Abstract
Our investigation includes the synthesis of new naphthalene-bis-triazole-bis-quinolin-2(1H)-ones 4a–e and 7a–e via Cu-catalyzed [3 + 2] cycloadditions of 4-azidoquinolin-2(1H)-ones 3a–e with 1,5-/or 1,8-bis(prop-2-yn-1-yloxy)naphthalene (2) or (6). All structures of the obtained products have been confirmed with different spectroscopic analyses. Additionally, a mild and versatile method based on copper-catalyzed [3 + 2] cycloaddition (Meldal–Sharpless reaction) was developed to tether quinolinones to O-atoms of 1,5- or 1,8-dinaphthols. The triazolo linkers could be considered as anti and syn products, which are interesting precursors for functionalized epidermal growth factor receptor (EGFR) inhibitors with potential apoptotic antiproliferative action. The antiproliferative activities of the 4a–e and 7a–e were evaluated. Compounds 4a–e and 7a–e demonstrated strong antiproliferative activity against the four tested cancer cell lines, with mean GI50 ranging from 34 nM to 134 nM compared to the reference erlotinib, which had a GI50 of 33 nM. The most potent derivatives as antiproliferative agents, compounds 4a, 4b, and 7d, were investigated for their efficacy as EGFR inhibitors, with IC50 values ranging from 64 nM to 97 nM. Compounds 4a, 4b, and 7d demonstrated potent apoptotic effects via their effects on caspases 3, 8, 9, Cytochrome C, Bax, and Bcl2. Finally, docking studies show the relevance of the free amino group of the quinoline moiety for antiproliferative action via hydrogen bond formation with essential amino acids.
Keywords: azide, naphthalene, click, quinolin-2-one, apoptosis, caspases, antiproliferative, reaction mechanism
1. Introduction
Over the past few decades, quinolones have transformed from a small and insignificant class of drugs primarily utilized for treating mild urinary tract infections to some of the most prescribed antibacterials globally [1,2,3,4]. An important different activity for quinolones has been investigated despite being well known as antibacterial. In the late 1980s, quinolone derivatives held significant potency against eukaryotic Type II topoisomerases (topoisomerase II) and demonstrated cytotoxic activity against cancer cell lines. Hence, quinolinone derivatives are promising candidates for cancer treatment [5,6,7]. Several quinolone derivatives exemplified by voreloxin, AT-3639, and quarfloxin have already been used in clinics or in clinical trials [8,9].
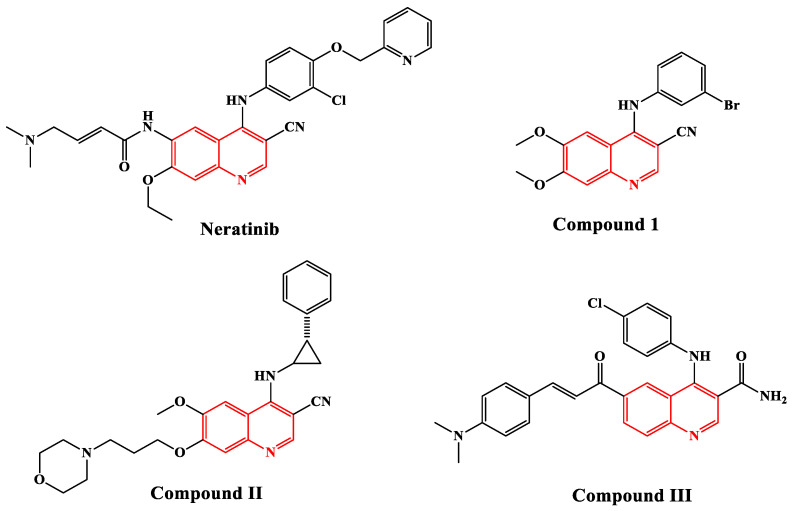
In addition, quinolinone is an intriguing fused heterocyclic scaffold that is found in several FDA-approved and commercialized anticancer medications [10], including Neratinib (Nerlynx®), an EGFR-TK inhibitor [11] (Figure 1). Moreover, several quinolinone scaffold-containing compounds are potent EGFR inhibitors. Compound I has an EGFR IC50 of 0.0075 µM [12], compound II has an EGFR IC50 of 5 nM [13], and compound III has an EGFR IC50 of 5.2 µM [14,15] (Figure 1).
Figure 1.
Structure of some quinoline-based EGFR inhibitors.
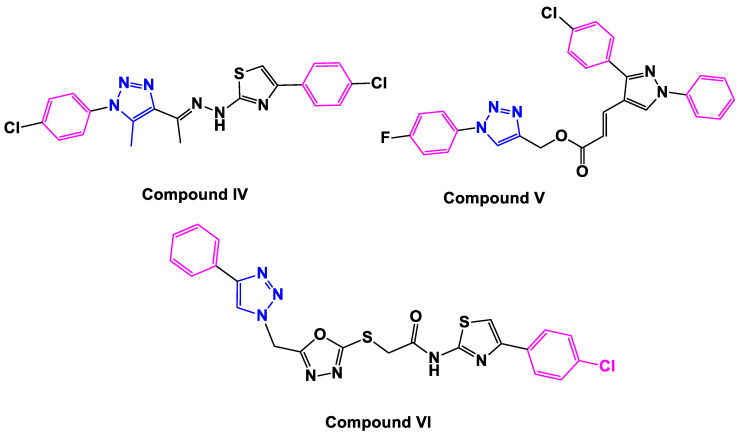
Currently, 1,2,3-triazoles exhibit a diverse set of biological actions [16,17,18,19]. They possess various pharmacological and biological features, including anticancer action [20]. Additionally, the importance and applications of 1,2,3-triazole compounds have increased [21,22], after development of the reactions of organic azides with terminal alkynes under mild conditions catalyzed by Cu(I). Further, the regioselective formation of 1,2,3-triazoles using Cu-catalyzed [3 + 2] cycloaddition has proved to be the best example of click chemistry with extensive applications in organic and medicinal chemistry [23]. However, several investigations have shown that their biological features are attributable to the triazole moiety’s making of hydrogen bonds, dipole–dipole, and stacking interactions, which warrant the development of stable complexes and, as a result, activate a cascade of metabolic activations such as apoptosis [24,25]. As a result, the antiproliferative activity of 1,2,3-triazole derivatives is explained by different mechanisms of action. Perihan and coworkers, for example, used a multi-target design technique to develop various 1,2,3-triazoles, one of which, compound IV (Figure 2), has been demonstrated to arrest the G2/M cell cycle and induce apoptosis in human cancer cells [26]. In addition, Khan and colleagues [27] described novel diphenyl-1H-pyrazole-based acrylates linked to 1,2,3-triazole V (Figure 2) as prospective apoptosis-inducing cytotoxic agents.
Figure 2.
Structure of 1,2,3-triazole-based apoptotic anti-cancer agents IV–VI.
We recently reported on compound VI’s design, synthesis, and antiproliferative activity (Figure 2) [28]. Compound VI displayed a substantial antiproliferative activity against four cancer cell lines, with a mean GI50 = 0.23 µM. Compound VI inhibited EGFR activity with an IC50 of 0.11 µM. The apoptotic mechanism demonstrated that compound VI increased Caspase-3, Caspase-9, and Cytochrome-C levels in human (Panc-1) cancer cells by 7.80, 19.30, and 13 times, respectively, compared to doxorubicin. Furthermore, VI elevated Bax levels to 40-fold greater than normal untreated cells while decreasing anti-apoptotic Bcl-2 levels 6.3-fold.
Hybridization has emerged as a promising strategy in developing new drugs with the potential to overcome cross-resistance and improve affinity and efficacy compared with the parent drugs [29,30]. Combining quinolinone with other anticancer pharmacophores may provide new candidates with great potency against drug-sensitive and drug-resistant cancers.
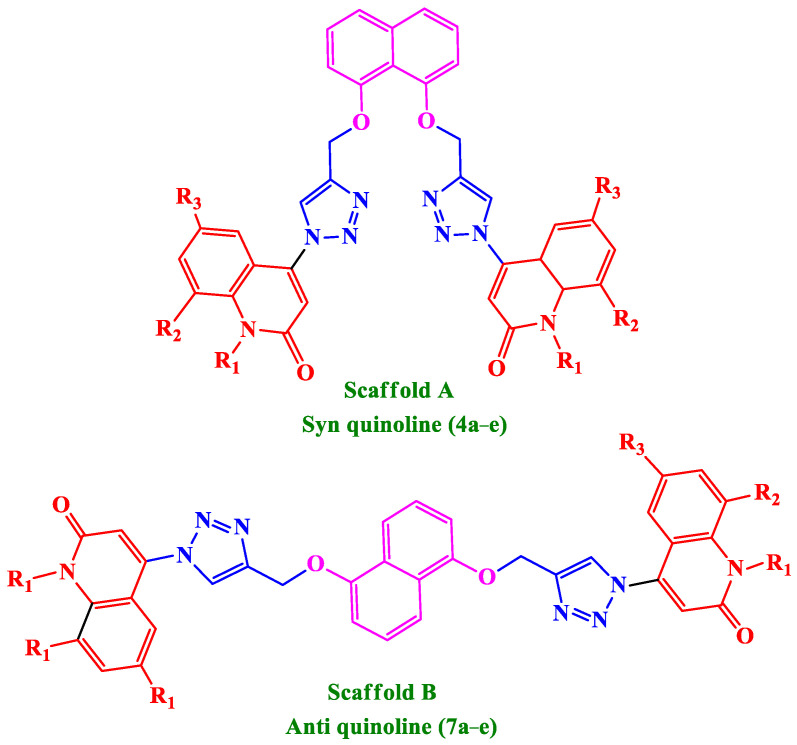
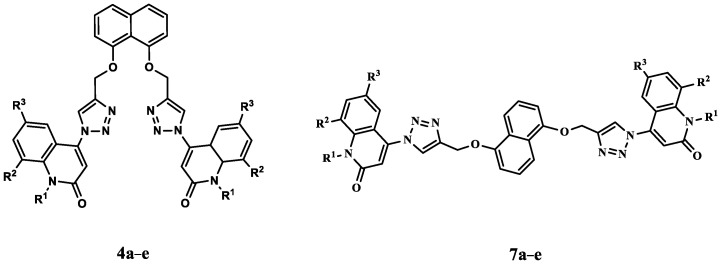
Motivated by the facts presented here, we present the synthesis of a small set of quinoline-1,2,3-triazole hybrids that will be evaluated for antiproliferative activity. The newly synthesized compounds consist of two scaffolds: Scaffold A (4a–e), which represents Syn-like-quinoline derivatives, and Scaffold B, which represents Anti-like-quinoline derivatives 7a–e, Figure 3. The two ligands of triazole and quinolinone moieties attached to the naphthalene core of Syn and Anti conformation-like represented diversity in the field of biology.
