Abstract
Basement membranes (BMs) are structured regions of the extracellular matrix that provide multiple functions including physical support and acting as a barrier, as a repository for nutrients and growth factors, and as biophysical signalling hubs. At the core of all BMs is the laminin (LM) family of proteins. These large heterotrimeric glycoproteins are essential for tissue integrity, and differences between LM family members represent a key nexus in dictating context and tissue-specific functions. These variations reflect genetic diversity within the family, which allows for multiple structurally and functionally distinct heterotrimers to be produced, each with different architectures and affinities for other matrix proteins and cell surface receptors. The ratios of these LM isoforms also influence the biophysical properties of a BM owing to differences in their relative ability to form polymers or networks. Intriguingly, the LM superfamily is further diversified through the related netrin family of proteins and through alternative splicing leading to the generation of non-LM short proteins known as the laminin N-terminus (LaNt) domain proteins. Both the netrins and LaNt proteins contain structural domains involved in LM-to-LM interaction and network assembly. Emerging findings indicate that one netrin and at least one LaNt protein can potently influence the structure and function of BMs, disrupting the networks, changing physical properties, and thereby influencing tissue function. These findings are altering the way that we think about LM polymerisation and, in the case of the LaNt proteins, suggest a hitherto unappreciated form of LM self-regulation.
Keywords: basement membrane, laminin, netrin
Introduction
Basement membranes (BMs) are specialised extracellular matrix (ECM) structures, underlying all epithelium, mesothelium and endothelium [1]. BMs provide physical support for cell attachment, act as semi-permeable barriers and influence cell behaviour through; direct signalling via interaction with cell surface receptors or through presenting tuneable biophysical interaction sites, indirectly influencing signalling via the sequestration and controlled release of signalling molecules. These overlapping but diverse functions mean that BMs are key modulators of a myriad of cell and tissue behaviours; these include: regulating cell migration during development, tissue remodelling and wound repair, controlling cellular movement during immune extravasation and tumour progression, and, via their signalling roles, defining differentiation and lineage specification [1–3]. Together these amount to critical functions and make BMs essential for multicellular life. Indeed, inherited or acquired disorders leading to dysfunction of individual BM components are often embryonic lethal or cause devastating human diseases, for example, major kidney problems in Alport syndrome and Pearson's syndrome, skin and other epithelial fragility in junctional epidermolysis bullosa, and severe muscular dystrophy in merosin-deficient congenital muscular dystrophy, as reviewed in [4–7].
At the core of every BM are two networks of proteins: a collagen IV network and a laminin (LM) network. These are connected via nidogens and heparan sulfate chains of the proteoglycans perlecan and agrin [3,8–11]. There are six different α-chain subunits of collagen IV, which form different heterotrimers, with α1α1α2 being the most common [1]. However, the LM family provides greater flexibility for tissue specialisation and is described in more detail below [2].
One of the most intriguing aspects of BM biology are the localised differences in structure and function. Whereas, historically, BM studies relied on electron microscopy approaches on dehydrated tissue; more recently atomic force microscopy (AFM), mass spectrometry and advanced light microscopy imaging have revealed complexities in biophysical properties including stiffness, in BM structure and protein composition that differ depending on the tissue. Indeed, BMs are no longer considered as static physical structures but rather undergo remodelling throughout life via the action of proteolytic processing and non-disruptive network modifying proteins [12,13], and/or through contextual differences in protein expression. Human and mouse studies focused on the inner limiting membrane (ILM) of the retina, Descemet's membrane supporting the corneal endothelium, lens capsule and retinal blood vessels have revealed a general trend that with ageing, BMs become thicker, more amorphous and change biochemical composition with the relative concentrations of collagen IV and agrin increased and LMs reduced [14–19].
AFM studies have revealed a distinct sided-ness to BMs, with the LM-enriched epithelial/endothelial side being considerably stiffer than the stromal-facing aspect [20], and ILM stiffness greater on the retinal side than the vitreous side [19,21]. Stochastic optical reconstruction microscopy (STORM) microscopy has identified that the glomerular BM, which also thickens with age, to be highly structured and laminar, not amorphous as previously thought [22,23]. Moreover, localised differences in biophysics have been established in drosophila egg elongation studies, where AFM measurements showed BM stiffness anisotropy between the centre and posterior regions. Within this context, collagen IV was central to defining the bulk properties of the BM whereas the LM mechanical contribution was more nuanced, with LM knockdown eggs showing increased stiffness in the centre region but decreased stiffness in the posterior region in comparison with controls [21]. These recent advances in appreciation of the biomechanical and structural features of BM places greater emphasis on understanding how the core proteins of LMs self-assemble to form the different types of BM.
Laminins
Each LM is a glycoprotein heterotrimer composed of three subunits: an α-, a β- and a γ-chain, with each chain derived from a distinct gene. In humans, there are five α-chain (LAMA1–5), three β-chain (LAMB1–3) and three γ-chain (LAMC1–3) genes [2,14]. A fourth β has been identified; however, this has not been found in any heterotrimers and may be a pseudogene. Additionally, LAMA3 contains two distinct promoters which generate functionally and structurally distinct subunits; a ‘full-length' form, LMα3B and a much shorter LMα3A [15,16]. The naming convention for the assembled LM heterotrimers reflects the subunit composition with the LM formed from α1, β1 and γ1 subunits being known as LM111 [17].
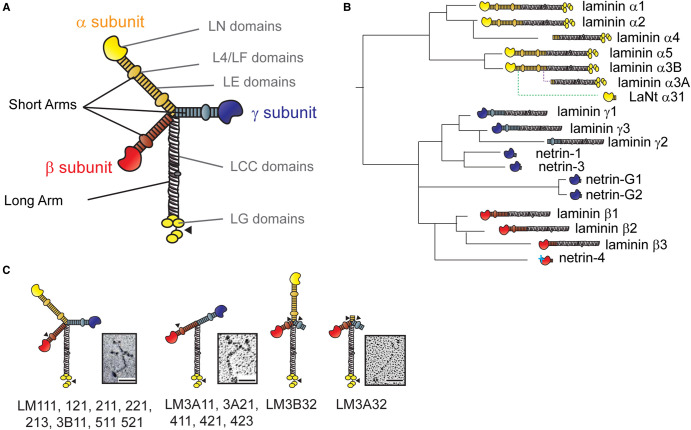
The archetypal LM heterotrimer structure is cross-shaped [24], formed of one long arm and three short arms (Figure 1A). The long arms comprise the defining feature of LMs; a 561-591 amino acid LM coiled-coil (LCC) domain through which the chains combine into an α-helical coiled-coil with disulfide bonds stabilising the structure [2]. The formation of the coiled-coil is dependent upon critical residues toward the C-terminal regions of the LCC, which means not all αβγ combinations are possible, with 16 having been purified in mammals [25–28]. In the α-chains, five LM globular (LG) domains are attached C-terminally to the LCC. These comprise a LG1-3 trio and a LG4-5 duo connected to one another by a linker region/hinge. The LG domains contain most of the high-affinity binding sites for LM cell surface receptors, including integrins, syndecans and α-dystroglycan [2,3]. Proteolytic processing can occur within the linker region, releasing the LG4-5 domain [29] and having implications for how cells interact with the LMs, and is associated, for example, with hemidesmosome maturation [30-32]. Indeed, like most multi-domain ECM proteins, multiple proteolytic processing events can potentially unmask matricryptic sites [33-36]. The LG domains also appear to contribute to the ability of BMs to sequester and control the release of growth factors, with vascular endothelial cell growth factor, platelet-derived growth factor, fibroblast growth factor, bone morphogenetic protein and neurotrophin families all capable of binding to LMs via the heparan-binding domains located on α-chain LG domains [37,38].
