 | 8 | ≤ 0.015 to ≥ 256 | ||||
| P. aeruginosa | 31.4 | MDR | 8 | 128 | 0.25 to ≥ 256 | |||
| P. aeruginosa | 96.1 | CR and MBL positive | 128 | ≥ 256 | 2 to ≥ 256 | |||
| European countries | P. aeruginosa | 7.7 | 2 | 8 | ≤ 0.015 to ≥ 256 | (Stone et al., 2020) | ||
| P. aeruginosa | 28.3 | MDR | 8 | 64 | 0.12 to ≥256 | |||
| P. aeruginosa | 82.7 | CR | 32 | ≥ 256 | 1 to ≥ 256 | |||
| P. aeruginosa | 95.3 | MBL‐positive | 64 | ≥ 256 | 1 to ≥ 256 | |||
| ATLAS (2016–2018) | Africa‐Middle East | Enterobacterales | 39.5 | CR | ≥ 256 | 0.12 to ≥256 | (Kiratisin et al., 2021) | |
| Enterobacterales | 98.4 | CR and MBL positive | ≥ 256 | 8 to ≥256 | ||||
| P. aeruginosa | 92.3 | CR | 128 | 2 to ≥256 | ||||
| P. aeruginosa | 100.0 | CR and MBL positive | 128 | 16 to ≥256 | ||||
| Asia/South Pacific | Enterobacterales | 66.6 | CR | ≥ 256 | 0.12 to ≥256 | (Kiratisin et al., 2021) | ||
| Enterobacterales | 99.5 | CR and MBL positive | ≥ 256 | 1 to ≥256 | ||||
| P. aeruginosa | 90.6 | CR | ≥ 256 | 2 to ≥256 | ||||
| P. aeruginosa | 95.6 | CR and MBL positive | ≥ 256 | 2 to ≥256 | ||||
| Europe | Enterobacterales | 28.0 | CR | ≥ 256 | ≤ 0.015 to ≥ 256 | (Kiratisin et al., 2021) | ||
| Enterobacterales | 96.9 | CR and MBL positive | ≥ 256 | 1 to ≥ 256 | ||||
| P. aeruginosa | 90.6 | CR | ≥ 256 | 1 to ≥256 | ||||
| P. aeruginosa | 95.6 | CR and MBL positive | ≥ 256 | 1 to ≥256 | ||||
| Latin American | Enterobacterales | 18.5 | CR | ≥ 256 | ≤ 0.015 to ≥ 256 | (Kiratisin et al., 2021) | ||
| Enterobacterales | 97.4 | CR and MBL positive | ≥ 256 | 0.25 to ≥256 | ||||
| P. aeruginosa | 82.9 | CR | ≥ 256 | 1 to ≥256 | ||||
| P. aeruginosa | 96.0 | CR and MBL positive | ≥ 256 | 4 to ≥256 | ||||
| All regions | P. aeruginosa | 35.7 | KPC‐positive | 64 | 1 to ≥256 | (Kiratisin et al., 2021) | ||
| P. aeruginosa | 97.0 | NDM‐positive | ≥ 256 | 2 to ≥256 | ||||
| P. aeruginosa | 37.0 | GES‐positive | 64 | 1 to ≥256 | ||||
| Enterobacterales | 1.5 | KPC‐positive | 4 | ≤0.015 to ≥256 | ||||
| Enterobacterales | 99.6 | NDM‐positive | ≥ 256 | 0.25 to ≥256 | ||||
| INFORM (2015–2017) | All regions | Enterobacterales | 27.0 | Meropenem‐NS | ≥ 256 | (Spiliopoulou et al., 2020) | ||
| Europe | Enterobacterales | 23.2 | Meropenem‐NS | ≥ 256 | ||||
| Latin American | Enterobacterales | 12.5 | Meropenem‐NS | 64 | ||||
| Asia | Enterobacterales | 51.7 | Meropenem‐NS | ≥ 256 | ||||
| Africa/Middle East | Enterobacterales | 49.2 | Meropenem‐NS | ≥ 256 | ||||
| Oceania | Enterobacterales | 88.9 | Meropenem‐NS | ≥ 256 | ||||
| INFORM (2015–2017) | Global | K. pneumoniae | 4.7 | MDR | 2 | ≤0.015 to ≥256 | (Rossolini & Stone, 2020) | |
| K. pneumoniae | 0.1 | MDR, ESBL‐positive, carbapenemase‐negative | 1 | ≤0.015 to ≥256 | ||||
| K. pneumoniae | 0.2 | MDR, CR, MBL‐negative | 2 | ≤0.015 to ≥16 | ||||
| K. pneumoniae | 98.0 | MDR, CR, MBL‐positive | ≥ 256 | 2 to ≥256 | ||||
| ATLAS (2015–2019) | Asia‐Pacific Region | P. aeruginosa | 8.5 | 2 | 8 | 0.015–256 | (Lee et al., 2022) | |
| P. aeruginosa | 40.3 | CR | 8 | 256 | 0.03–256 | |||
| P. aeruginosa | 39.1 | MDR | 8 | 256 | 0.25–256 | |||
| P. aeruginosa | 94.0 | MBL‐positive | 256 | 256 | ||||
| ATLAS (2017–2019) | Latin American | Enterobacterales | 1.9 | 0.12 | 0.5 | (Karlowsky et al., 2021) | ||
| Enterobacterales | 25.3 | Meropenem‐NS | 1 | >128 | ||||
| Enterobacterales | 8.2 | Multidrug‐resisstant | 0.5 | 4 | ||||
| Enterobacterales | 0.5 | KPC‐positive | 0.5 | 2 | ||||
| Enterobacterales | 98.6 | MBL‐positive | >128 | >128 | ||||
| P. aeruginosa | 13.1 | 2 | 32 | |||||
| P. aeruginosa | 38.1 | Meropenem‐NS | 8 | 64 | ||||
| P. aeruginosa | 51.0 | Multidrug‐resisstant | 16 | 128 | ||||
| P. aeruginosa | 35.3 | KPC‐positive | 8 | 32 | ||||
| P. aeruginosa | 95.8 | MBL‐positive | 32 | >128 |
Abbreviations: ATLAS, Antimicrobial Testing Leadership and Surveillance; CR, carbapenem‐resistant; INFORM, the International Network for Optimal Resistance Monitoring; KPC, Klebsiella pneumonia carbapenemase; MBL, metallo‐β‐lactamase; MDR, multidrug‐resistant.
2.2. Regional resistance surveillance of Enterobacteriaceae
The 2012–2015 INFORM (the International Network for Optimal Resistance Monitoring, INFORM) resistance monitoring system showed that the overall resistance rate of Enterobacteriaceae to CAZ/AVI was low in different regions of the world, with resistance rates in Latin America and Europe is only 0.3%, while the resistance rate in the Asia Pacific is relatively higher, at 1.0% (Karlowsky et al., 2018, 2019; K. M. Kazmierczak, de Jonge, et al., 2018a). By 2015–2017, the resistance rate of Enterobacteriaceae to CAZ/AVI in the Asia‐Pacific region increased to 1.9%, and further investigation of the resistance rate of carbapenem nonsusceptible Enterobacteriaceae to CAZ/AVI in different regions showed that Oceania had the highest CAZ/AVI resistance rate of 88.9%, followed by Asia (51.7%), Africa‐Middle East (49.2%), Europe (23.2%), and the lowest resistance rate was found in Latin America (12.5%; Spiliopoulou et al., 2020). By 2017–2019, carbapenem nonsusceptible Enterobacteriaceae bacteria in Latin America increased their resistance rate to CAZ/AVI nearly 1‐fold to 25.3% (Karlowsky et al., 2021). In addition, global resistance surveillance by the ATLAS project from 2012 to 2016 showed that carbapenem‐resistant Enterobacteriaceae had a resistance rate of 57.7% to CAZ/AVI, followed by CREC with a resistance rate of 27.7% to CAZ/AVI (H. Zhang, Xu, et al., 2020). The distribution of resistance rate of CRE to CAZ/AVI in different regions was as follows: the highest resistance rate to CAZ/AVI was 66.6% in the Asia‐Pacific region, followed by Africa‐Middle East (39.5%), Europe (28.0%), and Latin America (18.5%), whose trend was consistent with the 2015–2017 INFORM surveillance system showing the distribution of resistance rates of carbapenem nonsusceptible Enterobacteriaceae to CAZ/AVI (Table 1). These results suggest that CRE resistance to CAZ/AVI varies by region and that the differences in CAZ/AVI resistance rates are likely due to the different percentages of MBLs in different regions (Kiratisin et al., 2021; Spiliopoulou et al., 2020).
2.3. Regional resistance surveillance of P. aeruginosa
Pseudomonas aeruginosa has a higher resistance rate to CAZ/AVI compared to Enterobacteriaceae. INFORM resistance surveillance system in 2012–2015 showed the highest resistance rate to CAZ/AVI in Latin America at 12.6%, followed by Europe (7.6%) and the Asia Pacific (7.4%; Karlowsky et al., 2018, 2019; K. M. Kazmierczak, de Jonge, et al., 2018b). In recent years, the resistance rate of P. aeruginosa to CAZ/AVI has been relatively stable across continents, with 13.1% in Latin America, 7.7% in Europe, and 7.3% in the Asia Pacific (Karlowsky et al., 2021; K. M. Kazmierczak, de Jonge, et al., 2018b; Ko & Stone, 2020; Lee et al., 2022). Among carbapenemase‐producing P. aeruginosa, the resistance rate to CAZ/AVI was high in all continents: 92.3% in Africa‐Middle East, 90.6% in the Asia Pacific and Europe, and 82.9% in Latin America (Kiratisin et al., 2021), showing that carbapenemase production correlates strongly with CAZ/AVI resistance (Table 1).
2.4. Resistance surveillance in China
Evaluation of 10,661 Gram‐negative strains collected by the Blood Bacterial Resistant Investigation Collaborative System in China from January 2018 to December 2019 showed that 1% of E. coli strains were resistant to CAZ/AVI (MIC50, 0.25/4 mg/L; MIC90, 1/4 mg/L), including 60.7% of CREC strains. In addition, 1.6% of K. pneumoniae strains were resistant to CAZ/AVI (MIC50, 0.5/4 mg/L; MIC90, 4/4 mg/L), including 8.3% of CRKP strains (MIC50, 4/4 mg/L; MIC90, 8/4 mg/L). Also, 2.8% of P. aeruginosa strains were resistant to CAZ/AVI (MIC50, 4/4 mg/L; MIC90, 8/4 mg/L), including 11.6% of CRPA strains (MIC50, 4/4 mg/L. MIC90, 16/4 mg/L; T. Xu, Guo, et al., 2021). Furthermore, data from the China Antimicrobial Surveillance Network (CHINET) show that in 2017, the CAZ/AVI resistance rates of Enterobacteriaceae and P. aeruginosa were 5.4% (MIC50, ≤0.25/4 mg/L; MIC90, 2/4 mg/L) and 13.5% (MIC50, 2/4 mg/L; MIC90, 16/4 mg/L), respectively. The CAZ/AVI resistance rate was higher for CRE, at 24.7% (MIC50, 2/4 mg/L; MIC90, >32/4 mg/L), among which the resistance rate of CREC to CAZ/AVI was up to 71.4% (Yin et al., 2019). Resistance rates to CAZ/AVI were slightly higher in 2018, up to 6.0% (MIC50, 0.25/4 mg/L; MIC90, 2/4 mg/L) for Enterobacteriaceae, 35.7% (MIC50, 8/4 mg/L; MIC90, 64/4 mg/L) for CRPA and 90.9% for CREC (MIC50, >64/4 mg/L. MIC90, >64/4 mg/L; Yang et al., 2020).