Figure 3.
Structures of new quinoline-1,2,3-triazole hybrids 4a–e and 7a–e.
The antiproliferative activity of compounds 4a–e and 7a–e will be investigated using four cancer cell lines: A549 (epithelial cancer cell line), MCF-7 (breast cancer cell line), Panc-1 (pancreas cancer cell line), and HT-29 (colon cancer cell line). The most effective antiproliferative agents will be examined further for their potential inhibitory activity against EGFR as a target for their mechanistic action. Furthermore, the most potent derivatives will be tested for their ability to trigger apoptosis against caspases 3, 8, and 9, cytochrome c, Bax, and the anti-apoptotic Bcl2.
2. Results and Discussion
2.1. Chemistry
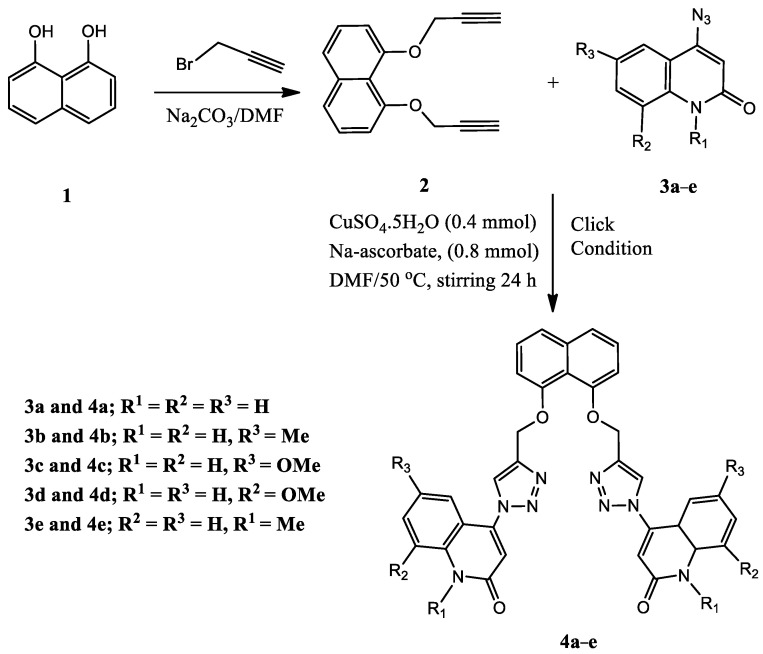
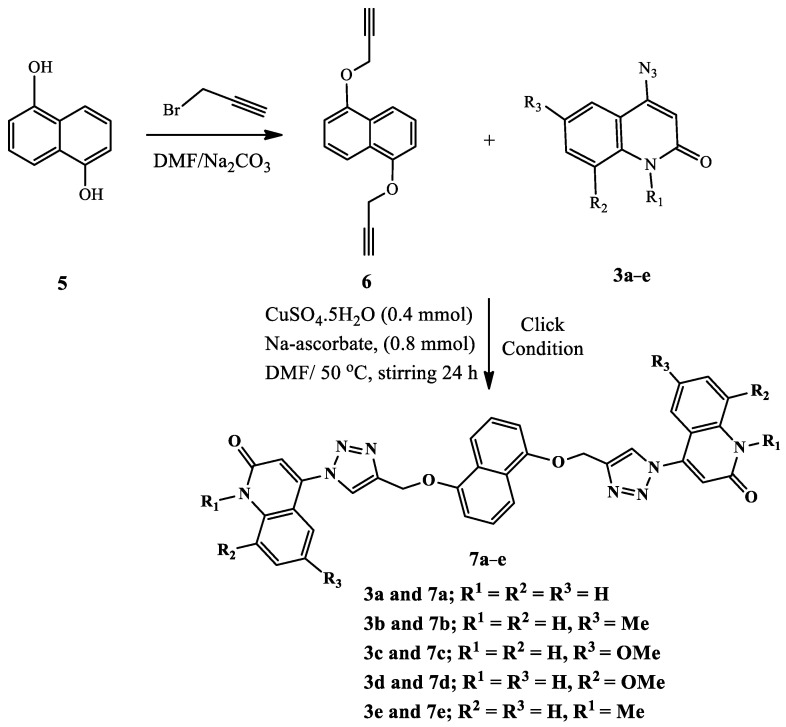
Scheme 1 and Scheme 2 depict the synthesis of compounds 4a–e and 7a–e via the cycloaddition reaction of the obtained 1,8-bis(prop-2-yn-1-yloxy)naphthalene (2) [31], or 1,5-bis(prop-2-yn-1-yloxy)naphthalene (6) with different readily prepared azides of quinolinones 3a–e [32,33,34,35,36]. The cycloaddition reaction of azide–alkyne was performed in DMF via click conditions to afford a series of five different Syn-like quinolinone-dioxo-naphthalene hybrids via triazole linker 4a–e. The characterization data, including 1H NMR, 13C NMR, 1H-1H COSY, HMBC, HSQC, and 15N NMR spectroscopy, mass spectrometry, and elemental analysis, confirmed the chemical structures of our new synthesized compounds 4a–e.
Scheme 1.
Synthesis of Syn-like quinolinone-triazole hybrids 4a–e.
Scheme 2.
Synthesis of Anti-like quinolinone-triazole hybrids 7a–e.
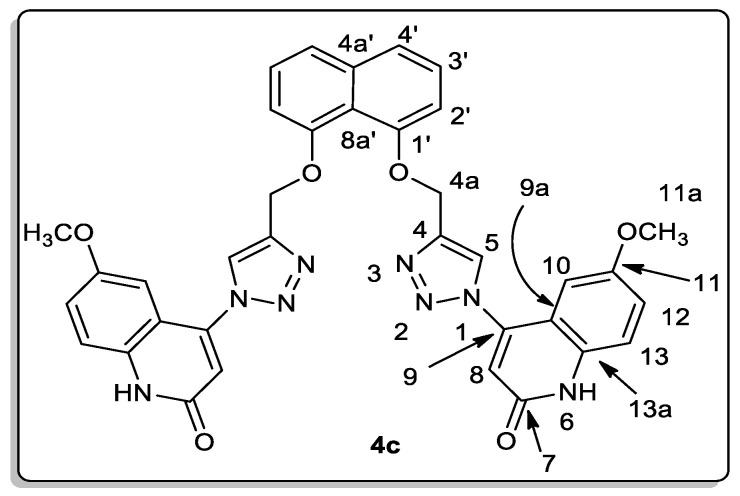
To confirm our obtained products, NMR, mass spectrometry, and elemental analysis were performed for all obtained products. All spectral data for synthesized products 4a–e, clarify that they are formed from two molecules of compounds 1a to e and one molecule of terminal alkyne 2 in a molar ratio (2:1) without any elimination. To rationalize our results, we choose compound 4c which assigned as [4,4′-(((naphthalene-1,8-diylbis(oxy))bis(methylene))-bis(1H-1,2,3-triazole-4,1-diyl))bis(6-methoxyquinolin-2(1H)-one)] (Figure 4).
Figure 4.
Structure for compound 4c.
Elemental analysis and mass spectrometry agreed with its general formula C36H28N8O6 and molecular weight (m/z = 668). The 1H NMR spectrum for compound 4c showed six-singlet signals at δH = 12.20 (2H), 9.03 (2H), 6.94 (2H), 6.90 (2H), 5.51 (4H), and 3.67 (6H) ppm., which are assigned as quinolinone-NH, H-5, H-10, H-8, H-4a (-O-CH2), and H-11a (OCH3) groups, respectively. Further, the methoxy protons H-11a are distinctive at δH 3.67 ppm; HMBC and HSQC correlation with the carbon appears at δC 55.33 ppm, which is assigned as C-11a, and other HMBC correlations with the non-protonated carbon appear at δC 154.51 ppm, which is assigned as C-11. The protons H-4a and H-5 also give HMBC correlation with the nitrogen at δN 247.6, assigned as N-1. The other sp3 nitrogen, at δN 151.3, gives only HSQC correlation with H-6; this nitrogen is assigned as N-6. Additionally, the carbon at δC 126.03 ppm gives three HMBC correlations with the protons appearing at δH 7.85 (d, J = 8.2 Hz; 2H), 7.45 (d, J = 8.2 Hz; 2H), and 7.30 (d, J = 7.4 Hz; 2H), ppm.; this carbon is assigned as C-4a’ and the signals were assigned as H-4′, H-3′ and H-2′, respectively. Furthermore, the carbon appeared at δC 125.47 ppm, giving one HSQC correlation with the proton at δH 7.45 and another HMBC correlation with the protons appearing at δH 7.85 and 7.30 ppm., and was assigned as C-3′.
On the other hand, the same synthetic methodology was used to produce a new series of bis-quinolinone-1,2,3-triazole hybrids as more analogs for biological testing. The cycloaddition reaction of 4-azido-2-quinolinones 3a–e with highly symmetrical di-o-propargylated compound 6, obtained by propargylation of 1,5-dinaphthol (5), was promoted under an above similar condition, resulting in 76–84% yield of the corresponding symmetrical bis-quinolinone triazole-based anti-like targets 7a–e (Scheme 2). The core architecture of these bis-quinolinone-triazoles and their diversity tempted us to study the anti-cancer properties of such compounds.
