Abstract
The main (Mpro) and papain-like (PLpro) proteases are highly conserved viral proteins essential for replication of the COVID-19 virus, SARS-COV-2. Therefore, a logical plan for producing new drugs against this pathogen is to discover inhibitors of these enzymes. Accordingly, the goal of the present work was to devise a computational approach to design, characterize, and select compounds predicted to be potent dual inhibitors – effective against both Mpro and PLpro. The first step employed LigDream, an artificial neural network, to create a virtual ligand library. Ligands with computed ADMET profiles indicating drug-like properties and low mammalian toxicity were selected for further study. Initial docking of these ligands into the active sites of Mpro and PLpro was done with GOLD, and the highest-scoring ligands were redocked with AutoDock Vina to determine binding free energies (ΔG). Compounds 89–00, 89–07, 89–32, and 89–38 exhibited favorable ΔG values for Mpro (−7.6 to −8.7 kcal/mol) and PLpro (−9.1 to −9.7 kcal/mol). Global docking of selected compounds with the Mpro dimer identified prospective allosteric inhibitors 89–00, 89–27, and 89–40 (ΔG -8.2 to −8.9 kcal/mol). Molecular dynamics simulations performed on Mpro and PLpro active site complexes with the four top-scoring ligands from Vina demonstrated that the most stable complexes were formed with compounds 89–32 and 89–38. Overall, the present computational strategy generated new compounds with predicted drug-like characteristics, low mammalian toxicity, and high inhibitory potencies against both target proteases to form stable complexes. Further preclinical studies will be required to validate the in silico findings before the lead compounds could be considered for clinical trials.
Keywords: COVID-19, Mpro/PLpro inhibitors, Nirmatrelvir, Molecular docking, Molecular dynamics simulation, In silico drug design, ADMET, Heterocyclic compounds, Pyrazolopyridazines, Tetrazoles
Graphical abstract
1. Introduction
The global pandemic of COVID-19 disease caused by the SARS-CoV-2 virus is one of the most challenging outbreaks the world has ever experienced. At the last update on September 30, 2022, the WHO reported 614, 385, 693 confirmed cases and 6,522,600 confirmed deaths [1]. This crushing pandemic continues to inflict major disruptions on national healthcare systems and the global economy. Furthermore, many countries have experienced second or third waves of COVID-19 outbreaks, mainly due to the emergence of mutant variants of the virus, such as the Omicron strain, which is the principal variant of SARS-CoV-2 that is currently circulating [2].
Fortunately, there are now vaccines to help protect against SARS-CoV-2 infection [3], and monoclonal antibodies to confer passive immunity against the virus [4]. Moreover, several small-molecule antiviral drug options have recently emerged to treat existing infection, including molnupiravir and remdesivir (nucleoside analogs that inhibit viral replication by different downstream mechanisms following their incorporation into the viral genome) [5,6] and nirmatrelvir (a dipeptide analog that blocks viral replication by covalently inhibiting an essential viral protease, Mpro) [7].
Nevertheless, current preventative and therapeutic advances have limitations. Although vaccines are regarded as the best safeguard against SARS-CoV-2 infection, they lose their effectiveness over time requiring periodic boosters and/or development of new vaccines in response to viral mutations [8]. Monoclonal antibodies are expensive to produce and subject to viral evasion arising from mutations [4,9]. Remdesivir must be administered by intravenous infusion, but it has higher efficacy than oral molnupiravir [10,11]. Nirmatrelvir is the most recent antiviral medication; it is given orally and has been shown to be highly efficacious [7], although as with remdesivir and molnupiravir, the target population consists of those with mild to moderate COVID-19 symptoms [10]. Thus, despite these encouraging prophylactic and therapeutic innovations, there is still an urgent need for the discovery and development of additional and highly selective antiviral agents [12].
Human coronaviruses contain a positive-sense single-stranded RNA genome and are among the fastest-evolving viruses owing to their high rates of nucleotide replacement and recombination [13]. SARS-CoV-2 belongs to the β-genus of the Coronaviridae family, and upon transcription of its genome, the virus produces a ∼800 kDa polypeptide that is proteolytically processed to yield non-structural proteins (Nsp) required for viral replication [14,15]. The specific cleavage of this long polypeptide chain is carried out by proteolytic enzymes: notably, the main protease (Mpro, also known as the 3-chymotrypsin-like cysteine protease 3CLpro, a domain within Nsp5), and the papain-like protease (PLpro, a domain within Nsp3). Despite the high mutation rate of SARS-CoV-2 and other coronaviruses, the sequences of essential viral proteins such as Mpro and PLpro tend to be preserved because mutations of such vital proteins are often lethal to the virus [16]. Moreover, compounds targeting Mpro can reduce the risk of mutation-mediated drug resistance in future deadly viral strains [17].
The Mpro enzyme of SARS-CoV-2 is a highly conserved homodimeric protease with >96% overall sequence identity (100% in the active site) to its counterpart in the SARS-CoV virus [18,19]. The active form of this enzyme is a homodimer; each monomer comprises 306 amino acid residues and three domains [20, PDB ID 7L0D]. The active site of Mpro has a non-classical His-Cys catalytic dyad (His41-Cys145), and it can be divided into four subsites, S1–S4. The following active site residues of Mpro are involved in substrate binding: His41, Met49, Glu143, Ser144, His163, His164, Met165, Glu166, Leu167, Asp187, Arg188, Gln189, Thr190, Ala191, and Gln192. The main protease plays pivotal roles for viral genome replication, transcription, and other processes that are vital for the continued survival of the virus. Therefore, Mpro is a promising target for the discovery and development of anti-coronavirus drugs, especially given that humans do not have a homologous protease [21]. Moreover, the recent success with the antiviral drug, nirmatrelvir, attests to the appropriateness of Mpro as a druggable target and the possibility of designing inhibitors with high selectivity against it [7]. However, some inhibitors of Mpro, including nirmatrelvir, can also inhibit human cysteine proteases such as cathepsin-L, although this cross-reactivity could be turned into an advantage because cathepsin-L is known to facilitate viral entry [19].
PLpro is a constituent of Nsp3, a large multidomain protein with a MW of ∼212 kDa and 1945 amino acid residues that is an essential component of the viral replicase-transcriptase complex [22]. The PLpro constituent is highly conserved among all coronaviruses, and it plays important roles in the proteolytic processing and maturation of viral polyproteins [23]. PLpro may also act on host cell proteins to disrupt host immune response machinery, thereby facilitating viral replication and proliferation. This viral protein is composed of 316 amino acid residues with a relatively high content of cysteine (3.5%) [24], PDB ID 7LBR]. The active site of PLpro has a canonical catalytic triad (Cys111, His272 and Asp286), unlike the Mpro protein [25]. Zinc ion binding is crucial for the structural integrity and protease activity of PLpro. The zinc ion is coordinated by four cysteine residues (Cys189, Cys192, Cys224 and Cys226), which are located on two loops of two β-hairpins. The following amino acid residues of PLpro are involved in substrate binding: Tyr268, Met208, Pro247, Pro248, Thr301, Tyr264, Asn267, Gln269, Leu162, Cys270, Gly271, and Tyr273 [26]. In view of these structural and functional characteristics of PLpro, this enzyme, along with Mpro, is a suitable target for developing inhibitors of its activity as therapeutic agents against SARS-CoV-2.
There are three broad functional classes of enzyme inhibitors that we considered when embarking on the present work: covalent irreversible inhibitors acting at the active site; noncovalent reversible inhibitors acting at the active site; and noncovalent reversible inhibitors acting at allosteric sites (ligand-binding at loci other than the active site that could modulate the activity of the active site) [27].
Covalent inhibitors were formerly shunned as viable drug candidates because of the perception of a greater toxicity risk associated with irreversible and non-selective binding of ligands to proteins [28,29]. However, covalent inhibitors have been gaining acceptance, owing to their potential for high potency and, when desired, a prolonged duration of action [30]. Nevertheless, covalent docking is still a relatively new approach that is more difficult to implement than reversible docking, especially in a high-throughput manner. Moreover, selectivity can be an issue with covalent inhibitors. For example, considering the design of covalent inhibitors of cysteine proteases such as Mpro, ligands would contain reactive electrophilic groups to form covalent bonds with the active site cysteine. However, such electrophiles could react indiscriminately with cysteine residues outside of the active site unless the ligand included specific groups equipped to recognize complementary residues in the active site, as was done with nirmatrelvir as a covalent inhibitor of Mpro [7]. For these reasons, the present work focused on noncovalent reversible inhibitors.
Recent studies aimed at discovering reversible inhibitors of Mpro identified ML188 (Table 1 ) as one of the most potent, with an IC50 of 2.5 μM [20]. Furthermore, in the same study, a crystal structure of this compound in complex with Mpro was solved to 2.39 Å resolution, thus furnishing a target structure for the present investigation (PDB ID 7L0D).
Table 1.
AutoDock Vina ΔG values for selected pyrazolopyridazine ligandsa docked into the active sites of Mpro and PLpro.
| Compound | Structure | ΔG Mpro, kcal/mol | ΔG PLpro, kcal/mol |
|---|---|---|---|
| 89–00b | 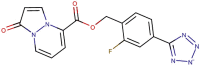 |
−7.8 | −9.1 |
| 89–03 | 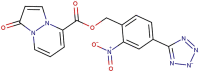 |
−8.2 | −8.7 |
| 89–07b | 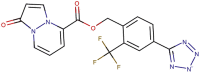 |
−8.7 | −9.2 |
| 89–10 | 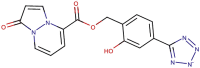 |
−7.8 | −8.9 |
| 89–22 | 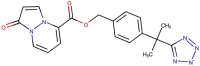 |
−8.3 | −8.5 |
| 89–24 | 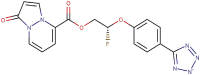 |
−7.5 | −8.4 |
| 89–25 | 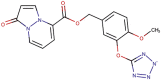 |
−7.6 | −8.3 |
| 89–27 | 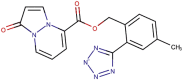 |
−7.6 | −9.2 |
| 89–30 | 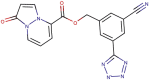 |
−8.1 | −8.5 |
| 89–31 | 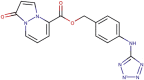 |
−8.1 | −8.3 |
| 89–32b | 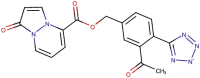 |
−8.1 | −9.2 |
| 89–33 | 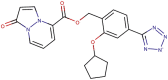 |
−8.2 | −8.5 |
| 89–37 | 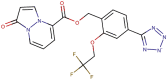 |
−7.8 | −8.3 |
| 89–38b | 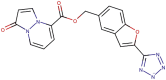 |
−8.7 | −9.7 |
| 89–40 | 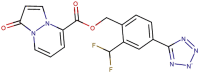 |
−7.7 | −8.3 |
| 89–42 | 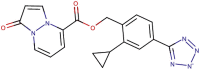 |
−8.1 | −8.5 |
| ML188 | 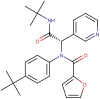 |
−7.9 | −6.7 |
| GRL0617 | 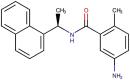 |
−7.4 | −9.9 |
| 94-00 (Azachalcone) | 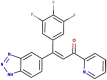 |
−8.5 | −9.3 |
In addition to the 16 compounds selected by their ChemPLP GOLD docking scores from the 43 ligands generated from compound 89–00 by LigDream, the known inhibitors ML188 (Mpro) [20] and GRL0617 (PLpro) [33] along with previously identified compound 94–00 [44] were included in the Vina docking studies for comparison.