3. MECHANISMS UNDERLYING RESISTANCE TO CAZ/AVI
Most MBLs have the metal ion Zn2+ at their active site; whereas other β‐lactamases have a serine (Ser70) at their active site; the latter are called serine β‐lactamases. The most common groups of MBLs are the Verona integrin‐encoded MBLs (VIMs), imipenemases (IMPs), and New Delhi MBLs (NDMs). MBL‐producing bacteria are naturally resistant to CAZ/AVI because their MBLs do not contain a serine at their active site (Centers for Disease Control and Prevention (CDC), 0). Evaluation of Enterobacteriaceae and P. aeruginosa collected from 42 medical centers in nine countries in the Asia‐Pacific region by the INFORM during the years 2012–2015 showed that 1% (91/9149) and 7.4% (151/2038), respectively, were resistant to CAZ/AVI, with MBL‐positive strains accounting for 80.2% (73/91) and 48.3% (73/151), respectively, of these CAZ/AVI‐resistant strains (Karlowsky et al., 2018). Of the 372 CRE strains collected by CHINET from more than 30 medical centers in 2017, 92 (24.7%) were resistant to CAZ/AVI, with 66 (71.7%) of these containing bla NDM. Of the 134 CAZ/AVI‐resistant strains collected in 2018, 57 (42.5%) contained bla NDM, whereas of the 30 CAZ/AVI‐resistant CREC, 21 (70%) were positive for NDM (Yang et al., 2020; Yin et al., 2019). Taken together, these findings indicate that MBL‐producing strains account for a high proportion of CAZ/AVI‐resistant strains.
By contrast, the main mechanisms underlying resistance to CAZ/AVI in Gram‐negative bacteria that do not produce MBL include: (1) β‐lactamase variants; (2) changes in bacterial membrane permeability; (3) increased expression of efflux pumps; and (4) mutation of PBPs (Shi et al., 2020).
3.1. β‐Lactamase variants
β‐Lactamase variants are an important mechanism of bacterial resistance to CAZ/AVI. Alteration of key residues at the active sites of serine β‐lactamases can increase their MIC values for CAZ/AVI significantly (Table 2). Serine β‐lactamases are dependent on several highly conserved sequences responsible for recognition of, and reactions with, antibiotics. These sequences include the Ω‐loop, consisting of amino acids 164–179. Glu166 and Asn170 are involved in the acylation and deacylation of antibiotics by β‐lactamases. The Ω‐loop acts as a structural domain unit that interacts with antibiotics, forming the binding cavity for β‐lactamase‐substrate action. This allows the carbonyl on the β‐lactam ring of the antibiotic to polarize, forming an acylated enzyme complex. The amino acid substitution in the KPC enzyme alters the structure of the binding cavity, thereby reducing resistance to the binding of CAZ and the active site of the enzyme, thereby increasing β‐lactamase binding and hydrolysis of CAZ, an important mechanism of bacterial resistance to CAZ/AVI. Individual amino acid substitutions, particularly at positions 164, 167, 169, and 179 of the Ω‐loop, increase the affinity of the β‐lactamase to CAZ and increase its hydrolysis (Winkler, Papp‐Wallace, & Bonomo, 2015). In addition, amino acid substitutions in the Ω‐loop can reduce the binding of β‐lactamase to AVI, weakening the inhibitory effect of AVI on β‐lactamase and increasing hydrolysis of CAZ (F. Compain et al., 2020a, 2020b). Alterations in β‐lactamase expression can also lead to CAZ/AVI resistance (Humphries et al., 2015).
TABLE 2.
CAZ/AVI resistance caused by β‐lactamase mutations
| β‐Lactamase | Variation | CAZ/AVI MIC | Possible mechanisms | Year/pathogen | Country | Notes | References |
|---|---|---|---|---|---|---|---|
| KPC‐3 | Asp179Tyr | >256/4 | Increased affinity for CAZ and restricted AVI binding | 2018/kpn | United States | Clinical strain | (Giddins et al., 2018) |
| Ala172Thr | 12/4 | Increased affinity for CAZ | 2021/kpn | France | Clinical strain | (Jousset et al., 2021) | |
| Asp179Tyr/Ala177Glu | 128/4 or 256/4 | Increased affinity for CAZ and restricted AVI binding | 2017/kpn | United States | Clinical strain | (Shields, Chen, et al., 2017) | |
| Asp179Tyr/Thr243Met | >256/4 | Increased affinity for CAZ and restricted AVI binding | 2017/kpn | United States | Clinical strain | (Shields, Chen, et al., 2017) | |
| Val240Gly | 32/4 | Increased affinity for CAZ and restricted AVI binding | 2017/kpn | United States | Clinical strain | (Shields, Chen, et al., 2017) | |
| 167–168Glu‐Leu del | 16/4 | Increased affinity for CAZ and restricted AVI binding | 2020/kpn | Italy | Clinical strain | (Antinori et al., 2020) | |
| 276–277 Glu‐Ala‐Val ins | >256/4 | Increased affinity for CAZ and restricted AVI binding | 2019/kpn | Switzerland | Clinical strain | (Poirel et al., 2020) | |
| 269–270 Pro‐Asn‐Lys ins | >128/4 | Increased affinity for CAZ and restricted AVI binding | 2019/kpn | Switzerland | Clinical strain | (Mueller et al., 2019) | |
| Val240Ala | 16/4 | Increased affinity for CAZ and restricted AVI binding | 2019/kpn | Greece | Clinical strain | (Galani et al., 2019) | |
| Ser181 ins + Thr244Ala | >256/4 | Increased affinity for CAZ and restricted AVI binding | 2020/kpn | Rome | Clinical strain | (Venditti et al., 2021) | |
| Asp163Gly | 32/4 | Increased affinity for CAZ and restricted AVI binding | 2015/kpn | United Kingdom | Laboratory selection strain | (Livermore et al., 2015) | |
| 180–181 Ser ins | 64/4 | Increased affinity for CAZ and restricted AVI binding | 2015/kpn | United Kingdom | Laboratory selection strain | (Livermore et al., 2015) | |
| 181–182 Ser‐Ser ins | 32/4 | Increased affinity for CAZ and restricted AVI binding | 2015/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2015) | |
| Thr243Pro | 32/4 | Increased affinity for CAZ and restricted AVI binding | 2015/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2015) | |
| 265–266 Ala‐Arg ins | 16/4 | Increased affinity for CAZ and restricted AVI binding | 2015/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2015) | |
| 183–184 Arg‐Ala‐Val‐Thr‐Thr‐Ser‐Ser‐Pro ins | 128/4 | Increased affinity for CAZ and restricted AVI binding | 2015/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2015) | |
| KPC‐2 | Asp179Asn | 64/4 | Increased affinity for CAZ and reduced inhibitory capacity of avibactam | 2015/eco | United States | Laboratory selection strain | (Winkler, Papp‐Wallace, & Bonomo, 2015) |
| 165–166 Gln‐Leu ins | 16/4 | Reduced inhibitory capacity of avibactam | 2021/kpn | China | Laboratory selection strain | (Guo et al., 2021) | |
| Leu169Pro plus Ser181ins | 128/4 | Reduced inhibitory capacity of avibactam | 2021/kpn | China | Laboratory selection strain | (Guo et al., 2021) | |
| 166–167 Gln‐Leu del | 32/4 | Reduced inhibitory capacity of avibactam | 2021/kpn | China | Laboratory selection strain | (Guo et al., 2021) | |
| Asp179Tyr | 32/4 | Increased affinity for CAZ and reduced inhibitory capacity of avibactam | 2020/kpn | China | Clinical strain | (P. Zhang, Shi, et al., 2020) | |
| Arg164Ala | 16/4 | Increased affinity for CAZ and restricted AVI binding | 2015/eco | United States | Laboratory selection strain | (Winkler, Papp‐Wallace, & Bonomo, 2015) | |
| Arg164Pro | 64/4 | Increased affinity for CAZ and restricted AVI binding | 2015/eco | United States | Laboratory selection strain | (Winkler, Papp‐Wallace, & Bonomo, 2015) | |
| Asp179Ala | 64/4 | Increased affinity for CAZ and restricted AVI binding | 2015/eco | United States | Laboratory selection strain | (Winkler, Papp‐Wallace, & Bonomo, 2015) | |
| Asp179Gln | 32/4 | Increased affinity for CAZ and restricted AVI binding | 2015/eco | United States | Laboratory selection strain | (Winkler, Papp‐Wallace, & Bonomo, 2015) | |
| 242–243Gly‐Thr del | >16/4 | Increased affinity for CAZ and restricted AVI binding | 2003/kpn | United States | Clinical strain | (Niu et al., 2020) | |
| CTX‐M‐15 | Gln169Leu + Gly130Ser | 16/4 | Increased affinity for CAZ and reduced inhibitory capacity for avibactam | 2018/kpn | France | Clinical strain | (Compain et al., 2018) |
| CTX‐M‐14 | Pro170Ser + Thr264Ile | 32/4 | Increased hydrolytic activity toward CAZ | 2017/kpn | Germany | Clinical strain | (Both et al., 2017) |
| AmpC | 245–249 (Asp245, Ala246, Glu247, Gly248 and Tyr249) del | 256/4 | Increased hydrolytic activity toward CAZ and decreased AVI inhibition | 2015/pae | United Kingdom | Laboratory selection strain | (Lahiri et al., 2015) |
| 238–244 (Arg238, Val239, Gly240, Pro241, Gly242, Pro243 and Leu244) del | 64/4 | Increased hydrolytic activity toward CAZ and reduced inhibitory capacity of avibactam | 2015/pae | United Kingdom | Laboratory selection strain | (Lahiri et al., 2015) | |
| Asn346Tyr | 16/4 | Loss of hydrogen interactions with AVI, reducing the inhibitory capacity of avibactam | 2018/ecl | United Kingdom | laboratory selection strain | (F. Compain et al., 2020a, 2020b) | |
| Gly176Arg | 64/4 | Increased affinity with CAZ | 2018/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2018) | |
| Arg168Pro | 16/4 | Decreased affinity with AVI | 2018/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2018) | |
| Asn366Tyr | 16/4 | Decreased affinity with AVI | 2018/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2018) | |
| Arg168His | 32/4 | Increased affinity with CAZ | 2018/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2018) | |
| Gly176Asp | 16/4 | Increased affinity with CAZ | 2018/ecl | United Kingdom | Laboratory selection strain | (Livermore et al., 2018) | |
| Gly183Asp | 32/4 | – | 2017/ecl | United States | Clinical strain | (MacVane et al., 2017) | |
| 290–292 Lys‐Val‐Ala del plus Asn346Ile | 128/4 | Increased hydrolytic activity toward CAZ and reduced inhibitory capacity of avibactam | 2021/kpn | China | Clinical strain | (M. Xu, Guo, et al., 2021) | |
| OXA‐2 | Asp149 dup | >32/4 | – | 2017/pae | Spain | Clinical strain | (Fraile‐Ribot et al., 2017) |
| OXA‐23405866 | – | ≥64/4 | Natural drug resistance | 2015/aba | Japan | clinical strain | (Yoshizumi et al., 2015)1 |
| VEB‐1 | Lys234Arg | 16/4 | Decreased ability to bond with AVI | 2019/kpn | Greece | Clinical strain | (Galani et al., 2020) |
Abbreviations: del: deletion; dup: duplication; ecl: Enterobacter cloacae; eco: Escherichia coli; ins: insertion; kpn: Klebsiella pneumoniae; pae: Pseudomonas aeruginosa.
