Abstract
Diabetes is a leading non-communicable disease and a risk factor for relapsing infections. The current study was aimed at investigating the prevalence and antibiotic susceptibility of carbapenem-resistant (CR) uropathogens of the family Enterobacteriaceae in diabetic patients. The data of 910 bacterial isolates was collected from diagnostic laboratories during January 2018 to December 2018. The bacterial isolates were identified using traditional methods including colonial characteristics, biochemical tests, and API (20E). Antimicrobial susceptibility and phenotypic characterization of ESBL, MBLs, and KPC was determined by utilizing CLSI recommended methods. The phenotypically positive isolates were further analyzed for resistance-encoding genes by manual PCR and Check-MDR CT103XL microarray. Susceptibility to colistin and cefiderocol was tested in accordance with CLSI guidelines. The data revealed that most of the patients were suffering from type 2 diabetes for a duration of more than a year and with uncontrolled blood sugar levels. Escherichia coli and Klebsiella pneumoniae were the most frequently encountered pathogens, followed by Enterobacter cloacae and Proteus mirabilis. More than 50% of the isolates showed resistance to 22 antibiotics, with the highest resistance (>80%) against tetracycline, ampicillin, and cefazolin. The uropathogens showed less resistance to non-β-lactam antibiotics, including amikacin, fosfomycin, and nitrofurantoin. In the phenotypic assays, 495 (54.3%) isolates were found to be ESBL producers, while ESBL-TEM and -PER were the most prevalent ESBL types. The resistance to carbapenems was slightly less (250; 27.5%) than ESBL producers, yet more common amongst E. coli isolates. MBL production was a common feature in carbapenem-resistant isolates (71.2%); genotypic characterization also validated this trend. The isolates were found to be sensitive against the new drugs, cefiderocol and eravacycline. with 7–28% resistance, except for P. mirabilis which had 100% resistance against eravacycline. This study concludes that a few types of ESBL and carbapenemases are common in the uropathogens isolated from the diabetic patients, and antibiotic stewardship programs need to be revisited, particularly to cure UTIs in diabetic patients.
Keywords: antimicrobial resistance, β-lactamase, E. coli, K. pneumoniae, type 2 diabetes
1. Introduction
Diabetes mellitus is one of the leading non-communicable diseases (NCD), ranked at number 5 among NCDs at a global level [1,2]. For the past 3–4 decades, a surge in diabetic cases has been witnessed. Due to a less established health care sector, the situation is worse in developing countries, and hence, a large population remains at risk of complications caused by diabetes. An unregulated and elevated level of blood glucose incurs damages to multiple organs, including the blood vessels, retinas, and kidneys, leading to various complications [3,4]. Neuropathy-related incomplete emptying of the bladder, poor metabolic control, and a weak immune system are amongst the risk factors causing repeated UTIs in diabetic patients [5]. The burden of UTIs and other complications in diabetic patients keep health care facilities under pressure, even in developed countries [6,7,8]. It is indeed reported that UTIs in diabetic patients exhibit more severe symptoms such as emphysematous cystitis, pyelonephritis, renal abscesses, and papillary necrosis [5] and result in longer hospital stays. Deplorably, the management of UTIs in diabetic patients is mainly concomitant with poor outcomes as compared to non-diabetic patients [9].
Bacteria commonly encountered in UTIs [10] include Escherichia coli and Klebsiella pneumoniae along with other Enterobacteriaceae members [11,12,13]. Indeed, diabetic patients are mainly infected by highly resistant pathogens, including extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae and carbapenem-resistant Enterobacteriaceae [5,14]. β-lactam antibiotics are frequently prescribed drugs because of their broad spectrum and safety profile [10,15,16]. Different resistance mechanisms against β-lactams include alterations in the drug target site, decreased membrane permeability, and the activity of the drug efflux pump, but β-lactamases, especially extended spectrum β-lactamases (ESBLs) and carbapenemases [16], are the most common features. ESBL producers are the leading cause of higher morbidity and mortality in UTI patients [17]. This may be attributed to the overuse of antibiotics or to the ability of the microbes to grow as biofilms, or caused by genetic means, including mutations and the horizontal transfer of resistance genes [11].
Several studies describe specific genotypes in ESBL-producing pathogens causing resistance to some antibiotics [17,18]. In India, TEM (temoniera) has frequently been reported among uropathogens [17]. Whilst in Europe, United States, and Africa, CTX-M (cefotaximase) has remained the most detected marker [19]. Carbapenems are the drug of choice for the treatment of infections caused by ESBL producers, but an increasing trend of resistance against these drugs has been reported in many Asian countries, including Pakistan [20]. The problem of emerging resistance in CRE along with poor prognosis in developing countries has resulted in a higher mortality rate of up to 40% [21]. The production of carbapenemases such as KPC, VIM, IPM, and NDM is a common feature in CRE isolates. blaNDM is the most prevalent marker in Asian countries [20], followed by blaKPC, blaIMP, blaVIM, and blaOXA-48 [21,22].
Cefiderocol (CFDC) and eravacycline (ERV) have been introduced lately as options against carbapenem-resistant (CR) isolates with an efficacy of 70% to 95% against MBL producers [23]. However, these drugs are not available in many developing countries, and hence the data related to resistance against these drugs has not been widely reported. It is also worthy to note that the prevalence of genetic variants of ESBLs and carbapenemases in Pakistan has not been studied extensively, particularly in uropathogens isolated from diabetic patients. Keeping this lag in view, the current study was aimed at understanding the basis of antibiotic resistance among Enterobacteriaceae uropathogens; the data will also help to improve antibiotic surveillance and stewardship guidelines.
2. Materials and Methods
2.1. Retrieval of Data, Bacterial Isolates, and Identification
The data related to the samples was obtained from several laboratories across the province of Khyber Pakhtunkhwa, Pakistan during the study period from January 2018 to December 2018. The data related to the patients’ age and sex, signs and symptoms of UTI, recurrence of UTI, duration, and type of diabetes was collected. Comorbidity other than diabetes, taking antibiotics, and refusal to provide consent were taken as exclusion criteria. In the case that the same patient appeared twice but with a different pathogen species or resistance type, it was considered as a new sample. Isolates other than Enterobacteriaceae were not included in this study. All the patients of UTI with diabetes of both sexes and without discrimination of age were included in this study. The laboratory data of 910 uropathogens was assessed to select Enterobacteriaceae isolates which belonged to E. coli, K. pneumoniae, P. mirabilis, and E. cloacae. The sample processing and bacterial isolation were carried out at the collaborating laboratories. Briefly, urine was inoculated on a cystine–lactose–electrolyte-deficient (CLED) medium and incubated at 37 °C for 24–48 h. The initial identification of the isolates was carried out by traditional methods, including Gram-staining, cultural characteristics, and biochemical tests (such as citrate utilization, oxidase, urea and indole test, and TSI). The identification was confirmed by API 20E (Biomerieux, Lyon, France).
