Abstract
Data suggesting that fecal indicating bacteria may persist and/or regrow in sand has raised concerns that fecal indicators may become uncoupled from sources of human fecal pollution. To investigate this possibility, wet and dry beach sand, beach water, riverine water, canal water, and raw sewage samples were screened by PCR for certain pathogenic microbes and molecular markers of human fecal pollution. The targets included in this study were human specific Bacteroides (HF8 marker), human-specific enterococci (esp gene), Staphylococcus aureus, Escherichia coli 0157:H7, Campylobacter jejuni, and adenovirus. Sewage samples were also tested for Salmonella species. The results were compared to concentrations of enterococci, Escherichia coli, and Bacteroides species, as determined by membrane filtration methods. Molecular analysis yielded positive results for human specific Bacteroides, and S. aureus, in samples of raw sewage. Two of the environmental samples were positive for human specific Bacteroides and one was positive for S. aureus. The PCR screen was negative for other samples and targets, despite exceedance of EPA single sample guidelines for recreational waters on several of the sample dates (5/11 dates). However, estimates of the number of cells delivered to the PCR reaction suggested that few of the samples met the detection limit of the PCR reaction due to a variety of factors. The analysis indicated a need to improve nucleic acid processing in order to enable better delivery of DNA to downstream molecular methods.
Introduction
The United States Environmental Protection Agency (EPA, 2003) and the Food and Drug Administration (FDA/ISSC, 2003) use fecal indicating bacteria to regulate the closure of recreational and shell fish harvesting waters. Fecal indicating bacteria are not necessarily human pathogens; instead, they are bacteria whose presence is supposed to indicate the existence of sewage-associated pathogens. Fecal indicating bacteria are employed because they are abundant in comparison to the actual pathogens that cause waterborne illness. In addition, measuring all potential pathogens in a given water sample would be technically and financially unfeasible. Therefore, fecal indicators are typically used to monitor and manage coastal water quality. In practice, a fecal indicator should be: (1) a member of the intestinal flora of warm blooded animals, (2) non-pathogenic, (3) present when pathogens are present and absent when they are not, (4) present in greater numbers than the pathogen, (5) unable to multiply in the environment, (6) at least equally resistant as the pathogen to environmental factors and to disinfection in water and wastewater treatment plants, and (7) detectable by means of rapid, easy, and inexpensive protocols (Bitton, 2005; National Research Council, 2004).
The risk of gastrointestinal illness is correlated to the concentration of fecal indicating bacteria when waters receive point sources of human fecal pollution (Cabelli et al., 1979; Dufour, 1984; Wade et al., 2003), and such illnesses cause negative economic impacts (Dwight et al. 2005). However, there is question whether concentrations of fecal indicators reliably predict the presence of human pathogens in regions that do not have point sources of human fecal contamination. For example, both E. coli and enterococci have been found in areas without apparent sewage contamination (Carillo et al., 1985; Rivera et al., 1988). Furthermore, there is evidence that fecal indicating bacteria may persist outside of their hosts (Solo-Gabriele et al., 2000; Wright, 1989). If fecal indicators are able to multiply in the environment, the theory of fecal indicators (described above) would be violated.
The role that sand plays in the survival of fecal indicating bacteria is an area of growing interest (Gerba and McLeod, 1976; Hood and Ness, 1982; Davies et al., 1995; Lee et al., 2006). Research suggests that sand may act a bacterial reservoir, providing a source of fecal indicators to adjacent waters (Yamahara et al. 2007; Whitman and Nevers, 2003; Craig et al., 2002; Goyal et al., 1977). However, only fecal indicator concentrations in water are currently monitored; therefore, there is growing concern that public exposure to human pathogens may be underestimated (Clean Beaches Council, 2005).
An underestimation of risk might occur if high concentrations of fecal indicators in sand signified the presence of pathogenic microbes. Conversely, an overestimation of risk might occur if persistence or growth occurred only for the indicators but not for the pathogens. Furthermore, it is possible that risk is estimated incorrectly by not considering the concentration of nonfecal pathogens such as Staphylococcus aureus.
To test the hypothesis that fecal indicating concentrations can be uncoupled from the presence of pathogenic organisms, samples from a variety of coastal environments were subjected to a suite of molecular assays in conjunction with traditional methods for detecting fecal indicting bacteria. The assays were designed to detect several human pathogens (Staphylococcus aureus, Campylobacter jejuni, E. coli O157:H7, Salmonella species, and adenovirus), and two markers of human fecal pollution (the esp gene for human specific Enterococcus faecium and the HF8 cluster for human specific Bacteroides species).
Materials and Methods
Site Description
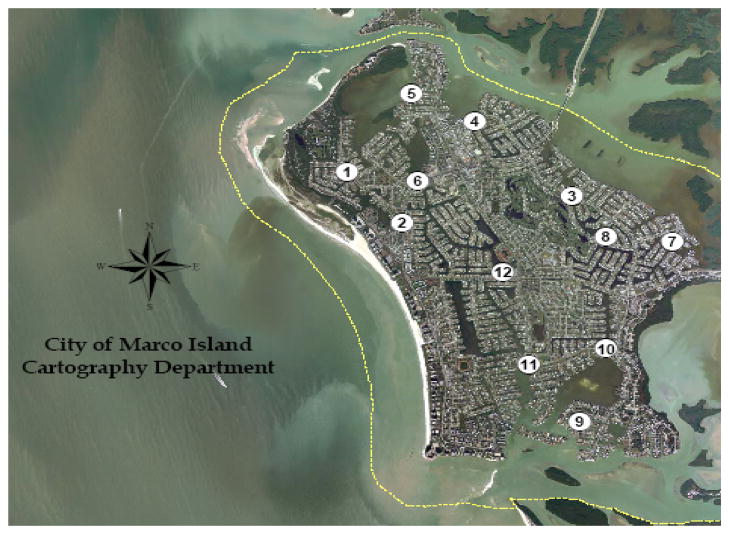
Samples were taken from several sites in southern Florida, USA, including: Hobie Beach, Marco Island, Wagner Creek, and the Virginia Key Waste Water Treatment Plant. Hobie Beach is a relatively shallow, semi-enclosed beach located in the southern portion of Biscayne Bay, Miami (Shibata et al., 2004). The Virginia Key Waste Water Treatment Plant is located near Hobie Beach. The outfall and the waters adjacent to Hobie Beach are not thought to have a significant hydrological connection; therefore, Hobie Beach is considered free of point sources of human fecal pollution. Wagner Creek is a tributary to the Miami River, which is an urbanized, tidally influenced river site that is located downstream from flood control gates. Marco Island (Figure 1) is located on the Gulf of Mexico in southwest Florida. Samples were collected from the following stations: Collier Bridge (N 25 56.726′, W 081 44.448′), Barfield Bridge (N 25 57.694′, W 081 43.345′), Perrine (N 25 57.943′, W 081 43.886′) JH Park (N 25 57.121′, W 081 43.830′) Hollyhock (N 25 56.607′, W 081 41.558′), Hummingbird (N 25 56.544′, W 081 42.189′), HC Center (N 25 56.263′, W 081 43.083′).
