Abstract
The C-terminal region of the tumor suppressor protein p53 contains three domains, nuclear localization signal (NLS), tetramerization domain (TET), and C-terminal regulatory domain (CTD), that are essential for p53 function. Characterization of the structure and interactions of these domains within full-length p53 has been limited by the overall size and flexibility of the p53 tetramer. Using trans-intein splicing, we have generated full-length p53 constructs in which the C-terminal region is isotopically labeled with 15N for NMR analysis, allowing us to obtain atomic-level information on the C-terminal domains in the context of the full-length protein. Resonances of NLS and CTD residues have narrow linewidths, showing that these regions are largely solvent exposed and dynamically disordered, whereas resonances from the folded tetramerization domain are broadened beyond detection. Two regions of the CTD, spanning residues 369-374 and 381-388 and with high lysine content, make dynamic and sequence-independent interactions with DNA in regions that flank the p53 recognition element. The population of DNA-bound states increases as the length of the flanking regions is extended up to approximately 20 base pairs on either side of the recognition element. Acetylation of K372, K373, and K382, using a construct of the transcriptional coactivator CBP containing the TAZ2 and acetyltransferase domains, inhibits interaction of the CTD with DNA. This work provides high resolution insights into the behavior of the intrinsically disordered C-terminal regions of p53 within the full-length tetramer and the molecular basis by which the CTD mediates DNA binding and specificity.
Introduction
The master tumor suppressor p53 performs a crucial role in determining the response to cellular stress and DNA damage.1,2 p53 is a multi-domain transcription factor consisting of an N-terminal transactivation domain (NTAD), Pro-rich domain (PRD), DNA-binding domain (DBD), nuclear localization signal (NLS, residues 316-325), tetramerization domain (TET, residues 326-356), and C-terminal regulatory domain (CTD, residues 364-393)2 (Figure 1). The DBD and TET are the only structured domains; the other regions of p53 are intrinsically disordered. P53 is regulated by a plethora of post-translational modifications, mostly within the disordered regions, that modulate p53 stability and its interactions with other proteins and with DNA.4-8 All three of the C-terminal regions are essential for p53 function, with the NLS region driving nuclear import.9-11, the TET domain regulating oligomeric state,12-14 and the CTD functioning in a multitude of roles.3,15
Figure 1.
Domain structure of the p53 protomer, showing the N-terminal domain (NTAD), proline-rich domain (PRD), DNA-binding domain, region containing the nuclear localization signal (NLS), tetramerization domain (TET), and the C-terminal regulatory domain (CTD). Intrinsically disordered domains are shown in yellow.
The CTD comprises the last 30 amino acids of p53 and is involved in protein and DNA interactions and overall regulation of p53.3,15 This domain is intrinsically disordered, but can fold upon binding to partner proteins to form localized structural elements, including α-helices (seen in binding to the S100B protein16), β-turns (seen with the bromodomain of CBP/p30017), or stable conformations lacking defined secondary structure (seen with 53BP1 18). The functional interactions of the CTD are modulated through numerous post-translational modifications, including acetylation, ubiquitination, phosphorylation, SUMOylation, and NEDDylation.15,19-22 The CTD is highly basic and binds non-specifically to DNA both as an isolated peptide and within the full-length p53 tetramer.23 It stabilizes DNA binding, enhances binding to long genomic DNA sequences, and enhances the specificity for different p53 response elements.24-26 The CTD is required for rapid linear diffusion of p53 on DNA,27 mediating fast sliding to facilitate the search for cognate p53 binding sites.28
Following DNA damage, p53 is activated by acetylation of multiple lysines in the CTD.29,30 The pattern of acetylation is directly involved in cell fate decisions. Differential acetylation of p53 by CBP/p300 and PCAF (p300/CBP-associated factor) regulates different subsets of target genes in response to cell stress. Under mild stress, p53 is acetylated at Lys320 by PCAF, which promotes activation of genes with high affinity p53 binding sites and leads to cell cycle arrest and survival.31 In contrast, severe and irreparable DNA damage results in CBP/p300-mediated acetylation of p53 at Lys373 and Lys382 and activation of pro-apoptotic genes and cell death.30,32,33 Acetylation of lysine residues (K372/K373/K381/K382) in an isolated CTD peptide results in decreased DNA-binding,34 consistent with the important role played by these residues in modulating interactions between p53 and DNA.
The molecular mechanism by which the CTD mediates site-specific DNA binding remains poorly understood. On the basis of biochemical experiments and molecular dynamics simulations, it has been suggested that long-range interactions between the CTD and DBD stabilize the tetramer and may promote conformational changes in the DBD that increase the stability of DNA complexes.24,35,36 In contrast, other work has presented evidence that the CTD binds directly to the N-terminal activation domain,26 an interaction that has been proposed as an organizing element of the p53 tetramer.37
High resolution studies of the CTD interactions have focused on isolated CTD peptides,34 leaving unanswered questions as to the nature of its interactions in full-length p53 both free and bound to DNA. While NMR can potentially provide detailed structural and dynamic information on the interactions of the CTD in full-length p53, such experiments are hampered by spectral overlap and by the unfavorable relaxation properties of a protein the size of the p53 tetramer.38,39 In the current work, we have used intein-based segmental isotopic labeling to simplify the NMR spectra of full-length p53, thereby allowing direct characterization of the structure and interactions of the C-terminal domain with atomic-level resolution.
Materials and Methods
Protein expression and purification
All DBD-containing p53 constructs contained the quadruple M133L/V203A/N239Y/N268D superstabilizing mutations.40 H6-GB1 fusion proteins containing a TEV cleavage site were expressed in BL21 (DE3) DNAY E. coli cells, while H6-SUMO fusions were expressed in BL21 (DE3) cells. For expression of H6-GB1-p53(1-393) and H6-GB1-p53(1-303)-Intn, cells were grown in LB or M9 medium for 16-20 hours at 16-18°C following induction with 0.5mM IPTG and addition of 150μM ZnSO4 (ZnSO4 was added only for constructs containing the p53 DBD). For expression of H6-GB1-p53(304-393), H6-GB1-p53(363-393), H6-GB1-Intc-p53(304-393), and H6-SUMO-Intc-p53(304-393), cells were grown for 4-5 hours at 37°C following induction with 0.1mM IPTG. 15N- and 15N/2H- labeled samples were prepared by growing cells in M9 medium containing 15N ammonium sulfate in H2O or D2O. Cell cultures were harvested by centrifugation and cell pellets were stored at −20°C. Cells were lysed by sonication in 40-60mL of 40mM Tris, pH 8.0, 1M NaCl, 5mM DTT (lysis buffer), with 1 EDTA-free protease tablet (Roche). The H6-GB1-tagged proteins were initially purified on His cOmplete Ni resin (Roche) and the fusion tag was removed by incubation with TEV protease as described previously.41 Full-length p53(1-393) was further purified by previously described methods.41 Following TEV cleavage, the p53(363-393) and p53(304-393) constructs were purified via HiTrap SP using a gradient from 40-700mM, eluting at ~350mM NaCl and ~450mM NaCl, respectively. The purified proteins were dialyzed into NMR buffer (see below) and concentrated to 150-300μM using Millipore Centriprep concentrators. For NMR assignments, a sample of 2H/13C/15N p53(304-393) was obtained by growing cells in M9 medium containing 15N ammonium sulfate and 13C glucose in D2O.