2.2. Antimicrobial Susceptibility Testing
To investigate antibiotic sensitivity, the isolates were grown on MacConkey agar plates for 24 h, and an inoculum standardized with 0.5 McFarland was transferred onto Mueller–Hinton agar plates to prepare a uniform bacterial lawn. Antibiotic discs were placed at appropriate distances. For antimicrobial sensitivity, the panel of antibiotics included amoxicillin/clavulanic acid; aztreonam; trimethoprim-sulphamethoxazole; cefepime; cefoxitin; ceftazidime; tetracycline: levofloxacin; gentamicin; nitrofurantoin; piperacillin/tazobactam; norfloxacin; ciprofloxacin; meropenem; imipenem; ampicillin; cefazolin; fosfomycin; amikacin; ceftriaxone; ertapenem; and doripenem. The isolates were categorized as sensitive, intermediate, or resistant according to the CLSI (2017) guidelines.
Susceptibility to colistin was performed by the colistin broth disk elution method following the protocol given in the CLSI (2021) guidelines. Briefly, Mueller–Hinton broth (MHB) with cation adjusted was arranged in four sets where each tube contained 10 mL MHB. The tubes were labeled as 0, 1, 2, and 4. One tube was kept as control (0), while in the remaining tubes, 1, 2, and 4 colistin discs (10 µg; OXOID, Basingstoke, UK) were added to obtain 0, 1, 2, and 4 µg mL−1 of the final concentration of colistin in the respective tubes. Colistin was eluted by vortexing and by keeping the tubes at room temperature for half an hour, following which the inoculum was added and the turbidity was adjusted to the 0.5 McFarland standard. The tubes were incubated at 37 °C for 24 h. The growth was inspected visually, and the minimum inhibitory concentration (MIC) was determined. The isolates were categorized as intermediate or resistant according to the CLSI guidelines.
In addition to routinely prescribed combinations, novel regimens of cefiderocol (FDC 30 µg; Liofilchem srl, Italy) and eravacycline (ERV 20 µg; Liofilchem, Italy) were also tested for antibacterial sensitivity by following the guidelines given by the Clinical and Laboratory Standards Institute and the Food and Drug Administration (FDA). The breakpoint criteria of ≤11 and ≤15 mm was used for FDC and ERV, respectively.
2.3. Phenotypic Characterization of ESBLs
The isolates having resistance to one or more cephalosporin or aztreonam were further tested for ESBL production through a double disc synergy test as described previously in the CLSI (2017) guidelines. The isolates were grown at 37 °C for 20 h on MHA plates containing discs of cephalosporin, aztreonam, and amoxicillin (with clavulanic acid). The zone of inhibition around cephalosporin or aztreonam was noted for any extension towards the other combination, and they were then categorized as ESBL producers.
2.4. Phenotypic Characterization of Carbapenemases
Initially, carbapenem resistance was assessed by the Kirby–Bauer disc diffusion method using MHA. The isolates were spread over the plates to make lawns, and four carbapenem discs (10 µg, each), including imipenem, meropenem, ertapenem, and doripenem were placed at appropriate distances. The plates were incubated at 37 °C for 20 h and the results were interpreted as per the CLSI guidelines.
The isolates exhibiting resistance to carbapenem were phenotypically assayed using EDTA and phenylboronic acid for MBL and KPC production. The cultures were inoculated on MHA by making lawns. Imipenem (IMP, 10 µg) and meropenem (MEM, 10 µg) discs were placed in three different sets; besides one set as control, the other two sets were either added with 0.5 mg of EDTA or 400 mg of phenylboronic acid. After cultivation at 37 °C for 20 h, an increase in the zone of inhibition (5–7 mm) around EDTA- or phenylboronic-acid-impregnated discs was compared with the control, and such isolates were marked as MBL/KPC producers.
2.5. Genotypic Characterization of ESBLs and Carbapenemases
Phenotypically screened ESBLs and carbapenemase producers were genotypically analyzed by two methods, including manual PCR and Check-MDR CT103XL microarray. The Check-MDR CT103XL microarray kit (15LTN0384) was used following the manufacturer’s instructions. DNA was extracted by the kit method (WizPrepTM gDNA Mini Kit, Seongnam-si, Korea). The relevant genes of the ESBLs and carbapenemases were also determined by manual PCR using the primers and conditions mentioned in the Table 1. The results of manual PCR were compared with microarray analysis.
Table 1.
Primers and details for the amplification of common ESBLs and carbapenemases.
| Primer | Sequence | Annealing Temperature °C | Product Size bp | Reference |
|---|---|---|---|---|
| TEM | F-ATGAGTATTCAACATTTCCG R-TTAATCAGTGAGGCACCTAT |
51 | 851 | Grimm et al., 2004 [24] |
| SHV | F-GCAAAACGCCGGGTTATTC R-GGTTAGCGTTGCCAGTGCT |
57.4 | 940 | Gröbner at el., 2009 [25] |
| PER | F-GCTCCGATAATGAAAGCGT R-TTCGGCTTGACTCGGCTGA |
53 | 520 | Uddin et al., 2022 [20] |
| GES | F-GTT AGA CGG GCG TAC AAA GAT AAT R-TGT CCG TGC TCA GGA TGA GT |
55 | 903 | Ryoo et al., 2005 [26] |
| CTX-M | F-CGCTTTGCGATGTGCAG R-ACCGCGATATCGTTGGT |
55.7 | 551 | Paterson et al., 2003 [27] |
| NDM | F-CACCTCATGTTTGAATTCGCC R-CTCTGT CACATCGAAATCGC |
60 | 984 bp | Kaase et al., 2011 [28] |
| KPC | F-CAGCTCATTCAAGGGCTTTC R-AGTCATTTGCCGTGCCATAC |
56 | 533 | Gröbner at el., 2009 [25] |
| VIM | F-GGTGTTTGGTCGCATATCGC CCATTCAGCCAGATCGGCATC |
65 | 503 bp | Wolter et al., 2009 [29] |
| IMP | F-GGAATAGAGTGGCTTAATTC R-CAACCAGTTTTG CCTTACC |
53 | 327 bp | Wolter et al., 2009 [29] |
2.6. Statistical Analysis
SPSS ver 26.0 was used to analyze the data and most of the values are given as frequencies and their percentages.