These ligands were selected for more detailed study based on their average docking scores and favorable ADMET properties.
Likewise, compound GRL0617 (Table 1) has been shown to be highly effective at reversibly inhibiting the in vitro activity of PLpro, with an IC50 of 1.6 μM [31,32]. However, the available crystal structures of PLpro in complex with GRL0617 had resolutions in the 2.5–3.2 Å range; therefore, a structure of PLpro in complex with XR8-89, an analog of GRL0617, with a resolution of 2.2 Å was chosen as a target structure for the present work (PDB ID 7LBR). Notwithstanding its favorable potency against PLpro, GRL0617 exhibited poor metabolic stability that prevented its potential clinical application [33]. In fact, despite intensive studies, efficacious PLpro inhibitors with low mammalian toxicity have still not been developed and subsequently approved by the US FDA [3].
Our preliminary investigations of Mpro included blind docking of ligands using a simulation cell that encompassed the entire dimer rather than being focused on the catalytic active sites in the constituent monomers. These studies revealed sites other than the catalytic active sites with favorable free energies of binding that overlapped the values obtained by docking the same ligands into the active sites. Furthermore, these results were in agreement with recent reports of allosteric binding sites in Mpro [34,35]. Because allosteric sites have been less explored than catalytic sites and allosteric binding could furnish new avenues for modulating the activity of Mpro [36], discovery of allosteric binding sites was incorporated into the present work.
A common approach for drug discovery and development in medicinal chemistry is a single-target strategy, i.e., a search for an inhibitor with high affinity for a single target. However, this approach does not always lead to success, and the number of single-target drugs discovered in recent years is quite small [37]. An alternative tactic for drug design is a multi-target methodology, which implies the partial inhibition of several targets simultaneously by one compound. In this case, the inhibitor should not necessarily demonstrate a high binding affinity for these targets. Indeed, surprising advances in multi-target drug discovery have demonstrated that partial inhibition of a small number of targets can be more effective than complete inhibition of a single target [37].
The goal of the present work was to discover a new class of dual inhibitors of Mpro and PLpro with predicted high potency and low mammalian toxicity using in silico original drug design encompassing a novel computational multistep approach similar in principle to a recent report by Yang et al. [38]. Starting with a compound predicted in our previous work to have high inhibitory potency against Mpro, we have applied our computational workflow to yield a series of novel structures exhibiting the desired inhibitory potencies and ADMET properties as assessed in silico. These compounds all contain heterocyclic rings (in particular, pyrazolopyridazine and tetrazole moieties) that are important organic building blocks for the construction of bioactive molecules. Pyrazolopyridazine and tetrazole derivatives have a wide range of biological activities, including antibacterial, antifungal, anticancer, analgesic, anti-inflammatory, and antidiabetic properties [39,40]. Moreover, these derivatives are synthetically accessible compounds with significant antiviral activity [[41], [42], [43]]. Therefore, our identified compounds, which contain pyrazolopyridazine and tetrazole moieties as organic scaffolds, were deemed suitable as starting points for continued discovery of novel SARS-CoV-2 Mpro/PLpro dual inhibitors and further development as potential therapeutic drugs for COVID-19.
2. Materials and methods
2.1. Creation and screening of the virtual ligand library
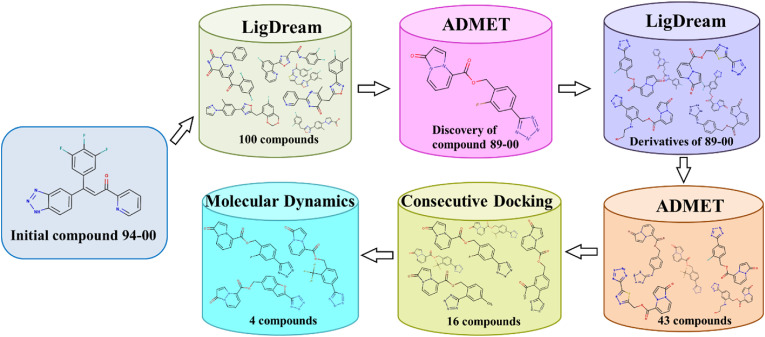
The workflow for creating and screening the virtual ligand library is shown in Fig. 1 . Starting with structure 94–00 (Table 1), which was identified in our previous study as a potential inhibitor of Mpro [44]. 2D structures of new ligands (100 compounds) were generated using an artificial neural network-driven platform, LigDream, as a shape decoding tool (Fig. 1; Fig. S1) [45,46]. The SMILE string of compound 94–00 was uploaded to the server and ran to generate 100 SMILE strings for new compounds. Default parameters of LigDream for generation of ligands were used.
Fig. 1.
Workflow for creation and screening of the virtual ligand library.
After filtering these structures for desirable ADMET properties using ADMETlab 2.0 [47] (Tables S4 and S5) (section 2.5), compound 89–00 (Table 1) was selected for a second round of ligand generation by LigDream to produce 2D structures of 43 novel 1-oxo-1H-pyrazolo[1,2-a]pyridazine derivatives (Table S1). The 2D structures were converted to 3D using MarvinSketch 21.10 with the addition of true ionization and tautomeric states at pH 7.4. All ligands were structurally optimized using the Universal Force Field [48] with a steepest descent algorithm (500 steps) in Avogadro 1.2.0 [49]. OpenBabel 3.0.0 [50] was used to convert the 3D structures (as mol files) to mol2 and pdbqt formats for use in consecutive molecular docking. For the first phase of docking, the 43 ligands generated by the second cycle of LigDream were docked into Mpro and PLpro by GOLD 5.3. The 16 highest-scoring compounds from GOLD were redocked with AutoDock Vina 1.1.2 along with three reference compounds to determine the affinity scores shown in Table 1 (sections 2.2 and 2.3). To gauge their intrinsic diversity, the structural and pharmacophoric Tanimoto similarities of the 16 new ligands and three reference compounds were quantified relative to compound 89–00 using ROCS 3.5.0.2: OpenEye Scientific Software, Santa Fe, NM, http://www.eyesopen.com (Fig. 2 ). Based on rankings of docking scores along with favorable ADMET characteristics, four compounds were then subjected to molecular dynamics (MD) simulations in YASARA 21.12.19 to assess the stability of the ligand-protein complexes (section 2.4).
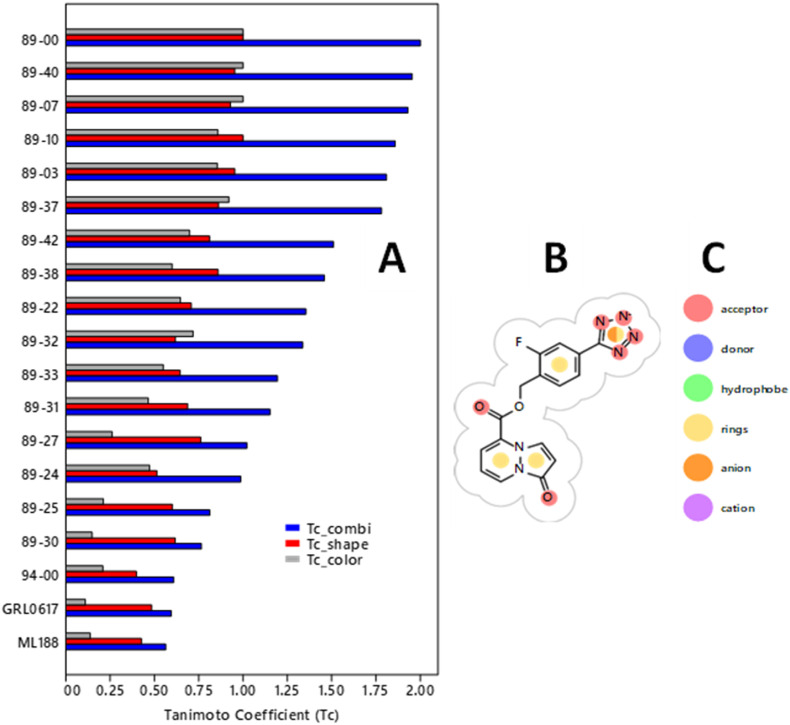
Fig. 2.
(A) Tanimoto similarity coefficients (Tc) for the 16 89-series compounds with the highest PLP docking scores from GOLD and 3 reference compounds (94–00, GRL0617, and ML188). Bar colors: gray = Tc_color (pharmacophore similarity); red = Tc_shape (shape similarity from overlap of molecular volumes); blue = Tc_combi (Tc_color + Tc_shape). Tc values are relative to compound 89–00. Tc_color and Tc_shape range from 0 (no similarity) to 1 (complete similarity). Tc_combi ranges from 0 (no similarity to 2 (complete similarity). (B) 2D pharmacophoric structure of 89–00 showing its 3 types of pharmacophores. (C) Color codes for the 6 pharmacophores assessed (red = acceptor, blue = donor, green = hydrophobe, yellow = rings, orange = anion, magenta = cation). Tc values were calculated using ROCS 3.5.0.2: OpenEye Scientific Software, Santa Fe, NM, http://www.eyesopen.com [77].
2.2. Preparation of protein docking targets
The X-ray crystal structures of SARS-CoV-2 main protease (PDB ID: 7L0D, resolution 2.39 Å) and SARS-CoV-2 papain-like protease (PDB ID: 7LBR, resolution 2.20 Å) were downloaded from the Protein Data Bank (PDB). Potential allosteric binding sites of the Mpro dimer (7L0D) were detected using the DoGSiteScorer tool [51,52] of the structure-based modeling server, ProteinsPlus [53,54]. For molecular docking, co-crystallized ligands, ions, and solvents were removed from the protein structures. The asymmetric unit of PLpro (7LBR) contained two chains, A and B; chain B was selected for use based on its higher-quality structure factors given in the PDB. The protonation states of proteins were adjusted at pH 7.4 using PROPKA 3.1 with addition of missing atoms and overall optimization of the hydrogen-bonding network using PDB2PQR 2.1, as provided by the ProteinPrepare tool on the PlayMolecule web server [55].
2.3. Consecutive molecular docking
Consecutive molecular docking was performed using CCDC GOLD Suite 5.3 software [56] followed by AutoDock Vina 1.1.2 on the graphical platform, MGL Tools 1.5.6 [57,58].