Figure 1.
Satellite image showing water quality collection sites for the city of Marco Island, Florida. For this study, samples were collected from the following stations: 2) Collier Bridge, 4) Barfield Bridge, 5) Perrine, 6) JH Park, 7) Hollyhock, 8) Hummingbird, 12) HC Center. Map provided courtesy of City of Marco Island, Florida Cartography Department.
Sample Collection
Samples of sand or water were collected in sterile containers between June 2004 and June 2007. Sand was collected from Hobie Beach above the high water mark (“dry sand”) or from the surf zone at knee-deep depth (“wet sand”). All samples were transported on ice and kept cool until processed within 6 hours of collection. Raw sewage samples were collected on 17 September 2005 from the liquid sewage of the primary settling tank from the Miami-Dade Water and Sewer wastewater treatment plant on Virginia Key, Miami, Florida.
Sample Processing and Enumeration
Samples for bacterial culture analysis were filtered onto 47 mm, 0.45 μm, cellulose nitrate membrane filters (Whatman) and rinsed with 20 ml phosphate buffered saline (PBS), according to standard membrane filtration protocols (EPA, 2002). Typically 3 to 5 dilutions were plated and values for plates with countable colonies (~6 to 100) were averaged to obtain the bacterial density for that sample. Samples were processed within 6 hours of collection. The filters were placed on selective media and incubated as outlined in Table 1. Anaerobic conditions for selection of Bacteroides were generated using the Gas Pak EZ-Anaerobe Container System with GasPak indicators (Becton Dickenson).
Table 1.
Summary of membrane filtration protocols used to enumerate bacteria in this study
| Target | Growth medium | Incubation | Target colony description | Reference |
|---|---|---|---|---|
| E. coli | Modified mTEC | 35 °C for 2 hr, then 44.5 °C for 22 hr | Red or magenta color | EPA method 1603 (EPA 2002) |
| Enterococcus | mEI | 41 °C for 24 hr | Colonies with blue halo (regardless of colony color) | EPA method 1600 (EPA 2002) |
| Fecal coliform | mFC | 44.5 °C for 24 hr | Blue colonies | Standard Method 9222D (American Water Works Association, 1999) |
| Bacteroides | BBE (Bacteroides Bile Esculin Agar) | Anaerobic, 35 °C for 22 – 48 hr | Brown or black colonies surrounded by a brown zone in the medium | Anaerobe Systems package insert |
| Bacteroides | BVSA (Bacteroides Vulgatus Selective Agar) | Anaerobic, 35 °C for 22 – 48 h | Brown or black colonies surrounded by a brown zone in the medium | Anaerobe Systems package insert |
| Total coliform | M-Endo-LES | 35 °C for 24h | Red colony with golden metallic sheen | Standard Method 9222B (American Water Works Association, 1999) |
Samples for nucleic acid analysis were filtered onto 47 mm, 0.2 μm, hydrophilic polyethersulfone membrane filters (Supor-200, Pall). Filters were placed in Analyslide® petri dishes (Pall) and frozen at −80 °C until used for DNA extraction.
Membrane filtration for sand samples was achieved by first vigorously hand shaking 2 g of sand into 80 ml of PBS for 2 min (Baums et al. 2007). This solution of sand and dislodged particles was vacuum filtered through a sterile, 30 μm, 47 mm nylon net filter (Millipore). Two additional 10 ml rinses with PBS were used to remove any remaining sand from the shaking container. This procedure was repeated until a sufficient volume of “sand water” was generated to satisfy the membrane filtration needs for that site. The sample was homogenized by hand mixing prior to filtration.
The water content of the sand was determined by weighing sand aliquots before and after overnight drying at 110°C. Concentrations of bacteria were calculated in terms of colony forming units (CFU) per 100 ml of water or CFU per gram dry sand. To estimate the number of bacterial cells available for DNA extraction, the CFU values were multiplied by the amount of sand processed onto the Supor-200 membrane filters used for DNA extraction.
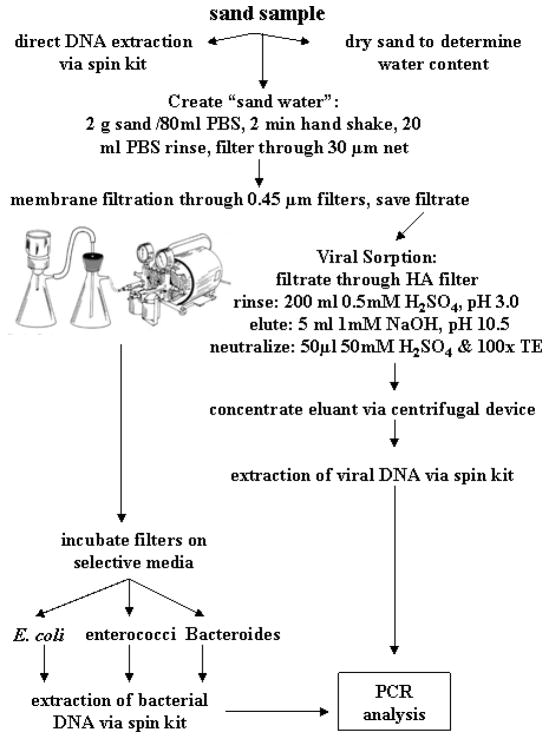
Viral analysis was performed on sand samples collected 1 February 2006 and on water samples collected on 14 March 2006 and 19 April 2007, as outlined in Figure 2. In these cases, samples for bacterial culture analysis were filtered through Durapore HV (Millipore HVLP04700) or through cellulose nitrate filters (Whatman 7141104) (47 mm, 0.45 μm). The filtrate was saved and combined and then filtered through a 90 mm, 0.45 μm HA filter (Millipore HAWP04700) using a custom-made filter holder (courtesy of Dr. H. Solo-Gabriele, University of Miami). The filter was rinsed with 200 ml of 0.5 mM H2SO4, pH 3.0. Elution was achieved with 5 ml of 1 mM NaOH, pH 10.5 (Katayama et al. 2002), and the eluant was neutralized with 50 μl of 50 mM H2SO4 and 100× TE buffer.
Figure 2.
Schematic of sample processing to perform PCR analyses on sand for bacterial and viral targets.
DNA Extraction
DNA was obtained from sand using the FastDNA Spin Kit for Soil (Q-BIOgene). Either the sand or the membrane filter used to process the sand was placed into the microcentrifuge tubes supplied by the kit. DNA from membrane filters used to process water or raw sewage was obtained either with the FastDNA Spin Kit for Soil or the FastDNA Spin Kit using the plant protocol (Q-BIOgene). In addition, some membrane filters were incubated on growth media to allow colony formation (Table 1). The goal was to filter enough water to achieve confluent growth; however, this was not achieved in all cases. Growth filters were placed into 50 ml conical tubes and the cells were dislodged by shaking the filters at 225 rpm for 1 hr in 25 ml 1× PBS. The solution was pelleted by centrifugation at 3000×g for 15 min. The pellets were resuspended in 600 μl of the lysis solution from the Wizard DNA Spin Kit (Promega) and the protocol for Gram (+) or Gram (−) bacteria was followed, depending on the growth medium.