For segmental labeling of the C-terminal domains the p53 N-split, p53(1-303), was expressed as a fusion protein attached at the N-terminus to His-tagged GB1 and at the C-terminus to the Intn fragment (Figure S1B). The C-split construct, p53(304-393), was expressed as a fusion to either an H6-GB1-Intc or H6-SUMO-Intc sequence (Figure S1B). After initial splicing attempts, the SUMO-fusion variant was used over the H6-GB1 construct as it gave a better yield of spliced protein. Splicing reactions were performed using a ratio of 1.2- to 1.4-fold excess H6-GB1-p53(1-303)-Intn precursor relative to 15N-labeled H6-SUMO-Intc-p53(304-393) precursor following a previously described protocol in which splicing and SUMO tag cleavage were performed simultaneously.41 After purification and TEV cleavage of the N-terminal H6-GB1 tag, the full-length protein, segmentally labeled with 15N at residues 304-393 (termed 15N(304-393)-p53) was dialyzed against NMR buffer overnight at 4°C and then concentrated to 150-300μM for use. Purified proteins were stored at −80°C after rapid freezing with liquid nitrogen. Purity was assessed by SDS-PAGE and identities of the purified products were confirmed by LC-MS or MALDI.
The CBP construct used in acetylation experiments (residues 1079-1855, spanning the BRD-PHD-RING-HAT-ZZ-TAZ2 domains) was expressed in a pET21 vector in E. coli BL21 (DE3) DNAY cells with a TEV-cleavable N-terminal H6 tag. Cells were grown to an OD of 0.6-0.8 before induction with 1mM IPTG at 15°C overnight. The expressed protein was purified as described previously42 and was used in acetylation reactions immediately after the final purification step.
Preparation of Duplex DNA
DNA was purchased from Integrated DNA Technologies. The DNA-S duplex used in NMR experiments was a palindromic sequence (5’-GAACATGTTCGAACATGTTC-3’) that contained an engineered p53 consensus site29. The DNA-L construct contained this same recognition element, with random base-pair extensions on both the 5’ and 3’ sides (5’-GCCTACAGAATCGCTCTACAGAACATGTTCGAACATGTTCGGGGACTGATGCTGGGGACT-3’). The 30-80 base-pair DNA duplexes contained a central 20 base-pair recognition element (the DNA-S sequence) flanked on either side with (ATG)n repeats. The 60-bp p21 DNA duplex contained the 20-bp recognition element found within the p21 gene and was flanked on either side by the native nucleotides. All DNA sequences are listed in Table S1.
Oligonucleotides were dissolved in 20 mM Tris, pH 7.0, and 150 mM NaCl before being mixed 1:1 with the complementary strand. After mixing, the DNA duplex was annealed by heating to 95°C for 5-10 minutes followed by cooling to room temperature over several hours. The annealed DNA constructs were then exchanged into NMR buffer.
NMR data collection, assignments, and analysis
All NMR data were collected in 20 mM Tris, pH 7.0, 150 mM NaCl, 2 mM DTT, and 5% D2O (NMR buffer) at 25°C. 1H-15N HSQC and 1H-15N TROSY-HSQC spectra were acquired on Bruker Avance 900MHz, DRX 800MHz, DRX 600MHz, and Avance 500MHz spectrometers. Spectra were processed using NMRPipe, and analyzed and visualized using NMR View.43,44 TROSY-HSQC45 experiments were performed on 2H labeled protein, while samples used in standard HSQC experiments were not 2H labeled. For all NMR experiments, protein concentrations ranged from 30-150 μM. All experiments involving the 20-80 base-pair DNA constructs used 30 μM protein. p53:DNA complexes were formed using a 4:1 ratio of p53 protomer to DNA, which represents a 1:1 tetramer:DNA ratio.
Backbone resonances of 2H/13C/15N-labeled p53(304-393) peptide were assigned using standard three-dimensional HNCACB and HN(CO)CACB46 experiments aided by comparisons to previously-published assignments.34 Backbone resonance assignments of p53(304-393) were readily transferable to 15N(304-393)-p53 as cross-peaks for residues in both constructs appear at similar chemical shifts. This was not the case for resonances from the TET domain, which are broadened beyond detection in the full-length protein. A 1H-15N TROSY spectrum of the isolated p53(304-393) peptide is shown in Figure S2 and assignments have been deposited in the BMRB under accession number 51613.
Acetylation of 15N(304-393)-p53
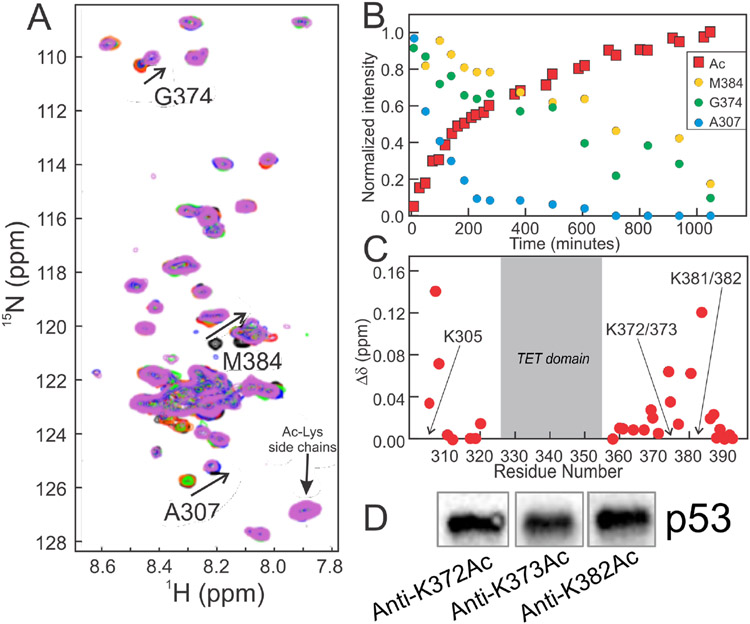
15N(304-393)-p53 was acetylated by adding CBP(1079-1855) directly to a p53 sample at a 100:1 (p53:CBP) molar ratio with addition of 1 mM acetyl-CoA (Ac-CoA). Typical acetylation reactions contained 50-100 μM p53 in NMR buffer. The progress of the reaction was monitored by recording sequential 1H-15N SOFAST HMQC spectra over the course of ~20 hours. Changes in chemical shift were observed for residues neighboring acetylation sites. The sites of acetylation in the CTD were confirmed by Western blot using antibodies specific for acetylation at K372 (Assay Biotech), K373 (Abcam), and K382 (BioLegend). Following acetylation, the reaction mixture was exchanged into fresh NMR buffer to remove the Ac-CoA and was either used directly in NMR experiments or frozen rapidly in liquid N2 for storage at −80°C.
Results
Segmental labeling of the C-terminal domain of tetrameric p53 by trans-intein splicing
The Npu DnaE trans-intein system47,48 was used for segmental labeling of full length p53 as previously described.41 The splice site for segmental labeling of the C-terminal region was placed in a disordered region approximately at the midpoint between the end of the C-terminal α-helix of the DBD and the start of the nuclear localization signal. The final spliced construct contains the non-native residues CFNG between S303/T304 (Figure S1A).
Production and purification of TET-CTD labeled p53
After completion of the ligation reaction, the mixture contained the desired product (full-length p53), unreacted precursors, and the IntN and IntC fragment byproducts. Separation of the ligated protein initially proved problematic due to the oligomeric nature of p53. At the protein concentrations required for efficient ligation (20-30μM), the tetrameric state of p53 was the predominant species in solution,49,50 with negligible amounts remaining in the dimer and monomer form. However, the desired full-length p53 readily oligomerized with TET domain containing unreacted precursors resulting in a hetero-tetramer containing full-length p53 and the IntC-p53(304-393) construct (the N-terminal H6-SUMO on this construct having already been removed). In addition, the large excess of correctly-ligated p53 over other components meant that any unreacted precursors containing the TET domain, and therefore capable of oligomer formation, were much more likely to exist in a tetramer with full-length p53, as opposed to a tetramer containing only the undesired construct. These ‘3:1’ tetramers, comprising 3 ligated full-length p53 and 1 precursor molecules, decreased overall yield and increased the complexity of the purification process. Although the homo-tetrameric and the 3:1 hetero-tetrameric species are similar in size and electrostatic properties, the proteins could be successfully separated through high-resolution anion-exchange chromatography, where the hetero-tetramer eluted at lower salt concentrations. Using this method, full-length p53, labeled segmentally in residues 304-393 with 15N, was obtained on the milligram scale with a purity of over 95%. This construct will be referred to as 15N(304-393)-p53. A comparison of 1H-15N TROSY-HSQC spectra of uniformly-labeled full-length p53 and 15N(304-393)-p53 is shown in Figure S3.