Figure 1. Laminin domain architecture, evolution and heterotrimer assemblies.
(A) Archetypal laminin structure. Yellow, red and blue regions indicate short arms generated from individual laminin chains, grey represents the long arm. LN — laminin N-terminus domain, LE — laminin-type epidermal growth factor-like domain, L4/LF — laminin domain IV globular domain, LCC — laminin coiled-coil domain, LG — laminin globular domain. (B) Derivation, relatedness and approximated order of appearance of laminin family members. Solid lines represent gene duplication and rearrangement events. Dotted lines indicate the evolution of an additional promoter (for LMα3A) and of intron-retention and alternative polyadenylation for LaNt α31. (C) Diagram of different laminin assemblies. Numbers represent chain composition e.g. LM111 is laminin α1β1γ1. Arrowheads indicate sites of proteolytic processing. Inset images are rotary shadowing electron microscopy images of purified LM111 (left) from [55], LM411 (middle) from [43] and LM332 (right) from [30]. Scale bars 50 mm. These images have been cropped from the original figures and are re-used in accordance with Creative Commons BY 4.0 license agreement.
The LM short arms are each comprised of the amino-terminal end of one individual chain [39,40]. At the end of most short arms are LM N-terminal (LN) globular domains, of 228–259 residues. These are node-forming globules involved in LM self-polymerisation and network formation, described below [3]. The majority of the short arms are formed of rod-like arrays of LE domains, which, with the LN domain, give a distinct ‘stalk and flowerhead' structure [41]. Each LE domain contains eight cysteine residues, paired together by disulfide bonds, forming interconnected loops [42]. In chains apart from LMα3A, α4 and β3, these LE arrays are interrupted by either an L4 domain inserted between residues three and four of one LE domain or an LF domain located between LE arrays [2,42]. Gene duplication and domain rearrangements have meant that not all the LM chains contain all structural domains (Figure 1B). Of particularly relevance when considering LM polymerisation is the existence of three chains that have much shorter N-terminal arms (LMα3a, α4 and γ2), with heterotrimers containing these chains taking on either a T-shape (LM3A11, 3A21, 411, 421, 423) [43] or more rod-like shape (LM3B32 and 3A32) [30,44] (Figure 1C).
The presence and distribution of the different LM types are tissue-specific and changes during development and tissue remodelling. In generalised terms, LM111 is ubiquitous during embryogenesis, but becomes tissue-specific in the adult as α5 containing LMs become more common [45,46]. As development continues, different LM heterotrimers become associated with certain tissue types, for example, LM332 is enriched in most epithelial basal lamina, LM411 and LM421 throughout the vasculature endothelium and LM211 and LM221 in muscle BMs [1–3,47–49]. However, most tissues contain a combination of different LMs that overlap or can be spatially or temporally distinct. These localised differences can be functionally important; for example, both LM511 and LM332 are abundant in developing and adult skin [50]; however, in the hair cycle LM511 induces hair growth with a steady increase in expression during anagen, while LM332 has a suppressing effect on hair growth and is down-regulated. Conversely, during catagen, LM332 is up-regulated, while LM511 is down-regulated [51,52]. BM LM composition changes with age are seen with decreased LM511 due to photoageing in epidermal BM [53], with differences also observed between infants and adult human corneal BMs [54].
LM network formation
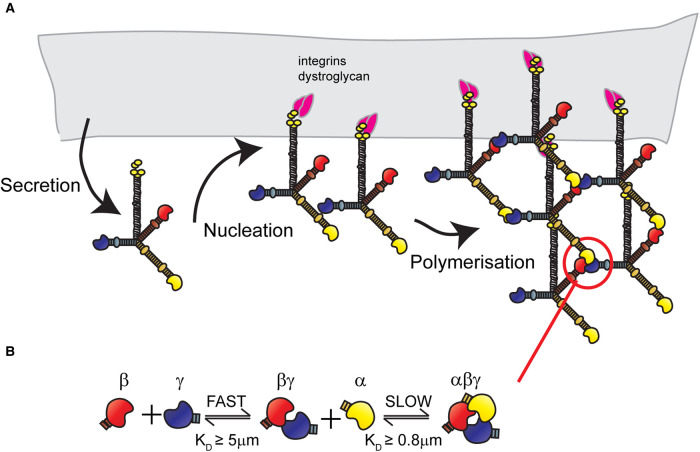
LM network self-assembly is LN domain dependant and occurs via a two-step process of nucleation, where the LMs bind to cell surface receptors, followed by propagation involving LN domain interaction [55,56] (Figure 2A). The network assembly steps can be further broken into a temperature-dependent oligomerisation step followed by a calcium-dependent polymerisation step [56]. For in vitro network assembly, all three short arms must contain a LN domain; leading to a ‘three-arm hypothesis’ [56,57]. Indeed, the arms must also be different, as the α, β and γ LN domains each play distinct roles [56-60] (Figure 2B). First, a fast but unstable interaction occurs between the β and γ LN domains. Then the βLN-γLN binary complex allows for a second, slower, stabilisation step to occur, involving an α LN binding to form an αβγ ternary node [61] (Figure 2B).
Figure 2. Laminin network assembly.
(A) Network assembly involves, first, secretion of the fully trimerised laminin protein. Then, in the ‘nucleation' step, the proteins bind to cell surfaces receptors including integrins α3β1, α6β1 and α6β4 or dystroglycans. Once sufficient local concentration is achieved through receptor clustering, the ‘polymerisation' step occurs via interaction between laminin N-terminal domains. (B) Laminin ternary node formation is a two-step process involving a rapid formation of relatively unstable βγ dimers, followed by slower interaction with an α laminin N-terminal domain to form a more stable αβγ ternary node.
The LN domain crystal structures have shed further light on the polymerisation process [41,61]. Each LN domain is centred around a β-sandwich jelly-roll motif with differing loop regions between the chains. While the surfaces are not conserved between the α, β and γ families, there is conservation within each chain family of β-sandwich residues on one face and of shielding glycans on the other [41]. The exposed residues on the non-glycosylated faces of the β and γ LN domains mediate binding with the α LN domain [41,61]. Whereas the α LN domains contain a conserved surface loop, involved in the stabilisation of the βγ dimer. Recent elegant studies have further established that the LN domains interact in a heel-to-toe fashion, forming triskelion-like structures [59] (Figure 2B). These new findings imply that two distinct interaction sites exist between each LN domain, and that functional conservation of both is required for the stabilisation of the network [59]. This becomes particularly important when considering netrin-4.
Netrins — widening the laminin superfamily
Phylogenetic analyses indicate that LM α-, β- and γ-chains and netrins were all generated from a shared common ancestor (Figure 1B). An early evolutionary duplication separated the LM α subunit ancestors from the β and γ subunits, then a second split occurred between the β and γ genes. Thereafter, the netrins evolved separately at least twice and possibly three times; generating netrins-1 and -3 from the γ-chain ancestor, netrin-G1 and -G2 from the βγ ancestor, and netrin-4 from the β-chain ancestor [62,63] (Figure 1B). The netrins lost the majority of the C-terminal LM domain, including the LCC domain, meaning netrins cannot form trimers. However, they contain LN domains and short stretches of LE repeats followed by a netrin-specific C-terminal domain [64].