Sources of Control DNA
A plasmid containing cloned DNA from the human-specific esp gene of E. faecium (Scott et al., 2005) was provided by Dr. Joan Rose (Michigan State University). A plasmid containing the PCR target for adenovirus (He and Jiang, 2005) was supplied by Dr. Sunny Jiang (University of California at Irvine). The human-specific HF8 cluster of Bacteroides (Bernhard and Field, 2000) was PCR amplified from human fecal DNA and cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA). DNA from E. coli (ATCC #25922) and E. faecalis (ATCC #29212) was isolated from cultures using the FastDNA SPIN Kit for Soil. DNA from Campylobacter jejuni, Salmonella typhi, and E. coli 0157:H7 were provided by Dr. Nick Cirino (New York State Department of Health). DNA from Staphylococcus aureus was obtained from ATCC (#700699D).
Target Amplification and Screening
The PCR primer sequences, thermal cycling conditions, amplicon sizes, and references for the procedures are given in Table 2. The PCR reactions for pathogens contained 5μl of 10x DyNAzyme II buffer (contains 1.5 mM MgCl2), 1.25 μl of 0.2 mM dNTP, primers (concentrations given in Table 2), 0.75 μl of DyNAzyme II DNA polymerase (Finnzymes), 1.50 μl BSA (10 mg/ml), 5 μl of DNA sample, and nuclease free water for a total reaction volume of 50 μl.
Table 2.
Summary of PCR and qPCR conditions used in this study
| Target | Gene | Primer labelb-name-sequence, 5′→3′ (μM per PCR reaction) | Cyclinga | based on Reference |
|---|---|---|---|---|
| Enterococcus | 23S rRNA | ECST748F-AGAAATTCCAAACGAACTTG (0.9) ENC854R-CAGTGGTCTACCTCCATCATT (0.3) |
94°C 30 s; 60°C 30 s; 72°C 30 s; 30 cycles | Haugland et al., 2005 |
| Human-specific Enterococcus faecium | esp | Biotin-espF-TATGAAAGCACAAGTT (0.3) FITC-espR-ACGTCGAAAGTTCGATTTCC (0.3) |
94°C 1 min; 58°C 1 min; 72°C 1 min; 40 cycles | Scott et al., 2005 |
| Human-specific Bacteroides | 16S rRNA | Biotin-HF183F-ATCATGAGTTCACATGTCCG (0.4) FITC-Bac708R-CAATCGGAGTTCTTCGTG (0.4) |
94°C 30 s; 59°C 30 s; 72°C 30 s; 40 cycles | Bernhard and Field, 2000 |
| Campylobacter jejuni | hipO | Biotin-CjF1-TGCTAGTGAGGTTGCAAAAGAATT (0.5) FITC-CjR1-TCATTTCGCAAAAAAATCCAAA (0.5) |
94°C 30 s; 60°C 30 s; 72°C 30 s; 40 cycles | LaGier et al., 2004 |
| Salmonella spp. | ipaB | Biotin-IpaBF-GGACTTTTTAAAAGCGGCGG (0.3) FITC-IpaBR-GCCTCTCCCAGAGCCGTCTGG (0.3) |
94°C 1 min; 62°C 1 min; 72°C 1 min; 35 cycles | Kong et al., 2002 |
| E. coli 0157:H7 | rfb | Biotin-0157PF8-CGTGATGATGTTGAGTTG (1.0) FITC-0157PR8-AGATTGGTTGGCATTACTG (1.0) |
94°C 30 s; 55°C 30 s; 72°C 30 s; 40 cycles | Maurer et al., 1999 |
| Human adenovirus | Hexon | Biotin-AD2F-CCCTGGTAKCCRATRTTGTA (0.3) FITC-AD3R-GACTCYTCWGTSAGYGGCC (0.3) |
94°C 30 s; 60°C 30 s; 72°C 30 s; 40 cycles | He and Jiang, 2005 |
| Staphylococcus aureus | clfA | Biotin-clfAF-GCAAAATCCAGCACAACAGGAAACGA (0.1) FITC-clfAR-CTTGATCTCCAGCCATAATTGGTGG (0.1) |
94°C 1 min; 55°C 1 min; 72°C 1 min; 40 cycles | Mason et al., 2001 |
In all cases, the initial heat denaturation step was 94°C for 10 min and the final extension step was 70°C for 8 min;
Biotin and FITC labels were included because the PCR product also was used for other projects that required these labels. FITC = fluorescein.
For the PCR of Salmonella, each 50 μl PCR reaction also contained 2% formamide (v/v) and an additional 1U of DNA polymerase. Positive and negative controls contained all of the PCR reagents but with 5 μl of isolated genomic DNA or nuclease free water, respectively.
DNA was amplified by standard PCR with an Eppendorf Mastercycler and PCR products were visualized using standard gel electrophoresis with 1% agarose gels (w/v). All of the samples were first run with 5 μl of sample. Negative samples were tested for PCR inhibition by adding 1 μl (10 ng) of the positive control genomic DNA to the 5 μl sample being tested and reanalyzed to determine possible inhibition. If grossly inhibited, the expected band of a specific amplicon size would not be seen or would be dim relative to the positive control. Inhibited samples were re-amplified using 1 μl of sample rather than 5μl, and in some cases the DNA was first diluted 1:10 prior to amplification.
DNA Sequencing of Colonies Picked from Agar Plates
Colonies were picked from BBE and BVSA agar plates and sequenced to determine the identity of the colonies that had grown. A single colony was placed in 25 μl sterile water and subjected to 95°C for 10 min in order to release the DNA from the cells. An aliquot of the solution (1 to 5 μl) was amplified for the 16S rRNA gene using a Bacteroides forward and a universal 16S rRNA reverse primer (Bacterfor/Unirev800) to increase the concentration of Bacteroides-like sequences. Amplification reactions contained 25 μl of HotStar MasterMix (Qiagen), 80 pmol of each primer, 3 μl genomic DNA (colony solution), and nuclease-free water for a final volume of 50 μl. A PTC-100 thermocycler (MJ Research) was used to amplify the DNA with the following PCR conditions: 94°C for 10 min; 40 cycles at 94°C for 1 min, 55°C for 1 min, 72°C for 1 min; and a final 8 min extension at 72°C. The amplicons were purified with the QIAquick PCR Purification Kit (Qiagen). Sequencing reactions were performed using the BigDye 3.1 sequencing kit (Applied Biosystems) in two directions, using forward and reverse primers. The two different reaction mixtures included the following: 1.075 × buffer, 0.32 pmol of Bacterfor or Unirev800 primer, 6 μl plasmid DNA, 1/16 dilution of BigDye 3.1 mix, and nuclease free water for a final volume of 10 μl. Reactions were sequenced on an ABI 3730 capillary sequencer (Applied Biosystems).