3.1.1. Alterations in KPC enzymes
KPC‐2 and KPC‐3, which are distributed widely worldwide, are not only present in K. pneumoniae, but have also been detected in other Enterobacteriaceae, including Acinetobacter baumannii and P. aeruginosa (Bush & Bradford, 2019). The main mechanisms of resistance associated with KPC are mutations in the bla KPC gene and increased bla KPC expression.
On the one hand, amino acid substitution in KPC caused by bla KPC mutation can lead to CAZ/AVI resistance. One of the most prevalent amino acid substitutions in KPC‐2 and KPC‐3 is Asp179Tyr (Giddins et al., 2018; Shields, Nguyen, et al., 2017b): this amino acid substitution, as well as others in KPC‐3, such as Val240Gly, Asp179Tyr/Ala177Glu, and Asp179Tyr/Thr243Met, leads to CAZ/AVI resistance (Gaibani et al., 2018; Galani et al., 2019; Giddins et al., 2018; Haidar et al., 2017; Shields et al., 2018; Shields, Nguyen, et al., 2017a). Moreover, amino acid substitutions at different sites are associated with varying degrees of increased MIC levels: Asp179Tyr/Thr243Met (MIC value 256/4 mg/L) ≥ Asp179Tyr/Ala177Glu (MIC value 128/4–256/4 mg/L) > Asp179Tyr (MIC value 128/4 mg/L) > Val240Gly (MIC value 32/4 mg/L; Haidar et al., 2017; Shields, Chen, et al., 2017; Shields, Nguyen, et al., 2017a).
Experimentally, mutations have been introduced into bla KPC by site‐directed mutagenesis, with the plasmids containing mutant bla KPC‐3 used to transform E. coli DH 5α by heat shock. The MIC value of native bla KPC toward CAZ/AVI is 0.5/4 mg/L, but is altered by mutation to 16/4 mg/L for Asp179Tyr/Thr243Met, 8/4 mg/L for Asp179Tyr and Thr243Met, and 4/4 mg/L for the 165–166 Glu‐Leu insertion (Haidar et al., 2017). The greater change in MIC value for the Asp179Tyr mutation than for the 165–166 Glu‐Leu insertion or the Thr243Met mutation in CAZ/AVI suggests that the Ω‐loop plays a central role in maintaining the stability of the KPC enzymes. The Ω‐loop is formed by a salt bridge between Asp at position 179 and Arg at position 164. The less stable Asp179Tyr mutant has increased affinity for CAZ, and restricted binding to AVI. The 167–168Glu‐Leu deletion in the Ω‐loop of KPC‐3 was also found to increase the MIC value of CAZ/AVI from 1/4 to 16/4 mg/L, resulting in a phenotypic shift from susceptible to resistant (Antinori et al., 2020).
Similarly, KPC‐2 mutants containing Arg164Ala, Arg164Pro, Asp179Ala, Asp179Gln, and Asp179Asn amino acid substitutions are all resistant to CAZ/AVI, with MIC values of 16/4, 64/4, 64/4, 32/4, and 64/4 mg/L, respectively. To further study the mechanism by which KPC mutants increase MIC values for CAZ/AVI, the β‐lactamase kinetics and the inhibitory activity of AVI were evaluated in the Arg164Ala and Asp179Asn mutants. Evaluation of their catalytic activities using nitrocefin kinetic assays showed that the Arg164Ala variant had a 10‐fold higher K m for nitrofecin than wild‐type KPC‐2. Rapid‐mixing stopped‐flow kinetic assays of KPC‐2 and the Asp179Asn variant on a 1.5 ms timescale revealed that the Asp179Asn variant revealed a phenomenon called “burst”. This “burst” contributed to the resistance of the enzyme to CAZ, and its inability to bind to penicillin binding proteins (PBPs). Evaluation of the K i app and K 2/K values of the Arg164Ala and Asp179Asn variants showed that similar concentrations of AVI were needed to fully inhibit both. These findings suggested that increased affinity for CAZ may prevent the binding of AVI to enzymes with Ω‐loop substitutions, thereby increasing MIC values for CAZ/AVI rather than reducing AVI inhibition (Winkler, Papp‐Wallace, & Bonomo, 2015). The Asp179Tyr variant of KPC‐2, called KPC‐33, which is induced by selective pressure asserted by CAZ/AVI, showed an increase in the MIC value for CAZ/AVI from 0.125/4 to 16/4 mg/L, while at the same time restoring susceptibility to carbapenem antibiotics, which is useful for treatment of infection by CAZ/AVI‐resistant strains (P. Zhang, Shi, et al., 2020). A strain containing the Ala172Thr variant of KPC‐3, called KPC‐39, was found to have a MIC value for CAZ/AVI of 12/4 mg/L. A study of its molecular structure showed that the Ala172Thr substitution enlarged the spatial structure of the Ω‐loop active site, enabling it to better accommodate CAZ and increase the affinity between CAZ and the variant enzyme, resulting in a resistant phenotype (Jousset et al., 2021).
In addition to the Ω‐loop amino acid substitutions, some other substitutions can alter the affinity of the enzyme for CAZ, leading to CAZ/AVI resistance. The Asp163Gly substitution adjacent to the Ω‐loop in KPC‐3 increased the MIC of K. pneumoniae clinical isolates for CAZ/AVI from 1/4 to 32/4 mg/L; the 181_182Ser‐Ser insertion increased the MIC from 1/4 to 64/4 mg/L; the 180_181Ser insertion increased the MIC of Enterobacter cloacae for CAZ/AVI from 0.25/4 to 32/4 mg/L, whereas the 183_184Arg‐Ala‐Val‐Thr‐Thr‐Ser‐Ser‐Pro insertion increased the MIC from 0.5/4 to 128/4 mg/L (Livermore et al., 2015). A novel CAZ/AVI‐resistant mutant, KPC‐14, containing a two‐amino acid deletion in KPC‐2 (242_243Gly‐Thr) and isolated from patients in New York in 2020, has a MIC value for CAZ/AVI of >16/4 mg/L. This enzyme was significantly more efficient than KPC‐2 at catalyzing CAZ, but did not affect AVI inhibitory properties, suggesting that CAZ/AVI resistance may be due to an increase in β‐lactamase hydrolysis of CAZ rather than a decrease in its inhibition by AVI (Niu et al., 2020). In addition, KPC‐3 variants from clinical isolates of K. pneumoniae in Switzerland were resistant to CAZ/AVI (Galani et al., 2019; Gregory et al., 2010; Mueller et al., 2019; Poirel et al., 2020). These included KPC‐41, with a 269_270 Pro‐Asn‐Lys insertion and a MIC >128/4 mg/L; KPC‐50, with a 276_277Glu‐Ala‐Val insertion and a MIC of 256/4 mg/L; KPC‐8, with a Val240Gly substitution and a MIC of 32/4 mg/L; and KPC‐23, with a Val240Ala substitution and a MIC of 16/4 mg/L. In addition, combinations of these alterations, such as KPC‐64, a KPC‐3 variant with a Ser181 insertion, and Tyr244Ala substitution had a MIC >256/4 mg/L and were resistant to CAZ/AVI (Venditti et al., 2021).
Bacterial resistance to CAZ/AVI is due not only to the altered affinity of KPC variants for CAZ, but also to the altered ability of AVI to inhibit these enzymes. A mutant containing a KPC‐2 variant with a Asp179Tyr substitution (Barnes et al., 2017; Compain & Arthur, 2017; Hemarajata & Humphries, 2019) was found to have a CAZ/AVI MIC of 32/4 mg/L, whereas the CAZ/AVI MIC for wild‐type KPC‐2 was 1/4 mg/L. The Asp179Tyr substitution reduced the AVI rate constant for KPC‐2 inhibition by approximately 70,000‐fold, suggesting that the resistance of this variant to CAZ/AVI was also partly due to a significant reduction in the ability of AVI to inhibit the enzyme.
Culture of strains susceptible to CAZ/AVI in LB medium containing CAZ/AVI for 50 generations resulted in the generation of several variants of KPC‐2, such as a 165_166 Gln‐Leu insertion, a Leu169Pro insertion plus a Ser181 insertion, and a 166_167 Gln‐Leu deletion. Introduction of these bla KPC mutations into a wild‐type strain susceptible to CAZ/AVI increased the MIC values for CAZ/AVI from 2/4, 4/4, and 0.25/4 mg/L to 16/4, 128/4, and 32/4 mg/L, respectively, but did not affect MIC values for CAZ significantly, suggesting that these variants mainly affect the inhibitory activity of AVI (Guo et al., 2021).
On other hand, increased expression of bla KPC can also contribute to CAZ/AVI resistance. The first KPC‐3‐producing K. pneumoniae strain resistant to CAZ/AVI did not contain mutations in the bla KPC‐3 gene (Humphries et al., 2015). Real‐time quantitative PCR analysis, however, found that expression of the bla KPC‐3 gene was 3.8 ± 0.2‐fold higher in this resistant strain than in susceptible strains (Humphries & Hemarajata, 2017). Moreover, bla KPC‐3 gene expression was found to be 2.5‐fold or 1.5‐fold higher in resistant strains than in wild‐type strains (Gaibani et al., 2020). A study comparing 12 strains resistant to CAZ/AVI carrying wild‐type bla KPC‐2 with five strains susceptible to CAZ/AVI, also carrying bla KPC‐2, found that the number of copies of bla KPC‐2 was 2.5‐fold higher, and its expression 2.7‐fold higher, in resistant than in susceptible strains (P. Zhang, Shi, et al., 2020). Evaluation of two CAZ/AVI‐resistant strains isolated from a patient who had not been treated with CAZ/AVI showed that both strains carried two copies of the transposon Tn4401a encoding the KPC‐3 enzyme, thereby increasing the number of bla KPC gene copies as well as its expression in the bacteria (Coppi et al., 2020). Moreover, increased expression of bla KPC resulted in the incorporation of structural changes in the outer membrane proteins, thereby reducing outer membrane protein pore size and permeability, leading (ultimately) to reduced resistance to CAZ/AVI (Coppi et al., 2020). In addition, the increase in bla KPC‐3 expression was due to the transposition of the Tn4401 transposon carrying bla KPC‐3 onto the plasmid plncX3, thereby increasing the number of bla KPC‐3 copies (Nelson et al., 2017). Taken together, these findings show that increases in the number of copies and expression of bla KPC are associated with CAZ/AVI resistance, and are most likely caused by an increase in the number of plasmids carrying bla KPC or an increase in the number of copies of bla KPC on the same plasmid.