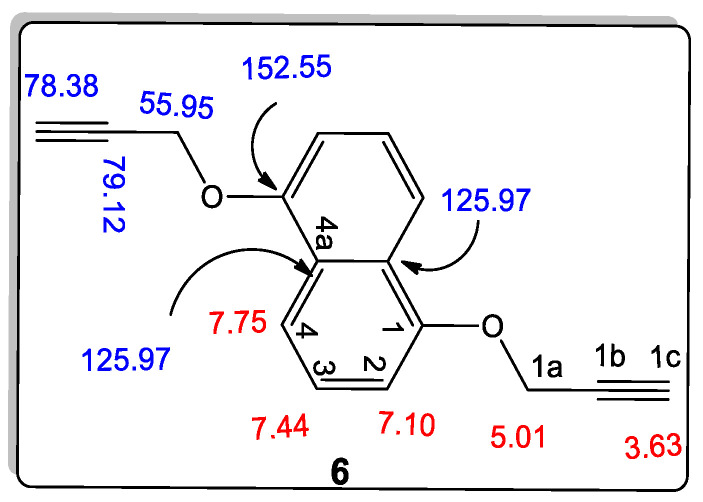
The structures of 1,5-bis(prop-2-yn-1-yloxy)naphthalene (6) were confirmed using different spectral data such as 1H NMR, 13C NMR, 2D-NMR spectrum, as well as mass spectrometry. For example, it exhibited a molecular formula of C16H12O2, which was compatible with its m/z = 236. Through the different data, we found that compound 6 fits perfectly with the previous compound 2 with a slight difference in the chemical shift’s values in its 1H NMR spectrum, and it also contains eight lines in this 13C NMR spectrum, while the previous compound 2 contains nine lines in this 13C NMR spectrum (Figure 5). In the 1H NMR spectrum of compound 6, three doublet signals appeared at δH 7.75, 7.10, and 5.01 ppm, assigned as H-4, H-2, and –OCH2, respectively. The doublet–doublet signal at δH 7.44 ppm gives HSQC and HMBC correlation with the carbon at δC 125.43 ppm., which is assigned as C-3, and this proton was assigned as H-3. This compound contains only one carbon signal at δC 125.97 ppm, which gives HMBC correlation with both signals at δH 7.75, 7.44, and 7.10 ppm., so it must be C-4a. Further, the 13C NMR spectrum showed other signals at δC 152.55, 106.74, 79.12, 78.38, and 55.95 ppm, which were assigned as C-1, C-2, C-1b, C-1c, and –O-CH2, respectively.
Figure 5.
Structure of 1,5-bis(prop-2-yn-1-yloxy)naphthalene (6).
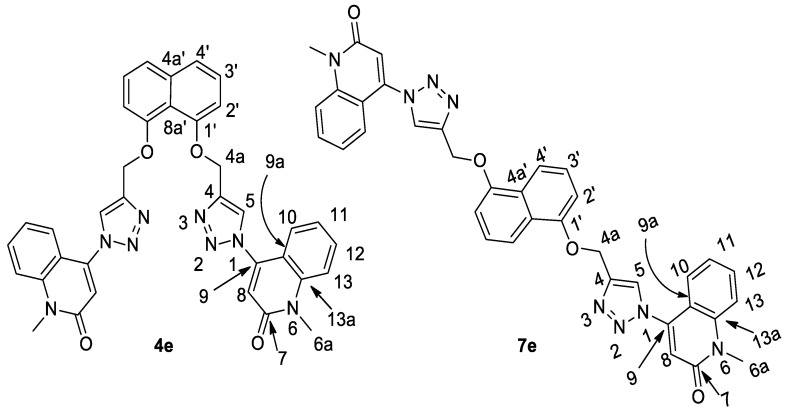
On the other hand, the novel synthesizing of naphthalene-bis-triazole-bis-quinolin-2(1H)-ones 7a–e was also confirmed on the bases of different spectral data. For example, we choose compound 7e which was assigned as [4,4′-(((naphthalene-1,5-diylbis(oxy))bis(methylene))bis-(1H-1,2,3-triazole-4,1-diyl))bis(1-methylquinolin-2(1H)-one)] (Figure 6).
Figure 6.
The similarity between structures of compounds 4e and 7e.
To confirm our results, we choose compound 7e which was assigned as [4,4′-(((naphthalene-1,5-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis(1-methylquinolin-2(1H)-one)]. Elemental analysis and all spectral data were acceptable with this proposed structure with chemical formula C36H28N8O4 (m/z = 636). Through the interpretation of the various analyses that were conducted for compound 7e and all the previous compounds, it is undoubtedly clear that the proposed chemical composition is correct, and also, to prove the composition in another way, it was compared with spectral data for its analogous compound 4e (Table 1). This is claimed to be the reaction product of 1,8-bis(propargyloxy)naphthalene (2) with 4-azido-1-methyl-1H-quinolin-2-one (3e). Additionally, the spectra are consistent with structure 7e but are very similar to those observed for sample 4e, for which 4e was assigned. The spectroscopic difference between these products is that, in 4e, C-4a’ and C-8a’ are nonequivalent, while in 7e, these two carbons are equivalent. However, in both compounds 4e and 2, C-8a’ was assigned as co-resonant with another carbon which could not be observed separately. Therefore, we suspect that the correct structure for both products is 4e; see Table 1. In addition, all spectroscopic analyses of all quinolin-2-one rings are completely identical to what was previously published [37,38,39].
Table 1.
Spectrum data for compounds 4e and 7e.
|
1H NMR
Compound 4e Compound 7e |
1H-1H COSY | Assignment | |
| 8.98 (s; 2H) 8.98 (s; 2H) |
5.50 5.50 |
H-5 H-5 |
|
| 7.85 (d, J = 8.3; 2H) 7.85 (d, J = 8.3; 2H) |
7.48 7.48 |
H-4′ H-4′ |
|
| 7.78 (dd, J = 8.4, 7.9; 2H) 7.79 (d, J = 8.0; 2H) |
7.73, 7.35 7.75 |
H-12 H-13 |
|
| 7.73 (d, J = 8.3; 2H) 7.75 (dd, J = 9.0, 8.3; 2H) |
7.78 7.79. 7.31 |
H-13 H-12 |
|
| 7.48 (dd, J = 7.6, 7.1; 2H) 7.48 (dd, J = 7.6, 6.5; 2H) |
7.85, 7.35 7.85 |
H-3′ H-3′ |
|
| 7.43 (d, J = 8.0; 2H) 7.43 (d, J = 8.2; 2H) |
H-10 H-10 |
||
| 7.35 (dd, J = 7.6, 7.3; 2H) 7.35 (dd, J = 7.7, 7.4; 2H) |
7.79, 7.48 | H-11 H-11 |
|
| 7.31 (d, J = 7.6; 2H) 7.31 (d, J = 7.7; 2H) |
7.48 7.48 |
H-2′ H-2′ |
|
| 7.04 (s; 2H) 7.03 (s; 2H) |
H-8 H-8 |
||
| 5.50 (s; 4H) 5.50 (s; 4H) |
8.98 8.98 |
H-4a H-4a |
|
| 3.74 (s; 6H) 3.74 (s; 6H) |
H-6a H-6a |
||
| 13C NMR | HSQC | HMBC | Assignment |
| 160.31 160.31 |
3.74 3.74 |
C-7 C-7 |
|
| 153.38 153.38 |
7.85, 7.43, 5.50 7.85, 7.43, 7.31, 5.50 |
C-1′ C-1′ |
|
| 143.44 143.44 |
8.98 8.98, 7.03, 5.50 |
C-4 C-4 |
|
| 142.64 142.64 |
7.48, 7.04, 5.50 7.48 |
C-9 C-9 |
|
| 140.14 140.14 |
7.78, 7.48, 3.74 7.48, 3.74 |
C-13a C-13a |
|
| 132.32 132.32 |
7.79 7.79 |
7.43 7.48, 7.35 |
C-12 C-12 |
| 126.68 126.66 |
8.98 8.98 |
8.98 8.98, 7.43, 7.31, 5.50 |
C-5 C-5 |
| 126.06 126.07 |
7.43 |
7.85, 7.43, 7.31, 5.50 7.85 |
C-4 C-10 |
| 125.51 125.50 |
7.43 7.48 |
C-10 C-3′ |
|
| 125.30 125.29 |
C-4a’ C-4a’ |
||
| 124.50 124.50 |
7.48 7.35 |
7.79, 7.35 |
C-3′ C-11 |
| 122.78 122.73 |
7.35 | 7.73 7.75 |
C-11 C-9a |
| 117.24 117.24 |
7.04 7.03 |
7.04 7.03 |
C-8 C-8 |
| 115.55 115.56 |
7.73 7.75 |
7.73, 7.35, 7.04, 3.74 7.75, 7.35, 7.04, 3.74 |
C-13 C-13 |
| 114.51 114.52 |
7.85 7.85 |
7.31 7.31 |
C-4a’,8a’,9a C-4′ |
| 106.84 106.85 |
7.31 7.31 |
7.85 7.85 |
C-2′ C-2′ |
| 61.69 61.70 |
5.50 5.50 |
5.50 |
C-4a C-4a |
| 29.59 29.59 |
3.74 3.74 |
3.74 3.74 |
C-6a C-6a |
| 15 N NMR | 1H-15N HSQC | 1H-15N HMBC | Assignment |
| 246.9 246.9 |
8.98, 7.04 8.98, 7.03 |
N-3 or N-1 N-1 |
|
| 146.9 146.9 |
7.04, 3.74 7.03, 3.74 |
N-6 N-6 |
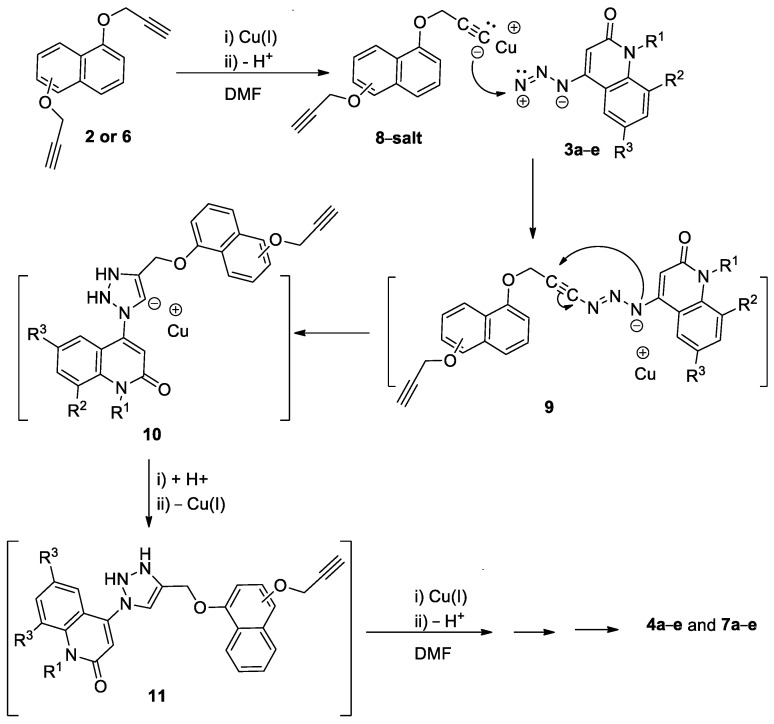
The mechanism for the obtained products 4a–e and 7a–e can be rationalized as, upon mixing (1 mmol) of terminal alkynes 2 and/or 6 with Cu(1), the salt of compound 8 would form. On the addition of (2 mmol) of azides 3a–e dissolved in DMF, a nucleophilic addition takes place to give the adduct 9, which undergoes nucleophilic attack to the N-negative charged on triple bond to give the adduct 10, which then further accepts H+ and forms the intermediate 11, which then reacts with Cu(1) to give the final products 4a–e and 7a–e via further repeating the above steps as shown in Scheme 3 [23,40].