Forward and reverse sequences were joined using the ContigExpress program of the Vector NTI 9.1 computer software (Invitrogen). Sequences were submitted to GenBank for BLAST analysis (http://www.ncbi.nlm.nih.gov/). The most homologous genes to the submitted sequences were included in a sequence alignment using the AlignX program of the Vector NTI 9.1 software (Invitrogen). Phylogenetic trees were produced to illustrate the percentage of similarities among the sequences. The tree was generated by PAUP* 4.0 software (Sinauer Associates) using the neighbor-joining algorithm with the Kimura 2-parameter correction factor.
Results
Bacterial Concentrations and Bacteria Available for DNA Extraction
Sand Samples
Bacterial concentrations in beach sand collected from Hobie Beach ranged from 77 to 2695 CFU/g dry weight for E. coli and from 73 to 445 CFU g/dry weight for enterococci (Table 3). Essentially no putative Bacteroides spp. colonies were observed on BBE or BVSA plates from dry or wet sand. The bacterial densities obtained from culturing were used to estimate the number of cells on membrane filters used for DNA extraction. This analysis (Table 3) showed that most of the membrane filters contained approximately 100 E. coli cells (76 – 128). Two of the samples had over 1000 E. coli cells and one sample contained over 66,000 E. coli cells prior to DNA extraction.
Table 3.
Average bacterial densities in sand (CFU/g dry wt) as determined by culturing, and the calculated cells available for DNA extraction per filter (Sample #) used for PCR analysis. NA = not available
| SAND
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Bacterial density (CFU/g dry wt ± STDa) | Sample parameters | # of cells on DNA extraction filters | |||||||
|
| |||||||||
| Sample date | E. coli | Enterococcus | Bacteroides | Sample # | Sand type | Amount processed (g dry wt) | E. coli | Enterococcus | Bacteroides |
| 14-Jul-04 | 77 | 73 ± 23 | <1 | 1 | wet sand | 0.99 | 76 | 72 | 0 |
| 2 | wet sand | 13.20 | 1214 | 660 | 0 | ||||
| 30-Jun-04 | 2695 ± 448 | 198 ± 175 | <2 | 3 | dry sand | 0.37 | 1001 | 74 | <1 |
| 4 | dry sand | 24.8 | 66781 | 4906 | <50 | ||||
| 14-Jul-04 | 79 | 445 ± 89 | <1 | 5 | dry sand | 1.45 | 114 | 644 | 0 |
| 1-Feb-06 | 270 ± 28 | 113 ± 40 | <1 | 9 | dry sand | 0.49 | 131 | 55 | 0 |
| 10 | dry sand | 0.49 | 131 | 55 | 0 | ||||
| 11 | dry sand | 0.49 | 131 | 55 | 0 | ||||
| 12 | dry sand | 0.49 | 131 | 55 | 0 | ||||
| 5-Jun-07b | NA | 335 ± 153 | NA | 36 | dry sand | 0.48 | NA | 161 | NA |
| 37 | dry sand | 0.56 | NA | 187 | NA | ||||
| 38 | dry sand | 5.2 | NA | 1740 | NA | ||||
performed in duplicate or triplicate, when no STD given n=1 countable plate;
taken from a natural drainage ditch that brings runoff to the beach during rain events.
The enterococci values were similar; most filters had about 100 enterococci cells (55–187), three samples had around 1000 cells (644 – 1740), and one sample had approximately 4900 cells. There did not appear to be a significant number of Bacteroides cells in the sand samples used for PCR analysis.
Water Samples
Lower bacterial concentrations were observed in seawater samples collected from Hobie Beach or Marco Island compared to river water from Wagner Creek (Tables 4 and 5). Concentrations of E. coli in seawater ranged from <1 – 83 CFU/100 ml and enterococci concentrations ranged from <1 – 61 CFU/100 ml. Few putative Bacteroides spp. were identified in the Hobie Beach samples and thus this parameter was not analyzed with the Marco Island samples (Table 4). In contrast, the river water samples ranged from 235 – 7600 CFU/100 ml E. coli and 47 – 5098 CFU/100 ml enterococci.
Table 4.
Average bacterial densities in seawater (CFU/100 ml) collected from Hobie Beach (knee-deep) or Marco Island (saltwater canal) as determined by growth on selective media and the calculated cells available for DNA extraction for each membrane filter (Sample #) used for PCR analysis. NA = not available
| SEAWATER
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Bacterial density (CFU/100 ml ± STDa) | Sample parameters | # of cells on DNA extraction filters | |||||||
|
| |||||||||
| Sample date | E. coli | Enterococcus | Bacteroides | Sample # | Water type (site #) | Amount processed (ml) | E. coli | Enterococcus | Bacteroides |
| 14-Jul-04 | 30 ± 14 | 61 ± 17 | <1 | 13 | beach | 700 | 210 | 427 | <1 |
| 14 | beach | 500 | 150 | 305 | <1 | ||||
| 14-Mar-06 | <1 | <1 | NA | 17,18a | canal (3) | 100 | <1 | <1 | NA |
| 14-Mar-06 | <1 | <1 | NA | 19, 20 | canal (7) | 100 | <1 | <1 | NA |
| 14-Mar-06 | <1 | <1 | NA | 21, 22 | canal (9) | 100 | <1 | <1 | NA |
| 14-Mar-06 | <1 | <1 | NA | 23, 24 | canal (10) | 100 | <1 | <1 | NA |
| 14-Mar-06 | <1 | <1 | NA | 25, 26 | canal (11) | 100 | <1 | <1 | NA |
| 14-Mar-06 | <1 | <1 | NA | 27, 28 | canal (12) | 100 | <1 | <1 | NA |
performed in duplicate.
Table 5.