GOLD (Genetic Optimization for Ligand Docking) is a genetic algorithm for docking flexible ligands into protein binding sites. GOLD was used for primary fast docking (calculation of fitness) and identification of possible Mpro/PLpro binders. ChemPLP [59] was selected as the scoring function with maximal search efficiency and number of operations. Default parameters were used for the genetic algorithm settings. The active sites of Mpro (7L0D) and PLpro (7LBR) were centered on the original co-crystallized ligands with a radius of 10 Å to define the search space. Input ligands with full protonation were used in mol2 format.
Subsequent to the initial docking with GOLD, AutoDock Vina was used for re-docking and prediction of the apparent free energies of binding (ΔG, also referred to as "affinity", in kcal/mol [57]) of prospective dual inhibitors of Mpro/PLpro. Kollman charges were added for proteins and Gasteiger charges were applied for ligands. The following docking parameters were used: number of XYZ grid points of the cubic simulation cell (60 × 60 × 60 Å), spacing (0.5 Å), XYZ coordinates of the grid center for the monomer of 7L0D (10.807, −17.259, 21.463), grid center for the monomer of 7L0D; and XYZ coordinates of the grid center for the B-chain of 7LBR (2.68, −63.737, 3.074). Exhaustiveness was set to 24 and other parameters were set to default values. Input ligands with polar hydrogens were used in pdbqt format. For docking of the 7L0D dimer, the following parameters were used: XYZ number of grid points of the cubic simulation cell (62 × 72 × 80 Å), spacing (1.0 Å), XYZ coordinates of the grid center (0.129, −11.308, −0.152), and exhaustiveness (100).
3D images of docking results were generated using UCSF Chimera 1.14 [60]. 2D images of protein-ligand interactions were constructed with BIOVIA Discovery Studio® Visualizer 21.1.0.20298 [61].
2.4. MD simulations
Each ligand-protein complex for MD simulations was obtained by molecular docking of the ligand into the known active site of the protein using AutoDock Vina as described in section 2.2.
All steps of the MD simulation were carried out using YASARA-Structure 21.12.19 for Linux [62,63]. The computer system was a desktop workstation equipped with an AMD 3970 × 32-core CPU @3.7 GHz accelerated by an Nvidia RTX 3090 GPU using OpenCL. The operating system was Linux Mint 20.3 Xfce, based on Ubuntu 20.04 LTS.
The ligand-protein complex was centered within a cuboid simulation cell to encompass the protein with an additional 10 Å clearance on all sides. The long axis of the protein was aligned parallel to the x-coordinate of the cell. Periodic boundary conditions were used. Explicit solvent consisted of TIP3 water [64] containing 0.15 M NaCl at pH 7.4 and additional Na or Cl ions as needed to balance the total charge of the system.
After steepest descent and simulated annealing minimizations to remove clashes followed by optimization of the hydrogen-bonding network [65,66], the simulation was run for 100.5 ns using the AMBER14 force field for protein [67] augmented by GAFF2 [68] and AM1BCC [69] for ligand components. The cutoff for van der Waals forces was 8 Å [70]. For electrostatic forces, the Particle Mesh Ewald (PME) algorithm was used [71]. Equations of motion were integrated with a multiple timestep of 1.25 fs (bonded interactions) and 2.50 fs (nonbonded interactions) at 310 K and 1 atm (modified NPT ensemble). Pressure was automatically regulated by isotropically resizing the simulation cell to achieve the target solvent density of 0.997 g/ml [72].
MD trajectories were saved every 100 ps as YASARA SIM files until 1005 files were collected, representing 100.5 ns of simulation time, in order to ensure that 100 ns durations were captured for each run. The trajectories were then processed in YASARA to yield data for assessing the characteristics and stability of the ligand-protein complexes, including the radius of gyration of the complex (Rg), root-mean-square-deviation of the C-alpha backbone of the protein (RMSD-Cα), root-mean-square-deviation of the ligand (RMSD-Ligand), and the root-mean-square-fluctuation per residue of the protein (RMSF).
Binding affinity (ΔG, no electrostatic contribution) of compounds was calculated for the last snapshot of each MD simulation using the PRODIGY-LIGAND web server [73,74].
2.5. In silico prediction of physicochemical properties and pharmacokinetic ADMET profiles
The following parameters were calculated for the pyrazolopyridazines using ADMETlab 2.0 [47]:
Physical and medicinal chemistry: MW, number of hydrogen bond acceptors, number of hydrogen bond donors, number of rotatable bonds, topological polar surface area, log of the octanol/water partition coefficient, logP at physiological pH 7.4, synthetic accessibility score, Lipinski rule, Pfizer Rule, Golden Triangle, PAINS, ALARM NMR, BMS, and Chelator Rule.
Pharmacokinetics, absorption: Caco-2 permeability, MDCK permeability, Pgp-inhibitor, Pgp-substrate, and human intestinal absorption.
Pharmacokinetics, distribution: plasma protein binding, volume of distribution, blood-brain barrier penetration, fraction unbound in plasma), and xenobiotic metabolism by cytochrome P450 (CYP) enzymes (CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 inhibitor and substrate potential).
Pharmacokinetics, excretion: drug clearance.
Toxicity: human ether-a-go-go related gene blockers, human hepatotoxicity, drug-induced liver injury, Ames test for mutagenicity, rat oral acute toxicity, carcinogenicity, respiratory toxicity, nuclear receptors (NR) or their ligand-binding domains (LBD) (NR-AR-LBD, NR-Aromatase, NR-ER-LBD, NR-PPAR-gamma), and stress response (SR) pathways (SR-ARE, SR-HSE, SR-MMP and SR-p53).
2.6. Statistical analyses of MD simulation data
Time-series data (Rg, RMSD-Cα, and RMSD-Ligand trajectories) from MD simulations were determined to be autocorrelated by applying the autocorrelation function (acf) in R version 4.2.1. Mean and standard error of the mean (SEM) values for Rg, RMSD-Ca, and RMSD-Ligand were calculated using block averaging to correct for autocorrelation [75] as implemented by the R function, block_average [76].
Statistical significance of the differences among all pairwise comparisons of mean values was determined by one-way ANOVA followed by the Tukey multiple comparisons test when the variances were statistically equivalent. For unequal variances, the Brown-Forsythe and Welch corrections were applied, and Dunnett's T3 multiple comparisons test was used. The alpha level for ANOVA was 0.05, and p < 0.05 was the level of significance for post-hoc tests. RMSF profiles were found to be not normally distributed by all four of the following tests for normality: D'Agostino and Pearson, Anderson-Darling, Sharpiro-Wilks, and Kolmogorov-Smirnov. Therefore, the non-parametric Spearman correlation coefficient was used to assess correlations among all pairs of RMSF profile data. All these tests were performed using GraphPad Prism version 9.4.1 for Windows 10 (GraphPad Software, San Diego, California USA, www.graphpad.com).
3. Results and discussion
3.1. Creation and screening of the virtual ligand library
As shown in Fig. 1, the azachalcone 94–00 was chosen as the initial seed structure for the computational design of dual inhibitors of Mpro and PLpro. This compound was identified in our previous in silico study as a potential inhibitor of Mpro (ΔG −8.5 kcal/mol by Autodock Vina) [44]. However, according to the ALARM NMR and BMC tools of ADMETlab 2.0, this ligand has a poor ADMET profile and possesses the potential to act as a thiol-reactive compound – an undesirable trait for a non-covalent inhibitor (Table S4). Therefore, through a generative and screening approach, we sought to discover novel dual inhibitors of Mpro and PLpro that exhibited favorable ADMET characteristics.
Thus, the first LigDream cycle (Fig. 1) generated 100 new structures (list in Fig. S1) that were subjected to a comprehensive in silico ADMET evaluation, resulting in the selection of a single compound (pyrazolopyridazine 89–00) with a favorable predicted pharmacokinetics profile. This molecule was used for a second generative cycle with LigDream to produce 43 novel pyrazolopyridazine structures (including compound 89–00; Table S1) for subsequent use as ligands in the consecutive docking and molecular dynamics steps.
3.2. Consecutive docking for identification of dual Mpro/PLpro inhibitors
To identify prospective high affinity dual inhibitors of Mpro and PLpro among the 43 newly generated pyrazolopyridazines (compound 89–00 and its derivatives), consecutive docking of the ligands into the catalytic active sites of the two proteases was performed with GOLD and AutoDock Vina.
Table S1 lists the GOLD ChemPLP docking scores for the 43 ligands. Of these, 16 compounds with the highest ChemPLP scores were selected for redocking with AutoDock Vina to determine the apparent free energies of binding (ΔG) [57]. In addition, compounds ML188, GRL0617, and 94–00 were included as reference compounds for comparison.
To quantitatively assess the intrinsic diversity of these 19 ligands, Tanimoto similarity coefficients (Tc) for shape (Tc_shape), pharmacophores ("color", Tc_color), and the combination of these two parameters (Tc_combi) were calculated as shown in Fig. 2. Compounds 89–00, 89–07, and 89–40 had the same Tc_color values but differed sufficiently in Tc_shape to yield noticeable differences in T_combi. Overall, there was a gradient in T_combi values throughout the 89-series of compounds ranging from 0.76 to 2.00. In particular, all of the 89-series of compounds had T_combi values greater than those of the three reference compounds (0.56–0.61), including the initial compound 94–00. These results serve to illustrate and confirm the ability of the workflow depicted in Fig. 1 to generate a series of structures distinct from the initial structure yet, as shown in Table 1, capable of producing favorable docking scores.
The Vina docking results are shown in Table 1. Excluding the three reference compounds, four ligands were selected for more detailed study based on their Mpro and PLpro docking scores and favorable ADMET properties: 89–00, 89–07, 89–32 and 89–38.
As shown in Table 1, ligand 89–00 had the same binding affinity as the reference inhibitor ML188. With respect to compounds 89–07 and 89–38, these ligands had the best inhibitory potential (ΔG – 8.7 kcal/mol) in comparison with 89–00 (ΔG – 7.8 kcal/mol) and 89–32 (ΔG – 8.1 kcal/mol). Moreover, 89–07 and 89–38 had higher in silico binding affinities for Mpro than ML188.
3.3. Docking interactions of selected ligands within the Mpro active site
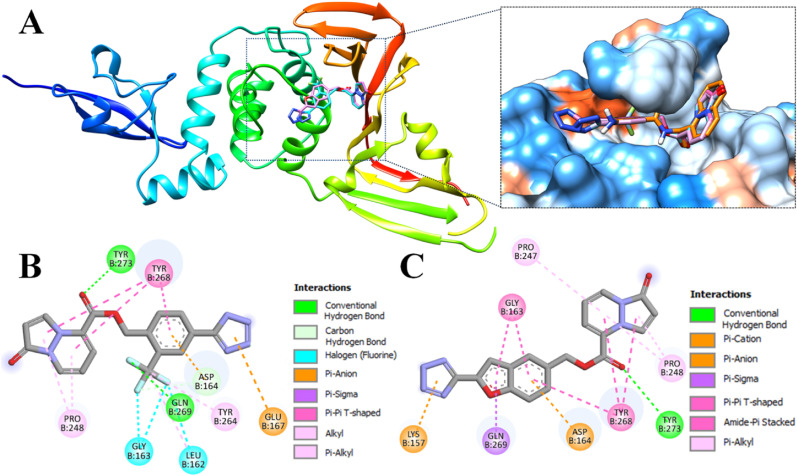
Ligands 89–00, 89–07, 89–32 and 89–38 were well accommodated within the active site of the Mpro monomer. Moreover, ligands 89–07 and 89–38 displayed a similar orientation (Fig. 3 A).