The NLS and CTD are disordered in full-length p53
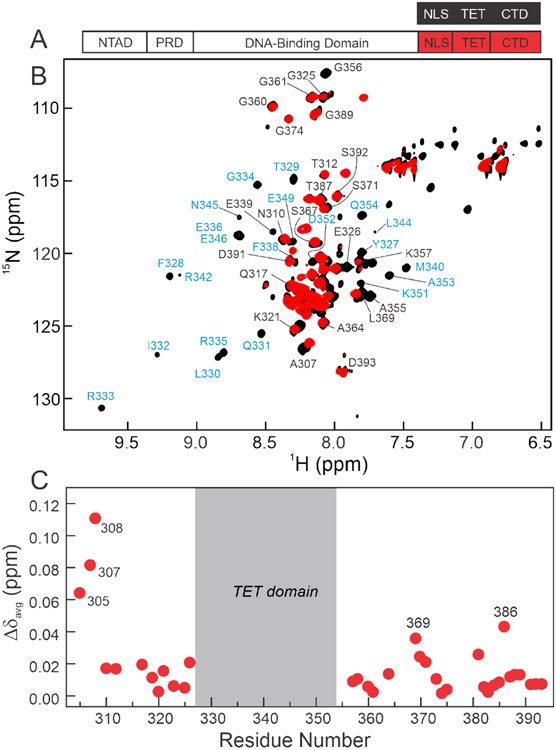
1H-15N TROSY-HSQC spectra of deuterated 15N(304-393)-p53 were recorded and compared with those of the isolated 15N/2H labeled peptide (residues 304-393, termed p53(304-393)) (Figure 2). The cross-peaks for residues 326 to 356, corresponding to the TET domain, 14,51-54 are unobservable in the HSQC spectrum of the segmentally labeled p53 tetramer but are visible and well dispersed in the spectrum of the p53(304-393) peptide (Figure 2B). The broadening and disappearance of TET resonances arises from the slow tumbling of the TET domain in the full-length p53 tetramer/39 Resonances arising from many of the residues in the disordered NLS (residues 304-325) and CTD (residues 357-393) domains are visible in the 1H-15N TROSY-HSQC spectrum of the segmentally labeled 15N(304-393)-p53, at similar chemical shifts as in the spectrum of an isolated p53(304-393) peptide. These data indicate that the NLS and CTD domains remain largely disordered in the full-length protein. A substantial number of the Ser and Thr residues within these regions do not have visible cross-peaks in 1H-15N spectra at pH 7.0, likely due to exchange of amide protons with H2O.
Figure 2.
A. Schematic representation of the isolated p53(304-393) peptide (top) and segmentally 15N-labeled full-length p53 (bottom, labeled at 304-393 through intein splicing) used for NMR experiments. B. 800MHz 1H-15N TROSY-HSQC spectra of isolated 15N/2H p53(304-393) peptide (black) and 15N/2H segmentally labeled tetrameric p53 (15N(304-393)-p53, red). Resonances appearing in the center of the spectra arise from the disordered NLS and CTD residues, while more dispersed cross-peaks in the spectrum of the p53(304-393) peptide correspond to the folded TET domain. Assignments labeled in black were obtained from triple resonance spectra of p53(304-393); assignments labeled in blue were mapped from published data for the isolated TET domain.55 Resonances of the TET domain residues are unobservable in segmentally labeled full-length p53 and do not give observable connectivities in triple resonance spectra of the p53(304-393) peptide. Cross peaks of residues 389 and 391-393 are split due to isomerism of Pro390. All samples were in 20mM Tris, pH7.0, 150mM NaCl, 5% D2O, and 2mM DTT and NMR spectra were acquired at 25°C. C. Weighted average 1H, 15N chemical shift differences between isolated p53(304-393) peptide and the full-length segmentally labeled p53 tetramer. Resonances of residues 305, 307, and 308 are shifted due to their proximity to the intein splice site. Residues in the CTD that exhibit significant chemical shift changes between spectra of 15N(304-393)-p53 and the p53(304-393) peptide are labeled. Weighted average chemical shift differences were calculated using the equation:
Despite the overall similarity of the spectra of the NLS and CTD domains in the isolated p53(304-393) peptide and full-length 15N(304-393)-p53, there are substantial chemical shift differences for residues adjacent to the splice site (S303/T304 in the native protein) (Figure 2C). These differences likely arise from the insertion of four non-native residues at the splice site. Several residues within the CTD of full-length p53 display small chemical shift differences relative to the p53(304-393) peptide that likely arise from transient intramolecular interactions within the p53 tetramer. Resonances of both the NLS and CTD are sharp, showing that these domains remain highly dynamic in the full-length protein.
The CTD contacts DNA outside the p53 recognition element
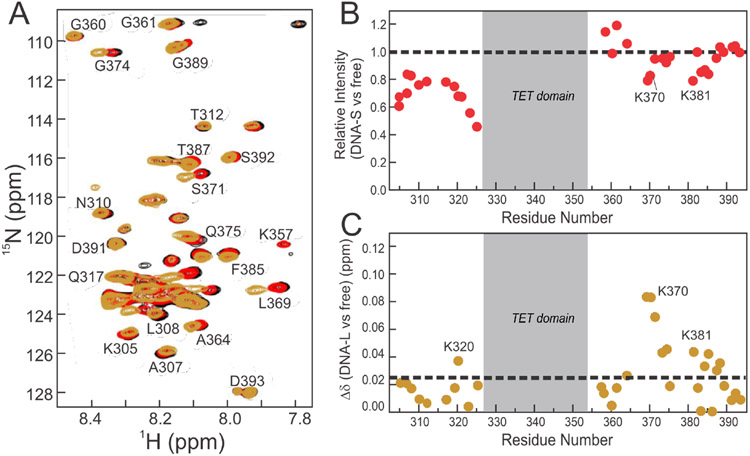
Complexes were formed between 15N/2H segmentally labeled p53 (15N(304-393)-p53) and either a 20 base-pair p53 target DNA (an engineered p53 consensus site59) or a 60 base-pair DNA that contained the same 20 base-pair consensus site flanked by 20 random base-pairs on each side (DNA-S and DNA-L respectively, sequences shown in Table S1). Upon binding to the DNA-S construct, only small chemical shift perturbations are observed in NLS and CTD cross peaks (Figure 3A). There is a substantial decrease in cross-peak intensity for residues 304-325 (NLS), suggesting that binding to DNA affects this region of p53, likely through increased motional restriction of the neighboring DBD and TET domains (Figure 3B). The loss of intensity is greatest for residues close to either the TET domain or the DBD. Cross peaks associated with CTD residues experience only minor intensity change upon binding to the DNA-S oligonucleotide (Figure 3B) which, coupled with the lack of chemical shift change, strongly suggests that the CTD interacts only weakly, if at all, with DNA within the p53 recognition element itself.
Figure 3.
A. 800MHz 1H-15N TROSY-HSQC spectra of 15N/2H segmentally labeled 15N(304-393)-p53 without DNA (black) and bound to the DNA-S (red) or the DNA-L (gold) constructs. Differences between the black and red spectra are difficult to discern as the two overlay closely. B. Intensity differences of 15N(304-393)-p53 cross peaks upon addition of DNA-S. C. Weighted average 1H,15N chemical shift differences upon addition of DNA-L. Dashed lines in (B) and (C) represent no change in intensity and average chemical shift difference, respectively. Spectra were recorded in NMR buffer (containing 150 mM NaCl) at 25°C. All p53:DNA complexes were at a ratio of 4:1, and as four p53 monomers bind 1 DNA, this represents a 1:1 DNA:p53 tetramer complex.