3. Results
3.1. Clinical and Other Variables
Considering the sharp increase in diabetes and related complications, the health sectors in developing countries are over-burdened. Repeated occurrence of UTIs caused by antibiotic-resistant bacteria is common in diabetic patients. This study was focused on the antibiotic-resistance characteristics of Enterobacteriaceae uropathogens isolated from diabetic patients. Table 2 indicates that most of the patients were suffering from type 2 diabetes with very poor control of the blood glucose level. Fasting blood sugar level, at least once during the active infection, exceeding 120 mg dL−1 was taken as a measure of control along with HbA1c > 6.5% [30]. Most of the patients were female, with a large proportion of them being middle aged (33.81%) to elderly women (50.64%), indicating the relationship between the reproductive stage of women with susceptibility to UTIs. A total of 910 isolates were collected from the 879 patients; 31 isolates obtained from previously included patients but were different either in their resistance profile or belonging to a different species. Such cases were considered as recurrent UTI. The samples showing the two or three pathogens including Gram-negative rods other than the required species and Gram-positive bacteria were not considered here. Only 209 isolates were from male patients. More than 90% of the patients had diabetes for a duration of more than a year and even for more than a decade.
Table 2.
Status of diabetes as informed by the patients (n = 910).
| Name of Variable | Occurrence |
|---|---|
| Type of DM | |
| Type 1 | 264 (29.02%) |
| Type 2 | 646 (70.98%) |
| Gender | |
| Male | 209 (22.96%) |
| Age (years) | |
| 1–20 | 1 (0.47%) |
| 20–40 | 32 (15.31%) |
| 40–60 | 107 (51.19%) |
| 60–80 | 61 (29.18%) |
| >80 | 8 (3.82%) |
| Female | 701 (77.03%) |
| Age (years) | |
| 1–20 | 3 (0.42%) |
| 20–40 | 49 (6.99%) |
| 40–60 | 237 (33.81%) |
| 60–80 | 355 (50.64%) |
| >80 | 57 (8.13%) |
| Blood glucose level | |
| Fasting Blood Sugar * | |
| ≥120 | 115 (12.63%) |
| <120 | 795 (87.36%) |
| HbA1c | |
| <6.5%) | 293 (32.2%) |
| >6.5% | 617 (67.8%) |
| Duration of DM | |
| <1 year | 72 (7.91%) |
| 1–10 years | 420 (46.15%) |
| >10 years | 418 (45.94%) |
* The data was obtained during the course of infection at least once per patient.
3.2. Prevalence of Major Uropathogens and Antimicrobial Susceptibility Pattern
Amongst Enterobacteriaceae, E. coli was the most frequently isolated pathogen (Table 3) followed by K. pneumoniae. While Enterobacter cloacae and P. mirabilis appeared less frequently.
Table 3.
Prevalence of enteric uropathogens from diabetic patients (n = 910).
| Pathogen | Prevalence |
|---|---|
| Escherichia coli | 536 (58.90%) |
| Klebsiella pneumoniae | 337 (37.03%) |
| Enterobacter cloacae | 19 (2.08%) |
| Proteus mirabilis | 18 (1.97%) |
The antimicrobial susceptibility testing presented a high level of resistance amongst the isolates, which can be a reason for the repeated episodes of UTI in diabetic patients. Piperacillin-tazobactam and tetracycline were found to be the most ineffective drugs with a resistance level of >80% (Table 4). The resistance to cephalosporins in E. coli was also alarmingly high (56–75%). More than two-thirds of the isolates showed resistance to amoxicillin-clavulanic acid, aztreonam, levofloxacin, and norfloxacin. The resistance to carbapenems was, however, lower (247; 27.14%) than the other antibiotics.
Table 4.
Antimicrobial resistance patterns of Enterobacteriaceae uropathogens.
| Antibiotics |
E. coli (n = 536) |
K. pneumoniae (n = 337) |
Enterobacter cloacae (n = 19) |
P. mirabilis (n = 18) |
Total (n = 910) |
|---|---|---|---|---|---|
| AMC | 458 (85.44%) | 208 (61.72%) | 18 (94.73%) | 16 (88.89%) | 700 (76.91%) |
| ATM | 382 (71.26%) | 203 (60.23%) | 16 (84.21%) | 15 (83.33%) | 616 (67.69%) |
| SXT | 324 (60.44%) | 241 (71.51%) | 11 (57.89%) | 12 (66.67%) | 588 (64.61%) |
| FEP | 402 (75.00%) | 210 (62.31%) | 13 (68.82%) | 11 (61.11%) | 636 (69.89%) |
| FOX | 301 (56.16%) | 177 (52.52%) | 19 (100%) | 10 (55.55%) | 507 (55.71%) |
| CAZ | 404 (75.37%) | 209 (62.01%) | 13 (68.82%) | 12 (66.67%) | 638 (70.10%) |
| TE | 415 (77.42%) | 283 (83.97%) | 16 (84.21%) | 18 (100%) | 732 (80.43%) |
| LEV | 406 (75.74%) | 239 (70.91%) | 16 (84.21%) | 14 (77.78%) | 675 (74.17%) |
| GN | 301 (56.16%) | 216 (64.09%) | 17 (89.47%) | 14 (77.78%) | 548 (60.21%) |
| F | 321 (59.88%) | 183 (54.30%) | 15 (78.94%) | 18 (100%) | 537 (59.01%) |
| TZP | 441 (82.27%) | 204 (60.53%) | 15 (78.94%) | 14 (77.78%) | 674 (74,06%) |
| NOR | 410 (76.49%) | 255 (75.66%) | 14 (73.68%) | 11 (61.11%) | 690 (75.82%) |
| CIP | 410 (76.49%) | 255 (75.66%) | 11 (57.89%) | 12 (66.67%) | 688 (75.60%) |
| MER | 138 (25.74%) | 91 (27.01%) | 04 (21.05%) | 05 (27.78%) | 238 (26.15%) |
| IMP | 142 (26.49%) | 88 (26.11%) | 06 (31.57%) | 05 (27.78%) | 241 (26.45%) |
| AMP | 465 (86.75%) | 263 (78.04%) | 19 (100%) | 16 (88.89%) | 763 (83.84%) |
| CFZ | 462 (86.19%) | 260 (77.15%) | 19 (100%) | 15 (83.33%) | 756 (83.07%) |
| FOS | 181 (33.76%) | 111 (32.93%) | 05 (26.31%) | 06 (33.33%) | 303 (33.29%) |
| AK | 176 (32.83%) | 177 (52.52%) | 13 (68.82%) | 10 (55.55%) | 376 (41.31) |
| CRO | 406 (75.74%) | 230 (68.24%) | 19 (100%) | 15 (83.33%) | 670 (73.62%) |
| ERT | 142 (26.49%) | 88 (26.11%) | 06 (31.57%) | 08 (44.44%) | 244 (26.81%) |
| DOR | 142 (26.49%) | 88 (26.11%) | 07 (36.84%) | 10 (55.55%) | 247 (27.14%) |
Abbreviations: AMC, amoxicillin/clavulanic acid; ATM, aztreonam; SXT, trimethoprim-sulphamethoxazole; FEP, cefepime; FOX, cefoxitin; CAZ, ceftazidime; TE, tetracycline: LEV, levofloxacin; GN, gentamicin; F, nitrofurantoin; TZP, piperacillin/tazobactam; NOR, norfloxacin; CIP, ciprofloxacin; MER, meropenem; IMP, imipenem; AMP, ampicillin; CFZ, cefazolin; FOS, fosfomycin; AK, amikacin; CRO, ceftriaxone; ERT, ertapenem; DOR, doripenem.