3.1.2. bla CTX‐M and bla SHV mutations lead to CAZ/AVI resistance
CTX‐M enzymes and SHV enzymes are not common in strains resistant to CAZ/AVI; however, they deserve attention. For example, a K. pneumoniae strain carrying wild‐type bla CTX‐M‐14 had a MIC value for CAZ/AVI of 1/4 mg/L, whereas a K. pneumoniae strain showing both high expression of bla OXA‐48 and a mutant bla CTX‐M‐14 △170△264 encoding a CTX‐M‐14 enzyme with Pro170Ser and Thr264Ile substitutions, showed an increase in the MIC value from 1/4 to 32/4 mg/L. To further confirm the role played by bla CTX‐M‐14 △170△264, its introduction into E. coli TOP 10 increased the MIC value for CAZ/AVI by 16‐fold, from 0.5/4 to 8/4 mg/L, and increased the MIC value for CAZ by 64‐fold, from 4/4 to 256/4 mg/L, suggesting that resistance of the mutant to CAZ/AVI is likely due to increased hydrolysis of CAZ. An Asp182Tyr substitution in the CTX‐M‐15 enzyme is also associated with decreased susceptibility to CAZ/AVI, with a 16‐fold increase in its MIC value for CAZ/AVI (Both et al., 2017; Dulyayangkul et al., 2020; Livermore et al., 2018). In addition, a variant of CTX‐M‐15 harboring two amino acid substitutions, Gln169Leu and Gly130Ser, induced resistance to CAZ/AVI in an E. coli strain susceptible to this combination (Compain et al., 2018). The Km value of WT CTX‐M‐15 was 10‐fold higher than that of the mutant enzyme, with the mechanism of resistance associated with increased affinity of the mutant enzyme for CAZ. CTX‐M‐15 includes several conserved sequences, such as 166_170 Glu‐Pro‐Thr‐Leu‐Asn on the 19‐residue Ω‐loop constituting the floor of active site. In addition, the conserved sequence 130_132 Ser‐Asp‐Asn is situated on a short loop in the all‐alpha domain, where it forms one side of the catalytic cavity (Compain et al., 2018). The structural features of CTX‐M‐15 suggest that the Pro170Ser substitution enhances the ability of the mutant enzyme to hydrolyze CAZ by altering the conformation of the Ω‐loop, thereby enabling CAZ to more easily enter the active site of the mutant enzyme. In addition, the Gly130Ser substitution may alter the structure of the catalytic cavity, enabling CAZ to more easily enter the catalytic cavity, as well as increasing the affinity between the variant enzyme and CAZ, leading ultimately to CAZ/AVI resistance.
The Ser130Gly substitution in the SHV‐1 enzyme was found to significantly reduce AVI‐induced carbamylation of β‐lactamase, with a 1700‐fold higher concentration of AVI required to inhibit the mutant than to inhibit wild‐type SHV‐1 β‐lactamase (Winkler, Papp‐Wallace, Taracila, & Bonomo, 2015).
3.1.3. ampC mutations cause CAZ/AVI resistance
Among the class C β‐lactamases, AmpC enzyme variants are the most common. The wild‐type AmpC enzyme has a MIC value for CAZ/AVI of 4/4 mg/L, whereas deletion of amino acid residues from its Ω‐loop, such as the 245_249 Asp‐Ala‐Glu‐Gly‐Tyr and 238_244 Arg‐Val‐Gly‐Pro‐Gly‐Pro‐Leu deletions, increase the MIC values for CAZ/AVI from 4/4 mg/L to 256/4 and 64/4 mg/L, respectively (Lahiri et al., 2015).
The Asn346Tyr amino acid substitution in AmpC cloacae, DHA‐1, and PDC‐5 affects the ability of AVI to inhibit these enzymes. In the wild‐type enzyme, the carboxamide at Asn346 forms hydrogen bonds with avibactam sulfonate. The amino acid substitution at this position results in loss of this interaction, which is likely reinforced by steric hindrance due to the bulky Tyr side chain, resulting in a significant reduction in carbamoylation activity (Compain et al., 2020a, 2020b). In addition, the Gly183Asp amino acid substitution, caused by a nonsynonymous G548A nucleotide mutation in bla PDC‐5, results in resistance to CAZ/AVI, thereby increasing the MIC of P. aeruginosa against CAZ/AVI from 4/4 to ≥32/4 mg/L (MacVane et al., 2017). Moreover, AmpC enzyme variants containing Gly176Arg, Gly176Asp, Arg168Pro, Asn366Tyr, and Arg168His amino acid substitutions, generated in Enterobacter cloacae by stepwise induction, were resistant to CAZ/AVI, with MIC values ranging from 0.5/4 mg/L in wild‐type to 64/4, 16/4, 16/4, and 32/4 mg/L, respectively, in the mutants. However, the mechanisms of resistance associated with these amino acid substitutions differed. Specifically, the Arg168Pro substitution reduced the affinity of the AmpC enzyme for AVI, and eliminated the synergy between CAZ and AVI, whereas the Arg168His and Gly176Arg/Asp substitutions increased the affinity of the enzyme for CAZ (Lahiri et al., 2014). Because the Asn346 residue is key to AVI binding, the Asn346Tyr substitution reduces the ability of AVI binding, resulting in resistance to CAZ/AVI (Lahiri et al., 2014). Moreover, CAZ/AVI resistance in ESBL‐producing mutants is associated mostly with changes in the efflux pump, membrane permeability, or β‐lactamase expression (Livermore et al., 2018). Compared with CYM‐2, CYM‐172 has a 290_292 Lys‐Val‐Ala deletion and a Asn346Ile substitution. Introduction of this mutant gene into E. coli DH5α through plasmid conjugation increases the MIC value for CAZ/AVI to 16/4 or 32/4 mg/L. CAZ/AVI resistance may be due to a change in CMY structure caused by the deletion of amino acids in the R2‐loop of the enzyme, which may increase CAZ hydrolysis. Additionally, the Asn346Ile substitution results in steric hindrance between the Ile side chain and the sulfate group of AVI, thereby altering the binding affinity of the latter for the enzyme and resulting in CAZ/AVI resistance (M. Xu, Zhao, et al., 2021).
In addition to mutations and increased expression of the ampC gene, AVI induces AmpC enzyme activity. To test the ability of AVI to induce AmpC enzyme activity, strains of E. cloacae, Citrobacter freundii, and P. aeruginosa were incubated with 1, 4, and 32 mg/L AVI. At a concentration of 32 mg/L, AVI strongly induced expression of the AmpC enzyme in two of five E. cloacae and in two of five P. aeruginosa strains, with the enzyme content being thousands‐fold higher than prior to induction. These findings suggest that the affinity of AmpC variants for AVI is reduced, but that these enzymes can be induced by AVI, resulting in CAZ/AVI resistance (D. M. Livermore, Jamrozy, et al., 2017).
3.1.4. bla OXA mutations lead to CAZ/AVI resistance
The MICs of 265 OXA‐48‐producing strains (collected by the International Optimal Resistance Monitoring Network between 2012 and 2015) for CAZ/AVI ranged from 0.03 to >128 mg/L, with 7.5% of these strains being resistant to CAZ/AVI (K. M. Kazmierczak, Bradford, et al., 2018). The presence of both Pro68Ala and Tyr211Ser amino acid substitutions in OXA‐48‐producing strains resulted in decreased susceptibility to CAZ/AVI. Enzyme kinetics showed that, compared with wild‐type OXA‐48, the variant enzyme had >20‐fold higher ability to hydrolyze CAZ, and a >5‐fold reduction in the inhibitory activity of AVI, suggesting that both are responsible for CAZ/AVI resistance. Molecular modeling showed that the hydrogen bond network mediated by water molecules was altered in the variant enzyme. In wild‐type enzymes, Tyr211OH and Thr234O, and Pro235O and Met236O, form hydrogen bonds with central water molecules. In the variant enzyme, however, Ser211 cannot participate in the same H‐bond network because of its less‐space‐filling properties than Tyr211. The loss of H‐bonds could enhance enzyme flexibility, which resulted in increased hydrolysis of CAZ. Moreover, the loss of aromatic stacking resulting from the Tyr211Ser amino acid substitution reduced AVI inhibitory activity (Fröhlich et al., 2019). In addition, OXA‐539, a variant of OXA‐2 with a Asp149 repeat, was found to be resistant to CAZ/AVI (Fraile‐Ribot et al., 2017), as were several Acinetobacter baumannii strains carrying class D β‐lactamases, such as OXA‐23, OXA‐40, OXA‐58, and OXA‐66. The inability of AVI to penetrate into their cell membranes prevents AVI from inhibiting these carbapenemases (Yoshizumi et al., 2015).
3.1.5. CAZ/AVI resistance caused by other β‐lactamase variants
In October 2019, an outbreak of CAZ/AVI‐resistant KPC‐2‐producing K. pneumoniae was reported in Athens, Greece. Resistance of the strain was due to a variant of the Vietnamese extended‐spectrum β‐lactamase‐1 (VEB‐1). Compared with VEB‐1, which had a MIC value for CAZ/AVI of 0.25/4 mg/L, this variant, VEB‐25, with a Lys234Arg amino acid substitution, had a MIC value of 16/4 mg/L. Resistance to CAZ/AVI was due to interference by Arg of the ability of AVI to bind to the active site of the enzyme, thereby attenuating the inhibitory effect of AVI (Galani et al., 2020).
3.2. CAZ/AVI resistance associated with changes in bacterial membrane permeability
The bacterial outer membrane proteins OmpF and OmpC, and their homologous proteins, play a role in CAZ/AVI resistance, although these proteins do not constitute the main pathway by which AVI passes through the cell walls of Enterobacteriaceae (Pagès et al., 2015).
A study investigating whether mutations in outer membrane proteins affect CAZ/AVI antibacterial activity found that the IS 5 insertion in ompK36, or the 134_135 Gly‐Asp insertion in OmpK36, when combined with ESBL such as SHV, TEM, and CTX‐M, are associated with decreased susceptibility of K. pneumoniae to CAZ/AVI (Shields et al., 2015). The decreased susceptibility of KPC‐producing K. pneumoniae to CAZ/AVI was not only related to high expression of the bla KPC gene, but also to loss of OmpK35, as confirmed by SDS‐PAGE. This indicates that the absence of OmpK35 contributes to the barrier of CAZ to permeate cells (Pagès et al., 2015). The genetic mechanism underlying OmpK35 deficiency varies by Sequence Type. For example, OmpK35 deficiency in ST11 strains may be due to early frameshift mutations and premature termination codons, whereas decreased OmpK35 expression in ST15 strains is associated with negative regulators of micF and ompR. MicF plays an important role in the transcriptional regulation of OmpK35 expression, whereas ompR negatively regulates the expression of OmpK35, either directly or through micF (Delihas & Forst, 2001).
The increased clinical use of CAZ/AVI has given rise to CAZ/AVI‐resistant strains of K. pneumoniae. The first CAZ/AVI‐resistant KPC‐3‐producing strain of K. pneumoniae, KP1244, was compared with the CAZ/AVI‐susceptible strain KP1245. Whole‐gene sequencing showed that both KP1244 and KP1245 have an Arg191Leu substitution in OmpK36, with KP1244 also having a Thr333Asn substitution in OmpK36. A frameshift mutation in ompK35 results in a truncated, 42‐amino acids OmpK35, resulting in a nonfunctional porin (Humphries & Hemarajata, 2017). Transfer of wild‐type ompK36 to KP1244 resulted in a 2‐fold decrease in its MIC value for CAZ/AVI, from 32/4 to 16/4 mg/L, whereas transfer of wild‐type ompK36 to KP1245 did not alter the MIC value. These findings indicate that although Arg191Leu does not affect CAZ/AVI MIC values, the Thr333Asn substitution alters susceptibility to CAZ/AVI. By contrast, transfer of wild‐type ompK35 to KP1244 and KP1245 reduces their MIC values for CAZ/AVI by 8‐fold, to 4/4 and 0.5 mg/L, respectively. A strain of Klebsiella oxytoca with a CAZ/AVI MIC value of 16/4 mg/L lacks the carbapenemase gene, whereas its OmpK36 has several amino acid substitutions (Ala192Pro and Asn229Ser) and deletions (Phe268 and 272_277GlyAspSerAspSerIle deletions; Aitken et al., 2016). Screening of two CAZ/AVI‐resistant strains showed decreased ompK36 gene expression and a premature stop codon in ompK35, resulting in a truncated porin (Castanheira et al., 2017). An alteration in the L3 region of OmpK36 in K. pneumoniae results in decreased susceptibility to CAZ/AVI (Castanheira et al., 2020), indicating that impaired cell membrane permeability is associated with elevated MIC values to CAZ/AVI. Resistance of A. baumannii strains to CAZ/AVI is due mainly to the failure of AVI to penetrate the outer membrane (Mushtaq et al., 2010). In addition to OmpC, OmpF, and their homologous proteins, the introduction of a wild‐type Lamb into resistant strains carrying a variant Lamb with Arg33His and Arg374Leu substitutions results in a 2‐fold reduction in the MIC values of these strains for CAZ/AVI, indicating that variant Lamb contributes to resistance to CAZ/AVI (Guo et al., 2021).