In contrast to DNA-S, binding to the longer DNA-L oligonucleotide results in substantial chemical shift changes for cross peaks associated with residues 369-374 and 381-388 in the CTD (Figure 3), suggesting that these regions are involved in interaction with the additional nucleotides in DNA-L that flank the consensus recognition element. These regions contain most of the positively charged lysine residues within the CTD, consistent with a role in DNA binding, and include acetylation sites (K372/K373/K381/K382) that are critical for p53 function.21,34,56 The chemical shift perturbations observed in these regions upon binding of p53 to DNA-L, but not to DNA-S, demonstrates that the CTD forms contacts outside the p53 recognition element, not with the same 20 base-pair binding site as the p53 DBD. Small chemical shift changes are also observed at K320, a PCAF acetylation site, at the center of a lysine cluster in the NLS.
Binding of the CTD to DNA is enhanced by increasing DNA length
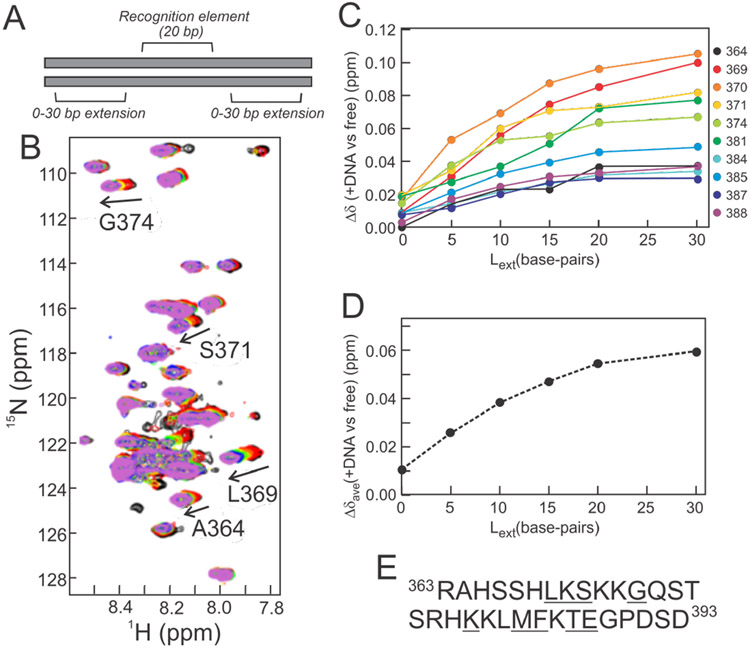
The experiments described in the previous section show clearly that the p53 CTD binds to regions of the DNA that lie outside of the p53 recognition element. However, these experiments are unable to provide information as to how far from the recognition element the interactions occur. CTD interactions were mapped by monitoring chemical shift changes upon binding to DNA constructs that ranged in length from 20 to 80 base-pairs. In each DNA, the central 20 nucleotides were the consensus p53 recognition element present in the DNA-S sequence, with (ATG)n repeats extending from this in both directions. These (ATG)n extensions resulted in ‘overhang’ sequences of 0, 5, 10, 15, 20, and 30 base-pairs for the 20, 30, 40, 50, 60, and 80 nucleotide-long sequences respectively (Figure 4A, Table S1). 1H-15N HSQC spectra were collected for full-length 15N(304-393)-p53 bound to each of the DNA sequences (Figure 4B). Addition of the 20 base-pair DNA results in an HSQC spectrum similar to the 1H-15N TROSY-HSQC spectrum of 15N(304-393)-p53 bound to DNA-S (Figure 3). As the length of the flanking regions increases, the chemical shift perturbations of CTD resonances relative to the unbound p53 tetramer increase proportionally (Figure 4C), but level off beyond ~20 flanking base-pairs (in the 60 and 80 base-pair oligonucleotides) (Figure 4C). Additionally, the CTD cross-peaks are sharp and clearly visible, even when the p53 is bound to the 80 base-pair DNA. These observations suggest that the CTD:DNA interactions are highly dynamic and that the population of bound states increases as the flanking regions are extended to ~20 base-pairs from the p53 recognition element. The gradual increase in chemical shift perturbations with increasing DNA length shows that the CTD does not interact with DNA at a specific distance from the recognition element, but transiently samples multiple sites on the DNA helix.
Figure 4.
DNA extension experiment. A. Schematic representation of DNA used in NMR experiments with 5’ and 3’ extensions consisting of ATG repeats. B. Superposition of 500MHz 1H-15N HSQC spectra of 15N(304-393)-p53 (black) with 20-bp (red), 30-bp (orange), 40-bp (yellow), 50-bp (green), 60-bp (blue), and 80-bp (violet) DNA constructs (sequences in Table S1). Selected resonances displaying readily observable shifts from the free form are labeled. C. Weighted average 1H, 15N chemical shift differences for resonances showing substantial changes in the presence of DNA of various lengths. D. Average chemical shift differences at each DNA extension length. The average was calculated from the Δδavg values for the residues underlined in panel E. E. Amino acid sequence of the p53 CTD (residues 363-393) with residues included in the analysis in C and D underlined (only assigned and well-resolved resonances were used). Spectra were recorded in NMR buffer (containing 150 mM NaCl) at 25°C. All protein concentrations were fixed to 30 μM monomer concentration and all samples used a protein:DNA ratio of 4:1.
DNA binding by the CTD is sequence independent
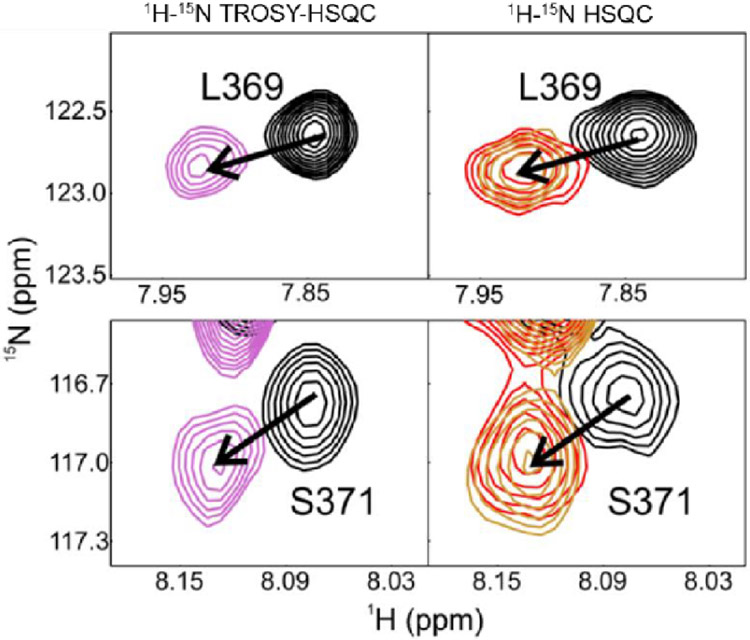
Complexes were formed with three different 60 base-pair DNA sequences. Two of these contained the same 20 base-pair p53 recognition element, but with flanking regions comprised of (ATG)n repeats or a random nucleotide sequence (Table S1, ’60-bp’ and DNA-L, respectively). The other DNA contained a high affinity p21 recognition element flanked on either side by 20 base-pairs of the native p21 sequence. Similar changes in the chemical shifts of CTD resonances were observed upon binding of full-length 15N(304-393)-p53 to each of the DNA sequences (Figure 5), suggesting that the CTD:DNA interactions are sequence independent, in accord with prior reports of non-specific interactions between the CTD and DNA.23
Figure 5.