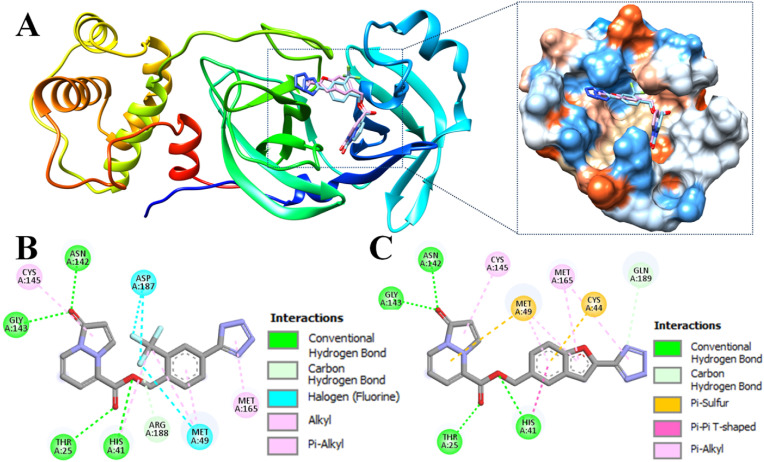
Fig. 3.
Docking poses of compounds 89–07 (cyan) and 89–38 (pink) within the active site of Mpro (PDB ID 7L0D). (A) Ribbon view depicted on the left; magnified hydrophobicity surface on the right (colors: blue = greatest polarity; red = greatest hydrophobicity; white = intermediate). (B) 2D interactions of 89–07. (C) 2D interactions of 89–38.
Despite the similar orientations of 89–07 and 89–38, it is important to note that 89–00, 89–07, 89–32, and 89–38 exhibited different types of interactions within the active site – especially with the catalytic dyad residues His41 and Cys145 of Mpro (Table 2 ). For example, ligand 89–00 formed strong hydrogen bonds with His41 and Gln189 as well as a halogen bond with His164. This compound occupied subsites S1–S3 of the active site [78]. Furthermore, ligands 89–07 and 89–38 engaged in many interactions with active site residues of Mpro (Fig. 1B and C). In particular, 89–07 and 89–38 were able to interact with Thr25, Asn142 and Gly143 to form conventional hydrogen bonds. Moreover, the fluorine atom of ligand 89–07 had strong halogen bonds with Met49 and Asp187. Ligand 89–38 displayed a T-shaped pi-pi interaction with His41 and hydrophobic interactions (pi-sulfur) with Cys44 and Met49. In general, pyrazolopyridazines 89–07 and 89–38 occupied subsites S1′, S2 and S3 of the Mpro active site.
Table 2.
Interactions of four selected ligands and a reference compound with the Mpro active site.
| Compound | Interacting residues (type and distance in Å) |
|---|---|
| 89–00 | His41a (HBb, 2.20, pi-pi T-shaped, 5.23), Met49 (pi-sulfur, 3.78, 4.04), His164 (halogen bond, 3.41), Met165 (pi-alkyl, 5.17), Glu166 (pi-donor HB, 2.54, pi-anion, 4.60), Gln189 (HB, 2.25) |
| 89–07 | Thr25 (HB, 2.53), His41 (carbon HB, 2.24, alkyl, 5.08), Met49 (halogen bond, 3.41, pi-alkyl, 4.26, alkyl, 4.93), Asn142 (HB, 2.27), Gly143 (HB, 2.29), Cys145 (pi-alkyl, 5.24), Met165 (pi-alkyl, 5.20), Asp187 (halogen bond, 3.01, 3.03), Arg188 (carbon HB, 2.18) |
| 89–32 | Cys44 (pi-sulfur, 5.64), Met49 (pi-sulfur, 3.51), Cys145 (pi-sulfur, 5.50), Met165 (pi-alkyl, 4.22, 4.49), Glu166 (carbon HB, 2.72, pi-donor HB, 3.11), Asp187 (carbon HB, 2.91) |
| 89–38 | Thr25 (HB, 2.20), His41 (carbon HB, 2.49, 2.93, pi-pi T-shaped, 5.06), Cys44 (pi-sulfur, 5.99), Met49 (pi-sulfur, 5.20, pi-alkyl, 4.53, 5.23), Asn142 (HB, 2.22), Gly143 (HB, 2.16), Cys145 (pi-alkyl, 5.11), Met165 (pi-alkyl, 4.91, 5.31), Gln189 (carbon HB, 2.64) |
| ML188 | Thr26 (carbon HB, 2.55), Leu27 (pi-alkyl, 5.32), His41 (pi-pi stacked, 4.65, pi-alkyl, 4.76), Met49 (pi-sulfur, 5.54, alkyl, 4.83), Phe140 (carbon HB, 2.56), Leu141 (amide pi-stacked, 4.05), Asn142 (HB, 2.57), Gly143 (HB, 2.10), Cys145 (pi-alkyl, 3.78), His163 (HB, 1.96), Met165 (amide pi-stacked, 4.96), Glu166 (HB, 2.59) |
Residues in boldface are members of the His41-Cys145 active site dyad.
HB = Conventional hydrogen bond.
3.4. Discovery of potential allosteric sites and modulators
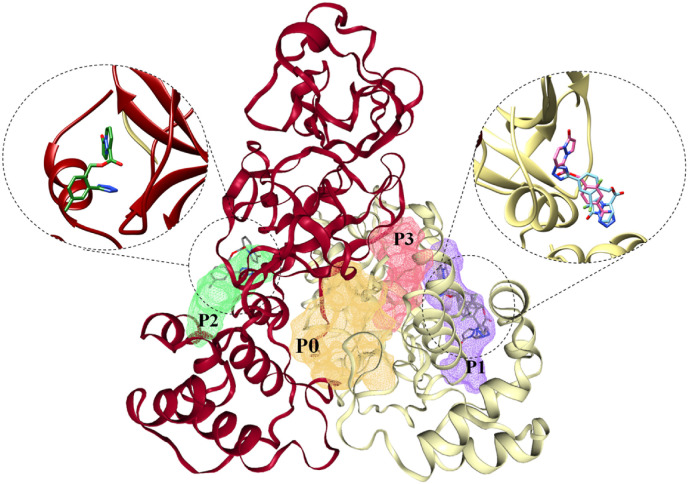
Using the ProteinPlus web server, four potential allosteric sites (pockets P0–P3) with high drug scores (>0.7) were located in the Mpro dimer (Fig. 4 ). It is important to note that the integrity of P1 and P2 is crucial for Mpro activity, which appears to be coupled with the conformational dynamics of the protease catalytic site [34,79]. Thus, when compounds bind to P1–P2, they can disrupt Mpro dimerization, resulting in inhibition of substrate cleavage via allosteric regulation [80]. Moreover, recently discovered inhibitors (e.g., AT7519 and pelitinib) of allosteric binding sites P1 (in the C-terminal dimerization domain) and P2 (between the catalytic domains and the dimerization domain) have demonstrated moderate to high antiviral activity in vitro [81]. Furthermore, the hydrophobic nature of the residues forming P1 is conserved for all human coronavirus Mpro enzymes. This means that allosteric modulators of this binding site in SARS-CoV-2 are likely to be effective against other coronaviruses as well [81].
Fig. 4.
The docking poses of compounds 89–00 (pink, right magnified view), 89–40 (dark blue, right magnified view) and 89–27 (forest green, left magnified view) within the Mpro dimer (chain A, dark red; chain B, khaki; drug pockets: P0 (yellow), P1 (purple), P2 (green), P3 (red).
Global blind docking with AutoDock Vina revealed that compounds 89–00, 89–27 and 89–40 occupied P1–P2 allosteric pockets, which were located outside of the catalytic active sites, thereby identifying potential allosteric sites and allosteric modulators.
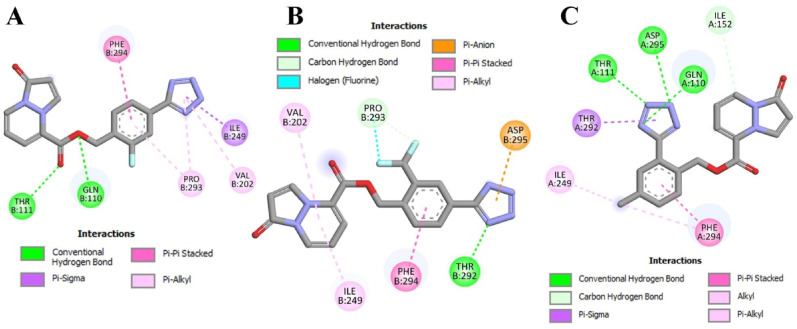
According to the docking data, the possible allosteric Mpro modulators 89–00 (ΔG –8.6 kcal/mol) and 89–40 (ΔG –8.2 kcal/mol) were accommodated in the P1 binding pocket of chain B protein (Fig. 4). Thus, ligand 89–00 formed a strong hydrogen bond with Thr111 (2.34 Å) and a weaker interaction of the same type with Gln110 (Fig. 5 A). Furthermore, compound 89–00 was able to interact with Pro293 and Val202 (pi-alkyl), and it had a pi-sigma contact with Ile249. Interestingly, the substituted benzene ring of pyrazolopyridazines 89–00 and 89–40 interacted with Phe294 (pi-pi stacked). For compound 89–40 (Fig. 5B), a hydrogen bond with Thr292 (2.94 Å) and a halogen bond (Pro293, 3.61 Å) were detected. Moreover, this ligand had an interaction with Asp295 (pi-anion, 3.56 Å).
Fig. 5.
2D interactions of 89–00 (A), 89–40 (B), and 89–27 (C) within potential allosteric sites of the Mpro dimer.
In the case of 89–27 (ΔG –8.9 kcal/mol), this ligand occupied the P2 drug pocket of chain A of the Mpro dimer (Fig. 5C). Interestingly, as with ligands 89–00 and 89–40, compound 89–27 interacted with Phe294 through pi-pi stacking (3.76 Å). Furthermore, the tetrazole ring of 89–27 was able to interact with Gln110 (2.29 Å), Thr111 (2.10 Å) and Asp295 (2.98 Å) to form hydrogen bonds. Moreover, weak interactions with Thr292, Ile152 and Ile249 were detected for pyrazolopyridazine 89–27.
3.5. Docking interactions of selected ligands within the PLpro active site
The selected ligands 89–00, 89–07, 89–32 and 89–38 had similar favorable docking scores and spatial orientations in the PLpro active site as the reference ligand, GRL0617 (Table 1 and Fig. 6 A).
Fig. 6.