3.3. CAZ/AVI resistance mechanisms associated with efflux pumps
Overexpression of efflux pumps can reduce intracellular concentration of antibiotics and strain susceptibility to antibiotics (Nikaido & Pagès, 2012). Addition of the efflux pump inhibitor phenylalanine–arginine β‐napthylamide (PaβN) and CAZ/AVI revealed that PaβN did not significantly affect the antibacterial activity of CAZ/AVI, showing that CAZ/AVI enters cells primarily through a passive barrier, with little contribution from active efflux AcrAB‐TolC (Pagès et al., 2015). In contrast to Enterobacteriaceae, the presence of PAβN or the efflux pump inhibitor carbonyl cyanide m‐chlorophenylhydrazone (CCCP) reduces the MIC values of some strains of P. aeruginosa for CAV/AZI, from >32/4 to <0.06/4 mg/L (M. L. Winkler, Papp‐Wallace, Hujer, et al., 2015).
Overexpression of the mexA, mexB, and oprM genes in CAZ/AVI‐resistant strains of P. aeruginosa isolated from patients with cystic fibrosis altered their MIC values to CAV/AZI from 0.5/4 to 2/4 mg/L. Further analysis showed that the efflux rate of the efflux pump was associated with the MIC value. For example, the MIC values for CAV/AVI of isolates with very slow and faster efflux rates of the efflux pump were 8/4 and 64/4 mg/L, respectively. The mechanism underlying the resistance of these bacteria to CAV/AVI was is due mainly to increased activity of the MexAB–OprM efflux system, resulting from overexpression of mexA and overproduction of AmpC (Chalhoub et al., 2018). These findings suggest that efflux pump‐related mutations play a non‐negligible role in the mechanism of CAZ/AVI resistance.
3.4. CAZ/AVI resistance mechanisms caused by variants of PBPs
The Leu169Pro amino acid substitution in PBP3 results in a significant increase in the MIC value for CAZ/AVI. Introduction of the wild‐type ftsl gene into the resistant strain reduces the MIC value for CAZ/AVI from 128/4 to 4/4 mg/L (Guo et al., 2021). Alterations in the PBP‐encoding gene ftsI are associated with CAZ/AVI resistance (Castanheira et al., 2019). A CAZ/AVI MIC of 8/4 μg/ml was reported against one E. coli isolate with an unusual TIPY insertion following Tyr344 in penicillin‐binding protein 3 (PBP 3) as the result of gene duplication, which could affect the entry of CAZ (Zhang et al., 2017). In addition, some studies showed that a strain of K. pneumoniae was continuously cultured in broth medium with CAZ/AVI concentration of 1/4 mg/L under antibiotic selective pressure. The results showed that amino acid substitution of Asp354Ala of mdrA gene encoding PBP2 eventually increased MIC of CAZ/AVI from 0.25/4 to 8/4 mg/L (Karlowsky et al., 2021; Livermore et al., 2018).
4. METHODS OF DETECTING CAZ/AVI RESISTANCE
Rapid detection of resistance to CAZ/AVI can be beneficial to the early implementation of appropriate therapeutic interventions. The rapid colorimetric method‐Andrade screening antimicrobial test (ASAT), involving a 3 h incubation with Andrade solution, can rapidly detect bacterial resistance to CAZ/AVI, thereby enabling timely treatment. The ASAT reagent contains NaCl, peptone, and Andrade indicator, with a pH adjusted to 7.4 ± 0.2. Each reaction contains 175 μl of ASAT reagent and a certain concentration of CAZ/AVI, to which is added 25 μl of bacterial suspension, followed by incubation at 37°C for 3 h. Gram‐negative bacteria are detected by fermentation of glucose medium. The presence of a purple‐red color indicates a CAZ/AVI‐resistant strain, whereas a light pink color indicates a CAZ/AVI‐susceptible strain. A selective medium called SuperCAZ/AVI has also been developed to screen for CAZ/AVI‐resistant Gram‐negative bacteria. This assay was based on findings showing that the resistance breakpoints of Enterobacteriaceae and P. aeruginosa to CAZ/AVI are 8/4 mg/L, with the medium optimized to a final CAZ/AVI concentration of 6/4 mg/L. A diluted suspension of bacteria is layered onto culture medium containing CAZ/AVI, and bacterial growth is evaluated after 18 h. SuperCAZ/AVI medium can be used to screen patients for the presence of potentially CAZ/AVI‐resistant strains, allowing the implementation of targeted measures to quickly limit the transmission of infection (Sadek et al., 2020). This culture medium can also be used in epidemiological investigations to assess the prevalence of CAZ/AVI‐resistant Gram‐negative bacteria in populations. The development of new methods of detecting CAZ/AVI resistance may facilitate more rapid detection and treatment of resistant strains. These methods to detect CAZ/AVI resistance are with excellent values of sensitivity and specificity. And these screening methods provide the opportunity to detect CAZ/AVI‐resistant isolates regardless of their resistance mechanisms. They are reliable and economical. However, the specific resistance mechanisms to CAZ/AVI are unclear, so it cannot guide medication concretely.
5. CONCLUSION
CAZ/AVI, a combination of β‐lactam and a novel β‐lactamase inhibitor (AVI), has been listed since 2015, which is an important drug used to treat complicated urinary tract infections and complicated intra‐abdominal infections caused by carbapenem‐resistant Gram‐negative bacterial infections. Increases in its clinical use have led to the development of bacterial resistance. According to the global resistance surveillance reports in recent years, the overall resistance rate of Gram‐negative bacteria to CAZ/AVI was low. The resistance rate of P. aeruginosa to CAZ/AVI was significantly higher than that of Enterobacteriaceae. In addition, the resistance rate of CAZ/AVI in carbapenem‐resistant Gram‐negative bacteria was significantly increased. And the resistance rate of CAZ/AVI was significantly different among different continents. By summarizing the mechanisms of resistance to CAZ/AVI, we find that β‐lactamase variants, especially those associated with bla KPC gene mutations, are the most widely reported. Other possible mechanisms associated with CAZ/AVI resistance include overexpression of efflux pumps, reduced expression of outer membrane proteins, and gene mutations leading to loss of outer membrane proteins. Finally, we summarized the detection methods of CAZ/AVI resistance.
These studies on the resistance mechanisms to CAZ/AVI will help guide clinical drug use. Currently, Combination drug therapy is mostly used to prevent and treat the emergence of resistant strains to CAZ/AVI in clinical practice, but the treatment lacks specificity. At present, broad‐spectrum metal β‐lactamase inhibitors have been developed for the strains which are natural resistance to CAZ/AVI. As for the treatment of CAZ/AVI non‐naturally resistant strains, research and development of specific medicine may be the trend of future development. Additionally, the development of new methods to detect CAZ/AVI resistance may be beneficial for the treatment and control of drug‐resistant bacterial infections.
AUTHOR CONTRIBUTIONS
Luying Xiong: Writing – original draft (lead); writing – review and editing (lead). Xueting Wang: Writing – original draft (supporting). Yuan Wang: Conceptualization (lead). Wei Yu: Writing – review and editing (supporting). Yanzi Zhou: Supervision (lead). Xiaohui Chi: Resources (lead). Tingting Xiao: Visualization (lead). Yonghong Xiao: Data curation (supporting); supervision (supporting).
FUNDING INFORMATION
This work was supported by the National Key Research and Development Program of China (2021YFC2300300) and the National Nature Science Foundation of China (No. 81971984) and the Key research and development program of Zhejiang Province (No. 2021C03068).
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLE
ACKNOWLEDGMENTS
We thank Chen Yunbo, Shen Ping, Ji Jinru, Ying Chaoqun, Liu Zhiying, and Chen Tao of the First Affiliated Hospital, College of Medicine, Zhejiang University for their helpful discussion.
Xiong, L. , Wang, X. , Wang, Y. , Yu, W. , Zhou, Y. , Chi, X. , Xiao, T. , & Xiao, Y. (2022). Molecular mechanisms underlying bacterial resistance to ceftazidime/avibactam. WIREs Mechanisms of Disease, 14(6), e1571. 10.1002/wsbm.1571
Edited by: Cui Hua Liu, Editor
Funding information Key Research and Development Program of Zhejiang Province, Grant/Award Number: 2021C03068; the National Key Research and Development Program of China, Grant/Award Number: 2021YFC2300300; the National Nature Science Foundation of China, Grant/Award Number: 81971984
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Aitken, S. L. , Tarrand, J. J. , Deshpande, L. M. , Tverdek, F. P. , Jones, A. L. , Shelburne, S. A. , Prince, R. A. , Bhatti, M. M. , Rolston, K. V. I. , Jones, R. N. , Castanheira, M. , & Chemaly, R. F. (2016). High rates of nonsusceptibility to Ceftazidime‐avibactam and identification of New Delhi Metallo‐β‐lactamase production in Enterobacteriaceae bloodstream infections at a major cancer center. Clinical Infectious Diseases, 63(7), 954–958. 10.1093/cid/ciw398 [DOI] [PubMed] [Google Scholar]
- Antinori, E. , Unali, I. , Bertoncelli, A. , & Mazzariol, A. (2020). Klebsiella pneumoniae carbapenemase (KPC) producer resistant to ceftazidime‐avibactam due to a deletion in the blaKPC3 gene. Clinical Microbiology and Infection, 26(7), 946.e941–946.e943. 10.1016/j.cmi.2020.02.007 [DOI] [PubMed] [Google Scholar]
- Barnes, M. D. , Winkler, M. L. , Taracila, M. A. , Page, M. G. , Desarbre, E. , Kreiswirth, B. N. , Shields, R. K. , Nguyen, M. H. , Clancy, C. , Spellberg, B. , Papp‐Wallace, K. M. , & Bonomo, R. A. (2017). Carbapenemase‐2 (KPC‐2), substitutions at ambler position Asp179, and resistance to Ceftazidime‐avibactam: Unique antibiotic‐resistant phenotypes emerge from β‐lactamase protein engineering. MBio, 8(5), e00528‐17. 10.1128/mBio.00528-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebrone, C. , Lassaux, P. , Vercheval, L. , Sohier, J.‐S. , Jehaes, A. , Sauvage, E. , & Galleni, M. (2010). Current challenges in antimicrobial chemotherapy: Focus on ß‐lactamase inhibition. Drugs, 70(6), 651–679. 10.2165/11318430-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Bonnefoy, A. , Dupuis‐Hamelin, C. , Steier, V. , Delachaume, C. , Seys, C. , Stachyra, T. , Fairley, M. , Guitton, M. , & Lampilas, M. (2004). In vitro activity of AVE1330A, an innovative broad‐spectrum non‐beta‐lactam beta‐lactamase inhibitor. The Journal of Antimicrobial Chemotherapy, 54(2), 410–417. [DOI] [PubMed] [Google Scholar]