3.3. Resistance to Colistin
Although the isolates did not exhibit a higher rate of resistance towards colistin compared to the other tested antibiotics (Table 5), the rate was more than reported earlier for E. coli and K. pneumoniae. It is also important to note that colistin is still not a routinely prescribed medicine to patients in this part of the world. However, its use in the veterinary sector has been stated by the concerned persons. Nonetheless, the emergence of multi-drug resistant bacteria necessitates that caution be taken in prescribing this antibiotic.
Table 5.
Resistance of the Enterobacteriaceae uropathogen against colistin. The isolates with MIC ≤ 2μg/ mL were considered as intermediate, and isolates with ≥4 µg/mL were taken as resistant. The criteria for sensitivity is not mentioned in CLSI (2021).
| Bacterial Isolate | Intermediate (≤2 µg/ mL) | Resistant (≥4 µg/mL) | Total n (%) |
|---|---|---|---|
| E. coli (n = 536) | 489 (91.24%) | 47 (8.76%) | 536(100) |
| K. pneumoniae (n = 337) | 302 (89.62%) | 35 (10.38%) | 337(100) |
| Enterobacter cloacae (n = 19) | 18 (94.74%) | 1 (5.26%) | 19(100) |
| P. mirabilis (n = 18) | 00 | 18 (100%) | 18(100) |
| Total (n = 910) | 809 (88.90%) | 101 (11.10%) | 910(100) |
3.4. Phenotypic Resistance Categories
The isolates were categorized as multidrug-resistant (MDR), extensively drug-resistant (XDR), and pan-drug resistant (PDR) by following guidelines given in Magiorakos et al. (2012) [31]. Out of 910 isolates, 421 (46.26%), 239 (26.26%), and 101 (11.09%) were found to be MDR, XDR, and PDR, respectively.
3.5. Phenotypic and Genotypic Characterization of Resistance Markers
In an attempt to characterize ESBL production by these uropathogens, phenotypic and genotypic assays were used. The isolates showing resistance to 3rd and 4th generation cephalosporin and/or aztreonam were considered as ESBL producers and were selected for phenotypic and genotypic analysis. Out of 910 isolates, 670 (73.62%) were resistant to 3rd and 4th generation cephalosporin. The double disc diffusion method presented 495 (54.39%) isolates as ESBL-positive. These phenotypically positive isolates were confirmed for the ESBL type by the Microarray technique. ESBL-PER was the most common in these isolates, except for P. mirabilis isolates which showed a lower rate (8.33%) for this marker. The data from the Microarray was affirmed by the manual PCR for ESBL types; however, the sensitivity of the PCR was lower than the Microarray (Table 6). Nonetheless, the pattern of ESBL types did not vary to a greater extent from the data that has been stated earlier for this geographic region, emphasizing the widespread transfer of the resistance genes among pathogens.
Table 6.
Detection of ESBL production using phenotypic method, manual PCR, and microarray assay.
| Isolates | Phenotypic Detection | Manual PCR (n = 495) (%) | Microarray Assay (n = 495) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TEM | PER | GES | CTX-M | Unrecognized | TEM | PER | GES | CTX-M | Unrecognized | ||
| E. coli | 312 (58.2) |
83 (26.6) |
134 (42.9) |
34 (10.8) |
39 (12.5) |
22(7.0) | 85 (27.2) |
139 (44.5) | 36 (11.5) |
41 (13.14) |
11 (3.52) |
| K. pneumoniae | 165 (48.9) |
55 (33.3) |
79 (47.8) |
5 (3.0) |
13 (7.8) |
13(7.8) | 59 (35.7) |
81 (49) | 6 (3.6) |
14 (8.48) |
05 (3.0) |
| Enterobacter cloacae | 08 (42.1) |
02 (25) |
02 (25) | 00 | 3 (37.5) |
1 (12.5) |
02 (25) |
02 (25) | 0 | 04 (50) |
Nil |
| P. mirabilis | 10 (55.5) |
02 (20) |
02 (20) | 02 (20) |
02 (20) |
Nil | 04 (40) |
02 (20) | 02 (20) |
02 (20) |
Nil |
| Total n (%) | 495 (54.3) |
134 (27) |
207 (41.8) |
36 (7.2) |
53 (10.7) |
36(7.2) | 150 (30.3) |
224 (45.2) | 44 (8.8) |
61 (12.32) |
16 (3.23) |
MBL production appeared as the most widely distributed feature in CR isolates (Table 7). Arguably, the level of MBL (65.71%) was even higher in K. pneumoniae isolates than the level of KPC (30.95%). The study indicated the endemic nature of MBL, particularly among CRE uropathogens. This finding can definitely aid physicians in prescribing antibiotics. The data from genotypic characterization revealed blaNDM as the predominant marker followed by the blaKPC encoding gene. blaIPM and blaVIM could not be detected in any CR isolate. The mechanism of resistance in a few isolates could not be recognized here; however, it can be attributed to the presence of chromosomal AmpCs, efflux pump, or change in permeability or target site. The results of microarray and manual PCR were almost similar except for the NDM in E. coli. Repeated investigations using manual PCR affirmed this finding.
Table 7.
Characterization of carbapenemases into MBL and KPC on phenotypic basis and detection of carbapenemases encoding genes (n = 250).