Docking poses of compounds 89–07 (cyan) and 89–38 (pink) within the active site of PLpro (PDB ID 7LBR) and hydrophobicity surface of PLpro substrate binding site (magnified view) with 89–07, 89–38 and GRL0617 (in orange) (A) Ribbon view depicted on the left; magnified hydrophobicity surface on the right (colors: blue = greatest polarity; red = greatest hydrophobicity; white = intermediate). (B) 2D interactions of 89–07. (C) 2D interactions of 89–38.
Interestingly, the pyrazolopyridazinone moieties of ligands 89–00, 89–07, 89–32 and 89–38 were located in the active site of PLpro, akin to the position of the naphthalene ring of GRL0617. Analogously, the substituted benzene moieties of these ligands (the benzofuran ring of 89–38) and GRL0617 were accommodated identically in the active site. Despite these similarities, it is noteworthy that compounds 89–00, 89–07, 89–32 and 89–38 had different interactions with key amino acid residues in the PLpro active site (Table 3 ; Fig. 6B and C).
Table 3.
Interactions of four selected ligands and a reference compound with the PLpro active site.
| Compound | Interacting residues (type and distance in Å) |
|---|---|
| 89–00 | Pro248a (pi-alkyl 4.21, 4.80), Tyr273 (HBb, 2.63), Gly163 (halogen bond, 3.47, pi-amide, 4.53), Lys157 (HB, 2.65), Asp164 (pi-anion, 4.13), Tyr268 (pi-pi T-shaped, 4.95, 5.25, 5.07) |
| 89–07 | Glu167 (pi-anion, 4.34), Gln269 (HB, 2.68), Asp164 (pi-anion, 4.02), Tyr264 (HB, 1.75, alkyl, 4.28), Gly163 (halogen bond, 2.78, 3.06), Leu162 (halogen bond, 2.96, alkyl, 4.23), Pro248 (pi-alkyl, 4.24, 4.73), Tyr273 (HB, 2.30), Tyr268 (pi-pi T-shaped, 4.81, 5.30, 4.93) |
| 89–32 | Lys157 (HB, 1.89), Gly163 (carbon HB, 2.66, pi-amide, 4.39), Asp164 (HB, 2.45, pi-anion, 4.00), Tyr273 (HB, 2.32), Pro248 (pi-alkyl, 4.29, 4.93), Tyr268 (pi-pi T-shaped, 5.13, 4.89, 5.20, HB, 1.99), Gln269 (HB, 1.84) |
| 89–38 | Pro248 (pi-alkyl, 4.24, 4.57), Pro247 (pi-alkyl, 5.48), Asp164 (pi-anion, 4.30), Gly163 (pi-amide, 4.14, 4.34), Lys157 (pi-cation, 3.03), Gln269 (pi-sigma, 2.68), Tyr273 (HB, 2.53), Tyr268 (pi-pi T-shaped, 4.96, 5.32, 5.49) |
| GRL0617 | Leu162 (alkyl, 4.15), Asp164 (HB, 1.93, pi-anion, 4.36), Pro247 (pi-alkyl, 5.28), Pro248 (pi-alkyl, 4.58, 4.36, alkyl, 4.52), Tyr264 (pi-alkyl, 3.91, 4.76), Tyr268 (pi-pi T-shaped, 5.09, 5.26, 5.30, carbon HB, 2.63), Gln269 (HB, 1.83, pi-sigma, 2.58), Tyr273 (pi-alkyl, 5.26) |
Residues in boldface are important for substrate binding with PLpro [33].
HB = Conventional hydrogen bond.
All selected ligands interacted with Pro248 (pi-alkyl), Tyr268 (pi-pi T-shaped) and Asp164 (pi-anion). Additionally, the ligand 89–32 formed a hydrogen bond with Asp164. The pyrazolopyridazines 89–07 and 89–32 each interacted with Gln269 (HB), while ligand 89–38 displayed a pi-sigma contact with Gln269. Furthermore, compound 89–07 also contacted Leu162 and formed a hydrogen bond with Tyr264. Moreover, ligand 89–38 additionally interacted with Pro247 (pi-alkyl) and this ligand had a more favorable calculated binding affinity to PLpro than pyrazolopyridazines 89–00, 89–07 and 89–32.
3.6. MD simulations for Mpro complexes
To evaluate the influence of ligand binding on dynamic properties of Mpro and PLpro and the stability of the ligand-protein complexes, MD simulations were performed. The following parameters were computed: radius of gyration of the complex (Rg), root-mean-square-deviation of the C-alpha backbone of the protein (RMSD-Cα), RMSD of ligand movement after superposing the protein on its reference structure (RMSD-Ligand), and the root-mean-square-fluctuation per residue of the protein (RMSF). In addition, the binding affinity of the ligands was calculated for the last snapshot of each MD simulation using the PRODIGY-LIGAND web server.
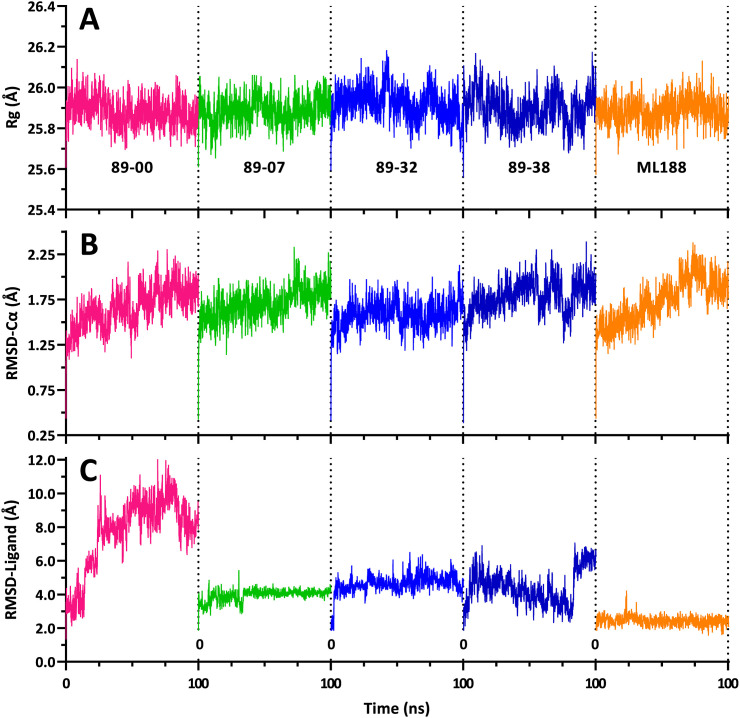
For the Mpro complexes, the biologically active dimer form of the protein in complex with each of the four selected ligands 89–00, 89–07, 89–32, and 89–38 as well as the reference compound ML188 was used for MD simulations. The MD trajectories are displayed in Fig. 7 , and the statistical analyses of the mean values of the Rg, RMSD-Cα, and RMSD-Ligand parameters are shown in Fig. 8 .
Fig. 7.
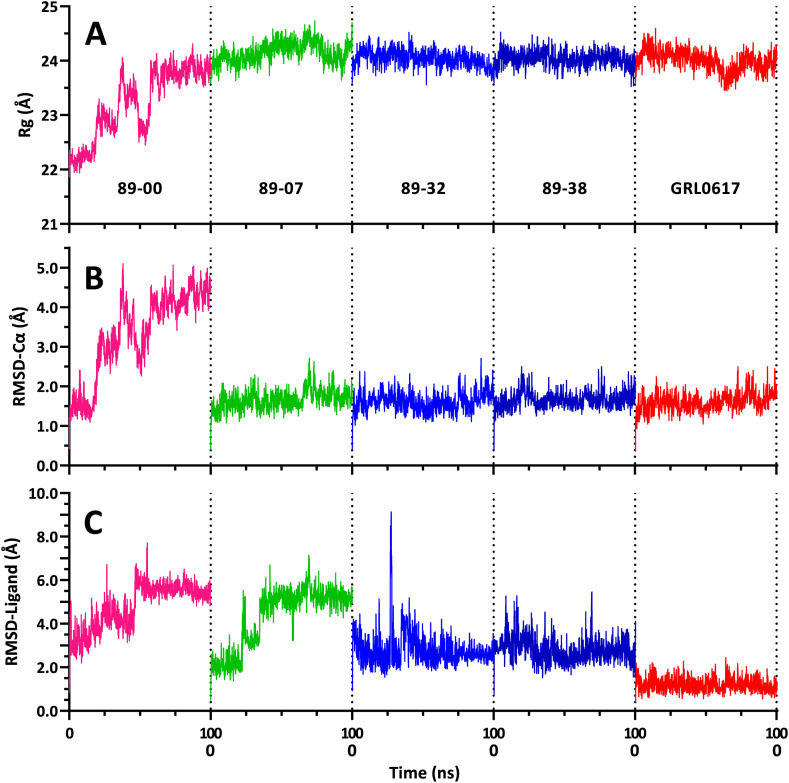
Trajectories (100 ns) from MD simulations of Mpro dimer complexes with ligands 89–00, 89–07, 89–32, 89–38, and reference compound ML188 bound to the active site of the dimer A-chain. (A) Rg of dimer complexes. (B) RMSD-Cα of A-chain complexes. (C) RMSD-Ligand: movement of the ligand relative to the A-chain. Trajectory colors: magenta, 89–00; green, 89–07; blue, 89–32; dark blue, 89–38; and orange, ML188.
Fig. 8.
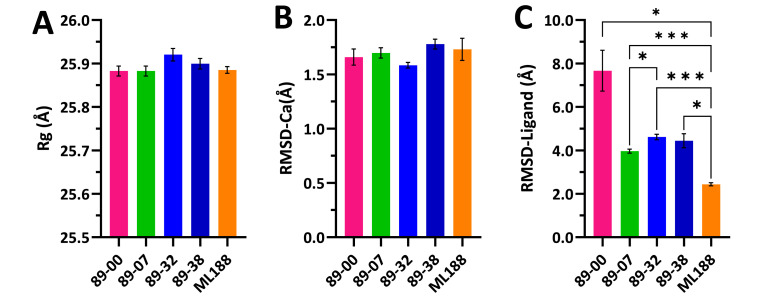
Statistical comparisons of mean-value parameters from 100 ns MD simulations of ligand complexes with Mpro dimers. (A) Rg of dimers. (B) RMSD-Cα of A-chain complexes. (C) RMSD-Ligand: movement of ligand relative to A-chain. Values are means ± SEM (n = 5 × 20 ns blocks) computed by block averaging to correct for time-series autocorrelation. There were no statistically significant differences among all pairwise comparisons of means for Rg or RMSD-Cα (one-way ANOVA, Tukey multiple comparisons test, p > 0.05). For RMSD-Ligand values, there were statistically significant differences between 5 pairs of means: *p < 0.05; ***p ≤ 0.0005 (one-way ANOVA with Brown-Forsythe and Welch corrections for unequal variances and Dunnett's T3 multiple comparisons test). Bar colors: magenta, 89–00; green, 89–07; blue, 89–32; dark blue, 89–38; and orange, ML188.