The γ LM-like netrins are considered primarily signalling proteins with important roles in axonal guidance; acting via cell surface receptors including deleted in colorectal cancer (DCC), neogenin and UNC5 [65]. Netrin-1 is known to gather and guide axons in the central nervous system, but has a dual role in neural migration acting as a chemorepellent for oligodendrocyte precursors and parallel fibres, but a chemoattractant for dopaminergic and pre-cerebellar neurons [66-68]. Netrin-3 is also involved in axonal guidance but is less effective in signalling than netrin-1; it has been suggested that netrin-3's weak affinity for DCC may be the reason for its relatively lower effectiveness [69]. Intriguingly, growth-cone attraction toward netrin-1 has been shown to be converted to repulsion when the neurons are plated on LM111, suggesting interplay between these protein families [70].
Netrin-4, the only LM β-type netrin, is different from the rest of the family. Although it also promotes axon outgrowth [71], it is mostly known as a regulator of angiogenesis. Specifically, netrin-4 localises to blood vessels and improves angiogenesis in the peri-infarct cortex after focal cerebral ischaemia [72], and absence of netrin-4 is associated with alteration of vascular structure and spontaneous leakage in the retina [73]. From a mechanistic perspective, netrin-4's influence upon angiogenesis requires Unc5B and neogenin as when either are silenced, the inhibitory effect of overexpressed netrin-4 are stopped and a direct effect is suggested as both proteins co-immunoprecipitate with netrin-4 [74]. However, although these data imply direct signalling, it has now also been established that netrin-4 is a potent modulator of LM network assembly and contributes to defining BM biophysical characteristics.
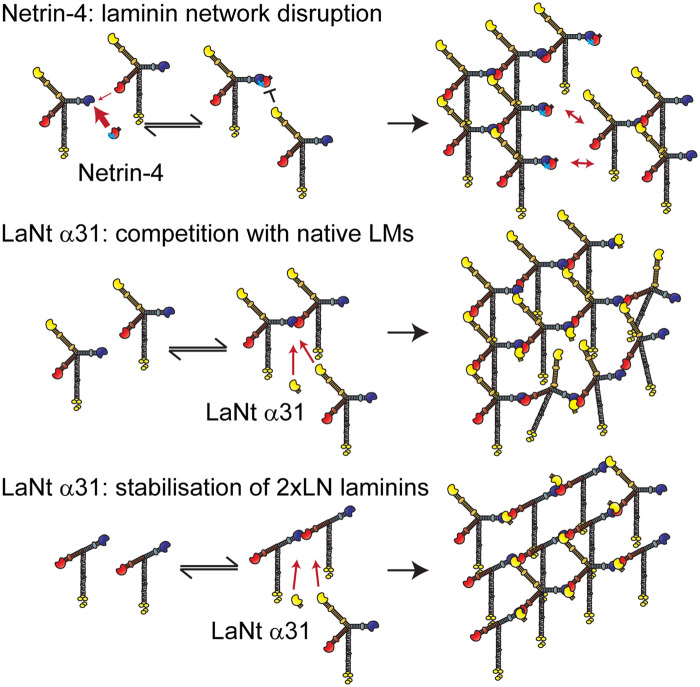
Central to the understanding of netrin-4 as a LM network disruptor is appreciating that although it contains a LM β-like LN domain, the domain itself is imperfect. The binding site for γ LN domains is conserved but the α LN binding site has been lost (Figure 3). Indeed, netrin-4 displays much higher affinity (2500× greater) than LMβ1 for LMγ1 LN domain [12]. In polymerisation assays, these differences mean that netrin-4 can not only prevent LM111 polymers from forming but can also disrupt preformed LM polymers through competing for the γ LN domain binding site [12,75], and this reduces the stiffness of LM containing hydrogels when netrin-4 is added as a recombinant protein [12]. In intact tissue, netrin-4 also may exhibit this non-enzymatic disruptive force on mature BM, with increased netrin-4 leading to larger pores and softening of BM mechanical properties [12,13]. These stiffness effects might be a key determinant of tumour invasion potential; softer tissues with higher netrin-4 to LM ratio display a resistance to metastasis formation [13]. This correlation between netrin-4 expression levels, tissue stiffness and tumour invasion does not preclude netrin-4 also acting through an alternative mechanism independently or in addition to its biophysical role.
Figure 3. Netrin-4 and LaNt α31 predicted effects on laminin networks.
(Top) Netrin-4 binding with high affinity to laminin γ-chains can outcompete the laminin β-chains. However, α-chains cannot bind to γ/netrin-4 complexes. Therefore, netrin-4 disrupts laminin networks leading to locally increased pore size and decreased basement membrane stiffness. (Middle) LaNt α31 has an identical laminin N-terminal domain as LMα3b and therefore LaNt α31 will compete with approximately equal affinity for α-chain binding sites. This will lead to a partially disrupted network with reduced stiffness, with the level of disruption proportional to the expression level. (Bottom) In basement membranes containing T-shaped laminins, where only the β- and γ-chains contain N-terminal domains (e.g. LM411), the LaNt α31 protein may stabilise transient βγ interactions allowing the formation of stable ternary nodes. This would lead to linear arrays of the two-arm laminins. These arrays could also be cross-linked via the integration of some three-arm laminins within the local structure. The mechanical properties of the formed network would depend on the ratio of the three-arm to two-arm laminins and the local LaNt α31 concentrations.
Laminin N-terminus proteins — the ‘missing' laminin superfamily members
While the netrins provide a source of β and γ type LN domains, alternative splicing from LM genes provides a means to produce α LN domain-containing protein fragments through a process of intron-retention and polyadenylation within the retained intron [76] (Figure 1B). At least one of these alternative splice isoforms from the LAMA3 gene has been confirmed at the protein level and is termed laminin N-terminus (LaNt) protein α31 (LaNt α31). The LaNt proteins are netrin-like in structure consisting of an α LN domain, LE repeats and a short unique C-terminus derived from the intronic sequence and could be considered the hitherto ‘missing' members of the LM/netrin family. Proteolytic processing of LM proteins can also lead to the release of LN domain-containing fragments and represent an additional mechanism to produce netrin-like proteins [60,77,78]. That multiple independent mechanisms exist to produce LN domain-containing proteins suggests important functional roles, and these are now beginning to become apparent.
LaNt α31 displays widespread expression at the mRNA and protein level (Figure 4), including being enriched in the basal layer of most epithelium, throughout the vascular system, around terminal ducts in the breast, and in specialised cell types in the brain [79]. Increased expression has been identified in invasive breast ductal carcinoma, with further increases in distant metastases and with high expression associated with a non-cohesive tumour phenotype [80]. The expression also increased in 2D scratch wound assays during the early stages of ex vivo burn wound closure and in epithelial stem cell activation assays [76,81]. Functional studies have demonstrated that both down-regulation and increased expression of LaNt α31 lead to defects in epithelial cell adhesion with reduced migration rates associated with changes to LM332 organisation and hemidesmosome maturation [76,81,82]. In breast cancer cells, the increased expression changed invasive behaviour from coherent multicellular streaming to individual cell invasion. This effect was only observed for cells invading into LM containing hydrogels, suggesting a LM dependency of function [80]. In vivo, ectopic transgenic overexpression during the late stages of development led to non-viable offspring with neonates displaying extensive leaky blood vessels most likely attributable to defective vessel BMs [83].
Figure 4. LaNt α31 distribution in human tissue.
Immunohistochemistry images of formalin-fixed paraffin-embedded human larynx, pancreas, kidney and breast tissue sections obtain from US Biomax (MBN481, US Biomax, Rockville, Maryland, U.S.A.) processed with mouse monoclonal antibodies against LaNt α31 [79]. Scale bar 200 µm.