Selected regions of 1H-15N TROSY-HSQC (left) and 1H-15N HSQC (right) spectra of full-length 15N(304-393)-p53 free (black) and bound to a 60-bp DNA with random nucleotide overhangs (violet, DNA-L sequence), with (ATG)n repeat overhangs (red) and with p21-native sequence overhangs (gold). Differences in cross-peak lineshapes between the left and right spectra are due to differences in experimental setup (TROSY vs non-TROSY) and different isotopic labeling schemes (2H labeling in TROSY-HSQC, 1H in HSQC). Spectra were recorded in NMR buffer (containing 150 mM NaCl) at 25°C, and all samples used a protein-DNA ratio of 4:1.
Multi-site acetylation of TET-CTD labeled p53
Acetylation plays a critical role in stabilization of p53 and modulation of DNA binding.24,34,56 The CTD of p53 is acetylated at multiple sites by CBP/p300.29,56-59 To probe how acetylation affects the CTD:DNA interaction within full-length p53, a CBP construct (residues 1079 to 1855, comprising the bromodomain, PHD, RING, ZZ, and TAZ2 domains) was used to acetylate 15N(304-393)-p53 in vitro. This construct contains both the TAZ2 domain, which recruits p53 through the disordered NTAD, and the acetyltransferase domain. Acetylation by shorter constructs lacking the TAZ2 domain is inefficient (Figure S4). Using 1H-15N SOFAST-HMQC experiments60 we were able to monitor the acetylation reaction in real time (Figure 6A). Acetylation of 15N(304-393)-p53 results in chemical shift changes for several resonances, and the progress of the reaction could be monitored by measuring the loss of peak intensity for resonances corresponding to the unmodified state as a function of time (Figure 6B). Additional chemical shift changes following acetylation were observed outside the CTD, near K305, which suggests this residue is also acetylated (Figure 6C). As acetylation results in the addition of an acetyl group onto the end of the lysine side-chain, the chemical shift difference observed for the backbone amides of the modified and neighboring residues are relatively small.61 Acetylation of K372, K373, and K382 was confirmed by western blot using site-specific antibodies (Figure 6D).
Figure 6.
Acetylation of full-length 15N(304-393)-p53. A. 500 MHz 1H-15N SOFAST HMQC spectra of unmodified 15N(304-393)-p53 (black) and ~2 hours (red), ~6 hours (orange), ~10 hours (green), ~13 hours (blue) and ~17 hours (violet) after mixing with CBP. Cross peaks used to determine rates of acetylation are labeled. Upon acetylation, these resonances decrease in intensity and reappear at new locations (indicated by arrows). Spectra were recorded in NMR buffer (containing 150 mM NaCl) at 25°C. B. Time-resolved intensity changes for resonances reporting on acetylation. Loss of intensity for M384 reports on K382/383 acetylation, G374 on K372/373 acetylation, A307 on K305 acetylation, and the increase in intensity for the acetyl-Lys resonance reports on overall modification. C. 1H, 15N weighted average chemical shift differences Δδavg between free 15N(304-393)-p53 and following acetylation of the indicated residues. D. Western blot confirming acetylation of p53 using acetylated-residue specific antibodies for K372Ac, K373Ac or K382Ac.
Acetylation of the p53 CTD inhibits interactions of the CTD with DNA
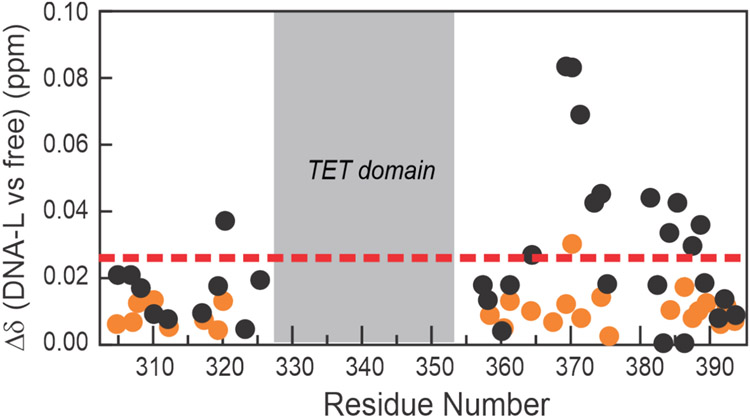
1H-15N HSQC spectra were acquired for the acetylated 15N(304-393)-p53 tetramer in the presence and absence of DNA-L. In contrast to the non-acetylated p53, which exhibits substantial chemical shift changes within the CTD upon addition of DNA-L, spectra of the acetylated protein bound to the same 60 base-pair DNA largely resemble those of the unbound protein (Figure S5). The changes in CTD chemical shifts upon DNA binding are substantially smaller than those for the non-acetylated protein (Figure 7), showing that acetylation of the CTD in the full-length p53 tetramer impairs but does not completely abrogate its interaction with DNA. Acetylation removes the positive charge of the Lys side chain, disrupting electrostatic interactions that are important for DNA binding.
Figure 7.
DNA binding to acetylated p53. 1H and 15N weighted average chemical shift change (Δδavg) upon addition of DNA-L to 15N(304-393)-p53 with (orange circles) and without (black circles) multi-site acetylation. The dashed line represents the average chemical shift change for the unacetylated protein after addition of DNA.
Discussion
The C-terminal domains in the context of full-length p53
The NMR experiments on full-length p53 were carried out with protein concentrations of at least 30 μM, far above the Kd for p53 tetramerization (50 nM).50,62 In 1H-15N TROSY-HSQC spectra of the segmentally 15N/2H labeled full-length p53 (15N(304-393)-p53), backbone amide resonances are not observed for residues Y327 to G356, which correspond to the TET domain boundaries in structures determined previously by both X-ray crystallography and NMR spectroscopy. 14,51-54 While the TET cross-peaks are present in spectra of the tetrameric p53(304-393) peptide, they are broadened in spectra of p53 due to slow tumbling of the TET domain within the full-length tetramer.59
In contrast to the broadened resonances of the TET domain, the narrow linewidths for residues in the NLS and CTD (Figure 2), and also residues in the N-terminal activation domain,63 show that these domains are highly dynamic in the full-length p53 tetramer. Apart from residues near the splice site, the backbone amide cross peaks of NLS and CTD residues occur at very similar 1H and 15N chemical shifts in spectra of full-length 15N(304-393)-p53 and the isolated p53(304-393) peptide. Small chemical shift differences are observed for K369 and K386 (Figure 2B) that might arise from transient interdomain interactions of the CTD within the p53 tetramer. Indeed, weak interactions between the C-terminal region of the CTD and the DBD have been reported previously55 and it has been suggested that the CTD modulates DNA binding by stabilizing interactions between the DBDs within the p53 tetramer.15,24
The observation that NTAD41, NLS, and CTD resonances are narrow in spectra of full-length p53 and are shifted only slightly from their chemical shifts in isolated peptides is inconsistent with an oligomerization model proposed on the basis of low-resolution (13.7 Å) cryo-electron microscopy data, in which the TET adopts a radically different structure compared to its isolated state and forms stable 3-helix bundle with the NTAD.37,64 In the event of a long-lived interaction between these the NTAD and TET domains, similar spectral qualities (i.e. strongly broadened cross-peaks) would be expected for both. Therefore, the sharp resonances of the NTAD, coupled with the extremely broad resonances of the TET, argues strongly against the NTAD:TET interactions proposed by Okorokov et al.37
Probing the CTD:DNA interaction with enhanced resolution