| Isolates | Phenotypic Methods | Total (%) | Manual PCR | Microarray Assay | ||||
|---|---|---|---|---|---|---|---|---|
| bla NDM | bla KPC | Unrecognized | bla NDM | bla KPC | Unrecognized | |||
|
E. coli (n = 142) |
MBL | 107 (75.35%) | 109 (76.7%) |
24 (16.90%) |
9 (6.33%) |
111 (78.16%) | 24 (16.90%) |
7 (4.92%) |
| KPC | 21 (14.78%) |
|||||||
| Unrecognized | 14 (8.85%) | |||||||
|
K. pneumoniae (n = 91) |
MBL | 61 (67.03%) | 64 (70.3%) |
27 (29.67%) |
00 | 64 (70.3%) |
27 (29.67%) |
0 |
| KPC | 27 (29.67%) | |||||||
| Unrecognized | 3 (3.29%) |
|||||||
|
E. cloacae (n = 7) |
MBL | 5 (71.42%) | 5 (71.42%) | 2 (28.58%) | 00 | 5 (71.42%) | 2 (28.58%) | 0 |
| KPC | 2 (28.58%) | |||||||
| Unrecognized | - | |||||||
|
P. mirabilis (n = 10) |
MBL | 5 (50.0%) | 5 (50.0%) | 5 (50%) | 2 (20%) | 5 (50%) | 5 (50%) | 0 |
| KPC | 3 (30.0%) | |||||||
| Unrecognized | 2 (20%) | |||||||
| Total (n = 250) | MBL | 178 (71.2%) | 183 (73.2%) | 55 (22%) | 11 (4.4%) | 188 (75.2%) | 55 (22%) | 7 (2.8%) |
| KPC | 53 (21.2%) | |||||||
| Unrecognized | 19 (7.6%) | |||||||
3.6. Co-Existence of Resistance-Encoding Genes
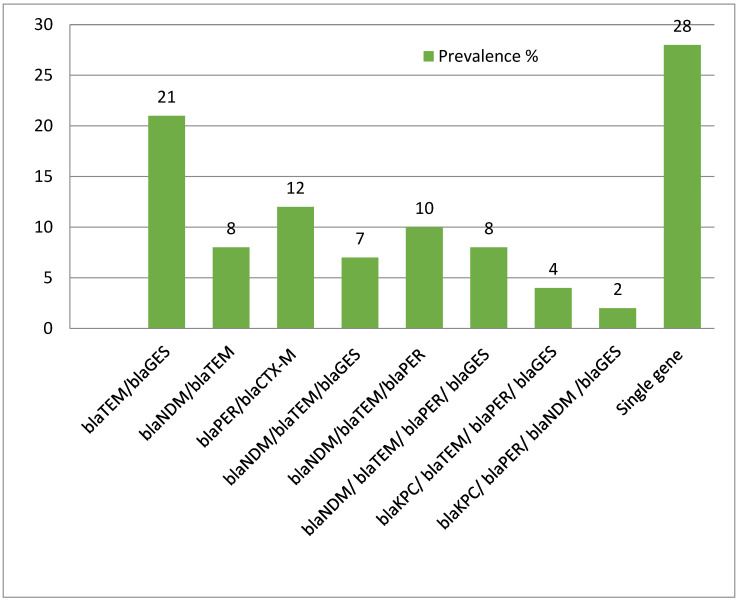
Out of 495, most of the isolates exhibited a co-existence of resistance genes (Figure 1). Indeed, the antimicrobial resistance spectrum was significantly increased with the co-existing genes (p < 0.01). blaTEM and blaGES were the most frequent pair of resistance markers in these isolates.
Figure 1.
Prevalence of co-existing resistance genes in isolates. (y-axis represents % of the isolates).
3.7. Resistance to Cefiderocol and Eravacycline
The resistance to new regimens, cefiderocol and eravacycline, was conceivably lower except for P. mirabilis which was completely resistant to eravacycline (Table 8). These regimens are not available in developing countries and hence it was presumed that these drugs will exhibit good sensitivity. However, the presence of complete resistance in P. mirabilis necessitates the assessment of the effectiveness of the new regimen in the near future, particularly with larger datasets.
Table 8.
Resistance to a new regimen, cefiderocol (FDC) and eravacycline (ERV), in CRE isolates (n = 250).
| Isolate | Resistant to | |
|---|---|---|
| FDC | ERV | |
| E. coli (n = 142) | 10 (7.04) | 19 (13.38) |
| K. pneumoniae (n = 91) | 7 (7.69) | 9 (9.89) |
| Enterobacter cloacae (n = 7) | 2 (28.57) | 1 (14.28) |
| P. mirabilis (n = 10) | 1 (10.0) | 10 (100) |
| Total (n = 250) | 20 (8.0) | 39 (15.6) |
4. Discussion
Diabetes mellitus has been recognized as a predisposing factor for UTIs [32,33]. The incidence of UTIs in women is higher and can cause more complications if the infection reaches the kidney [8]. The findings in the current study also corroborate that women, particularly those suffering from type 2 diabetes, are more susceptible to UTIs. It is generally believed that sexually active women are less susceptible to UTIs [34], which was also supported by the current findings as a large number of patients were older women.
E. coli has been described as a leading cause of UTI among all the bacteria [35] and the most common uropathogen in diabetic patients [32,36]. The findings of this study, despite being skewed towards Enterobacteriaceae, also corroborated it. The involvement of Enterobacteriaceae isolates in UTIs has widely been reported [37]; however, the data specifically obtained from diabetic patients is scarcely available. Antimicrobial resistance is a global issue and a challenge for clinicians because of the rapid increase in resistance. In the present study, the majority of the isolates were found to be resistant to commonly used antibiotics, including ampicillin, cefazolin, ceftriaxone, and amoxicillin-clavulanic acid. A considerable proportion was found to be MDR, while XDR and PDR were less common. The prevalence of these drug-resistant categories reportedly varies in different parts of the world. Pakistan, being a South Asian developing country, contributes a significant burden in AMR [38], with a high level (63–100%) of MDR and XDR bacteria [38]. Abbas et al. (2020) [39] reported a higher level of XDR (56%) strains of E. coli and Klebsiella species than the current findings. The resistance to colistin in Enterobacteriaceae is previously not reported from Peshawar, Pakistan [40]. However, in the present study, high values of MICs were obtained for the isolates from this region
In Pakistan, ESBLs have widely been reported in MDR bacterial strains with an overall frequency of 40% [41]. Here, it appeared higher (54.3%) in the isolates from diabetic patients. It can be linked with the presence of resistant bacteria in such patients, particularly isolates from the Enterobacteriaceae family. Abrar et al. (2018) [41] performed a meta-analysis where CTX-M appeared as a common ESBL variant in this region. The other variants reported from Pakistan included SHV and TEM. Here, PER appeared as a dominant marker. Previously, PER-1-type ESBLs have been reported in the P. aeruginosa and Acinetobacter species from Pakistan [42]. The current finding needs more investigation to ascertain this change in genotypic marker. Nonetheless, the commonly found ESBL genes, including CTX-M, GES, and TEM [7,43], also appeared in this study. The current findings regarding the absence of SHV confirmed a previous report from this region [44].
Antibiotic resistance is a global threat especially in immunocompromised or susceptible individuals such as chronic diabetic patients. The data showed that the resistance to reserve or less-prescribed drugs, including carbapenems and colistin, is higher [20]. Indeed, the resistance to carbapenems was higher as compared to a previous study from Karachi, Pakistan [45]. CREs have also been linked with high mortality and morbidity. Likewise, the resistance to colistin has also been increased compared to an earlier finding [20]. Colistin is considered as a last resort for the treatment of CR bacteria. Various factors are associated with the increased resistance to antibiotics, including over-the-counter sale, the irrational use of antibiotics, and lack of antibiotic stewardship programs. Enterobacteriaceae isolates, notably E. coli, K. pneumoniae and E. cloacae, harbor resistance to a variety of antibiotics, particularly by elaborating β-lactamases [20]. The incidence of as high as 32% carbapenem resistance has also been reported in these isolates, leading to an expansion of the list of PDR bacteria [46].