Average bacterial densities in river water (CFU/100 ml) collected from Wagner Creek as determined by growth on selective media and the calculated cells available for DNA extraction for each membrane filter (Sample #) used for PCR analysis
| RIVERWATER
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial density (CFU/100 ml ± STD) | Sample parameters | # of cells on DNA extraction filters | ||||||
|
| ||||||||
| Sample date | E. coli | Enterococcus | Bacteroides | Sample # | Amount processed (ml) | E. coli | Enterococcus | Bacteroides |
| 11-Feb-04 | 7600 ± 5657 | 94 ± 37 | 215 ± 7 | 29 | 500 | 3.8 × 105 | 470 | 1075 |
| 29-Mar-04 | 1158 ± 706 | 5098 ± 5958 | 115 ± 27 | 30 | 500 | 5790 | 2.5×104 | 575 |
| 10-Feb-05 | 2050 ± 919 | 1381 ± 680 | 62 ± 60 | 31 | 700 | 1.4 × 104 | 9667 | 434 |
| 32 | 700 | 1.4 × 104 | 9667 | 434 | ||||
| 19-Apr-06 | 235 ± 50 | 47 ± 33 | 850 ± 212 | 33 | 100 | 235 | 47 | 850 |
| 34 | 100 | 235 | 47 | 850 | ||||
| 35 | 100 | 235 | 47 | 850 | ||||
Unlike the other tested sites, a significant number of putative Bacteroides spp. colonies were obtained (62 – 850 CFU/100 ml). The estimated number of cells on the membrane filters used for DNA extraction was correspondingly higher in the river samples than for the seawater samples. As expected, the raw sewage samples contained high concentrations of fecal indicating bacteria, with concentrations ranging from 4×106 to 6×106 CFU/100 ml for E. coli, enterococci, and Bacteroides spp. (Table 6).
Table 6.
Average bacterial densities (CFU/100 ml) in raw sewage samples collected from a wastewater treatment plant as determined by growth on selective media and the calculated cells available for DNA extraction for each membrane filter (Sample #) used for PCR analysis
| RAW SEWAGE
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial density (CFU/100 ml ± STD) | Sample parameters | # of cells available for DNA extraction | ||||||
|
| ||||||||
| Sample date | E. coli | Enterococcus | Bacteroides | Sample # | Amount processed (ml) | E. coli | Enterococcus | Bacteroides |
| 17-Sep-05 | 6×106 ± 8×105 | 1.4×106 ± 2×106 | 3×106 ± 2×106 | 50 | 8 | 4.8×105 | 1.1 ×105 | 2.4 ×105 |
| 54 | 12 | 7.2 ×105 | 1.7 ×105 | 3.6 ×105 | ||||
| 60 | 2 | 1.2 ×105 | 2.8 ×104 | 6.0 ×104 | ||||
Molecular Detection of Source Tracking Markers and Bacterial and Viral Pathogens
The marker for human-specific Bacteroides spp. (HF8 human cluster) and the pathogen S. aureus were positively detected by PCR in the samples of raw sewage. The human-specific enterococci marker (esp gene) and the pathogens E. coli O157:H7, Salmonella spp., C. jejuni, and adenovirus were not detected (Table 7).
Table 7.
Results of PCR screening of raw sewage for a variety of fecal indicators, source tracking markers, and pathogens. Sample numbers are described in Table 6. Positive (+) or negative (−) result based on visual inspection of electrophoresis gel
| Sample # | Enterococcus | Enterococcus human marker | Bacteroides human marker | S. aureus | C. jejuni | E. coli 0157:H7 | Salmonella spp. | Adenovirus |
|---|---|---|---|---|---|---|---|---|
| 50 | + | − | + | + | − | − | − | − |
| 54 | + | − | + | + | − | − | − | − |
| 60 | + | − | + | + | − | − | − | − |
Two environmental samples were positive for human-specific Bacteroides spp. (HF8 human cluster) and one was positive for S. aureus. Samples 31 and 35 were positive for human specific Bacteroides and sample 31 was positive for S. aureus (Table 8). It is interesting to note that the replicate of sample 31 (sample 32) was negative for both S. aureus and the human Bacteroides marker. These samples differed only in the treatment of the filter, in that sample 32 was cut into pieces before being placed into the microfuge tube used for DNA extraction, whereas sample 31 was left intact (Table 8).
Table 8.
Results of PCR screening of sand and water samples for a variety of fecal indicators, source tracking markers, and pathogens. Sample numbers are described in Tables 3 – 5. Positive (+) or negative (−) result based on visual inspection of electrophoresis gel
| Sample # | Sample type | E. coli | Enterococci | Bacteroides human marker | enterococci human marker | E. coli 0157:H7 | Adenovirusb | S. aureus |
|---|---|---|---|---|---|---|---|---|
| 1 – 2 | wet sand | + | + | − | − | − | ||
| 3 – 4 | dry sand | + | + | − | − | − | ||
| 9 – 12 | dry sand | + | + | − | − | − | − | |
| 36 – 38 | dry sand | + | + | − | − | − | − | |
| 39, 41G | dry sand, enriched | − | ||||||
| 13 – 14 | beach water | + | + | − | − | − | ||
| 17 – 28 | canal water | + | − | − | ||||
| 29 | river water | + | + | − | − | − | ||
| 30 | river water | + | + | − | − | − | ||
| 31 – 32 | river water | + | +/−c | + | − | +/−c | ||
| 33 – 35a | river water | + | + | − | − | − | ||
| 40G | river water, enriched | − |
DNA extracted from growth filter (MeI agar);
these samples were also negative for C. jejuni;
only samples 9–12, 17–28, and 33–35 were processed for viral analysis;
sample 31 was positive, but sample 32 was negative. These samples were replicates except that sample 32 was cut during processing and sample 31 was left intact.
Many of these samples contained low amounts of fecal indicating bacteria (Tables 3–5); therefore it is not surprising that source tracking markers and pathogens were not detected. However, a few samples did contain high numbers of fecal indicating bacteria (samples 3, 29, 30, 31, 32). Nonetheless, source tracking markers and pathogens were not detected in these samples (Table 8).
A two-part enrichment step is recommended to achieve detection of the esp gene (Scott, 2005), but it was not used here for the sewage samples or for dry sand samples (runoff ditch) collected 5 June 2007 (#36, 37, 38). Three of the environmental samples (#39, 40, 41) did receive an incubation step. Filters were incubated on MeI agar plates to select for enterococci (Table 1), and DNA was extracted from the growth filters. DNA from dry sand collected 10 February 2006 (sample #39) was extracted from 69 CFU of enterococci. DNA from river water collected 19 April 2006 (sample #40) was extracted from 24 CFU of enterococci. DNA from dry sand collected 5 June 2007 (sample #41) was extracted from 883 CFU. None of these samples tested positive for the esp gene (Table 8).
DNA Sequence Results
Plate culture results indicated the presence of a high number of Bacteroides spp. in Wagner Creek river water (Table 5) in comparison to seawater samples. Two Wagner Creek samples were positive for human specific Bacteroides; whereas, none of the seawater samples were positive. In contrast, other Wagner Creek samples with high Bacteroides counts were not positive for human-specific marker.