The Rg values of each of the Mpro-ligand complexes remained essentially constant throughout the 100 ns simulations (Fig. 7A) indicating that the compactness of the proteins did not substantially change, suggesting that the ligands did not dissociate from the complexes. Moreover, the mean Rg values of the five complexes were statistically identical (Fig. 8A), varying over a small range from 25.88 ± 0.011 Å (mean ± SEM) for the 89–00 and 89–07 complexes to 25.92 ± 0.014 Å (mean ± SEM) for the 89–32 complex. These values of Rg for Mpro dimers in complex with small molecule ligands at 310 K are in excellent agreement with previous reports of 26.2 ± 0.15 Å for the apo dimeric protein at 300 K [82] and 26.1 ± 0.2 Å at 310 K [82], respectively, as well as 25.7 ± 0.3 Å to 26.1 ± 0.2 Å for the Mpro dimer in complex with 17 different small-molecule ligands at 310 K [82].
In like manner to the Rg trajectories, the RMSD-Cα profiles of the Mpro complexes were similar throughout the 100 ns simulations (Fig. 7B) and had statistically equivalent mean values ranging from 1.58 ± 0.03 to 1.79 ± 0.04 Å (Fig. 8B). These results are in accord with a comprehensive MD study of 62 Mpro complexes with reversible ligands that exhibited an average RMSD-Cα of 1.79 ± 0.04 Å, whereas the apo protein had an RMSD-Ca of 1.81 Å [83]. Thus, the binding of these reversible ligands to Mpro had little or no effect on the backbone dynamics of this protein.
In contrast to the Rg and RMSD-Cα results, the RMSD-Ligand trajectories of the pyrazolopyridazine ligands were different from that of inhibitor ML188, which maintained consistently low RMSD-Ligand values throughout the simulation (Fig. 7, Fig. 8C). The highest RMSD values were detected for compound 89–00 after ∼25 ns of MD simulation. Although this ligand was still located within the active site of Mpro with a final ΔG value of −5.7 kcal/mol and exhibited interactions with residues Met49, Met165, Arg188 and Gln189, it did not display any post-MD interactions with the active site dyad residues, His41 and Cys145 (Table S2). Compound 89–38 appeared to have reached a stable state after ∼40 ns, but at ∼85 ns, its RMSD rose abruptly from 3.0 to 6.9 Å. Nevertheless, 89–38 remained within the active site and made sufficient residue contacts (including His41 but not Cys145 of the catalytic dyad) to yield the second-best binding affinity (ΔG –7.6 kcal/mol) of the four test ligands. Ligands 89–07 and 89–32 maintained stable RMSD-Ligand values during 100 ns of MD, and their final snapshots revealed contacts with both residues of the active site dyad. Moreover, the final calculated post-MD binding affinity of 89–07 was the most favorable of the four test ligands (ΔG –9.1 kcal/mol) (Table S2). These impressions of the RMSD-Ligand trajectories coupled with post-MD binding affinities were in agreement with the rank order of the mean RMSD-Ligand values: ML188 < 89–07 < 89–38 < 89–32 < 89–00 (Fig. 8C).
The per-residue RMSF profiles of the A and B chains of Mpro dimer ligand complexes are depicted in Fig. S2. Each chain has 305 amino acid residues, and the highest RMSF values were observed for the C-terminal residue Phe305 of MPro complexes with ML188A (8.40 Å), ML188B (5.53 Å), and 89–38B (7.33 Å). Nevertheless, large fluctuations of Phe305 are not unusual given that this residue is located at the end of a flexible 5-residue C-terminal loop. Otherwise, the overall RMSF profiles were highly similar to each other with Spearman correlation coefficients between corresponding A and B chains of 0.85–0.93 and between A or B chains of different complexes ranging from 0.85 to 0.95 (Fig. S3).
However, the ligand RMSF values are of particular interest. The ligand was bound to the A-chain of the dimer and assigned residue number 306. Ligand RMSF values for the Mpro complexes were as follows (in descending order): 89–00 (4.90 Å), 89–32 (3.00 Å), 89–38 (2.08 Å), 89–07 (1.43 Å), and ML188 (1.27 Å). Thus, it is noteworthy that the rank order of the ligand RMSF values mirrored that of the RMSD-Ligand mean values shown in Fig. 8C. Furthermore, as with RMSD-Ligand values, smaller ligand RMSF values are better [84].
Considering all of the MD simulation results for the complexes with Mpro, the best candidates among the four test ligands for further consideration as inhibitors of this protease are 89–07, 89–32, and 89–38.
3.7. MD simulations for PLpro complexes
Complexes of the biologically active monomer form of PLpro with each of the four selected ligands (89–00, 89–07, 89–32, and 89–38) along with the reference inhibitor GRL0617 were used for MD simulations. The MD trajectories are displayed in Fig. 9 , and the statistical analyses of the mean values of the Rg, RMSD-Cα, and RMSD-Ligand parameters are shown in Fig. 10 .
Fig. 9.
MD trajectories of PLpro monomer active site complexes with selected ligands 89–00, 89–07, 89–32, 89–38, and reference compound GRL0617 during 100 ns. (A) Rg of complexes. (B) RMSD-Cα of complexes: movement of the protein backbone. (C) RMSD-Ligand: movement of the ligand relative to the protein. Trajectory colors: magenta, 89–00; green, 89–07; blue, 89–32; dark blue, 89–38; and red, GRL0617.
Fig. 10.
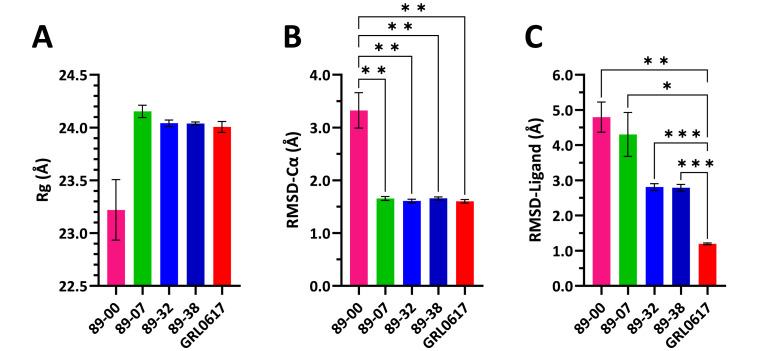
Statistical comparisons of mean-value parameters from 100 ns MD simulations of ligand complexes with monomeric PLpro. (A) Rg. (B) RMSD-Cα of complexes. (C) RMSD-Ligand: movement of ligand relative to the protein. Values are means ± SEM (n = 5 × 20 ns blocks for (A) and (C); n = 10 × 10 ns blocks for (B) computed by block averaging to correct for time-series autocorrelation. Because of the similar mean values and the high variance for 89–00, there were no statistically significant differences among all pairwise comparisons of means for Rg (p > 0.05). For RMSD-Ca and RMSD-Ligand values, there were statistically significant differences between 4 pairs of means: *p < 0.05; **p < 0.01; ***p = 0.0001 (one-way ANOVA with Brown-Forsythe and Welch corrections for unequal variances and Dunnett's T3 multiple comparisons test). Bar colors: magenta, 89–00; green, 89–07; blue, 89–32; dark blue, 89–38; and red, GRL0617.
As can be seen in Fig. 9A, the Rg of the PLpro complex with 89–00 increased during the 100 ns simulation from 22.3 to 23.8 Å. An increase in the Rg of a ligand-protein complex tends to be interpreted as dissociation of the ligand from the protein, although both increases and decreases in Rg have been reported in comparisons between proteins with and without bound ligands [85,86]. Moreover, the Rg value of the 89–00 complex that was attained toward the end of the simulation was within the range of Rg values seen with the other four complexes. In addition, the final snapshot of the simulation revealed that the ligand was still within the active site of the protein. Furthermore, Fig. 10A shows that the mean Rg value for the 89–00 complex was not statistically different from the mean Rg values of the other complexes, although this was because of the comparatively high variance of the 89–00 complex and the relatively small differences between the means. Nevertheless, the Rg trajectory of the 89–00 complex with PLpro was qualitatively distinct from those of the other complexes, and the reason for this discrepancy remains unexplained.
Fig. 9B shows that RMSD-Cα values for the PLpro complex with 89–00 increased from 1.5 to 4.4 Å during the 100 ns simulation, while this parameter maintained a steady average of 1.6 Å for the other complexes. Indeed, the mean RMSD-Cα value for the 89–00 complex was statistically different from the mean values for the other complexes, which did not differ from each other (Fig. 10B). A recent study of changes in protein flexibility from ligand binding found that the increase in RMSD-Cα values between apo and holo states was less than 0.5 Å for 90% of the 305 proteins investigated, but 10% of the proteins exhibited changes of greater than 2 Å [87]. However, MD simulations of PLpro have found RMSD-Cα increases from 2 to 4 Å for the native protein and increases from 2 to 5 Å in PLpro-ligand complexes during 100 ns [88]. Nonetheless, the erratic RMSD-Cα trajectory of the 89–00 complex compared with the others would tend to weigh against selecting this ligand for further consideration as a candidate for a PLpro inhibitor.
The RMSD-Ligand trajectories are shown in Fig. 9C. Both the 89–00 and 89–07 complexes displayed increases of 2.5 and 3.0 A, respectively, during 100 ns. In contrast, apart from a spike in the 89–32 complex at ∼27 ns, the 89–32 and 89–38 complexes maintained relatively steady RMSD-Ligand values of 2.8 A. However, the complex with the reference compound GRL0617 kept the lowest and most stable RMSD-Ligand value of 1.2 A throughout the 100 ns simulation, which compares favorably with an RMSD-Ligand value of 2.3 for this complex obtained in a recent MD study [89].
These observations of the trajectories in Fig. 9C were borne out by the statistical comparisons of mean values shown in Fig. 10C, wherein the RMSD-Ligand mean value of the GRL0617 complex was statistically different from all the other mean values. It was also evident that the mean values for the 89–32 and 89–38 complexes were essentially identical and less than the values for the 80–00 and 89–07 complexes, although these two pairs were not statistically distinct owing to the large variances in the 89–00 and 89–07 values.
Fig. S4 shows the RMSF profiles for the ligand complexes with PLpro. This protein has N-terminal and C-terminal loops that would be expected to exhibit a high degree of flexibility, and this is readily apparent from the RMSF plots. Other residues with relatively high flexibility in all of the complexes included Lys191, Thr226, Gly228, Lys229, and Ile315. Overall, the RMSF profiles were remarkably similar, with Spearman correlation coefficients ranging from 0.92 to 0.95 (Fig. S5).
Apart from the global similarities among the RMSF profiles of the complexes, it is important to focus on the ligand RMSF because it would be desirable to select compounds with low values of this parameter [90]. The ligand RMSF values for the PLpro complexes were as follows (in descending order): 89–07 (2.58 Å), 89–32 (1.82 Å), 89–38 (1.76 Å), 89–00 (1.65 Å), and GRL0617 (0.97 Å). In this case, the rank order of the ligand RMSF values did not coincide with that of the RMSD-Ligand mean values shown in Fig. 10C because 89–00 was ranked fourth rather than first, as occurred in the complex of this ligand with dimeric Mpro.