Through its interactions with DNA, the CTD mediates binding of p53 to non-specific DNA sites, enables rapid diffusion on DNA to facilitate the search for cognate target sites, and enhances the specificity for specific p53 recognition elements.15,27,28,65-67 The enhanced resolution of the NMR spectrum of the full-length p53 tetramer afforded by segmental 15N labeling provided new insights into the interaction of the CTD with DNA. Our experiments confirm that interactions between the CTD and DNA are sequence independent; the CTD binds equally well to a random nucleotide sequence, ATG repeats, and genomic DNA flanking the p21 recognition element (Figure 5). Binding occurs through two neighboring regions of the CTD, centered on residues 370 and 385, that contain multiple positively charged lysine residues (Figure 3). These regions have been identified previously in studies focused on binding of isolated CTD peptides to DNA,23,34 and in molecular simulations of full-length p53.65
Interestingly, our data show that the CTD makes only minimal interactions within the 20 base-pair p53 recognition element, but instead undergoes dynamic interactions with flanking nucleotides. Binding of full-length p53 to a 20 base-pair consensus site produces only small changes in backbone amide resonances of the CTD but leads to broadening of cross-peaks corresponding to residues 304-326, with residues closest to the DBD and TET domains the most affected (Figure 3B). This broadening most likely reflects restricted motions of the neighboring DBD and TET domains as the tetramer assembles onto DNA. In contrast to the 20 base-pair DNA, binding to the longer 60 base-pair oligonucleotide leads to changes in chemical shift for several residues within the CTD (Figure 3C). These results suggest that upon binding of the DBDs to the recognition element, the DNA is occluded in such a way as to impair direct interactions with the CTD, which instead binds to flanking regions of the double helix. By systematically extending the length of the flanking sequence, we showed that the CTD:DNA interactions primarily occur over approximately 20 base-pairs (~70Å) on either side of the p53 recognition element (Figure 4). Increasing the length of the flanking regions further, to 30 base-pairs, causes little further change in the chemical shifts. As the CTD resonances remain sharp for all DNA lengths tested, the CTD-DNA interaction must be highly dynamic. Our data are consistent with electron microscopy and single molecule FRET studies which show that the p53 tetramer can adopt a closed conformation on DNA in which the CTD and DBD approach more closely than in the unbound state.66 They are also consistent with long time scale molecular dynamics simulations of full-length p53 bound to p21 and Puma promoter sequences, in which K370, K372, K373, R379, K381, K382, and K386, located in the regions of the CTD that exhibit above average chemical shift changes (Figure 3), interact dynamically with DNA backbone atoms flanking the cognate recognition elements.65
Overall, our work supports a model in which the four DBDs anchor the p53 tetramer to the recognition element through base-specific contacts while positively charged residues in the CTD make secondary, non-specific contacts with flanking DNA. Neutralization of the positive charge by acetylation of K372, K373, and K382 impairs, but does not completely abrogate, the interaction of the CTD with the flanking DNA (Figure 7). Inhibition of binding by acetylation of short peptides derived from the CTD has been reported previously;34 however, it is noteworthy that even in the full-length protein, where binding of the DBD to a cognate recognition element anchors p53 tightly to the DNA and thus brings the CTD close to the DNA, acetylation still has a strong inhibitory effect on CTD interactions with DNA.
As backbone amide resonances of the TET domain (residues 326-356) are broadened beyond detection in the full-length p53 tetramer, we are unable to comment on potential interactions of this domain with DNA. While the TET domain is expected to be positioned on the opposite side of the DNA relative to the four DBDs,65,66,68 the nature of any potential TET:DNA interactions remains uncharacterized. The larger than average chemical shift perturbation experienced by K320 upon binding to DNA (Figure 3), which is substantially diminished by acetylation of K305 and lysine residues in the CTD (Figure 7), likely arises from transient interactions of the DBD-TET linker with the DNA backbone. Acetylation of K320 modulates the activity of p53 and its interactions with DNA.31 and non-specific contacts between the DBD-TET linker and DNA have been identified previously in single molecule fluorescence experiments.69
Role of disordered N- and C-terminal regions in p53 target site search
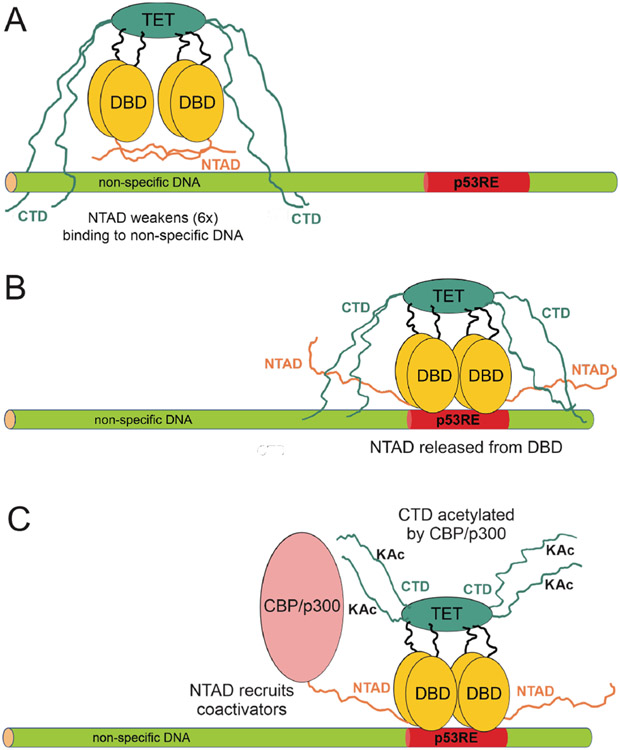
The CTD plays an essential role in facilitating the search for p53 target sites by promoting rapid one-dimensional sliding along the DNA helix and fast intersegmental transfer.28,67,70,71 In search mode, the DBDs interact with the DNA via repetitive dissociation and association steps, while the CTD loosely tethers the p53 to the DNA through weak and highly dynamic contacts with the phosphate backbone. A schematic model summarizing the role of the disordered NTAD and CTD regions in target site search is shown in Figure 8. The CTD makes only minimal contact with the DNA within or immediately adjacent to the 20-basepair site occupied by the DBD, but makes optimal contacts over a span of 20-30 base pairs away from the DBD binding site (Figure 4). Binding of the DBDs to non-specific DNA, but not to cognate recognition elements, is autoinhibited by dynamic intramolecular interactions between the N-terminal activation domain (NTAD) and the DNA binding surface (Figure 8A).41,72,73 The NTAD thus functions synergistically with the CTD to facilitate diffusion by weakening binding of the DBDs to non-specific DNA sites. Once a cognate sequence is encountered during target search, favorable interactions between the DBD and the DNA would displace the NTAD and enhance the specificity for p53 recognition elements in the promoters of p53-mediated genes (Figure 8B). Subsequent acetylation of lysine residues in the CTD by recruitment of CBP or p300 to the promoter would impair competing interactions between the CTD and flanking DNA sequences that could cause p53 to diffuse away from the recognition element (Figure 8C).
Figure 8.
Schematic representation of the interaction of p53 with DNA. A. Interaction with non-specific DNA. The NTAD interacts with the DBD,41 lowering the affinity of p53 for the DNA. Only two NTADs are shown for clarity. The CTD makes contact with DNA outside the immediate vicinity of the DBD, maintaining proximity to the DNA while allowing sliding and hopping for target site search. B. When p53 encounters a recognition element, the NTAD is competed off the DBD and acts to recruit transcriptional coactivators such as CBP/p300. The CTD continues to interact with the DNA flanking the recognition element. C. The CTD is acetylated by the coactivator, weakening its interactions with flanking DNA and thereby removing competing interactions that could cause p53 to diffuse away from the cognate recognition site.