CRE commonly and frequently produce KPC and NDM as modes of action against antibiotics [20]. In this study, the resistance to β-lactam antibiotics was found higher, which could be attributed to the production of ESBLs and modification in surface receptors [47]. Among carbapenemase producers, NDM was the prevalent marker that confirmed an earlier finding from the same geographic region [20]. Habib et al. (2022) [48] also reported the widespread nature of NDM in CRE isolates from this region. Here, the majority of cases of CRE were of carbapenemase-producing Enterobacteriaceae (CPE), as reported in a recent report [48].
Lately, the survey of data available regarding eravacycline showed a cumulative (%) degree of MIC for non-MDR Enterobacteriaceae in the range of 0.5–1 µg mL−1, while for MDR isolates, it was 2–8 µg mL−1 [49]. The data was particularly encouraging as this fluorocycline drug was developed to treat infections caused by MDR bugs [50]. The drug is not available in this part of the world; however, the resistance pattern indicated the extent of the global antimicrobial resistance pattern, and therefore, the introduction of new drugs in any region should be based on a larger dataset (that should also be reviewed periodically). Considering this lag in view, a study published from China in 2022 investigated the efficacy of cefiderocol against CRE isolates [51], and deciphered the role of cirA, pbp3, and blaNDM-5 in the resistance to this drug. The siderophore-related gene cirA was of particular interest as cefiderocol is a catecholamine-siderophore-derived cephalosporin [52]. This study affirmed the hypothesis that even before the introduction of a drug, resistance can be found in the local isolates as resistance against cefiderocol and eravacycline was found in ~8 and 15% of the isolates, respectively. The isolates with the NDM and PER combination exhibited resistance to cefiderocol as previously reported [53].
5. Conclusions
Urinary tract infections are common amongst patients of type 2 diabetes. The uropathogens, particularly the members of Enterobacteriaceae, exhibit a high level of drug resistance. Antibiotic resistance relies on various mechanisms, including the production of extended spectrum β-lactamases (ESBLs) and carbapenemases. The isolates investigated in this study exhibited PER and TEM variants as the prevalent ESBLs. The presence of high levels of PER needs further investigation as it has not been reported earlier from this geographic region. Among carbapenem-resistant isolates, blaNDM was the common mode of resistance; a large proportion of such isolates showed the co-existence of β-lactamases with carbapenemases. The mechanisms of resistance other than ESBLs and carbapenemases need to be explored in future studies. Colistin along with relatively new drugs, cefiderocol and eravacycline, appeared effective, and hence can be regarded as the drugs of choice for the treatment of UTIs in diabetic patients. This study provides a basis for improving antibiotic surveillance and stewardship programs.
Acknowledgments
The author acknowledge the financial support from Taif University Researchers Supporting Project number (TURSP-2020/262), Taif University, Taif, Saudi Arabia. The assistance of Bakht Jahan is acknowledged in acquiring relevant information.
Author Contributions
O.M.A., data analysis, funding; F.U., methodology, writing initial draft; S.F.M., data curation, analysis; A.S.A., investigation, data curation; M.S., editing final draft, data curation; M.Y., data analysis, investigation. All authors approved the final draft. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical approval was taken from Kuwait Teaching Hospital, Khyber Pakhtunkhwa through IRB/KTH/135/2011718.
Informed Consent Statement
The data from the diagnostic labs was obtained with oral and written informed consent from the relevant patients.
Data Availability Statement
Data associated with this manuscript can be obtained from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Funding was obtained from Taif University, Saudia Arabia, through TURSP-2020/262.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mendenhall E., Kohrt B.A., Norris S.A., Ndetei D., Prabhakaran D. Non-communicable disease syndemics: Poverty, depression, and diabetes among low-income populations. Lancet. 2017;389:951–963. doi: 10.1016/S0140-6736(17)30402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng Y., Ley S.H., Hu F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018;14:88–98. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 3.Hicks C.W., Canner J.K., Mathioudakis N., Lippincott C., Sherman R.L., Abularrage C.J. Incidence and Risk Factors Associated With Ulcer Recurrence Among Patients With Diabetic Foot Ulcers Treated in a Multidisciplinary Setting. J. Surg. Res. 2020;246:243–250. doi: 10.1016/j.jss.2019.09.025. [DOI] [PubMed] [Google Scholar]
- 4.Sherwani S.I., Khan H.A., Ekhzaimy A., Masood A., Sakharkar M.K. Significance of HbA1c test in diagnosis and prognosis of diabetic patients. Biomark. Insights. 2016;11:95–104. doi: 10.4137/BMI.S38440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nitzan O., Elias M., Chazan B., Saliba W. Urinary tract infections in patients with type 2 diabetes mellitus: Review of prevalence, diagnosis, and management. Diabetes. Metab. Syndr. Obes. Targets Ther. 2015;8:129–136. doi: 10.2147/DMSO.S51792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Percival S.L., Suleman L., Vuotto C., Donelli G. Healthcare-Associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015;64:323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- 7.Wyres K.L., Hawkey J., Hetland M.A.K., Fostervold A., Wick R.R., Judd L.M., Hamidian M., Howden B.P., Löhr I.H., Holt K.E. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J. Antimicrob. Chemother. 2019;74:577–581. doi: 10.1093/jac/dky492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu S., Fu A.Z., Qiu Y., Engel S.S., Shankar R., Brodovicz K.G., Rajpathak S., Radican L. Disease burden of urinary tract infections among type 2 diabetes mellitus patients in the U.S. J. Diabetes Complicat. 2014;28:621–626. doi: 10.1016/j.jdiacomp.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Okwume C.C., Onyemelukwe N.F., Abdullahi I.N., Okoyeocha O.E., Asamota S.D. Prevalence of symptomatic urinary tract infection and bacterial spectrum of diabetic and non-diabetic patients at the two teaching hospitals in Enugu, Nigeria. Afr. J. Clin. Exp. Microbiol. 2021;22:480–488. doi: 10.4314/ajcem.v22i4.8. [DOI] [Google Scholar]