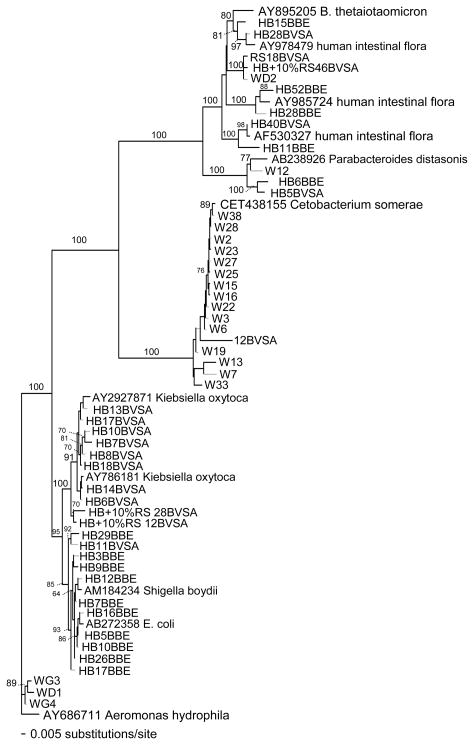
Bacteroides counts could have been overestimated if the media used were not selective. Colony sequencing was used to investigate the specificity of BBE and BVSA plates when used with environmental samples. A portion of the 16S rRNA gene was sequenced for colonies grown from samples collected from Hobie Beach, Wagner Creek, and sewage samples. The analysis showed that some of the picked colonies contained sequences closely related to those of known Bacteroides species (Figures 3 and 4), although few of the sequences obtained from environmental samples were exact matches to sequences available in Genbank.
Figure 3.
Portion of an alignment of partial 16S rRNA gene sequences from colonies grown on BBE or BVSA plates from samples of river or beach water.
Figure 4.
Phylogenetic relationships of partial 16S rRNA gene sequences (654 bp) from putative Bacteroides colonies grown on BBE or BVSA media. Aeromonas hydrophila was used as an outgroup. Bootstrap values for 1000 trees generated by PAUP* 4.0 are shown for nodes >60.
Phylogenetic relationships were determined for partial 16S rRNA gene sequences (654 base pairs) from colonies grown from river or beach water and picked from BBE or BVSA plates designed to select for Bacteroides species (Figure 4). Some sequences (9/52) were related to named Bacteroides species or were closely related to various uncultured human intestinal flora, but few were exact matches (Figure 4).
Many colonies were not related to Bacteroides. Sequences related to Cetobacterium somerae, Parabacteroides distasonis, Klebsiella oxytoca, E. coli, and Aeromonas hydrophila were obtained (Figure 4). This data demonstrates that the BBE and BVSA plates were not perfectly selective for Bacteroides spp. when used for environmental samples. Overall, the paucity of exact matches to species previously identified in GenBank illustrated the molecular diversity present in these samples.
Discussion
Several studies support the idea that sand acts as a reservoir for fecal indicating bacteria (Alm et al., 2006; Lee et al., 2006; Yamahara et al. 2007). Sand may protect adsorbed bacteria by reducing exposure to stressors such as ultraviolet radiation, high salinity, high temperatures, and wave action. Studies also have suggested that the persistence of bacteria in sediments may result from a balance between the rates of bacterial growth versus predation. (Davies et al., 1995). The survival of enteric bacteria on dry sand has been hypothesized to be minimal due to environmental stresses including a lack of adequate moisture and nutrients (World Health Organization, 2003). However, dry sand in this study harbored a significant number of fecal indicating bacteria (Table 3).
The possibility that fecal indicators can grow and persist in sand and become a source of these bacteria to adjacent waters raises the possibility that the fecal indicators can be independent of human feces and thus not indicative of the presence of pathogens. An alternative hypothesis is that if sand is a reservoir for fecal indicators it may also be a reservoir for pathogens. This preliminary study performed PCR screening of samples collected from rivers, canals, beach water, and beach sand in order to look at the relationship between fecal indicator concentrations and several pathogens and source tracking markers.
Raw sewage was used as a positive control to compare against sand and water results. As expected, sewage showed the most bacterial colony growth and positive hits in the PCR screen. S. aureus and the HF8 human marker for Bacteroides spp. were detected in the samples (Table 7). Salmonella spp., E. coli O157:H7, C. jejuni, and adenovirus were not detected. Unlike S. aureus, which is carried by 30–50% of the population (Youmans et al., 1985), these other pathogens are not expected in healthy individuals; therefore, lack of detection may have been due to the lack of their presence at the time of sampling. In addition, the sewage samples were not processed by the viral sorption method (Fig. 2), making it less likely to achieve viral detection in those samples.
Although only a subpopulation is expected to carry the human specific enterococci marker (Shankar et al., 1999), the lack of detection of the esp gene in the sewage samples was unexpected. However, this analysis did not utilize a preincubation step. The published protocol suggested a two-stage culture step in which filtered cells were grown at 41 °C on MeI agar for 48 hr and then the filter was incubated at 41 °C in tryptic soy broth for 3 hr (Scott et al., 2005). The lack of enrichment step for the sewage samples may have accounted for the lack of detection of the esp gene. Three samples (#39, 40, 41) were preincubated on MeI agar, and DNA was extracted from the growth filters. None of these samples tested positive for human specific enterococci (Table 8). However, two of the samples (# 39, #40) had few enterococci colonies from which to extact DNA (see results section); therefore, negative results were not surprising. For the third sample, it is possible that the PCR was inhibited due to carry over of inhibitory substances in the MeI agar.
On 45% of the sampling dates (5/11), the water exceeded EPA standards for recreational water quality (EPA, 2003) using the single sample standard of ≥104 CFU of enterococci/100 ml or ≥235 CFU of E. coli/100 ml (Tables 4 and 5). On 80% of the sampling dates (4/5), the sand had relatively high concentrations of fecal indicating bacteria, as defined here by ≥ 100 CFU/g dry weight sand of enterococci or E. coli (Table 4). How these values translated to the number of cells available for molecular analysis greatly depended on the processing procedures used (Tables 3–5). In some cases, less cells were available for molecular detection than was implied by the bacterial density because of the small amount of sample processed (Table 3). Six of the 27 samples had no cells on the DNA extraction filters, despite filtering 100 ml of canal water (Table 4). Nine of the 27 samples had <100 cells available on the DNA extraction filter, and 10 of the samples had >100 cells available. The remaining samples (3/27), had >9500 cells (Table 5), and one of those had on the order of 104 enterococci cells, similar to what was seen in sewage samples. Only two samples returned positive detects in the PCR screen. Sample 35 was positive for human specific Bacteroides and sample 31 was positive for HF8 and S. aureus. Both of these samples had high numbers of cells available for DNA extraction (Table 5); although other samples that were negative in the screen contained even higher numbers of cells (Table 5).
The detection limits for some of the molecular targets used in this study have been previously determined. The detection limit for the Bacteroides spp. human marker (HF8) was found to be 1 plasmid per PCR reaction, and the detection limit for the human specific enterococci marker (esp gene) was found to be 10 plasmid copies per PCR reaction (LaGier et al., 2007). Other research found that the Bacteroides marker could be detected by PCR in sewage samples diluted to 1:150,000 (Bower et al., 2005), which would equate to ~3 Bacteroides cells in the PCR reaction for the samples used in this study. For the esp gene, approximately 100 CFU of enterococci was needed on a growth plate to achieve detection, and this detection limit included a two-step enrichment procedure, as described above (Scott et al. 2005).