Indeed, single molecule fluorescence studies in living cells show that acetylation of the CTD stabilizes p53 binding by increasing the residence time at cognate sites.74 Acetylation of the p53 CTD has been linked to differential gene activation.15,24,31,56 Specifically, acetylation at K372/K373 is required for activation of lower affinity p53 promoters that regulate pro-apoptotic genes.31 Our data show that within full-length p53, the dominant CTD interactions with DNA are mediated by this region and are impaired by lysine acetylation (Figure 7). By inhibiting the CTD:DNA interaction, acetylation at K372 and K373 would increase the lifetime of p53:DNA complexes with weak apoptotic promoters, leading to their activation.74
Supplementary Material
Acknowledgments
We thank Gerard Kroon for help with NMR experiments and Euvel Manlapaz for technical assistance.
Funding
This work was supported by grants GM075995 and CA096865 (P.E.W.) and GM131693 (H.J.D.) from the National Institutes of Health and by the Skaggs Institute for Chemical Biology.
Footnotes
References
- [1].Levine AJ, and Oren M (2009) The first 30 years of p53: growing ever more complex, Nat. Rev. Cancer 9, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Joerger AC, and Fersht AR (2010) The tumor suppressor p53: from structures to drug discovery, Cold Spring Harb. Persp. Biol 2, a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Joerger AC, and Fersht AR (2008) Structural biology of the tumor suppressor p53, Ann. Rev. Biochem 77, 557–582. [DOI] [PubMed] [Google Scholar]
- [4].Bode AM, and Dong Z (2004) Post-translational modification of p53 in tumorigenesis, Nat. Rev. Cancer 4, 793–805. [DOI] [PubMed] [Google Scholar]
- [5].Lavin MF, and Gueven N (2006) The complexity of p53 stabilization and activation, Cell Death Differ. 13, 941–950. [DOI] [PubMed] [Google Scholar]
- [6].Dai C, and Gu W (2010) p53 post-translational modification: deregulated in tumorigenesis, Trends Mol. Med 16, 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kruse JP, and Gu W (2009) Modes of p53 regulation, Cell 137, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Murray-Zmijewski F, Slee EA, and Lu X (2008) A complex barcode underlies the heterogeneous response of p53 to stress, Nat. Rev. Mol. Cell Biol 9, 702–712. [DOI] [PubMed] [Google Scholar]
- [9].Shaulsky G, Goldfinger N, Ben-Ze'ev A, and Rotter V (1990) Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis, Mol Cell Biol 10, 6565–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dang CV, and Lee WM (1989) Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins, J. Biol. Chem 264, 18019–18023. [PubMed] [Google Scholar]
- [11].Addison C, Jenkins JR, and Sturzbecher HW (1990) The p53 nuclear localisation signal is structurally linked to a p34cdc2 kinase motif, Oncogene 5, 423–426. [PubMed] [Google Scholar]
- [12].Sturzbecher HW, Brain R, Addison C, Rudge K, Remm M, Grimaldi M, Keenan E, and Jenkins JR (1992) A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization, Oncogene 7, 1513–1523. [PubMed] [Google Scholar]
- [13].Sakamoto H, Lewis MS, Kodama H, Appella E, and Sakaguchi K (1994) Specific sequences from the carboxyl terminus of human p53 gene product form anti-parallel tetramers in solution, Proc Natl Acad Sci U S A 91, 8974–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee W, Harvey TS, Yin Y, Yau P, Litchfield D, and Arrowsmith CH (1994) Solution structure of the tetrameric minimum transforming domain of p53, Nature Struct. Biol 1, 877–890. [DOI] [PubMed] [Google Scholar]
- [15].Laptenko O, Tong DR, Manfredi J, and Prives C (2016) The tail that wags the dog: how the disordered C-terminal domain controls the transcriptional activities of the p53 tumor-suppressor protein, Trends Biochem. Sci 41, 1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rustandi RR, Baldisseri DM, and Weber DJ (2000) Structure of the negative regulatory domain of p53 bound to S100B(bb), Nature Struct. Biol 7, 570–574. [DOI] [PubMed] [Google Scholar]
- [17].Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik L, Ronai Z, and Zhou MM (2004) Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation, Mol. Cell 13, 251–263. [DOI] [PubMed] [Google Scholar]
- [18].Tong Q, Mazur SJ, Rincon-Arano H, Rothbart SB, Kuznetsov DM, Cui G, Liu WH, Gete Y, Klein BJ, Jenkins L, Mer G, Kutateladze AG, Strahl BD, Groudine M, Appella E, and Kutateladze TG (2015) An acetyl-methyl switch drives a conformational change in p53, Structure 23, 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Boehme KA, and Blattner C (2009) Regulation of p53 - insights into a complex process, CRC Crit. Rev. Biochem. Mol. Biol 44, 367–392. [DOI] [PubMed] [Google Scholar]
- [20].Kruse JP, and Gu W (2008) SnapShot: p53 posttranslational modifications, Cell 133, 930–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gu B, and Zhu W-G (2012) Surf the post-translational modification network of p53 regulation, Int. J. Biol. Sci 8, 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brooks CL, and Gu W (2011) p53 regulation by ubiquitin, FEBS Lett. 585, 2803–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Weinberg RL, Freund SM, Veprintsev DB, Bycroft M, and Fersht AR (2004) Regulation of DNA binding of p53 by its C-terminal domain, J. Mol. Biol 342, 801–811. [DOI] [PubMed] [Google Scholar]
- [24].Laptenko O, Shiff I, Freed-Pastor W, Zupnick A, Mattia M, Freulich E, Shamir I, Kadouri N, Kahan T, Manfredi J, Simon I, and Prives C (2015) The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain, Mol. Cell 57, 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hamard PJ, Lukin DJ, and Manfredi JJ (2012) p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes, J. Biol. Chem 287, 22397–22407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim H, Kim K, Choi J, Heo K, Baek HJ, Roeder RG, and An W (2012) p53 requires an intact C-terminal domain for DNA binding and transactivation, J. Mol. Biol 415, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].McKinney K, Mattia M, Gottifredi V, and Prives C (2004) p53 linear diffusion along DNA requires its C terminus, Mol. Cell 16, 413–424. [DOI] [PubMed] [Google Scholar]
- [28].Tafvizi A, Huang F, Fersht AR, Mirny LA, and van Oijen AM (2011) A single-molecule characterization of p53 search on DNA, Proc. Natl. Acad. Sci. USA 108, 563–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gu W, and Roeder RG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain, Cell 90, 595–606. [DOI] [PubMed] [Google Scholar]
- [30].Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, and Appella E (1998) DNA damage activates p53 through a phosphorylation-acetylation cascade, Genes Devel. 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Knights CD, Catania J, Giovanni SD, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, and Avantaggiati ML (2006) Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate, J. Cell. Biol 173, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, and Berger SL (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage, Mol. Cell. Biol 19, 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, and Gu W (2004) Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo, Proc. Natl. Acad. Sci. USA 101, 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Friedler A, Veprintsev DB, Freund SMV, von Glos KI, and Fersht AR (2005) Modulation of binding of DNA to the C-terminal domain of p53 by acetylation, Structure 13, 629–636. [DOI] [PubMed] [Google Scholar]
- [35].Retzlaff M, Rohrberg J, Küpper NJ, Lagleder S, Bepperling A, Manzenrieder F, Peschek J, Kessler H, and Buchner J (2013) The regulatory domain stabilizes the p53 tetramer by intersubunit contacts with the DNA binding domain, J. Mol. Biol 425, 144–155. [DOI] [PubMed] [Google Scholar]