- 10.Seifu W.D., Gebissa A.D. Prevalence and antibiotic susceptibility of Uropathogens from cases of urinary tract infections (UTI) in Shashemene referral hospital, Ethiopia. BMC Infect. Dis. 2018;18:1–9. doi: 10.1186/s12879-017-2911-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albu S., Voidazan S., Bilca D., Badiu M., Truta A., Ciorea M., Ichim A., Luca D., Moldovan G. Bacteriuria and asymptomatic infection in chronic patients with indwelling urinary catheter the incidence of ESBL bacteria. Medicine. 2018;97:e11796. doi: 10.1097/MD.0000000000011796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Oliveira D.M.P., Forde B.M., Kidd T.J., Harris P.N.A., Schembri M.A., Beatson S.A., Paeterson D.L., Walker M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020;33:e00181-19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Díaz Álvarez M., Acosta Batista B., Pérez Córdova R., Hernández Robledo E. Urinary tract infection caused by Enterobacteriaceae and its relationship with vesicoureteral reflux. Bol. Méd. Hosp. Infant. México Engl. Ed. 2017;74:34–40. doi: 10.1016/j.bmhimx.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Vatan A., Saltoglu N., Yemisen M., Balkan I.I., Surme S., Demiray T., Mete B., Tabak F. Association between biofilm and multi/extensive drug resistance in diabetic foot infection. Int. J. Clin. Pract. 2018;72:e13060. doi: 10.1111/ijcp.13060. [DOI] [PubMed] [Google Scholar]
- 15.De Lastours V., Foxman B. Urinary tract infection in diabetes: Epidemiologic considerations topical collection on genitourinary infections. Curr. Infect. Dis. Rep. 2014;16:389. doi: 10.1007/s11908-013-0389-2. [DOI] [PubMed] [Google Scholar]
- 16.Khalifa S.M., Abd El-Aziz A.M., Hassan R., Abdelmegeed E.S. β-lactam resistance associated with β-lactamase production and porin alteration in clinical isolates of E. coli and K. pneumoniae. PLoS ONE. 2021;16:e0251594. doi: 10.1371/journal.pone.0251594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajpai T., Pandey M., Varma M., Bhatambare G.S. Prevalence of TEM, SHV, and CTX-M Beta-Lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna J. Med. 2017;7:12–16. doi: 10.4103/2231-0770.197508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reid R., Al-bayati M., Samarasinghe S. Genotypic Identification of Extended-Spectrum β-Lactamase (ESBL)- Producing Enterobacteriaceae from Urinary Tract Infections in the Leicestershire Area, United Kingdom: A One Health Prospective. J. Infect. Dis. Diagnosis. 2018;3:1000122. doi: 10.4172/2576-389X.1000122. [DOI] [Google Scholar]
- 19.Zeynudin A., Pritsch M., Schubert S., Messerer M., Liegl G., Hoelscher M., Belachew T., Wieser A. Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum β-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infect. Dis. 2018;18:1–10. doi: 10.1186/s12879-018-3436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uddin F., Imam S.H., Khan S., Khan T.A., Ahmed Z., Sohail M., Elnaggar A.Y., Fallatah A.M., El-Bahy Z.M. NDM Production as a Dominant Feature in Carbapenem-Resistant Enterobacteriaceae Isolates from a Tertiary Care Hospital. Antibiotics. 2022;11:48. doi: 10.3390/antibiotics11010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Q., Wang Y., Yu J., Li S., Zhang Y., Wang H., Lai X., Liu D., Mao L., Luo Y., et al. Bacterial characteristics of carbapenem-resistant Enterobacteriaceae (CRE) colonized strains and their correlation with subsequent infection. BMC Infect. Dis. 2021;21:638. doi: 10.1186/s12879-021-06315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe M., Kock M.M., Coetzee J., Hoosien E., Peirano G., Strydom K., Ehlers M.M., Mbelle N.M., Shashkina E., Haslam D.B., et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014–2016. Emerg. Infect. Dis. 2019;25:739–747. doi: 10.3201/eid2504.181482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston B.D., Thuras P., Porter S.B., Anacker M., VonBank B., Snippes V.P., Witwer M., Castanheira M., Johnson J.R. Activity of Cefiderocol, Ceftazidime-Avibactam, and Eravacycline against Carbapenem-Resistant Escherichia coli Isolates from the United States and International Sites in Relation to Clonal Background, Resistance Genes, Coresistance, and Region. Antimicrob. Agents Chemother. 2020;64:e00797-20. doi: 10.1128/AAC.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm V., Ezaki S., Susa M., Knabbe C., Schmid R.D., Bachmann T.T. Use of DNA microarrays for rapid genotyping of TEM beta-lactamases that confer resistance. J. Clin. Microbiol. 2004;42:3766–3774. doi: 10.1128/JCM.42.8.3766-3774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gröbner S., Linke D., Schütz W., Fladerer C., Madlung J., Autenrieth I.B., Witte W., Pfeifer Y. Emergence of carbapenem-non-susceptible extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tübingen, Germany. J. Med. Microbiol. 2009;58:912–922. doi: 10.1099/jmm.0.005850-0. [DOI] [PubMed] [Google Scholar]
- 26.Ryoo N.H., Kim E.C., Hong S.G., Park Y.J., Lee K., Bae I.K., Song E.H., Jeong S.H. Dissemination of SHV-12 and CTX-M-type extended-spectrum β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J. Antimicrob. Chemother. 2005;56:698–702. doi: 10.1093/jac/dki324. [DOI] [PubMed] [Google Scholar]
- 27.Paterson D.L., Hujer K.M., Hujer A.M., Yeiser B., Bonomo M.D., Rice L.B., Bonomo R.A., International Klebsiella Study Group Extended-spectrum b-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: Dominance and widespread prevalence of SHV- and CTX-M-type b-lactamases. Antimicrob. Agents Chemother. 2003;47:3554–3560. doi: 10.1128/AAC.47.11.3554-3560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaase M., Nordmann P., Wichelhaus T.A., Gatermann S.G., Bonnin R.A., Poirel L. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 2011;66:1260–1262. doi: 10.1093/jac/dkr135. [DOI] [PubMed] [Google Scholar]
- 29.Wolter D.J., Khalaf N., Robledo I.E., Vázquez G.J., Santé M.I., Aquino E.E., Goering R.V., Hanson N.D. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rican Medical Center Hospitals: Dissemination of KPC and IMP-18 β-lactamases. Antimicrob. Agents Chemother. 2009;53:1660–1664. doi: 10.1128/AAC.01172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaiafa G., Veneti S., Polychronopoulos G., Pilalas D., Daios S., Kanellos I., Didangelos T., Pagoni S., Savopoulos C. Is HbA1c an ideal biomarker of well-controlled diabetes? Postgrad. Med. J. 2021;97:380–383. doi: 10.1136/postgradmedj-2020-138756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbath S., Hindler J.F., Kahlmeter G., Olsson-Lijequist B., et al. Monnet, Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 32.Ozawa K., Takai M., Taniguchi T., Kawase M., Takeuchi S., Kawase K. Diabetes Mellitus as a Predictive Factor for Urinary Tract Infection for Patients Treated with Kidney Transplantation. Medicina. 2022;58:1488. doi: 10.3390/medicina58101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stapleton A. Urinary tract infections in patients with diabetes. Am. J. Med. 2002;113:80–84. doi: 10.1016/S0002-9343(02)01062-8. [DOI] [PubMed] [Google Scholar]