Despite the anomalous behavior of the PLpro complex with 89–00 in the three trajectories shown in Fig. 9A–C and that of the PLpro complex with 89–07 in Fig. 9C, these and the other two ligands were found in the active site in the last snapshot from the MD simulation. Moreover, all four test ligands had comparable post-MD binding affinities as the reference compound GRL0617 (Table S3). Nevertheless, in view of all the results from the MD simulations of PLpro complexes, the best of the four test ligands for further consideration as inhibitors of this protease are 89–32 and 89–38.
3.8. ADMET results for selected compounds
It is noteworthy that the predicted ADMET profiles of prospective dual Mpro/PLpro inhibitors (89–00, 89–07, 89–32 and 89–38) and allosteric modulators of Mpro dimer (89–00, 89–27 and 89–40) were significantly better than the profiles calculated for the reference inhibitors ML188 and GRL0617 (Tables S4 and S5). In particular, compounds 89–00, 89–07, 89–32 and 89–38 satisfied fundamental medicinal chemistry rules (Lipinski Rule, Pfizer Rule, and Golden Triangle) and did not exhibit any undesirable reactivity (PAINS, ALARM NMR, BMS, and Chelator Rule) in comparison with 94–00, ML188 and GRL0617 (Tables S4 and S5). Moreover, hERG blockade, mutagenicity (AMES toxicity), rat oral acute toxicity, and carcinogenicity were not predicted for these ligands. However, all compounds (89–00, 89–07, 89–32, and 89–38) together with the reference inhibitors (ML188 and GRL0617) displayed a potential risk of drug-induced liver injury (DILI) and human hepatotoxicity (H-HT). DILI and H-HT are common safety problems for many marketed drugs, including amoxicillin, diclofenac, isoniazid, and methotrexate [91]. Nevertheless, some degree of drug toxicity during COVID-19 treatment might be considered acceptable so that high doses of antiviral agents could be used for short period of time in view of recommendations that proliferation of the SARS-CoV-2 virus should be halted within the first few days of infection [12].
4. Conclusion
We have applied a computational workflow that combined an artificial neural network, LigDream, comprehensive ADMET evaluation, quantification of ligand diversity, consecutive molecular docking, and MD simulations to generate and characterize new compounds as potential dual inhibitors of the SARS-CoV-2 targets, Mpro and PLpro. An important feature of our protocol is that it was designed to find reversible noncovalent inhibitors of the viral cysteine proteases rather than irreversible covalent inhibitors. This approach identified four lead compounds, 89–00, 89–07, 89–32, and 89–38 with predicted high potency directed against the catalytic active sites of the target enzymes, favorable drug-like properties, and low mammalian toxicity. Moreover, affinities (ΔG) of our compounds determined by AutoDock Vina were in the range −8.1 to −8.7 kcal/mol for Mpro, which was more favorable than the reference inhibitor, ML188 (−7.9 kcal/mol). Our procedure also found three potential allosteric modulators of the Mpro dimer, 89–00, 89–27 and 89–40, as biocompatible compounds with high predicted affinities to Mpro in the range −8.2 to −8.9 kcal/mol. Similarly, the affinities of 89–00, 89–07, 89–32, and 89–38 for PLpro were in the range −9.1 to −9.7 kcal/mol, which closely approached that of the reference inhibitor GRL0617 (−9.9 kcal/mol). Data from 100 ns all-atom MD simulations demonstrated that the most stable complexes of Mpro and PLpro were afforded by compounds 89–32 and 89–38. These promising in silico results will require validation with in vitro and in vivo tests before the lead compounds could be advanced to clinical trials.
Author contributions
Roman S. Tumskiy: Conceptualization, Methodology, Writing - original draft, Writing - review & editing. Anastasiia V. Tumskaia: Writing - original draft, Writing - Review & Editing, Visualization. Iraida N. Klochkova: Conceptualization, Supervision. Rudy J. Richardson: Conceptualization, Data curation, Formal analysis, Methodology, Writing - review & editing, Investigation, Resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced, or appear to have influenced, the work reported in this paper.
Acknowledgements
This work was supported, in part, by a research grant to the University of Michigan from the Alternatives Research and Development Foundation (ARDF) to support research on computational drug discovery for prevention and treatment of SARS-CoV-2 infections (Rudy J. Richardson, PI). We also thank Dr. P. Y. Huais (National University of Córdoba, Argentina) for assistance with the R-function: block_average. The sponsor had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.106449.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.World Health Organization 2022. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 2.World Health Organization 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 3.US FDA 2022. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines
- 4.Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byléhn F., Menéndez C.A., Perez-Lemus G.R., Alvarado W., De Pablo J.J. Modeling the binding mechanism of remdesivir, favilavir, and ribavirin to SARS-CoV-2 RNA-dependent RNA polymerase. ACS Cent. Sci. 2021;7:164–174. doi: 10.1021/acscentsci.0c01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian L., Pang Z., Li M., Lou F., An X., Zhu S., Song L., Tong Y., Fan H., Fan J. Molnupiravir and its antiviral activity against COVID-19. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.855496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duveau Y., Thomas C.J. The remarkable selectivity of nirmatrelvir. ACS Pharmacol. Transl. Sci. 2022;5:445–447. doi: 10.1021/acsptsci.2c00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLean G., Kamil J., Lee B., Moore P., Schulz T.F., Muik A., Sahin U., Tureci O., Pather S. The impact of evolving SARS-CoV-2 mutations and variants on COVID-19 vaccines. mBio. 2022;13 doi: 10.1128/mbio.02979-21. e02979-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corti D., Purcell L.A., Snell G., Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184:3086–3108. doi: 10.1016/j.cell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ASPR 2022. https://aspr.hhs.gov/COVID-19/Therapeutics/Documents/side-by-side-overview.pdf
- 11.Lee T.C., Morris A.M., Grover S.A., Murthy S., McDonald E.G. Outpatient therapies for COVID-19: how do we choose? Open Forum Infect. Dis. 2022;9 doi: 10.1093/ofid/ofac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez M.A. Efficacy of repurposed antiviral drugs: lessons from COVID-19. Drug Discov. Today. 2022;27(7):1954–1960. doi: 10.1016/j.drudis.2022.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wit de E., Doremalen van N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehr A.R., Perlman S. In: Maier H., Bickerton E., Britton P., editors. vol. 1282. Humana Press; New York, NY: 2015. Coronaviruses: an overview of their replication and pathogenesis. (Coronaviruses. Methods in Molecular Biology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jade D., Ayyamperumal S., Tallapaneni V., Nanjan C.M.J., Barge S., Mohan S., Nanjan M.J. Virtual high throughput screening: potential inhibitors for SARS-CoV-2 PLPRO and 3CLPRO proteases. Eur. J. Pharmacol. 2021;901 doi: 10.1016/j.ejphar.2021.174082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L.C., Zhao H.L., Liu J., He L., Yu R.L., Kang C.M. Design of SARS-CoV-2 Mpro, PLpro dual-target inhibitors based on deep reinforcement learning and virtual screening. Future Med. Chem. 2022;14:393–405. doi: 10.4155/fmc-2021-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22(6):297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 18.Amin S.A., Banerjee S., Ghosh K., Gayen S., Jha T. Protease targeted COVID-19 drug discovery and its challenges: insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 2021;29 doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Z., Ratia K., Cooper L., Kong D., Lee H., Kwon Y., Li Y., Alqarni S., Huang F., Dubrovskyi O., Rong L., Thatcher G.R.J., Xiong R. Design of SARS-CoV-2 PLpro inhibitors for COVID-19 antiviral therapy leveraging binding cooperativity. J. Med. Chem. 2022;65:2940–2955. doi: 10.1021/acs.jmedchem.1c01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockbaum G.J., Reyes A.C., Lee J.M., Tilvawala R., Nalivaika E.A., Ali A., Yilmaz N.K., Thompson P.R., Schiffer C.A. Crystal structure of SARS-CoV-2 main protease in complex with the non-covalent inhibitor ML188. Viruses. 2021;13(2):174. doi: 10.3390/v13020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Liang C., Xin L., Ren X., Tian L., Ju X., Li H., Wang Y., Zhao Q., Liu H., Cao W., Jian Y. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189–15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osipiuk J., Azizi S.A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R.S., Kim Y., Lisnyak V.G., Maki S.L., Nicolaescu V., Taylor C.A., Tesar C., Zhang Y.-A., Zhou Z., Randall G., Michalska K., Snyder S.A., Dickinson B.C., Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12(1):743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Copeland R.A. second ed. John Wiley & Sons, Inc.; Hoboken, NJ: 2013. Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists; p. 538. [PubMed] [Google Scholar]
- 28.Ghosh A.K., Samanta I., Mondal A., Liu W.R. Covalent inhibition in drug discovery. ChemMedChem. 2019;14:889–906. doi: 10.1002/cmdc.201900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macchiagodena M., Pagliai M., Procacci P. Identification of potential binders of the main protease 3CLpro of the COVID-19 via structure-based ligand design and molecular modeling. Chem. Phys. Lett. 2020;750 doi: 10.1016/j.cplett.2020.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strelow J.M. A perspective on the kinetics of covalent and irreversible inhibition. SLAS Discov. 2017;22(1):3–20. doi: 10.1177/1087057116671509. [DOI] [PubMed] [Google Scholar]
- 31.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B.S., Johnson M.E., Baker S.C., Ghosh A.K., Mesecar A.D. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. USA. 2008;105(42):16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., Wang M., Cui S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2021;11(1):237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Z., Ratia K., Cooper L., Kong D., Lee H., Kwon Y., Li Y., Alqarni S., Huang F., Dubrovskyi O., Rong L., Thatcher G.R.J., Xiong R. Design of SARS-CoV-2 PLpro inhibitors for COVID-19 antiviral therapy leveraging binding cooperativity. J. Med. Chem. 2022;65(4):2940–2955. doi: 10.1021/acs.jmedchem.1c01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhat Z.A., Chitara D., Iqbal J., Sanjeev B.S., Madhumalar A. Targeting allosteric pockets of SARS-CoV-2 main protease Mpro. J. Biomol. Struct. Dyn. 2021;40(14):6603–6618. doi: 10.1080/07391102.2021.1891141. [DOI] [PubMed] [Google Scholar]