The understanding of LM network formation and LN-LN interaction leads to a hypothesis where LaNt α31 could elicit its functional effects by competing with α LN domains of the LMs in a similar but distinct way to netrin-4. Unlike netrin-4's competition with β LN domains (Figure 3, Top), the LaNt proteins would compete with the α-chains during the stabilisation step of network assembly. Indeed, as LaNt LN domains are identical with the native LM, any competitive effect would be much weaker than netrin-4 at equivalent concentrations. Moreover, this LN domain conservation raises a very different potential situation. As they are perfect matches to the LMs, LaNt proteins could actually be integrated into the ternary node in the same way as the native LM α-chain. LaNt integration would still change the network structure and mechanics, but one could predict the effects to be subtler than the full localised disruption induced by netrin-4 (Figure 3, Middle). The perfect conservation and putative integration of the LaNt-βγ ternary node raises a further potential situation, where LaNt proteins could play a stabilising role in those BMs where the predominant LMs present are normally considered unable to self-assemble.
As LMα3A, α4- and γ2-chains all lack LN domains, any heterotrimers containing these chains are unable to independently polymerise under the ‘three-arm hypothesis' [56,57,78,84]; this includes LM311, LM321, LM411, LM421, LM423, all of which lack an α LN domain. A biological rationale for this scenario is that networks containing LMs with missing LN domains have different tissue biomechanical properties in terms of flexibility and stability than those with fully polymerised networks [44]. A good example of differential polymerisation status potential can be seen in blood vessels, where the 3xLN domain LM511 and 2xLN domain LM411 are the main LMs. These two LMs are found at different ratios dependent on the blood vessel type and their maturation state; lower expression of LM511 is observed in post-capillary venules relative to venules and relative to veins [85]. This leads distinct areas of defined LM411 to LM511 ratio. Intriguingly high LM511 sites have higher endothelial-to-endothelial cell junctional adhesion strength, and the low LM511 are preferred for leukocyte extravasation; although the reasons behind this later effect also reflect T-cell interactions with LM511 that inhibit migration [85-87].
The LaNt proteins raise a possibility where the additional free α LN domain allows for partial network assembly between non-network forming LMs, compensating for the missing α LN domain. This stabilisation effect would lead to a different assembly state; rather than robust hexagonal arrays that would be self-supporting, the 2xLN + LaNt nodes would be predicted to assemble into rod-like linear arrays (Figure 3, Bottom). These arrays could be cross-linked by any three-arm LMs integrated into the network. Indeed, most biological tissues contain a mixture of two and three LN domain LMs; therefore, the relative abundance of the three LN LMs would determine the level of cross-linking between these LaNt/LM rods and provide a mechanism to fine-tune biomechanical properties. Within this model, excess LaNt α31 would disrupt cross-links and could explain the vessel disruption observed in the LaNt α31 transgenic animals [83].
While LaNt proteins are likely to be capable of influencing network assembly, they could be involved in direct cell signalling. Cell surface receptor binding sites have been identified in LM α-chain LN domains. Specifically, α-chain LN domains can bind heparan sulfate chains of perlecan [88], and a fragment produced from LMα3B N-terminal proteolytic cleavage supported cell attachment in a α3β1 integrin-dependent manner [89], while an LN domain fragment from LMβ1 has also been implicated in the process of epithelial-mesenchymal transition [90]. However, it is important to note that the relative affinities of LN domains for cell surface receptors are orders of magnitude lower than LM LG domain binding. Whether any of these putative signalling pathways are physiologically relevant is yet to robustly be determined but is an important unresolved question. When considering signalling effects, LaNt proteins and netrin-4 are very likely to have indirect effects via their biophysical effects. As BMs sequester growth factors, and as LMs are involved in this process [37,38], one might envisage that modulation of the LM network could modify the bind/release rate of these growth factors. Changing LM network status could also conceivably affect the ways in which the LMs are presented to cells [82]. This, in turn, would influence the clustering or localisation of cell surface receptors and impact upon downstream signalling cascades.
Dissecting the direct vs indirect pathways will not be a trivial task but will shed light on how these relatively new proteins elicit their potent effects and more fundamentally to understand the potentially diverse ways through which LMs can influence cell and tissue behaviour.
Do the LaNt proteins provide two layers of LM regulation?
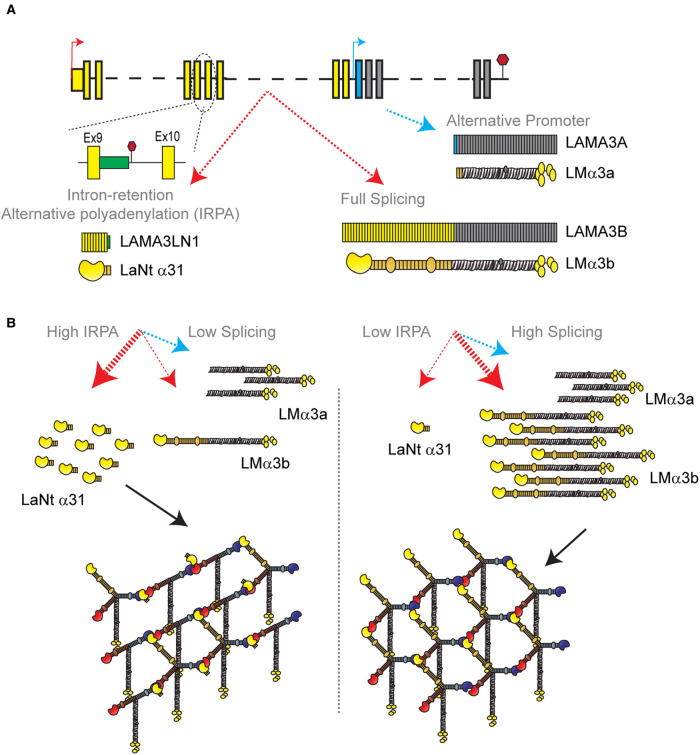
The findings to date point toward LaNt proteins being potent mediators of BM function. However, their genetic derivation raises the potential for a second layer of LM regulation, at the transcript level. As the LaNt transcripts are derived from LM genes, they share a promoter with their parent gene. Each time transcription is initiated from that promoter, a proportion of the transcripts will make the LaNt instead of the LM. When the efficiency of splicing or the rate at which the intronic cleavage and polyadenylation events are processed changes, then the relative abundance of the LM and LaNt transcripts will also change. Changing this intron-retention ‘splicing switch' so that more LaNt is made, would effectively decrease protein production of the ‘full-length' LM and simultaneously, by increasing LaNt production, produce a protein that disrupts LM networks.
This concept of dual-layer intron-retention control becomes particularly intriguing when one considers the LAMA3 gene. LAMA3 has two distinct promoters; one which produces the ‘full-length' LMα3b and the LaNt α31 encoding transcripts, and a second promoter that produces the N-terminally truncated LMα3a-chain [76,91] (Figure 5A). Although these two promoters are independently regulated [92], the evolution of the splicing switch allows for the reduction in the ‘full-length' LMα3b by a secondary means (Figure 5B). As LMα3b contains a LN domain and LMα3a does not, reducing the relative production rate of LMα3b would influence the proportion of network-forming LMs being deposited by cells and the mechanical properties of the network. This might be particularly important in tumour situations where the ratio of the LAMA3A to LAMA3B transcripts has been shown to be a better predictor of patient outcome in head and neck squamous cell carcinoma than the expression of either transcript alone [93]. Splicing and polyadenylation rates are frequently dysregulated in cancers but also change in numerous normal situations, including during development and tissue morphogenesis. Determining the contribution of alternative splicing to regulating LM and LaNt production during these processes and their influences on LM networks and biomechanics represents an exciting new area for BM research.