- Both, A. , Büttner, H. , Huang, J. , Perbandt, M. , Belmar Campos, C. , Christner, M. , Maurer, F. P. , Kluge, S. , König, C. , Aepfelbacher, M. , Wichmann, D. , & Rohde, H. (2017). Emergence of ceftazidime/avibactam non‐susceptibility in an MDR Klebsiella pneumoniae isolate. The Journal of Antimicrobial Chemotherapy, 72(9), 2483–2488. 10.1093/jac/dkx179 [DOI] [PubMed] [Google Scholar]
- Bush, K. , & Bradford, P. A. (2019). Interplay between β‐lactamases and new β‐lactamase inhibitors. Nature Reviews. Microbiology, 17(5), 295–306. 10.1038/s41579-019-0159-8 [DOI] [PubMed] [Google Scholar]
- Castanheira, M. , Doyle, T. B. , Hubler, C. , Sader, H. S. , & Mendes, R. E. (2020). Ceftazidime‐avibactam activity against a challenge set of carbapenem‐resistant Enterobacterales: Ompk36 L3 alterations and β‐lactamases with ceftazidime hydrolytic activity lead to elevated MIC values. International Journal of Antimicrobial Agents, 56(1), 106011. 10.1016/j.ijantimicag.2020.106011 [DOI] [PubMed] [Google Scholar]
- Castanheira, M. , Doyle, T. B. , Smith, C. J. , Mendes, R. E. , & Sader, H. S. (2019). Combination of MexAB‐OprM overexpression and mutations in efflux regulators, PBPs and chaperone proteins is responsible for ceftazidime/avibactam resistance in Pseudomonas aeruginosa clinical isolates from US hospitals. The Journal of Antimicrobial Chemotherapy, 74(9), 2588–2595. 10.1093/jac/dkz243 [DOI] [PubMed] [Google Scholar]
- Castanheira, M. , Mendes, R. E. , & Sader, H. S. (2017). Low frequency of Ceftazidime‐avibactam resistance among Enterobacteriaceae isolates carrying collected in U.S. hospitals from 2012 to 2015. Antimicrobial Agents and Chemotherapy, 61(3), e02369‐16. 10.1128/AAC.02369-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira, M. , Rhomberg, P. R. , Flamm, R. K. , & Jones, R. N. (2016). Effect of the β‐lactamase inhibitor Vaborbactam combined with Meropenem against serine Carbapenemase‐producing Enterobacteriaceae . Antimicrobial Agents and Chemotherapy, 60(9), 5454–5458. 10.1128/aac.00711-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub, H. , Sáenz, Y. , Nichols, W. W. , Tulkens, P. M. , & Van Bambeke, F. (2018). Loss of activity of ceftazidime‐avibactam due to MexAB‐OprM efflux and overproduction of AmpC cephalosporinase in Pseudomonas aeruginosa isolated from patients suffering from cystic fibrosis. International Journal of Antimicrobial Agents, 52(5), 697–701. 10.1016/j.ijantimicag.2018.07.027 [DOI] [PubMed] [Google Scholar]
- Compain, F. , & Arthur, M. (2017). Impaired inhibition by avibactam and resistance to the Ceftazidime‐avibactam combination due to the DY substitution in the KPC‐2 β‐lactamase. Antimicrobial Agents and Chemotherapy, 61(7), e00451‐17. 10.1128/AAC.00451-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compain, F. , Debray, A. , Adjadj, P. , Dorchêne, D. , & Arthur, M. (2020a). Ceftazidime‐avibactam resistance mediated by the N(346)Y substitution in various AmpC β‐lactamases. Antimicrobial Agents and Chemotherapy, 64(6), e02311‐19. 10.1128/aac.02311-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compain, F. , Debray, A. , Adjadj, P. , Dorchêne, D. , & Arthur, M. (2020b). Ceftazidime‐avibactam resistance mediated by the NY substitution in various AmpC β‐lactamases. Antimicrobial Agents and Chemotherapy, 64(6), e02311‐19. 10.1128/AAC.02311-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compain, F. , Dorchène, D. , & Arthur, M. (2018). Combination of amino acid substitutions leading to CTX‐M‐15‐mediated resistance to the Ceftazidime‐avibactam combination. Antimicrobial Agents and Chemotherapy, 62(9), e00357‐18. 10.1128/AAC.00357-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi, M. , Di Pilato, V. , Monaco, F. , Giani, T. , Conaldi, P. G. , & Rossolini, G. M. (2020). Ceftazidime‐avibactam resistance associated with increased gene copy number mediated by pKpQIL plasmid derivatives in sequence type 258. Antimicrobial Agents and Chemotherapy, 64(4), e01816‐19. 10.1128/AAC.01816-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikos, G. L. , & Markogiannakis, A. (2011). Carbapenemase‐producing Klebsiella pneumoniae: (when) might we still consider treating with carbapenems? Clinical Microbiology and Infection, 17(8), 1135–1141. 10.1111/j.1469-0691.2011.03553.x [DOI] [PubMed] [Google Scholar]
- Delihas, N. , & Forst, S. (2001). MicF: An antisense RNA gene involved in response of Escherichia coli to global stress factors. Journal of Molecular Biology, 313(1), 1–12. https://pubmed.ncbi.nlm.nih.gov/11601842 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) . (2010). Detection of Enterobacteriaceae isolates carrying metallo‐beta‐lactamase ‐ United States, 2010. MMWR Morbidity and Mortality Weekly Report, 59(24), 750. [PubMed] [Google Scholar]
- Drawz, S. M. , Papp‐Wallace, K. M. , & Bonomo, R. A. (2014). New β‐lactamase inhibitors: A therapeutic renaissance in an MDR world. Antimicrobial Agents and Chemotherapy, 58(4), 1835–1846. 10.1128/aac.00826-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulyayangkul, P. , Douglas, E. J. A. , Lastovka, F. , & Avison, M. B. (2020). Resistance to Ceftazidime/avibactam plus Meropenem/Vaborbactam when Both are used together is achieved in four steps in Metallo‐β‐lactamase‐negative Klebsiella pneumoniae . Antimicrobial Agents and Chemotherapy, 64(10), e00409‐20. 10.1128/AAC.00409-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann, D. E. , Jahić, H. , Ross, P. L. , Gu, R.‐F. , Hu, J. , Kern, G. , Walkup, G. K. , & Fisher, S. L. (2012). Avibactam is a covalent, reversible, non‐β‐lactam β‐lactamase inhibitor. Proceedings of the National Academy of Sciences of the United States of America, 109(29), 11663–11668. 10.1073/pnas.1205073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile‐Ribot, P. A. , Mulet, X. , Cabot, G. , Del Barrio‐Tofiño, E. , Juan, C. , Pérez, J. L. , & Oliver, A. (2017). Emergence of resistance to novel cephalosporin‐β‐lactamase inhibitor combinations through the duplication of amino acid D149 from OXA‐2 β‐lactamase (OXA‐539) in sequence type 235 Pseudomonas aeruginosa . Antimicrobial Agents and Chemotherapy, 61(9), e01117‐17. 10.1128/AAC.01117-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich, C. , Sørum, V. , Thomassen, A. M. , Johnsen, P. J. , Leiros, H.‐K. S. , & Samuelsen, Ø. (2019). OXA‐48‐mediated Ceftazidime‐avibactam resistance is associated with evolutionary trade‐offs. mSphere, 4(2), e00024‐19. 10.1128/mSphere.00024-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaibani, P. , Campoli, C. , Lewis, R. E. , Volpe, S. L. , Scaltriti, E. , Giannella, M. , Pongolini, S. , Berlingeri, A. , Cristini, F. , Bartoletti, M. , Tedeschi, S. , & Ambretti, S. (2018). In vivo evolution of resistant subpopulations of KPC‐producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. The Journal of Antimicrobial Chemotherapy, 73(6), 1525–1529. 10.1093/jac/dky082 [DOI] [PubMed] [Google Scholar]
- Gaibani, P. , Re, M. C. , Campoli, C. , Viale, P. L. , & Ambretti, S. (2020). Bloodstream infection caused by KPC‐producing Klebsiella pneumoniae resistant to ceftazidime/avibactam: Epidemiology and genomic characterization. Clinical Microbiology and Infection, 26(4), 516.e511–516.e514. 10.1016/j.cmi.2019.11.011 [DOI] [PubMed] [Google Scholar]
- Galani, I. , Antoniadou, A. , Karaiskos, I. , Kontopoulou, K. , Giamarellou, H. , & Souli, M. (2019). Genomic characterization of a KPC‐23‐producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime‐avibactam. Clinical Microbiology and Infection, 25(6), 763.e765–763.e768. 10.1016/j.cmi.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Galani, I. , Karaiskos, I. , Souli, M. , Papoutsaki, V. , Galani, L. , Gkoufa, A. , Antoniadou, A. , & Giamarellou, H. (2020). Outbreak of KPC‐2‐producing Klebsiella pneumoniae endowed with ceftazidime‐avibactam resistance mediated through a VEB‐1‐mutant (VEB‐25), Greece, September to October 2019. Euro Surveillance, 25(3). 10.2807/1560-7917.Es.2020.25.3.2000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddins, M. J. , Macesic, N. , Annavajhala, M. K. , Stump, S. , Khan, S. , McConville, T. H. , Mehta, M. , Gomez‐Simmonds, A. , & Uhlemann, A.‐C. (2018). Successive emergence of Ceftazidime‐avibactam resistance through distinct genomic adaptations in ‐harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrobial Agents and Chemotherapy, 62(3), e02101‐17. 10.1128/AAC.02101-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, C. J. , Llata, E. , Stine, N. , Gould, C. , Santiago, L. M. , Vazquez, G. J. , Robledo, I. E. , Srinivasan, A. , Goering, R. V. , & Tomashek, K. M. (2010). Outbreak of carbapenem‐resistant Klebsiella pneumoniae in Puerto Rico associated with a novel carbapenemase variant. Infection Control and Hospital Epidemiology, 31(5), 476–484. 10.1086/651670 [DOI] [PubMed] [Google Scholar]
- Guh, A. Y. , Bulens, S. N. , Mu, Y. , Jacob, J. T. , Reno, J. , Scott, J. , Wilson, L. E. , Vaeth, E. , Lynfield, R. , Shaw, K. M. , Vagnone, P. M. S. , Bamberg, W. M. , Janelle, S. J. , Dumyati, G. , Concannon, C. , Beldavs, Z. , Cunningham, M. , Cassidy, P. M. , Phipps, E. C. , … Kallen, A. J. (2015). Epidemiology of Carbapenem‐resistant Enterobacteriaceae in 7 US communities, 2012‐2013. JAMA, 314(14), 1479–1487. 10.1001/jama.2015.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. , Liu, N. , Lin, Z. , Ba, X. , Zhuo, C. , Li, F. , Wang, J. , Li, Y. , Yao, L. , Liu, B. , Xiao, S. , Jiang, Y. , & Zhuo, C. (2021). Mutations in porin LamB contribute to ceftazidime‐avibactam resistance in KPC‐producing Klebsiella pneumoniae . Emerging Microbes & Infections, 10(1), 2042–2051. 10.1080/22221751.2021.1984182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar, G. , Clancy, C. J. , Shields, R. K. , Hao, B. , Cheng, S. , & Nguyen, M. H. (2017). Mutations in that confer Ceftazidime‐avibactam resistance encode novel KPC‐3 variants that function as extended‐Spectrum β‐lactamases. Antimicrobial Agents and Chemotherapy, 61(5), e02534‐16. 10.1128/AAC.02534-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker, S. J. , Reddy, K. R. , Totrov, M. , Hirst, G. C. , Lomovskaya, O. , Griffith, D. C. , King, P. , Tsivkovski, R. , Sun, D. , Sabet, M. , Tarazi, Z. , Clifton, M. C. , Atkins, K. , Raymond, A. , Potts, K. T. , Abendroth, J. , Boyer, S. H. , Loutit, J. S. , Morgan, E. E. , … Dudley, M. N. (2015). Discovery of a cyclic Boronic acid β‐lactamase inhibitor (RPX7009) with utility vs class A serine Carbapenemases. Journal of Medicinal Chemistry, 58(9), 3682–3692. 10.1021/acs.jmedchem.5b00127 [DOI] [PubMed] [Google Scholar]