Scheme 3.
Suggested mechanism for the formation of products 4a–e and 7a–e.
2.2. Biology
2.2.1. Antiproliferative Action
Cell Viability Assay
A cell viability test was performed using MCF-10A (human mammary gland epithelial) cell line [41,42,43] to investigate the effect of 4a–e and 7a–e on normal cell lines. In this investigation, a concentration of 50 µM of the studied compound is employed for four days, after which cell viability is assessed. The results showed that compounds 4a–e and 7a–e have no toxic effect and have more than 86% cell viability, as shown in Table 2.
Table 2.
IC50 of tested compounds 4a–e, 7a–e, and Erlotinib.
| Comp. | Cell Viability % | Antiproliferative Activity IC50 ± SEM (nM) | ||||
|---|---|---|---|---|---|---|
| A-549 | MCF-7 | Panc-1 | HT-29 | Average (GI50) | ||
| 4a | 91 | 31 ± 3 | 33 ± 3 | 36 ± 3 | 36 ± 3 | 34 |
| 4b | 89 | 41 ± 4 | 43 ± 4 | 46 ± 4 | 48 ± 4 | 45 |
| 4c | 87 | 78 ± 8 | 79 ± 8 | 83 ± 8 | 81 ± 8 | 80 |
| 4d | 86 | 90 ± 9 | 95 ± 9 | 98 ± 9 | 97 ± 9 | 95 |
| 4e | 90 | 123 ± 12 | 127 ± 12 | 135 ± 13 | 137 ± 13 | 130 |
| 7a | 89 | 57 ± 6 | 59 ± 6 | 62 ± 6 | 65 ± 6 | 61 |
| 7b | 93 | 63 ± 6 | 65 ± 6 | 67 ± 6 | 67 ± 6 | 65 |
| 7c | 91 | 70 ± 7 | 74 ± 7 | 75 ± 7 | 76 ± 7 | 74 |
| 7d | 89 | 50 ± 5 | 53 ± 5 | 57 ± 5 | 57 ± 5 | 54 |
| 7e | 85 | 126 ± 13 | 130 ± 13 | 136 ± 13 | 143 ± 14 | 134 |
| Erlotinib | - | 30 ± 3 | 40 ± 3 | 30 ± 3 | 30 ± 3 | 33 |
Antiproliferative Assay
The newly synthesized compounds were tested for antiproliferative activity against four different types of cancer cells [44,45]: A-549 (epithelial cancer cell line), MCF-7 (breast cancer cell line), Panc-1 (pancreas cancer cell line), and HT-29 (colon cancer cell line). Erlotinib was used as the reference, and Table 2 displays the results of calculating the IC50 of each compound.
Generally, the newly evaluated compounds 4a–e and 7a–e displayed significant antiproliferative activity, with mean GI50 ranging from 34 nM to 134 nM compared to the reference erlotinib with a GI50 of 33 nM against the four tested cancer cell lines. Three compounds with the highest antiproliferative activity were identified: 4a and 4b with a Syn quinoline backbone structure (Scaffold A), and 7d with an Anti-quinoline backbone structure (Scaffold B), with GI50 values ranging from 34 nM to 54 nM.
Compound 4a (R1 = R2 = R3 = H) was the most potent synthetic derivative, with a mean GI50 of 34 nM compared to the reference erlotinib’s GI50 of 33 nM. Compound 4a inhibited the MCF-7 (breast cancer) cell line more effectively than erlotinib, with an IC50 of 33 nM versus 40 nM. The antiproliferative effects of compounds 4e (R1 = CH3, R2 = R3 = H, Syn quinoline backbone) and compound 7e (R1 = CH3, R2 = R3 = H, Anti quinoline backbone), which have GI50 of 130 and 134 nM and are approximately 4-fold less potent than 4a, were greatly diminished when the free NH group in compound 4a was replaced with a methyl group. This finding highlights the significance of the free amino group at position-1 of the quinoline moiety for the antiproliferative action.
Compound 4b (R1 = R2 = H, R3 = CH3) ranks second in efficacy against the cancer cell lines tested, with a GI50 of 45 nM, which is 1.4-fold less potent than erlotinib (GI50 = 33 nM). Compound 7b (R1 = R2 = H, R3 = CH3) shares the same substitution pattern as compound 4b, but was found to be 1.4 times less potent due to its anti-quinoline backbone structure. This finding emphasizes the impact of stereochemistry in the action of this class of organic compounds, with the syn derivatives being more active than the anti-derivatives. The same pattern becomes apparent when 4a (GI50 = 34 nM) and 7a (GI50 = 130 nM) are compared.
The nature and position of the substitution on the quinoline moiety were also studied. The GI50 of the 6-methyl quinoline derivative 4b (R1 = R2 = H, R3 = CH3) was found to be 1.8-fold more potent than that of the 6-methoxy derivative 4c (R1 = R2 = H, R3 = OCH3), indicating that the methyl group was better tolerated than the methoxy group. Finally, 8-methyl quinoline derivative 4d (R1 = R3 = H, R2 = CH3) was found to be at least twofold less potent than that of the 6-methyl derivative 4b (R1 = R2 = H, R3 = CH3), demonstrating that the 6-position on the quinoline moiety was more tolerated than the 8-position.
2.2.2. EGFR Inhibitory Assay
Compounds 4a, 4b, and 7d, the most potent derivatives as antiproliferative agents, were tested for their efficiency as EGFR inhibitors [46,47] to understand how these substances affected the EGFR enzyme. According to Table 3, compounds 4a, 4b, and 7d significantly inhibited the activity of the EGFR enzyme, with IC50 values ranging from 64 nM to 97 nM. Compound 4a, the most effective antiproliferative of all synthetic derivatives, showed higher potency than the standard drug erlotinib, with an IC50 of 64 nM as opposed to erlotinib’s IC50 of 70 nM. Compounds 4b and 7d significantly inhibited EGFR, with IC50 values of 93 and 97 nM, respectively, which were roughly 1.3-fold less effective than erlotinib. The outcomes of this assay supplemented cancer cell-based assay results, suggesting that EGFR-TK may be a viable target for these drugs’ antiproliferative effects.
Table 3.
IC50 of compounds 4a, 4b, and 7d against EGFR.
| Code No. | EGFR IC50 ± SEM (nM) |
|---|---|
| 4a | 64 ± 6 |
| 4b | 93 ± 9 |
| 7d | 97 ± 9 |
| Erlotinib | 70 ± 5 |
2.2.3. Apoptosis Assays
Effect of Compounds 4a, 4b, and 7d on Caspases Cascade
The effects of derivatives 4a, 4b, and 7d on caspase-3 were studied using human epithelial cancer cell line (A-549) and compared to the reference drug doxorubicin [39,48]. The results showed that 4a, 4b, and 7d increased the level of active caspase-3 by 7–9 times when compared to control untreated cells, and that 4a, 4b, and 7d had remarkable overexpression of caspase-3 protein level (587.50 ± 4.50, 535.50 ± 4.50, and 485.50 ± 4.25 pg/mL, respectively) when compared to the reference doxorubicin (503.2 ± 4.50 pg/mL), as shown in Table 4. Compared with control untreated cells, the most active derivatives 4a and 4b increased the level of active caspase-3 by 9 and 8 times, respectively, and activated caspase-3 higher than doxorubicin, Table 4.
Table 4.
Effect of compounds 4a, 4b, and 7d on caspases 3, 8, 9 and Cytochrome C.
| Comp. No | Caspase-3 | Caspase-8 | Caspase-9 | Cytochrome C | ||||
|---|---|---|---|---|---|---|---|---|
| Conc (pg/mL) | Fold Change | Conc (ng/mL) | Fold Change | Conc (ng/mL) | Fold Change | Conc (ng/mL) | Fold Change | |
| 4a | 587.50 ± 4.50 | 8.95 | 1.20 | 6 | 17.80 | 18.75 | 0.80 | 16 |
| 4b | 535.50 ± 4.50 | 8.15 | 0.80 | 4 | 16.60 | 17.50 | 0.70 | 14 |
| 7d | 485.50 ± 4.25 | 7.40 | -- | -- | -- | -- | -- | -- |
| Doxorubicin | 503.25 ± 4.50 | 7.70 | 1.80 | 9 | 16.25 | 17.00 | 0.60 | 12 |
| Control | 65.65 | 1 | 0.20 | 1 | 0.95 | 1 | 0.05 | 1 |
The impact of compounds 4a and 4b on caspase-8 and caspase-9 was also assessed to clarify how compounds 4a and 4b induce apoptosis by activating the intrinsic or extrinsic route. The results showed that compound 4a increased caspase-8 and 9 levels by 6 and 19 times, respectively, while compound 4b showed a 4- and 18-fold increase in levels, respectively, compared to control cells. This indicates that both the intrinsic and extrinsic pathways were activated, with an effect that was more noticeable on the intrinsic pathway, because caspase-9 levels were higher, as shown in Table 4.
Effect of Compounds 4a, 4b, and 7d on Cytochrome C Level
The concentration of Cytochrome c in a cell is crucial for activating caspases and starting the intrinsic apoptosis process [49]. The evaluation of hybrids 4a and 4b as inducers of Cytochrome c is summarized in Table 4. In the A-549 epithelial cancer cell line, hybrids 4a and 4b result in a 16- and 14-fold overexpression of Cytochrome c compared to the control. Accordingly, the results presented above show that Cytochrome c overexpression and the activation of the intrinsic apoptotic pathway by the investigated hybrids may be responsible for apoptosis.