Figure 5. Intron-retention and alternative polyadenylation as a two-layer laminin regulation mechanism.
(A) Schematic of LAMA3 gene indicating three distinct transcripts and proteins produced. Red arrow — LAMA3B promoter, blue arrow — LAMA3A promoter. Yellow boxes — LAMA3B exons, grey boxes — LAMA3A and B shared exons, blue box — LAMA3A specific exon, green box — protein coding intronic sequence included in LAMA3LN1 but not present in LAMA3B transcript. (B) Potential effects of changing the intron-retention/splicing rates on protein production and laminin network assembly.
Conclusions
The central importance of LMs to normal and disease processes has long been established. The LaNt proteins not only expand the LM superfamily but opens up new ways in which LMs are regulated at both the transcript and protein level; they suggest hitherto unknown methods of defining and remodelling BM organisation, biomechanics and signalling, and thereby influencing cell and tissue function in normal and disease situations.
Perspectives
The LM family of ECM proteins are essential for normal tissue function with dysfunction associated with inherited and acquired diseases. Central to LM function is their ability to form a network or polymer, with that polymer determining matrix biophysical features, signalling and barrier functions.
LM polymerisation involves interaction between laminin N-terminal domains; with stable networks requiring a ternary interaction between an α, β and γ laminin chain. The LM family has been extended by additional proteins that contain laminin N-terminal domains but which are not LMs. Emerging results indicate β-type netrins and the LaNt proteins can influence LM network assembly with implications for development and tumour progression.
The new LaNt proteins and netrin findings generate scenarios where localised and contextually regulated modification of LM networks and BM can occur. Questions remain about the regulation of these modifications and their mechanisms, particularly for situations where LMs are not known to polymerise.
Abbreviations
- BM
basement membrane
- DCC
deleted in colorectal cancer
- ECM
extracellular matrix
- LaNt
laminin N-terminus
- LCC, laminin coiled coil; LE repeats
laminin-type epidermal growth factor-like repeats
- LM
laminin
- LN
laminin N-terminal
- STORM
stochastic optical reconstruction microscopy
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
UKRI Newcastle, Liverpool, Durham Biotechnology and Biological Sciences Research Council Doctoral Training Partnership. International Graduate Education Scholarship (YLYS), the Ministry of National Education, Republic of Turkey. The Ministry of Education of the Kingdom of Saudi Arabia, University of Bisha.
Open Access
Open access for this article was enabled by the participation of the University of Liverpool in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Author Contribution
All authors contributed to the preparation of the initial draft and editing of the manuscript text and figures.
References
- 1.Pozzi, A., Yurchenco, P.D. and Iozzo, R.V. (2017) The nature and biology of basement membranes. Matrix Biol. 57-58, 1–11 10.1016/j.matbio.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aumailley, M. (2013) The laminin family. Cell Adh. Migr. 7, 48–55 10.4161/cam.22826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hohenester, E. and Yurchenco, P.D. (2013) Laminins in basement membrane assembly. Cell Adh. Migr. 7, 56–63 10.4161/cam.21831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardhan, A., Bruckner-Tuderman, L., Chapple, I.L.C., Fine, J.D., Harper, N., Has, C.et al. (2020) Epidermolysis bullosa. Nat. Rev. Dis. Primers 6, 78 10.1038/s41572-020-0210-0 [DOI] [PubMed] [Google Scholar]
- 5.Barraza-Flores, P., Bates, C.R., Oliveira-Santos, A. and Burkin, D.J. (2020) Laminin and integrin in LAMA2-related congenital muscular dystrophy: from disease to therapeutics. Front. Mol. Neurosci. 13, 1 10.3389/fnmol.2020.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chew, C. and Lennon, R. (2018) Basement membrane defects in genetic kidney diseases. Front. Pediatr. 6, 11 10.3389/fped.2018.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGowan, K.A. and Marinkovich, M.P. (2000) Laminins and human disease. Microsc. Res. Tech. 51, 262–279 [DOI] [PubMed] [Google Scholar]
- 8.Groffen, A.J., Ruegg, M.A., Dijkman, H., van de Velden, T.J., Buskens, C.A., van den Born, J.et al. (1998) Agrin is a major heparan sulfate proteoglycan in the human glomerular basement membrane. J. Histochem. Cytochem. 46, 19–27 10.1177/002215549804600104 [DOI] [PubMed] [Google Scholar]
- 9.Noonan, D.M., Fulle, A., Valente, P., Cai, S., Horigan, E., Sasaki, M.et al. (1991) The complete sequence of perlecan, a basement membrane heparan sulfate proteoglycan, reveals extensive similarity with laminin A chain, low density lipoprotein-receptor, and the neural cell adhesion molecule. J. Biol. Chem. 266, 22939–22947 10.1016/S0021-9258(18)54445-8 [DOI] [PubMed] [Google Scholar]
- 10.Yurchenco, P.D., Cheng, Y.S. and Schittny, J.C. (1990) Heparin modulation of laminin polymerization. J. Biol. Chem. 265, 3981–3991 10.1016/S0021-9258(19)39691-7 [DOI] [PubMed] [Google Scholar]
- 11.Ghohestani, R.F., Li, K., Rousselle, P. and Uitto, J. (2001) Molecular organization of the cutaneous basement membrane zone. Clin. Dermatol. 19, 551–562 10.1016/s0738-081x(00)00175-9 [DOI] [PubMed] [Google Scholar]
- 12.Reuten, R., Patel, T.R., McDougall, M., Rama, N., Nikodemus, D., Gibert, B.et al. (2016) Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes. Nat. Commun. 7, 13515 10.1038/ncomms13515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuten, R., Zendehroud, S., Nicolau, M., Fleischhauer, L., Laitala, A., Kiderlen, S.et al. (2021) Basement membrane stiffness determines metastases formation. Nat. Mater. 20, 892–903 10.1038/s41563-020-00894-0 [DOI] [PubMed] [Google Scholar]
- 14.Candiello, J., Cole, G.J. and Halfter, W. (2010) Age-dependent changes in the structure, composition and biophysical properties of a human basement membrane. Matrix Biol. 29, 402–410 10.1016/j.matbio.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 15.Halfter, W., Oertle, P., Monnier, C.A., Camenzind, L., Reyes-Lua, M., Hu, H.et al. (2015) New concepts in basement membrane biology. FEBS J. 282, 4466–4479 10.1111/febs.13495 [DOI] [PubMed] [Google Scholar]
- 16.Murphy, C., Alvarado, J. and Juster, R. (1984) Prenatal and postnatal growth of the human Descemet's membrane. Invest. Ophthalmol. Vis. Sci. 25, 1402–1415 https://iovs.arvojournals.org/article.aspx?articleid=2176847 [PubMed] [Google Scholar]