Although the detection limits mentioned above appear low, there are several reasons that concerns remain about false negative results possibly arising from an inability to meet the detection limit of the PCR assays. First, previous work and the analysis here have shown that the media were not completely selective when used with environmental samples. For Bacteroides, the per cent match ranged from 6–39% (Table 9) (Baums et al. 2007). In the work presented here, sequencing of putative Bacteroides colonies (Figures 3 and 4) confirmed that many of the sequences (40/52) were not closely related to Bacteroides spp.; therefore, the number of Bacteroides actually available for DNA detection was less than that estimated from the colony counts. In addition, the source tracking markers are expected to be present in only a subset of the Bacteroides or enterococci colonies because not every human is a carrier of these markers. These factors act to reduce the number of targets available for molecular detection as compared to the number of cells estimated to be available on the DNA filters (Tables 3–5).
Of even more concern with regard to meeting the detection limit of a PCR assay is the fact that few of the cells on a DNA extraction filter are likely to reach the PCR reaction. The number of cells delivered to the PCR reaction depends on several factors such as 1) the DNA extraction efficiency, 2) the final volume of the eluant or lysate, 3) the amount of the eluant or lysate put into the PCR reaction, 4) the amount of dilution necessary to overcome PCR inhibition, and 5) the amplification efficiency. For example, if one assumes a DNA extraction efficiency of 30% (Mumy and Findlay, 2004), an eluant volume of 100 μl, the use of 1 μl of template in the PCR reaction, no dilution necessary to achieve PCR (which was not always the case here), and an amplification efficiency of 1 (although less is expected in reality), it would take 1000 cells on the DNA extraction filter in order to deliver 3 cells into the PCR reaction. Only one environmental sample is estimated to have met this criteria for Bacteroides (Table 5). In comparison, ~3,350 cells would be needed to deliver the 10 cells required for the esp assay. This criteria for enterococci was met for 4 samples, and 5 samples met this criteria for E. coli. Out of these samples, none returned positive detections for any of the tested targets.
This analysis points to the need to find better methods of extracting nucleic acids from environmental samples in order to reduce the possibility of false negative results. Pre-incubation is a strategy, but that does not enable rapid detection. Overall, this study could not fully support or deny the hypothesis that the concentrations of fecal indicating bacteria can be uncoupled from markers of human fecal pollution or with human pathogens. This analysis illustrated the need to improve the current standard practices of sample concentration and DNA extraction.
Conclusion
The PCR screen returned positive results for human specific Bacteroides and S. aureus for the raw sewage samples. In addition, positive results were obtained for two of the environmental samples (sample 31 and 35 for human specific Bacteroides and sample 31 for S. aureus). This preliminary investigation did not find a correlation between the concentration of fecal indicators and detection of a variety of human pathogens and source tracking markers. However, the possibility of false negative results made it difficult to properly test the hypothesis that fecal indicating bacteria can become uncoupled from human fecal pollution. This analysis suggested that most of the samples may not have been able to achieve the detection limit of the PCR reaction, despite the fact that several of the water samples exceeded single sample standards for fecal indicators and many of the sand samples contained relatively high counts of fecal indicating bacteria. Failure to meet the detection limit likely could arise from a combination of factors that effectively reduce the number of cells that reach the PCR reaction tube from the DNA extraction filter. These factors include the DNA extraction efficiency, the amount of sample dilution due to the volume of eluant or the need to remove PCR inhibition, the amount of template in the PCR reaction and the overall amplification efficiency (which will be reduced by PCR inhibitors present in the sample).
Overall, this analysis pointed to the need to find better methods of extracting nucleic acids from environmental samples in order to reduce the possibility of false negative results. High quality nucleic acids need to be consistently and efficiently delivered to the detector system if the relationship between fecal indicators and human pathogens and human source tracking markers is to be elucidated.
Acknowledgments
We thank the Rookery Bay National Estuarine Research Reserve and the City of Marco Island for help with sample collection. We thank H. Spencer, a NOAA Ernest F. Hollings Undergraduate Scholar, for field and laboratory work with the June 2007 sand samples. Some of the research was conducted in partial fulfillment of the University of Miami requirements for summa cum laude for L. Matragrano. We thank I. Baums (U. Miami) for aid with sequencing. Financial support is gratefully acknowledged from the Cooperative Institute of Estuarine and Environmental Technology (CICEET) and from a pilot project supported through the National Science Foundation and National Institute of Environmental Health Sciences Oceans and Human Health Center at the University of Miami Rosenstiel School (NSF 0CE0432368; NIEHS P50 ES12736). This research was carried out in part under the auspices of the Cooperative Institute for Marine and Atmospheric Studies (CIMAS), a joint institute of the University of Miami and the National Oceanic and Atmospheric Administration, cooperative agreement #NA17RJ1226.
References
- Alm EW, Burke J, Hagan E. Persistence and potential growth of the fecal indicator bacteria, Escherichia coli, in shoreline sand at Lake Huron. J Great Lakes Res. 2006;32:401–405. [Google Scholar]
- American Water Works Association. Standard Methods for the Examination of Water and Wastewater. 2 1999. [Google Scholar]
- Baums IB, Goodwin KD, Kiesling T, Wanless D, Fell JW. Luminex detection of fecal indicators in river samples, marine recreational water, and beach sand. Mar Pollut Bullet. 2007;54:521–536. doi: 10.1016/j.marpolbul.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard AE, Field KG. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol. 2000;66:4571–4574. doi: 10.1128/aem.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton G. Wastewater Microbiology. 3. Wiley–Liss; Hoboken: 2005. [Google Scholar]
- Bower PA, Scopel CO, Jensen ET, Depas MM, McLellan SL. Detection of genetic markers of fecal indicator bacteria in Lake Michigan and determination of their relationship to Escherichia coli densities using standard microbiological methods. Appl Environ Microbiol. 2005;71:8305–8313. doi: 10.1128/AEM.71.12.8305-8313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelli VJ, Dufour AP, Levin MA, McCabe J, Haberman PW. Relationship of microbial indicators to health effects at bathing beaches. Am J Pub Health. 1979;69:690–696. doi: 10.2105/ajph.69.7.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelli VJ, Dufour AP, McCabe LJ, Levin MA. Swimming-associated gastroenteritis and water quality. Am J Epidemol. 1982;115:606–616. doi: 10.1093/oxfordjournals.aje.a113342. [DOI] [PubMed] [Google Scholar]
- Carillo J, Estrada E, Hazen TC. Survival and enumeration of the fecal indicators Bifidobacterium adolescentis and Escherichia coli in a tropical rain forest watershed. Appl Environ Microbiol. 1985;50:468–476. doi: 10.1128/aem.50.2.468-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clean Beaches Council. 2005 State of the Beach Report: Bacteria and Sand, A National Call to Action. 2005. [Online] http://www.cleanbeaches.org/ewebeditpro/items/O31F6450.pdf.