Effect of Compounds 4a, 4b, and 7d on BaX and Bcl2 Levels
The most effective hybrids 4a and 4b were further investigated for their impact on Bax and Bacl-2 levels against the A-549 epithelial cancer cell line, as shown in Table 5 [50]. The findings demonstrated that, compared to doxorubicin, 4a and 4b evoked a notable increase in Bax level. Compound 4a induction of Bax (298 pg/mL) was comparable to doxorubicin (276 pg/mL), 36 times higher than control untreated A-549 cancer cells, followed by compound 4b (284 pg/mL and 34-fold change). Finally, compound 4a (1.05 ng/mL), compound 4b (1.17 ng/mL), and doxorubicin (1.98 ng/mL) all reduced the level of the anti-apoptotic Bcl-2 protein in the A-549 cell line.
Table 5.
Bax and Bcl-2 levels for compounds 4a and 4b.
| Comp. No | Bax | Bcl-2 | ||
|---|---|---|---|---|
| Conc (pg/mL) | Fold Change | Conc (ng/mL) | Fold Reduction | |
| 4a | 298.60 | 36 | 1.05 | 5 |
| 4b | 284.50 | 34 | 1.17 | 4 |
| Doxorubicin | 276.20 | 33 | 1.98 | 2.5 |
| Control | 8.30 | 1 | 5.10 | 1 |
2.3. Molecular Docking Simulations
The EGFR is a recognized receptor that binds to the EGF outside of the cell membrane and is activated, leading to the receptor’s phosphorylation. Cell survival, proliferation, and metabolism are mediated by phosphorylated EGFR. Dysfunction of the EGFR promotes uncontrolled cell development, which results in cell overgrowth and, ultimately, oncogenesis [51]. As a result, EGFR has been considered as a potential target for cancer therapy. Molecular docking analyses were carried out using the Glide software to better understand the interactions of promising compounds (4a, 4b, and 7d) with the EGFR target protein. In this methodology, Glide docking score, emodel, and Molecular mechanics with generalized Born and surface area solvation (MMGBSA) binding free energy (ΔG Bind) were kept as support for the present work. ΔG Bind is a popular method to calculate the free energy of the binding of ligands to proteins. The minimal docking score and ΔG Bind needed for complex formation between ligand and protein show good binding affinity. More negative values suggest that the ligand is buried in the receptor cavity. The mean docking score for all three compounds is −6.98 kcal/mol, and the ΔG Bind is −67 kcal/mol, as shown in Table 6.
Table 6.
Glide XP dock score and binding free energy of promising compounds with human EGFR (PDB ID: 1M17) protein.
| Compounds | XP Docking Score (kcal/mol) |
Glide Emodel (kcal/mol) |
MMGBSA ΔG Bind (kcal/mol) |
Interacting Residues with Distance |
|---|---|---|---|---|
| 4a | −7.20 | −102.60 | −75.62 | Arg817(2.21 A°) a, Lys721(4.08 A°) d |
| 4b | −6.80 | −107.55 | −72.35 | Lys721(2.31 A°) d |
| 7d | −6.94 | −85.55 | −61.43 | Met769(1.77 A°) a, Phe771(4.90 A°) b, Lys405(4.52 A°) c |
| Erlotinib | −9.07 | −74.22 | −84.86 | Met769(1.98 A°) a,Cys773(1.43 A°) a |
a hydrogen bonding, b hydrophobic (π-π interaction), c salt bridge, d π-cation.
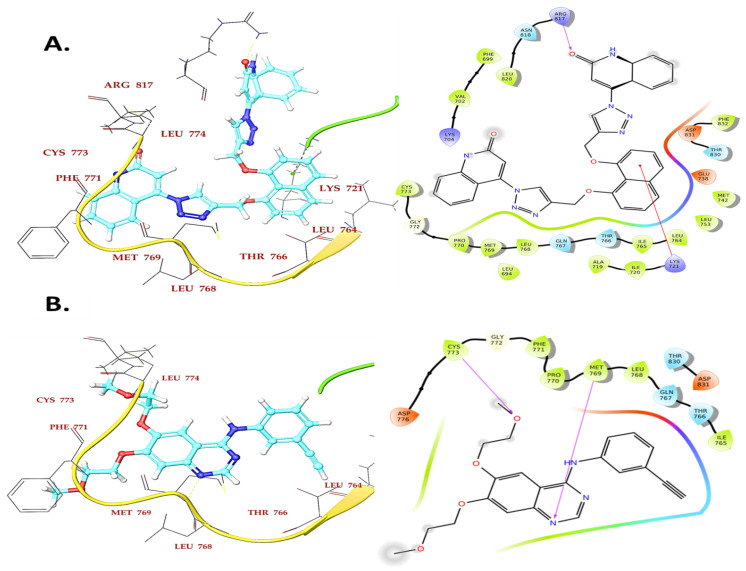
Compound 4a, the most active EGFR inhibitor among the investigated compounds, also had the most significant docking score of −7.20 kcal/mol and a ΔG Bind of −75.62 kcal/mol when contrasted to Erlotinib (−9.07 and −84.86 kcal/mol, respectively). The binding interaction showed that compound 4a formed one hydrogen bond with Arg817 at a 2.21 A° bond length, while the central naphthyl ring formed a π-cation interaction with Lys721(4.08 A°) in the EGFR kinase domain (Figure 7A). The quinolin-2(1H)-one and triazole portions of compound 4a were shown to have substantial van der Waals contacts with Gly772 (−3.09 kcal/mol), Val702 (−3.95 kcal/mol), and Leu694 (−4.14 kcal/mol), which demonstrated that the molecule is entrenched within the active site.
Figure 7.
Binding interaction of compound 4a (A) and reference compound Erlotinib (B) in the ATP binding cavity of EGFR kinase domain.
On the other hand, Compounds 4b and 7d demonstrated hydrogen and hydrophobic interactions with Lys721, Met769, Phe771, and Lys405 residues in the EGFR kinase domain that were comparable to erlotinib, supporting its inhibitory activity towards EGFR.
3. Conclusions
In conclusion, a straightforward way to tether different quinolinones derivatives to 1,5-dinaphthol and 1,8-dinaphthol via 1,2,3-triazole linkers was elaborated based on Cu-catalyzed [3 + 2] cycloaddition of o-propargyl units. In the course of the introduction of the propargyl groups, an interesting dependence of the regioselectivity on the substituent found at the OH groups of the naphthalene rings was observed as Syn and Anti isomers-like. By sequential [3 + 2] cycloadditions, it was possible to link two quinolinone moieties to the naphthalene skeleton. Remarkably, this cycloaddition occurred, and all products obtained were hitherto unknown. Pharmacological screening of novel products showed interesting and promising results as EGFR inhibitors with potential apoptotic antiproliferative action.
4. Experimental Section
4.1. Chemistry
General Details: See Appendix SA.
4.1.1. Starting Materials
The materials 4-Azido-2-quinoline-(1H)-ones 3a–e, naphthalene-1,8-diol (1) and naphthalene-1,5-diol (5) (Aldrich) were used as received. The 1,8-Bis(prop-2-yn-1-yloxy)naphthalene (2) and 1,5-bis(prop-2-yn-1-yloxy)naphthalene (6) were synthesized according to the literature.
4.1.2. General Procedure for the Synthesis of Compounds 4a–e and 7a–e
A mixture of 1,8-bis(prop-2-yn-1-yloxy)naphthalene (2) or 1,5-bis(prop-2-yn-1-yloxy)-naphthalene (6) (1.1 mmol) in 20 mL DMF, CuSO4.5H2O (0.4 mmol) and (0.4 mmol) of sodium ascorbate was stirred for 10 min at room temperature. To the above mixture, 4-azido compounds 3a–e (1.0 mmol) in 20 mL DMF were added dropwise. The reaction mixture was stirred at 50 °C for 24 h. After 14 hr, another portion of sodium ascorbate (0.4 mmol) was added to the reaction mixture to prevent the reversible process for Cu (+1). The reaction mixtures were monitored with TLC. After completion, the mixture was diluted with 100 gm ice, and the formed precipitate was filtered off and washed four times with cold water to give compounds 4a–e and 7a–e in excellent yields.
1,8-Bis(prop-2-yn-1-yloxy)naphthalene (2) m.p 190–192 °C [31].
[4,4′-(((naphthalene-1,8-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(quinolin-2(1H)-one)] (4a). This compound was obtained as colorless powder, (85%), m.p. > 360 °C. 1H NMR (DMSO-d6): δH = 12.29 (s, 2H; NH-6), 9.00 (s, 2H; H-5), 7.85 (d, J = 8.3, 2H; H-4′), 7.66 (dd, J = 7.7, 7.0, 2H; H-12), 7.49 (d, J = 7.7, 2H; H-13), 7.48 (dd, J = 7.7, 6.8, 2H; H-3′), 7.45 (d, J = 8.2, 2H; H-10), 7.31 (d, J = 7.8, 2H; H-2′), 7.26 (dd, J = 7.5, 7.2, 2H; H-11), 6.91 (s, 2H; H-8), 5.50 ppm (s, 4H; H-4a), 13C NMR (DMSO-d6): δC = 160.99 (C-7), 153.37 (C-1′), 143.64 (C-4), 143.42 (C-9), 139.43 (C-13a), 131.88 (C-12), 126.46 (C-5), 126.03 (C-4a’), 125.51 (C-3′), 124.04 (C-13), 122.59 (C-11), 117.76 (C-8), 115.92 (C-10), 114.48 (C-4′, 8a’, 9a), 106.80 (C-2′), 61.67 ppm (C-4a), 15N NMR (DMSO-d6): δN = 247.4 (N-3), 152.3 ppm (N-6), N-1 and N-2 n/o. m/z = 608 (M+, 8). Anl. Calcd. for C34H24N8O4: C, 67.10; H, 3.97; N, 18.41; Found: C, 67.19; H, 4.11; N, 18.55.