- 17.Xi, Y.P., Nette, E.G.. King, D.W. and Rosen, M. (1982) Age-related changes in Normal human basement membrane. Mech. Ageing Dev. 19, 315–324 10.1016/0047-6374(82)90015-x [DOI] [PubMed] [Google Scholar]
- 18.To, M., Goz, A., Camenzind, L., Oertle, P., Candiello, J., Sullivan, M.et al. (2013) Diabetes-induced morphological, biomechanical, and compositional changes in ocular basement membranes. Exp. Eye Res. 116, 298–307 10.1016/j.exer.2013.09.011 [DOI] [PubMed] [Google Scholar]
- 19.Henrich, P.B., Monnier, C.A., Halfter, W., Haritoglou, C., Strauss, R.W., Lim, R.Y.et al. (2012) Nanoscale topographic and biomechanical studies of the human internal limiting membrane. Invest. Ophthalmol. Vis. Sci. 53, 2561–2570 10.1167/iovs.11-8502 [DOI] [PubMed] [Google Scholar]
- 20.Halfter, W., Monnier, C., Muller, D., Oertle, P., Uechi, G., Balasubramani, M.et al. (2013) The bi-functional organization of human basement membranes. PLoS One 8, e67660 10.1371/journal.pone.0067660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topfer, U., Guerra Santillan, K.Y., Fischer-Friedrich, E. and Dahmann, C. (2022) Distinct contributions of ECM proteins to basement membrane mechanical properties in Drosophila. Development 149, dev200456 10.1242/dev.200456 [DOI] [PubMed] [Google Scholar]
- 22.Suleiman, H., Zhang, L., Roth, R., Heuser, J.E., Miner, J.H., Shaw, A.S.et al. (2013) Nanoscale protein architecture of the kidney glomerular basement membrane. eLife 2, e01149 10.7554/eLife.01149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann, K.H., Kellner, C., Kuhn, K., Stolte, H. and Schurek, H.J. (2004) Age-dependent thickening of glomerular basement membrane has no major effect on glomerular hydraulic conductivity. Nephrol. Dial. Transplant. 19, 805–811 10.1093/ndt/gfh067 [DOI] [PubMed] [Google Scholar]
- 24.Yurchenco, P.D. and Schittny, J.C. (1990) Molecular architecture of basement membranes. FASEB J. 4, 1577–1590 10.1096/fasebj.4.6.2180767 [DOI] [PubMed] [Google Scholar]
- 25.Paulsson, M., Deutzmann, R., Timpl, R., Dalzoppo, D., Odermatt, E. and Engel, J. (1985) Evidence for coiled-coil alpha-helical regions in the long arm of laminin. EMBO J. 4, 309–316 10.1002/j.1460-2075.1985.tb03630.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engel, J., Hunter, I., Schulthess, T., Beck, K., Dixon, T.W. and Parry, D.A. (1991) Assembly of laminin isoforms by triple- and double-stranded coiled-coil structures. Biochem. Soc. Trans. 19, 839–843 10.1042/bst0190839 [DOI] [PubMed] [Google Scholar]
- 27.Nissinen, M., Vuolteenaho, R., Boot-Handford, R., Kallunki, P. and Tryggvason, K. (1991) Primary structure of the human laminin A chain. Limited expression in human tissues. Biochem. J. 276(Pt 2), 369–379 10.1042/bj2760369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utani, A., Nomizu, M., Timpl, R., Roller, P.P. and Yamada, Y. (1994) Laminin chain assembly. Specific sequences at the C terminus of the long arm are required for the formation of specific double- and triple-stranded coiled-coil structures. J. Biol. Chem. 269, 19167–19175 10.1016/S0021-9258(17)32290-1 [DOI] [PubMed] [Google Scholar]
- 29.Senyurek, I., Kempf, W.E., Klein, G., Maurer, A., Kalbacher, H., Schafer, L.et al. (2014) Processing of laminin alpha chains generates peptides involved in wound healing and host defense. J. Innate Immun. 6, 467–484 10.1159/000357032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker, S.E., Hopkinson, S.B., Fitchmun, M., Andreason, G.L., Frasier, F., Plopper, G.et al. (1996) Laminin-5 and hemidesmosomes: role of the alpha 3 chain subunit in hemidesmosome stability and assembly. J. Cell Sci. 109 (Pt 10), 2509–2520 10.1242/jcs.109.10.2509 [DOI] [PubMed] [Google Scholar]
- 31.Jones, J.C., Hopkinson, S.B. and Goldfinger, L.E. (1998) Structure and assembly of hemidesmosomes. Bioessays 20, 488–494 [DOI] [PubMed] [Google Scholar]
- 32.O'Toole, E.A., Marinkovich, M.P., Hoeffler, W.K., Furthmayr, H. and Woodley, D.T. (1997) Laminin-5 inhibits human keratinocyte migration. Exp. Cell Res. 233, 330–339 10.1006/excr.1997.3586 [DOI] [PubMed] [Google Scholar]
- 33.Davis, G.E., Bayless, K.J., Davis, M.J. and Meininger, G.A. (2000) Regulation of tissue injury responses by the exposure of matricryptic sites within extracellular matrix molecules. Am. J. Pathol. 156, 1489–1498 10.1016/S0002-9440(10)65020-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirila, E., Sharabi, A., Salo, T., Quaranta, V., Tu, H., Heljasvaara, R.et al. (2003) Matrix metalloproteinases process the laminin-5 gamma 2-chain and regulate epithelial cell migration. Biochem. Biophys. Res. Commun. 303, 1012–1017 10.1016/s0006-291x(03)00452-2 [DOI] [PubMed] [Google Scholar]
- 35.Giannelli, G., Falk-Marzillier, J., Schiraldi, O., Stetler-Stevenson, W.G. and Quaranta, V. (1997) Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science 277, 225–228 10.1126/science.277.5323.225 [DOI] [PubMed] [Google Scholar]
- 36.Yamashita, H., Goto, C., Tajima, R., Koparal, A.T., Kobori, M., Ohki, Y.et al. (2008) Cryptic fragment alpha4 LG4-5 derived from laminin alpha4 chain inhibits de novo adipogenesis by modulating the effect of fibroblast growth factor-2. Dev. Growth Differ. 50, 97–107 10.1111/j.1440-169X.2007.00979.x [DOI] [PubMed] [Google Scholar]
- 37.Sekiguchi, R. and Yamada, K.M. (2018) Basement membranes in development and disease. Curr. Top. Dev. Biol. 130, 143–191 10.1016/bs.ctdb.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishihara, J., Ishihara, A., Fukunaga, K., Sasaki, K., White, M.J.V., Briquez, P.S.et al. (2018) Laminin heparin-binding peptides bind to several growth factors and enhance diabetic wound healing. Nat. Commun. 9, 2163 10.1038/s41467-018-04525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tzu, J. and Marinkovich, M.P. (2008) Bridging structure with function: structural, regulatory, and developmental role of laminins. Int. J. Biochem. Cell Biol. 40, 199–214 10.1016/j.biocel.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamill, K.J., Kligys, K., Hopkinson, S.B. and Jones, J.C. (2009) Laminin deposition in the extracellular matrix: a complex picture emerges. J. Cell Sci. 122(Pt 24), 4409–4417 10.1242/jcs.041095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carafoli, F., Hussain, S.A. and Hohenester, E. (2012) Crystal structures of the network-forming short-arm tips of the laminin beta1 and gamma1 chains. PLoS One 7, e42473 10.1371/journal.pone.0042473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stetefeld, J., Mayer, U., Timpl, R. and Huber, R. (1996) Crystal structure of three consecutive laminin-type epidermal growth factor-like (LE) modules of laminin gamma1 chain harboring the nidogen binding site. J. Mol. Biol. 257, 644–657 10.1006/jmbi.1996.0191 [DOI] [PubMed] [Google Scholar]