- Craig DL, Fallowfield HJ, Cromar NJ. Enumeration of faecal coliforms from recreational coastal sites: evaluation of techniques for the separation of bacteria from sediments. J Appl Microbiol. 2002;93:557–565. doi: 10.1046/j.1365-2672.2002.01730.x. [DOI] [PubMed] [Google Scholar]
- Davies CM, Long JAH, Donald M, Ashbolt NJ. Survival of fecal microorganisms in marine and freshwater sediments. Appl Environ Microbiol. 1995;61:1888–1896. doi: 10.1128/aem.61.5.1888-1896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour AP. Research and Development EPA-600/1-84/004. USEPA, Health Effects Research Laboratory; NC: 1984. Health effects criteria for fresh recreational waters. [Google Scholar]
- Dwight RH, Fernandez LM, Baker DB, Semenza JC, Olson BH. Estimating the economic burden from illnesses associated with recreational coastal water pollution--a case study in Orange County, California. Journal of Environmental Management. 2005;76:95–103. doi: 10.1016/j.jenvman.2004.11.017. [DOI] [PubMed] [Google Scholar]
- EPA. Method 1600: Enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-B-D-glucoside agar (mEI). EPA-821-R-02-022. United States Environmental Protection Agency; 2002. [Google Scholar]
- EPA. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli Agar (modified mTEC). EPA-821-R-02-023. United States Environmental Protection Agency; 2002. [Google Scholar]
- EPA. Guidelines establishing test procedures for the analysis of pollutants; Analytical methods for biological pollutants in ambient water; Final Rule. Federal Register. 2003;68:40. CFR Part 136. [Google Scholar]
- FDA/ISSC. National Shellfish Sanitation Program Guide for the Control of Molluscan Shellfish 2003. US Food and Drug Administration and the Interstate Shellfish Sanitation Conference.2003. [Google Scholar]
- Gerba CP, McLeod JS. Effect of sediments on the survival of Escherichia coli in marine waters. Appl Environ Microbiol. 1976;32:114–120. doi: 10.1128/aem.32.1.114-120.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal SM, Gerba CP, Melnick JL. Occurrence and distribution of bacterial indicators and pathogens in canal communities along the Texas coast. Am Soc Microbiol. 1977;34:139–149. doi: 10.1128/aem.34.2.139-149.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Wat Res. 2005;39:559–568. doi: 10.1016/j.watres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- He JW, Jiang S. Quantification of Enterococci and human adenoviruses in environmental samples by real-time PCR. Appl Environ Microbiol. 2005;71:2250–2255. doi: 10.1128/AEM.71.5.2250-2255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MA, Ness GE. Survival of Vibrio cholerae and Escherichia coli in estuarine waters and sediments. Appl Environ Microbiol. 1982;43:578–584. doi: 10.1128/aem.43.3.578-584.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Shimasaki A, Ohgaki S. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl Environ Microbiol. 2002;68:1033–1039. doi: 10.1128/AEM.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong RYC, Lee SKY, Lee TFW, Law SHW, Wu RSS. Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Wat Res. 2002;36:2802–2812. doi: 10.1016/s0043-1354(01)00503-6. [DOI] [PubMed] [Google Scholar]
- LaGier MJ, Joseph LA, Passaretti TV, Musser KA, Cirino NM. A real-time multiplexed PCR assay for rapid detection and differentiation of Campylobacter jejuni and Campylobacter coli. Mol Cell Probes. 2004;18:275–282. doi: 10.1016/j.mcp.2004.04.002. [DOI] [PubMed] [Google Scholar]
- LaGier MJ, Fell JW, Goodwin KD. Electrochemical detection of harmful algae and other microbial contaminants in coastal waters using hand-held biosensors. Mar Pollut Bullet. 2007;54:757–770. doi: 10.1016/j.marpolbul.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Lin TY, Lin C, Kohbodi GA, Bhatt A, Lee R, Jay JA. Persistence of fecal indicator bacteria in Santa Monica Bay beach sediments. Wat Res. 2006;40:2593–2602. doi: 10.1016/j.watres.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Mason WJ, Blevins JS, Beenken K, Wibowo N, Ojha N, Smeltzer MS. Multiplex PCR protocol for the diagnosis of Staphylococcal infection. J Clin Microbiol. 2001;39:3332–3338. doi: 10.1128/JCM.39.9.3332-3338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer JJ, Schmidt D, Petrosko P, Sanchez S, Bolton L, Lee MD. Development of primers to O-antigen biosynthesis genes for specific detection of Escherichia coli O157 by PCR. Appl Environ Microbiol. 1999;65:2954–2960. doi: 10.1128/aem.65.7.2954-2960.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumy KL, Findlay RH. Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative-competitive PCR. J Microbiol Methods. 2004;57:259–268. doi: 10.1016/j.mimet.2004.01.013. [DOI] [PubMed] [Google Scholar]
- National Research Council. Committee on Indicators for Waterborne Pathogens. National Academy of Sciences; Washington, DC: 2004. Indicators for Waterborne Pathogens. [Google Scholar]
- Rivera SC, Hazen TC, Toranzos GA. Isolation of fecal coliforms from pristine sites in a tropical rain forest. Appl Environ Microbiol. 1988;54:513–517. doi: 10.1128/aem.54.2.513-517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TM, Jenkins TM, Lukasik J, Rose JB. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ Sci Technol. 2005;39:283–287. [PubMed] [Google Scholar]
- Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Solo-Gabriele HM, Fleming LE, Elmir S. Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Wat Res. 2004;38:3119–3131. doi: 10.1016/j.watres.2004.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solo-Gabriele HM, Wolfert MA, Desmarais TR, Palmer CJ. Sources of Escherichia coli in a coastal subtropical environment. Appl Environ Microbiol. 2000;66:230–237. doi: 10.1128/aem.66.1.230-237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade TJ, Pai N, Eisenberg JNS, Colford JM., Jr Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environ Health Perspect. 2003;111:1102–1109. doi: 10.1289/ehp.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, Nevers MB. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl Environ Microbiol. 2003;69:5555–5562. doi: 10.1128/AEM.69.9.5555-5562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Gutell R, Gupta R, Noller HF. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47:621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Coastal and Fresh Waters. Vol. 1. World Health Organization; Geneva, Switzerland: 2003. Guidelines for safe recreational water environments. [Google Scholar]
- Wright RC. The survival patterns of selected faecal bacteria in tropical fresh water. Epidemiol Infect. 1989;103:603–611. doi: 10.1017/s0950268800031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamahara KM, Layton BA, Santoro AE, Boehm AB. Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters. Environ Sci Technol. 2007;41:4515–4521. doi: 10.1021/es062822n. [DOI] [PubMed] [Google Scholar]
- Youmans GP, Paterson PY, Sommers HM. The Biological and Clinical Basis of Infectious Diseases. 3. The W. B. Saunders Company; Philadelphia: 1985. [Google Scholar]