- [36].D'Abramo M, Besker N, Desideri A, Levine AJ, Melino G, and Chillemi G (2016) The p53 tetramer shows an induced-fit interaction of the C-terminal domain with the DNA-binding domain, Oncogene 35, 3272–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Okorokov AL, Sherman MB, Plisson C, Grinkevich V, Sigmundsson K, Selivanova G, Milner J, and Orlova EV (2006) The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity, EMBO J. 25, 5191–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Veprintsev DB, Freund SMV, Andreeva A, Rutledge SE, Tidow H, Cañadillas JMP, Blair CM, and Fersht AR (2006) Core domain interactions in full-length p53 in solution, Proc. Natl. Acad. Sci. USA 103, 2115–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bista M, Freund SM, and Fersht AR (2012) Domain-domain interactions in full-length p53 and a specific DNA complex probed by methyl NMR spectroscopy, Proc. Natl. Acad. Sci. USA 109, 15752–15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nikolova PV, Henckel J, Lane DP, and Fersht AR (1998) Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability, Proc. Natl. Acad. Sci. USA 95, 14675–14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Krois AS, Dyson HJ, and Wright PE (2018) Long-range regulation of p53 DNA binding by its intrinsically disordered N-terminal transactivation domain, Proc. Natl. Acad. Sci. USA 115, E11302–E11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Park S, Stanfield RL, Martinez-Yamout MA, Dyson HJ, Wilson IA, and Wright PE (2017) Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation, Proc. Natl. Acad. Sci. USA 114, E5335–E5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Delaglio F, Grzesiek S, Vuister GW, Guang Z, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- [44].Johnson BA, and Blevins RA (1994) NMRView: A computer program for the visualization and analysis of NMR data, J. Biomol. NMR 4, 603–614. [DOI] [PubMed] [Google Scholar]
- [45].Pervushin K, Riek R, Wider G, and Wüthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution, Proc. Natl. Acad. Sci. USA 94, 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wittekind M, and Mueller L (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins, J. Magn. Res 101, 201–205. [Google Scholar]
- [47].Iwai H, Züger S, Jin J, and Tam PH (2006) Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme, FEBS Lett. 580, 1853–1858. [DOI] [PubMed] [Google Scholar]
- [48].Aranko AS, Züger S, Buchinger E, and Iwai H (2009) In vivo and in vitro protein ligation by naturally occurring and engineered split DnaE inteins, PLoS One 4, e5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Weinberg RL, Veprintsev DB, and Fersht AR (2004) Cooperative binding of tetrameric p53 to DNA, J. Mol. Biol 341, 1145–1159. [DOI] [PubMed] [Google Scholar]
- [50].Rajagopalan S, Huang F, and Fersht AR (2011) Single-molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53, Nucl. Acids Res 39, 2294–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jeffrey PD, Gorina S, and Pavletich NP (1995) Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms, Science 267, 1498–1502. [DOI] [PubMed] [Google Scholar]
- [52].Mittl PR, Chene P, and Grutter MG (1998) Crystallization and structure solution of p53 (residues 326-356) by molecular replacement using an NMR model as template, Acta Crystallogr D Biol Crystallogr 54, 86–89. [DOI] [PubMed] [Google Scholar]
- [53].Clore GM, Ernst J, Clubb R, Omichinski JG, Kennedy WMP, Sakaguchi K, Appella E, and Gronenborn AM (1995) Refined solution structure of the oligomerization domain of the tumour suppressor p53, Nature Struct. Biol 2, 321–333. [DOI] [PubMed] [Google Scholar]
- [54].Emamzadah S, Tropia L, and Halazonetis TD (2011) Crystal structure of a multidomain human p53 tetramer bound to the natural CDKN1A (p21) p53-response element, Mol. Cancer Res 9, 1493–1499. [DOI] [PubMed] [Google Scholar]
- [55].Clubb RT, Omichinski JG, Sakaguchi K, Appella E, Gronenborn AM, and Clore GM (1995) Backbone dynamics of the oligomerization domain of p53 determined from 15N NMR relaxation measurements, Protein Sci. 4, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Reed S, and Quelle D (2014) p53 acetylation: regulation and consequences, Cancers 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine A, and Kelly K (1997) Recruitment of p300/CBP in p53-dependent signal pathways, Cell 89, 1175–1184. [DOI] [PubMed] [Google Scholar]
- [58].Tang Y, Zhao W, Chen Y, Zhao Y, and Gu W (2008) Acetylation is indispensable for p53 activation, Cell 133, 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wright DG, Marchal C, Hoang K, Ankney JA, Nguyen ST, Rushing AW, Polakowski N, Miotto B, and Lemasson I (2015) Human T-cell leukemia virus type-1-encoded protein HBZ represses p53 function by inhibiting the acetyltransferase activity of p300/CBP and HBO1, Oncotarget 7, 1687–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Schanda P, Kupce E, and Brutscher B (2005) SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds, J. Biomol. NMR 33, 199–211. [DOI] [PubMed] [Google Scholar]
- [61].Lee JH, Ying J, and Bax A (2016) Nuclear Magnetic Resonance Observation of α-Synuclein Membrane Interaction by Monitoring the Acetylation Reactivity of Its Lysine Side Chains, Biochemistry 55, 4949–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Brandt T, Petrovich M, Joerger AC, and Veprintsev DB (2009) Conservation of DNA-binding specificity and oligomerisation properties within the p53 family, BMC Genom. 10, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Krois AS, Ferreon JC, Martinez-Yamout MA, Dyson HJ, and Wright PE (2016) Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein, Proc. Natl. Acad. Sci. USA 113, E1853–E1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Aramayo R, Sherman MB, Brownless K, Lurz R, Okorokov AL, and Orlova EV (2011) Quaternary structure of the specific p53–DNA complex reveals the mechanism of p53 mutant dominance, Nucl. Acids Res 39, 8960–8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Demir O, Ieong PU, and Amaro RE (2017) Full-length p53 tetramer bound to DNA and its quaternary dynamics, Oncogene 36, 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Melero R, Rajagopalan S, Lázaro M, Joerger AC, Brandt T, Veprintsev DB, Lasso G, Gil D, Scheres SHW, Carazo JM, Fersht AR, and Valle M (2011) Electron microscopy studies on the quaternary structure of p53 reveal different binding modes for p53 tetramers in complex with DNA, Proc. Natl. Acad. Sci. USA 108, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Terakawa T, Kenzaki H, and Takada S (2012) p53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains, J. Am. Chem. Soc 134, 14555–14562. [DOI] [PubMed] [Google Scholar]
- [68].Petty TJ, Emamzadah S, Costantino L, Petkova I, Stavridi ES, Saven JG, Vauthey E, and Halazonetis TD (2011) An induced fit mechanism regulates p53 DNA binding kinetics to confer sequence specificity, EMBO J. 30, 2167–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Subekti DRG, Murata A, Itoh Y, Fukuchi S, Takahashi H, Kanbayashi S, Takahashi S, and Kamagata K (2017) The disordered linker in p53 participates in nonspecific binding to and one-dimensional sliding along DNA revealed by single-molecule fluorescence measurements, Biochemistry 56, 4134–4144. [DOI] [PubMed] [Google Scholar]
- [70].Kamagata K, Itoh Y, and Subekti DRG (2020) How p53 molecules solve the target DNA search problem: A review, Int. J. Mol. Sci 21, 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Subekti DRG, Murata A, Itoh Y, Takahashi S, and Kamagata K (2020) Transient binding and jumping dynamics of p53 along DNA revealed by sub-millisecond resolved single-molecule fluorescence tracking, Sci. Rep 10, 13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].He F, Borcherds W, Song T, Wei X, Das M, Chen L, Daughdrill GW, and Chen J (2019) Interaction between p53 N terminus and core domain regulates specific and nonspecific DNA binding, Proc. Natl. Acad. Sci. USA 116, 8859–8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sun X, Dyson HJ, and Wright PE (2021) A phosphorylation-dependent switch in the disordered p53 transactivation domain regulates DNA binding, Proc. Natl. Acad. Sci. USA 118, e2021456118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Loffreda A, Jacchetti E, Antunes S, Rainone P, Daniele T, Morisaki T, Bianchi ME, Tacchetti C, and Mazza D (2017) Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity, Nat. Comm 8, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.