- 35.DasGupta D., Chan W.K.B., Carlson H.A. Computational identification of possible allosteric sites and modulators of the SARS-CoV-2 main protease. J. Chem. Inf. Model. 2022;22:618–626. doi: 10.1021/acs.jcim.1c01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubanevics I., McLeish T.C.B. Computational analysis of dynamic allostery and control in the SARS-CoV-2 main protease. J. R. Soc. Interface. 2021;18 doi: 10.1098/rsif.2020.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Csermely P., Agoston V., Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol. Sci. 2005;26(4):178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Yang J., Lin X., Xing N., Zhang Z., Zhang H., Wu H., Xue W. Structure-based discovery of novel nonpeptide inhibitors targeting SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021;61(8):3917–3926. doi: 10.1021/acs.jcim.1c00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaushik N., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A., Chen X., Hou T., Cao D. Tetrazoles: synthesis and biological activity, immun., endoc. & metab. Agents in Med. Chem. 2018;18(1):3–21. doi: 10.2174/1871522218666180525100850. [DOI] [Google Scholar]
- 40.Ghareb N., Elshihawy H.A., Abdel-Daim M.M., Helal M.A. Novel pyrazoles and pyrazolo [1, 2-a] pyridazines as selective COX-2 inhibitors; Ultrasound-assisted synthesis, biological evaluation, and DFT calculations. Bioorg. Med. Chem. Lett. 2017;27(11):2377–2383. doi: 10.1016/j.bmcl.2017.04.020. [DOI] [PubMed] [Google Scholar]
- 41.Mikolaichuk O.V., Zarubaev V.V., Muryleva A.А., Esaulkova Y.L., Spasibenko D.V., Batyrenko A.А., Kornyakov I.V., Trifonov R.Е. Synthesis, structure, and antiviral properties of novel 2-adamantyl-5-aryl-2H-tetrazoles. Chem. Heterocycl. Compd. (N. Y., NY, U. S.) 2021;57(4):442–447. doi: 10.1007/s10593-021-02931-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y., Kitamura N., Musharrafieh R., Wang J. Discovery of potent and broad-spectrum pyrazolopyridine-containing antivirals against enteroviruses D68, A71, and coxsackievirus B3 by targeting the viral 2C protein. J. Med. Chem. 2021;64(12):8755–8774. doi: 10.1021/acs.jmedchem.1c00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamdy N.A., El-Senousy W.M. Synthesis and antiviral evalution of some novel pyrazoles and pyrazolo [3,4-d] pyridazines bearing 5,6,7,8-tetrahydronaphthalene. Acta Pol. Pharm. 2013;70(1):99–110. [PubMed] [Google Scholar]
- 44.Tumskiy R.S., Tumskaia A.V. Multistep rational molecular design and combined docking for discovery of novel classes of inhibitors of SARS-CoV-2 main protease 3CLpro. Chem. Phys. Lett. 2021;780 doi: 10.1016/j.cplett.2021.138894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skalic M., Martínez-Rosell G., Jiménez J., De Fabritiis G. PlayMolecule BindScope: large scale CNN-based virtual screening on the web. Bioinformatics. 2019;35(7):1237–1238. doi: 10.1093/bioinformatics/bty758. [DOI] [PubMed] [Google Scholar]
- 46.Umar H.I., Ajayi A., Bello R.O., Alabere H.O., Sanusi A.A., Awolaja O.O., Alshehri M.M., Chukwuemeka P.O. Novel molecules derived from 3-O-(6-galloylglucoside) inhibit main protease of SARS-CoV 2 in silico. Chem. Zvesti. 2022;76:785–796. doi: 10.1007/s11696-021-01899-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A., Chen X., Hou T., Cao D. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49(W1):W5–W14. doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casewit C.J., Colwell K.S., Rappe A.K. Application of a universal force field to organic molecules. J. Am. Chem. Soc. 1992;114(25):10035–10046. doi: 10.1021/ja00051a041. [DOI] [Google Scholar]
- 49.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminf. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volkamer A., Griewel A., Grombacher T., Rarey M. Analyzing the topology of active sites: on the prediction of pockets and subpockets. J. Chem. Inf. Model. 2010;50(11):2041–2052. doi: 10.1021/ci100241y. [DOI] [PubMed] [Google Scholar]
- 52.Volkamer A., Kuhn D., Grombacher T., Rippmann F., Rarey M. Combining global and local measures for structure-based druggability predictions. J. Chem. Inf. Model. 2012;52(2):360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 53.Schöning-Stierand K., Diedrich K., Fährrolfes R., Flachsenberg F., Meyder A., Nittinger E., Steinegger R., Rarey, ProteinsPlus M. Interactive analysis of protein-ligand binding interfaces. Nucleic Acids Res. 2020;48:W48–W53. doi: 10.1093/nar/gkaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fährrolfes R., Bietz S., Flachsenberg F., Meyder A., Nittinger E., Otto T., Volkamer A., Rarey M. ProteinsPlus: a web portal for structure analysis of macromolecules. Nucleic Acids Res. 2017;45:W337–W343. doi: 10.1093/nar/gkx333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martínez-Rosell G., Giorgino T., De Fabritiis G. PlayMolecule ProteinPrepare: a web application for protein preparation for molecular dynamics simulations. J. Chem. Inf. Model. 2017;57(7):1511–1516. doi: 10.1021/acs.jcim.7b00190. [DOI] [PubMed] [Google Scholar]
- 56.Cole J.C., Nissink J.W.M., Taylor R. Virtual Screening in Drug Discovery. Taylor & Francis. CRC Press; Boca Raton: 2005. Protein–ligand docking and virtual screening with GOLD. [Google Scholar]
- 57.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korb O., Stutzle T., Exner T.E. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 2009;49(1):84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- 60.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 61.BIOVIA. Systèmes Dassault. Dassault Systèmes; San Diego: 2022. Studio® Visualizer 21.1.0.20298. [Google Scholar]
- 62.Krieger E., Vriend G. YASARA View – molecular graphics for all devices – from smartphones to workstations. Bioinformatics. 2014;30 doi: 10.1093/bioinformatics/btu426. 2991-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozvoldik K., Stockner T., Rammner B., Krieger E. Assembly of biomolecular gigastructures and visualization with the Vulkan graphics API. J. Chem. Inf. Model. 2021;61:5293–5503. doi: 10.1021/acs.jcim.1c00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jorgensen W.L., Chandrasekhar J., Madura J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 65.Krieger E., Dunbrack R.L., Hooft R.W., Krieger B. Assignment of protonation states in proteins and ligands: combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012;819:405–421. doi: 10.1007/978-1-61779-465-0_25. [DOI] [PubMed] [Google Scholar]
- 66.Krieger E., Nielsen J.E., Spronk C.A., Vriend G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006;25:481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 67.Maier J., Martinez C., Kasavajhala K., Wickstrom L., Hauser K., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general AMBER force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 69.Jakalian A., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 70.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple AMBER force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Essman U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. B. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 72.Krieger E., Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kurkcuoglu Z., Koukos P.I., Citro N., Trellet M.E., Rodrigues J.P.G.L.M., Moreira I.S., Roel-Touris J., Melquiond A.S.J., Geng C., Schaarschmidt J., Xue L.C., Vangone A., Bonvin A.M. Performance of HADDOCK and a simple contact-based protein-ligand binding affinity predictor in the D3R Grand Challenge 2. J. Comput. Aided Mol. Des. 2018;32:175–185. doi: 10.1007/s10822-017-0049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vangone A., Schaarschmidt J., Koukos P., Geng C., Citro N., Trellet M.E., Xue L.C., Bonvin A.M. Large-scale prediction of binding affinity in protein-small ligand complexes: the PRODIGY-LIG web server. Bioinformatics. 2018;35(9):1585–1587. doi: 10.1093/bioinformatics/bty816. [DOI] [PubMed] [Google Scholar]
- 75.Flyvbjerg H., Petersen H.G. Error estimates on averages of correlated data. J. Chem. Phys. 1989;91:461–466. doi: 10.1063/1.457480. [DOI] [Google Scholar]
- 76.Huais P.Y. R-function: block_average. 2022. https://github.com/phuais/block_average
- 77.Hawkins P.C.D., Skillman A.G., Nicholls A. Comparison of shape-matching and docking as virtual screening tools. J. Med. Chem. 2007;50:74–82. doi: 10.1021/jm0603365. [DOI] [PubMed] [Google Scholar]
- 78.Liu Y., Liang C., Xin L., Ren X., Tian L., Ju X., Li H., Wang Y., Zhao Q., Liu H., Cao W., Xie X., Zhang D., Wang Y., Jian Y. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan J., Verschueren K.H., Anand K., Shen J., Yang M., Xu Y., Hilgenfeld R. pH-dependent conformational flexibility of the SARS-CoV main proteinase (Mpro) dimer: molecular dynamics simulations and multiple X-ray structure analyses. J. Mol. Biol. 2005;354(1):25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El‐Baba T.J., Lutomski C.A., Kantsadi A.L., Malla T.R., John T., Mikhailov V., Robinson C.V. Allosteric inhibition of the SARS‐CoV‐2 main protease: insights from mass spectrometry based assays. Angew, Chem. Int. 2020;59(52):23544–23548. doi: 10.1002/anie.202010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Günther S., et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372(6542):642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kneller D.W., Phillips G., O'Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bellow M. Prediction of potential inhibitors of the dimeric SARS-CoV2 main proteinase through the MM/GBSA approach. J. Mol. Graph. Model. 2020;101 doi: 10.1016/j.jmgm.2020.107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weng Y.L., Naik S.R., Dingelstad N., Lugo M.R., Kalyaanamoorthy S., Ganesan A. Molecular dynamics and in silico mutagenesis on the reversible inhibitor-bound SARS-CoV-2 main protease complexes reveal the role of lateral pocket in enhancing the ligand affinity. Sci. Rep. 2021;11:7429. doi: 10.1038/s41598-021-86471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhowmick S., Saha A., Osman S.M., Alasmary F.A., Almutairi T.M., Islam M.A. Structure-based identification of SARS-CoV-2 main protease inhibitors from anti-viral specific libraries: an exhaustive computational screening approach. Mol. Divers. 2021;25:1979–1997. doi: 10.1007/s11030-021-10214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Durchschlag H., Zipper P., Wilfing R., Purr G. Detection of small conformational changes of proteins by small-angle scattering. J. Appl. Crystallogr. 1991;24:822–831. doi: 10.1107/S0021889891004831. [DOI] [Google Scholar]
- 87.Zhang Y., Hess H. Enhanced diffusion of catalytically active enzymes. ACS Cent. Sci. 2019;5 doi: 10.1021/acscentsci.9b00228. 939-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clark J.J., Benson M.L., Smith R.D., Carlson H.A. Inherent versus induced protein flexibility: comparisons within and between apo and holo structures. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh E., Jha R.K., Khan R.J., Kumar A., Jain M., Kuthukumaran J., Singh A.K. A computational essential dynamics approach to investigate structural influences of ligand binding on Papain like protease from SARS-CoV-2, Comput. Biol. Chem. 2022;99 doi: 10.1016/j.compbiolchem.2022.107721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prajapati J., Patel R., Rao P., Saraf M., Rawal R., Goswami D. Perceiving SARS-CoV-2 Mpro and PLpro dual inhibitors from pool of recognized antiviral compounds of endophytic microbes: an in silico simulation study. Struct. Chem. 2022;33:1619–1643. doi: 10.1007/s11224-022-01932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Björnsson E.S. Hepatotoxicity by drugs: the most common implicated agents. Int. J. Mol. Sci. 2016;17(2):224. doi: 10.3390/ijms17020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.