[4,4′-(((naphthalene-1,8-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis(6-methylquinolin-2(1H)-one)] (4b). This compound was obtained as colorless powder (80%), m.p > 360 °C. 1H NMR (DMSO-d6): δH = 12.21 (s, 2H; H-6), 8.98 (s, 2H; H-5), 7.86 (d, J = 8.1, 2H; H-4′), 7.48 (dd, J = 8.3, 8.3, 4H; H-12, 3′), 7.39 (d, J = 8.2, 2H; H-13), 7.31 (d, J = 7.7, 2H; H-2′), 7.26 (s, 2H; H-10), 6.86 (s, 2H; H-8), 5.50 (s, 4H; H-4a), 2.31 ppm (s, 6H; H-11a), 13C NMR (DMSO-d6): δC = 160.81 (C-7), 153.37 (C-1′), 143.44 (C-4,9), 137.53 (C-13a), 133.18 (C-12), 131.74 (C-11), 126.45 (C-5), 125.63 (C-4a’), 125.29 (C-3′), 123.20 (C-10), 117.78 (C-8), 115.91 (C-13), 114.42 (C-4′, 8a’, 9a), 106.77 (C-2′), 61.67 (C-4a), ppm 20.55 (C-11a), 15N NMR (DMSO-d6): δN = 247.8 (N-3), 151.8 ppm (N-6). N-1 and N-2 n/o. m/z = 636 (M+, 100). Anl. Calcd. for C36H28N8O4: C, 67.91; H, 4.43; N, 17.60; Found: C, 68.07; H, 4.52; N, 17.44.
[4,4′-(((naphthalene-1,8-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(6-methoxyquinolin-2(1H)-one)] (4c). This compound was obtained as colorless powder (77%), m.p > 360 °C. 1H NMR (DMSO-d6): δH = 12.20 (s, 2H; NH-6), 9.03 (s, 2H; H-5), 7.85 (d, J = 8.2, 2H; H-4′), 7.45 (dd, J = 8.2, 6.8, 2H; H-3′), 7.44 (d, J = 8.2, 2H; H-13), 7.34 (d, J = 8.7, 2H; H-12), 7.30 (d, J = 7.4, 2H; H-2′), 6.94 (s, 2H; H-10), 6.90 (s, 2H; H-8), 5.51 (s, 4H; H-4a), 3.67 ppm (s, 6H; H-11a), 13C NMR (DMSO-d6): δC = 160.52 (C-7), 154.51 (C-11), 153.32 (C-1′), 143.55 (C-4), 143.11 (C-9), 134.09 (C-13a), 126.35 (C-5), 126.03 (C-4a’), 125.47 (C-3′), 121.07 (C-12), 118.05 (C-8), 117.44 (C-13), 114.88 (C-4′, 9a), 114.44 (C-8a’), 106.69 (C-2′), 105.49 (C-10), 61.62 (C-4a), 55.33 ppm (C-11a), 15N NMR (DMSO-d6): δN = 247.6 (N-1), 151.3 ppm (N-6). N-2 and N-3 n/o. m/z = 668 (M+, 41). Anal. Calcd. for C36H28N8O6: C, 64.66; H, 4.22; N, 16.76; Found: C, 64.59; H, 4.31; N, 16.88.
[4,4′-(((naphthalene-1,8-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(8-methylquinolin-2(1H)-one)] (4d). This compound was obtained as colorless powder (81%), m.p > 360 °C. 1H NMR (DMSO-d6): δH = 11.40 (bs, 2H; NH-6), 8.97 (s, 2H; H-5), 7.85 (d, J = 8.1, 2H; H-4′), 7.52 (d, J = 6.8, 2H; H-12), 7.46 (dd, J = 8.0, 7.4, 2H; H-3′), 7.31 (d, J = 7.4, 2H; H-2′), 7.22 (d, J = 7.2, 2H; H-10), 7.16 (dd, J = 7.4, 6.9, 2H; H-11), 6.91 (s, 2H; H-8), 5.50 (s, 4H; H-4a), 2.51 ppm (s, 6H; H-13b), 13C NMR (DMSO-d6): δC = 161.34 (C-7), 153.38 (C-1′), 144.22 (C-4), 143.39 (C-9), 137.82 (C-13a), 133.10 (C-12), 126.63 (C-5), 126.06 (C-4a’), 125.51 (C-3′), 124.42 (C-13), 122.32 (C-11), 121.80 (C-10), 117.93 (C-8), 114.85 (C-4′), 114.52 (C-8a’, 9a), 106.85 (C-2′), 61.69 (C-4a), 17.53 ppm (C-13b), 15N NMR (DMSO-d6): δN = 247.4 (N-3 or N-1), 149.3 ppm (N-6). N-2 and (N-1 or N-3) n/o. m/z = 636 (M+, 31). Anl. Calcd. for C36H28N8O4: C, 67.91; H, 4.43; N, 17.60; Found: C, 68.10; H, 4.39; N, 17.49.
[4,4′-(((naphthalene-1,8-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(1-methylquinolin-2(1H)-one)] (4e). This compound was obtained as colorless powder (79%), m.p > 360 °C. NMR (DMSO-d6) (See Table 1) m/z = 636 (M+, 12). Anl. Calcd. for C36H28N8O4: C, 67.91; H, 4.43; N, 17.60; Found: C, 67.88; H, 4.55; N, 17.79.
1,5-Bis(prop-2-yn-1-yloxy)naphthalene (6). This compound was obtained as colorless powder (90%), m.p 150–152 °C. 1H NMR (DMSO-d6): δH = 7.75 (d, J = 8.4, 2H; H-4), 7.44 (dd, J = 8.2, 8.0, 2H; H-3), 7.10 (d, J = 7.7, 2H; H-2), 5.01 (d, J = 2.0, 4H; -OCH2), 3.34 (s, 2H; H-1c), 13C NMR (DMSO-d6): δC = 152.55 (C-1), 125.97 (C-4a), 125.43 (C-3), 114.23 (C-4), 106.74 (C-2), 79.12 (C-1b), 78.38 (C-1c), 55.99 (-OCH2). M/z = 236 (M+, 35). Anl. Calcd. for C16H12O2: C, 81.34; H, 5.12; Found: C, 81.44; H, 4.97.
[4,4′-(((naphthalene-1,5-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(quinolin-2(1H)-one)] (7a). This compound was obtained as colorless powder (86%), m.p > 360 °C. 1H NMR (DMSO-d6): δH = 12.21 (s, 2H; NH-6), 8.98 (s, 2H; H-5), 7.84 (d, J = 8.4 Hz, 2H; H-4′), 7.77 (d, J = 8.5 Hz, 2H; H-13), 7.51–7.26 (m, 9H; H-12,3′,10,11,2′,8), 6.86 (s, 2H; H-2′), 5.50 (s, 4H; H-4a), 13C NMR (DMSO-d6): δC = 160.84 (C-7), 153.11 (C-1′), 143.42 (C-4), 142.64 (C-9), 140.11 (C-13a), 133.18 (C-12), 126.45 (C-5), 126.07 (C-10), 125.50 (C-3′), 125.29 (C-4a’), 124.50 (C-11), 123.20 (C-9a), 117.24 (C-8), 115.91 (C-13), 114.42 (C-4′), 106.77 (C-2′), 61.88 (C-4a). m/z = 608 (M+, 20). Anl. Calcd. for C34H24N8O4: C, 67.10; H, 3.97; N, 18.41; Found: C, 66.95; H, 3.88; N, 18.58.
[4,4′-(((naphthalene-1,5-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(6-methylquinolin-2(1H)-one)] (7b). This compound was obtained as colorless powder (73%), m.p > 360 °C. 1H NMR (DMSO-d6): δH = 12.22 (s, 2H; H-6), 8.96 (s, 2H; H-5), 7.96 (d, J = 8.0, 2H; H-4′), 7.85 (d, J = 8.4, 4H; H-12, 3′), 7.51–7.26 (m, 6H; H-13,2′,10), 6.86 (s, 2H; H-8), 5.50 (s, 4H; H-4a), 2.09 ppm (s, 6H; H-11a). m/z = 636 (M+, 31). Anl. Calcd. for C36H28N8O4: C, 67.91; H, 4.43; N, 17.60; Found: C, 67.83; H, 4.38; N, 17.78.
[4,4′-(((naphthalene-1,5-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(6-methoxyquinolin-2(1H)-one)] (7c). This compound was obtained as colorless powder (78%), m.p > 360 °C. 1H NMR (DMSO-d6): δH = 12.19 (s, 2H; NH-6), 9.02 (s, 2H; H-5), 7.84 (d, J = 6.2, 2H; H-4′), 7.45 (d, J = 6.0, 2H; H-13), 7.31 (dd, J = 9.1, 8.2, 2H; H-12), 6.94 (d, J = 8.0, 2H; H-3′), 7.30 (d, J = 7.4, 2H; H-10), 6.94 (s, 2H; H-11), 6.70 (s, 2H; H-8), 5.51 (s, 4H; H-4a), 3.32 ppm (s, 6H; H-11a), 13C NMR (DMSO-d6): δC = 160.52 (C-7), 154.51 (C-11), 153.32 (C-1′), 143.55 (C-4), 143.11 (C-9), 134.09 (C-13a), 126.35 (C-5), 126.03 (C-4a’), 125.47 (C-3′), 121.07 (C-12), 118.05 (C-8), 117.44 (C-13), 114.88 (C-4′, 9a), 114.44 (C-8a’), 106.69 (C-2′), 105.49 (C-10), 61.62 (C-4a), 55.33 ppm (C-11a), 15N NMR (DMSO-d6): δN = 151.4 ppm (N-6). N-1, N-2 and N-3 n/o. m/z = 668 (M+, 8). Anl. Calcd. for C36H28N8O6: C, 64.66; H, 4.22; N, 16.76; Found: C, 64.79; H, 4.18; N, 16.59.
[4,4′-(((naphthalene-1,5-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(8-methylquinolin-2(1H)-one)] (7d). This compound was obtained as colorless powder (88%), m.p > 360 °C. 1H NMR (DMSO-d6): δH =11.38 (bs, 2H; NH-6), 8.96 (s, 2H; H-5), 7.96–690 (m, 12H; H-4′,12,3′,2′,10,8), 5.50 (s, 4H; H-4a), 2.50 ppm (s, 6H; H-13b). m/z = 636 (M+, 23). Anl. Calcd. for C36H28N8O4: C, 67.91; H, 4.43; N, 17.60; Found: C, 68.06; H, 4.52; N, 17.79.
[(4,4′-(((naphthalene-1,5-diylbis(oxy))bis(methylene))bis(1H-1,2,3-triazole-4,1-diyl))-bis-(1-methylquinolin-2(1H)-one)] (7e). This compound was obtained as colorless powder (74%), m.p > 360 °C. 1H NMR (DMSO-d6) (See Table 1). m/z = 636 (M+, 58). Anl. Calcd. for C36H28N8O4: C, 67.91; H, 4.43; N, 17.60; Found: C, 67.78; H, 4.39; N, 17.74.
4.2. Biology
Appendix SA (Supplementary File) contains information on all biological experimental tests.