- Hemarajata, P. , & Humphries, R. M. (2019). Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC‐2. The Journal of Antimicrobial Chemotherapy, 74(5), 1241–1243. 10.1093/jac/dkz026 [DOI] [PubMed] [Google Scholar]
- Hirsch, E. B. , Ledesma, K. R. , Chang, K. T. , Schwartz, M. S. , Motyl, M. R. , & Tam, V. H. (2012). In vitro activity of MK‐7655, a novel β‐lactamase inhibitor, in combination with imipenem against carbapenem‐resistant gram‐negative bacteria. Antimicrobial Agents and Chemotherapy, 56(7), 3753–3757. 10.1128/aac.05927-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries, R. M. , & Hemarajata, P. (2017). Resistance to Ceftazidime‐avibactam in Klebsiella pneumoniae due to Porin mutations and the increased expression of KPC‐3. Antimicrobial Agents and Chemotherapy, 61(6), e00537‐17. 10.1128/aac.00537-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries, R. M. , Yang, S. , Hemarajata, P. , Ward, K. W. , Hindler, J. A. , Miller, S. A. , & Gregson, A. (2015). First report of Ceftazidime‐avibactam resistance in a KPC‐3‐expressing Klebsiella pneumoniae isolate. Antimicrobial Agents and Chemotherapy, 59(10), 6605–6607. 10.1128/AAC.01165-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset, A. B. , Oueslati, S. , Emeraud, C. , Bonnin, R. A. , Dortet, L. , Iorga, B. I. , & Naas, T. (2021). KPC‐39‐mediated resistance to Ceftazidime‐avibactam in a Klebsiella pneumoniae ST307 clinical isolate. Antimicrobial Agents and Chemotherapy, 65(12), e0116021. 10.1128/aac.01160-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky, J. A. , Kazmierczak, K. M. , Bouchillon, S. K. , de Jonge, B. L. M. , Stone, G. G. , & Sahm, D. F. (2018). In vitro activity of Ceftazidime‐avibactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa collected in Asia‐Pacific countries: Results from the INFORM global surveillance program, 2012 to 2015. Antimicrobial Agents and Chemotherapy, 62(7), e02569‐17. 10.1128/aac.02569-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky, J. A. , Kazmierczak, K. M. , Bouchillon, S. K. , de Jonge, B. L. M. , Stone, G. G. , & Sahm, D. F. (2019). In vitro activity of Ceftazidime‐avibactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa collected in Latin American countries: Results from the INFORM global surveillance program, 2012 to 2015. Antimicrobial Agents and Chemotherapy, 63(4), e01814‐18. 10.1128/aac.01814-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky, J. A. , Kazmierczak, K. M. , Valente, M. , Luengas, E. L. , Baudrit, M. , Quintana, A. , Irani, P. , Stone, G. G. , & Sahm, D. F. (2021). In vitro activity of ceftazidime‐avibactam against Enterobacterales and Pseudomonas aeruginosa isolates collected in Latin America as part of the ATLAS global surveillance program, 2017‐2019. The Brazilian Journal of Infectious Diseases, 25(6), 101647. 10.1016/j.bjid.2021.101647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak, K. M. , Bradford, P. A. , Stone, G. G. , de Jonge, B. L. M. , & Sahm, D. F. (2018). Activity of Ceftazidime‐avibactam and Aztreonam‐avibactam against OXA‐48‐carrying Enterobacteriaceae isolated as part of the international network for optimal resistance monitoring (INFORM) global surveillance program from 2012 to 2015. Antimicrobial Agents and Chemotherapy, 62(12), e00592‐18. 10.1128/AAC.00592-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak, K. M. , de Jonge, B. L. M. , Stone, G. G. , & Sahm, D. F. (2018a). In vitro activity of ceftazidime/avibactam against isolates of Enterobacteriaceae collected in European countries: INFORM global surveillance 2012‐15. The Journal of Antimicrobial Chemotherapy, 73(10), 2782–2788. 10.1093/jac/dky266 [DOI] [PubMed] [Google Scholar]
- Kazmierczak, K. M. , de Jonge, B. L. M. , Stone, G. G. , & Sahm, D. F. (2018b). In vitro activity of ceftazidime/avibactam against isolates of Pseudomonas aeruginosa collected in European countries: INFORM global surveillance 2012‐15. The Journal of Antimicrobial Chemotherapy, 73(10), 2777–2781. 10.1093/jac/dky267 [DOI] [PubMed] [Google Scholar]
- Kiratisin, P. , Kazmierczak, K. , & Stone, G. G. (2021). In vitro activity of ceftazidime/avibactam and comparators against carbapenemase‐producing Enterobacterales and Pseudomonas aeruginosa isolates collected globally between 2016 and 2018. Journal of Global Antimicrobial Resistance, 27, 132–141. 10.1016/j.jgar.2021.08.010 [DOI] [PubMed] [Google Scholar]
- Ko, W. C. , & Stone, G. G. (2020). In vitro activity of ceftazidime‐avibactam and comparators against gram‐negative bacterial isolates collected in the Asia‐Pacific region as part of the INFORM program (2015‐2017). Annals of Clinical Microbiology and Antimicrobials, 19(1), 14. 10.1186/s12941-020-00355-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri, S. D. , Johnstone, M. R. , Ross, P. L. , McLaughlin, R. E. , Olivier, N. B. , & Alm, R. A. (2014). Avibactam and class C β‐lactamases: Mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrobial Agents and Chemotherapy, 58(10), 5704–5713. 10.1128/aac.03057-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri, S. D. , Walkup, G. K. , Whiteaker, J. D. , Palmer, T. , McCormack, K. , Tanudra, M. A. , Nash, T. J. , Thresher, J. , Johnstone, M. R. , Hajec, L. , Livchak, S. , McLaughlin, R. E. , & Alm, R. A. (2015). Selection and molecular characterization of ceftazidime/avibactam‐resistant mutants in Pseudomonas aeruginosa strains containing derepressed AmpC. The Journal of Antimicrobial Chemotherapy, 70(6), 1650–1658. 10.1093/jac/dkv004 [DOI] [PubMed] [Google Scholar]
- Lapuebla, A. , Abdallah, M. , Olafisoye, O. , Cortes, C. , Urban, C. , Landman, D. , & Quale, J. (2015). Activity of Imipenem with Relebactam against gram‐negative Pathogens from new York City. Antimicrobial Agents and Chemotherapy, 59(8), 5029–5031. 10.1128/aac.00830-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. L. , Ko, W. C. , & Hsueh, P. R. (2022). Geographic patterns of Carbapenem‐resistant Pseudomonas aeruginosa in the Asia‐Pacific region: Results from the antimicrobial testing leadership and surveillance (ATLAS) program, 2015‐2019. Antimicrobial Agents and Chemotherapy, 66(2), e0200021. 10.1128/aac.02000-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore, D. M. , Jamrozy, D. , Mushtaq, S. , Nichols, W. W. , Young, K. , & Woodford, N. (2017). AmpC β‐lactamase induction by avibactam and relebactam. The Journal of Antimicrobial Chemotherapy, 72(12), 3342–3348. 10.1093/jac/dkx298 [DOI] [PubMed] [Google Scholar]
- Livermore, D. M. , Mushtaq, S. , Doumith, M. , Jamrozy, D. , Nichols, W. W. , & Woodford, N. (2018). Selection of mutants with resistance or diminished susceptibility to ceftazidime/avibactam from ESBL‐ and AmpC‐producing Enterobacteriaceae . The Journal of Antimicrobial Chemotherapy, 73(12), 3336–3345. 10.1093/jac/dky363 [DOI] [PubMed] [Google Scholar]
- Livermore, D. M. , Mushtaq, S. , Warner, M. , Vickers, A. , & Woodford, N. (2017). In vitro activity of cefepime/zidebactam (WCK 5222) against gram‐negative bacteria. The Journal of Antimicrobial Chemotherapy, 72(5), 1373–1385. 10.1093/jac/dkw593 [DOI] [PubMed] [Google Scholar]
- Livermore, D. M. , Warner, M. , Jamrozy, D. , Mushtaq, S. , Nichols, W. W. , Mustafa, N. , & Woodford, N. (2015). In vitro selection of ceftazidime‐avibactam resistance in Enterobacteriaceae with KPC‐3 carbapenemase. Antimicrobial Agents and Chemotherapy, 59(9), 5324–5330. 10.1128/AAC.00678-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore, D. M. , Warner, M. , & Mushtaq, S. (2013). Activity of MK‐7655 combined with imipenem against Enterobacteriaceae and Pseudomonas aeruginosa . The Journal of Antimicrobial Chemotherapy, 68(10), 2286–2290. 10.1093/jac/dkt178 [DOI] [PubMed] [Google Scholar]
- Lomovskaya, O. , Sun, D. , Rubio‐Aparicio, D. , Nelson, K. , Tsivkovski, R. , Griffith, D. C. , & Dudley, M. N. (2017). Vaborbactam: Spectrum of Beta‐lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae . Antimicrobial Agents and Chemotherapy, 61(11), e01443‐17. 10.1128/aac.01443-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVane, S. H. , Pandey, R. , Steed, L. L. , Kreiswirth, B. N. , & Chen, L. (2017). Emergence of Ceftolozane‐Tazobactam‐resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrobial Agents and Chemotherapy, 61(12), e01183‐17. 10.1128/AAC.01183-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka, A. , Tsutsumi, Y. , Yamada, M. , Suzuki, K. , Watanabe, T. , Abe, T. , Furuuchi, T. , Inamura, S. , Sakamaki, Y. , Mitsuhashi, N. , Ida, T. , & Livermore, D. M. (2015). OP0595, a new diazabicyclooctane: Mode of action as a serine β‐lactamase inhibitor, antibiotic and β‐lactam ‘enhancer’. The Journal of Antimicrobial Chemotherapy, 70(10), 2779–2786. 10.1093/jac/dkv166 [DOI] [PubMed] [Google Scholar]
- Mueller, L. , Masseron, A. , Prod'Hom, G. , Galperine, T. , Greub, G. , Poirel, L. , & Nordmann, P. (2019). Phenotypic, biochemical and genetic analysis of KPC‐41, a KPC‐3 variant conferring resistance to ceftazidime‐avibactam and exhibiting reduced carbapenemase activity. Antimicrobial Agents and Chemotherapy, 63(12), e01111‐19. 10.1128/aac.01111-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq, S. , Warner, M. , & Livermore, D. M. (2010). In vitro activity of ceftazidime+NXL104 against Pseudomonas aeruginosa and other non‐fermenters. The Journal of Antimicrobial Chemotherapy, 65(11), 2376–2381. 10.1093/jac/dkq306 [DOI] [PubMed] [Google Scholar]