- 34.Giesen L.G., Cousins G., Dimitrov B.D., Van De Laar F.A., Fahey T. Predicting acute uncomplicated urinary tract infection in women: A systematic review of the diagnostic accuracy of symptoms and signs. BMC Fam. Pract. 2010;11:78. doi: 10.1186/1471-2296-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magill S.S., O’Leary E., Janelle S.J., Thompson D.L., Dumyati G., Nadle J., Wilson L.E., Kainer M.A., Lynfield R., Greissman S., et al. Changes in Prevalence of Health Care–Associated Infections in U.S. Hospitals. N. Engl. J. Med. 2018;379:1732–1744. doi: 10.1056/NEJMoa1801550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salari N., Karami M.M., Bokaee S., Chaleshgar M., Shohaimi S., Akbari H., Mohammadi M. The prevalence of urinary tract infections in type 2 diabetic patients: A systematic review and meta-analysis. Eur. J. Med. Res. 2022;27:1–13. doi: 10.1186/s40001-022-00644-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayakaran J., Soundararajan N., Shanmugam P. Phenotypic and genotypic characterization of multidrug-resistant isolates from patients with catheter-associated urinary tract infection in a tertiary care hospital. J. Lab. Physicians. 2019;11:206–211. doi: 10.4103/JLP.JLP_22_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bilal H., Khan M.N., Rehman T., Hameed M.F., Yang X. Antibiotic resistance in Pakistan: A systematic review of past decade. BMC Infect. Dis. 2021;21:244. doi: 10.1186/s12879-021-05906-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbas S., Sabir A.U., Khalid N., Sabir S., Khalid S., Haseeb S., Khan N.M., Ajmal W.M., Azhar F., Saeed M.T. Frequency of Extensively Drug-Resistant Gram-Negative Pathogens in a Tertiary Care Hospital in Pakistan. Cureus. 2020;12:e11914. doi: 10.7759/cureus.11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masseron A., Poirel L., Ali B.J., Syed M.A., Nordmann P. Molecular characterization of multidrug-resistance in Gram-negative bacteria from the Peshawar teaching hospital, Pakistan. New Microbes New Infect. 2019;32:100605. doi: 10.1016/j.nmni.2019.100605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrar S., Hussain S., Khan R.A., Ain N.U., Haider H., Riaz S. Prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae: First systematic meta-analysis report from Pakistan. Antimicrob. Resist. Infect. Control. 2018;7:26. doi: 10.1186/s13756-018-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uddin F., McHugh T.D., Roulston K., Platt G., Khan T.A., Sohail M. Detection of carbapenemases, AmpC and ESBL genes in Acinetobacter isolates from ICUs by DNA microarray. J. Microbiol. Methods. 2018;155:19–23. doi: 10.1016/j.mimet.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Ghenea A.E., Zlatian O.M., Cristea O.M., Ungureanu A., Mititelu R.R., Balasoiu A.T., Vasile C.M., Salan A.I., Iliuta D., Popescu M. TEM,CTX-M,SHV Genes in ESBL-Producing Escherichia coli and Klebsiella pneumoniae Isolated from Clinical Samples in a County Clinical Emergency Hospital Romania-Predominance of CTX-M-15. Antibiotics. 2022;11:503. doi: 10.3390/antibiotics11040503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahad A., Salman M., Ikram A., Ashraf Z., Amir A., Saeed A., Ahmad A. Prevalence and molecular Characterization of ESBL-producing Escherichia coli in waste water samples from Pakistan. Int. J. Infect. Dis. 2020;101:33. doi: 10.1016/j.ijid.2020.09.119. [DOI] [Google Scholar]
- 45.Nasir F., Khan M.I., Kashif S., Uddin F., Naseer A., Masood S. Prevalence of ESBLs secreting and carbapenem-resistant E. coli from urinary tract infection. Rawal Med. J. 2021;46:518–521. [Google Scholar]
- 46.Nordmann P., Poirel L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019;69:521–528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohsin M., Brekhna H., Khan A.U., Ali A., Swedberg G., Hasan B. Genomic characterization of high-risk Escherichia coli and Enterobacter hormaechei clones recovered from a single care hospital in Pakistan. J. Appl. Microbiol. 2022;132:3907–3914. doi: 10.1111/jam.15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habib A., Lo S., Villageois-Tran K., Petitjean M., Malik S.A., Armand-Lefèvre L. Dissemination of carbapenemase-producing Enterobacterales in the community of Rawalpindi, Pakistan. PLoS ONE. 2022;17:e0270707. doi: 10.1371/journal.pone.0270707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrissey I., Olesky M., Hawser S., Lob S.H., Karlowsky J.A., Corey G.R., Bassetti M. In Vitro Activity of Eravacycline against Gram-Negative Bacilli Isolated in Clinical Laboratories Worldwide from 2013 to 2017. Antimicrob. Agents Chemother. 2017;64:01715–01719. doi: 10.1128/AAC.01699-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhanel G.G., Cheung D., Adam H., Zelenitsky S., Golden A., Schweizer F., Gorityala B., Lagacé-Wiens P.R.S., Walkty A., Gin A.S., et al. Review of Eravacycline, a Novel Fluorocycline Antibacterial Agent. Drugs. 2016;76:567–588. doi: 10.1007/s40265-016-0545-8. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q., Jin L., Sun S., Yin Y., Wang R., Chen F., Wang X., Zhang Y., Hou J. Occurrence of High Levels of Ce fi derocol Resistance in Carbapenem-Resistant Escherichia coli before Its Approval in China: A Report from China CRE-Network. Microbiol. Spectr. 2022;10:e02670-21. doi: 10.1128/spectrum.02670-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji C., Juárez-Hernández R.E., Miller M.J. Exploiting bacterial iron acquisition: Siderophore conjugates. Future Med. Chem. 2012;4:297–313. doi: 10.4155/fmc.11.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohira N., Hackel M.A., Ishioka Y., Kuroiwa M., Sahm D.F., Sato T., Maki H., Yamano Y. Reduced susceptibility mechanism to cefiderocol, a siderophore cephalosporin, among clinical isolates from a global surveillance programme (SIDERO-WT-2014) J. Glob. Antimicrob. Resist. 2020;22:738–741. doi: 10.1016/j.jgar.2020.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this manuscript can be obtained from the corresponding author upon reasonable request.