- 43.Kortesmaa, J., Yurchenco, P. and Tryggvason, K. (2000) Recombinant laminin-8 (alpha(4)beta(1)gamma(1)). Production, purification,and interactions with integrins. J. Biol. Chem. 275, 14853–14859 10.1074/jbc.275.20.14853 [DOI] [PubMed] [Google Scholar]
- 44.Aumailley, M. (2021) Laminins and interaction partners in the architecture of the basement membrane at the dermal-epidermal junction. Exp. Dermatol. 30, 17–24 10.1111/exd.14239 [DOI] [PubMed] [Google Scholar]
- 45.Miner, J.H., Patton, B.L., Lentz, S.I., Gilbert, D.J., Snider, W.D., Jenkins, N.A.et al. (1997) The laminin alpha chains: expression, developmental transitions, and chromosomal locations of alpha1-5, identification of heterotrimeric laminins 8-11, and cloning of a novel alpha3 isoform. J. Cell Biol. 137, 685–701 10.1083/jcb.137.3.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falk, M., Ferletta, M., Forsberg, E. and Ekblom, P. (1999) Restricted distribution of laminin alpha1 chain in normal adult mouse tissues. Matrix Biol. 18, 557–568 10.1016/s0945-053x(99)00047-5 [DOI] [PubMed] [Google Scholar]
- 47.Domogatskaya, A., Rodin, S. and Tryggvason, K. (2012) Functional diversity of laminins. Annu. Rev. Cell Dev. Biol. 28, 523–553 10.1146/annurev-cellbio-101011-155750 [DOI] [PubMed] [Google Scholar]
- 48.Spenle, C., Simon-Assmann, P., Orend, G. and Miner, J.H. (2013) Laminin alpha5 guides tissue patterning and organogenesis. Cell Adh. Migr. 7, 90–100 10.4161/cam.22236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timpl, R. and Brown, J.C. (1996) Supramolecular assembly of basement membranes. Bioessays 18, 123–132 10.1002/bies.950180208 [DOI] [PubMed] [Google Scholar]
- 50.DeRouen, M.C., Zhen, H., Tan, S.H., Williams, S., Marinkovich, M.P. and Oro, A.E. (2010) Laminin-511 and integrin beta-1 in hair follicle development and basal cell carcinoma formation. BMC Dev. Biol. 10, 112 10.1186/1471-213X-10-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imanishi, H., Tsuruta, D., Tateishi, C., Sugawara, K., Paus, R., Tsuji, T.et al. (2010) Laminin-511, inducer of hair growth, is down-regulated and its suppressor in hair growth, laminin-332 up-regulated in chemotherapy-induced alopecia. J. Dermatol. Sci. 58, 43–54 10.1016/j.jdermsci.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugawara, K., Tsuruta, D., Kobayashi, H., Ikeda, K., Hopkinson, S.B., Jones, J.C.et al. (2007) Spatial and temporal control of laminin-332 (5) and -511 (10) expression during induction of anagen hair growth. J. Histochem. Cytochem. 55, 43–55 10.1369/jhc.6A6920.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iriyama, S., Yasuda, M., Nishikawa, S., Takai, E., Hosoi, J. and Amano, S. (2020) Decrease of laminin-511 in the basement membrane due to photoaging reduces epidermal stem/progenitor cells. Sci. Rep. 10, 12592 10.1038/s41598-020-69558-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabosova, A., Azar, D.T., Bannikov, G.A., Campbell, K.P., Durbeej, M., Ghohestani, R.F.et al. (2007) Compositional differences between infant and adult human corneal basement membranes. Invest. Ophthalmol. Vis. Sci. 48, 4989–4999 10.1167/iovs.07-0654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schittny, J.C. and Yurchenco, P.D. (1990) Terminal short arm domains of basement membrane laminin are critical for its self-assembly. J. Cell Biol. 110, 825–832 10.1083/jcb.110.3.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng, Y.S., Champliaud, M.F., Burgeson, R.E., Marinkovich, M.P. and Yurchenco, P.D. (1997) Self-assembly of laminin isoforms. J. Biol. Chem. 272, 31525–31532 10.1074/jbc.272.50.31525 [DOI] [PubMed] [Google Scholar]
- 57.Yurchenco, P.D. and Cheng, Y.S. (1994) Laminin self-assembly: a three-arm interaction hypothesis for the formation of a network in basement membranes. Contrib. Nephrol. 107, 47–56 10.1159/000422960 [DOI] [PubMed] [Google Scholar]
- 58.McKee, K.K., Harrison, D., Capizzi, S. and Yurchenco, P.D. (2007) Role of laminin terminal globular domains in basement membrane assembly. J. Biol. Chem. 282, 21437–21447 10.1074/jbc.M702963200 [DOI] [PubMed] [Google Scholar]
- 59.McKee, K.K., Hohenester, E., Aleksandrova, M. and Yurchenco, P.D. (2021) Organization of the laminin polymer node. Matrix Biol. 98, 49–63 10.1016/j.matbio.2021.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Odenthal, U., Haehn, S., Tunggal, P., Merkl, B., Schomburg, D., Frie, C.et al. (2004) Molecular analysis of laminin N-terminal domains mediating self-interactions. J. Biol. Chem. 279, 44504–44512 10.1074/jbc.M402455200 [DOI] [PubMed] [Google Scholar]
- 61.Hussain, S.A., Carafoli, F. and Hohenester, E. (2011) Determinants of laminin polymerization revealed by the structure of the alpha5 chain amino-terminal region. EMBO Rep. 12, 276–282 10.1038/embor.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fahey, B. and Degnan, B.M. (2012) Origin and evolution of laminin gene family diversity. Mol. Biol. Evol. 29, 1823–1836 10.1093/molbev/mss060 [DOI] [PubMed] [Google Scholar]
- 63.Rajasekharan, S. and Kennedy, T.E. (2009) The netrin protein family. Genome Biol. 10, 239 10.1186/gb-2009-10-9-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, H., Vreeken, D., Leuning, D.G., Bruikman, C.S., Junaid, A., Stam, W.et al. (2021) Netrin-4 expression by human endothelial cells inhibits endothelial inflammation and senescence. Int. J. Biochem. Cell Biol. 134, 105960 10.1016/j.biocel.2021.105960 [DOI] [PubMed] [Google Scholar]
- 65.Ishii, N., Wadsworth, W.G., Stern, B.D., Culotti, J.G. and Hedgecock, E.M. (1992) UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron 9, 873–881 10.1016/0896-6273(92)90240-e [DOI] [PubMed] [Google Scholar]
- 66.Jarjour, A.A. and Kennedy, T.E. (2004) Oligodendrocyte precursors on the move: mechanisms directing migration. Neuroscientist 10, 99–105 10.1177/1073858403260751 [DOI] [PubMed] [Google Scholar]
- 67.Alcantara, S., Ruiz, M., De Castro, F., Soriano, E. and Sotelo, C. (2000) Netrin 1 acts as an attractive or as a repulsive cue for distinct migrating neurons during the development of the cerebellar system. Development 127, 1359–1372 10.1242/dev.127.7.1359 [DOI] [PubMed] [Google Scholar]
- 68.Lin, L., Rao, Y. and Isacson, O. (2005) Netrin-1 and slit-2 regulate and direct neurite growth of ventral midbrain dopaminergic neurons. Mol. Cell. Neurosci. 28, 547–555 10.1016/j.mcn.2004.11.009 [DOI] [PubMed] [Google Scholar]
- 69.Wang, H., Copeland, N.G., Gilbert, D.J., Jenkins, N.A. and Tessier-Lavigne, M. (1999) Netrin-3, a mouse homolog of human NTN2L, is highly expressed in sensory ganglia and shows differential binding to netrin receptors. J. Neurosci. 19, 4938–4947 10.1523/JNEUROSCI.19-12-04938.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hopker, V.H., Shewan, D., Tessier-Lavigne, M., Poo, M. and Holt, C. (1999) Growth-cone attraction to netrin-1 is converted to repulsion by laminin-1. Nature 401, 69–73 10.1038/43441 [DOI] [PubMed] [Google Scholar]