- Nelson, K. , Hemarajata, P. , Sun, D. , Rubio‐Aparicio, D. , Tsivkovski, R. , Yang, S. , Sebra, R. , Kasarskis, A. , Nguyen, H. , Hanson, B. M. , Leopold, S. , Weinstock, G. , Lomovskaya, O. , & Humphries, R. M. (2017). Resistance to Ceftazidime‐avibactam is due to transposition of KPC in a Porin‐deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrobial Agents and Chemotherapy, 61(10), e00989‐17. 10.1128/AAC.00989-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, H. , & Pagès, J.‐M. (2012). Broad‐specificity efflux pumps and their role in multidrug resistance of gram‐negative bacteria. FEMS Microbiology Reviews, 36(2), 340–363. 10.1111/j.1574-6976.2011.00290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, S. , Chavda, K. D. , Wei, J. , Zou, C. , Marshall, S. H. , Dhawan, P. , Wang, D. , Bonomo, R. A. , Kreiswirth, B. N. , & Chen, L. (2020). A ceftazidime‐avibactam‐resistant and Carbapenem‐susceptible Klebsiella pneumoniae strain harboring Bla KPC‐14 isolated in New York City. mSphere, 5(4), e00775‐20. 10.1128/mSphere.00775-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi, G. B. , Falcone, M. , & Venditti, M. (2011). Surveillance and management of multidrug‐resistant microorganisms. Expert Review of Anti‐Infective Therapy, 9(8), 653–679. 10.1586/eri.11.77 [DOI] [PubMed] [Google Scholar]
- Pagès, J.‐M. , Peslier, S. , Keating, T. A. , Lavigne, J.‐P. , & Nichols, W. W. (2015). Role of the outer membrane and Porins in susceptibility of β‐lactamase‐producing Enterobacteriaceae to Ceftazidime‐avibactam. Antimicrobial Agents and Chemotherapy, 60(3), 1349–1359. 10.1128/AAC.01585-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel, L. , Vuillemin, X. , Juhas, M. , Masseron, A. , Bechtel‐Grosch, U. , Tiziani, S. , Mancini, S. , & Nordmann, P. (2020). KPC‐50 confers resistance to Ceftazidime‐avibactam associated with reduced Carbapenemase activity. Antimicrobial Agents and Chemotherapy, 64(8), e00321‐20. 10.1128/AAC.00321-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossolini, G. M. , & Stone, G. G. (2020). Assessment of the in vitro activity of ceftazidime/avibactam against a global collection of multidrug‐resistant Klebsiella spp. from the INFORM surveillance programme (2015‐2017). International Journal of Antimicrobial Agents, 56(3), 106111. 10.1016/j.ijantimicag.2020.106111 [DOI] [PubMed] [Google Scholar]
- Sadek, M. , Poirel, L. , Tinguely, C. , & Nordmann, P. (2020). A selective culture medium for screening Ceftazidime‐avibactam resistance in and Pseudomonas aeruginosa . Journal of Clinical Microbiology, 58(9). 10.1128/JCM.00965-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Q. , Yin, D. , Han, R. , Guo, Y. , Zheng, Y. , Wu, S. , Yang, Y. , Li, S. , Zhang, R. , & Hu, F. (2020). Emergence and recovery of Ceftazidime‐avibactam resistance in blaKPC‐33‐harboring Klebsiella pneumoniae sequence type 11 isolates in China. Clinical Infectious Diseases, 71(Suppl 4), S436–S439. 10.1093/cid/ciaa1521 [DOI] [PubMed] [Google Scholar]
- Shields, R. K. , Chen, L. , Cheng, S. , Chavda, K. D. , Press, E. G. , Snyder, A. , Pandey, R. , Doi, Y. , Kreiswirth, B. N. , Nguyen, M. H. , & Clancy, C. J. (2017). Emergence of Ceftazidime‐avibactam resistance due to plasmid‐borne mutations during treatment of Carbapenem‐resistant Klebsiella pneumoniae infections. Antimicrobial Agents and Chemotherapy, 61(3), e02097‐16. 10.1128/AAC.02097-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields, R. K. , Clancy, C. J. , Hao, B. , Chen, L. , Press, E. G. , Iovine, N. M. , Kreiswirth, B. N. , & Nguyen, M. H. (2015). Effects of Klebsiella pneumoniae carbapenemase subtypes, extended‐spectrum β‐lactamases, and porin mutations on the in vitro activity of ceftazidime‐avibactam against carbapenem‐resistant K. pneumoniae. Antimicrobial Agents and Chemotherapy, 59(9), 5793–5797. 10.1128/AAC.00548-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields, R. K. , Nguyen, M. H. , Chen, L. , Press, E. G. , Kreiswirth, B. N. , & Clancy, C. J. (2018). Pneumonia and renal replacement therapy are risk factors for Ceftazidime‐avibactam treatment failures and resistance among patients with Carbapenem‐resistant Enterobacteriaceae infections. Antimicrobial Agents and Chemotherapy, 62(5), e02497‐17. 10.1128/AAC.02497-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields, R. K. , Nguyen, M. H. , Press, E. G. , Chen, L. , Kreiswirth, B. N. , & Clancy, C. J. (2017a). Emergence of Ceftazidime‐avibactam resistance and restoration of Carbapenem susceptibility in Carbapenemase‐producing: A case report and review of literature. Open Forum Infectious Diseases, 4(3), ofx101. 10.1093/ofid/ofx101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields, R. K. , Nguyen, M. H. , Press, E. G. , Chen, L. , Kreiswirth, B. N. , & Clancy, C. J. (2017b). Selection of Meropenem resistance among Ceftazidime‐avibactam‐resistant, Meropenem‐susceptible Klebsiella pneumoniae isolates with variant KPC‐3 Carbapenemases. Antimicrobial Agents and Chemotherapy, 61(5), e00079‐17. 10.1128/AAC.00079-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, M. (2018). Ceftazidime‐avibactam: A review in the treatment of serious gram‐negative bacterial infections. Drugs, 78(6), 675–692. 10.1007/s40265-018-0902-x [DOI] [PubMed] [Google Scholar]
- Spiliopoulou, I. , Kazmierczak, K. , & Stone, G. G. (2020). In vitro activity of ceftazidime/avibactam against isolates of carbapenem‐non‐susceptible Enterobacteriaceae collected during the INFORM global surveillance programme (2015‐17). The Journal of Antimicrobial Chemotherapy, 75(2), 384–391. 10.1093/jac/dkz456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, G. G. , Seifert, H. , & Nord, C. E. (2020). In vitro activity of ceftazidime‐avibactam against gram‐negative isolates collected in 18 European countries, 2015‐2017. International Journal of Antimicrobial Agents, 56(3), 106045. 10.1016/j.ijantimicag.2020.106045 [DOI] [PubMed] [Google Scholar]
- Tamma, P. D. , Goodman, K. E. , Harris, A. D. , Tekle, T. , Roberts, A. , Taiwo, A. , & Simner, P. J. (2017). Comparing the outcomes of patients with Carbapenemase‐producing and non‐Carbapenemase‐producing Carbapenem‐resistant Enterobacteriaceae bacteremia. Clinical Infectious Diseases, 64(3), 257–264. 10.1093/cid/ciw741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke, C. L. , Hinchliffe, P. , Bragginton, E. C. , Colenso, C. K. , Hirvonen, V. H. A. , Takebayashi, Y. , & Spencer, J. (2019). β‐Lactamases and β‐lactamase inhibitors in the 21st century. Journal of Molecular Biology, 431(18), 3472–3500. 10.1016/j.jmb.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti, C. , Butera, O. , Meledandri, M. , Balice, M. P. , Cocciolillo, G. C. , Fontana, C. , D'Arezzo, S. , de Giuli, C. , Antonini, M. , Capone, A. , Messina, F. , Nisii, C. , & di Caro, A. (2021). Molecular analysis of clinical isolates of ceftazidime‐avibactam‐resistant Klebsiella pneumoniae . Clinical Microbiology and Infection, 27, 1040.e1–1040.e6. 10.1016/j.cmi.2021.03.001 [DOI] [PubMed] [Google Scholar]
- Winkler, M. L. , Papp‐Wallace, K. M. , & Bonomo, R. A. (2015). Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV β‐lactamases with single amino acid substitutions in the Ω‐loop. The Journal of Antimicrobial Chemotherapy, 70(8), 2279–2286. 10.1093/jac/dkv094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, M. L. , Papp‐Wallace, K. M. , Hujer, A. M. , Domitrovic, T. N. , Hujer, K. M. , Hurless, K. N. , Tuohy, M. , Hall, G. , & Bonomo, R. A. (2015). Unexpected challenges in treating multidrug‐resistant gram‐negative bacteria: Resistance to ceftazidime‐avibactam in archived isolates of Pseudomonas aeruginosa . Antimicrobial Agents and Chemotherapy, 59(2), 1020–1029. 10.1128/aac.04238-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, M. L. , Papp‐Wallace, K. M. , Taracila, M. A. , & Bonomo, R. A. (2015). Avibactam and inhibitor‐resistant SHV β‐lactamases. Antimicrobial Agents and Chemotherapy, 59(7), 3700–3709. 10.1128/AAC.04405-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M. , Zhao, J. , Xu, L. , Yang, Q. , Xu, H. , Kong, H. , Zhou, J. , & Fu, Y. (2021). Emergence of transferable ceftazidime‐avibactam resistance in KPC‐producing Klebsiella pneumoniae due to a novel CMY AmpC β‐lactamase in China. Clinical Microbiology and Infection, 28, 136.e1–136.e6. 10.1016/j.cmi.2021.05.026 [DOI] [PubMed] [Google Scholar]
- Xu, T. , Guo, Y. , Ji, Y. , Wang, B. , & Zhou, K. J. E. (2021). Epidemiology and mechanisms of Ceftazidime‐avibactam resistance in gram‐negative bacteria. Engineering, 11, 138–145. [Google Scholar]
- Yang, Y. , Guo, Y. , Yin, D. , Zheng, Y. , Wu, S. , Zhu, D. , & Hu, F. (2020). In vitro activity of Cefepime‐Zidebactam, Ceftazidime‐avibactam, and other comparators against clinical isolates of Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii: Results from China antimicrobial surveillance network (CHINET) in 2018. Antimicrobial Agents and Chemotherapy, 65(1), e01726‐20. 10.1128/aac.01726-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, D. , Wu, S. , Yang, Y. , Shi, Q. , Dong, D. , Zhu, D. , & Hu, F. (2019). Results from the China antimicrobial surveillance network (CHINET) in 2017 of the in vitro activities of Ceftazidime‐avibactam and Ceftolozane‐Tazobactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa . Antimicrobial Agents and Chemotherapy, 63(4), e02431‐18. 10.1128/aac.02431-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi, A. , Ishii, Y. , Aoki, K. , Testa, R. , Nichols, W. W. , & Tateda, K. (2015). In vitro susceptibility of characterized β‐lactamase‐producing gram‐negative bacteria isolated in Japan to ceftazidime‐, ceftaroline‐, and aztreonam‐avibactam combinations. Journal of Infection and Chemotherapy, 21(2), 148–151. 10.1016/j.jiac.2014.08.028 [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Xu, Y. , Jia, P. , Zhu, Y. , Zhang, G. , Zhang, J. , Duan, S. , Kang, W. , Wang, T. , Jing, R. , Cheng, J. , Liu, Y. , & Yang, Q. (2020). Global trends of antimicrobial susceptibility to ceftaroline and ceftazidime‐avibactam: A surveillance study from the ATLAS program (2012‐2016). Antimicrobial Resistance and Infection Control, 9(1), 166. 10.1186/s13756-020-00829-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Shi, Q. , Hu, H. , Hong, B. , Wu, X. , du, X. , Akova, M. , & Yu, Y. (2020). Emergence of ceftazidime/avibactam resistance in carbapenem‐resistant Klebsiella pneumoniae in China. Clinical Microbiology and Infection, 26(1), 124.e121–124.e124. 10.1016/j.cmi.2019.08.020 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Kashikar, A. , Brown, C. A. , Denys, G. , & Bush, K. (2017). Unusual Escherichia coli PBP 3 insertion sequence identified from a collection of Carbapenem‐resistant Enterobacteriaceae tested in vitro with a combination of Ceftazidime‐, Ceftaroline‐, or Aztreonam‐avibactam. Antimicrobial Agents and Chemotherapy, 61(8), e00389‐17. 10.1128/aac.00389-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