4.3. Docking Study
Molecular docking simulations were performed using MOE® software within EGFR protein crystal structure with erlotinib as a co-crystallized ligand (PDB ID: 1M17). Docking protocol and other experimental details were used exactly as reported elsewhere [52,53,54]. See Appendix SA (Supplementary Files).
Acknowledgments
We acknowledge support by the KIT-Publication Fund of the Karlsruhe Institute of Technology, Karlsruhe, Germany.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/molecules27248765/s1, Figure S1–S85: Spectral data for compound; Appendix SA.
Author Contributions
Credit authorship contribution statement. E.M.E.-S., M.A.A., K.M.E.-S. and F.F.A.-L.: Conceptualization, writing, and editing; A.I.A.-n.: editing and revision; H.A.M.G.: Biology, editing; B.G.M.Y.: Biology, methodology, writing, and editing; A.H.M. and H.M.F.: writing the draft, editing; A.B.B. and S.B.: Editing and revision; I.A. and H.P.: Modeling, writing and editing; A.H.M.: Conceptualization, methodology, writing the draft and editing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The study did not report any data.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the all new compounds are available from the authors up on request.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Emmerson A.M., Jones A.M. The quinolones: Decades of development and use. J. Antimicrob. Chemother. 2003;51((Suppl. S1)):13–20. doi: 10.1093/jac/dkg208. [DOI] [PubMed] [Google Scholar]
- 2.Mitscher L.A. Bacterial topoisomerase inhibitors: Quinolone and pyridone antibacterial agents. Chem. Rev. 2005;105:559–592. doi: 10.1021/cr030101q. [DOI] [PubMed] [Google Scholar]
- 3.Linder J.A., Huang E.S., Steinman M.A., Gonzales R., Stafford R.S. Fluoroquinolone prescribing in the United States: 1995 to 2002. Am. J. Med. 2005;118:259–268. doi: 10.1016/j.amjmed.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Chao Y.S., Farrah K. Cost-Effectiveness, and Guidelines. Canadian Agency for Drugs and Technologies in Health; Ottawa, Canada: 2019. Fluoroquinolones for the treatment of urinary tract infection: A review of clinical effectiveness. [PubMed] [Google Scholar]
- 5.Sharma P.C., Chaudhary M., Sharma A., Piplani M., Rajak H., Prakash O. Insight view on possible role of fluoroquinolones in cancer therapy. Curr. Top. Med. Chem. 2013;13:2076–2096. doi: 10.2174/15680266113139990133. [DOI] [PubMed] [Google Scholar]
- 6.Kan J.Y., Hsu Y.L., Chen Y.H., Chen T.C., Wang J.Y., Kuo P.L. Gemifloxacin, a fluoroquinolone antimicrobial drug, inhibits migration and invasion of human colon cancer cells. Bio. Med. Res. Int. 2013;2013:159786–159797. doi: 10.1155/2013/159786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batalha P.N., Souza M.C.B., Pena-Cabrera E., Cruz D.C., Boechat F.C.S. Quinolone in the search for new anti-cancer agents. Curr. Pharmaceut. Des. 2016;22:6009–6020. doi: 10.2174/1381612822666160715115025. [DOI] [PubMed] [Google Scholar]
- 8.Makhanya T.R., Gengan R.M., Pandian P., Chuturgoon A.A., Tiloke C., Atar A. Phosphotungstic acid catalyzed one pot synthesis of 4,8,8-trimethyl-5-phenyl-5,5a,8,9-tetrahydrobenzo[b][1,8]naphthyridin-6(7H)-one derivatives and their biological evaluation against A549 lung cancer cells. J. Heterocycl. Chem. 2018;55:1193–1204. doi: 10.1002/jhet.3153. [DOI] [Google Scholar]
- 9.Musiol R. An overview of quinoline as a privileged scaffold in cancer drug discovery. Expert Opin. Drug Discov. 2017;12:583–597. doi: 10.1080/17460441.2017.1319357. [DOI] [PubMed] [Google Scholar]
- 10.Afzal O., Afzal O., Kumar S., Haider R.M., Ali R.M., Kumar R., Jaggi M., Bawa S. A review on anti-cancer potential of bioactive heterocycle quinoline. Eur. J. Med. Chem. 2015;97:871–910. doi: 10.1016/j.ejmech.2014.07.044. [DOI] [PubMed] [Google Scholar]
- 11.Cherian M.A., Ma C.X. The role of neratinib in HER2-driven breast cancer. Future Oncol. 2017;13:1931–1943. doi: 10.2217/fon-2017-0186. [DOI] [PubMed] [Google Scholar]
- 12.Wissner A., Berger D., Boschelli D., Floyd M., Greenberger L., Gruber B., Johnson B., Mamuya N., Nilakantan R., Reich M., et al. 4-Anilino-6,7-dialkoxyquinoline-3-carbonitrile inhibitors of epidermal growth factor receptor kinase and their bioisosteric relationship to the 4-anilino-6,7-dialkoxy-quinazoline inhibitors. J. Med. Chem. 2000;43:3244–3256. doi: 10.1021/jm000206a. [DOI] [PubMed] [Google Scholar]
- 13.Pannala M., Kher S., Wilson N., Gaudette J., Sircar I., Zhang S., Bakhirev A., Yang G., Yuen P., Gorcsan F., et al. Synthesis and structure–activity relationship of 4-(2-aryl-cyclopropylamino)-quinoline-3-carbonitriles as EGFR tyrosine kinase inhibitors. Bioorg. Med. Chem. Lett. 2007;17:5978–5982. doi: 10.1016/j.bmcl.2007.07.071. [DOI] [PubMed] [Google Scholar]
- 14.Ibrahim D., El Ella D.A., El-Motwally A., Aly R. Molecular design and synthesis of certain new quinoline derivatives having potential anti-cancer activity. Eur. J. Med. Chem. 2015;102:115–131. doi: 10.1016/j.ejmech.2015.07.030. [DOI] [PubMed] [Google Scholar]
- 15.Aly R., Serya R., El-Motwally A., Al-Ansary G., El Ella D.A. Review: Quinoline-based small molecules as effective protein kinases inhibitors. J. Am. Sci. 2016;12:10–32. [Google Scholar]
- 16.Romagnoli R., Baraldi P.G., Prencipe F., Oliva P., Baraldi S., Salvador M.K., Lopez-Cara L.C., Brancale A., Ferla S., Hamel E., et al. Synthesis and Biological Evaluation of 2-Methyl-4,5- Disubstituted Oxazoles as a Novel Class of Highly Potent Antitubulin Agents. Sci. Rep. 2017;7:46356–46375. doi: 10.1038/srep46356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youssif B.G.M., Elshaier Y.A.M.M., Abdu-Allah H.H.M., Salim M.T.A., Inagak F., Mukai C. Synthesis of some benzimidazole derivatives endowed with 1,2,3-triazole as potential inhibitors of hepatitis C virus. Acta Pharm. 2016;66:219–231. doi: 10.1515/acph-2016-0014. [DOI] [PubMed] [Google Scholar]
- 18.Li Y.-S., Hu D.-K., Zhao D.-S., Liu X.-Y., Jin H.-W., Song G.-P., Cui Z.-N., Zhang L.-H. Design, synthesis and biological evaluation of 2,4-disubstituted oxazole derivatives as potential PDE4 inhibitors. Bioorg. Med. Chem. 2017;25:1852–1859. doi: 10.1016/j.bmc.2017.01.047. [DOI] [PubMed] [Google Scholar]
- 19.Ansari M.F., Siddiqui S.M., Agarwal S.M., Vikramdeo K.S., Mondal N., Azam A. Metronidazole hydrazone conjugates: Design, synthesis, antiamoebic and molecular docking studies. Bioorg. Med. Chem. Lett. 2015;25:3545–3549. doi: 10.1016/j.bmcl.2015.06.091. [DOI] [PubMed] [Google Scholar]
- 20.Dheer D., Singh V., Shankar R. Medicinal attributes of 1,2,3-triazoles: Current developments. Bioorg. Chem. 2017;71:30–54. doi: 10.1016/j.bioorg.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Meldal M., Tornøe C.W. Cu-catalyzed azide-alkyne cycloaddition. Chem. Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 22.Kolb H.C., Finn M.G., Sharpless K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Rostovtsev V.V., Green L.G., Fokin V.V., Sharpless K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Mohammed I., Kummetha I.R., Singh G., Sharova N., Lichinchi G., Dang J., Stevenson M., Rana T.M. 1,2,3-Triazoles as amide bioisosteres: Discovery of a new class of potent HIV-1 Vif antagonists. J. Med. Chem. 2016;16:7677–7682. doi: 10.1021/acs.jmedchem.6b00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonandi E., Christodoulou M.S., Fumagalli G., Perdicchia D., Rastelli G., Passarella D. The 1,2,3-triazole ring as a bioisostere in medicinal chemistry. Drug Discov. Today. 2017;22:1572–1581. doi: 10.1016/j.drudis.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Sun J., Yang Y.-S., Li W., Zhang Y.-B., Wang X.-L., Tang J.-F., Zhu H.-L. Synthesis, biological evaluation and molecular docking studies of 1,3,4-thiadiazole derivatives containing 1,4-benzodioxan as potential antitumor agents. Bioorg. Med. Chem. Lett. 2011;21:6116–6121. doi: 10.1016/j.bmcl.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 27.Khan M.F., Anwer T., Bakht A., Verma G., Akhtar W., Alam M.M., Rizvi M.A., Akhter M., Shaquiquzzaman M. Unveiling novel diphenyl-1H-pyrazole based acrylates tethered to 1,2,3-triazole as promising apoptosis inducing cytotoxic and anti-inflammatory agents. Bioorg. Chem. 2019;87:667–678. doi: 10.1016/j.bioorg.2019.03.071. [DOI] [PubMed] [Google Scholar]
- 28.Mahmoud M.A., Mohammed A.F., Salem O.I.A., Gomaa H.A.M., Youssif B.G.M. New 1,3,4-oxadiazoles linked 1,2,3-triazole moiety as antiproliferative agents targeting EGFR-TK. Arch. Der Pharm. 2022;355:2200009–2200023. doi: 10.1002/ardp.202200009. [DOI] [PubMed] [Google Scholar]