- 71.Qin, S., Yu, L., Gao, Y., Zhou, R. and Zhang, C. (2007) Characterization of the receptors for axon guidance factor netrin-4 and identification of the binding domains. Mol. Cell. Neurosci. 34, 243–250 10.1016/j.mcn.2006.11.002 [DOI] [PubMed] [Google Scholar]
- 72.Hoang, S., Liauw, J., Choi, M., Choi, M., Guzman, R.G. and Steinberg, G.K. (2009) Netrin-4 enhances angiogenesis and neurologic outcome after cerebral ischemia. J. Cereb. Blood Flow Metab. 29, 385–397 10.1038/jcbfm.2008.128 [DOI] [PubMed] [Google Scholar]
- 73.Crespo-Garcia, S., Reichhart, N., Wigdahl, J., Skosyrski, S., Kociok, N., Strauss, O.et al. (2019) Lack of netrin-4 alters vascular remodeling in the retina. Graefes Arch. Clin. Exp. Ophthalmol. 257, 2179–2184 10.1007/s00417-019-04447-3 [DOI] [PubMed] [Google Scholar]
- 74.Lejmi, E., Leconte, L., Pedron-Mazoyer, S., Ropert, S., Raoul, W., Lavalette, S.et al. (2008) Netrin-4 inhibits angiogenesis via binding to neogenin and recruitment of Unc5B. Proc. Natl Acad. Sci. U.S.A. 105, 12491–12496 10.1073/pnas.0804008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneiders, F.I., Maertens, B., Bose, K., Li, Y., Brunken, W.J., Paulsson, M.et al. (2007) Binding of netrin-4 to laminin short arms regulates basement membrane assembly. J. Biol. Chem. 282, 23750–23758 10.1074/jbc.M703137200 [DOI] [PubMed] [Google Scholar]
- 76.Hamill, K.J., Langbein, L., Jones, J.C. and McLean, W.H. (2009) Identification of a novel family of laminin N-terminal alternate splice isoforms: structural and functional characterization. J. Biol. Chem. 284, 35588–35596 10.1074/jbc.M109.052811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kariya, Y., Yasuda, C., Nakashima, Y., Ishida, K., Tsubota, Y. and Miyazaki, K. (2004) Characterization of laminin 5B and NH2-terminal proteolytic fragment of its alpha3B chain: promotion of cellular adhesion, migration, and proliferation. J. Biol. Chem. 279, 24774–24784 10.1074/jbc.M400670200 [DOI] [PubMed] [Google Scholar]
- 78.Yurchenco, P.D. and Cheng, Y.S. (1993) Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J. Biol. Chem. 268, 17286–17299 10.1016/S0021-9258(19)85334-6 [DOI] [PubMed] [Google Scholar]
- 79.Troughton, L.D., Reuten, R., Sugden, C.J. and Hamill, K.J. (2020) Laminin N-terminus alpha31 protein distribution in adult human tissues. PLoS One 15, e0239889 10.1371/journal.pone.0239889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Troughton, L.D., O'Loughlin, D.A., Zech, T. and Hamill, K.J. (2022) Laminin N-terminus alpha31 is upregulated in invasive ductal breast cancer and changes the mode of tumour invasion. PLoS One 17, e0264430 10.1371/journal.pone.0264430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barrera, V., Troughton, L.D., Iorio, V., Liu, S., Oyewole, O., Sheridan, C.M.et al. (2018) Differential distribution of laminin N-terminus alpha31 across the ocular surface: Implications for corneal wound repair. Invest. Ophthalmol. Vis. Sci. 59, 4082–4093 10.1167/iovs.18-24037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Troughton, L.D., Iorio, V., Shaw, L., Sugden, C.J., Yamamoto, K. and Hamill, K.J. (2020) Laminin N-terminus α31 regulates keratinocyte adhesion and migration through modifying the organisation and proteolytic processing of laminin 332. bioRxiv 10.1101/617597 [DOI] [Google Scholar]
- 83.Sugden, C.J., Iorio, V., Troughton, L.D., Liu, K., Morais, M., Lennon, R.et al. (2022) Laminin N-terminus alpha31 expression during development is lethal and causes widespread tissue-specific defects in a transgenic mouse model. FASEB J. 36, e22318 10.1096/fj.202002588RRR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yurchenco, P.D., Cheng, Y.S. and Colognato, H. (1992) Laminin forms an independent network in basement membranes. J. Cell Biol. 117, 1119–1133 10.1083/jcb.117.5.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yousif, L.F., Di Russo, J. and Sorokin, L. (2013) Laminin isoforms in endothelial and perivascular basement membranes. Cell Adh. Migr. 7, 101–110 10.4161/cam.22680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song, J., Zhang, X., Buscher, K., Wang, Y., Wang, H., Di Russo, J.et al. (2017) Endothelial basement membrane laminin 511 contributes to endothelial junctional tightness and thereby inhibits leukocyte transmigration. Cell Rep. 18, 1256–1269 10.1016/j.celrep.2016.12.092 [DOI] [PubMed] [Google Scholar]
- 87.Li, L., Song, J., Chuquisana, O., Hannocks, M.J., Loismann, S., Vogl, T.et al. (2020) Endothelial basement membrane laminins as an environmental cue in monocyte differentiation to macrophages. Front. Immunol. 11, 584229 10.3389/fimmu.2020.584229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garbe, J.H., Gohring, W., Mann, K., Timpl, R. and Sasaki, T. (2002) Complete sequence, recombinant analysis and binding to laminins and sulphated ligands of the N-terminal domains of laminin alpha3B and alpha5 chains. Biochem. J. 362(Pt 1), 213–221 10.1042/bj3620213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kikkawa, Y., Sasaki, T., Nguyen, M.T., Nomizu, M., Mitaka, T. and Miner, J.H. (2007) The LG1-3 tandem of laminin alpha5 harbors the binding sites of lutheran/basal cell adhesion molecule and alpha3beta1/alpha6beta1 integrins. J. Biol. Chem. 282, 14853–14860 10.1074/jbc.M611706200 [DOI] [PubMed] [Google Scholar]
- 90.Horejs, C.M., Serio, A., Purvis, A., Gormley, A.J., Bertazzo, S., Poliniewicz, A.et al. (2014) Biologically-active laminin-111 fragment that modulates the epithelial-to-mesenchymal transition in embryonic stem cells. Proc. Natl Acad. Sci. U.S.A. 111, 5908–5913 10.1073/pnas.1403139111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ryan, M.C., Tizard, R., VanDevanter, D.R. and Carter, W.G. (1994) Cloning of the LamA3 gene encoding the alpha 3 chain of the adhesive ligand epiligrin. Expression in wound repair. J. Biol. Chem. 269, 22779–22787 10.1016/S0021-9258(17)31713-1 [DOI] [PubMed] [Google Scholar]
- 92.Ferrigno, O., Virolle, T., Galliano, M.F., Chauvin, N., Ortonne, J.P., Meneguzzi, G.et al. (1997) Murine laminin alpha3A and alpha3B isoform chains are generated by usage of two promoters and alternative splicing. J. Biol. Chem. 272, 20502–20507 10.1074/jbc.272.33.20502 [DOI] [PubMed] [Google Scholar]
- 93.Moller-Levet, C.S., Betts, G.N., Harris, A.L., Homer, J.J., West, C.M. and Miller, C.J. (2009) Exon array analysis of head and neck cancers identifies a hypoxia related splice variant of LAMA3 associated with a poor prognosis. PLoS Comput. Biol. 5, e1000571 10.1371/journal.pcbi.1000571 [DOI] [PMC free article] [PubMed] [Google Scholar]