- 29.Xu J.H., Fan Y.L., Zhou J. Quinolone-triazole hybrids and their biological activities. J. Heterocycl. Chem. 2018;55:1854–1862. doi: 10.1002/jhet.3234. [DOI] [Google Scholar]
- 30.Ren Q.C., Gao C., Xu Z., Feng L.S., Liu M.L., Wu X., Zhao F. Bis-coumarin derivatives and their biological activities. Curr. Top. Med. Chem. 2018;18:101–113. doi: 10.2174/1568026618666180221114515. [DOI] [PubMed] [Google Scholar]
- 31.Hemamalini A., Das M.T. Design and synthesis of sugar-triazole low molecular weight gels as mercury ion sensor. New. J. Chem. 2013;37:2419–2425. doi: 10.1039/c3nj00072a. [DOI] [Google Scholar]
- 32.Steinschifter W., Fiala W., Stadlbauer W. Synthesis of oxazolo[4,5-c]quinolones by thermolytic degradation of 4-azido-2(1H)-quinolones. J. Heterocycl. Chem. 1994;31:1647–1652. doi: 10.1002/jhet.5570310662. [DOI] [Google Scholar]
- 33.Nayl A.A., Aly A.A., Arafa W.A.A., Ahmed I.M., Abd-Elhamid A.I., El-Fakharany E.M., Abdelgawad M.A., Tawfeek H.N., Bräse S. Azides in the Synthesis of Various Heterocycles. Molecules. 2022;27:3716. doi: 10.3390/molecules27123716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bräse S., Gil C., Knepper K., Zimmermann V. Organic Azides: An Exploding Diversity of a Unique Class of Compounds. Angew. Chem. 2005;117:5320–5374. doi: 10.1002/anie.200400657. [DOI] [PubMed] [Google Scholar]
- 35.Stadlbauer W., Laschober R., Kappe T. Potential non-steroidal estrogens and antiestrogens, IV Organicazides in heterocyclic synthesis, part 13: Synthesis of aza-and diazacoumestrols via azido derivatives. Monathefte Chem. 1991;122:853–861. doi: 10.1007/BF00815924. [DOI] [Google Scholar]
- 36.Aizikovich A., Kuznetsov V., Gorohovsky S., Levy A., Meir S., Byk G., Gellerman G. A new application of diphenylphosphorylazide (DPPA) reagent: Convenient transformations of quinolin-4-one, pyridin-4-one and quinazolin-4-one derivatives into the 4-azido and 4-amino counterparts. Tetrahedron Lett. 2004;45:4241–4243. doi: 10.1016/j.tetlet.2004.04.032. [DOI] [Google Scholar]
- 37.El-Sheref E.M., Aly A.A., Alshammari M.B., Brown A.B., Abdel-Hafez S.M.N., Abdelzaher W.Y., Bräse S., Abdelhafez E.M.N. Design, Synthesis, Molecular Docking, Anti-apoptotic and Caspase-3 Inhibition of New 1,2,3-Triazole/Bis-2(1H)-Quinolinone Hybrids. Molecules. 2020;25:5057. doi: 10.3390/molecules25215057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aly A.A., El-Sheref E.M., Mourad A.E., Bakheet M.E.M., Bräse S. 4-Hydroxy-2-quinolones: Syntheses, reactions and fused heterocycles. Mol. Divers. 2020;24:477–524. doi: 10.1007/s11030-019-09952-5. [DOI] [PubMed] [Google Scholar]
- 39.El-Sheref E.M., Elbastawesy M.A.I., Brown A.B., Shawky A.M., Gomaa H.A.M., Bräse S., Youssif B.G.M. Design and synthesis of (2-oxo-1,2-dihydroquinolin-4-yl)-1,2,3-triazole derivatives via click reaction: Potential apoptotic antiproliferative agents. Molecules. 2021;26:6798. doi: 10.3390/molecules26226798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Himo F., Lovell T., Hilgraf R., Rostovtsev V.V., Noodleman L., Sharpless K.B., Fokin V.V. Copper(I)-Catalyzed Synthesis of Azoles. DFT Study Predicts Unprecedented Reactivity and Intermediates. J. Am. Chem. Soc. 2005;127:210–216. doi: 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
- 41.Gomaa H.A.M., Shaker M.E., Alzarea S.I., Hendawy O.M., Mohamed F.A.M., Gouda A.M., Ali A.T., Morcoss M.M., Abdelrahman M.H., Trembleau L., et al. Optimization and SAR investigation of novel 2,3-dihydropyrazino[1,2-a]indole-1,4-dione derivatives as EGFR and BRAFV600E dual inhibitors with potent antiproliferative and antioxidant activities. Bioorg. Chem. 2022;120:105616–105629. doi: 10.1016/j.bioorg.2022.105616. [DOI] [PubMed] [Google Scholar]
- 42.Youssif B.G.M., Gouda A.M., Moustafa A.H., Abdelhamid A.A., Gomaa H.A.M., Kamal I., Marzouk A.A. Design and synthesis of new triarylimidazole derivatives as dual inhibitors of BRAFV600E/p38α with potential antiproliferative activity. J. Mol. Struct. 2022;1253:132218. doi: 10.1016/j.molstruc.2021.132218. [DOI] [Google Scholar]
- 43.Lamya H., Al-Wahaibi L.H., Gouda A.M., Abou-Ghadir O.F., Salem O.I.A., Ali A.T., Farghaly H.S., Abdelrahman M.H., Trembleau L., Abdu-Allah H.H.M., et al. Design and synthesis of novel 2,3-dihydropyrazino[1,2-a]indole-1,4-dione derivatives as antiproliferative EGFR and BRAFV600E dual inhibitors. Bioorg. Chem. 2020;104:104260–104276. doi: 10.1016/j.bioorg.2020.104260. [DOI] [PubMed] [Google Scholar]
- 44.Zha G.-F., Qin H.-L., Youssif B.G.M., Amjad M.W., Raja M.A., Abdelazeem A.H., Bukhari S.N.A. Discovery of potential anti-cancer multi-targeted ligustrazine based cyclohexanone and oxime analogs overcoming the cancer multidrug resistance. Eur. J. Med. Chem. 2017;135:34–48. doi: 10.1016/j.ejmech.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 45.Mohamed F.A.M., Gomaa H.A.M., OM H., Ali A.T., Farghaly H.S., Gouda A.M., Abdelazeem A.H., Abdelrahman M.H., Trembleau L., Youssif B.G.M. Design, synthesis, and biological evaluation of novel EGFR inhibitors containing 5-chloro-3-hydroxymethyl-indole-2-carboxamide scaffold with apoptotic antiproliferative activity. Bioorg. Chem. 2021;112:104960–104973. doi: 10.1016/j.bioorg.2021.104960. [DOI] [PubMed] [Google Scholar]
- 46.Hisham M., Hassan H.A., Gomaa H.A.M., Youssif B.G.M., Hayallah A.M., Abdel-Aziz M. Structure-based design, synthesis and antiproliferative action of new quinazoline-4-one/chalcone hybrids as EGFR inhibitors. J. Mol. Struct. 2022;1254:132422. doi: 10.1016/j.molstruc.2022.132422. [DOI] [Google Scholar]
- 47.Mohassab A.M., Hassan H.A., Abdelhamid D., Gouda A.M., Youssif B.G.M., Tateishi H., Fujita M., Otsuka M., Abdel-Aziz M. Design and Synthesis of Novel quinoline/chalcone/1,2,4-triazole hybrids as potent antiproliferative agent targeting EGFR and BRAFV600E kinases. Bioorg. Chem. 2021;106:104510–104521. doi: 10.1016/j.bioorg.2020.104510. [DOI] [PubMed] [Google Scholar]
- 48.AL-Mahmoudy A.M.M., Hassan A.N., Ibrahim T.S., Youssif B.G.M., Taher E.S., Tantawy M.A., Abdel-Aal E.H., Osman N.A. Novel benzyloxyphenyl pyrimidine-5-carbonitrile derivatives as potential apoptotic antiproliferative agents. Anti-Cancer Agent. Med. Chem. 2021;21:1–7. doi: 10.2174/1871520621666210612043812. [DOI] [PubMed] [Google Scholar]
- 49.Abdelbaset M.S., Abdel-Aziz M., Abuo-Rahma G.E.A., Abdelrahman M.H., Ramadan M., Youssif B.G.M. Novel quinoline derivatives carrying nitrones/oximes nitric oxide donors: Design, synthesis, antiproliferative and caspase-3 activation activities. Arch. Der Pharm. 2019;352:1800270–1800283. doi: 10.1002/ardp.201800270. [DOI] [PubMed] [Google Scholar]
- 50.Hisham M., Youssif B.G.M., Osman E.E.A., Hayallah A.M., Abdel-Aziz M. Synthesis and biological evaluation of novel xanthine derivatives as potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019;176:117–128. doi: 10.1016/j.ejmech.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 51.Shaikh M., Shinde Y., Pawara R., Noolvi M., Surana S., Ahmad I., Patel H. Emerging Approaches to Overcome Acquired Drug Resistance Obstacles to Osimertinib in Non-Small-Cell Lung Cancer. J. Med. Chem. 2022;65:1008–1046. doi: 10.1021/acs.jmedchem.1c00876. [DOI] [PubMed] [Google Scholar]
- 52.Abou-Zied H.A., Youssif B.G.M., Mohamed M.F.A., Hayallah A.M., Abdel-Aziz M. EGFR inhibitors and apoptotic inducers: Design, synthesis, anti-cancer activity and docking studies of novel xanthine derivatives carrying chalcone moiety as hybrid molecules. Bioorg. Chem. 2019;89:102997–103007. doi: 10.1016/j.bioorg.2019.102997. [DOI] [PubMed] [Google Scholar]
- 53.Al-Sanea M.M., Gotina L., Mohamed M.F.A., Parambi D.G.T., Gomaa H.A.M., Mathew B., Youssif B.G.M., Alharbi K.S., Elsayed Z.M., Abdelgawad M.A., et al. Design, Synthesis and Biological Evaluation of New HDAC1 and HDAC2 Inhibitors Endowed with Ligustrazine as a Novel Cap Moiety. Drug Des. Dev. Ther. 2020;14:497–508. doi: 10.2147/DDDT.S237957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elbastawesy M.A.I., Aly A.A., Ramadan M., Elshaier Y.A.M.M., Youssif B.G.M., Brown A.B., Abuo-Rahma G.E.A. Novel Pyrazoloquinolin-2-ones: Design, Synthesis, Docking Studies, and Biological Evaluation as Antiproliferative EGFR- TK Inhibitors. Bioorg. Chem. 2019;90:103045–103060. doi: 10.1016/j.bioorg.2019.103045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The study did not report any data.